Abstract
A previously published clinical trial demonstrated the benefit of autologous CD34+ cells transduced with a self-inactivating lentiviral vector (HPV569) containing an engineered β-globin gene (βA-T87Q-globin) in a subject with β-thalassemia major. This vector has been modified to increase transduction efficacy without compromising safety. In vitro analyses indicated that the changes resulted in both increased vector titers (3 to 4 fold) and increased transduction efficacy (2 to 3 fold). An in vivo study in which 58 β-thalassemic mice were transplanted with vector- or mock-transduced syngenic bone marrow cells indicated sustained therapeutic efficacy. Secondary transplantations involving 108 recipients were performed to evaluate long-term safety. The six month study showed no hematological or biochemical toxicity. Integration site (IS) profile revealed an oligo/polyclonal hematopoietic reconstitution in the primary transplants and reduced clonality in secondary transplants. Tumor cells were detected in the secondary transplant mice in all treatment groups (including the control group), without statistical differences in the tumor incidence. Immunohistochemistry and quantitative PCR demonstrated that tumor cells were not derived from transduced donor cells. This comprehensive efficacy and safety data provided the basis for initiating two clinical trials with this second generation vector (BB305) in Europe and in the USA in patients with β-thalassemia major and sickle cell disease.
Keywords: β-hemoglobinopathy, β-thalassemia, gene therapy, lentiviral vector, mouse model
INTRODUCTION
Human genetic diseases have been treated successfully by gene therapy with lentiviral vectors, as shown for the first time in the treatment of subjects with childhood cerebral adrenoleukodystrophy (CCALD) [1, 2] and β-thalassemia major [3, 4]. The subjects treated in these studies experienced clinically beneficial outcomes even though only 10% to 20% of their blood cells carried the transgene sequences. More recently positive clinical data have been reported for the treatment of metachromatic leukodystrophy [5] and Wiscott-Aldrich syndrome [6] with substantially higher gene marking and expression.
β-thalassemia is a rare hereditary blood disorder caused by the absence or reduced synthesis of the beta chains of hemoglobin A (HbA). Reduction in β-globin production results in an accumulation of excess uncomplexed α-globin in erythroblasts, leading to premature death of the cells. The ineffective erythropoiesis and hemolysis causes the anemia, characteristic of patients with β-thalassemia [7]. Allogeneic hematopoietic stem cell transplantation (HSCT) is a proper treatment for patients with severe hemoglobin disorders [8, 9]. However, because of the significant risk of transplant related mortality, graft versus host disease (GVHD) and graft rejection with allogeneic HSCT, transplants are offered primarily to patients with available human leukocyte antigen (HLA) matched sibling donors (below 25% of cases) [10, 11]. In adults, HSCT is not a commonly prescribed treatment and only 50-65% of β-thalassemic patients survive beyond 35 years in high income countries [12-14].
The safety and efficacy of autologous CD34+ hematopoietic stem cells transduced with LentiGlobin HPV569 lentiviral vectors were investigated in 3 subjects with β-thalassemia major in a Phase 1/2 clinical study in France [3]. Clinical benefit was demonstrated in 1 of 3 subjects treated, who became transfusion-independent 12 months after treatment with LentiGlobin HPV569 lentiviral vector and remains transfusion-independent 5 years post-transplant [4].
The LentiGlobin HPV569 lentiviral vector is a self-inactivating (SIN) vector, containing two copies of the 250 bp core element of the chicken 1.2 kb hypersensitive site-4 (cHS4) chromatin insulator in the 3’ long terminal repeat (LTR), that are duplicated in the 5’LTR of the provirus upon cell transduction [3, 15]. Tandem copies of the cHS4 elements were shown to provide partial protection against oncogene activation by oncoretroviral long-terminal repeats (LTR) in cell lines [16] and chromatin barrier activities to the surrounded genes [16, 17]. Thus, cHS4 elements were believed to provide additional safety features to the SIN design of lentiviral vectors.
However, recent results suggest that the cHS4 elements may not be as beneficial as previously believed. The protective effect was shown to be variable [18], limited in efficacy and dependent on their position in the chromatin [19]. Furthermore, their insertion into the 3’LTR reduces the functional titer of lentiviral vectors [20-22] due to impaired reverse transcription at the post-entry step and/or integration of incomplete viral sequences. In addition, the tandem cHS4 cassette is prone to rearrange into a single cHS4 element upon vector insertion [3, 20, 23], which does not protect against gene activation when placed between enhancer and promoter regions [3]. Therefore, it seemed likely that removing the cHS4 elements would result in a lentiviral vector with higher transduction capabilities and an equivalent level of safety.
The LentiGlobin HPV569 lentiviral vector used in the clinical study for the treatment of patients with β-thalassemia major had a relatively low titer and modest transduction efficiency. Therefore, modifications were made to the HPV569 vector with the aim to increase titers and transduction efficiency without compromising safety. The 5’LTR of the HPV569 vector was modified by replacing the wild-type HIV-1 U3 region with the strong and constitutive CMV promoter to drive higher expression levels of packageable RNA in producer cells. This change also resulted in removing the need for the Tat gene, which reduced the number of plasmids required for lentiviral vector productions and improved vector yield. The resulting new, improved second generation globin vector construct, is called LentiGlobin BB305 lentiviral vector.
We have compared the lentiviral vectors HPV569 and BB305 in vitro for vector production and transduction efficiencies in human CD34+ hematopoietic cells. In addition, the efficacy and safety of both vectors were assessed in mouse bone marrow transplants using β-thalassemia mice (Hbbth1/th1) in primary and C57BL/6J mice in secondary bone marrow transplants. The efficacy was demonstrated by the correction of the thalassemic phenotype in the primary transplants and the safety was assessed by in life observation, blood chemistry, macroscopic and microscopic observation and histopathology of selected organs in both primary and secondary transplant animals. Integration site (IS) analyses were carried out using linear amplification-mediated polymerase chain reaction (LAM PCR) and the genomic integration profiles of both vectors were evaluated from >7,000 unique insertion sites. Overall, the data from the in vitro and in vivo nonclinical studies indicate a better efficacy of the LentiGlobin BB305 compared to the LentiGlobin HPV569 lentiviral vector with equivalent safety. Results from the studies described in this report supported the initiation of clinical trials using autologous CD34+ hematopoietic stem cells transduced with the LentiGlobin BB305 lentiviral vector for treatment of β-thalassemia in France and the USA.
MATERIAL AND METHODS
Lentiviral Vector Design, Production, Titration and CD34+ Cell Transduction
The HPV569 vector has been described previously [3, 15]. “It is a self-inactivating (SIN), Tat-dependent vector, containing two copies of the 250-base-pair (bp) core element of the cHS4 chromatin insulator in the U3 region of the 3’ LTR. It encodes a mutated adult βA-T87Q-globin” [3]. The SIN vector BB305 contains a Cytomegalovirus (CMV) promoter and enhancer instead of the HIV U3 region at the 5’ LTR, and a 3’ deleted U3 region (Fig. 1A). Clinical-grade vesicular stomatitis virus glycoprotein pseudotyped lentiviral particles of the two vectors were produced by a plasmid based co-transfection method. Purification was done by ion exchange chromatography and buffer was exchanged for SCGM medium (CellGenix) by ultrafiltration prior to final filtration according to published protocols [24, 25].
Fig. (1).
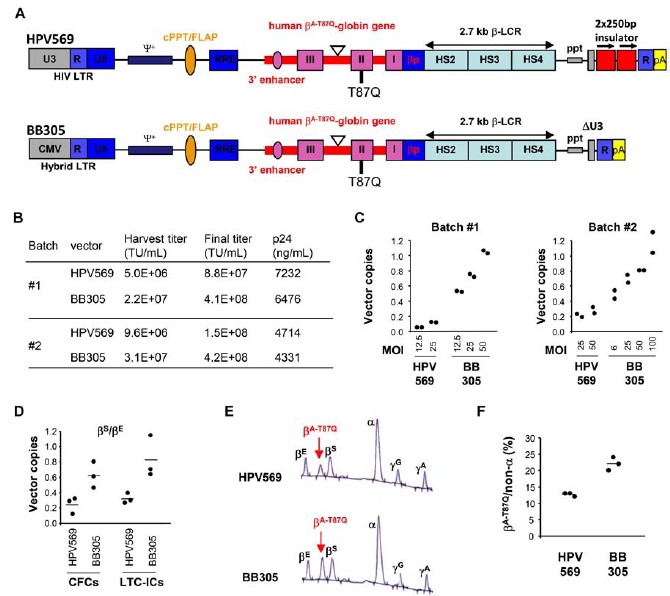
In vitro evaluation of LentiGlobin lentiviral vectors. A) Diagram of the LentiGlobin HPV569 and BB305 lentiviral vectors. The 3′ β-globin enhancer, the 372 base pairs (bp) IVS2 deletion in intron 2 (triangle), the βA-T87Q mutation (ACA [Thr] to CAG [Gln]) and DNase I hypersensitive sites (HS) 2, HS3, and HS4 of the human β-globin locus control region (LCR) are indicated. Safety modifications including the 400 bp deletion in the U3 of the right HIV LTR, the rabbit β-globin polyA signal and the 2 × 250 bp cHS4 chromatin insulators are indicated. βp, human β-globin promoter; cPPT/flap, central polypurine tract; HIV LTR, human immunodeficiency type-1 virus long-terminal repeat; ppt, polypurine tract; RRE, Rev-responsive element; Ψ+, packaging signal. B) Functional (transducing unit/mL) and physical (ng p24/mL) titers of lentiviral vectors produced in HEK293T cells, before (harvest) and after purification/concentration (final). C) Y axis: Vector copies = Average vector copy number per cell in transduced human CD34+ cells. D) Y axis: Vector copies = Average vector copy numbers per cell in transduced progenitor cells, using either short-term cultures to determine colony forming cells (CFCs) or long-term cultures to determine long-term culture initiating cells (LTC-ICs). MOI was 50. E) HPLC profile of globin chains from pooled erythroid colonies obtained from HPV569 (top) and BB305 (bottom) lentiviral vector-transduced CD34+ cells. F) Y axis: βA-T87Q globin chains as a percentage of all non-α globin chains as detected by HPLC.
The infectious titer was determined by transducing NIH3T3 cells as previously described [26]. CD34+ cells were grown 24 hours in SCGM medium containing human cytokines fms-related tyrosine kinase 3 ligand (Flt3L), stem cell factor (SCF), thrombopoietin (TPO) at 100 ng/mL and IL-3 (at 60 ng/mL) and transduced another 24 hours in medium containing protamine sulfate (8 µg/mL). The liquid culture and progenitor assays were performed as previously described [26]. DNA was prepared from liquid culture or pooled colonies and amplified by quantitative PCR for vector copy number determination, as previously described [23]. For individual colonies and determination of the percentage of vector bearing progenitors, DNA was prepared and amplified by quantitative PCR using the TaqMan Sample-to-SNP kit (Life Technologies).
Insertional Genotoxic Assay
Aliquots of the test and control vectors were used to transduce primary murine hematopoietic cells. Fresh lineage-negative (Lin-) cells were isolated from complete bone marrow of young adult C57BL/6J mice using lineage specific antibodies and magnetic beads (Miltenyi Biotec). Cells were prestimulated in Stem Span medium (Stemcell Technologies) containing mouse SCF and IL-3, human Flt3L, and interleukin 11 (IL-11), (all at 100 ng/mL and from Pepro-
tech), penicillin/streptomycin (PAN-Biotech) and glutamine (Biochrom). Lin- cells were transduced on days 2 and 3 at a multiplicity of infection (MOI) between 10 and 100. On day 9, transduction efficiency was monitored by flow cytometry for GFP vectors and qPCR for BB305 and HPV569. Following transduction, bone marrow cells were expanded for two weeks in IMDM medium (Biochrom) containing 10% fetal calf serum (PAA Laboratories) and cytokines as above. Then 100 cells per well were plated in 96-well plates. The wells containing cells were scored two weeks later and the frequencies of replating cells were determined as described [27]. Six independent assays were performed per sample. As a positive control, an LTR-driven gammaretroviral vector expressing GFP (RSF91) was used. It induces a replating phenotype with a penetrance close to 100%. We also used a SIN-lentiviral vector containing the same SFFV promoter/enhancer to control the expression of GFP. As a negative control, mock-transduced cells were used. Based on these data, the replating frequency, normalized to the mean vector copy number, was calculated.
Animals, Bone Marrow Transduction and Transplantation
“All animal experiments were approved by the ethical committee of Ile-de-France and were in accordance with the French decree 2001-464. The β-Thal intermedia mouse model Hbbth-1/th-1 [28] referred to as β-thal mice, bear a homozygous deletion of the mouse β major-globin gene and manifest clinical and biological features similar to those observed in human β-Thal [29]. The β-thal mice, originally provided by F Constantini (Columbia University, New York, NY), were backcrossed in C57BL/6J mice and maintained in our animal facility” [23]. C57BL/6-Ly5.1 (B6.SJL-PtprcaPepcb/BoyCrl) mice were purchased from Charles River Laboratory (L’Abresle, France).
Bone marrow cells from β-thalassemic Hbbth1/th1 mice were harvested from femurs and tibias and lineage depleted (Lin-) with a cocktail of biotinylated antibodies against a panel of lineage antigens and anti-biotin micro-beads (Mouse Lineage Cell Depletion kit, Miltenyi Biotec). Lin- β-thal cells were transduced in SCGM medium containing protamine sulfate (8µg/mL), mouse SCF (100ng/mL), mouse IL3 (6 ng/mL) and mouse interleukin 6 (IL-6) (10 ng/mL) with lentiviral vectors at a MOI of 50 (or mock-transduced) and infused intravenously (2.8x105 cells/mouse) in syngenic β-thalassemic Hbbth1/th1 mice submitted to total body irradiation (TBI) of 11 Grays (two doses of 5.5 Gy three hours apart). MOI calculation was based on functional transduction units (TU), as determined on NIH3T3 cells. Before transplantation, 30,000 transduced bone marrow cells were grown in liquid culture for 10 days and seeded in triplicate in methyl cellulose medium (MethoCult M3434; Stem Cell Technologies). Colonies were scored on day 7 and DNA was prepared for vector insertion analyses. Groups, sexes and numbers of animals are indicated in Fig. (2A). Four months after transplantation, mice were sacrificed and necropsied. Twelve millions bone marrow cells of each sacrificed primary transplanted β-thal mice were injected in two secondary C57BL/6-Ly5.1 mice.
Fig. (2).
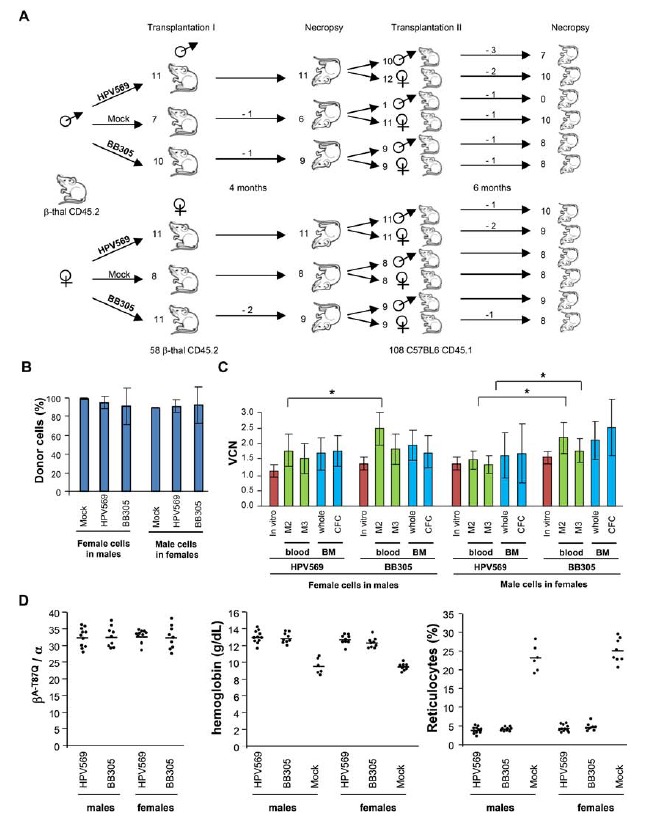
Design of in vivo transplant studies and primary transplant results. A) Study design. Lin- bone marrow cells from β-thalassemia mice (Hbbth1/th1 ; β-thal CD45.2) mice were transduced with lentiviral vectors and infused in 58 syngenic recipients. Four months after primary transplantation, the remaining 54 animals were sacrificed and bone marrow cells were injected in 108 secondary normal mice. The numbers of prematurely dead mice are indicated on top of the arrows. B) Mean (± SD) percentage chimerism estimated in primary transplant recipients. C) Mean VCN (± SD) pre-transplantation (in vitro), and in blood at 2 and 3 months after transplantation (2M, 3M), and in bone marrow after sacrifice (whole bone marrow and after culture for CFCs). D) Proportion of therapeutic β-globin chains and correction of the β-thalassemic phenotype (hemoglobin concentration and reticulocyte percentage) per group of mice. Horizontal bar represents the mean.
Post-Transplant Monitoring
Mortality and morbidity were checked five times a week. Animal were observed individually twice a week, for the recording of clinical changes including skin ulcerations, changes in the fur, unusual respiratory pattern, piloerection, stereotypes (e.g. repetitive cycling). The body weight of each animal was recorded on the first day of treatment and then once a week until the end of the study. During in vivo follow-up, hemoglobin and blood cell counts were determined using an automated cell counter (Cell Dyn 3700, Abbott Diagnostic). Hematocrit values were obtained by standard manual centrifugation method. Reticulocytes were determined after Thiazole orange staining and flow cytometry[23]. Genomic DNA was extracted from blood for determination of vector copy number and donor chimerism in primary transplants as described previously [30]. In secondary transplant, engraftment was assessed by measuring the proportion of CD45.2 donor cells in blood by flow cytometry.
Necropsy
The following organs were taken and weighted: adrenal gland, brain including medulla / pons, cerebellar and cerebral cortex, epididymis, heart, kidneys, large intestine (cecum, colon, rectum), liver, lungs with bronchi, lymph nodes, mesenteric and mandibular, ovaries with oviduct, pancreas, skeletal muscle, skin, small intestine (duodenum, ileum, jejunum), spleen, sternum with bone marrow, stomach with forestomach, testes, thymus, uterus, vagina. They were fixed in 10% buffered formalin or modified Davidson’s fixative for testes and epididymis, trimmed according to the RITA guidelines when possible [31-33], embedded in paraffin wax, sectioned at a thickness of approximately four microns and stained with hematoxylin-eosin. Immunohistochemical staining of formalin-fixed paraffin embedded (FFPE) thymus and few other organs with anti-CD3 (rat IgG1, AbD Serotec), CD45RA (rat IgG2a, Pharmingen), CD45.1 (mouse IgG2a, Southern Biotech), and CD45.2 (mouse IgG2a, Southern Biotech), were realized. Microscopic examination was performed by a board-certified veterinary pathologist. A peer review was performed on all animals with significant histological finding, including all neoplasia and thymic hyperplasia, in a randomized way.
DNA extraction from FFPE thymus was performed using the QIAamp® DNA FFPE Tissue kit (Qiagen). DNA samples were quantified by UV spectrophotometry using a NanoDrop™ and by a PicoGreen® dsDNA quantitation assay (Life Technologies). Forty nanograms (based on the concentration obtained by the PicoGreen® method) were amplified by qPCR. Eighteen out of the nineteen processed samples returned a similar Ct value for the two endogenous controls tested (GAPDH and 18S). One sample (FFPE block from animal 15) returned a slightly higher Ct. When the Ct values were below a threshold of 30 cycles and the coefficients of variation were low, the samples were considered to display satisfactory quality for qPCR analysis.
“Vector copy numbers per cell were determined by the delta-delta-Ct method (ΔΔCt) by comparison with those obtained after serial dilutions of genomic DNA from a cell line containing one copy of the integrated globin lentiviral vector per haploid genome as described” [30]. Two series of primers and probes were used to detect the vector. The set 1 made of GAGF (5’-ggagctagaacgattcgcagtta-3’), GAGR (5’-ggttgtagctgtcccagtatttgtc-3’) and GAGP (5’FAM-acagccttctgatgtctctaaaaggccagg-3’TAMRA) and the set 2 made of LTRF (5’-tgtgtgcccgtctgttgtgt-3’), LTRR (5’-cgagtcctgcgtcgagaga-3’) and LTRP, (5’FAM-cagtggcgcccgaacagga-3'TAMRA). The mouse beta actin gene was used to normalize for DNA quantity using primers mbActF1 (5’-acggccaggtcatcactattg-3’), mbActR1 (5’-caagaaggaaggctggaaaaga-3’) and mbActP2 (5’Yakima Yellow-caacgagcggttccgatgccct-3’BHQ1). All primers and probes were from Eurogentec.
Bone marrow cells were harvested from tibias and femurs. The relative percentages of leukocyte subsets were determined using the anti-mouse antibodies directed against CD45.2, CD45.1, CD11b, Gr1, CD3, B220, CD71, Ter119, their corresponding isotypes (eBioscience), flow cytometry (Becton–Dickinson FACSCanto II flow cytometer) and the Diva software (Becton-Dickinson).
Blood samples for biochemistry analyses were collected in lithium-heparin tubes. At least 100 µL of plasma were aliquoted for each animal. Sodium, Potassium, Chloride, Calcium, Inorganic phosphorus, Glucose, Urea, Creatinine, Total bilirubin, Total cholesterol, Triglycerides, Total protein, Albumin, Albumin/Globulin ratio, Alkaline phosphatase, Alanine Aminotransferase, Aspartate Aminotransferase were measured in plasma using the ADVIA 1650 blood biochemistry analyzer (Siemens).
Blood samples for hematology analyses were collected in EDTA tubes (250 µL per animal). Erythrocyte count, mean cell volume, packed cell volume, hemoglobin, mean cell hemoglobin concentration, mean cell hemoglobin, thrombocyte count, leucocyte count, differential white cell count with cell morphology, reticulocyte count were determined using the ADVIA 120 hematology analyzer (Siemens). In case the samples were not accepted by the machine, a blood smear was prepared and stained with May Grünwald Giemsa for determination of the differential white cell count. A blood smear was also prepared for manual determination of the reticulocyte count (stained with blue cresyl) if necessary.
High-Performance Liquid Chromatography
Percentage of the therapeutic globin βA-T87Q chain in human and mouse samples was determined by high-performance liquid chromatography with a Prominence chromatograph (Shimadzu). Cells lysates were injected onto a C4 column EC 250/4 Nucleosil 300-5 (Macherey-Nagel) and elution was achieved with a gradient of Milli-Q water (Millipore), acetonitril (Carlo Erba), trifluoroacetic (Fluka) acid and heptafluorobutyric acid (Fluka). The detection wavelength was 220 nm. Data acquisition was performed with the LC Solution software (Shimadzu).
Integration Site (IS) Analysis by Standard LAM-PCR or nrLAM-PCR and Deep Sequencing
LAM-PCR and/or nrLAM-PCR were performed as described previously [34, 35]. PCR amplicons of pretransplant and primary mice were sequenced with 454 pyrosequencing (Roche) as previously described [34, 36] whereas those of secondary recipients were sequenced on the MiSeq instrument (Illumina) after sample preparation for high-throughput sequencing. Therefore, an additional PCR with special fusion-primers carrying MiSeq specific sequencing adaptors was performed. DNA barcoding was used for sequencing of multiple samples in a single run.
Bioinformatical Analyses of Sequenced LAM-PCR and nrLAM-PCR Amplicons
Raw sequence data were trimmed according to sequence quality. Only sequences carrying correct (100% sequence identity) sequences in both molecular barcodes (linker cassette barcode, sequencing barcodes) were retained and analysed. Our (semi-) automated bioinformatical data mining pipeline was used to analyse the data [37]. In brief, sequences were trimmed (vector- and linker cassette-specific parts removed) and aligned to the mouse genome using UCSC BLAT (mm10), while nearby genes and other integrating features were annotated as previously described [34]. Only sequences which showed at least 18 nucleotides of vector specific sequence after MiSeq specific fusion-primer sequences were analysed further to ensure the analysis of specific PCR products and vector-genome junctions, respectively.
Determination of Common Integration Sites (CIS)
Standard mathematical programs to analyse CIS formation [38] were used by applying the following definition for CIS: 2nd order CIS: 2 IS in 30kb; 3rd order CIS: 3 IS in 50kb; 4th order CIS: 4 IS in 100 kb; ≥5th order CIS: 5 or more IS in 200 kb. RefSeq genes which have been detected next to the IS were analysed for their presence within three different cancer gene databases: CGC (Cancer Gene Census Database; http://www.sanger.ac.uk/genetics/CGP/Census/), RTCGD (Ret-rovirus and Transposon tagged Cancer Gene Database; http://variation.osu.edu/rtcgd/index.html and CBioPortal (cBio Cancer Genomics Portal; http://www.cbioportal.org/public-portal/). The frequency of IS detected next to cancer genes was assessed in comparison to an in silico generated data set of 7362 unique exactly mappable IS.
Diversity Index Calculation
The Shannon index is defined by SA = -∑i (ni / N) log (ni / N) where ni is the number of sequence reads belonging to the ith IS and N the total number of sequence counts in a given group and provides information on both the richness of the population and the homogeneity of the distribution. The first component is defined by the total number of species (unique integration sites) and the homogeneity measures the distribution of the individuals in the population (concept of dominance).
Statistics
SigmaPlot, GraphPad prism 6 and CiToxLAB softwares were used to perform the statistical analysis.
RESULTS
LentiGlobin BB305 Lentiviral Vector Titer and In vitro Transduction Efficiency
We redesigned the LentiGlobin HPV569 lentiviral vector by replacing the 5' HIV U3 LTR promoter/enhancer with the CMV promoter/enhancer. We also removed the 2 copies of 250 base pair (bp) core chicken hypersensitivity site 4 (cHS4) insulators imbedded in the SIN U3 LTR of LentiGlobin HPV569, which resulted in the construction of LentiGlobin BB305 lentiviral vector (Fig. 1A). The sequences of the integrated provirus containing the βA-T87Q-globin gene expression cassette the cPPT/cTS, RRE, and SIN U3 are identical in both lentiviral vectors and the internal globin promoter together with the LCR sequence driving the transgene expression exclusively in the erythroid lineage, remained unchanged.
Functional titers of harvested vector supernatants and the concentrated BB305 and HPV569 lentiviral vector preparations were measured on NIH3T3 cells (Fig. 1B). Harvest titers were approximately 3 to 4 fold higher for BB305 than for HPV569, with similar differences in titers after the purification and concentration processes. The concentrations of p24 were similar for both lentiviral vectors indicating that the proportion of functional to physical particles was higher for BB305 than for HPV569 (Fig. 1B).
The respective lentiviral vectors transduction efficiency as expressed by the average vector copy number (VCN) per cell was evaluated on human CD34+ hematopoietic stem cells with varying multiplicities of infection (MOI). At equivalent MOIs the VCN was consistently at least 2- to 3-fold higher for BB305 compared to HPV569 in CD34+ cells following 7 days in liquid culture (Fig. 1C), in short-term (day 14 methyl cellulose culture) culture colony forming cells (CFCs; Fig. 1D), and in long-term culture initiating cells (LTC-ICs; Fig. 1D). Consequently, therapeutic globin βA-T87Q-chain was produced at a higher level with BB305 (Figs. 1E and 1F) as measured in pooled erythroid colonies from CFCs. In order to understand whether VCN improvement resulted from higher transduction of cells and/or higher percentage of transduced cells, we also compared the percentages of transduced colonies. We observed a moderate but significant increase (one-tailed p-value = 0.0072) of vector bearing colonies with BB305 compared to HPV569, indicating that probably both the cell transduction and the level of vector integration increased with the Lentiglobin BB305 lentiviral vector (Supp. Fig. 1).
We also produced a vector without insulator but with the 5’ HIV-derived LTR. Harvest titers on 3T3 cells and transduction potency on CD34+ cells were within the intermediate range between those of HPV569 and BB305 vectors (data not shown). As the BB305 vector displayed the highest efficacy, all subsequent experiments were performed with the Lentiglobin BB305 lentiviral vector and compared to the Lentiglobin HPV569 vector.
Low Genotoxic Potential of LentiGlobin Vectors In vitro
Two independent in vitro insertional genotoxic assays [39] were completed to evaluate the genotoxic potential of LentiGlobin BB305 and HPV569 lentiviral vectors relative to a gamma-retroviral control. High transduction efficiency was observed with both lentiviral vectors with a mean VCN of 4.2 per cell for HPV569 and a mean VCN of 7.4 per cell for BB305. The control RSF91 gamma-retroviral vector was capable of immortalization of clonal outgrowth at 3-fold lower VCN level. In comparison to the positive control gamma-retroviral vectors RSF91 and lentiviral vector LV-SF with known in vivo and in vitro mutagenic potential, the two lentiviral vectors LentiGlobin BB305 and HPV569 showed a strongly reduced risk of growth advantage for transduced murine hematopoietic stem cells and are hence believed to be significantly less genotoxic. Furthermore, there was no significant cytotoxicity associated with high titer virus transduction with LentiGlobin HPV569 and BB305 lentiviral vectors (Supp. Fig. 2).
Efficient Engraftment and Correction of Phenotype in Primary Transplant β-Thalassemia Mice (Hbbth1/th1)
Lin- bone marrow cells from male and female β-thalassemia mice (Hbbth1/th1) were transduced with LentiGlobin HPV569 or LentiGlobin BB305 lentiviral vector at a MOI of 50, or mock-transduced (no vector) and transplanted into 58 lethally irradiated Hbbth1/th1 animals (female donor cells to male recipients and vice versa) according to the procedure schematically represented in Fig. (2A). Four months after transplantation, 54 mice were sacrificed and necropsied. Four mice died before sacrifice (3 in the BB305 group and 1 in the Mock group).
VCNs were measured in vitro after transduction in blood samples at 2 and 3 months after transplantation (M2 and M3) and in bone marrow after sacrifice. After transduction, the average VCN per cell was measured by qPCR on cells grown for 10 days in liquid cultures (1.50 and 1.57 for HPV569 transduced female and male cells and 1.96 and 2.05 for BB305 transduced female and male cells respectively) and on cells plated for 7 days in methylcellulose (1.13 and 1.36 for HPV569 transduced female and male cells and 1.35 and 1.56 for BB305 transduced female and male cells respectively). In vivo, donor chimerism, verified by quantitative male PCR, was close to 90% in all groups (Fig. 2B). Overall, VCN were slightly higher in the BB305 than in the HPV569-transduced cells (1.1 to 1.5-fold), but with statistical significance in blood samples (M2) only (Fig. 2C). The ratio of β-A-T87Q-globin /α-globin was above 30% and was similar in the groups engrafted with the LentiGlobin HPV569 and the LentiGlobin BB305 transduced cells (Fig. 2D). Anemia was corrected in all mice that received vector-transduced cells at three months post-transplantation (Fig. 2D) and at terminal sacrifice (Table 1). “A significant improvement of hemoglobin concentration, hematocrit level, red blood cell counts and reticulocyte percentage was observed in mice transplanted with bone marrow cells transduced with HPV569 or BB305 compared to the group transplanted with untransduced cells” [40]. As expected, the proportion of differentiated Ter119+ CD71- erythroid cell was significantly higher in the bone marrow of mice transplanted with LentiGlobin transduced cells than in the control group, indicative of corrected dyserythropoiesis (Supp. Fig. 3). “Improvement of the β-thalassemic phenotype was similar between mice transplanted with cells transduced with both the HPV569 and with the BB305 lentiviral vector” [40]. These results were expected, as the levels of expression per vector copy are similar between the two lentiviral vectors (BB305 and HPV569) as demonstrated in previous in vitro studies performed in human CD34+ cells (Supp. Fig. 4). White blood cell counts based on flow cytometry analysis of cell subsets of peripheral blood and bone marrow did not show differences in white blood cell homeostasis between the three groups (Supp. Fig. 3).
Table 1. Hematological parameters in the blood of primary recipients at terminal sacrifice (ADVIA 120).
| Sex | Male | Female | ||||
|---|---|---|---|---|---|---|
| Vector | HPV569 | BB305 | Mock | HPV569 | BB305 | Mock |
| Animals (Examined/Alive) | 11/11 | 7/9(2) | 5/6(1) | 10/11(1) | 9/9 | 5/8(1) |
| Red blood cell parameters | ||||||
| Erythrocytes (x1012/L) | 10.16** | 10.30** | 7.90 | 9.94** | 10.23** | 7.54 |
| Hemoglobin (g/dL) | 13.0** | 13.1** | 8.6 | 12.8** | 13.0** | 8.2 |
| Hematocrit (%) | 46.0** | 47.0** | 33.0 | 46.0** | 46.0** | 31.0 |
| MCV (fL) | 45.3 | 45.5 | 41.9 | 45.7** | 45.4** | 41.2 |
| MCH (pg) | 12.8** | 12.7** | 10.9 | 12.9** | 12.7** | 10.9 |
| MCHC (g/dL) | 28.3** | 28.0** | 26.0 | 28.2** | 28.0** | 26.6 |
| Reticulocyte (%) | 3.54** | 3.89** | 14.88 | 4.30** | 3.95** | 19.04 |
| White blood cell counts | ||||||
| WBC (x109/L) | 3.86 | 4.24 | 4.31 | 4.44 | 6.74* | 5.00 |
| Neutrophiles (x109/L) | 0.71 | 0.73 | 0.63 | 0.56 | 0.89* | 0.44 |
| Eosinophiles (x109/L) | 0.06 | 0.07* | 0.03 | 0.05 | 0.09 | 0.05 |
| Monocytes (x109/L) | 0.09 | 0.14* | 0.06 | 0.10 | 0.16 | 0.09 |
* p < 0.05; ** p < 0.01
MCV (Mean Corpuscular Volume); MCH (Mean Corpuscular Hemoglobin); MCHC (Mean Corpuscular Hemoglobin Concentration)
Notes: (1) five samples had to be diluted; they were not included in the statistical analysis; (2) insufficient sample for hematological analysis of two animals
Safety Assessment of LentiGlobin BB305 Vector in Primary Transplant Recipient Mice
Safety was evaluated in the transplanted Hbb(th1/th1) mice based on hematology analysis (Table 1), blood biochemistry (Supp. Table 1), bone marrow cytology and histopathology of selected organs four months after the bone marrow transplantation with Lin- cells untransduced (mock) or transduced with LentiGlobin BB305 or HPV569 lentiviral vectors.
The minor changes observed in blood chemistry and white blood cell counts were considered to be incidental and not vector related except the lower mean total bilirubin levels observed in the blood of mice transplanted with the LentiGlobin HPV569 and BB305 lentiviral vectors, which is expected as treatment induces lower destruction of red blood cells.
The microscopic histological examination was performed by a board-certified veterinary pathologist on all tissues of the study animals (58 mice). Out of the four prematurely
dead animals, two males died at 27 and 34 days after transplantation (BB305 and mock groups, respectively). These mice had a marked lymphoid atrophy of all compartments with decreased erythroid cell numbers in the bone marrow. These observations might indicate the reason for their death and were most likely related to the pre-transplant irradiation. Another two females of the BB305 group died at 49 and 65 days after transplantation and based on the microscopic observation the cause of death for these mice could not be clearly determined.
Some microscopic findings were observed in the bone marrow and spleen of the mock group. They consisted of a marked to severe increase in extramedullary hematopoiesis in the spleen associated with minimal to moderate increase in erythroid cell numbers in the bone marrow and are part of the normal background changes in the Hbbth1/th1 mice [41]. A lower severity of extramedullar hematopoiesis was observed in spleens from the HPV569 or BB305 groups, indicating a correction of the β-thalassemic splenic phenotype (Supp. Table 2). There was no difference in the severity of the extramedullar hematopoiesis in the spleen between the HPV569 or BB305 groups. The decreased severity of hematopoiesis in treated groups correlated with the absence of macroscopic enlargement of the spleen noted at necropsy. Other microscopic findings observed in treated animals were considered incidental changes, as they also occurred in the mock group, were of low incidence and/or are common background findings for the mouse species and for the age of the animals.
Insertion Site Analyses of Primary Recipient Mice Bone Marrow Cells
Insertion site (IS) analysis was performed by both restriction and non-restriction based-linear amplification-mediated PCR and sequencing more than 700,000 DNA fragments. Pre-transplant and 4 months post-transplant samples from the four transduction groups (males and females transduced with HPV569 or BB305) were analyzed. Only a few sites were found duplicated in mice of the same transduction group (6%) and in the pre- and post-transplant samples (1% for HPV569 and 0.2% for BB305). A total of 7767 unique sites were identified, including 4258 for the HPV569 groups and 3509 for the BB305 groups (Fig. 3A). Among the unique sites, 2605 and 1372 were identified in pre-transplant cells samples of the HPV569 and the BB305 groups respectively. Post-transplantation, 1699 and 2143 unique IS were identified in mice receiving HPV569 and BB305 transduced cells respectively. IS analyzes in transplanted mice revealed an oligo/polyclonal hematopoietic repopulation in all four groups, with a mean number of 73 (male) and 94 (females) vector integration per mouse receiving HPV569-transduced cells, and 86 in males and 127 in females for BB305 vector transduced group. The 10 most prominent IS in all four tested groups, pre- and post-transplantation, are given in Fig. (3B). No specific clone was selected in vivo, each representing less than 10% of all detected sequences. Individual mouse data are shown (Supp. Fig. 5).
Fig. (3).
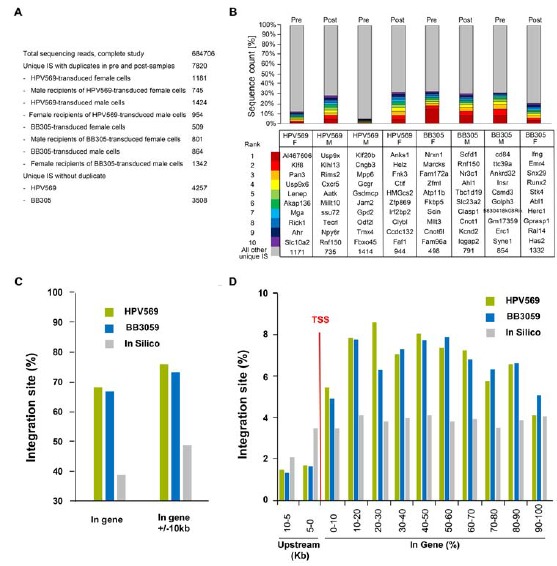
Integration site analysis in primary transplants. A) Total sequencing reads, numbers of unique IS with duplicates in pre- and post-transplant samples and unique IS without duplicates combining male and female data. B) Top ten IS pre- and post-transplantation in all groups of mice receiving transduced cells. C) Percentage of IS within genes and in genes +/- 10kb. D) Percentage of IS upstream and downstream of the transcription start site (TSS).
As expected for lentiviral vectors, around 70% of the IS were localized in genes (Fig. 3C). 1552 and 1146 IS (36% and 33%) were clustered in common insertion sites (CIS) for HPV569 and BB305, respectively (combining in vitro and in vivo data, males and females). CIS were classified as follows: 2 IS within 30 kb for 2nd order, 3 IS within 50 kb for 3rd order, 4 IS within 100 kb for 4th order, and n IS within 200 kb for the nth order [42]. Computer simulations based on an assumed lentiviral vector distribution using the CISLENTIc program [38] showed that the occurrence of CIS for both vectors is significantly higher than expected for a distribution that takes into account the spatial structure of 19513 transcribed RefSeq gene regions of the mouse genome and
the preference of lentiviral vectors to insert into gene regions. Combining all the IS obtained with the two lentiviral vectors (7767 unique IS), 4028 IS (51.9%) were clustered in CIS regions which is in line with previously published data [43, 44] and reflects the differential affinity of individual cellular genes to serve as a target for lentiviral integration. The data suggest that no specific region of the mouse genome was targeted by the LentiGlobin lentiviral vectors overall although the IS were not randomly distributed. Extracting IS from in vivo data (combining male and females) and comparison with in vitro results indicated that departure from semi-random integration did not occur through in vivo reconstitution. In order to further study the risk of in vivo selection, we compared the proportion of IS near oncogenes before and after in vivo repopulation. There was no significant increase of IS near oncogenes in post-transplantation samples compared to the in vitro IS for both lentiviral vectors (Table 2).
Long-Term Safety of LentiGlobin Lentiviral Vectors in Secondary Transplant Mice
In an effort to fully assess the potential impact of insertional mutagenesis on clonal imbalance and/or leukemogenesis, we performed secondary bone marrow transplantation, an alternative assay for oncogenic event detection [45]. Twelve million bone marrow cells of each of the 54 necropsied primary recipients were isolated and transplanted into 108 (one donor bone marrow into 2 recipients) lethally irradiated C57BL/6J CD45.1 mice (Fig. 2A).
The mean donor chimerism as the percentage of CD45.2 cells in blood was determined four and five months after transplantation and was above 85% in all six groups of mice (Supp. Fig. 6). Percentages of myeloid (Gr1/Mac1+) and lymphoid (B220+, CD3+) peripheral blood cells were similar in all groups (Supp. Fig. 6). As observed in primary transplant animals, the percentages of myeloid and lymphoid B cells were slightly but statistically different between males and females (p=0.006 and p=0.01 for myeloid and lymphoid B cells respectively four months post-transplantation) and were independent of the vector. The mean VCN (± SD) in peripheral blood was higher (p = 0.02) for the BB305 groups (2.31 ± 1.40) than for the HPV569 groups (1.63 ± 1.02) at five months post-secondary transplantation. As expected, red blood cell parameters were normalized in both the BB305 and the HPV569 groups but not in the mock-transduced groups (not shown).
Thirteen mice died before sacrifice (n = 8, 3, and 2 in HPV569, BB305, and Mock groups, respectively). Organs and bone marrow cells from these mice were collected for histopathology. Kaplan-Meier survival curves (Supp. Fig. 7) did not show any statistically significant difference in mean survival between mock and LentiGlobin groups (p = 0.27). The 95 surviving mice were sacrificed at 6.8 months after transplantation. Consistent with correction of the thalassemia phenotype, there was a decreased hematopoiesis in the spleen of necropsied animals.
All animals including the prematurely dead were necropsied and microscopic findings are reported per sex and group and individually (data not shown). The most probable causes of premature death are summarized in supplementary Table 3. Systemic malignant lymphoma was seen in 2 animals from the HPV569 groups and 1 animal from the BB305 groups and unclassified leukemia was found in 2 animals from the HPV569 groups. Cutaneous ulcer contributed or was the most probable cause of death for 3 animals (all in the HPV569 group), including two that were prematurely sacrificed. The cause of death could not be determined for the other five mice.
Malignant T cell lymphomas occurred in mice that received cells from donors that were primarily transplanted with either mock transduced or lentiviral vector-transduced cells (Table 3). The lymphomas were either thymic or systemic. For systemic lymphoma, based on the severity of the infiltration, the most likely primary site was the thymus, except for one mouse (F48) for which it could have been thymus or spleen. T-cell origin of the lymphoma cells was confirmed by CD3 immunostaining of the thymus (Supp. Fig. 8). Two mice (M104 and F15) had leukemias that could not be characterized as CD3 positive, possibly due to high autolysis level in those prematurely dead animals. However, the donor cells injected in M104 and F15 were also transplanted in animals F42 and M76 respectively, which also developed malignant lymphomas suggesting that leukemic cells were of the same origin (F16 primary transplant into F15 and M76 secondary transplant; M21 primary transplant into M104 and F42 secondary transplant). Overall, lymphomas/leukemia occurred in 7/44, 6/36, 2/28 animals in HPV569, BB305, and Mock groups, which is statistically not significantly different by Chi-square (p=0.4848). Unilateral lymphoid hyperplasia, involving a single lobe of the thymus, characterized by a loss of the normal cortico-medullary demarcation, also occurred in 4 mice (Table 3). There was no extension beyond the capsule and cells were CD3+CD45RA-.
Table 3. Malignant lymphomas, unclassified leukemias and thymus hyperplasia.
| Vector | Primary Animal | Secondary Animal | Premature Death (Days) or TS | HLP Change | Staining CD3+CD45RA- | Staining CD45.2/CD45.1 |
|---|---|---|---|---|---|---|
| Malignant cells | ||||||
| HPV569 | M21 | M104 | 140 | LEUK | - | equivocal |
| F42 | 203 | ML(S) | + | equivocal | ||
| M25 | M108 | 186 | ML(S) | + | equivocal | |
| F10 | F9 | TS | ML(T2) | + | equivocal | |
| F13 | F12 | TS | ML(T1) | + | CD45.2 | |
| F16 | M76 | TS | ML(T1) | + | CD45.2 | |
| F15 | 142 | LEUK | - | equivocal | ||
| BB305 | M14 | M101 | TS | ML(S) | + | equivocal |
| M24 | M107 | TS | ML(S) | + | CD45.1 | |
| M27 | F48 | TS | ML(S) | + | CD45.1 | |
| F14 | F13 | 192 | ML(S) | + | equivocal | |
| F17 | M77 | TS | ML(S) | + | CD45.2 | |
| F22 | F22 | TS | ML(T2) | + | equivocal | |
| Mock | M18 | F58 | TS | ML(S) | + | equivocal |
| F12 | F11 | TS | ML(S) | + | CD45.2 | |
| Hyperplasia | ||||||
| HPV569 | M7 | F34 | TS | TLH | + | nd |
| BB305 | M22 | M105 | TS | TLH | + | nd |
| F26 | M86 | TS | TLH | + | nd | |
| F28 | F26 | TS | TLH | + | nd | |
TS: Terminal sacrifice (6.8 months post-transplantation); HLP: Hemato-lymphopoietic change; +: CD3+CD45RA-cells; -: CD3-CD45RA-cells; equivocal: either no or faint staining of a low proportion of lymphomatous cells, LEUK: Leukemia, not otherwise specified; ML: Malignant lymphoma (S: systemic; T1: thymic, 1 lobe; T2: thymic, 2 lobes); TLH: Thymic lymphoid hyperplasia; nd: not done.
In order to identify the origin of the proliferative lesions as secondary or non-secondary recipient origin, immunohistochemical staining with anti CD45.2 antibodies (specific of transplanted cells and potentially gene modified) and CD45.1 (specific of the hematopoietic cells of the host) were performed (Table 3 and Supp. Fig. 9). Two animals of the BB305 group had lymphomatous cells of secondary recipient (M107 and F48), whereas four lymphomas were of non-secondary origin (M76 and F12 from the HPV569 group, M77 from BB305 and F11 from the control group). The origins of the nine other lymphoma/leukemia were equivocal, as no or very faint staining of a small proportion of lymphomatous cells could be detected. Normal thymus from two animals (M63 and F2 of the control group) which served as a control showed strong positive CD45.2 staining. It is possible that equivocal staining for lymphomas may be related to poor differentiation of tumor cells and consequently low expression of panleukocytic marker CD45. Cytometric analysis of bone marrow cells led to congruent conclusions as total donor chimerism was lower for mice F48, M107 and M101 as compared to other animals (Supp. Fig. 10). The donor chimerism of bone marrow T-cells in those mice was also very low, whereas the proportion of the overall T cells fraction was higher in mice M101 and M107 than in others. Lymphoma cells were detected in bone marrow of those three mice at a grade level between three and four.
In conclusion, the frequencies of mice with lymphoma/leukemia possibly coming from transduced donor cells were 7/44 (M76, M104, M108, F9, F12, F15, F42), 4/36 (M77, M101, F13, F22) and 2/28 (F11, F58) for HPV569, BB305 and control mice, respectively. The numbers of animals with unclassified leukemia, malignant lymphoma and lymphoid hyperplasia were compiled per group for each sex and pooled sexes (Supp. Table 4). There was no statistical difference as detected by Chi-square and Fisher exact tests between tumor occurrence and transduction with the LentiGlobin lentiviral vectors even when the mice with tumor cells from recipient origin were included (Supp. Table 5).
Table 4. Vector copy number in thymus of mice with malignant tumors.
| Vector Group | Sex | Animal Number | Tumor | Tumor Cells (%) | VCN |
|---|---|---|---|---|---|
| HPV569 | Male | 73 | - | NA | 0.054 |
| 76 | ML | ≈70% | 0.490 | ||
| 108 | ML | >90% | 0.389 | ||
| Female | 9 | ML | >95% | 0.076 | |
| 12 | ML | ≈70% | 0.017 | ||
| 15 | LEUK | >90% | 0.056 | ||
| 42 | ML | >90% | 0.004 | ||
| BB305 | Male | 77 | ML | >95% | 0.071 |
| 83 | - | NA | 5.511 | ||
| 101 | ML | >95% | 0.004 | ||
| 107 | ML | >95% | 0.101 | ||
| Female | 13 | ML | >95% | 0.125 | |
| 16 | - | NA | 1.389 | ||
| 22 | ML | >95% | 0.261 | ||
| 48 | ML | >95% | 0.059 | ||
| Mock | Male | 11 | ML | >95% | <0.001 |
| 5 | - | NA | <0.001 | ||
| 8 | - | NA | <0.001 | ||
| Female | 17 | - | NA | <0.001 | |
| 58 | ML | >95% | <0.001 |
*ML: Malignant lymphoma; LEUK: Leukemia of unknown origin; -: no tumor; NA: not applicable
Determination of VCN in Secondary Transplant Mice
In order to answer the key question as to whether the integration of the vectors could be involved in tumor generation, vector quantification was performed on DNA extracted from sections of fixed thymuses with high proportion of tumor cells. Microscopic analysis of tissue section by a board-certified pathologist showed that more than 90% of the cells were malignant in tissue sections of mice with malignant lymphomas and unclassified leukemias except mice F12 and M76 from the HPV569 group (Table 4). The thymus of mouse 104 was not included in this study as the tissue procured at necropsy from the thymic location was skeletal muscle infiltrated by tumor cells. Each extracted DNA sample was quantified and amplified by qPCR using endogenous controls for assessment of DNA quality (not shown). Vector copy number determination is shown in Table 4 and compared to the percentage of tumor cells. Although we cannot rule out that tumor cells arose from more than one clone in the same mouse (one of which containing the vector), we considered this event to be very unlikely. We then assume that if vector mediated insertional mutagenesis was the tumor initiating event(s), all tumor cells would contain at least one copy of the vector. None of the DNA extracted from tumor samples had a mean VCN at or above this threshold (equal to the fraction of tumor cells in the tissue section), indicating that vector insertion was not the cause of tumor development (Table 4). In order to affirm that no recombination event may have led to inaccurate vector quantification, two sets of primers and probe were used for qPCR, which gave very similar results.
Integration Site Analysis in Secondary Transplant Mice
IS profiles of the vectors were assessed in bone marrow cells of all mice together with prematurely dead animals. A total of 3068 unique sites, including 1353 and 1715 for HPV569 and BB305, respectively, were identified. As expected, vectors integrated preferentially within gene coding region (Figs. 4A and 4), though less pronounced than in primary transplant. Again, computer simulation using the CISLENTIc program established that numbers of CIS (27.4% and 32.3% for samples transduced with HPV569 and BB305, respectively) were higher than expected (data not shown). Comparison of the ten most prominent IS in primary and secondary recipients is given in Fig. (4C). Some of the top ten IS found in primary recipients were also detected among the most frequent clones of secondary transplanted animals but were not more preponderant. Reconstitution clonality was similar to that observed in primary transplanted mice (Supp. Fig. 5 and Supp. Fig. 11). Interestingly, none of the mice whose BM was invaded with lymphomatous cells (animals F15 and F42 from HPV569 group and M101, M107, F13 and F48 from BB305 group) had a dominant clone. As expected due to selection of long-term reconstituting cells in primary and secondary transplant, clonal diversity decreased between pre- and post-transplant samples (Fig. 4D). Injecting a limited number of hematopoietic cells in secondary transplants and extra selection for long-term stem cells further reduced the clonal diversity in secondary transplanted animals. The decreased clonal diversity from primary to secondary animals was independent from the vector used (Fig. 4D).
Fig. (4).
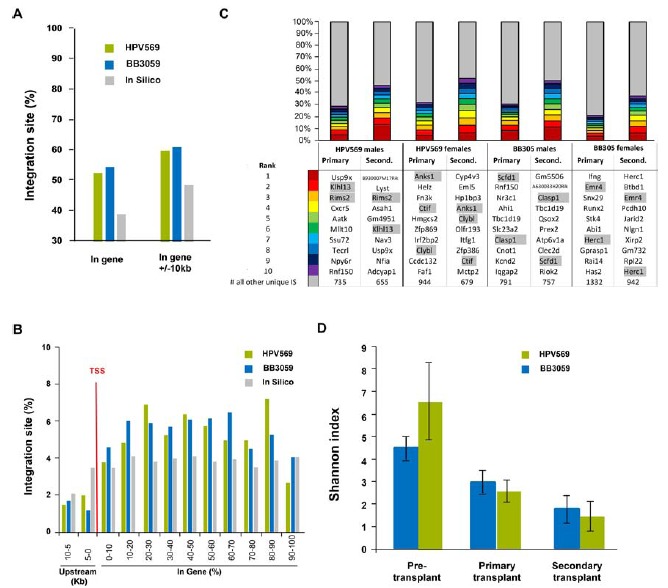
Integration site analysis in secondary transplants. A) Percentage of IS within genes and in genes +/- 10kb. B) Percentage of IS upstream and downstream of the transcription start site (TSS). C) Top ten IS comparing primary and secondary transplanted animals. D) Diversity index in pre- and post-transplant samples.
No enrichment for genes listed in oncogene databases were observed between samples obtained from primary and secondary transplants (Table 2). No enrichment for genes listed in RTCGD and CGC database was observed, but the data set with the HPV569 and BB305 lentiviral vector showed a statistically significant enrichment for genes listed in the cBio Portal as compared to in silico data. Since the CGC and cBio represent human genes and this integration study was conducted in mouse BM cells, the RTCGD database that represents mouse genes might be more relevant. There was no preferred integration in the MECOM (MDS1-EVI1), in LMO2 or in HMGA2 gene loci.
DISCUSSION
We previously reported that a patient with β-thalassemia major (βE/β0) became transfusion independent after autologous transplantation with CD34+ cells transduced with the LentiGlobin HPV569 lentiviral vector [3]. However, long-term VCNs in subsets of blood cells of this patient were relatively low (0.1-0.2 copy per cell), and analysis of some IS showed partial deletion of the cHS4 element [23] with abnormal splicing and transcriptional activation of the HMGA2 gene [3].
In order to improve the vector titers and stability, the new LentiGlobin vector (BB305) was constructed by removing the cHS4 sequences and incorporating a chimeric CMV/HIV LTR sequence to drive high level expression of the viral transcript. Results showed that the LentiGlobin BB305 lentiviral vector could be produced with approximately 3- to 4-fold higher functional titers than the LentiGlobin HPV569 lentiviral vector with a better ratio of functional to physical particles. This higher titer was expected from earlier studies where insulator sequences were removed [20-22]. We show here that additional modification of the lentiviral enhancer to a CMV promoter further increased the vector production. Additionally, at an equivalent MOI, LentiGlobin BB305 transduced human hematopoietic cells more effectively than the LentiGlobin HPV569, with the average VCN approximately 3-fold higher in late and early human progenitors. In mouse hematopoietic progenitors (Lin- cells), relative transduction efficacy was slightly but not highly different, suggesting that mouse cells were less sensitive that human progenitors to the functional to physical particle ratio. In vitro immortalization (IVIM) assays [27, 39] in mouse cells showed equivalent safety for both lentiviral vectors, indicating a strongly reduced risk of genotoxicity on murine hematopoietic cells as compared to control retroviral vectors and lentiviral vectors containing the strong SFFV viral promoter.
Based on these encouraging results, we designed an in vivo study aimed to evaluate and compare the safety and efficacy of the new LentiGlobin BB305 to the LentiGlobin HPV569 to support the use of LentiGlobin BB305 in clinical studies for the treatment of β-thalassemia major and severe sickle cell disease. Bone marrow cells from male and female β-thalassemia (Hbbth1/th1) mice were transduced and transplanted into β-thalassemic Hbbth1/th1 mice. Engraftment and
transduction were efficient and the β-thalassemic phenotype was fully corrected. Careful microscopic observation of organs, blood biochemistry, and hematology did not show any vector-transduced cell-related safety concerns.
“IS analyses of the pre-transplant samples showed a highly polyclonal vector integration profile for both lentiviral vectors” [40]. In primary recipients, a total of 3509 and 4258 unique IS were identified for BB305 and HPV569, respectively. As expected from previous studies [46] the IS profile demonstrated a preferred integration (68.2% and 66.8% for HPV569 and BB305, respectively) in gene coding regions. Host factors (i.e. Lens-Epithelium Growth Factor, LEDGF), gene expression state, chromatin state and other so far unknown factors seem to contribute to this target site selection [46-48].
Despite a general tendency of lentiviral vectors to insert in clusters, this integration profile has not been reported to be accompanied by severe adverse events in patients [1-6]. In contrast, integration of gamma retroviral vectors into preferred IS has been associated with upregulation of MDS1-EVI1 in patients treated for chronic granulomatous disease [49] or LMO2 in patients with X-chromosomal linked severe combined immunodeficiency and Wiskott Aldrich syndrome [50-52] leading to malignant transformation in the affected patients.
Neither LentiGlobin HPV569 nor LentiGlobin BB305 lentiviral vectors showed preferred integration in either of the two “dangerous gene loci” MDS1-EVI1 or LMO2. One sample showed an integration site 74 kb upstream to CCND2 and three samples showed three unique IS within the fourth intron of HMGA2, but with a low clonal contribution ranging between 0.12% and 1.10%. The transplantation of 2.8x105 lineage negative mouse bone marrow cells regularly led to an oligo- to polyclonal hematopoietic repopulation for both vectors. Insertions near oncogenes were not favored.
In order to get insight into the potential impact of insertional mutagenesis on leukemogenesis, secondary bone marrow transplantation was performed. The efficacy of engraftment was high (≥ 85%) and the average VCN was similar to that found in primary transplants. Macroscopic findings (spleen enlargement, tumor mass, abnormal tumor size) seen in premature decedents and at terminal sacrifice in animals from all groups correlated with malignant lymphoma microscopically. However, in view of the presence of malignancies in the mock group, a relationship to the treatment could not be established for these findings. With the aim to determine whether the tumors could have been induced by insertional mutagenesis, the average VCN per cell and the proportion of tumor cells were assessed in the thymuses of affected animals. The VCN detected in the tumor were below the threshold of one copy per tumor cell, arguing against LentiGlobin BB305 or HPV569 lentiviral vector integration as a tumor initiating event.
“Lymphomas are among the most common tumors in many strains of mice, especially those used in safety assessment. CD-1, C57BL/6, B6C3F1 and B6; 129 mice develop 10–50% incidences of lymphomas in aging mice. Lymphomas and leukemias may be induced by chemicals, retroviruses or irradiation. Genetics also play a major role in mouse lymphomagenesis and leukemogenesis” [53-57]. Our findings are consistent with numerous published reports describing spontaneous lymphomas in mice receiving genetically modified cells and undergoing radiation. Will et al. [56] reported lymphoma in mice transplanted with ex vivo manipulated bone marrow cells to assess the long-term effects of the transduction of hematopoietic cells with the retroviral vector MSCV-MGMTP140Kwc. In this study, six of 38 test animals developed a malignancy, whereas no malignancies developed in control animals. Malignant lymphomas were observed at a much higher frequency than expected (as high as 28.6%) after 12 months observation and it was determined that at least five of the six lymphomas were vector negative, arguing against vector integration or ectopic MGMTP140K expression as a cause of tumor development. Furthermore, Ginn et al [58] reported that 4 of 14 common gamma-chain (γc) deficient mice reconstituted with a “vector expressing the γc from an elongation factor-1-α (EF1α) promoter subsequently developed lymphoma. Extensive analyses failed to implicate insertional mutagenesis or γc overexpression as the underlying mechanism, highlighting the need for detailed mechanistic analysis of tumor readouts in preclinical animal models assessing vector safety, and suggesting the existence of other ill-defined risk factors for oncogenesis, including replicative stress, in gene therapy protocols targeting the hematopoietic compartment” [58].
The IS analyses in bone marrow samples derived from secondary transplant mice revealed an oligoclonal hematopoietic repopulation. However, we found that the proportion of integrations within genes decreased over time, maybe reflecting that “integration of lentiviral vectors into genes more often leads to a fitness cost than a fitness advantage”, as we previously suggested [23]. The reduction of clonality in the secondary transplant animals was expected and is due to the reduced clonal diversity present in the bone marrow used from the primary recipients (twelve million cells injected in two mice out of the total bone marrow). Progressive selection of surviving stem cells may also contribute to this phenomenon. As for primary transplants, there was no preferred integration in the MECOM, in LMO2 or in HMGA2 genes. One sample showed an integration site next to MECOM, but with a low clonal contribution of 0.02%. The identified integration profiles showed no substantial differences between both groups.
The detected clustering of lentiviral IS in a small number of genomic sites is as expected from previously described IS profiles in mouse and human hematopoietic cells. Notably, there were no recurrent integrations found in the secondary animals that i) were located near oncogenes that previously triggered malignant transformation in individual patients taking part in gamma-retroviral gene therapy trials to treat immunodeficiencies, ii) were already present in primary transplants and showing elevated frequencies of clonal contribution that may point to in vivo clonal dominance and iii) were found in genes recurrently listed in known oncogene databases.
conclusion
In conclusion, overall phenotype correction of β-thalassemic mice was observed with both LentiGlobin HPV569 and BB305 lentiviral vectors with no alteration of bone marrow homeostasis in the primary transplant animals. No evidence of toxic effect related to transplantation with Lin- bone marrow cells transduced with either HPV569 of BB305 was observed. The integration patterns of the two vectors BB305 and HPV569 were similar, mostly within or close to RefSeq genes, without any sign of clonal outgrowth or in vivo selection. Leukemias and thymic lymphomas were observed in several secondary transplants animals. With the new LentiGlobin BB305 lentiviral vector, comprehensive analyses of thymuses containing tumor cells by immunohistochemistry and quantitative PCR, showed that tumor cells were derived from either primary or secondary recipient and not from transduced donor cells. The high baseline rate of malignancy observed in the secondary transplant model is in agreement with the published literature and questioning the usefulness of secondary bone marrow transplantation to evaluate lentiviral vector safety.
In summary we have developed a second generation Lentiglobin vector (BB305) with improved transduction efficiency and manufacturing characteristics. In both, in vitro assays and in vivo animal models, BB305 has demonstrated improved efficacy that was not associated with insertional oncogenesis. Clinical trials of BB305 are underway in both β-thalassemia and sickle cell disease in Europe and for β-thalassemia in the USA. Preliminary clinical results showing the advantage of the new Lentiglobin BB305 lentiviral vector over the previous HPV569 vector in two β-thalassemia patients were reported at the 2014 annual meeting of the European Hematology Association and the 2014 annual meeting of the American Society of Hematology and can be read at http://investor.bluebirdbio.com/phoenix.zhtml?c=251820&p=irol-newsArticle&ID=1939867 and at http://investor.bluebirdbio.com/phoenix.zhtml?c=251820&p=irol-newsArticle&ID=1995913, respectively.
Patient Consent
Declared none.
Table 2. Distribution of IS near oncogenes in pre-transplanted cells and in vivo (primary and secondary transplanted animals).
| Genes of Cancer Gene Census | ||||||
|---|---|---|---|---|---|---|
| Vector | Pre / Post-Transplant |
Unique IS
Number |
Frequency of IS Next to CGC Genes (%) |
p-value
Compared to In silico Data Set* |
p-value Comparing Pre Transplant Samples and
Primary Mice* |
p-value
Comparing Primary and Secondary Mice* |
| HPV569 + BB305 | Pre | 3978 | 4.7 | 1.7 x 10-7 | 0.1 | 0.95 |
| Primary | 3842 | 4.0 | 6.0 x 10-4 | |||
| Secondary | 3068 | 3.9 | 2.0 x 10-3 | |||
| HPV569 | Pre | 2605 | 4.4 | 7.0 x 10-5 | 0.6 | 0.71 |
| Primary | 1699 | 4.1 | 7.0 x 10-2 | |||
| Secondary | 1353 | 3.8 | 5.2 x 10-2 | |||
| BB305 | Pre | 1372 | 5.2 | 6.0 x 10-6 | 0.07 | 0.8 |
| Primary | 2143 | 3.9 | 8.0 x 10-2 | |||
| Secondary | 1715 | 4.1 | 6.0 x 10-3 | |||
| In silico | 7108 | 2.8 | ||||
| Genes of RTCGD Database | ||||||
| HPV569 + BB305 | Pre | 3978 | 2.5 | 0.4 | 0.7 | 0.13 |
| Primary | 3842 | 2.3 | 0.8 | |||
| Secondary | 3068 | 1.8 | 0.15 | |||
| HPV569 | Pre | 2605 | 2.9 | 0.06 | 0.09 | 1 |
| Primary | 1699 | 2.1 | 0.7 | |||
| Secondary | 1353 | 2.1 | 0.7 | |||
| BB305 | Pre | 1372 | 1.7 | 0.2 | 0.1 | 0.04 |
| Primary | 2143 | 2.6 | 0.4 | |||
| Secondary | 1715 | 1.6 | 0.09 | |||
| In Silico | 7108 | 2.3 | ||||
| Genes of cBio Cancer Genomics Portal | ||||||
| HPV569 + BB305 | Pre | 3978 | 23.9 | 1.2 x 10-7 | 0.05 | 0.09 |
| Primary | 3842 | 22.1 | 2.0 x 10-3 | |||
| Secondary | 3068 | 20.4 | 0.39 | |||
| HPV569 | Pre | 2605 | 24.0 | 3.0 x 10-6 | 0.03 | 0.23 |
| Primary | 1699 | 21.1 | 0.2 | |||
| Secondary | 1353 | 19.4 | 0.85 | |||
| BB305 | Pre | 1372 | 23.7 | 7.0 x 10-4 | 0.6 | 0.21 |
| Primary | 2143 | 22.9 | 1.0 x 10-3 | |||
| Secondary | 1715 | 21.2 | 0.15 | |||
| In Silico | 7108 | 19.6 | ||||
*Fisher Exact Test, two-tailed
ACKNOWLEDGEMENTs
This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM) and an ANR Chaire Industrielle to P.L.
Olivier Negre, Gabor Veres, Byoung Ryu, Emmanuel Payen, Michael Rothe, Raffaele Fronza and Manfred Schmidt contributed to the design, performance, analysis and reporting of the work.
Christof von Kalle, Marina Cavazzana, Yves Beuzard, Mitchell Finer and Philippe Leboulch contributed to the design and the reporting of the work.
Cynthia Bartholomae, Lauryn Christiansen, Anaïs Paulard, Céline Courne, Annette Deichmann, Béatrix Gillet-Legrand, Francis J. Pierciey Jr, Christophe Joubert and Robert Kutner contributed to the performance and analysis of the work.
Maria Denaro, Edouard de Dreuzy and Leïla Maouche contributed to the analysis and reporting of the work.
We are grateful to Véronique Neuville for her expertise in animal care and Jean-Baptiste Lahaye for mouse irradiations.
We thank Anne-Virginie Eggimann and Holly Horton from bluebird bio for suggestions and help in preparation of the manuscript; Cécile Sobry, Fleurance Renaud and Frédéric Gervais from CiToxLAB for contributing to the in vivo mouse study.
LIST OF ABBREVIATIONS
- ANR
Agence nationale de la recherche
- CCALD
Childhood cerebral adrenoleukodystrophy
- CCND2
Cyclin D2
- CD
Cluster of differentiation
- CEA
Commissariat à l’énergie atomique et aux énergies alternatives
- CFC
Colony forming cell
- CIS
Common insertion site
- CMV
Cytomegalovirus
- Ct
Cycle threshold
- DNA
Deoxyribonucleic acid
- dsDNA
Double stranded DNA
- EDTA
Ethylenediaminetetraacetic acid
- EVI1
Ecotropic viral integration site 1
- FFPE
Formalin –fixed paraffin embedded
- FLT3-L
Fms-like tyrosine kinase 3-ligand
- GVHD
Graft-versus-host disease
- HIV
Human immunodeficiency virus
- HLA
Human leukocyte antigen
- HMGA2
High mobility group A2
- HS4
Hypersensitive site-4
- HSCT
Hematopoietic stem cell transplantation
- IL-11
Interleukin-11
- IL-3
Interleukin-3
- IL-6
Interleukin-6
- INSERM
Institut national de la santé et de la recherche médicale
- IS
Integration site
- IVIM
In vitro immortalization assay
- LAM PCR
Linear amplification mediated polymerase chain reaction
- LEDGF
Lens-epithelium growth factor
- LMO-2
LIM domain only 2
- LTC-IC
Long-term culture initiating cell
- LTR
Long terminal repeat
- MDS1
Myelodysplastic syndrome 1
- MECOM
MDS1 and EVI1 complex locus
- MOI
Multiplicity of infection
- nr LAM-PCR
Non-restrictive LAM-PCR
- PPT
Polypurine tract
- qPCR
Quantitative polymerase chain reaction
- RRE
Rev-responsive element
- RTCGD
Retrovirus and transposon tagged cancer gene database
- SA
Splice acceptor site
- SCF
Stem cell factor
- SD
Standard deviation
- SIN
Self-inactivating
- TBI
Total body irradiation
- TPO
Thrombopoietin
- TSS
Transcription start site
- UV
Ultra violet
- VCN
Vector copy number
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publishers web site along with the published article.
CONFLICT OF INTEREST
Olivier Negre, Lauryn Christiansen, Céline Courne, Maria Denaro, Mitchell Finer, Béatrix Gillet-Legrand, Robert Kutner, Anaïs Paulard, Francis J. Pierciey Jr., Byoung Ryu and Gabor Veres are employees of bluebird bio Inc. and receive salary and other compensations from the company.
Philippe Leboulch, Emmanuel Payen, Manfred Schmidt and Christof von Kalle are consultants for bluebird bio and receive financial compensation.
REFERENCES
- 1.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., et al. Lentiviral hematopoietic cell gene therapy for X-linked adrenoleukodystrophy. Methods Enzymol. 2012;507:187–198. doi: 10.1016/B978-0-12-386509-0.00010-7. [DOI] [PubMed] [Google Scholar]
- 2.Cartier N., Hacein-Bey-Abina S., Bartholomae C.C., et al. Hematopoietic stem cell gene therapy with a lentiviral vector in X-linked adrenoleukodystrophy. Science. 2009;326:818–823. doi: 10.1126/science.1171242. [DOI] [PubMed] [Google Scholar]
- 3.Cavazzana-Calvo M., Payen E., Negre O., et al. Transfusion independence and HMGA2 activation after gene therapy of human beta-thalassaemia. Nature. 2010;467:318–322. doi: 10.1038/nature09328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Payen E, Leboulch P. Advances in stem cell transplantation and gene therapy in the beta-hemoglobinopathies. 2012. [DOI] [PubMed]
- 5.Biffi A., Montini E., Lorioli L., et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 6.Aiuti A., Biasco L., Scaramuzza S., et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science. 2013;341:1233151. doi: 10.1126/science.1233151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galanello R., Origa R. Beta-thalassemia. Orphanet J. Rare Dis. 2010;5:11. doi: 10.1186/1750-1172-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isgro A., Gaziev J., Sodani P., et al. Progress in hematopoietic stem cell transplantation as allogeneic cellular gene therapy in thalassemia. Ann. N. Y. Acad. Sci. 2010;1202:149–154. doi: 10.1111/j.1749-6632.2010.05543.x. [DOI] [PubMed] [Google Scholar]
- 9.Lucarelli G., Galimberti M., Polchi P., et al. Marrow transplantation in patients with advanced thalassemia. N. Engl. J. Med. 1987;316:1050–1055. doi: 10.1056/NEJM198704233161703. [DOI] [PubMed] [Google Scholar]
- 10.Caocci G., Efficace F., Ciotti F., et al. Prospective assessment of health-related quality of life in pediatric patients with beta-thalassemia following hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2011;17:861–866. doi: 10.1016/j.bbmt.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 11.Luznik L., Jones R.J., Fuchs E.J. High-dose cyclophosphamide for graft-versus-host disease prevention. Curr. Opin. Hematol. 2010;17:493–499. doi: 10.1097/MOH.0b013e32833eaf1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borgna-Pignatti C. The life of patients with thalassemia major. Haematologica. 2010;95:345–348. doi: 10.3324/haematol.2009.017228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Modell B., Khan M., Darlison M., et al. Improved survival of thalassaemia major in the UK and relation to T2*. Cardiovasc Magn Reson. 2008;10:42. doi: 10.1186/1532-429X-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thuret I., Pondarre C., Loundou A., et al. Complications and treatment of patients with beta-thalassemia in France: results of the National Registry. Haematologica. 2010;95:724–729. doi: 10.3324/haematol.2009.018051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bank A., Dorazio R., Leboulch P. A phase I/II clinical trial of beta-globin gene therapy for beta-thalassemia. Ann. N. Y. Acad. Sci. 2005;1054:308–316. doi: 10.1196/annals.1345.007. [DOI] [PubMed] [Google Scholar]
- 16.Recillas-Targa F., Pikaart M.J., Burgess-Beusse B., et al. Position-effect protection and enhancer blocking by the chicken beta-globin insulator are separable activities. Proc. Natl. Acad. Sci. USA. 2002;99:6883–6888. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell A.C., West A.G., Felsenfeld G. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999;98:387–396. doi: 10.1016/s0092-8674(00)81967-4. [DOI] [PubMed] [Google Scholar]
- 18.Ryu B.Y., Evans-Galea M.V., Gray J.T., et al. An experimental system for the evaluation of retroviral vector design to diminish the risk for proto-oncogene activation. Blood. 2008;111:1866–1875. doi: 10.1182/blood-2007-04-085506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Desprat R., Bouhassira E.E. Gene specificity of suppression of transgene-mediated insertional transcriptional activation by the chicken HS4 insulator. PLoS One. 2009;4:e5956. doi: 10.1371/journal.pone.0005956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanawa H., Yamamoto M., Zhao H., et al. Optimized lentiviral vector design improves titer and transgene expression of vectors containing the chicken beta-globin locus HS4 insulator element. Mol. Ther. 2009;17:667–674. doi: 10.1038/mt.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jakobsson J., Rosenqvist N., Thompson L., et al. Dynamics of transgene expression in a neural stem cell line transduced with lentiviral vectors incorporating the cHS4 insulator. Exp. Cell Res. 2004;298:611–623. doi: 10.1016/j.yexcr.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 22.Nielsen T.T., Jakobsson J., Rosenqvist N., et al. Incorporating double copies of a chromatin insulator into lentiviral vectors results in less viral integrants. BMC Biotechnol. 2009;9:13. doi: 10.1186/1472-6750-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ronen K., Negre O., Roth S., et al. Distribution of lentiviral vector integration sites in mice following therapeutic gene transfer to treat beta-thalassemia. Mol. Ther. 2011;19:1273–1286. doi: 10.1038/mt.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kutner R.H., Puthli S., Marino M.P., et al. Simplified production and concentration of HIV-1-based lentiviral vectors using HYPERFlask vessels and anion exchange membrane chromatography. BMC Biotechnol. 2009;9:10. doi: 10.1186/1472-6750-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutner R.H., Zhang X.Y., Reiser J. Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors. Nat. Protoc. 2009;4:495–505. doi: 10.1038/nprot.2009.22. [DOI] [PubMed] [Google Scholar]
- 26.Payen E., Colomb C., Negre O., et al. Lentivirus vectors in beta-thalassemia. Methods Enzymol. 2012;507:109–124. doi: 10.1016/B978-0-12-386509-0.00006-5. [DOI] [PubMed] [Google Scholar]
- 27.Modlich U., Navarro S., Zychlinski D., et al. Insertional transformation of hematopoietic cells by self-inactivating lentiviral and gammaretroviral vectors. Mol. Ther. 2009;17:1919–1928. doi: 10.1038/mt.2009.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Skow L.C., Burkhart B.A., Johnson F.M., et al. A mouse model for beta-thalassemia. Cell. 1983;34:1043–1052. doi: 10.1016/0092-8674(83)90562-7. [DOI] [PubMed] [Google Scholar]
- 29.Rouyer-Fessard P., Leroy-Viard K., Domenget C., et al. Mouse beta thalassemia, a model for the membrane defects of erythrocytes in the human disease. J. Biol. Chem. 1990;265:20247–20251. [PubMed] [Google Scholar]
- 30.Negre O., Fusil F., Colomb C., et al. Correction of murine beta-thalassemia after minimal lentiviral gene transfer and homeostatic in vivo erythroid expansion. Blood. 2011;117:5321–5331. doi: 10.1182/blood-2010-01-263582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kittel B., Ruehl-Fehlert C., Morawietz G., et al. Revised guides for organ sampling and trimming in rats and mice--Part 2. A joint publication of the RITA and NACAD groups. Exp. Toxicol. Pathol. 2004;55:413–431. doi: 10.1078/0940-2993-00349. [DOI] [PubMed] [Google Scholar]
- 32.Morawietz G., Ruehl-Fehlert C., Kittel B., et al. Revised guides for organ sampling and trimming in rats and mice--Part 3. A joint publication of the RITA and NACAD groups. Exp. Toxicol. Pathol. 2004;55:433–449. doi: 10.1078/0940-2993-00350. [DOI] [PubMed] [Google Scholar]
- 33.Ruehl-Fehlert C., Kittel B., Morawietz G., et al. Revised guides for organ sampling and trimming in rats and mice--part 1. Exp. Toxicol. Pathol. 2003;55:91–106. [PubMed] [Google Scholar]
- 34.Paruzynski A., Arens A., Gabriel R., et al. Genome-wide high-throughput integrome analyses by nrLAM-PCR and next-generation sequencing. Nat. Protoc. 2010;5:1379–1395. doi: 10.1038/nprot.2010.87. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt M., Schwarzwaelder K., Bartholomae C., et al. High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR). Nat. Methods. 2007;4:1051–1057. doi: 10.1038/nmeth1103. [DOI] [PubMed] [Google Scholar]
- 36.Gabriel R., Eckenberg R., Paruzynski A., et al. Comprehensive genomic access to vector integration in clinical gene therapy. Nat. Med. 2009;15:1431–1436. doi: 10.1038/nm.2057. [DOI] [PubMed] [Google Scholar]
- 37.Arens A., Appelt J.U., Bartholomae C.C., et al. Bioinformatic clonality analysis of next-generation sequencing-derived viral vector integration sites. Hum. Gene Ther. Methods. 2012;23:111–118. doi: 10.1089/hgtb.2011.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abel U., Deichmann A., Nowrouzi A., et al. Analyzing the number of common integration sites of viral vectors--new methods and computer programs. PLoS One. 2011;6:e24247. doi: 10.1371/journal.pone.0024247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Modlich U., Bohne J., Schmidt M., et al. Cell-culture assays reveal the importance of retroviral vector design for insertional genotoxicity. Blood. 2006;108:2545–2553. doi: 10.1182/blood-2005-08-024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Negre O., Bartholoma C., Kutner R., et al. Abstract 674 of the American Society of Gene & Cell Therapy (ASGGT) 16th Annual Meeting. May 15-18, 2013. Salt Lake City, Utah, USA. Mol. Ther. 2013;21(Suppl. 1):s257. doi: 10.1038/mt.2013.82. [DOI] [PubMed] [Google Scholar]
- 41.Imren S., Payen E., Westerman K.A., et al. Permanent and panerythroid correction of murine beta thalassemia by multiple lentiviral integration in hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 2002;99:14380–14385. doi: 10.1073/pnas.212507099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deichmann A., Hacein-Bey-Abina S., Schmidt M., et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 2007;117:2225–2232. doi: 10.1172/JCI31659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biffi A., Bartolomae C.C., Cesana D., et al. Lentiviral vector common integration sites in preclinical models and a clinical trial reflect a benign integration bias and not oncogenic selection. Blood. 2011;117:5332–5339. doi: 10.1182/blood-2010-09-306761. [DOI] [PubMed] [Google Scholar]
- 44.Tubsuwan A., Abed S., Deichmann A., et al. Parallel assessment of globin lentiviral transfer in induced pluripotent stem cells and adult hematopoietic stem cells derived from the same transplanted beta-thalassemia patient. Stem Cells. 2013;31:1785–1794. doi: 10.1002/stem.1436. [DOI] [PubMed] [Google Scholar]
- 45.Kustikova O., Fehse B., Modlich U., et al. Clonal dominance of hematopoietic stem cells triggered by retroviral gene marking. Science. 2005;308:1171–1174. doi: 10.1126/science.1105063. [DOI] [PubMed] [Google Scholar]
- 46.Schroder A.R., Shinn P., Chen H., et al. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 47.Ciuffi A., Llano M., Poeschla E., et al. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 48.Marshall H.M., Ronen K., Berry C., et al. Role of PSIP1/LEDGF/p75 in lentiviral infectivity and integration targeting. PLoS One. 2007;2:e1340. doi: 10.1371/journal.pone.0001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ott M.G., Schmidt M., Schwarzwaelder K., et al. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- 50.Avedillo Diez I., Zychlinski D., Coci E.G., et al. Development of novel efficient SIN vectors with improved safety features for Wiskott-Aldrich syndrome stem cell based gene therapy. Mol. Pharm. 2011;8:1525–1537. doi: 10.1021/mp200132u. [DOI] [PubMed] [Google Scholar]
- 51.Boztug K., Schmidt M., Schwarzer A., et al. Stem-cell gene therapy for the Wiskott-Aldrich syndrome. N. Engl. J. Med. 2010;363:1918–1927. doi: 10.1056/NEJMoa1003548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 53.Gold L.S., Manley N.B., Slone T.H., et al. Compendium of chemical carcinogens by target organ: results of chronic bioassays in rats, mice, hamsters, dogs, and monkeys. Toxicol. Pathol. 2001;29:639–652. doi: 10.1080/019262301753385979. [DOI] [PubMed] [Google Scholar]
- 54.Taddesse-Heath L., Chattopadhyay S.K., Dillehay D.L., et al. Lymphomas and high-level expression of murine leukemia viruses in CFW mice. J. Virol. 2000;74:6832–6837. doi: 10.1128/jvi.74.15.6832-6837.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ward J.M. Lymphomas and leukemias in mice. Exp. Toxicol. Pathol. 2006;57:377–381. doi: 10.1016/j.etp.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 56.Will E., Bailey J., Schuesler T., et al. Importance of murine study design for testing toxicity of retroviral vectors in support of phase I trials. Mol. Ther. 2007;15:782–791. doi: 10.1038/sj.mt.6300083. [DOI] [PubMed] [Google Scholar]
- 57.Boorman G.A., Rafferty C.N., Ward J.M., et al. Leukemia and lymphoma incidence in rodents exposed to low-frequency magnetic fields. Radiat. Res. 2000;153:627–636. doi: 10.1667/0033-7587(2000)153[0627:laliir]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 58.Ginn S.L., Liao S.H., Dane A.P., et al. Lymphomagenesis in SCID-X1 mice following lentivirus-mediated phenotype correction independent of insertional mutagenesis and gammac overexpression. Mol. Ther. 2010;18:965–976. doi: 10.1038/mt.2010.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publishers web site along with the published article.


