Abstract
Background and study aims: Biopsy sampling error can be a problem for the diagnosis of certain gastrointestinal tract diseases. Spectrally-encoded confocal microscopy (SECM) is a high-speed reflectance confocal microscopy technology that has the potential to overcome sampling error by imaging large regions of gastrointestinal tract tissues. The aim of this study was to test a recently developed SECM endoscopic probe for comprehensively imaging large segments of the esophagus at the microscopic level in vivo.
Methods: Topical acetic acid was endoscopically applied to the esophagus of a normal living swine. The 7 mm diameter SECM endoscopic probe was transorally introduced into the esophagus over a wire. Optics within the SECM probe were helically scanned over a 5 cm length of the esophagus. Confocal microscopy data was displayed and stored in real time.
Results: Very large confocal microscopy images (length = 5 cm; circumference = 2.2 cm) of swine esophagus from three imaging depths, spanning a total area of 33 cm2, were obtained in about 2 minutes. SECM images enabled the visualization of cellular morphology of the swine esophagus, including stratified squamous cell nuclei, basal cells, and collagen within the lamina propria.
Conclusions: The results from this study suggest that the SECM technology can rapidly provide large, contiguous confocal microscopy images of the esophagus in vivo. When applied to human subjects, the unique comprehensive, microscopic imaging capabilities of this technology may be utilized for improving the screening and surveillance of various esophageal diseases.
Introduction
Standard of care endoscopic biopsy suffers from sampling error, which can limit the diagnosis of many esophageal diseases. Confocal laser endomicroscopy (CLE) is a recently introduced imaging technique where a microscopy probe is inserted into the gastrointestinal tract to obtain microscopic images of tissue without biopsy excision 1 2 3 4 5. While CLE has the potential to improve tissue sampling 6, the speeds and modes of operation of CLE devices prohibit automatic and comprehensive acquisition of microscopic image data over large segments of luminal gastrointestinal organs.
Spectrally-encoded confocal microscopy (SECM) is a new reflectance CLE technology that uses a diffraction grating and a wavelength-swept laser to image tissues at very high speeds (> 10 × video rate) 7. Because of its high-speed capabilities, SECM has a potential to image the entire distal esophagus in an acceptable procedural time. Differences between CLE and SECM are summarized in Table 1. Previously, SECM, implemented using a bench top microscope, has been shown to be capable of visualizing key histomorphologic features associated with various esophageal diseases ex vivo 8 9. Recently, we have developed a miniature endoscopic SECM probe that contains high-resolution optics 10, can be placed into the gastrointestinal tract endoscopically over a guide wire, and automatically images over large swatches of gastrointestinal tissues using a helical scan pattern. The aim of this study was to test this SECM endoscopic probe for imaging large areas of the esophagus in vivo.
Table 1. Comparison of eCLE (endoscope-based CLE), pCLE (probe-based CLE) and SECM.
| eCLE | pCLE | SECM | |
| Image acquisition speed | 0.8 frames/sec1 | 12 frames/sec2 | 100 frames/sec |
| Contrast mechanism | Fluorescence | Fluorescence | Reflectance |
| Contrast agent | Intravenous fluorescein | Intravenous fluorescein | Topical acetic acid |
| Implementation | Integrated in endoscope | Though an accessary port | Over a guide wire |
| Laser wavelength | 488 nm1 | 488 nm2 | 1300 nm |
| Resolution | Lateral: 0.7 µm1 Axial: 7 µm1 | Lateral: 1 µm2 Axial: 3 µm2 | Lateral: 2 µmAxial: 17 µm |
| Imaging depth | 0 – 250 µm1 | 0 – 50 µm3 | 86 – 114 µm4 |
| Field of view (FOV) | 500 × 500 µm2 1 | 240 × 200 µm2 2 | 280 × 1300 µm2 |
| Contiguous mosaic size | No published value available | 8 mm2 5 | 1,100 mm2 6 |
| Large-area imaging method | Manual scanning | Manual scanning | Automatic pullback |
These values were taken from Kiesslich et al. [1].
These values were taken from Becker et al. [6].
Imaging depth of pCLE can be set as a fixed value between 0 and 50 µm [16].
Multiple depth levels are imaged simultaneously over this range.
This value was taken from De Palma et al. [17].
This value is for a 5-cm-long pullback of the probe.
Materials and methods
SECM endoscopic imaging setup
We conducted comprehensive confocal microscopy using an SECM endoscopic imaging setup shown in Fig. 1. Near infrared light from the laser was delivered to the 7 mm diameter SECM probe inside the esophagus through a 2-m-long optical fiber. The light was then focused inside the tissue by the SECM probe optics. The reflected light from the tissue was collected by the same SECM probe optics and delivered back to the SECM system. The SECM endoscopic probe had a lateral resolution of 2.3 µm, an axial resolution of 17 µm, and a field size of 280 µm. The focal plane was located 100 µm below the tissue surface, and was slightly tilted to image multiple depths during a single scan 11. The line image of the tissue was acquired at the rate of 100 kHz, which resulted in the 1024 × 1024-pixel frame rate of 100 frames/sec. The SECM endoscopic probe was helically scanned at a rotation rate of 360 rpm and a pullback rate of 0.4 mm/sec to automatically obtain large, contiguous confocal microscopy images of the esophagus.
Fig. 1.
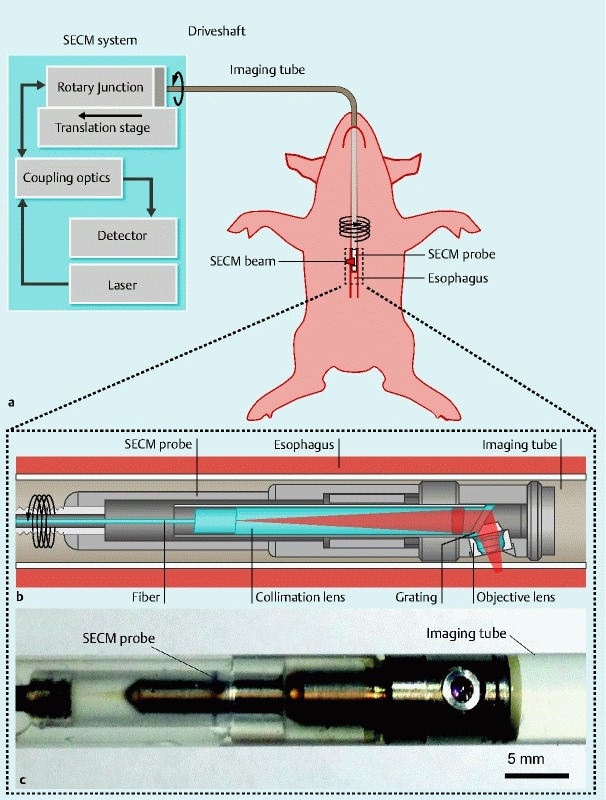
Schematics and photo of the SECM endoscopic imaging setup. a Overview of the SECM endoscopic imaging setup. An SECM endoscopic probe is inserted into an imaging tube, which is a transparent semi-flexible plastic tube. The imaging tube is introduced to the esophagus transorally. A SECM beam is focused into the esophageal tissue. While SECM images are continuously acquired, the SECM probe is helically scanned by a rotary junction and translation stage to image a large area of the esophagus. b Detailed schematic of the SECM probe optics. In the probe optics, light from the fiber is collimated by a collimation lens and diffracted by a grating. The diffracted light is focused by an objective lens (water immersion; numerical aperture = 0.5) into a 280-µm-long line. c Photo of the SECM optical probe inside the imaging tube. The diameter of the probe is 5.9 mm, and the rigid length is 30 mm. The outer diameter of the imaging tube is 7.0 mm.
In vivo swine imaging
SECM imaging of swine esophagus in vivo was conducted according to a study protocol approved by the Massachusetts General Hospital Subcommittee on Research Animal Care (Protocol# 2011N000115). Imaging was performed in a 45 kg female Yorkshire swine. After sedation and intubation, a videoendoscope (EPM-3300, Pentax) was advanced transorally to the gastroesophageal junction, and the initial evaluation of the esophagus was conducted. To increase the contrast of squamous nuclei, 12 a low-concentration of acetic acid (6 % concentration by volume) was topically applied to the esophagus using a spray catheter. A guidewire was then introduced to the stomach through the auxiliary channel. The videoendoscope was then removed while leaving the guidewire in place. The SECM endoscopic probe was introduced over the guidewire to approximately 5 cm proximal from the gastroesophageal junction. Once the probe was in place, the optics automatically rotated and pulled back within the transparent imaging tube. Light returned from the esophagus was detected and stitched together to form contiguous confocal microscopy images in 3 planes over a 5-cm-long esophageal segment. The animal was euthanized immediately following the procedure. At necropsy, histological examination of the esophagus was performed to conduct a comparative analysis.
Results
Comprehensive confocal image of swine esophagus in vivo
Contiguous 5 cm (length) by 2.2 cm (circumference) reflectance confocal microscopy images were acquired from three depth planes in 2.1 minutes. A large-area confocal image from the middle depth is shown in Fig. 2 a. The horizontal axis of the image corresponds to the longitudinal direction of the esophagus, and the vertical axis is the circumferential axis. Approximately 22 cm2 (65 %) of the imaged area contains microscopic tissue information. At a low-magnification view (Fig. 2 a), gross morphology of the esophagus can be seen. The epithelium (E) and lamina propria (LP) are visualized, and numerous papillae are seen as circular features with low SECM signal (arrows). Portions of the SECM image (~ 35 %) are artifactual; in the top portion of the image, the imaging tube lost contact with the esophagus (NC) and the focal plane of the probe was not inside the tissue. At the bottom of the image, a segment of the image reveals high reflectance at the border between the liquid-air interface (LA).
Fig. 2.
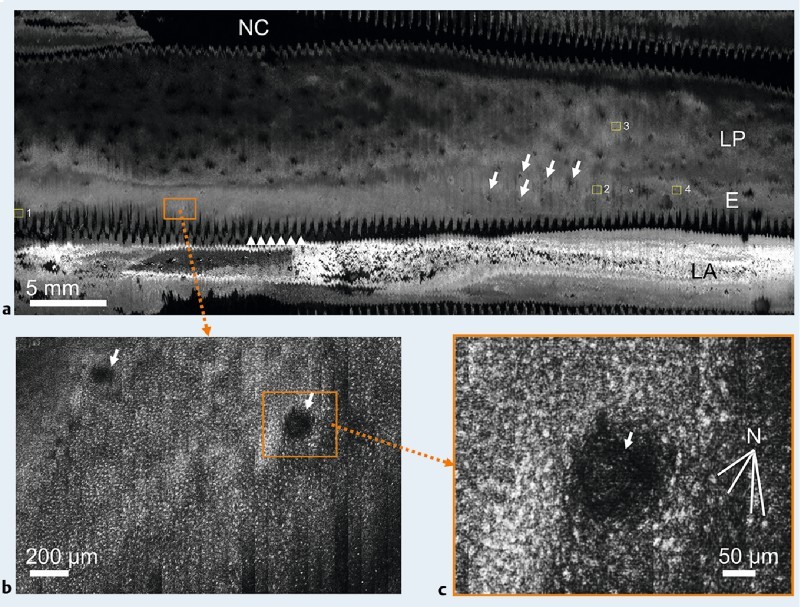
SECM images of the swine esophagus in vivo. a Low-magnification image; b magnified view of Fig. 2 a; c magnified view of Fig. 2 b. NC , no contact between the tissue and imaging tube; LP, lamina propria; E, epithelium; LA, reflected light from the air-liquid boundary; N , nuclei; arrows , papillae; and arrowheads, zig-zag pattern on the tissue surface.
As shown in Fig. 2 b, the large-area image can be zoomed in to show a small region of interest at a higher magnification. The size of Fig. 2 b is 2.0 mm by 1.3 mm, which is similar in size to a low-power microscopic field used in histologic analysis. At this magnification, cellular features of the tissue can be clearly seen, including numerous nuclei (seen as bright, highly reflecting dots) surrounding two papillae (shown as dark circular regions of low SECM signal marked by arrows). The image can be zoomed in further to produce a view (Fig. 2 c) that is similar in size to a high-power field (40 ×). At this high magnification, numerous nuclei (N) can be seen encircling a papilla.
Cellular features were visualized in SECM images at various depths within the epithelium. Fig. 3 shows high-magnification SECM images at three distinctive locations and en face histologic images of the swine esophagus. Fig. 3 a, a SECM image taken from a superficial plane within the epithelium (box 1 in Fig. 2 a), shows scattered nuclei (bright dots) similar to that seen in a histologic image of representative, nearby superficial squamous epithelium (Fig. 3 d). Fig. 3 b, a SECM image taken from a deeper region of the epithelium (box 2 in Fig. 2 a), demonstrates numerous nuclei (bright dots) with a higher density than Fig. 3 a and multiple papillae (dark circular regions) surrounded by the nuclei. Similar cellular features are observed in a representative histologic image of the basal cell layer (Fig. 3 e). Fig. 3 c, an SECM region from the LP (box 3 in Fig. 2 a), enables the visualization of microstructures with high SECM signals that are similar in appearance to collagen seen in a representative histologic image of the LP (Fig. 3 f).
Fig. 3.
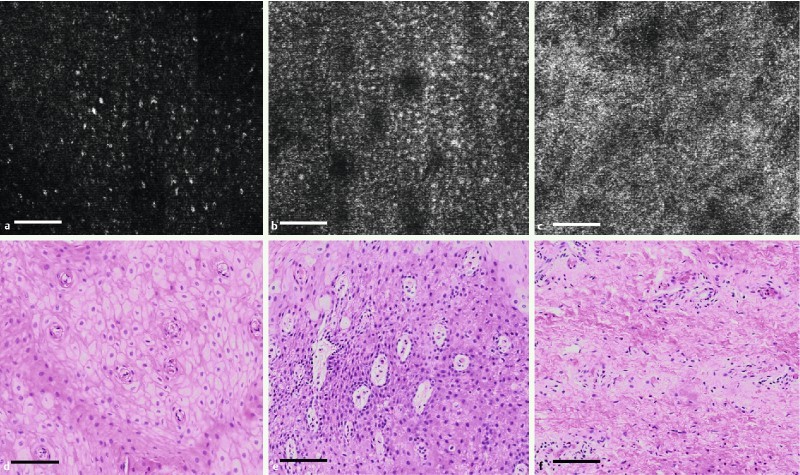
High-magnification SECM images (a, b, c) and representative, nearby en face histologic images (d, e, f) of swine esophagus. SECM images were taken from the regions marked by the boxes 1, 2, and 3 in Fig. 2 a. a and d Stratified squamous epithelium; b and e basal cell layer; c and f lamina propria (scale bar = 100 µm).
Imaging multiple depths in a single scan
SECM images from multiple imaging depths were obtained through post-operation image processing of the volumetric data that was acquired in a single helical scan of the probe 11. Fig. 4 shows SECM and en face histologic images obtained from the same respective transverse locations but at different imaging depths. The imaging depth changes from superficial (Fig. 4 a and d) to deep (Fig. 4 c and f) with a depth interval of 14 µm between images. The SECM images were taken from the box 4 in Fig. 2 a. In the SECM images, the papillae (dark circular regions) become bigger as the imaging depth increases from Fig. 4 a to 4 c, and a similar morphologic change is observed in the histologic images. Nuclear density increases with the imaging depth in the SECM images, and a similar trend is shown in the histologic images.
Fig. 4.
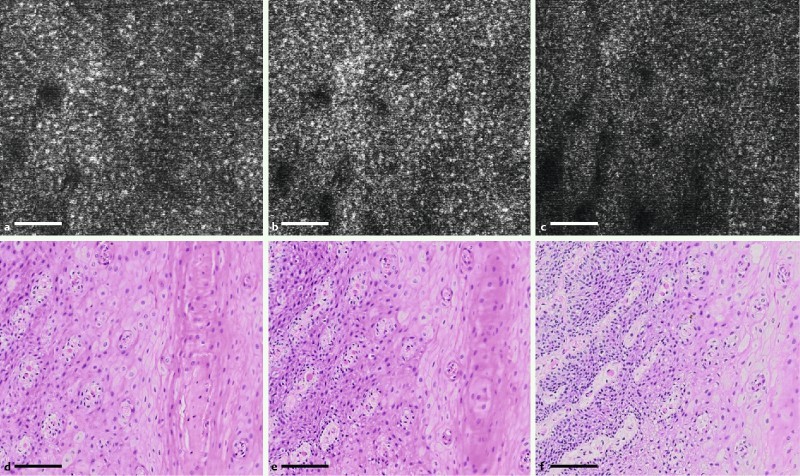
SECM and representative, nearby en face histologic images taken at the same respective transverse locations but at different imaging depths. The imaging depth changes from superficial (a and d) to deep (c and f), with a depth difference of 14 µm between images. The SECM images were taken from the region marked by box 4 in Fig. 2 a (scale bar = 100 µm).
Discussion
We demonstrated the feasibility of conducting automatic, comprehensive confocal microscopy of the esophagus in vivo using SECM. Very large confocal images (11 cm2) of the swine esophagus from three imaging depths were obtained in about 2 minutes. Low-magnification views enabled the visualization of the gross morphology of the tissue, while high-magnification views revealed cellular features. This technology can also be modified to be used to image large portions of other gastrointestinal tract organs, including the stomach, small intestines, colon, and rectum.
There were, however, areas where the technology needs to be improved further. Artifacts were present in portions of the SECM images. The first artifact was likely caused by peristalsis, where tissue motion relative to the SECM probe resulted in a zig-zag pattern in some regions (Fig. 2 a, arrowheads). A second artifact was loss of contact between the probe and the tissue (Fig. 2 a, NC). Adding a suction tube on the side of the probe increases and stabilizes tissue contact 13, which will likely reduce these two artifacts. The processing time to generate a large-area image such as Fig. 2 a (file size = 1.3 Gbytes) was around 4 minutes. This amount of processing time is acceptable for post-operation image analysis. However, a shorter processing time will be needed if we want to conduct intraprocedural visualization of the entire confocal microscopy dataset. Because of the current imaging depth of 100 µm, SECM was capable of visualizing the epithelium and LP but was unable to be used to image deeper tissue regions such as muscularis mucosa and submucosa. Optical coherence tomography (OCT) has been successfully used to visualize architectural features of these deeper tissue regions 14. Because conditions such as Barrett's esophagus (BE) and esophageal squamous dysplasia are primarily limited to the superficial mucosa, we expect that the cellular features visualized by SECM will provide comprehensive diagnostic information for most screening and surveillance cases.
As the next step of this research, we will utilize this probe to obtain comprehensive confocal microscopy images of the human esophagus in vivo. When imaging the human esophagus, we will use a lower concentration of acetic acid (1 – 3 %) than the concentration used for the swine esophagus (6 %), since our previous study of imaging human esophageal biopsy tissues showed that 0.6 % acetic acid provides sufficient nuclear contrast in human tissues 8. The transparent imaging tube will be disposed after each imaging session, while the internal SECM probe optics will be used for multiple imaging sessions. The costs of the imaging probe and disposable transparent imaging tube will likely be comparable to the costs of other imaging devices that are used in conjunction with endoscopy.
SECM images could enable targeted biopsy based on microscopic tissue assessment of the entire distal esophagus. During the imaging procedure, SECM images will be displayed in real time while the SECM probe automatically scans a long segment of the esophagus. An endoscopist or pathologist will review the SECM images and will determine regions that need further examination by histopathology. In conjunction with the laser marking functionality 15, the SECM probe can generate visible marks around the identified tissue regions and guide the endoscopist to biopsy tissues at these regions. The targeted biopsy enabled by SECM imaging could be helpful for improving the sampling yield of biopsy procedures such as those performed for Barrett’s surveillance. For endoscopists or pathologists to be able to read SECM images, we will develop and validate diagnostic criteria for SECM interpretation. Operators will undergo training sessions to gain expertise in reviewing and interpreting SECM videos and images.
Several challenges, however, are expected during SECM imaging of the human esophagus in vivo, including following four. First, certain shapes of the distal esophagus might pose difficulties for SECM imaging, including esophageal stricture or tortuosities. For these cases, it may be difficult to pass the probe as it would be for conventional endoscopy. These geometrical distortions can also bend the probe, which can bind the driveshaft, causing rotational artifacts. If the patient has an esophagus that is larger than normal, the SECM probe may not come into contact with a large portion of the esophagus. This situation may be mitigated by obtaining one pullback SECM scan and then repositioning the probe to acquire images from another region of the esophagus. Second, luminal contents, which are occasionally present in patients with gastroesophageal reflux disease (GERD) or BE, might make the SECM imaging difficult. The luminal contents could enter the space between the SECM probe and tissue, which can attenuate the light and change the focal plane so that it is no longer within the tissue itself. To mitigate this factor, we ensure that luminal contents are cleared during the initial endoscopic examination. As a result, we do not expect that luminal debris will interfere with SECM imaging. Third, the current SECM endoscopic probe always requires video endoscopy to introduce a guide wire. Conducting SECM imaging in conjunction with video endoscopy is beneficial since the esophagus can be cleaned during the endoscopy procedure, acetic acid can be applied in a controlled manner, and the location of the gastroesophageal junction (GEJ) can be determined, which guides SECM probe placement. Fourth, due to its current diameter of 7 mm, the SECM probe cannot be introduced through the accessary port of a standard video endoscope. Compared to the current over-the-guide wire introduction of the SECM probe, through-the-scope SECM could facilitate good spatial registration between SECM images and endoscopic observation. To utilize the SECM probe through the endoscopic accessory port, the diameter of the probe will need to be reduced.
Abbreviations
- BE
Barrett’s esophagus
- CLE
Confocal laser endomicroscopy
- GEJ
gastroesophageal junction
- GERD
gastroesophageal reflux disease
- LP
Lamina propria
- OCT
Optical coherence tomography
- SECM
Spectrally-encoded confocal microscopy
Acknowledgment
The authors thank William Puricelli, RN for his help during the animal study. This research has been sponsored by Nine Point Medical, Cambridge, MA. S. C. S is currently with Nine Point Medical. K. W. is currently with Emory University Hospital, Atlanta, GA. J. L. is currently with Tufts Medical Center, Boston, MA.
Footnotes
Competing interests: G. J. Tearney received sponsored research from Nine Point Medical. M. Shishkov, K. Woods and G. J. Tearney consult for Nine Point Medical. Massachusetts General Hospital has a licensing arrangement with Nine Point Medical. D. Kang, S. C. Schlachter, R. W. Carruth, N. Tabatabaei, M. Shishkov, J. S. Sauk, N. S. Nishioka, G. J. Tearney have the rights to receive royalty payments as part of this licensing arrangement.
References
- 1.Kiesslich R, Gossner L, Goetz M. et al. In vivo histology of Barrett’s esophagus and associated neoplasia by confocal laser endomicroscopy. Clin Gastroenterol Hepatol. 2006;4:979–987. doi: 10.1016/j.cgh.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Kitabatake S, Niwa Y, Miyahara R. et al. Confocal endomicroscopy for the diagnosis of gastric cancer in vivo. Endoscopy. 2006;38:1110–1114. doi: 10.1055/s-2006-944855. [DOI] [PubMed] [Google Scholar]
- 3.Buchner A M, Shahid M W, Heckman M G. et al. Comparison of Probe-Based Confocal Laser Endomicroscopy With Virtual Chromoendoscopy for Classification of Colon Polyps. Gastroenterology. 2010;138:834–842. doi: 10.1053/j.gastro.2009.10.053. [DOI] [PubMed] [Google Scholar]
- 4.Meining A, Chen Y K, Pleskow D. et al. Direct visualization of indeterminate pancreaticobiliary strictures with probe-based confocal laser endomicroscopy: a multicenter experience. Gastrointestinal endoscopy. 2011;74:961–968. doi: 10.1016/j.gie.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Konda V J, Aslanian H R, Wallace M B. et al. First assessment of needle-based confocal laser endomicroscopy during EUS-FNA procedures of the pancreas (with videos) Gastrointestinal endoscopy. 2011;74:1049–1060. doi: 10.1016/j.gie.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 6.Becker V, Vercauteren T, von Weyhern C H. et al. High-resolution miniprobe-based confocal microscopy in combination with video mosaicing (with video) Gastrointest Endosc. 2007;66:1001–1007. doi: 10.1016/j.gie.2007.04.015. [DOI] [PubMed] [Google Scholar]
- 7.Tearney G J, Webb R H, Bouma B E. Spectrally encoded confocal microscopy. Opt Lett. 1998;23:1152–1154. doi: 10.1364/ol.23.001152. [DOI] [PubMed] [Google Scholar]
- 8.Kang D, Suter M J, Boudoux C. et al. Comprehensive imaging of gastroesophageal biopsy samples by spectrally encoded confocal microscopy. Gastrointest Endosc. 2010;71:35–43. doi: 10.1016/j.gie.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoo H, Kang D, Katz A J. et al. Reflectance confocal microscopy for the diagnosis of eosinophilic esophagitis: a pilot study conducted on biopsy specimens. Gastrointest Endosc. 2011;74:992–1000. doi: 10.1016/j.gie.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kang D, Carruth R W, Kim M. et al. Endoscopic probe optics for spectrally encoded confocal microscopy. Biomed Opt Express. 2013;4:1925–1936. doi: 10.1364/BOE.4.001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang D, Yoo H, Jillella P. et al. Comprehensive volumetric confocal microscopy with adaptive focusing. Biomed Opt Express. 2011;2:1412–1422. doi: 10.1364/BOE.2.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carlson K, Pavlova I, Collier T. et al. Confocal microscopy: Imaging cervical precancerous lesions. Gynecologic oncology. 2005;99:84–88. doi: 10.1016/j.ygyno.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y-M, Bergman J J, Weusten B. et al. Endoscopy Innovation Forum: Radiofrequency Ablation for Early Esophageal Squamous Cell Neoplasia. Endoscopy. 2010;42:327–333. doi: 10.1055/s-0029-1244017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suter M J, Vakoc B J, Yachimski P S. et al. Comprehensive microscopy of the esophagus in human patients with optical frequency domain imaging. Gastrointest Endosc. 2008;68:745–753. doi: 10.1016/j.gie.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suter M J, Jillella P A, Vakoc B J. et al. Image-guided biopsy in the esophagus through comprehensive optical frequency domain imaging and laser marking: a study in living swine. Gastrointestinal Endoscopy. 2010;71:346–353. doi: 10.1016/j.gie.2009.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiberville L, Salaün M, Lachkar S. et al. Confocal fluorescence endomicroscopy of the human airways. Proceedings of the American Thoracic Society. 2009;6:444–449. doi: 10.1513/pats.200902-009AW. [DOI] [PubMed] [Google Scholar]
- 17.De Palma G D, Staibano S, Siciliano S. et al. In vivo characterisation of superficial colorectal neoplastic lesions with high-resolution probe-based confocal laser endomicroscopy in combination with video-mosaicing: A feasibility study to enhance routine endoscopy. Digestive and Liver Disease. 2010;42:791–797. doi: 10.1016/j.dld.2010.03.009. [DOI] [PubMed] [Google Scholar]


