Abstract
Background
Skeletal muscle is a major contributor to whole-body metabolism as it serves as a depot for both glucose and amino acids, and is a highly metabolically active tissue. Within skeletal muscle exists an intrinsic molecular clock mechanism that regulates the timing of physiological processes. A key function of the clock is to regulate the timing of metabolic processes to anticipate time of day changes in environmental conditions. The purpose of this study was to identify metabolic genes that are expressed in a circadian manner and determine if these genes are regulated downstream of the intrinsic molecular clock by assaying gene expression in an inducible skeletal muscle-specific Bmal1 knockout mouse model (iMS-Bmal1−/−).
Methods
We used circadian statistics to analyze a publicly available, high-resolution time-course skeletal muscle expression dataset. Gene ontology analysis was utilized to identify enriched biological processes in the skeletal muscle circadian transcriptome. We generated a tamoxifen-inducible skeletal muscle-specific Bmal1 knockout mouse model and performed a time-course microarray experiment to identify gene expression changes downstream of the molecular clock. Wheel activity monitoring was used to assess circadian behavioral rhythms in iMS-Bmal1−/− and control iMS-Bmal1+/+ mice.
Results
The skeletal muscle circadian transcriptome was highly enriched for metabolic processes. Acrophase analysis of circadian metabolic genes revealed a temporal separation of genes involved in substrate utilization and storage over a 24-h period. A number of circadian metabolic genes were differentially expressed in the skeletal muscle of the iMS-Bmal1−/− mice. The iMS-Bmal1−/− mice displayed circadian behavioral rhythms indistinguishable from iMS-Bmal1+/+ mice. We also observed a gene signature indicative of a fast to slow fiber-type shift and a more oxidative skeletal muscle in the iMS-Bmal1−/− model.
Conclusions
These data provide evidence that the intrinsic molecular clock in skeletal muscle temporally regulates genes involved in the utilization and storage of substrates independent of circadian activity. Disruption of this mechanism caused by phase shifts (that is, social jetlag) or night eating may ultimately diminish skeletal muscle’s ability to efficiently maintain metabolic homeostasis over a 24-h period.
Electronic supplementary material
The online version of this article (doi:10.1186/s13395-015-0039-5) contains supplementary material, which is available to authorized users.
Keywords: Circadian, Molecular clock, Skeletal muscle, Metabolism, Temporal separation, Anabolic, Catabolic, Bmal1, Rev-erbα
Background
Skeletal muscle plays a large role in whole-body metabolism as it constitutes approximately 40% of body mass and is a highly metabolically active tissue [1,2]. Basal metabolic rate is dependent on both the size and activity of skeletal muscle as cross-bridge cycling and calcium handling associated with contraction are energetically expensive processes [2-5]. Skeletal muscle is a principle contributor to whole-body glucose handling as it is responsible for approximately 80% of postprandial glucose uptake [6,7]. It has been widely reported that skeletal muscle has regulatory mechanisms that modulate substrate utilization and storage in response to varying metabolic demands and environmental conditions (for example, nutrient status) [3,8-12]. For instance, skeletal muscle rapidly modulates rates of glucose uptake and utilization in response to contraction and/or insulin stimulation [13-15]. While the fluctuations in the role for muscle to store vs. use is commonly linked with the fed/fasted and active/inactive behaviors, these changes in storage and use are also aligned with the 24-h (circadian) light/dark cycles attributed to the rising and setting of the sun and feeding/activity behavior [16].
At the core of circadian rhythms is a mechanism known as the molecular clock. In the last 15 years, many researchers have shown that the clock mechanism exists in virtually all cell types in the body including skeletal muscle [17,18]. The intrinsic molecular clock is most known for its role in regulating cellular metabolism even under constant lighting or feeding conditions [19-26]. These studies have shown that the molecular clock temporally regulates the rhythmic activation or repression of rate-limiting metabolic genes to help the cell anticipate changes in environmental conditions and metabolic demand [27]. The molecular clock comprises a transcriptional-translational feedback mechanism driven by the rhythmic expression of the PAS-bHLH family of transcription factors BMAL1:CLOCK, which reach maximal activity during the inactive phase (that is, light phase for mice) [28-31]. Direct targets of BMAL1:CLOCK typically reach peak expression (acrophase) prior to the beginning of the active phase of the day (that is, dark phase for mice). The capacity of the molecular clock in regulating metabolism is highlighted by the metabolic phenotypes observed in genetic core-clock mutant models [32-38]. On-going studies are aimed at utilizing organ-specific molecular clock mutant models to determine the function of the clock in each tissue as well as assessing the role the peripheral clocks play in regulating whole-body metabolism [39-43].
Utilizing high-resolution temporal transcriptome data coupled with circadian statistics has proved to be an effective method for identifying genes expressed in a circadian manner [44,45]. In the present study, we employ a bioinformatics approach with a publically available high-resolution circadian data set collected under constant dark conditions to analyze the skeletal muscle circadian transcriptome (gastrocnemius muscle) with a focus on the temporal phase of gene expression. We reveal that skeletal muscle circadian genes are highly enriched for metabolic processes, and furthermore, we identify the temporal pattern of peak expression for different key metabolic genes separating catabolic vs. anabolic processes over 24 h. To identify which circadian-metabolic genes are regulated downstream of the intrinsic molecular clock, we generated an inducible muscle-specific Bmal1 knockout (iMS-Bmal1−/−) mouse and performed a time series transcriptome analysis. Mice lacking Bmal1 in skeletal muscle displayed no apparent changes in circadian behavior, yet we observed significant decreases in the expression of circadian genes involved in glucose utilization and adrenergic signaling, while observing significant increases in lipogenic genes. Consistent with a substrate shift from carbohydrate to lipid utilization, we observed a concomitant shift from a fast to slow fiber-type gene expression profile indicative of a more oxidative muscle in iMS-Bmal1−/−. These findings demonstrate that the endogenous molecular clock in skeletal muscle contributes significantly to the time of day shifts in carbohydrate/lipid metabolism.
Methods
High-resolution circadian microarray
Microarray data for the high-resolution circadian time-course are from gastrocnemius muscles of male C57Bl6 mice collected every 2 h for 48 h under constant dark conditions and ad libitum food availability [46]. The data were downloaded from NCBI GEO datasets (GSE54652) and consist of 24 individual arrays, one for each time point from circadian time 18 to 64 [45,46]. Expression intensities from the series matrix file for all probesets at all time points were used as input for JTK_CYCLE analysis, with period length set to 24 h [47]. We defined circadian genes as having a JTK_CYCLE adjusted P value of less than 0.05. We utilized the Bioconductor package to identify mapped probesets on the Affymetrix Mouse Gene 1.0 ST chip that represent unique genes, thus eliminating control probesets from further analyses. Genes with median expression intensities of at least 100 were considered as expressed in skeletal muscle. We entered the list of circadian genes into Gene Ontology Consortium online tools to identify enriched biological processes [48,49]. Enrichment P values were adjusted for multiple testing using Bonferroni correction.
Inducible skeletal muscle-specific Bmal1 inactivation mouse model
All animal procedures were conducted in accordance with institutional guidelines for the care and use of laboratory animals as approved by the University of Kentucky Institutional Animal Care and Use Committee. The floxed Bmal1 mouse [B6.129S4(Cg)-Arntltm1Weit/J] was purchased from The Jackson Laboratory and has no reported breeding, physical, or behavioral abnormalities [50]. The skeletal muscle-specific Cre-recombinase mouse, [human skeletal actin (HSA)-MerCreMer] has been previously characterized [51]. The floxed Bmal1 mouse has loxP sites flanking exon 8 and is indistinguishable from wild-type littermates. Breeding with the skeletal muscle-specific Cre-recombinase mouse generates offspring in which selective deletion of the bHLH domain of Bmal1 in skeletal muscle can be induced upon tamoxifen administration. Inducible skeletal muscle-specific Bmal1 knockout mice were generated as follows: the Bmal1flox/flox female was crossed with the skeletal muscle-specific Cre-recombinase male. This yielded an F1 generation of skeletal muscle-specific Cre+/−;Bmal1+/flox mice. Breeding the F1 generation males to the Bmal1flox/flox females resulted in the skeletal muscle-specific Cre+/−;Bmal1flox/flox mice (referred to as iMS-Bmal1flox/flox) needed for this study. Mouse genotypes were determined by PCR using genomic DNA isolated from tail snips. Activation of Cre-recombination was done by intraperitoneal injections of tamoxifen (Sigma-Aldrich, St. Louis, MO, USA; Cat. No. T5648) (2 mg/day) for five consecutive days when the mice reached 12 weeks of age. This age was chosen to eliminate any effects that the lack of Bmal1 might have on skeletal muscle development and postnatal maturation. Controls were vehicle (15% ethanol in sunflower seed oil)-treated iMS-Bmal1flox/flox mice.
Recombination specificity
The iMS-Bmal1 mice were injected (intraperitoneal) with either vehicle (iMS-Bmal1+/+) or tamoxifen (iMS-Bmal1−/−) between 12 and 16 weeks of age. Five weeks post injection, mice were anesthetized with isoflurane, and the heart, diaphragm, liver, lung, abdominal aorta, brain, tibialis anterior, soleus, gastrocnemius, brown fat, white fat, and cartilage were collected and immediately frozen in liquid nitrogen for DNA analysis. Genomic DNA was extracted from the tissues using the DNeasy Blood and Tissue Kit (Qiagen, Venlo, Netherlands). To assess recombination specificity, PCR was performed with tissue DNA and primers for the recombined and non-recombined alleles as described in Storch et al. [50]. The forward and reverse primers for the non-recombined allele were the same as the genotyping primers and yielded a 431-bp product. A second forward primer 5′-CTCCTAACTTGGTTTTTGTCTGT-3′ was included to detect the recombined allele, which showed a band at 572 bp [50]. The PCR reaction was run on a 1.5% agarose gel (0.0005% ethidium bromide) to visualize the DNA products.
RNA isolation and real-time PCR
Total RNA was prepared from frozen gastrocnemius tissue samples using TRIzol (Invitrogen) according to the manufacturer’s directions. RNA samples were treated with TURBO DNase (Ambion, Austin, TX, USA) to remove genomic DNA contamination. Isolated RNA was quantified by spectrophotometry (λ = 260 nm). First-strand cDNA synthesis from total RNA was performed with a mixture of oligo(dT) primer and random hexamers using SuperScript III First-Strand Synthesis SuperMix (Invitrogen, Waltham, MA, USA). All isolated RNA and cDNA samples were stored at −80°C until further analysis. Real-time quantitative PCR using TaqMan (Applied Biosystems, Waltham, MA, USA) assays was used to examine the gene expression of Bmal1 (Mm00500226_m1), Rev-erbα (Mm00520708_m1), Dbp (Mm00497539_m1), Hk2 (Mm00443385_m1), Pdp1 (Mm01217532_m1), Fabp3 (Mm02342495), and Pnpla3 (Mm00504420_m1). The ΔΔCT method was used for the quantification of real-time PCR data in the circadian collections.
Wheel activity monitoring
One cohort of mice was used for analysis of circadian behavior (gene expression not analyzed in this cohort). A total of 20 mice (mixed genders) were analyzed with 11 receiving tamoxifen treatment and the remaining 9 receiving vehicle treatment. The mice were maintained in individual cages with a running wheel under 12L:12D (LD) conditions for 4 weeks. The wheel running of the vehicle (iMS-Bmal1+/+) or tamoxifen (iMS-Bmal1−/−) mice were continuously recorded and monitored throughout the experiment using ClockLab software [52]. To determine the free-running period of the mice, we released them into total darkness (DD) for 3 weeks. Activity was evaluated using voluntary running wheel rotations plotted in 1-min bins. The free-running period (tau) during the 3-week DD period was calculated using periodogram analysis in the ClockLab software.
Circadian collections
Forty-eight iMS-Bmal1flox/flox mice were housed in individual cages in light boxes, entrained to a 12-h LD cycle for 14 days, and had ad libitum access to food and water. Following the 2-week entrainment period, 24 mice were injected with vehicle and 24 with tamoxifen for five consecutive days, generating 24 iMS-Bmal1+/+ and 24 iMS-Bmal1−/− mice, respectively. The light schedule was kept the same during injections and for the subsequent 5 weeks. Five weeks after the last day of injections, mice were released into constant darkness for 30 h following protocols established in the circadian field [46,53]. Mice were sacrificed in darkness (dim red light), and skeletal muscles were collected every 4 h for 20 h (six time points) and frozen for RNA and protein analysis.
Western blot
Whole cell lysates were prepared from the liver and gastrocnemius of iMS-Bmal1+/+ and iMS-Bmal1−/− mice (n = 3/strain). SDS-PAGE (4-15% separating gel, Bio-Rad, Hercules, CA, USA) and immunoblotting were carried out with routine protocols. Affinity-purified Bmal1 polyclonal antibody (Sigma-Aldrich, SAB4300614) was visualized with IRDye-conjugated secondary antibody using the Odyssey system (Li-Cor, Lincoln, NE, USA). Each lane contained 50 μg total protein.
Microarray analysis of iMS-Bmal1+/+, iMS-Bmal1−/−, and MKO (Dyar et al.)
We pooled equivalent amounts of total RNA from four mice for each time point (circadian time 18, 22, 26, 30, 34, 38) and treatment (vehicle or tamoxifen). Pooled RNA samples were used to construct cDNA libraries that were hybridized to Affymetrix Mouse Gene 1.0 ST microarrays (Affymetrix, Santa Clara, CA, USA) (1 sample/time point). Intensity data for iMS-Bmal1+/+ and iMS-Bmal1−/− gastrocnemius muscles are quantile normalized, and a low pass median intensity filter of greater than or equal to 100 is applied to both iMS-Bmal1+/+ and iMS-Bmal1−/− datasets separately. Nine thousand one hundred eighty-four non-redundant, mapped genes (9,988 probesets) are considered to be expressed in one or both datasets. Gene expression changes in iMS-Bmal1−/− muscle tissue were calculated by averaging the change in expression for each gene throughout the circadian time course (CT18-38, n = 6). Tibialis anterior and soleus gene expression values for control and muscle-specific knockout model (MKO) from Dyar et al. [43] were downloaded from NCBI GEO datasets (GSE43071) and consists of 18 individual arrays, three for each time point from circadian time 0 to 20. To compare temporal gene expression changes for the TA and SOL, we averaged Affymetrix ST 1.0 expression values for each gene at circadian times 0, 4, 8, 12, 16, and 20. Student’s t test was used to identify differentially expressed probesets at a significance of P ≤ 0.05.
Results and discussion
Cellular metabolic processes are highly enriched in the circadian transcriptome of skeletal muscle
To identify circadian gene expression in skeletal muscle, we used a publicly available, high-resolution, circadian time-course microarray dataset from gastrocnemius muscles of male C57BL/6 mice [45,46]. These mice were housed in constant darkness, and food was provided ad libitum to eliminate the influence of external environmental cues. We chose this dataset because it has double the sampling frequency of previously published circadian muscle transcriptomes, and this allows for greater precision for circadian analysis [46,54]. Using the JTK_CYCLE statistical algorithm [47] for the reliable detection of oscillating transcripts with a 24-h periodicity, we identified 1,628 circadian mRNAs (adjusted P < 0.05). An unbiased Gene Ontology enrichment analysis of these circadian genes revealed a significant overrepresentation of cellular metabolic processes, with approximately 1,004 (62%) genes directly involved in skeletal muscle metabolic processes as well as the regulation of metabolism (Figure 1).
Figure 1.
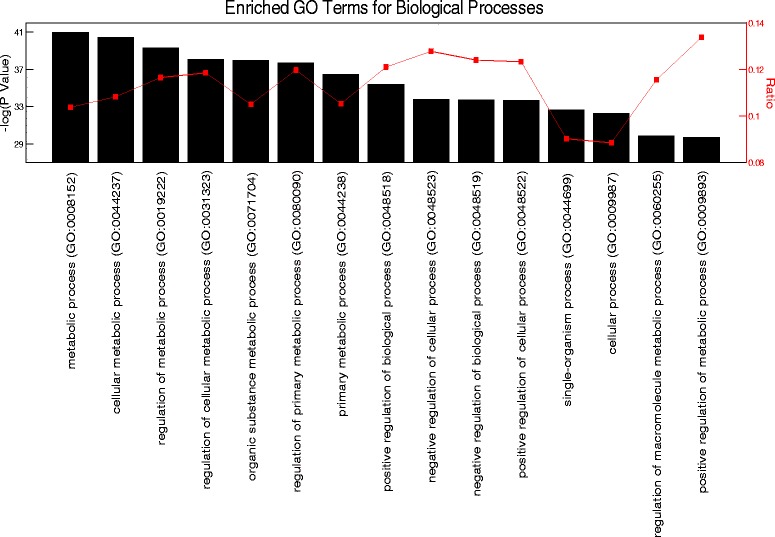
Gene ontology analysis of the skeletal muscle circadian transcriptome. Top 15 enriched GO processes listed from left to right in order of significance.
An additional benefit of using the JTK_CYCLE algorithm is its ability to determine the acrophase, or time of peak expression, of each circadian probeset. Identifying the acrophase of genes that have common ontologies may help to predict the potential timing of cellular and physiological processes. Herein, we report the acrophase according to their respective circadian times (CT), which is standardized to the free-running period of the mice under constant conditions. For the array studies, the mice were in DD for 30 h so CT 0 denotes the start of the inactive period, while CT 12 denotes the start of the active period. To identify the timing of gene expression and its relationship to metabolic processes in skeletal muscle, we annotated a subset of circadian genes by their known functions, timing of peak expression, and involvement in key metabolic pathways. We focused our analysis on metabolic functions that involve substrate (carbohydrate and lipid) utilization as well as storage and biosynthetic processes.
Lipid metabolism: genes involved in fatty-acid uptake and β-oxidation peak in the mid-inactive/light phase
Skeletal muscle expresses specialized membrane transporters to facilitate the transport of lipids into the cell [55-57]. Two lipid transport genes that encode for fatty-acid binding proteins, Fabp4 (CT 24.0) and Fabp3 (heart/muscle isoform, CT 6.0), are expressed in a circadian manner with the highest mRNA expression in the early- and mid-inactive periods, respectively. Acrophase of circadian genes involved in lipid metabolism are illustrated in Figure 2. Normalized expression traces for each gene are located in Additional files 1, 2, and 3. Previous studies have demonstrated oscillations in plasma fatty acid concentrations in mice with peak levels occurring during the inactive/light period [58-60]. Further functional analysis is required to validate the predition that the rate of fatty-acid uptake in skeletal muscle peaks during the mid-late inactive period. Upon uptake into the cell, fatty acids can be stored as triglycerides or be converted to acetyl-CoA through β-oxidation [61]. Slc25a20 encodes for an acyl-carnitine translocase that transfers fatty acids into the inner-mitochondrial matrix and reaches peak expression in the middle of the inactive period (CT 7.5) [62]. We identified multiple genes that encode for β-oxidation enzymes to be circadian and also reach peak expression around the mid-inactive phase. These include the enoyl CoA hydratase Ech1 (CT 7.0), the tri-functional enzyme subunits Hadha (CT 8.0) and Hadhb (CT 8.0), and the acetyl-CoA acyltransferase Acaa2 (CT 9.0). Malonyl-CoA, an intermediate formed during de novo fatty acid synthesis, is a potent inhibitor of β-oxidation. The striated muscle enriched gene Mlycd (CT 7.5) encodes for the malonyl-CoA decarboxylase that promotes β-oxidation by reducing cytosolic concentrations of malonyl-CoA and reaches peak expression during the mid-inactive period similar to that of the circadian β-oxidation genes. These observations suggest that rates of β-oxidation are modulated over time of day and potentially through the endogenous molecular clock in skeletal muscle [10,63,64].
Figure 2.
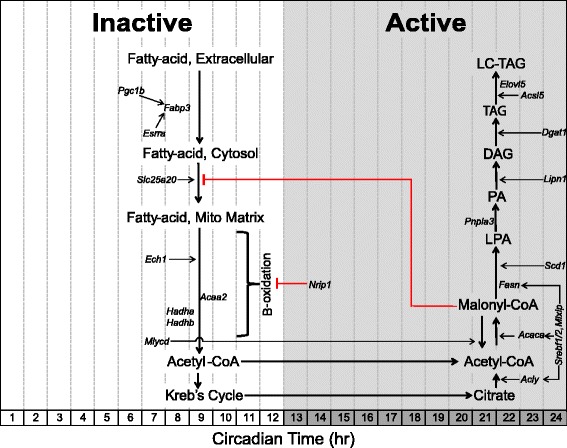
Schematic acrophase diagram of circadian genes involved in lipid metabolic processes. The relative location of the circadian genes (italicized) in respect to the x-axis indicates acrophase or time of peak expression calculated by the JTK_CYCLE algorithm. Location of substrates and pathways does not represent peak substrate concentrations and/or rates of individual pathways as these were not measured in our analysis. White/grey shading is representative of the inactive and active phases, respectively.
Nuclear receptors are known to be potent transcriptional regulators of metabolism as they sense changes in environmental conditions and induce appropriate changes in the expression of metabolic genes [65-69]. The nuclear receptor Estrogen-related receptor alpha (Esrra, CT 7.5) and the nuclear co-activator PPARγ coactivartor-1 beta (Ppargc1b, CT 7.0) are both circadian genes in skeletal muscle with peak expression occurring at the mid-inactive phase. These factors have been shown to promote mitochondrial biogenesis, fatty-acid uptake (targets Fabp3), and β-oxidation [70,71]. The nuclear co-repressor Nrip1, also known as Rip140, is a potent negative regulator of skeletal muscle oxidative metabolism and has been shown to suppress expression of the fatty-acid transporter, Fabp3, in skeletal muscle [72-74]. NRIP1 suppresses gene expression by binding nuclear receptors (including PPARs and estrogen-related receptors) and recruiting histone deacetylases [75]. Interestingly, peak expression of Nrip1 occurs during the beginning of the active period (CT 13.0) and may therefore act as a molecular brake to oxidative metabolism as the muscle transitions from lipid to carbohydrate utilization during the early active phase.
Lipid metabolism: lipogenic genes reach peak expression at the end of the active/dark phase
The lipogenic genes Acly (CT23.0), Acaca (CT 23.0), and Fasn (CT 22.5) involved in de novo fatty-acid synthesis, or the conversion of excess carbohydrates into fatty acids, reach peak expression at the end of the active phase (Figure 2) [61,76]. Scd1 (CT 24.0) encodes the enzyme that catalyzes the rate-limiting reaction of monounsaturated fatty-acid formation to promote lipid bilayer fluidity and lipogenesis [77,78]. The genes Srebf1 (CT 24.5), Srebf2 (CT 24.0), and Mlxip (CT 23.5) encode transcription factors that target carbohydrate response elements within lipogenic gene promoter regions (Acly, Acaca, and Fasn) and are also circadian with peak expression at the end of the active phase [79,80]. Consistent with our results, Srebf1 oscillations have been reported in the liver and genome-wide binding studies have shown a circadian recruitment pattern of SREBF1 to the promoters of lipogenic genes with maximal binding during the active (fed) stage [81-84].
The gene Pnpla3 (CT 21.0), also known as adiponutrin, promotes lipogenesis by converting LPA to phosphatidic acid (PA) [85]. The gene Lpin1 (CT 24.0) which encodes for the lipin-1 enzyme is responsible for converting phosphatidic acid (PA) to diacylglycerol (DAG), the upstream metabolite required in phospholipid biosynthesis [86,87]. The highly regulated, committing step in triacylglycerol (TG) synthesis, addition of a fatty-acyl-CoA to DAG, is performed by the product encoded by Dgat1 (CT 24.5), which is also expressed in a circadian manner [88]. Once a TG molecule is formed, it can be elongated by enzymes encoded by Acsl5 (CT 23.0) or Elovl5 (CT 22.5) [89,90]. The observation that circadian lipogenic genes reach peak expression levels around the end of the active phase suggests that skeletal muscle promotes storage of excess energy at the end of the active/absorptive phase.
Carbohydrate metabolism: genes involved in carbohydrate catabolism peak in the early active/dark phase
Glycolysis, the breakdown of glucose to form pyruvate, is primarily regulated at two enzymatic reactions catalyzed by the hexokinase and phosphofructokinase enzymes [91]. We observe that the hexokinase-2 (Hk2) gene is circadian with peak expression occurring at the beginning of the active phase (CT 12.0). Acrophase of circadian genes involved in carbohydrate metabolism are illustrated in Figure 3. Normalized expression traces for each gene are located in Additional files 1, 2, and 3. Hk2 is responsible for the first step in glycolysis by phosphorylating glucose to make glucose-6-phosphate, thereby trapping glucose within the cell [92]. The rate-limiting step of glycolysis involves the catalysis of fructose-6-phosphate to the highly unstable fructose-1,6-bisphosphate by the enzyme phosphofructokinase-1 (PFKM) [93,94]. A potent allosteric activator of PFKM is fructose-2,6-bisphosphate, which is the product of the other phosphofructokinase isozyme phosphofructokinase-2 (PFK2) [95]. Three genes (Pfkfb-1,3,4) that encode phosphofructokinase-2 subunits are circadian with peak expression occurring during the mid- and late-inactive phases (CT 10.0, CT 4.5, and CT 12.0, respectively).
Figure 3.
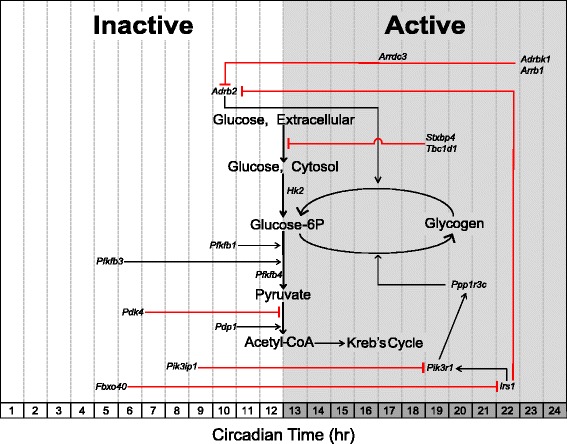
Schematic acrophase diagram of circadian genes involved in carbohydrate metabolic processes. The relative location of the circadian genes (italicized) in respect to the x-axis indicates acrophase or time of peak expression calculated by the JTK_CYCLE algorithm. Location of substrates and pathways does not represent peak substrate concentrations and/or rates of individual pathways as these were not measured in our analysis. White/grey shading is representative of the inactive and active phases, respectively.
Glycolytic flux through the Kreb’s cycle is controlled by pyruvate dehydrogenase complex (PDH) [96,97]. PDH decarboxylates pyruvate to form acetyl-CoA, which is a substrate for the Kreb’s cycle. The activity of PDH is regulated at the posttranslational level. Phosphorylation by kinases (PDKs) inhibits PDH activity, while dephosphorylation by phosphatases (PDPs) activates the complex [98,99]. The Pdk4 gene, which encodes for a PDH kinase that inhibits PDH, reaches maximal expression at the mid-inactive phase (CT 6.0). This expression pattern is similar to that of the β-oxidation genes and suggests that skeletal muscle substrate preference is pushed toward utilization of lipids over carbohydrates during the mid- to late-inactive phase. Conversely, the PDH phosphatase gene, Pdp1, peaks at the beginning of the active phase (CT 10.0) in a similar temporal fashion compared to the glycolytic enzymes described above. This temporal regulation of Pdp1 may therefore help increase glycolytic flux during the active phase. Dyar et al. observed similar expression patterns of Pdk4 and Pdp1 in skeletal muscle and were first to report a shift to carbohydrate utilization at the beginning of the active phase [43].
Adrb2 encodes for the β2-adrenergic receptor (β2AR) involved in the fight-or-flight response in peripheral tissues [100,101]. Agonist (that is, catecholamine) binding is well established to evoke a cell-signaling cascade that promotes glucose uptake, glycogenolysis, and lipolysis to provide a readily available source of energy for skeletal muscle [102-104]. Adrb2 is expressed in a similar pattern to that of the glycolytic activating genes as it peaks at the beginning of the active phase. Interestingly, the expression of Adrb2 coincides with that of oscillating epinephrine concentrations in mammals, which has previously been identified as peaking at the beginning of the active phase in mouse models [105]. The G-protein receptor kinase, encoded by Adrbk1, phosphorylates the β2AR, thereby rendering it susceptible to receptor-mediated endocytosis via β-arrestin proteins encoded by Arrdc3 and Arrb1 [106-108]. Adrbk1, Arrdc3, and Arrb1 are all expressed in a circadian manner and antiphasic to the expression of Adrb2. These observations suggest there is a time of day difference in adrenergic signaling and that sensitivity to epinephrine may be highest in skeletal muscle during the active period while being desensitized prior to the inactive period.
Carbohydrate metabolism: genes involved in carbohydrate storage peak at the mid-active/dark phase
Excess carbohydrates are stored as glycogen in skeletal muscle which accounts for approximately 70 to 80% of whole body stores [109]. Unlike the liver, skeletal muscle glycogen content is not responsible for maintaining blood glucose concentrations but serves as a rapidly accessible energy depot for active contractions [110]. Glycogenesis is regulated by both glucose-6P concentrations and the enzymatic activity of glycogen synthase [111,112]. The gene Ppp1r3c (CT 20.0) reaches peak expression around the mid-inactive phase and encodes a regulatory subunit of the protein phosphatase-1 (PP-1) responsible for activating glycogen synthase while also inhibiting glycogen breakdown (Figure 3) [113]. Enzymatic activity of PP-1, and subsequent activation of glycogen synthase, is regulated downstream of the insulin signaling pathway [114].
Insulin promotes an anabolic signaling cascade that works in opposition to that of adrenergic signaling to drive glycogen and lipid storage. Previous reports have identified a ‘counter-regulatory’ role of the insulin receptor to selectively inhibit β2AR signaling through phosphorylation and subsequent internalization of the receptor [101,115]. Interestingly, the genes that encode the insulin receptor substrate-1, Irs1 (CT 22.0), and its downstream PI3-kinase target, Pik3r1 (CT 19.0), are both circadian with peak expression occurring at the late-active phase while the genes involved in suppressing PI3-kinase, Pik3ip1 (CT 8.0), and the insulin-receptor substrate-1, Fbxo40 (CT 5.0), reach peak expression during the inactive phase [116,117]. These data suggest that the molecular clock may act to prime skeletal muscle to store excess glucose during the mid- to late-active phase. This prediction is further supported by previous studies that report skeletal muscle glycogen content as having a diurnal rhythm with the highest levels occurring during the mid-active phase [118-120]. Skeletal muscle glucose uptake is primarily controlled via the presence/absence of the glucose transporter GLUT4/Slc2a4 in the plasma membrane (sarcolemma) and transverse tubules. A t-SNARE syntaxin-4 interacting protein, encoded by Stxbp4, has previously been shown to repress GLUT4 insertion into the plasma membrane in the absence of insulin signaling [121-123]. The gene Tbc1d1 encodes for Rab-GTPase that represses GLUT4 translocation in the absence of insulin- or contraction-induced signaling cascades [124-126]. Interestingly, Tbc1d1 and Stxbp4 are both expressed in a circadian manner and reach peak expression in the middle of the active phase (CT 19.0). Previous reports have identified Tbc1d1 as a circadian gene in skeletal muscle and other tissues [43,127]. Together, these gene products may play a role in reducing glucose uptake at the end of the active phase by repressing GLUT4 translocation and/or insertion into the plasma membrane. This temporal separation of anabolic and catabolic signaling processes in skeletal muscle may be vital for maintaining a tight regulation of serum glucose levels, and disruption of which may contribute to the metabolic phenotypes often reported in clock-mutant mice models.
Generation of an inducible skeletal muscle-specific mouse model of Bmal1 inactivation
Use of the high-resolution microarray data set allowed for the identification of mRNAs expressed in a circadian pattern, but this could be due to the intrinsic molecular clock or could be a response to external behavioral (feeding/activity) or neural/humoral cues [24,128,129]. To determine the role of the intrinsic skeletal muscle molecular clock in the temporal regulation of metabolic gene expression, we generated an inducible mouse model to inactivate Bmal1 specifically in adult skeletal muscles. Upon treatment with tamoxifen in 12-week-old adult mice, we detect recombination of exon-8 (that is, DNA binding region) of the Bmal1 gene specifically in skeletal muscle (Figure 4A), confirming the tissue specificity of the mouse model. We waited until 12 weeks of age to limit possible developmental effects as BMAL1 has been shown to promote myogenesis [20,130]. As seen in Figure 4A, recombination was not detected in the skeletal muscle or non-muscle tissues of vehicle-treated mice (iMS-Bmal1+/+). Western blot analysis confirmed the depletion of BMAL1 protein in the skeletal muscle of the iMS-Bmal1−/− mice with no effect on the liver (Figure 4B). Tamoxifen-induced loss of Bmal1 in adult skeletal muscle resulted in significant and expected gene expression changes of genes involved in the core clock mechanism. In particular, genes directly activated by the BMAL1/CLOCK heterodimer, such as Rev-erbα and Dbp, are markedly downregulated in iMS-Bmal1−/− but not in the iMS-Bmal1+/+ samples (Figure 4C). Collectively, these results demonstrate the effective loss of BMAL1 protein and disruption of core-clock gene expression in the iMS-Bmal1−/− muscle tissue.
Figure 4.
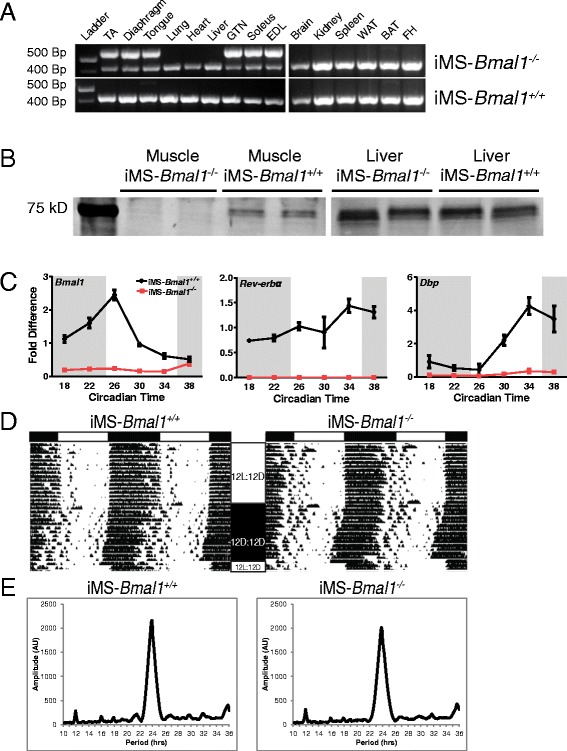
Characterization of iMS-Bmal1 −/− mice. Recombination assay (A) of genomic DNA isolated from muscle and non-muscle tissues from tamoxifen-treated (iMS-Bmal1 −/−) and vehicle-treated (iMS-Bmal1 +/+) mice at 17 to 18 weeks of age (5 weeks postinjection). Recombination of the Bmal1 gene (exon 8) yields a 572-bp PCR product. The non-recombined allele is detected at 431 bp. Western blot (B) analysis of BMAL1 expression in iMS-Bmal1 −/− and iMS-Bmal1 +/+ liver and gastrocnemius samples. Note that the original blot containing both muscle and liver samples was cut, and brightness/contrast was altered to enhance the visibility of Bmal1 in the muscle samples. (C) Real-time PCR results of time-course expression values for Bmal1, Rev-erbα, and Dbp in the iMS-Bmal1 +/+ (black) and iMS-Bmal1 −/− (red). Representative wheel running rhythms (D) of iMS-Bmal1 −/− and iMS-Bmal1 +/+ mice. White and black bars (top) indicate light and dark phases. 12 L/12D represents the 12-h light/12-h dark cycle. 12D/12D represents constant darkness conditions. Tick marks indicate wheel running activity. Representative chi-squared periodograms (E) of iMS-Bmal1 −/− and iMS-Bmal1 +/+ mice indicating approximate 24-h period lengths in both mice.
iMS-Bmal1−/− display normal circadian activity rhythms
We used voluntary wheel running to assess circadian behavior in the iMS-Bmal1 mice 22 to 29 weeks posttreatment. We did not detect any significant differences in entrainment to light under 12-h light/12-h dark conditions between iMS-Bmal1+/+ and iMS-Bmal1−/−, and analysis of activity rhythms under constant darkness did not reveal any changes in circadian behavior (Figure 4D,E). Clock-lab analysis indicates that both iMS-Bmal1+/+ and iMS-Bmal1−/− exhibit approximate 24-h period lengths (23.85 ± 0.083 and 23.77 ± 0.138 h, respectively) with no differences in amplitude, the relative strength of the rhythm. These data are consistent with other studies and confirm that inactivation of BMAL1 in skeletal muscle does not directly alter circadian activity patterns [43,131]. Therefore, gene expression changes observed in this model are more likely to be downstream of the endogenous molecular clock mechanism in skeletal muscle.
Expression of key circadian metabolic genes are significantly altered in iMS-Bmal1−/− skeletal muscle
Gene expression analysis of iMS-Bmal1+/+ and iMS-Bmal1−/− muscle tissue reveals that the intrinsic molecular clock, even in constant conditions, plays a role in temporally regulating carbohydrate and lipid metabolism. We performed our transcriptome analysis at 5 weeks postrecombination to identify early gene expression changes caused by the loss of the clock mechanism in skeletal muscle. Analyzing gene expression at this time point also limits potential off-target effects of tamoxifen treatment by allowing for a sufficient wash-out period. We found that the circadian genes involved in carbohydrate metabolism were most affected by loss of Bmal1. The expression of the glycolytic enzymes, Pfkfb1, Pfkfb3, and Hk2 as well as the PDH phosphatase, Pdp1 were all significantly downregulated in the gastrocnemius (Figure 5A). In addition, expression of the adrenergic receptor, Adrb2, was also significantly decreased. These genes are convincing clock-controlled candidates in skeletal muscle as they have circadian expression patterns similar to that of known clock-controlled genes (peak expression during inactive to active phase transition), and their loss of expression following Bmal1 inactivation is indicative of direct transcriptional regulation by the clock. By targeting these genes, the molecular clock mechanism can precisely regulate the timing of carbohydrate utilization to occur during the active phase. The observation that circadian genes involved in glucose utilization are diminished in our model is in agreement with the muscle-specific Bmal1 knockout model generated by Dyar et al. in which they report significant decreases in glucose oxidation and insulin stimulated glucose uptake in their muscle tissues [43].
Figure 5.
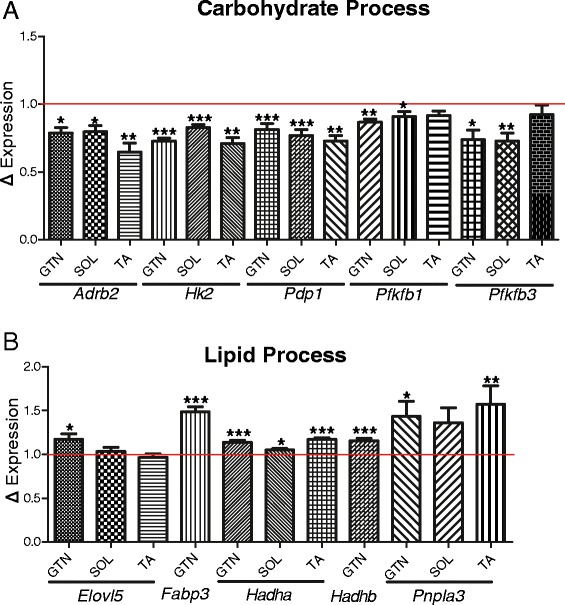
Differentially expressed circadian, metabolic genes in iMS-Bmal1−/− skeletal muscle. Average expression changes of the circadian carbohydrate (A) and lipid (B) genes in iMS-Bmal1−/− gastrocnemius averaged over circadian times 18, 22, 26, 30, 34, and 38. Tibialis anterior and soleus gene expression changes (Dyar et al.) averaged over circadian times 0, 4, 8, 12, 16, and 20. The red line denotes control (iMS-Bmal1 +/+) gene expression values. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Lipid metabolic processes appear to be elevated as the nuclear co-repressor, Nrip1, involved in repressing β-oxidation was significantly decreased with loss of Bmal1 (approximately 21% decrease, Student’s t test P value = 0.019). Previous studies have shown that knockout of Nrip1 results in an increase in succinate dehydrogenase staining of gastrocnemius muscle consistent with a shift to slow oxidative fiber types [72]. Interestingly, the fatty-acid transporter, Fabp3, and the β-oxidation genes, Hadha and Hadhb, were significantly elevated in the iMS-Bmal1−/− gastrocnemius tissues (Figure 5B). Two circadian genes involved in triacylglycerol elongation, Pnpla3 and Elovl5, were also increased in the iMS-Bmal1−/−. Altogether, we report significant expression changes in circadian genes that are key regulators of metabolism in skeletal muscle. We think that the gene changes observed in iMS-Bmal1−/− are either directly or indirectly regulated downstream of BMAL1/molecular clock in skeletal muscle and not due to changes in external cues as circadian activity patterns in iMS-Bmal1−/− are indistinguishable from vehicle-treated controls. The observation that circadian genes involved in carbohydrate and lipid metabolism are disrupted in iMS-Bmal1−/− highlights a fundamental importance of the intrinsic molecular clock in temporal regulation of substrate utilization and storage in skeletal muscle in the absence of external cues.
iMS-Bmal1−/− gene expression changes reveal a fast to slow fiber-type shift
Skeletal muscle comprises different fiber types that are differentiated based on contractile function as well as predominant substrate utilization [132-135]. For example, fast-type skeletal muscles (type IIX/IIB) primarily rely on ATP generated from anaerobic metabolism (glycolysis/lactic-acid fermentation) to provide quick energy sources required for short bursts of activity, while slow-type skeletal muscles and fast-type IIA muscles rely on oxidative metabolism to promote a more sustained and less fatigable bout of contractions. We analyzed changes in gene expression related to fiber type following Bmal1 ablation in adult skeletal muscle and included both circadian and non-circadian transcripts. We identified a selective increase in slow-type sarcomeric genes in the gastrocnemius muscles with a limited effect on fast-type sarcomeric genes (Figure 6A,B). We chose the list of ‘slow’ and ‘fast’ sarcomeric genes, because these have been shown to be significantly enriched in either slow-soleus or fast-EDL myofiber preparations [136]. Additionally, calcium handling genes and nuclear receptors common in slow-fiber muscles (for example, Casq2, Atp2a2, Ankrd2, Csrp3.) were significantly increased in iMS-Bmal1−/− (Table 1). Similar to the changes observed for the circadian metabolic genes, we see that non-circadian metabolic genes involved in carbohydrate metabolism are significantly decreased, while genes involved in lipid metabolism are increased (Tables 2 and 3). This switch from a fast to a slow fiber type mRNA profile is in agreement with the observed metabolic changes as slow fiber type muscles rely more heavily on oxidative metabolism compared to fast-type skeletal muscle.
Figure 6.

Increase in slow type sarcomeric genes in iMS-Bmal1 −/−. Average gene expression changes of slow (A) and fast (B) type sarcomeric genes in iMS-Bmal1 −/− compared to control values (red line). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Table 1.
Fiber-type specific gene expression changes in iMS- Bmal1 −/−
| Gene symbol | Fast or slow | Gene description | ΔExpression (Tam/Veh) | Student’s t test |
|---|---|---|---|---|
| Atp2a1 | Fast | Calcium handling | 0.99 | ns |
| Atp2a2 | Slow | Calcium handling | 1.06 | ns |
| Calm3 | Fast | Calcium handling | 0.84 | *** |
| Casq1 | Fast | Calcium handling | 1.00 | ns |
| Casq2 | Slow | Calcium handling | 2.89 | *** |
| Itpr1 | Fast | Calcium handling | 1.05 | ns |
| Pvalb | Fast | Calcium handling | 1.02 | ns |
| Ankrd2 | Slow | Nuclear receptor | 1.66 | * |
| Csrp3 | Slow | Nuclear receptor | 2.13 | ** |
| Fhl1 | Slow | Nuclear receptor | 1.28 | ** |
| Nfatc2 | Slow | Nuclear receptor | 0.88 | ns |
| Pdlim1 | Slow | Nuclear receptor | 1.51 | *** |
| Ppara | Slow | Nuclear receptor | 1.23 | * |
| Ppargc1a | Fast | Nuclear receptor | 0.83 | * |
| Sos2 | Fast | Nuclear receptor | 0.84 | *** |
Average gene expression changes of calcium handling and nuclear receptor genes in iMS-Bmal1 −/−. ns, non-significant; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Table 2.
Metabolic genes upregulated in iMS- Bmal1 −/−
| Gene symbol | Gene function | ΔExpression (Tam/Veh) | Student’s t test |
|---|---|---|---|
| Agpat3 | Lipogenesis | 1.59 | *** |
| Acadm | Lipolysis | 1.31 | *** |
| Acot7 | Lipolysis | 1.18 | *** |
| Acot9 | Lipolysis | 1.44 | ** |
| Acsl1 | Lipolysis | 1.24 | ** |
| Cd36 | Lipid transport | 1.18 | ** |
| Cox5a | Electron transport chain | 1.24 | *** |
| Cox6a1 | Electron transport chain | 1.30 | * |
| Cpt2 | Lipolysis | 1.11 | * |
| Fabp1 | Lipid transport | 1.28 | * |
| Fabp5 | Lipid transport | 1.29 | ** |
| Fads2 | Lipogenesis | 1.29 | * |
| Ldhb | Lactate metabolism | 1.33 | *** |
| Ndufa8 | Electron transport chain | 1.24 | *** |
| Ndufb8 | Electron transport chain | 1.18 | ** |
| Plin5 | Lipogenesis | 1.41 | *** |
| Sdhc | Electron transport chain | 1.18 | *** |
| Sdhd | Electron transport chain | 1.21 | ** |
| Uqcr10 | Electron transport chain | 1.14 | ** |
Average gene expression changes of metabolic genes that are significantly upregulated in iMS-Bmal1 −/− skeletal muscle. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Table 3.
Metabolic genes downregulated in iMS- Bmal1 −/−
| Gene symbol | Gene function | ΔExpression (Tam/Veh) | Student’s t test |
|---|---|---|---|
| Agl | Glycogenolysis | 0.83 | *** |
| Akt1 | Glucose uptake | 0.84 | ** |
| Il15 | Glucose uptake | 0.86 | * |
| Pak1 | Glucose uptake | 0.79 | * |
| Pfkm | Glycolysis | 0.81 | *** |
| Pgm2 | Glycogenolysis | 0.87 | *** |
| Phka1 | Glycogenolysis | 0.81 | ** |
| Prkab2 | Glucose uptake | 0.85 | * |
| Prkag2 | Glucose uptake | 0.83 | *** |
| Prkag3 | Glucose uptake | 0.71 | ** |
| Rab10 | Glucose uptake | 0.86 | ** |
| Slc2a3 | Glucose uptake | 0.35 | *** |
Average gene expression changes of metabolic genes that are significantly downregulated in iMS-Bmal1 −/− skeletal muscle. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
Conclusions
Here, we report that the intrinsic molecular clock regulates the timing of genes involved in substrate catabolic and anabolic processes in skeletal muscle. We have identified the mid-inactive period as the time of peak expression of genes involved in fatty-acid breakdown, possibly serving as the main energy source to skeletal muscle during the overnight fasting period. The temporal expression pattern of genes that regulate glycolysis and glycolytic flux into the Kreb’s cycle suggests a shift in substrate utilization during the early active period from lipids to carbohydrates, which has previously been documented in other muscle-specific Bmal1 knockout models [43]. Genes involved in glucose and lipid storage were observed as reaching peak expression toward the end of the active phase, where we predict excess energy is stored for usage during the postabsorptive phase. Expression analysis of time-course data from iMS-Bmal1−/− skeletal muscle revealed the differential expression of a number of key circadian metabolic genes in the absence of BMAL1. These finding suggests that the temporal regulation and circadian rhythmicity of these genes is directly downstream of the intrinsic skeletal muscle molecular clock mechanism. Lastly, we observe a gene expression profile that is indicative of a glycolytic to oxidative fiber type shift with loss of Bmal1 in adult muscle tissue. These findings suggest a potential unidentified role of Bmal1 in the maintenance of fast-type muscle fibers, possibly via direct transcriptional regulation of glucose handling. It is widely reported that aging is associated with a selective loss of fast-type skeletal muscle fibers [137,138]. In addition, aging is also associated with decreases in the robustness of the molecular clock [139,140]. These observations raise the possibility that fast to slow fiber-type shifts may be a result of dampening of the molecular clock with age.
Acknowledgements
The authors would wish to thank John Hogenesch for providing the skeletal muscle circadian transcriptome data. We would like to thank John McCarthy for his intellectual support in the analysis of the data. We would like to thank Tanya Seward for the breeding and maintaining of mouse colonies. We would like to thank Trusha Mehta for contributing with the collection and analysis of the mouse activity data. We would like to thank Donna Wall and the Microarray Core Facility at the University of Kentucky for performing the microarray experiments. This work was supported by funding from the National Institutes of Health, NIH AR066082.
Abbreviations
- Acaa2
acetyl-Coenzyme A acyltransferase 2
- Acaca
acetyl-Coenzyme A carboxylase alpha
- Acadm
acyl-Coenzyme A dehydrogenase, medium chain
- Acly
ATP citrate lyase
- Acot7
acyl-CoA thioesterase 7
- Acot9
acyl-CoA thioesterase 9
- Acsl1
acyl-CoA synthetase long-chain family member 1
- Acsl5
acyl-CoA synthetase long-chain family member 5
- Actn2
actinin alpha 2
- Actn3
actinin alpha 3
- Adrb2
β2-adrenergic receptor
- Adrbk1
adrenergic receptor kinase, beta 1
- Agl
amylo-1,6-glucosidase, 4-alpha-glucanotransferase
- Agpat3
1-acylglycerol-3-phosphate O-acyltransferase 3
- Akt1
thymoma viral proto-oncogene 1
- Ankrd2
ankyrin repeat domain 2 (stretch-responsive muscle)
- Arrb1
arrestin, beta 1
- Arrdc3
arrestin domain containing 3
- Atp2a1
ATPase, Ca++ transporting, cardiac muscle, fast twitch 1
- Atp2a2
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
- BAT
brown adipose tissue
- bHLH
basic helix-loop-helix
- Bhlhe40
basic helix-loop-helix family, member e40
- Bmal1
brain and muscle ARNT-like 1
- Calm3
calmodulin 3
- Casq1
calsequestrin 1
- Casq2
calsequestrin 2
- Cd36
(FAT) fatty acid translocase
- cDNA
complementary DNA
- Clock
Circadian Locomotor Output Cycles Kaput
- CoA
coenzyme A
- Cox5a
cytochrome c oxidase subunit Va
- Cox6a1
cytochrome c oxidase subunit VIa polypeptide 1
- Cpt2
carnitine palmitoyltransferase 2
- Csrp3
cysteine and glycine-rich protein 3
- CT
circadian time
- DAG
diacylglycerol
- Dbp
D site albumin promoter binding protein
- DD
dark/dark
- Dgat1
diacylglycerol O-acyltransferase 1
- Ech1
enoyl coenzyme A hydratase 1
- EDL
extensor digitorum longus
- Elovl5
ELOVL family member 5, elongation of long chain fatty acids
- Esrra
estrogen-related receptor, alpha
- Fabp1
fatty acid binding protein 1
- Fabp3
fatty acid binding protein 3
- Fabp4
fatty acid binding protein 4
- Fabp5
fatty acid binding protein 5
- Fads2
fatty acid desaturase 2
- Fasn
fatty acid synthase
- Fbxo40
F-box protein 40
- FH
femoral head
- GTN
gastrocnemius
- Fhl1
four and a half LIM domains 1
- Hadha
enoyl-Coenzyme A hydratase (trifunctional protein), alpha subunit
- Hadhb
enoyl-Coenzyme A hydratase (trifunctional protein), beta subunit
- HDAC3
histone deacetylase 3
- Hk2
hexokinase-2
- Il15
interleukin 15
- iMS-Bmal1
inducible skeletal muscle-specific Bmal1
- Irs1
insulin receptor substrate-1
- Itpr1
inositol 1,4,5-trisphosphate receptor 1
- JTK_CYCLE
Jonckheer-Terpstra-Kendall Cycle Algorithm
- LD
light/dark
- Ldhb
lactate dehydrogenase B
- Ndufa8
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8
- LPA
lysophosphatidic acid
- Lpin1
lipin 1
- Mlxip
MLX interacting protein
- Mlycd
malonyl-CoA decarboxylase
- Murc
muscle-related coiled-coil protein
- Myh1
myosin, heavy polypeptide 1, skeletal muscle, adult
- Myh2
myosin, heavy polypeptide 2, skeletal muscle, adult
- Myh3
myosin, heavy polypeptide 3, skeletal muscle, embryonic
- Myh4
myosin, heavy polypeptide 4, skeletal muscle
- Myh6
myosin, heavy polypeptide 6, cardiac muscle, alpha
- Myh7
myosin, heavy polypeptide 7, cardiac muscle, beta
- Myh8
myosin, heavy polypeptide 8, skeletal muscle
- Myl2
myosin, light polypeptide 2, regulatory, cardiac, slow
- Myl3
myosin, light polypeptide 3
- Myl7
myosin, light polypeptide 7, regulatory
- Myom2
myomesin 2
- Myoz1
myozenin 1
- Myoz2
myozenin 2
- NCBI GEO
National Center for Biotechnology Information Gene Expression Omnibus
- Ndufb8
NADH dehydrogenase (ubiquinone) 1 beta subcomplex 8
- Nfatc2
nuclear factor of activated T cells, cytoplasmic, calcineurin-dependent 2
- Nrip1
nuclear receptor interacting protein 1
- PA
phosphatidic acid
- Pak1
p21 protein (Cdc42/Rac)-activated kinase 1
- PCR
polymerase chain reaction
- PDH
pyruvate dehydrogenase complex
- PDK
pyruvate dehydrogenase kinase
- Pdk4
pyruvate dehydrogenase kinase, isoenzyme 4
- Pdlim1
PDZ and LIM domain 1
- PDP
pyruvate dehydrogenase phosphatase
- Pdp1
pyruvate dehydrogenase phosphatase catalytic subunit 1
- PFK2
phosphofructokinase-2
- Pfkfb1
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 1
- Pfkfb3
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
- Pfkfb4
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4
- Pfkm
phosphofructokinase-1
- Pgm2
phosphoglucomutase 2
- Phka1
phosphorylase kinase alpha 1
- Pik3ip1
phosphoinositide-3-kinase interacting protein 1
- Pik3r1
phosphatidylinositol 3-kinase, regulatory subunit, polypeptide 1 (p85 alpha)
- Plin5
perilipin 5
- Pnpla3
patatin-like phospholipase domain containing 3
- PP-1
protein phosphatase-1
- Ppargc1b
peroxisome proliferative activated receptor, gamma, coactivator 1 beta
- Ppara
peroxisome proliferator activated receptor alpha
- Pparδ
peroxisome proliferator activator receptor delta
- Ppargc1a
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
- Ppp1r3c
protein phosphatase 1, regulatory (inhibitor) subunit 3C
- Prkab2
protein kinase, AMP-activated, beta 2 non-catalytic subunit
- Prkag2
protein kinase, AMP-activated, gamma 2 non-catalytic subunit
- Prkag3
protein kinase, AMP-activated, gamma 3 non-catatlytic subunit
- Pvalb
parvalbumin
- Rab10
RAB10, member RAS oncogene family
- Rev-erbα
nuclear receptor subfamily 1, group D, member 1
- RORE
REV-ERB response element
- Scd1
stearoyl-Coenzyme A desaturase 1
- Sdhc
succinate dehydrogenase complex, subunit C, integral membrane protein
- Sdhd
succinate dehydrogenase complex, subunit D, integral membrane protein
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- Slc25a20
solute carrier family 25 (mitochondrial carnitine/acylcarnitine translocase)
- Slc2a3
solute carrier family 2 (facilitated glucose transporter), member 3
- Smtnl1
smoothelin-like 1
- Sos2
son of sevenless homolog 2
- Srebf1
sterol regulatory element binding transcription factor 1
- Srebf2
sterol regulatory element binding factor 2
- Stxbp4
syntaxin binding protein 4
- TA
tibialis anterior
- TAG
triacylglycerol
- Tbc1d1
TBC1 domain family, member 1
- Tmod1
tropomodulin 1
- Tnnc1
troponin C, cardiac/slow skeletal
- Tnnc2
troponin C2, fast
- Tnni1
troponin I, skeletal, slow 1
- Tnni2
troponin I, skeletal, fast 2
- Tnnt1
troponin T1, skeletal, slow
- Tnnt3
troponin T3, skeletal, fast
- Uqcr10
ubiquinol-cytochrome c reductase, complex III subunit X
- Vcl
vinculin
- WAT
white adipose tissue
- Xirp1
xin actin-binding repeat containing 1
Additional files
Normalized gene expression traces of circadian metabolic genes. Normalized expression traces of the circadian metabolic genes from the high-resolution skeletal muscle time-course transcriptome (data were downloaded from NCBI GEO datasets-GSE54652). Grey bars indicated the active period and white bars indicate the inactive period. Note that mice were in constant darkness during the time-course collection. Red lines indicate the acrophase (time of peak expression) calculated by JTK_CYCLE algorithm. A 6° polynomial was fitted to the data to highlight the temporal expression pattern (black line). The genes are categorized by function and listed in the following order: lipid breakdown, lipid storage, carbohydrate breakdown, and carbohydrate storage.
Temporal gene expression traces of circadian metabolic genes. Gene expression traces for circadian metabolic genes from the Mouse ST 1.0 Affymetrix gene array for gastrocnemius tissue collected at circadian times 18 to 38. iMS-Bmal1 +/+ control values are indicated as black diamonds and iMS-Bmal1 −/− are indicated as red squares. Grey bars indicated the active period, and white bars indicate the inactive period. Note that mice were in constant darkness during the time-course collection.
Real-time PCR results for circadian metabolic genes. Real-time PCR results (C) of time-course expression values for Fabp3, Pnpla3, Hk2, and Pdp1 in the iMS-Bmal1 +/+ (black) and iMS-Bmal1 −/− (red). Paired t test of Fabp3 (P value = 0.02), Pnpla3 (P value = 0.4), Hk2 (P value = 0.001), and Pdp1 (P value = 0.15).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
BAH participated in the time-course collection and RNA extraction from iMS-Bmal1 muscle tissue, contributed to the recombination assay, carried out the bioinformatics analysis, and drafted the manuscript. YW participated in the bioinformatics analysis, contributed to the interpretation of the data, and helped with the writing of the manuscript. LAR helped with the bioinformatics analysis and figure production. JHE participated in the time-course collection and preparation of the RNA used in the microarray assays. BDH and EAS participated in the time-course collection and completed the mouse activity data collection and analysis. EAS also carried out the real-time PCR analysis and participated in the recombination assay. XZ participated in the time-course collection and performed the Western blot analysis. KAE was involved in the design and coordination of the study and helped with the drafting of the manuscript. The final manuscript was read and approved by all authors.
Contributor Information
Brian A Hodge, Email: baho223@uky.edu.
Yuan Wen, Email: yuan.wen@uky.edu.
Lance A Riley, Email: lance.riley@uky.edu.
Xiping Zhang, Email: xiping.zhang@uky.edu.
Jonathan H England, Email: jonathan.england@uky.edu.
Brianna D Harfmann, Email: harfmannbd@uky.edu.
Elizabeth A Schroder, Email: eschr0@uky.edu.
Karyn A Esser, Email: karyn.esser@gmail.com.
References
- 1.Hoppeler H, Fluck M. Normal mammalian skeletal muscle and its phenotypic plasticity. J Exp Biol. 2002;205:2143–52. doi: 10.1242/jeb.205.15.2143. [DOI] [PubMed] [Google Scholar]
- 2.Zurlo F, Larson K, Bogardus C, Ravussin E. Skeletal muscle metabolism is a major determinant of resting energy expenditure. J Clin Invest. 1990;86:1423–7. doi: 10.1172/JCI114857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egan B, Zierath JR. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17:162–84. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Frontera WR, Ochala J. Skeletal muscle, a brief review of structure and function: Calcif Tissue Int. 2014. [DOI] [PubMed] [Google Scholar]
- 5.Rall JA. Energetic aspects of skeletal muscle contraction: implications of fiber types. Exerc Sport Sci Rev. 1985;13:33–74. [PubMed] [Google Scholar]
- 6.Defronzo RA, Simonson D, Ferrannini E, Barrett E. Insulin resistance: a universal finding in diabetic states. Bull Schweiz Akad Med Wiss. 1981-1982:223–238. [PubMed]
- 7.Ferrannini E, Simonson DC, Katz LD, Reichard G, Jr, Bevilacqua S, Barrett EJ, et al. The disposal of an oral glucose load in patients with non-insulin-dependent diabetes. Metabolism. 1988;37:79–85. doi: 10.1016/0026-0495(88)90033-9. [DOI] [PubMed] [Google Scholar]
- 8.Green HJ, Helyar R, Ball-Burnett M, Kowalchuk N, Symon S, Farrance B. Metabolic adaptations to training precede changes in muscle mitochondrial capacity. J Appl Physiol. 1992;72:484–91. doi: 10.1152/jappl.1992.72.2.484. [DOI] [PubMed] [Google Scholar]
- 9.Benziane B, Burton TJ, Scanlan B, Galuska D, Canny BJ, Chibalin AV, et al. Divergent cell signaling after short-term intensified endurance training in human skeletal muscle. Am J Physiol Endocrinol Metab. 2008;295:E1427–38. doi: 10.1152/ajpendo.90428.2008. [DOI] [PubMed] [Google Scholar]
- 10.Jeukendrup AE. Regulation of fat metabolism in skeletal muscle. Ann N Y Acad Sci. 2002;967:217–35. doi: 10.1111/j.1749-6632.2002.tb04278.x. [DOI] [PubMed] [Google Scholar]
- 11.McCarthy JJ, Esser KA. Anabolic and catabolic pathways regulating skeletal muscle mass. Curr Opin Clin Nutr Metab Care. 2010;13:230–5. doi: 10.1097/MCO.0b013e32833781b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pilegaard H, Saltin B, Neufer PD. Effect of short-term fasting and refeeding on transcriptional regulation of metabolic genes in human skeletal muscle. Diabetes. 2003;52:657–62. doi: 10.2337/diabetes.52.3.657. [DOI] [PubMed] [Google Scholar]
- 13.Romijn JA, Coyle EF, Sidossis LS, Gastaldelli A, Horowitz JF, Endert E, et al. Regulation of endogenous fat and carbohydrate metabolism in relation to exercise intensity and duration. Am J Physiol. 1993;265:E380–91. doi: 10.1152/ajpendo.1993.265.3.E380. [DOI] [PubMed] [Google Scholar]
- 14.van Loon LJ, Greenhaff PL, Constantin-Teodosiu D, Saris WH, Wagenmakers AJ. The effects of increasing exercise intensity on muscle fuel utilisation in humans. J Physiol. 2001;536:295–304. doi: 10.1111/j.1469-7793.2001.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdulla H, Phillips B, Smith K, Wilkinson D, Atherton PJ, Idris I. Physiological mechanisms of action of incretin and insulin in regulating skeletal muscle metabolism. Curr Diabetes Rev. 2014;10:327–35. doi: 10.2174/1573399810666141017153749. [DOI] [PubMed] [Google Scholar]
- 16.Gaitanos GC, Williams C, Boobis LH, Brooks S. Human muscle metabolism during intermittent maximal exercise. J Appl Physiol. 1993;75:712–9. doi: 10.1152/jappl.1993.75.2.712. [DOI] [PubMed] [Google Scholar]
- 17.Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15 Spec No 2:R271–7. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- 18.Fukuhara C, Tosini G. Peripheral circadian oscillators and their rhythmic regulation. Front Biosci. 2003;8:d642–51. doi: 10.2741/1042. [DOI] [PubMed] [Google Scholar]
- 19.Amaral IP, Johnston IA. Circadian expression of clock and putative clock-controlled genes in skeletal muscle of the zebrafish. Am J Physiol Regul Integr Comp Physiol. 2012;302:R193–206. doi: 10.1152/ajpregu.00367.2011. [DOI] [PubMed] [Google Scholar]
- 20.Andrews JL, Zhang X, McCarthy JJ, McDearmon EL, Hornberger TA, Russell B, et al. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc Natl Acad Sci U S A. 2010;107:19090–5. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dudek M, Meng QJ. Running on time: the role of circadian clocks in the musculoskeletal system. Biochem J. 2014;463:1–8. doi: 10.1042/BJ20140700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazado CC, Kumaratunga HP, Nagasawa K, Babiak I, Giannetto A, Fernandes JM. Daily rhythmicity of clock gene transcripts in Atlantic cod fast skeletal muscle. PLoS One. 2014;9 doi: 10.1371/journal.pone.0099172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy BA, Wagner AL, McGlynn OF, Kharazyan F, Browne JA, Elliott JA. Exercise influences circadian gene expression in equine skeletal muscle. Vet J. 2014;201:39–45. doi: 10.1016/j.tvjl.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 24.Schroder EA, Esser KA. Circadian rhythms, skeletal muscle molecular clocks, and exercise. Exerc Sport Sci Rev. 2013;41:224–9. doi: 10.1097/JES.0b013e3182a58a70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Squarcini CF, Pires ML, Lopes C, Benedito-Silva AA, Esteves AM, Cornelissen-Guillaume G, et al. Free-running circadian rhythms of muscle strength, reaction time, and body temperature in totally blind people. Eur J Appl Physiol. 2013;113:157–65. doi: 10.1007/s00421-012-2415-8. [DOI] [PubMed] [Google Scholar]
- 26.Wolff G, Duncan MJ, Esser KA. Chronic phase advance alters circadian physiological rhythms and peripheral molecular clocks. J Appl Physiol. 2013;115:373–82. doi: 10.1152/japplphysiol.01139.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bass J. Circadian topology of metabolism. Nature. 2012;491:348–56. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- 28.Korencic A, Kosir R, Bordyugov G, Lehmann R, Rozman D, Herzel H. Timing of circadian genes in mammalian tissues. Sci Rep. 2014;4:5782. doi: 10.1038/srep05782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998;95:5474–9. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–67. doi: 10.1016/S0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 31.Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–41. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, et al. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shimba S, Ishii N, Ohta Y, Ohno T, Watabe Y, Hayashi M, et al. Brain and muscle Arnt-like protein-1 (BMAL1), a component of the molecular clock, regulates adipogenesis. Proc Natl Acad Sci U S A. 2005;102:12071–6. doi: 10.1073/pnas.0502383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science. 2005;308:1043–5. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan X, Zhang Y, Wang L, Hussain MM. Diurnal regulation of MTP and plasma triglyceride by CLOCK is mediated by SHP. Cell Metab. 2010;12:174–86. doi: 10.1016/j.cmet.2010.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimba S, Ogawa T, Hitosugi S, Ichihashi Y, Nakadaira Y, Kobayashi M, et al. Deficient of a clock gene, brain and muscle Arnt-like protein-1 (BMAL1), induces dyslipidemia and ectopic fat formation. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bugge A, Feng D, Everett LJ, Briggs ER, Mullican SE, Wang F, et al. Rev-erbalpha and Rev-erbbeta coordinately protect the circadian clock and normal metabolic function. Genes Dev. 2012;26:657–67. doi: 10.1101/gad.186858.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, et al. Regulation of circadian behaviour and metabolism by REV-ERB-alpha and REV-ERB-beta. Nature. 2012;485:123–7. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–7. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bray MS, Young ME. The role of cell-specific circadian clocks in metabolism and disease. Obes Rev. 2009;10(Suppl 2):6–13. doi: 10.1111/j.1467-789X.2009.00684.x. [DOI] [PubMed] [Google Scholar]
- 41.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–31. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paschos GK, Ibrahim S, Song WL, Kunieda T, Grant G, Reyes TM, et al. Obesity in mice with adipocyte-specific deletion of clock component Arntl. Nat Med. 2012;18:1768–77. doi: 10.1038/nm.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dyar KA, Ciciliot S, Wright LE, Bienso RS, Tagliazucchi GM, Patel VR, et al. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol Metab. 2014;3:29–41. doi: 10.1016/j.molmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu G, Zhu J, Yu J, Zhou L, Huang JZ, Zhang Z. Evaluation of five methods for genome-wide circadian gene identification. J Biol Rhythms. 2014;29:231–42. doi: 10.1177/0748730414537788. [DOI] [PubMed] [Google Scholar]
- 45.Pizarro A, Hayer K, Lahens NF, Hogenesch JB. CircaDB: a database of mammalian circadian gene expression profiles. Nucleic Acids Res. 2013;41:D1009–13. doi: 10.1093/nar/gks1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang R, Lahens NF, Ballance HI, Hughes ME, Hogenesch JB. A circadian gene expression atlas in mammals: implications for biology and medicine. Proc Natl Acad Sci U S A. 2014;111:16219–24. doi: 10.1073/pnas.1408886111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–80. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–10. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, et al. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–41. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCarthy JJ, Srikuea R, Kirby TJ, Peterson CA, Esser KA. Inducible Cre transgenic mouse strain for skeletal muscle-specific gene targeting. Skelet Muscle. 2012;2:8. doi: 10.1186/2044-5040-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Processing circadian data collected from the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc. 2010;2010:pdb.prot5519. doi: 10.1101/pdb.prot5519. [DOI] [PubMed] [Google Scholar]
- 53.McCarthy JJ, Andrews JL, McDearmon EL, Campbell KS, Barber BK, Miller BH, et al. Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol Genomics. 2007;31:86–95. doi: 10.1152/physiolgenomics.00066.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hughes M, Deharo L, Pulivarthy SR, Gu J, Hayes K, Panda S, et al. High-resolution time course analysis of gene expression from pituitary. Cold Spring Harb Symp Quant Biol. 2007;72:381–6. doi: 10.1101/sqb.2007.72.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang L, Li L, Jiang J, Wang Y, Zhong T, Chen Y, et al. Molecular characterization and different expression patterns of the FABP gene family during goat skeletal muscle development: Mol Biol Rep. 2014. [DOI] [PubMed] [Google Scholar]
- 56.Malik ZA, Cobley JN, Morton JP, Close GL, Edwards BJ, Koch LG, et al. Label-free LC-MS profiling of skeletal muscle reveals heart-type fatty acid binding protein as a candidate biomarker of aerobic capacity. Proc Natl Acad Sci U S A. 2013;1:290–308. doi: 10.3390/proteomes1030290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Syamsunarno MR, Iso T, Hanaoka H, Yamaguchi A, Obokata M, Koitabashi N, et al. A critical role of fatty acid binding protein 4 and 5 (FABP4/5) in the systemic response to fasting. PLoS One. 2013;8 doi: 10.1371/journal.pone.0079386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shostak A, Husse J, Oster H. Circadian regulation of adipose function. Adipocyte. 2013;2:201–6. doi: 10.4161/adip.26007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gooley JJ, Chua EC. Diurnal regulation of lipid metabolism and applications of circadian lipidomics. J Genet Genomics. 2014;41:231–50. doi: 10.1016/j.jgg.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 60.Minami Y, Kasukawa T, Kakazu Y, Iigo M, Sugimoto M, Ikeda S, et al. Measurement of internal body time by blood metabolomics. Proc Natl Acad Sci U S A. 2009;106:9890–5. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ameer F, Scandiuzzi L, Hasnain S, Kalbacher H, Zaidi N. De novo lipogenesis in health and disease. Metabolism. 2014;63:895–902. doi: 10.1016/j.metabol.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 62.Indiveri C, Iacobazzi V, Tonazzi A, Giangregorio N, Infantino V, Convertini P, et al. The mitochondrial carnitine/acylcarnitine carrier: function, structure and physiopathology. Mol Aspects Med. 2011;32:223–33. doi: 10.1016/j.mam.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt I, Herpin P. Carnitine palmitoyltransferase I (CPT I) activity and its regulation by malonyl-CoA are modulated by age and cold exposure in skeletal muscle mitochondria from newborn pigs. J Nutr. 1998;128:886–93. doi: 10.1093/jn/128.5.886. [DOI] [PubMed] [Google Scholar]
- 64.Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu Rev Nutr. 2008;28:253–72. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]
- 65.Francis GA, Fayard E, Picard F, Auwerx J. Nuclear receptors and the control of metabolism. Annu Rev Physiol. 2003;65:261–311. doi: 10.1146/annurev.physiol.65.092101.142528. [DOI] [PubMed] [Google Scholar]
- 66.Fan W, Evans R. PPARs and ERRs: molecular mediators of mitochondrial metabolism. Curr Opin Cell Biol. 2014;33C:49–54. doi: 10.1016/j.ceb.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang X, Downes M, Yu RT, Bookout AL, He W, Straume M, et al. Nuclear receptor expression links the circadian clock to metabolism. Cell. 2006;126:801–10. doi: 10.1016/j.cell.2006.06.050. [DOI] [PubMed] [Google Scholar]
- 68.Sonoda MT, Martinez L, Webb P, Skaf MS, Polikarpov I. Ligand dissociation from estrogen receptor is mediated by receptor dimerization: evidence from molecular dynamics simulations. Mol Endocrinol. 2008;22:1565–78. doi: 10.1210/me.2007-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yang X, Lamia KA, Evans RM. Nuclear receptors, metabolism, and the circadian clock. Cold Spring Harb Symp Quant Biol. 2007;72:387–94. doi: 10.1101/sqb.2007.72.058. [DOI] [PubMed] [Google Scholar]
- 70.Kressler D, Schreiber SN, Knutti D, Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J Biol Chem. 2002;277:13918–25. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 71.Finck BN, Kelly DP. PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–22. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Seth A, Steel JH, Nichol D, Pocock V, Kumaran MK, Fritah A, et al. The transcriptional corepressor RIP140 regulates oxidative metabolism in skeletal muscle. Cell Metab. 2007;6:236–45. doi: 10.1016/j.cmet.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Constantinescu S, Turcotte LP. Genetic downregulation of receptor-interacting protein 140 uncovers the central role of Akt signalling in the regulation of fatty acid oxidation in skeletal muscle cells. Exp Physiol. 2013;98:514–25. doi: 10.1113/expphysiol.2012.068833. [DOI] [PubMed] [Google Scholar]
- 74.Fritah A, Christian M, Parker MG. The metabolic coregulator RIP140: an update. Am J Physiol Endocrinol Metab. 2010;299:E335–40. doi: 10.1152/ajpendo.00243.2010. [DOI] [PubMed] [Google Scholar]
- 75.Fritah A. Control of skeletal muscle metabolic properties by the nuclear receptor corepressor RIP140. Appl Physiol Nutr Metab. 2009;34:362–7. doi: 10.1139/H09-026. [DOI] [PubMed] [Google Scholar]
- 76.Funai K, Semenkovich CF. Skeletal muscle lipid flux: running water carries no poison. Am J Physiol Endocrinol Metab. 2011;301:E245–51. doi: 10.1152/ajpendo.00152.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobrzyn A, Dobrzyn P. Stearoyl-CoA desaturase—a new player in skeletal muscle metabolism regulation. J Physiol Pharmacol. 2006;57(Suppl 10):31–42. [PubMed] [Google Scholar]
- 78.Ntambi JM, Miyazaki M. Recent insights into stearoyl-CoA desaturase-1. Curr Opin Lipidol. 2003;14:255–61. doi: 10.1097/00041433-200306000-00005. [DOI] [PubMed] [Google Scholar]
- 79.Rui L. Energy metabolism in the liver. Compr Physiol. 2014;4:177–97. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dentin R, Girard J, Postic C. Carbohydrate responsive element binding protein (ChREBP) and sterol regulatory element binding protein-1c (SREBP-1c): two key regulators of glucose metabolism and lipid synthesis in liver. Biochimie. 2005;87:81–6. doi: 10.1016/j.biochi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 81.Brewer M, Lange D, Baler R, Anzulovich A. SREBP-1 as a transcriptional integrator of circadian and nutritional cues in the liver. J Biol Rhythms. 2005;20:195–205. doi: 10.1177/0748730405275952. [DOI] [PubMed] [Google Scholar]
- 82.Matsumoto E, Ishihara A, Tamai S, Nemoto A, Iwase K, Hiwasa T, et al. Time of day and nutrients in feeding govern daily expression rhythms of the gene for sterol regulatory element-binding protein (SREBP)-1 in the mouse liver. J Biol Chem. 2010;285:33028–36. doi: 10.1074/jbc.M109.089391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Matsumoto K, Yokoyama S. Gene expression analysis on the liver of cholestyramine-treated type 2 diabetic model mice. Biomed Pharmacother. 2010;64:373–8. doi: 10.1016/j.biopha.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 84.Gilardi F, Migliavacca E, Naldi A, Baruchet M, Canella D, Le Martelot G, et al. Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kumari M, Schoiswohl G, Chitraju C, Paar M, Cornaciu I, Rangrez AY, et al. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metab. 2012;15:691–702. doi: 10.1016/j.cmet.2012.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ishimoto K. Lipin 1 in lipid metabolism. Yakugaku Zasshi. 2011;131:1189–94. doi: 10.1248/yakushi.131.1189. [DOI] [PubMed] [Google Scholar]
- 87.Takeuchi K, Reue K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipin enzymes in triglyceride synthesis. Am J Physiol Endocrinol Metab. 2009;296:E1195–209. doi: 10.1152/ajpendo.90958.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jakobsson A, Westerberg R, Jacobsson A. Fatty acid elongases in mammals: their regulation and roles in metabolism. Prog Lipid Res. 2006;45:237–49. doi: 10.1016/j.plipres.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 90.Bu SY, Mashek DG. Hepatic long-chain acyl-CoA synthetase 5 mediates fatty acid channeling between anabolic and catabolic pathways. J Lipid Res. 2010;51:3270–80. doi: 10.1194/jlr.M009407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dashty M. A quick look at biochemistry: carbohydrate metabolism. Clin Biochem. 2013;46:1339–52. doi: 10.1016/j.clinbiochem.2013.04.027. [DOI] [PubMed] [Google Scholar]
- 92.Vogt C, Ardehali H, Iozzo P, Yki-Jarvinen H, Koval J, Maezono K, et al. Regulation of hexokinase II expression in human skeletal muscle in vivo. Metabolism. 2000;49:814–8. doi: 10.1053/meta.2000.6245. [DOI] [PubMed] [Google Scholar]
- 93.Wegener G, Krause U. Different modes of activating phosphofructokinase, a key regulatory enzyme of glycolysis, in working vertebrate muscle. Biochem Soc Trans. 2002;30:264–70. doi: 10.1042/BST0300264. [DOI] [PubMed] [Google Scholar]
- 94.Wegener G, Krause U, Thuy M. Fructose 2,6-bisphosphate and glycolytic flux in skeletal muscle of swimming frog. FEBS Lett. 1990;267:257–60. doi: 10.1016/0014-5793(90)80939-G. [DOI] [PubMed] [Google Scholar]
- 95.Tornheim K. Fructose 2,6-bisphosphate and glycolytic oscillations in skeletal muscle extracts. J Biol Chem. 1988;263:2619–24. [PubMed] [Google Scholar]
- 96.Spriet LL, Heigenhauser GJ. Regulation of pyruvate dehydrogenase (PDH) activity in human skeletal muscle during exercise. Exerc Sport Sci Rev. 2002;30:91–5. doi: 10.1097/00003677-200204000-00009. [DOI] [PubMed] [Google Scholar]
- 97.Harris RA, Bowker-Kinley MM, Huang B, Wu P. Regulation of the activity of the pyruvate dehydrogenase complex. Adv Enzyme Regul. 2002;42:249–59. doi: 10.1016/S0065-2571(01)00061-9. [DOI] [PubMed] [Google Scholar]
- 98.Holness MJ, Sugden MC. Regulation of pyruvate dehydrogenase complex activity by reversible phosphorylation. Biochem Soc Trans. 2003;31:1143–51. doi: 10.1042/BST0311143. [DOI] [PubMed] [Google Scholar]
- 99.Fan J, Kang HB, Shan C, Elf S, Lin R, Xie J, et al. Tyr-301 phosphorylation inhibits pyruvate dehydrogenase by blocking substrate binding and promotes the Warburg effect. J Biol Chem. 2014;289:26533–41. doi: 10.1074/jbc.M114.593970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Emrick MA, Sadilek M, Konoki K, Catterall WA. Beta-adrenergic-regulated phosphorylation of the skeletal muscle Ca(V)1.1 channel in the fight-or-flight response. Proc Natl Acad Sci U S A. 2010;107:18712–7. doi: 10.1073/pnas.1012384107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mandic M, Drinovec L, Glisic S, Veljkovic N, Nohr J, Vrecl M. Demonstration of a direct interaction between beta2-adrenergic receptor and insulin receptor by BRET and bioinformatics. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jensen J, Lai YC. Regulation of muscle glycogen synthase phosphorylation and kinetic properties by insulin, exercise, adrenaline and role in insulin resistance. Arch Physiol Biochem. 2009;115:13–21. doi: 10.1080/13813450902778171. [DOI] [PubMed] [Google Scholar]
- 103.Dehvari N, Hutchinson DS, Nevzorova J, Dallner OS, Sato M, Kocan M, et al. β(2)-Adrenoceptors increase translocation of GLUT4 via GPCR kinase sites in the receptor C-terminal tail. Br J Pharmacol. 2012;165:1442–56. doi: 10.1111/j.1476-5381.2011.01647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boyda HN, Procyshyn RM, Pang CC, Barr AM. Peripheral adrenoceptors: the impetus behind glucose dysregulation and insulin resistance. J Neuroendocrinol. 2013;25:217–28. doi: 10.1111/jne.12002. [DOI] [PubMed] [Google Scholar]
- 105.Lee S, Donehower LA, Herron AJ, Moore DD, Fu L. Disrupting circadian homeostasis of sympathetic signaling promotes tumor development in mice. PLoS One. 2010;5 doi: 10.1371/journal.pone.0010995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Usui I, Imamura T, Satoh H, Huang J, Babendure JL, Hupfeld CJ, et al. GRK2 is an endogenous protein inhibitor of the insulin signaling pathway for glucose transport stimulation. EMBO J. 2004;23:2821–9. doi: 10.1038/sj.emboj.7600297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Patwari P, Lee RT. An expanded family of arrestins regulate metabolism. Trends Endocrinol Metab. 2012;23:216–22. doi: 10.1016/j.tem.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Han S-O, Kommaddi RP, Shenoy SK. Distinct roles for β-arrestin2 and arrestin-domain-containing proteins in β2 adrenergic receptor trafficking. EMBO Rep. 2013;14:164–71. doi: 10.1038/embor.2012.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang SC, Muscat GE. Nuclear receptors and epigenetic signaling: novel regulators of glycogen metabolism in skeletal muscle. IUBMB Life. 2013;65:657–64. doi: 10.1002/iub.1181. [DOI] [PubMed] [Google Scholar]
- 110.Jensen J, Rustad PI, Kolnes AJ, Lai YC. The role of skeletal muscle glycogen breakdown for regulation of insulin sensitivity by exercise. Front Physiol. 2011;2:112. doi: 10.3389/fphys.2011.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Villar-Palasi C, Guinovart JJ. The role of glucose 6-phosphate in the control of glycogen synthase. FASEB J. 1997;11:544–58. [PubMed] [Google Scholar]
- 112.Halse R, Bonavaud SM, Armstrong JL, McCormack JG, Yeaman SJ. Control of glycogen synthesis by glucose, glycogen, and insulin in cultured human muscle cells. Diabetes. 2001;50:720–6. doi: 10.2337/diabetes.50.4.720. [DOI] [PubMed] [Google Scholar]
- 113.Montori-Grau M, Guitart M, Garcia-Martinez C, Orozco A, Gomez-Foix AM. Differential pattern of glycogen accumulation after protein phosphatase 1 glycogen-targeting subunit PPP1R6 overexpression, compared to PPP1R3C and PPP1R3A, in skeletal muscle cells. BMC Biochem. 2011;12:57. doi: 10.1186/1471-2091-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Brady MJ, Saltiel AR. The role of protein phosphatase-1 in insulin action. Recent Prog Horm Res. 2001;56:157–73. doi: 10.1210/rp.56.1.157. [DOI] [PubMed] [Google Scholar]
- 115.Baltensperger K, Karoor V, Paul H, Ruoho A, Czech MP, Malbon CC. The beta-adrenergic receptor is a substrate for the insulin receptor tyrosine kinase. J Biol Chem. 1996;271:1061–4. doi: 10.1074/jbc.271.2.1061. [DOI] [PubMed] [Google Scholar]
- 116.Palus S, von Haehling S, Springer J. Muscle wasting: an overview of recent developments in basic research. Int J Cardiol. 2014;176:640–4. doi: 10.1016/j.ijcard.2014.08.086. [DOI] [PubMed] [Google Scholar]
- 117.Shi J, Luo L, Eash J, Ibebunjo C, Glass DJ. The SCF-Fbxo40 complex induces IRS1 ubiquitination in skeletal muscle, limiting IGF1 signaling. Dev Cell. 2011;21:835–47. doi: 10.1016/j.devcel.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 118.Leighton B, Cooper GJ. Pancreatic amylin and calcitonin gene-related peptide cause resistance to insulin in skeletal muscle in vitro. Nature. 1988;335:632–5. doi: 10.1038/335632a0. [DOI] [PubMed] [Google Scholar]
- 119.Leighton B, Kowalchuk JM, Challiss RA, Newsholme EA. Circadian rhythm in sensitivity of glucose metabolism to insulin in rat soleus muscle. Am J Physiol. 1988;255:E41–5. doi: 10.1152/ajpendo.1988.255.1.E41. [DOI] [PubMed] [Google Scholar]
- 120.Leighton B, Lozeman FJ, Vlachonikolis IG, Challiss RA, Pitcher JA, Newsholme EA. Effects of adenosine deaminase on the sensitivity of glucose transport, glycolysis and glycogen synthesis to insulin in muscles of the rat. Int J Biochem. 1988;20:23–7. doi: 10.1016/0020-711X(88)90005-5. [DOI] [PubMed] [Google Scholar]
- 121.Min J, Okada S, Kanzaki M, Elmendorf JS, Coker KJ, Ceresa BP, et al. Synip: a novel insulin-regulated syntaxin 4-binding protein mediating GLUT4 translocation in adipocytes. Mol Cell. 1999;3:751–60. doi: 10.1016/S1097-2765(01)80007-1. [DOI] [PubMed] [Google Scholar]
- 122.Okada S, Ohshima K, Uehara Y, Shimizu H, Hashimoto K, Yamada M, et al. Synip phosphorylation is required for insulin-stimulated Glut4 translocation. Biochem Biophys Res Commun. 2007;356:102–6. doi: 10.1016/j.bbrc.2007.02.095. [DOI] [PubMed] [Google Scholar]
- 123.Saito T, Okada S, Nohara A, Tagaya Y, Osaki A, Oh IS, et al. Syntaxin4 interacting protein (Synip) binds phosphatidylinositol (3,4,5) triphosphate. PLoS One. 2012;7 doi: 10.1371/journal.pone.0042782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.An D, Toyoda T, Taylor EB, Yu H, Fujii N, Hirshman MF, et al. TBC1D1 regulates insulin- and contraction-induced glucose transport in mouse skeletal muscle. Diabetes. 2010;59:1358–65. doi: 10.2337/db09-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Cartee GD. Roles of TBC1D1 and TBC1D4 in insulin- and exercise-stimulated glucose transport of skeletal muscle. Diabetologia. 2015;58:19–30. doi: 10.1007/s00125-014-3395-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Szekeres F, Chadt A, Tom RZ, Deshmukh AS, Chibalin AV, Bjornholm M, et al. The Rab-GTPase-activating protein TBC1D1 regulates skeletal muscle glucose metabolism. Am J Physiol Endocrinol Metab. 2012;303:E524–33. doi: 10.1152/ajpendo.00605.2011. [DOI] [PubMed] [Google Scholar]
- 127.Yan J, Wang H, Liu Y, Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLoS Comput Biol. 2008;4 doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Harfmann BD, Schroder EA, Esser KA. Circadian rhythms, the molecular clock, and skeletal muscle. J Biol Rhythms. 2014. [DOI] [PMC free article] [PubMed]
- 129.Cermakian N, Sassone-Corsi P. Multilevel regulation of the circadian clock. Nat Rev Mol Cell Biol. 2000;1:59–67. doi: 10.1038/35036078. [DOI] [PubMed] [Google Scholar]
- 130.Chatterjee S, Nam D, Guo B, Kim JM, Winnier GE, Lee J, et al. Brain and muscle Arnt-like 1 is a key regulator of myogenesis. J Cell Sci. 2013;126:2213–24. doi: 10.1242/jcs.120519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–8. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev. 2011;91:1447–531. doi: 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 133.Blaauw B, Schiaffino S, Reggiani C. Mechanisms modulating skeletal muscle phenotype. Compr Physiol. 2013;3:1645–87. doi: 10.1002/cphy.c130009. [DOI] [PubMed] [Google Scholar]
- 134.Pette D. The adaptive potential of skeletal muscle fibers. Can J Appl Physiol. 2002;27:423–48. doi: 10.1139/h02-023. [DOI] [PubMed] [Google Scholar]
- 135.Zierath JR, Hawley JA. Skeletal muscle fiber type: influence on contractile and metabolic properties. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chemello F, Bean C, Cancellara P, Laveder P, Reggiani C, Lanfranchi G. Microgenomic analysis in skeletal muscle: expression signatures of individual fast and slow myofibers. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brunner F, Schmid A, Sheikhzadeh A, Nordin M, Yoon J, Frankel V. Effects of aging on type II muscle fibers: a systematic review of the literature. J Aging Phys Act. 2007;15:336–48. doi: 10.1123/japa.15.3.336. [DOI] [PubMed] [Google Scholar]
- 138.Purves-Smith FM, Sgarioto N, Hepple RT. Fiber typing in aging muscle. Exerc Sport Sci Rev. 2014;42:45–52. doi: 10.1249/JES.0000000000000012. [DOI] [PubMed] [Google Scholar]
- 139.Wyse CA, Coogan AN. Impact of aging on diurnal expression patterns of CLOCK and BMAL1 in the mouse brain. Brain Res. 2010;1337:21–31. doi: 10.1016/j.brainres.2010.03.113. [DOI] [PubMed] [Google Scholar]
- 140.Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–73. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]


