Abstract
We investigate how the coulombic Gibbs free energy and salt ion association per phosphate charge of DNA oligomers vary with oligomer size (i.e. number of charged residues ∣ZD∣) at 0.15 M univalent salt by non-linear Poisson Boltzmann (NLPB) analysis of all-atom DNA models. Calculations of these quantities (, ) are performed for short and long double-stranded (ds) and single-stranded (ss) DNA oligomers, ranging from 4 to 118 phosphates (ds) and from 2 to 59 phosphates (ss). Behaviors of and as functions of ∣ZD∣ provide a measure of the range of the coulombic end effect and determine the size of an oligomer at which an interior region with the properties (per charge) of the infinite-length polyelectrolyte first appears. This size (10–11 phosphates at each end for ds DNA and 6–9 for ss DNA at 0.15 M salt) is in close agreement with values obtained previously by Monte Carlo and NLPB calculations for cylindrical models of polyions, and by analysis of binding of oligocations to DNA oligomers. Differences in and in between ss and ds DNA are used to predict effects of oligomeric size and salt concentration on duplex stability in the vicinity of 0.15 M salt. Results of all-atom calculations are compared with results of less structurally detailed models and with experimental data.
Introduction
DNA and RNA molecules in aqueous salt solutions are oligo- or polyanions with high axial densities of negatively charged phosphate groups: 2 phosphates per 3.4 Å for double-stranded (ds) DNA, 1 phosphate per 3.4 Å (or >3.4 Å) for stacked (or unstacked) single-stranded (ss) DNA. As a consequence of these high axial charge densities, steep gradients in concentrations of salt cations and anions extend radially for approximately 100 Å from the nucleic acid surface at low to moderate salt concentration ([salt]). Even at very low [salt], the local salt cation concentration near the surface of interior regions of duplex DNA or RNA is in the molar range and the local salt anion concentration is negligibly small. Local concentrations of both salt cations and anions increase with increasing bulk [salt] and the concentration gradients for both salt ions are reduced, reducing this source of thermodynamic nonideality. At low [salt], the thermodynamic consequences of the coulombic interactions of the phosphate charges on one nucleic acid molecule with each other and with the surrounding atmosphere of salt ions are accurately described by the solution of the nonlinear Poisson–Boltzmann equation for the cylindrical cell model, and to a good approximation by counterion condensation theory.1,2
In each of these approaches, the salt ion distribution is evaluated around a DNA model and and/or is calculated from this distribution. Detailed comparisons have been made between NLPB and CC thermodynamic predictions,2–4 and between NLPB and canonical or grand canonical Monte Carlo (MC) predictions for the same (cylindrical) model of the polyion.5–7 These comparisons revealed (1) that analytical CC limiting law thermodynamic expressions are obtained directly from NLPB without the assumption of counterion condensation, (2) that NLPB ion distributions and thermodynamics are in quantitative agreement with results of MC simulations for the cylinder model of nucleic acids over a wide range of univalent salt concentrations (<1 mM to approximately 1 M), and (3) that even for salt solutions containing both divalent and univalent cations, trends in NLPB results with [salt] agree with those from MC predictions.
All high-charge-density oligo- and polyelectrolytes, including ds and ss nucleic acids, exhibit significant coulombic end effects (CEE). Radial salt ion concentration gradients near the ends of the nucleic acid are predicted by MC8 and NLPB9 calculations to be less steep than those characteristic of the interior and, as a consequence, strong axial gradients in salt ion concentration at and near the surface of the nucleic acid are predicted to exist over ~10 phosphates at each of its ends. The salt cation (anion) concentration at the surface of the nucleic acid is predicted to decrease (increase) at each end of the nucleic acid, relative to that in the central region. 23 Na NMR experiments comparing local Na+ accumulation for 20 bp and 160 bp DNA oligomers are in agreement with NLPB predictions for the cylindrical model.10 The [salt]-dependent thermodynamic behavior of oligomeric nucleic acids with less than approximately 20–30 phosphate charges is dominated by this coulombic end effect,8,9,11–15 which is also important for analysis of [salt]-dependent thermodynamic properties and interactions of the ends of polymeric nucleic acids.
Unambiguous experimental evidence of CEE is obtained from analysis of effects of [salt] on helix–coil transition temperatures Tm of hairpin helices formed by DNA oligomers of different ∣ZD∣16 and on equilibrium constants Kobs for binding of oligocationic ligands (Lz+) to DNA oligomers of different ∣ZD∣.11,15 For example, the derivative of Tm with respect to ln[salt] (STm = dTm/dln[salt]) is only half as large for a 22 residue (9 bp) dAT hairpin as for polymeric dAT.16 Values of the standard free energy change at 0.1 M salt and the salt derivative SKobs = ∂ln Kobs/∂ln[salt] for binding of L8+ ligand to dT(dT)10 are only two-thirds as large in magnitude as for polymeric DNA.15 The corresponding [salt] derivatives STm for melting of oligomeric DNA duplexes do not differ greatly from the polymeric value except for very short oligomers (e.g. 70% of polymeric STm for 6 bp duplex9). This does not mean there is no CEE in these case; in fact it means just the opposite, in that the CEEs for the two smaller ss DNA oligomer formed from one ds DNA oligomer are sufficiently large so that the overall salt ion release in these transitions is as large as for polymeric DNA, even though the amounts of salt ion accumulation for both the ds and ss oligomers are predicted to be much less than the polymeric values.8,9 These computational analyses used the cylindrical model of nucleic acids. What are the thermodynamic consequences of replacing the cylinder model of DNA by a detailed all-atom model in a NLPB analysis of the role of coulombic end effects on the [salt]-dependence of DNA helix formation or melting?
In this study we report NLPB calculations of the coulombic contribution to the Gibbs free energy for all atom models of double and single stranded nucleic acid oligoanions in the vicinity of 0.15 M salt for a wide range of oligomer lengths from 4 to 118 (ds) or from 2 to 59 (ss) phosphate charges. Numerical results are analyzed to obtain a NLPB predictions of the thermodynamic extent of salt ion association for any length ds or ss nucleic acid oligomer at 0.15 M salt. These results provide basis for analysis of experimental values of STm and SKobs as a function of oligomer size (∣ZD∣) for formation or melting of nucleic acid oligomers in the vicinity of 0.15 M salt.
NLPB analysis of DNA model
Coulombic free energy and extent of salt ion association per phosphate charge of oligomeric NA
The cylindrical model of polymeric nucleic acids (an infinite cylinder with a uniform axial charge density) is well known to capture the essence of the extraordinary strong interactions of these polyions with salt ions. The model requires knowledge of only two structural quantities of the nucleic acid, the axial charge separation b and the cylinder radius a (distance of closest approach of salt ions to the cylinder axis). Thermodynamic properties such as the per charge Gibbs coulombic free energy (), obtained from the NLPB free energy by subtracting the high [salt] limit, and the thermodynamic extent of salt ion association (), obtained from analysis of free energy differences or derivatives as a function of [salt], are calculated for any choice of a, b and [salt] (ref. 17 and references therein). For oligomeric nucleic acids, the number of charged residues ∣ZD∣ is an additional variable that very significantly affects the (per charge) values of and . For nucleic acid oligomers, it is therefore necessary to consider ([salt],∣ZD∣). An extended set of numerical coefficients is needed to describe the function , including the derivatives of and with respect to ∣ZD∣ evaluated at the same reference salt concentration of 0.15 M and at the limits ∣ZD∣ = 0 (for short oligomers) or at ∣ZD∣ = ∞ (for long oligomers). The expansions for coulombic free energy in these two limits were developed18 and yield two equations
| (1) |
| (2) |
Here is the per charge free energy of a short oligomer, and is its first derivative with respect to ∣ZD∣, both are taken at ∣ZD∣ = 0. In eqn (2), Gu,∞/RT is the polymeric per charge free energy, and . The changeover between the two different regimes of ∣ZD∣-dependence specified in eqn (1) and (2) occurs in the vicinity of a characteristic value of ∣ZD∣, which we define as the length of coulombic end effect Ne. At this length, we require the function and its first derivative with respect to ∣ZD∣ to be continuous, yielding following two equations for the coefficients and the characteristic length Ne:18
| (3) |
| (4) |
Eqn (1) and (2) are given in the Table 1 as eqn (1a) and (2a). Eqn (1b) and (2b) in Table 1 for the salt ion association are obtained by replacing Gu,∞/RT by nu,∞, Gu,0/RT by nu,0, by , and g/RT by γ. Coefficients for long oligomers, Gu,∞/RT, g/RT, and Ne, depend on DNA structural quantities but coefficients for short oligomers, Gu,0/RT and , do not.
Table 1.
NLPB analysis of all-atom DNA: coulombic free energy and salt ion association per charge of short and long ds and ss oligomersa
| (A) Short ds and ss DNA oligomers | ||||
|---|---|---|---|---|
|
| ||||
| Eqn (1a) |
Eqn (1b) |
|||
| DNA | Gu,0/RT | n u,0 | ||
| ss1 | 0.801 | 0.0437 | 0.125 | 0.0198 |
| ss2 | 0.764 | 0.0455 | 0.114 | 0.0209 |
| ds | 0.806 | 0.0506 | 0.115 | 0.0213 |
| Averageb | 0.790 | 0.0466 | 0.118 | 0.0207 |
|
| ||||
| (B) Long ds and ss DNA oligomers | ||||
|
| ||||
| Eqn (2a) |
Eqn (2b) |
|||
| DNA | Gu,∞/RT | 2g/RT | n u,∞ | 2γ |
|
| ||||
| ss1 | 1.268 | 1.407 | 0.400 | 0.939 |
| ss2 | 1.271 | 1.323 | 0.399 | 0.924 |
| ds | 1.819 | 5.015 | 0.590 | 2.576 |
Error estimation for coefficients of Table 1 is performed as described in Methods. Error for Gu,∞/RT and nu,∞ is the smallest and does not exceed 2%. Error for g and γ is between 3 and 5%. Short oligomer coefficients have larger error: 2 to 6% for linear coefficients Gu,0/RT and nu,0, and 10% for quadratic coefficients and .
Since short oligo coefficients are expected to be independent on DNA type, the average number is assumed to apply to ss and ds oligomer
Analysis of coulombic effects of salt on the melting transition of a nucleic acid duplex (ds → ss1 + ss2)
For the melting transition ds → ss1 + ss2 the number of phosphate charges in ds and ss oligomers are related as ∣ZD,ss∣ = ∣ZD,ds∣/2, where ∣ZD,ds∣ is the number of phosphate charges in the ds form. Then, the per charge free energy change for the transition is related to the per charge free energies of ds and ss oligomers as
| (5) |
For the situation where both ds and ss nucleic acids are long oligomers (i.e. where both ds and ss lengths are larger than characteristic length Ne, then ∣ZD,ds∣ ≥ Ne,ds and ∣ZD,ss∣ ≥ Ne,ss) coulombic free energies per phosphate charge of ss and ds oligomers are described by eqn (2a) from Table 1. Expressed per phosphate on the ds form, the free energy change for the transition of a two-stranded duplex (ds) to two strands (ss) is given by
| (6) |
where ΔGu,∞/RT = Gu,∞,ss/RT − Gu,∞,ds/RT, and Δg/RT = 2gss/RT − gds/RT. Therefore, the coulombic free energy difference per phosphate for the ds to ss transition of an oligomer is smaller in magnitude than for the polymeric case. Similar equations for the thermodynamic extent of salt ion association per DNA phosphate are obtained by substituting coefficients of eqn (2a) from Table 1 to eqn (6). The equation for free energy change eqn (6) can be presented in alternative forms by eliminating all coefficients except , Ne,ss, and Ne,ds:
| (7) |
The long-oligomer range of ∣ZD∣ is characterized by a linear dependence of on 1/∣ZD∣. These eqn (6) were derived and tested in ref. 9 for the cylindrical model.
An intermediate length regime exists where the ds oligomer is longer than Ne,ds, but when each ss oligomer is shorter than Ne,ss: ∣ZD,ds∣ ≥ Ne,ds but ∣ZD,ss∣ = ∣ZD,ds∣/2 ≤ Ne,ss or as a single inequality Ne,ds ≤ ∣ZD,ds∣ ≤ 2Ne,ss. The expression for the free energy change per phosphate for the transition of a long ds oligomer to two short ss oligomers is obtained from Table 1 using eqn (2a) for the ds oligomer and eqn (1a) for the ss oligomer. Two alternative forms of this equation are:
| (8) |
| (9) |
Similarly, an equation for the change in the thermodynamic extent of salt ion association per DNA phosphate in the transition is obtained by replacing coefficient by and coefficient g/RT by γ.
If both ss and ds oligomers are shorter than corresponding Ne, ∣ZD,ds∣ ≤ Ne,ds and ∣ZD,ss∣ ≤ Ne,ss, the per-charge coulombic free energies of ss and ds oligomers are described by eqn (1a) of Table 1. The per charge coulombic free energy change for the melting of a short nucleic acid duplex to two strands is
| (10) |
Methods
Oligomer model
An all-atom model of 12 bp B-DNA oligomer with strands 5′-CGATTAGATAGC-3′ (ss1) and 5′-GCTATCTAATCG-3′ (ss2) was built with the Nucleic Acid Builder (NAB) program19 via web-server http://casegroup.rutgers.edu/. Melting studies on this oligomer were reported by Pegram et al.20 To calculate properties of longer oligomers (approaching the polyelectrolyte limit), five repeats of the above sequence were used to make a 60 bp oligomer (118 phosphates, Fig. 1, top). This oligomer was then shortened in 2 bp (4 phosphates) steps by removing one bp from each end. This model is used to investigate the range of DNA oligomer length from ∣ZD,ds∣ = 118 charges (60 bp) to ∣ZD,ds∣ = 2 (2 bp). Calculations for 12 bp oligomer are compared to calculations for other 12 bp duplexes. Models of stacked single strands (ss1 and ss2) are obtained from ds oligo model by separating corresponding atoms from ds file (or deleting the other strand from the coordinate file). All Protein Data Bank (PDB)21 files were then converted to files with charges and radii for all-atom calculations (PQR format) by online converter22,23 and used as input file for Adaptive Poisson–Boltzmann Solver (APBS).24 Free energy and its salt dependence is calculated by APBS with Nonlinear Poisson–Boltzmann equation as described previously.17 Calculations are performed with grid sizes and length specified in Table S1 in ESI.†
Fig. 1.
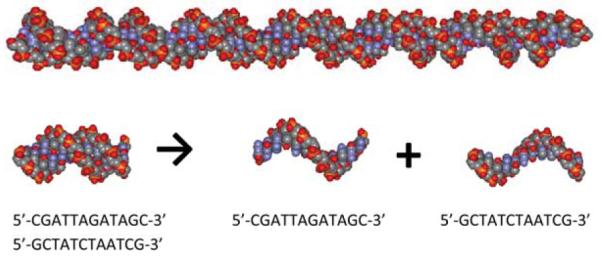
Image of 60 bp ds DNA oligomer built as five repeats of the sequence 5′-CGATTAGATAGC-3′ as described in Methods (top) and schematics of duplex melting transition ds → ss1 + ss2 with images of 12 bp ds oligomer of the above sequences and two fully stacked single strands ss1 and ss2 (bottom).
Other duplex models
Two all-atom models of 12 bp oligo were obtained from PDB files 1CS2.pdb25 and 1DUF.pdb26 and are duplex B-DNA structures. We chose to use NMR solution structures where they are available, and tested structures with different GC content. File 1DUF is the NMR structure of the dodecamer for which the X-ray crystal structure was obtained by Drew and Dickerson27 with the sequence 5′-CGCGAATTCGCG-3′. There is no structure with the exact sequence match in the PDB database for 5′-CGATTAGATAGC-3′ duplex. Therefore, we choose the file 1CS2 with the sequence 5′-CTACTGCTTTAG-3′ and the same GC content as 5′-CGATTAGATAGC-3′ duplex (41% GC). GC-content of duplex from the file 1DUF is 67% allowing to test the effect of sequence in ΔGcoul.
Treatment of computational results for Gcoul(∣ZD∣) in the vicinity of 0.15 M salt and ncoul(∣ZD∣) at 0.15 M salt
NLPB calculations on these oligomers yield Gcoul as function of ∣ZD∣ between 2 ≤ ∣ZD∣ ≤ 118 for ds DNA and 1 ≤ ∣ZD∣ ≤ 59 for ss DNA. The extent of salt ion association ncoul(0.15) is obtained by evaluating for intervals 0.1–0.15 M and 0.15–0.2 M, and then averaged to obtain . Uncertainty was estimated from the difference in these determinations. Uncertainties in oligomeric values of salt ion association ncoul(0.15) are less than 1%. Uncertainties in values of Δncoul(0.15) of melting an oligomer are larger but do not exceed 2%. For small ∣ZD∣ data were fit with ∣ZD∣ ≤ Ne where Ne = 6 for ss and 12 for ds DNA to eqn (1a) and Ne = 7 for ss and 12 for ds DNA to eqn (1b) of Table 1. The choice of fitting range for ss data is determined by the point where ss data start to deviate from the common with ds DNA trend of the length dependence. For values of ∣ZD∣ in long oligomer range, values of and per DNA phosphate were fit as an inverse function of ∣ZD∣ ≥ Ne to eqn (2a) and (2b). Uncertainties for coefficients of fitting equations are estimated by varying range of ∣ZD∣ by one unit.
Results and discussion
NLPB predictions of dependences of coulombic free energy and salt ion association per phosphate on ds and ss DNA oligomer length at 0.15 M salt
All-atom models of a B-DNA oligomer and its two fully stacked single strands (ss1, ss2) were built as five repeats of the sequence 5′-CGATTAGATAGC-3′ (ss1) and 5′-GCTATCTAATCG-3′ (ss2) and truncated in a step-wise fashion to create the family of ds and ss oligomers investigated here, as described in Methods. While this fully stacked state of the individual strands is convenient for all-atom NLPB calculations, it is only approached in solution at low temperature (typically <0 °C).29 Unstacking occurs as a broad noncooperative transition extending over the full temperature range to 100 °C. The stacked-to-unstacked transition in single stranded polynucleotides is only slightly favored by a reduction in salt concentration (see ref. 30 and references therein), indicating that coulombic properties of unstacked or partially stacked single-stranded nucleic acids are not very different from those of the fully stacked strand, and that unstacking involves only a small increase in the average distance between adjacent phosphates.
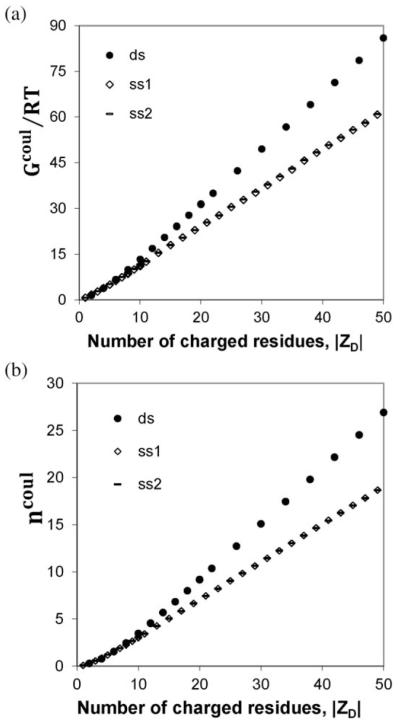
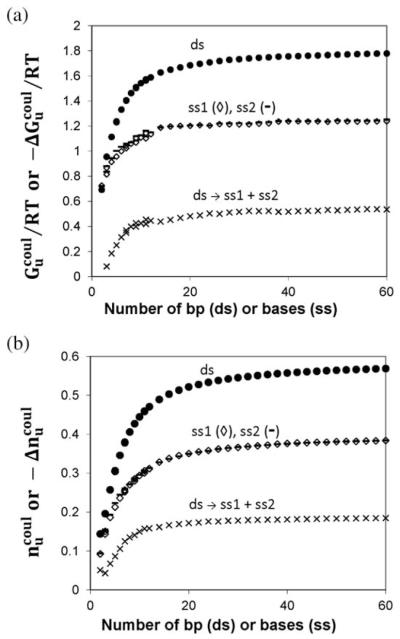
Numerical results for the dependences on ∣ZD∣ of the coulombic free energy of the oligomer Gcoul(0.15)/RT and the extent of salt ion association with the oligomer ncoul(0.15) are plotted in Fig. 2. For sufficiently long oligomers, these all-atom calculations predict that both Gcoul(0.15)/RT (A) and ncoul(0.15) (B) increase approximately linearly with increasing ∣ZD∣, as predicted by NLPB calculations for the cylindrical model.9
Fig. 2.
Coulombic (NLPB) results for an all-atom model of nucleic acid oligomers. NLPB results for the reduced coulombic free energy (Gcoul/RT; panel A) and the thermodynamic extent of salt ion association (ncoul = −δ(Gcoul/RT)/δln C; panel B), both at a 1 : 1 salt concentration C = 0.15 M, are plotted as a function of the number of charged residues (phosphates) ∣ZD∣ on the oligomer. Calculations were performed for a duplex oligomer (solid dot symbol) built from 5 repeats of the 12-mer sequence 5′-CGATTAGATAGC-3′, for the individual stacked helical strands of this oligomer ss1 (diamond), ss2 (dash) as described in the text.
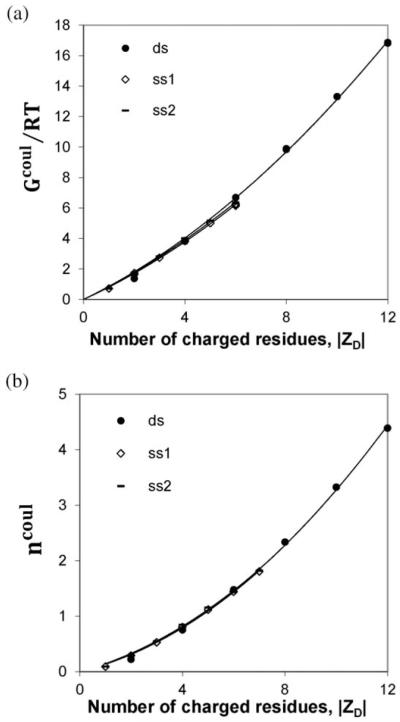
For short oligomers, Fig. 3A and B show that all-atom values of Gcoul(0.15)/RT and ncoul(0.15) are highly nonlinear functions of ∣ZD∣, as observed for the cylindrical model. As shown on an expanded scale in these figures, values of Gcoul(0.15)/RT and of ncoul(0.15) for ss and ds DNA of the same ∣ZD∣ converge for oligomers shorter than the length of the coulombic end effect Ne. Fits to the oligomer model of all-atom NLPB results for the coulombic free energy Gcoul(0.15)/RT (eqn (1a)) and of the extent of salt ion association ncoul(0.15) (eqn (2a)) are shown, and coefficients obtained from these fits are listed in Table 1. These coefficients (Gu,0/RT, , nu,0, and ) are approximately equal for both stacked strands (ss1 and ss2 and for ds DNA), indicating that arrangement of charges in a short oligomer does not greatly affect either its coulombic free energy or its extent of salt ion association. Intercepts of both Gcoul(0.15)/RT and ncoul(0.15) for ss and ds DNA are zero to a good approximation, indicating that the distribution of partial charges on the net-uncharged nucleoside unit(s) at the end make only a small contribution to Gcoul(0.15)/RT and ncoul(0.15). Setting the intercept equal to zero in these fits makes no significant difference for ∣ZD∣ > 5. For ∣ZD∣ = 6 or larger, agreement between fitted and calculated values is within 1% for both Gcoul/RT and ncoul. Neglecting the small differences between ds and ss results, average values of fitting coefficients are , , , and (Table 1).
Fig. 3.
Coulombic behavior of short oligomers. Nonlinear regions of Gcoul/RT (panel A) and ncoul (panel B) from all-atom NLPB calculations on short ds (∣ZD∣ ≤ 12) and ss (∣ZD∣ ≤ 7) DNA oligomers at 0.15 M salt (cf. Fig. 2) are plotted vs. ∣ZD∣. Fits shown are to quadratic equations in ∣ZD∣, the functional form derived for short oligomers (ref. 18 and eqn (1a) and (1b) in Table 1); fitting coefficients are listed in Table 1.
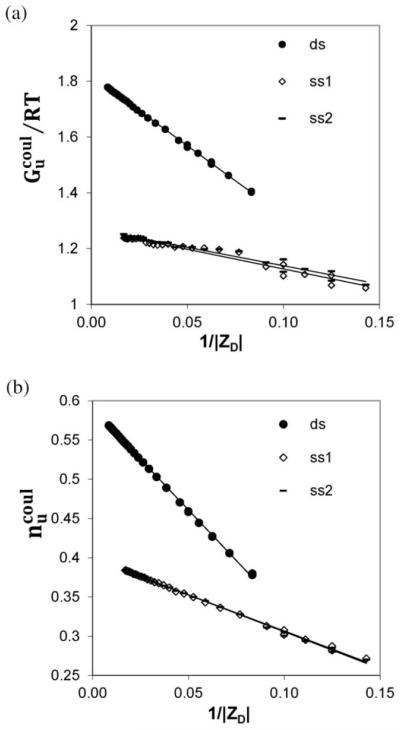
Values of Gcoul(0.15)/RT and ncoul(0.15) for oligomers with chain lengths that exceed the length of the CEE (i.e. ∣ZD∣ ≥ 12 (ds) and ∣ZD∣ ≥ 7 (ss) increase almost linearly with ∣ZD∣ (Fig. 2). To eliminate this primary dependence on ∣ZD∣, we analyze the values of these quantities normalized per DNA phosphate: and . It is straightforward to show for any kind of an end effect that the per-residue property of interest in a molecule of N residues approaches the polymeric value linearly in 1/N. It is therefore not surprising that, as shown in Fig. 4, both (Fig. 4A) and (Fig. 4B) are linear functions of 1/∣ZD∣ for long oligomers, as predicted for the oligomer CEE model (cf. eqn (2a) and (2b)). Equations and fit coefficients are given in Table 1. Unlike the situation reported above for short oligomers, in the long oligomer regime the coefficients describing the dependences of and on ∣ZD∣ (cf. Table 1B) are very different for ds and ss DNA, though very similar for ss1 and ss2 strands.
Fig. 4.
Coulombic behavior of long oligomers. Linear regions of Gcoul/RT (Fig. 2A) and ncoul (Fig. 2B) vs. ∣ZD∣ from all-atom NLPB calculations on ds (∣ZD∣ ≥ 12) and ss (∣ZD∣ ≥ 7) DNA oligomers at 0.15 M salt (cf. Fig. 2) are replotted. Values of , the coulombic free energy per charged residue of a DNA oligomer and of , the corresponding thermodynamic extent of salt ion association per charged residue, are plotted in panels A and B vs. 1/∣ZD∣. Fits to linear dependences on 1/∣ZD∣ (eqn (2a) and (2b)) in Table 1), expected for a situation in which a significant end effect is present and in which the end effect is independent of the number of interior residues8 are shown; fitting coefficients are listed in Table 1.
The [salt] dependence of the coulombic free energy per phosphate of ds and ss nucleic acid oligomers
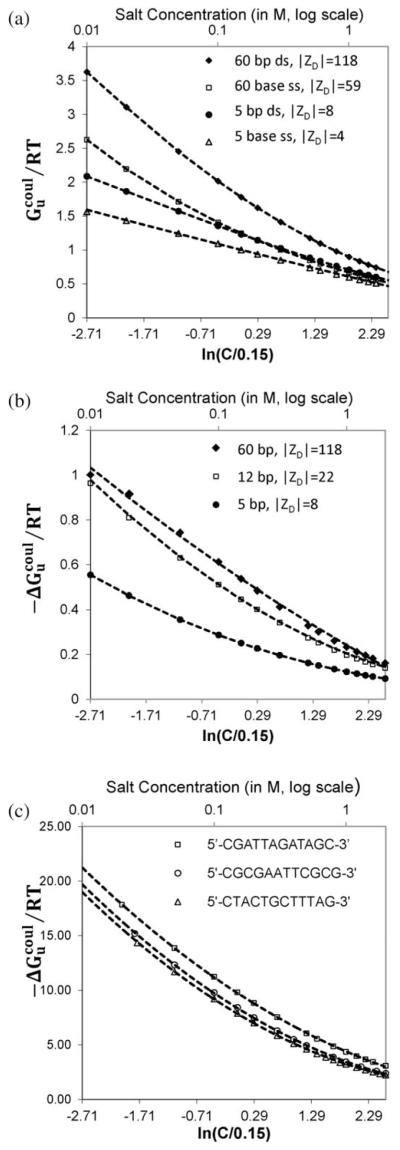
All-atom NLPB predictions of the per charge coulombic free energy of long and short ds and ss oligomers as functions of [salt] between 0.01 M and 2 M are shown in Fig. 5A. These results are well-fit by a second order polynomial in the logarithm of [salt], referenced to the mid-range [salt] of 0.15 M. Two important findings of NLPB analyses of both the all-atom model (Fig. 5A) and the cylindrical model9 of nucleic acids are that (1) per charge coulombic free energies of long ds and ss nucleic acid oligomers (and the corresponding polyanions) are nonlinear functions of ln [salt], while (2) per charge coulombic free energies of short ds and ss nucleic acid oligomers are approximately linear functions of ln[salt] in the range examined.
Fig. 5.
Salt dependence of per-charge coulombic free energy () from all-atom NLPB calculations. In panel A, values of for representative long (60 bp; ∣ZD∣ = 118) and short (5 bp; ∣ZD∣ 8) ds oligomers and for representative long (60 base; ∣ZD∣ 59) and short (5 base; ∣ZD∣ = 4) ss oligomers, calculated over the range 0.01 M to 2 M salt, are plotted vs. the natural logarithm of the relative salt concentration C/0.15 (i.e. the salt concentration normalized to the reference salt concentration of 0.15 M). In panel B, predicted values of (expressed per nucleic acid phosphate) for melting of 60 bp, 12 bp and 5 bp oligomers to stacked single strands are plotted vs. the logarithm of the salt concentration (expressed relative to 0.15 M salt). Panel C compares all-atom NLPB predictions of for melting of the 12 bp duplex of panel C with analogous predictions for two other 12 bp duplexes over the salt concentration range 0.01 to 2 M.
Changes in coulombic free energy and salt ion association per phosphate in melting nucleic acid oligomer duplexes
Fig. 5B plots for the melting transition of the 60 bp and 5 bp duplexes of Fig. 5A to two 60 base or 5 base stacked strands. All-atom NLPB results for for melting a 12 bp duplex to two 12 base stacked strands are also shown. Fig. 5B shows that, while is nonlinear in ln[salt] for long oligomers, as exemplified by the 60 bp ds results and 60 base ss results in Fig. 5A, for the transition of 60 bp ds DNA to two 60 base stacked ss DNAs is relatively linear in ln[salt] below 0.3 M. Moreover, while is linear in ln[salt] for short oligomers, as exemplified by the 5 bp ds results and 5 base ss results in Fig. 5A, for the transition of 5 bp ds DNA to two 5 base stacked ss DNAs is predicted to be quite nonlinear in ln [salt] over the entire range investigated.
Fig. 5C displays the predictions of all-atom NLPB calculations for the effects of [salt] on the coulombic part of the free energy change for the melting transitions of three different 12 bp duplex sequences to the corresponding 12 base stacked strands. Two of these dodecamers have the same base composition (5 GC and 7 AT base pairs) but different sequences; the third (8 GC and 4 AT base pairs) differs in composition from the other two.
Fig. 5C shows that per-charge values of the reduced coulombic free energy change () for duplex melting to fully stacked strands are quite similar for the two 12 bp duplexes with the same base composition , 7.90, and are systematically larger in magnitude (more negative) by approximately 1.6RT ~ 1 kcal per mole of DNA phosphates for melting of the more GC-rich dodecamer. This is a large difference, of the same magnitude as the net stability of the duplex relative to the melted strands at 37 °C and 0.15 M salt. Further research is needed to determine if this apparent GC-composition-dependent coulombic destabilization of a duplex is general, and what its molecular origins might be.
Length dependences of the coulombic free energy per phosphate residue in all-atom and cylindrical DNA models
Numerical coefficients summarizing the NLPB predictions of the length (∣ZD∣) dependence of the coulombic free energy per charged residue (phosphate) of DNA for cylindrical and all-atom models are listed in Table 2, and compared with previously published values derived from analysis of experimental data on and SKobs as function of DNA length for binding of L8+ ligand to ss DNA oligomers of 7 to 169 T bases and for melting of duplex and hairpin helices. Comparison of these coefficients for all-atom and cylinder models shows they are in general quite similar to each other and to the experimentally derived values, with a few notable exceptions. Differences in coefficients for the different models are larger in relative magnitude for than for , as expected.
Table 2.
Comparison of NLPB coulombic free energy and salt ion association coefficients for all-atom and cylindrical DNA models and with fits to experimental data
| (A) Salt ion association coefficients | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| DNA | n u,0 | n u,∞ | γ | nu,∞ − nu,0 | N e | ||
| All-atom NLPB, 0.15 M | ss | 0.118 | 0.0207 | 0.400 | 0.466 | 0.282 | 6.8 |
| Exp, DNA + L8+ → complex 0.1–0.2 M18 | ss | 0.019 | 0.77 | 0.35 | 9.1 | ||
| Cyl NLPBa & exp, ds → k ss9 | ss | 0.123 | 0.022 | 0.454 | 0.60 | 0.331 | 7.5 |
| All-atom NLPB, 0.15 M | ds | 0.118 | 0.0207 | 0.590 | 1.29 | 0.472 | 11.4 |
| Cyl NLPBa & exp, ds → k ss9 | ds | 0.123 | 0.022 | 0.612 | 1.42 | 0.464 | 11.1 |
|
| |||||||
| (B) Free energy coefficients | |||||||
|
| |||||||
| DNA | G u,0 b | G u,∞ | g | Gu,∞ − Gu,0 | N e | ||
|
| |||||||
| All-atom NLPB, 0.15 M | ss | 0.468 | 0.0276 | 0.752 | 0.404 | 0.284 | 5.2 |
| Exp, DNA + L8+ → complex 0.1 M18 | ss | 0.0193 | 0.78 | 0.35 | 9.0 | ||
| Exp, DNA + L8+ → complex 0.2 M18 | ss | 0.0126 | 0.51 | 0.23 | 9.1 | ||
| Cyl NLPBa & exp, ds → k ss9 | ss | 0.316 | 0.044 | 0.879 | 0.849 | 0.563 | 6.4 |
| All-atom NLPB, 0.15 M | ds | 0.468 | 0.0276 | 1.08 | 1.485 | 0.609 | 11.1 |
| Cyl NLPBa & exp, ds → k ss9 | ds | 0.372 | 0.041 | 1.257 | 2.415 | 0.885 | 10.7 |
Structural parameters of cylindrical model of DNA: ass = 7 Å, bss = 3.4 Å, ads = 9.4 Å, bds = 1.7 Å.
All coefficients for Gcoul except Ne are given in kcal mol−1 for comparison with Shkel et al. 2006,18 Ne has no units.
All-atom and cylinder models of nucleic acids differ in details of charge distributions and molecular shape. Cylindrical model represents polymeric DNA as an infinite cylinder of radius a with uniformly charged axis with electron charge e per length b. The radius a is the distance of the closest approach of mobile ion center to the cylinder axis. The oligomer DNA model considers finite charged cylinder of length ∣ZD∣b, which has uncharged cylindrical caps of 4 Å on both ends. The all-atom model starts from a specific DNA structure, X-ray or NMR, and assigns charges and radii to each atom according to a selected force field. Each charged residue has net charge of e. The surface of the molecule is built by rolling over atomic surfaces a small sphere representing a water molecule. Mobile ions are modeled as charged spheres. The average axial distance between phosphates is b = 1.7 Å for the various ds B-DNA structures. A previous comparison for polymeric DNA found that the [salt] dependence of the coulombic free energy predicted for an all-atom model with a detailed assignment of partial charges matched that of the cylinder model, calculated for an axial charge density corresponding to the preaveraged charge per residue and for a realistic choice of cylinder radius.17
The coefficients G0,u, , n0,u and describe short oligomers, and therefore are expected to be the same for any DNA type, ds or ss. Coefficients for show more variation than coefficient for : n0,u and are remarkably similar for both DNAs and all three determinations. Larger coefficient for all-atom model suggests larger non-linearity in reflecting difference in charge distribution of two models. The coefficients of long oligomer (G∞,u, n∞,u, g, and γ) are expected to depend on structural parameters of polymeric DNA (a and b). The axial charge separation b has much stronger effect on these coefficients then the radius a.17,31 The polymeric values G∞,u and n∞,u are smaller for all-atom model because they correspond to a larger effective cylinder radius a. The coefficients that describe the deviation from the polymeric limiting values (g and γ) may also depend on details of charge distribution and show larger variation between determinations. For cylindrical model calculation with other radii a than in Table 2 (data not shown), values of g and γ increase with increasing in a. The trend in g and γ is consistent with expectation that CEE contribution for simple charged cylinder is proportional to its radius. However, the all-atom model coefficient g is significantly smaller (g is 0.404 for all-atom vs. 0.85 for cylindrical model for ss DNA and 1.5 vs. 2.4 for ds DNA). Therefore this should be an effect of all-atom representation of DNA charges.
The differences Gu,∞ − Gu,0 and nu,∞ − nu,0 appear in expressions for ΔGcoul and Δncoul for oligolectrolyte processes like melting and ligand binding. Therefore these differences may be more important that the individual values. These differences all are quite consistent except that cylindrical model value for ds DNA (Gu,∞,ds − Gu,0) is significantly larger then for the all-atom case. The salt dependence coefficients nu,∞ − nu,0 again are much more consistent, which allows accurate application of developed equations and coefficients for experimental analysis.
The characteristic length of CEE Ne for ss DNA in the vicinity of 0.15 M salt is estimated to be between 5.2 and 9 phosphates charges. The smaller estimate is obtained from the all-atom model, and the larger is the experimental determination. This is outside of the uncertainty ±1 estimated previously.18 For ds DNA predictions of all-atom and cylindrical models are more consistent, giving Ne in the range 10.7–11.4 at 0.15 M salt, which are within the uncertainty of the determination. As discussed in the next section, the characteristic length Ne provides a lower bound on the oligomer size required for it to behave thermodynamically as a long oligomer, with values of and that approach the polymeric value linearly in 1/∣ZD∣ (see eqn (2a) and (2b)). Predicted values of Ne decrease modestly with increasing [salt], MC yields Ne of 18 residues at 1.76 mM salt.8 Values from Table 2 are given at 0.15 M of 1 : 1 salt and can be used in a range between 0.05 M and 0.3 M where variations in Ne with [salt] do not exceed uncertainty of ±1, as follows from calculations with cylindrical model of DNA.9
Coulombic end effect in melting of duplex DNA oligomers
The coulombic end effect length Ne is significant on several levels. Oligomers with less than Ne phosphates exhibit a linear dependence of (free energy per DNA phosphate) on ∣ZD∣. Conversely, DNA oligomers with more than Ne phosphates exhibit a linear dependence of on 1/∣ZD∣. Axial profiles of surface salt cation concentration are very different for short and long DNA oligomers. Critical change in axial gradients of surface concentration occurs for oligomer length 2Ne. Short oligomers exhibit parabolic axial cation concentration profiles where the maximum cation concentration in the center is much less than the salt cation concentration at the surface of polymeric DNA. As the number of phosphate charges on the oligomer increases, the surface cation concentration at the center of the oligomer increases attaining the concentration predicted for the interior of polymeric DNA when ∣ZD∣ > 2Ne. At larger oligomer sizes this interior region with uniform (polymeric) surface concentration extends over ∣ZD∣ − 2Ne residues. Surface cation concentration profiles in the two end regions of Ne charged residues do not change as oligomer size increases beyond 2Ne, which results in a linear dependence of thermodynamic properties like and on 1/∣ZD∣.
At what DNA oligomer size ∣ZD∣ does a process like helix melting or oligocation binding exhibit the same salt concentration dependence (SKobs or STm) as that characteristic of the same process with polymeric DNA? There is no one answer to this question; the answer is in fact very different for different processes.
Binding of a protein or cationic ligand to a site in the interior of a DNA oligomer is predicted to exhibit the same salt dependence SKobs as for binding to polymeric DNA if the oligomer is long enough so that the ends of the binding site are more than Ne phosphates from the ends of the oligomer. SKobs for binding to shorter oligomers will be smaller in magnitude. If the binding site were less than Ne phosphates in length and located at the end of the DNA, then SKobs is predicted to be smaller than for binding to the interior site, and to be the same for long DNA oligomers as for polymeric DNA. For nonspecific binding of a protein or ligand to a DNA oligomer, SKobs is smaller in magnitude than for nonspecific binding of the same ligand to polymeric DNA, and approaches the polymeric value linearly in 1/∣ZD∣.15
Melting of an oligomeric DNA hairpin helix formed from a self-complementary strand (like alternating dTA oligomers) exhibits a smaller STm than polymeric DNA; values of STm for long enough oligomers approach the polymeric value approximately linearly in 1/∣ZD∣. For melting of a duplex DNA oligomer to two strands (ds to ss1 + ss2), the situation is more complicated. Oligomers as short as 10 bp exhibit values of STm or SKobs of melting that are as large in magnitude as those for polymeric DNA. In this case, ∣ZD∣ for each ss oligomer is only one-half as large as for the duplex, and each ss oligomer exhibits coulombic end effects. In Fig. 6, all-atom NLPB results for (Fig. 6A) and (Fig. 6B) are plotted together with the corresponding values of (Fig. 6A) and (Fig. 6B) as a function of oligomer size in base pairs for ds DNA and in bases for ss DNA. On these plots, direct vertical comparison can be made of the contributions of ss and ds forms to and . For oligomeric duplexes in the range 10–30 bp, melting to strands with 10–30 bases each, the individual values of and for ds and ss conformations are significantly less than the polymeric values and are strong functions of the number of base pairs or bases (i.e. of ∣ZD∣), but the differences and are quite close to the polymeric values. A recent experimental study determined STm and for melting of 92 duplex DNA oligomers (10–30 bp).28 No systematic dependence of either STm or on oligomer size was observed, and values of these quantities were close to the polymeric values, as expected from the analysis of Fig. 6.
Fig. 6.
Length dependences of change in coulombic free energy () and salt ion association () in duplex melting transition. The per charge free energy (panel A) and salt ion association (panel B) both at 0.15 M concentration of 1 : 1 salt from all-atom calculations plotted as function of the number of bases (ss DNA oligomer) of base pairs (ds DNA oligomer). The change in free energy and in salt ion association calculated per charge of ds oligomer are plotted as function of the number of base pair.
Fig. 7 compares the dependences of for duplex melting (ds to ss1 + ss2) on the number of DNA phosphates in the ds form (∣ZD∣) calculated by NLPB for both cylindrical and all-atom models. The all-atom model predicts a somewhat larger magnitude of the polymeric value of (−0.185) than the cylindrical model (−0.158) for the same average axial charge densities as for the all-atom model (2 charged phosphates per 3.4 Å for ds and 1 charged phosphates per 3.4 Å for ss) and for a choice of cylinder radii (9.4 Å ds and 7.0 Å ss) that optimizes the fit to normalized duplex and hairpin melting data.9 The ds radius in the cylindrical model is less than the average distance of closest approach of mobile ions to the duplex axis in the all atom model; increasing this radius in the cylindrical model of ds DNA would improve the agreement of the NLPB predictions for the two models, but introduce an offset from the experimental data. Additional considerations for refinement of these analyses include a more realistic description of residual stacking in the ss form, which varies with temperature, base composition, and sequence. Unstacking of bases probably increases b of the ss form slightly, thereby increasing the predicted of transition, so introduction of ss unstacking into the model for ss oligomers would not improve agreement between all atom NLPB and experiment.
Fig. 7.
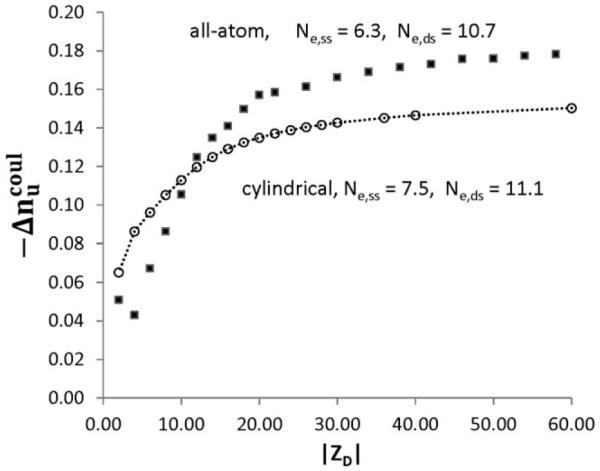
Comparison of per-charge change in salt ion association for duplex melting transition from all-atom and cylindrical models. All-atom (Fig. 6, symbols) and cylindrical model (Table 2, circles) predictions are plotted as a function of duplex charge. Calculations of for cylindrical model of ds and ss DNA are performed with radii ads = 9.4 Å, ass = 7 Å, and charge separations bds = 1.7 Å, bss = 3.4 Å.9
Conclusions
As shown previously, the [salt]-dependence of the polymeric coulombic free energy per phosphate () of both all-atom and cylindrical models of DNA is well described by a quadratic polynomial in ln C/0.15 in the range from 0.01 M to 2 M of univalent salt. Here we determine and analyze the dependence of and on oligomer size (number of phosphate charges ∣ZD∣) for all-atom models of ds DNA and fully stacked ss DNA for a wide range of oligomer lengths, for ds shorter than 60 bp and ss shorter than 60 bases. NLPB predictions of the coulombic free energy and salt ion association per DNA phosphate using all-atom DNA models exhibit very similar functional dependences on ∣ZD∣ to those previously observed for the cylindrical (preaveraged) model. Both and are linear functions in of the number of charged residues ∣ZD∣ for short oligomers and are linear functions of the reciprocal of the number of charged residues for long oligomers. NLPB results for the all-atom model are compared with those for the cylindrical model and with results obtained from analysis of experimental data in the vicinity of 0.15 M univalent salt. While predictions of the all-atom model and the cylinder model are very analogous, the ability to adjust the radius of the cylinder model provides better agreement with experiment. Once calibrated in this way, better predictive capability is obtained using the cylindrical model than with the all-atom model.
Supplementary Material
Appendix.
Abbreviation
| [salt], C | Salt concentration, concentration of 1 : 1 electrolyte |
| bp | Base pair |
| CEE | Coulombic end effect |
| ss | Single stranded |
| ds | Double stranded |
| NLPB | Nonlinear Poisson Boltzmann |
| CC | Counterion condensation |
| MC | Monte Carlo |
| MD | Molecular dynamics |
| APBS | Adaptive Poisson–Boltzmann solver |
| PDB | Protein data bank |
| NAB | Nucleic acid builder. |
| Symbol Equation | ||
|---|---|---|
| a | Radius of DNA in cylindrical model | |
| b | Axial charge separation in cylindrical model of DNA | |
| R | Gas constant, 1.9859 (cal K−1 mol−1) | |
| T | Temperature | |
| ∣ZD∣ | Number of phosphate charges in DNA oligomer | |
| Gcoul | Coulombic Gibbs free energy | |
| Gcoul/∣ZD∣ | Coulombic Gibbs free energy per phosphate charge |
|
| Polymeric limit of Coulombic Gibbs free energy |
||
| ncoul | −(RT)−1∂Gcoul/∂ln C | Salt ion association |
| Tm | Transition temperature | |
| STm | dTm/dln C | Salt-dependence of transition temperature |
| Kobs | Observed equilibrium constant | |
| −RTln Kobs | Standard free energy change | |
| SKobs | ∂ln Kobs/∂ln C | Salt-dependence of equilibrium constant |
| Coefficients of DNA oligomeric model at C = 0.15 M | ||
| Ne | Characteristic length of CEE | |
| Gu,0 | Per phosphate free energy of oligomer at ∣ZD∣ = 0 |
|
| First ∣ZD∣-derivative of per phosphate oligomer free energy at ∣ZD∣ = 0 |
||
| Gu,∞ | Per phosphate free energy of polymer | |
| 2g | Negative first derivative of per phosphate free energy with respect to 1/∣ZD∣ at ∣ZD∣ → ∞ |
|
| nu,0 | Per phosphate salt ion association of oligomer at ∣ZD∣ = 0 |
|
| First ∣ZD∣-derivative of per phosphate salt ion association at ∣ZD∣ = 0 |
||
| nu,∞ | Per phosphate salt ion association of polymer |
|
| 2γ | Negative first derivative of per phosphate salt ion association with respect to 1/∣ZD∣ at ∣ZD∣ → ∞ |
|
Footnotes
Electronic supplementary information (ESI) available. See DOI: 10.1039/c2sm25607j
References
- 1.Manning GS. Molecular theory of polyelectrolyte solutions with applications to electrostatic properties of polynucleotides, Q. Rev. Biophys. 1978;11:179–246. doi: 10.1017/s0033583500002031. [DOI] [PubMed] [Google Scholar]
- 2.Klein BK, Anderson CF, Record MT. Comparison of Poisson–Boltzmann and condensation model expressions for the colligative properties of cylindrical polyions. Biopolymers. 1981;20(10):2263–2280. [Google Scholar]
- 3.Anderson CF, Record MT. Poly-electrolyte theories and their applications to DNA, Annu. Rev. Phys. Chem. 1982;33:191–222. [Google Scholar]
- 4.Anderson CF, Record MT. Salt nucleic-acid interactions, Annu. Rev. Phys. Chem. 1995;46:657–700. doi: 10.1146/annurev.pc.46.100195.003301. [DOI] [PubMed] [Google Scholar]
- 5.Mills P, Anderson CF, Record MT. Monte-Carlo studies of counterion DNA interactions – comparison of the radial-distribution of counterions with predictions of other poly-electrolyte theories. J. Phys. Chem. 1985;89:3984–3994. [Google Scholar]
- 6.Mills P, Anderson CF, Record MT. Grand canonical Monte-Carlo calculations of thermodynamic coefficients for a primitive model of DNA salt-solutions. J. Phys. Chem. 1986;90:6541–6548. [Google Scholar]
- 7.Ni HH, Anderson CF, Record MT. Quantifying the thermodynamic consequences of cation (M2+, M+) accumulation and anion (X−) exclusion in mixed salt solutions of polyanionic DNA using Monte Carlo and Poisson–Boltzmann calculations of ion–polyion preferential interaction coefficients. J. Phys. Chem. B. 1999;103(17):3489–3504. [Google Scholar]
- 8.Olmsted MC, Anderson CF, Record MT. Monte-Carlo description of oligoelectrolyte properties of DNA oligomers – range of the end effect and the approach of molecular and thermodynamic properties to the poly-electrolyte limits. Proc. Natl. Acad. Sci. U. S. A. 1989;86:7766–7770. doi: 10.1073/pnas.86.20.7766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shkel IA, Record MT. Effect of the number of nucleic acid oligomer charges on the salt dependence of stability (ΔG°37) and melting temperature (Tm): NLPB analysis of experimental data. Biochemistry. 2004;43:7090–7101. doi: 10.1021/bi036225e. [DOI] [PubMed] [Google Scholar]
- 10.Stein VM, Bond JP, et al. Importance of coulombic end effects on cation accumulation near oligoelectrolyte B-DNA – a demonstration using NA-23 NMR. Biophys. J. 1995;68:1063–1072. doi: 10.1016/S0006-3495(95)80281-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang W, Bond JP, Anderson CF, Lohman TM, Record MT. Large electrostatic differences in the binding thermodynamics of a cationic peptide to oligomeric and polymeric DNA. Proc. Natl. Acad. Sci. U. S. A. 1996;93:2511–2516. doi: 10.1073/pnas.93.6.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olmsted MC, Bond JP, Anderson CF, Record MT. Grand-canonical Monte-Carlo molecular and thermodynamic predictions of ion effects on binding of an oligocation (L8+) to the center of DNA oligomers. Biophys. J. 1995;68:634–647. doi: 10.1016/S0006-3495(95)80224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Record MT, Lohman TM. Semi-empirical Extension of polyelectrolyte theory to treatment of oligoelectrolytes – application to oligonucleotide helix–coil transitions. Biopolymers. 1978;17:159–166. [Google Scholar]
- 14.Zhang WT, Ni HH, Capp MW, Anderson CF, Lohman TM, Record MT. The importance of coulombic end effects: experimental characterization of the effects of oligonucleotide flanking charges on the strength and salt dependence of oligocation (L8+) binding to single-stranded DNA oligomers. Biophys. J. 1999;76:1008–1017. doi: 10.1016/S0006-3495(99)77265-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballin JD, Shkel IA, Record MT. Interactions of the KWK6 cationic peptide with short nucleic acid oligomers: demonstration of large coulombic end effects on binding at 0.1–0.2 M salt. Nucleic Acids Res. 2004;32(11):3271–3281. doi: 10.1093/nar/gkh646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elson EL, Scheffler IE, Baldwin RL. Helix formation by d(TA) oligomers. III. Electrostatic effects. J. Mol. Biol. 1970;54:401–415. doi: 10.1016/0022-2836(70)90118-x. [DOI] [PubMed] [Google Scholar]
- 17.Shkel IA. Coulombic free energy of polymeric nucleic acid: low- and high-salt analytical approximations for the cylindrical Poisson–Boltzmann model. J. Phys. Chem. 2010;114:10793–10803. doi: 10.1021/jp908267c. [DOI] [PubMed] [Google Scholar]
- 18.Shkel IA, Ballin JD, Record M. Thomas. Interactions of cationic ligands and proteins with small nucleic acids: analytic treatment of the large coulombic end effect on binding free energy as a function of salt concentration. Biochemistry. 2006;45:8411–8426. doi: 10.1021/bi0520434. [DOI] [PubMed] [Google Scholar]
- 19. http://casegroup.rutgers.edu/
- 20.Pegram LM, Wendorff T, Erdmann R, Shkel I, Bellissimo D, Felitsky DJ, Record MT., Jr Why Hofmeister effects of many salts favor protein folding but not DNA helix formation. Proc. Natl. Acad. Sci. U. S. A. 2010;107:7716–7721. doi: 10.1073/pnas.0913376107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–W667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baker NA. Poisson–Boltzmann methods for biomolecular electrostatics. Methods Enzymol. 2004;383:94–118. doi: 10.1016/S0076-6879(04)83005-2. [DOI] [PubMed] [Google Scholar]
- 25.Leporc S, Mauffret O, Tevanian G, Lescot E, Monnot M, Fermandjian S. An NMR and molecular modelling analysis of d(CTACTGCTTTAG) d(CTAAAGCAGTAG) reveals that the particular behavior of TpA steps is related to edge-to-edge contacts of their base-pairs in the major groove. Nucleic Acids Res. 1999;27:4759–4767. doi: 10.1093/nar/27.24.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tjandra N, Tate S, Ono A, Kainosho M, Bax A. The NMR structure of a DNA dodecamer in an aqueous dilute liquid crystalline phase. J. Am. Chem. Soc. 2000;122:6190–6200. [Google Scholar]
- 27.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: conformation and dynamics. Proc. Natl. Acad. Sci. U. S. A. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owczarzy R, You Y, Moreira BG, Manthey JA, Huang LY, Behlke MA, Walder JA. Effects of sodium ions on DNA duplex oligomers: improved predictions of melting temperatures. Biochemistry. 2004;43:3537–3554. doi: 10.1021/bi034621r. [DOI] [PubMed] [Google Scholar]
- 29.Davis RC, Tinoco I. Temperature-dependent properties of dinucleoside phosphates. Biopolymers. 1968;6:223. doi: 10.1002/bip.1968.360060206. [DOI] [PubMed] [Google Scholar]
- 30.Electrostatic effects on polynucleotide transitions. 2. behavior of titrated systems. Biopolymers. 1967;5:993. doi: 10.1002/bip.1967.360051011. [DOI] [PubMed] [Google Scholar]
- 31.Bond JP, Anderson CF, Record M. Thomas., Jr Conformational transitions of duplex and triplex nucleic acid helices: thermodynamic analysis of effects of salt concentration on stability using preferential interaction coefficients. Biophys. J. 1994;67:825–836. doi: 10.1016/S0006-3495(94)80542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.