Abstract
Various studies have assessed the clinicopathological and prognostic value of Notch1 and Notch3 expression in Non-small cell lung cancer (NSCLC), but their results remain controversial. This meta-analysis was conducted to address the above issues by using a total of 19 studies involving 3663 patients. The correlations between Notch1 and Notch3 expression and clinicopathological features and NSCLC prognosis were analyzed. The meta-analysis indicated that higher expression of Notch1 was associated with greater possibility of lymph node metastasis and higher TNM stages. Moreover, patients with Notch1 overexpression and Notch3 overexpression showed significantly poor overall survival (Notch1: HR, 1.29; 95% CI, 1.06–1.57, p = 0.468 and I2 = 0.0%; Notch3: HR, 1.57; 95%CI, 1.04-2.36, p = 0.445 and I2 = 0.0%). Furthermore, there are statistically significant association between overall survival of NSCLC patients and the expression of Notch signaling ligand DLL3 and target gene HES1. Our meta-analysis supports that Notch signaling is a valuable bio-marker to predict progression and targeting Notch signaling could benefit subpopulation of NSCLC patients.
Non-small-cell lung cancer (NSCLC) is the most common cause of cancer-related deaths in the world and results in a serious problem to global health1. Despite advances in cancer biology as well as in diagnosis and treatment, progress in NSCLC therapy has been slow, leading to about 5% improvements in 5-year survival rates for the last 20 years2. Therefore, identifying the biomarkers is urgent to screen out high risk patients and predict outcome of NSCLC, in addition to the traditional clinicopathological features. Overwhelming evidence has proven that Notch signaling, comprised of four receptor isoforms (Notch1, Notch2, Notch3, Notch4) and five ligands members (Delta-like 1, Delta-like 3, Delta-like 4, Jagged-1 and Jagged-2), is crucial for cell proliferation, differentiation and apoptosis3. Earlier studies also discovered that deregulated Notch signaling led to various diseases, such as T cell leukemia and lung cancer4,5. In a transgenic mouse model, enhanced expression of Notch1 was found in the alveolar epithelium and increased abundance of Notch1 correlated with progressive from alveolar hyperplasia to pulmonary adenomas6. Subsequent study demonstrated that Notch signaling drove stemness and tumorigenicity of NSCLC7. The role of different Notch isoforms in NSCLC development is not clear, but Notch1 and Notch3 are believed to play a vital role8,9,10,11. Currently, one of the key points is to evaluate the value of Notch signaling as a molecular indicator of clinical features in NSCLC patients12,13,14,15.
A number of studies have examined the relationship between Notch1 and Notch3 expression and survival in patients with NSCLC. However, the prognostic value of Notch1 and Notch3 for NSCLC has yet to be confirmed. Some studies suggested that Notch1 and Notch3 overexpression was associated with poor prognosis in NSCLC16,17,18,19, but other researchers reported different results20. Besides, the ligands (Delta-like 1, Delta-like 3, Delta-like 4) and target genes, primarily involving in two families of helix-loop-helix transcription factors: HES (Hairy enhance of split) and HEY (Hairy/ enhancer of spit related with YRPW motif) targeting genes, were also used as biomarkers for the assessment of survival21,22,23. Given the uncertainty of causality and inconsistencies among studies, we performed a systematic review and a meta-analysis to evaluate the expression of Notch signaling members as predictive markers for clinicopathological parameters and outcomes of NSCLC.
Results
Search results
The study flow diagram for the identification of eligible studies is shown in Fig. 1. 111 articles were found by our search strategy. After the article titles, abstracts and main texts were checked, 19 articles with 3663 cases were subjected to the criteria for this analysis. The features of the eligible studies are presented in Table 1. These studies mainly evaluated the correlation between the expression of Notch1 and Notch3 with clinical parameters for NSCLC, based on either clinicopathological features or prognostic factors. In the meantime, we conducted an analysis of the association between other members of Notch signaling with prognosis of NSCLC patients, including DLL1, DLL3, DLL4, HES1 and HEY1. Six of the 19 studies were from USA, 5 were from China, 2 were from Germany, 1 was from Canada, 1 was from Norway, 1 was from Netherland, 1 was from Japan, 1 was from Italy and 1 was from Korea. TNM stages I and II were classified as low stage, III and IV as high stage.
Figure 1.
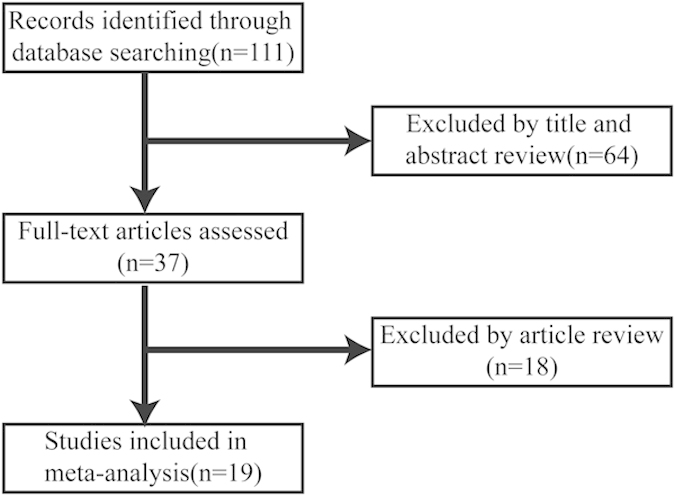
Flow diagram of article selection
Table 1. Characteristics of studies included in the meta-analysis.
| First author | year | Country or area | Duration Month(s) | Histology | Stage | Patients n | Quality score | Detection | Cutoff values |
|---|---|---|---|---|---|---|---|---|---|
| Andersen16 | 2011 | Norway | 216 | NSCLC | I-IIIA | 335 | 9 | IHC (Notch1, DLL4) | Staining of H-score 0–1 vs. 2–3 |
| Baumgart8 | 2010 | Germany | NR | NSCLC | NR | 46 | 9 | IHC(Notch1) | Staining of H-score 0–1 vs. 2–3 |
| Bild17 | 2005 | USA | 90 | NSCLC | NR | 111 | 9 | RT-PCR(Notch1, DLL4, HES1) | median expression |
| Cao9 | 2012 | China | NR | NSCLC | I-IIIA | 131 | 8 | IHC(Notch1) | Staining of H-score0–1 vs. 2–3 |
| Hassan23 | 2013 | USA | 60 | ADC | I-III | 441 | 9 | IHC(HES1) | median H-score |
| Hou20 | 2010 | Netherland | 130 | NSCLC | NR | 156 | 9 | RT-PCR (Notch1, HES1) | median expression |
| Lee24 | 2008 | Korea | NR | NSCLC | I-III | 158 | 8 | IHC(Notch1) | >10% |
| Li10 | 2009 | USA | NR | NSCLC | NR | 127 | 9 | IHC(Notch1) | median H-score |
| Li11 | 2010 | USA | NR | NSCLC | I-IV | 395 | 8 | IHC(Notch1) | Staining of H-score 0–1 vs. 2–3 |
| Okayama18 | 2011 | Japan | 120 | ADC | NR | 226 | 9 | RT-PCR(Notch1, DLL4, HES1) | median expression |
| Peng12 | 2011 | China | NR | NSCLC | NR | 55 | 7 | IHC(Notch1) | median H-score |
| Pesch13 | 2012 | Germany | NR | NSCLC | NR | 68 | 7 | IHC(Notch1) | Staining of H-score 0–1 vs. 2–3 |
| Raponi22 | 2006 | USA | 60 | SCC | I-IIIA | 20 | 7 | RT-PCR(HES1) | median expression |
| Shi28 | 2014 | China | 24 | NSCLC | III-IV | 594 | 8 | IHC(Notch3) | median H-score |
| Westhoff19 | 2009 | Italy | 60 | NSCLC | I-IV | 420 | 9 | IHC(Notch1) | median H-score |
| Xin14 | 2007 | China | NR | NSCLC | I-III | 80 | 8 | IHC(Notch1) | Staining of H-score 0–1 vs. 2–3 |
| Ye27 | 2013 | China | 60 | NSCLC | I-IV | 131 | 8 | IHC(Notch3) | median H-score |
| Zhang15 | 2013 | USA | NR | NSCLC | I-IV | 41 | 8 | IHC(Notch1) | Staining of H-score 0–1 vs. 2–3 |
| Zhu21 | 2010 | Canada | 120 | NSCLC | I-II | 133 | 9 | RT-PCR(HES1) | median expression |
Abbreviation: NR, not reporting; IHC, immunohistochemistry; RT-PCR, real-time reverse transcription polymerase chain reaction; ADC, lung adenocarcinoma; SCC, lung squamous cell carcinoma.
Notch1 and Notch3 expression positively correlated with NSCLC progression
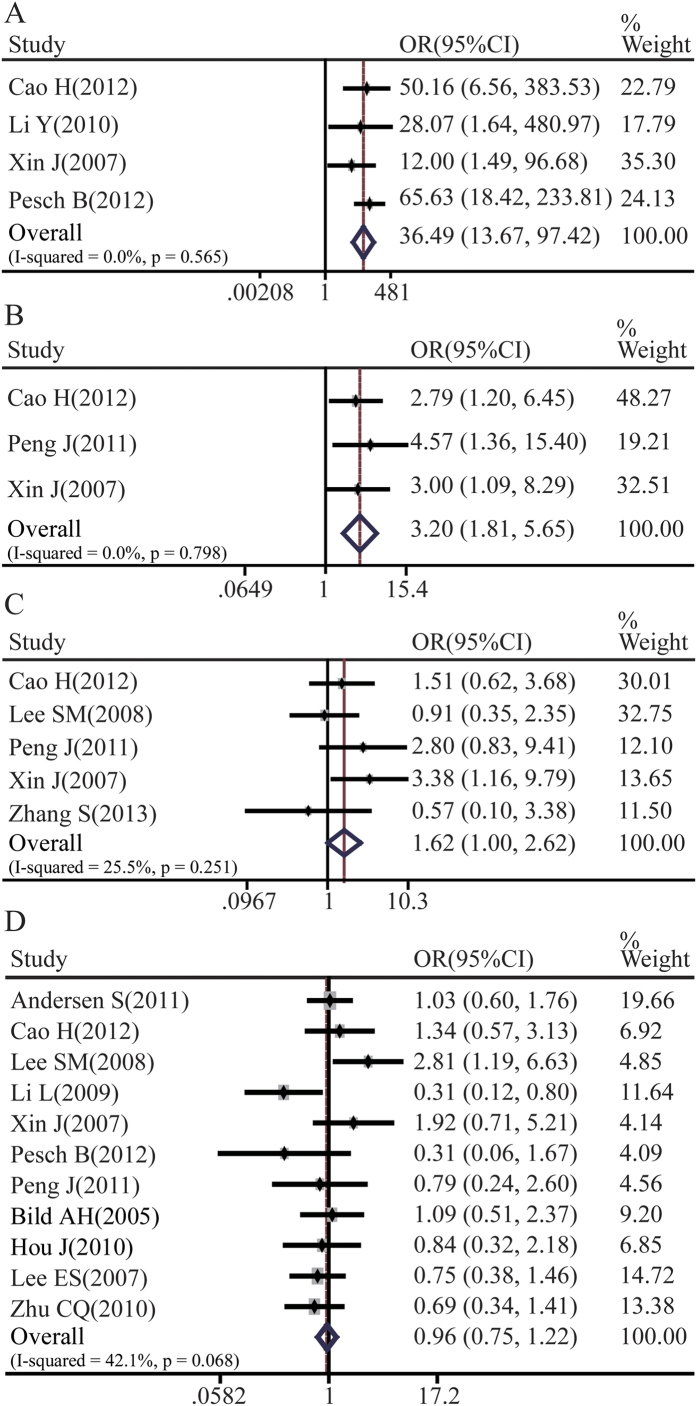
There were a total of 13 references that assessed the correlation between Notch1 expression and tumor clinicopathological parameters. Our meta-analysis demonstrated that Notch1 expressions in NSCLC tissues were significantly higher than normal lung tissues (pooled OR = 36.49, 95%CI: 13.67-97.42, p = 0.565 and I2 = 0.0%) (Fig. 2A). Moreover, greater possibility of lymph node metastasis (LNM) and higher tumor stages were linked with high Notch1 expression in NSCLC (pooled OR = 3.20, 95%CI: 1.81-5.65, p = 0.798 and I2 = 0.0%; pooled OR = 1.62, 95%CI: 1.00-2.62, p = 0.251 and I2 = 25.5%) (Fig. 2B,C). However, there was no statistical association between Notch1 expression and two main histological types of NSCLC: adenocarcinoma (ADC) and squamous cell carcinoma (SCC) (pooled OR = 0.96, 95%CI: 0.75-1.22, p = 0.068 and I2 = 42.1%) (Fig. 2D). Although Notch 3 expression associated with lymph node metastasis, there was no correlation between Notch3 and tumor size. Similarly, there was no statistical difference between adenocarcinoma and squamous carcinoma (Supplementary figure).
Figure 2.
Forest plot of odds ratio (OR). CI, confidence interval. A. Relative Notch1 abundance of NSCLC in comparison to normal tissues. B. Association between Notch1 expression and NSCLC lymph node metastasis. C. Association between Notch1 abundance and NSCLC clinical stages. D. Relative expression of Notch1 in ADC compared to SCC.
Impact of Notch1 expression on survival for NSCLC
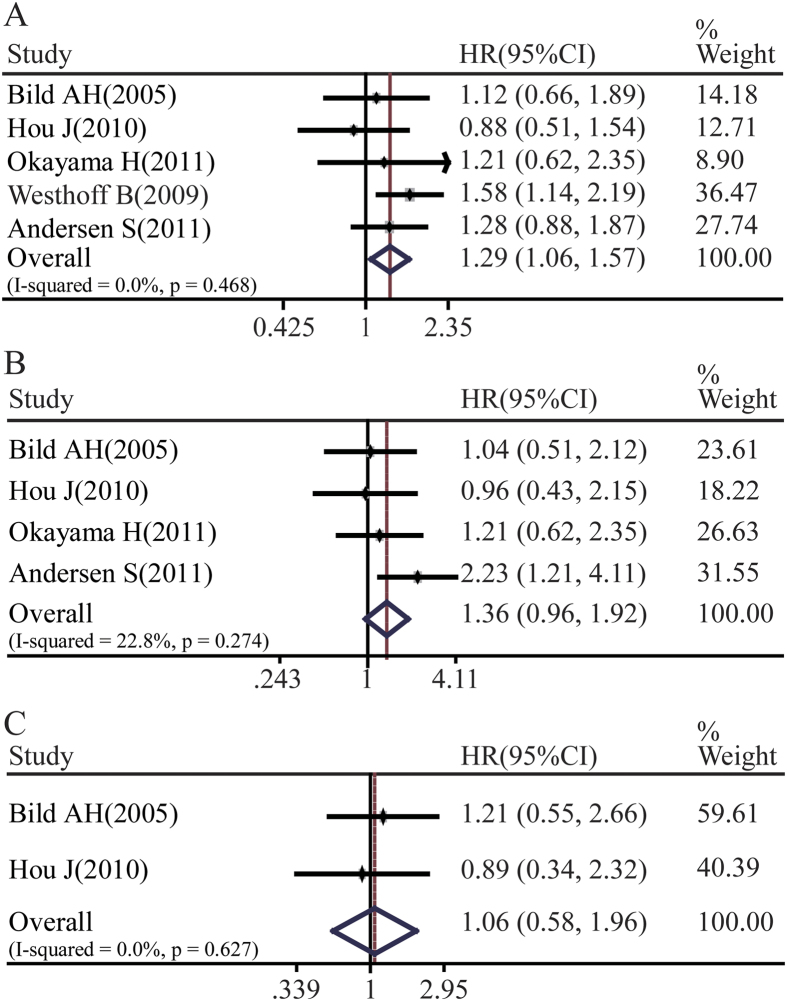
The association between Notch1 expression and NSCLC prognosis was also analyzed. We found that Notch1 was correlated with the overall survival rate of NSCLC patients (pooled HR = 1.29, 95%CI: 1.06-1.57, p = 0.468 and I2 = 0.0%) (Fig. 3A). Although there was a tendency indicating Notch 1 expression associated with poor OS of adenocarcinoma, however there was no significant difference (pooled OR = 1.36, 95%CI: 0.96-1.92, p = 0.274 and I2 = 22.8%) (Fig. 3B). There was no significant relationship between Notch1 expression with lung squamous cell carcinoma overall survival rate (pooled OR = 1.06, 95%CI: 0.58-1.96, p = 0.627 and I2 = 0.0%) (Fig. 3C).
Figure 3.
Forest plot of hazard ratio (HR). CI, confidence interval. A. Association between Notch1 and NSCLC OS. B. Association between Notch1 and ADC OS. C. Association between Notch1 and SCC OS.
Impact of Notch3 expression on survival for NSCLC
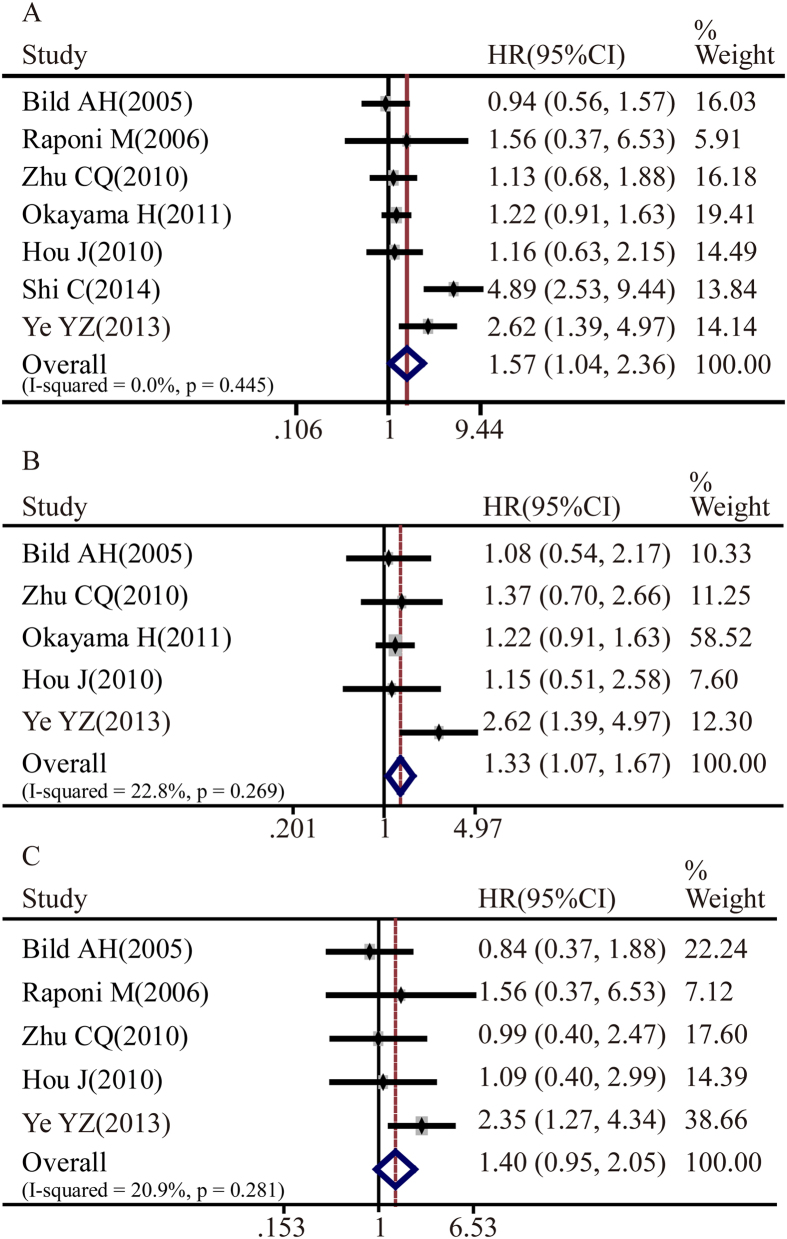
There were a total of 9 studies that assessed the correlation between Notch3 expression and tumor clinicopathological parameters. Our meta-analysis demonstrated that Notch3 expressions in NSCLC tissues were significantly correlated with the overall survival rate of NSCLC patients (pooled HR, 1.57; 95%CI, 1.04-2.36, p = 0.445 and I2 = 0.0%) (Fig. 4A). In addition, Notch3 was also associated with the overall survival rate of lung adenocarcinoma patients (pooled HR = 1.33, 95%CI: 1.07-1.67, p = 0.269 and I2 = 22.8%) (Fig. 4B). There was no significant relationship between Notch3 expression with lung squamous cell carcinoma overall survival rate (pooled HR = 1.40, 95%CI: 0.95-2.05, p = 0.281 and I2 = 20.9%) (Fig. 4C).
Figure 4.
Forest plot of hazard ratio (HR). CI, confidence interval. A. Association between Notch3 and NSCLC OS. B. Association between Notch3 and ADC OS. C. Association between Notch3 and SCC OS.
Impact of the expression of other members of Notch signaling on survival for NSCLC
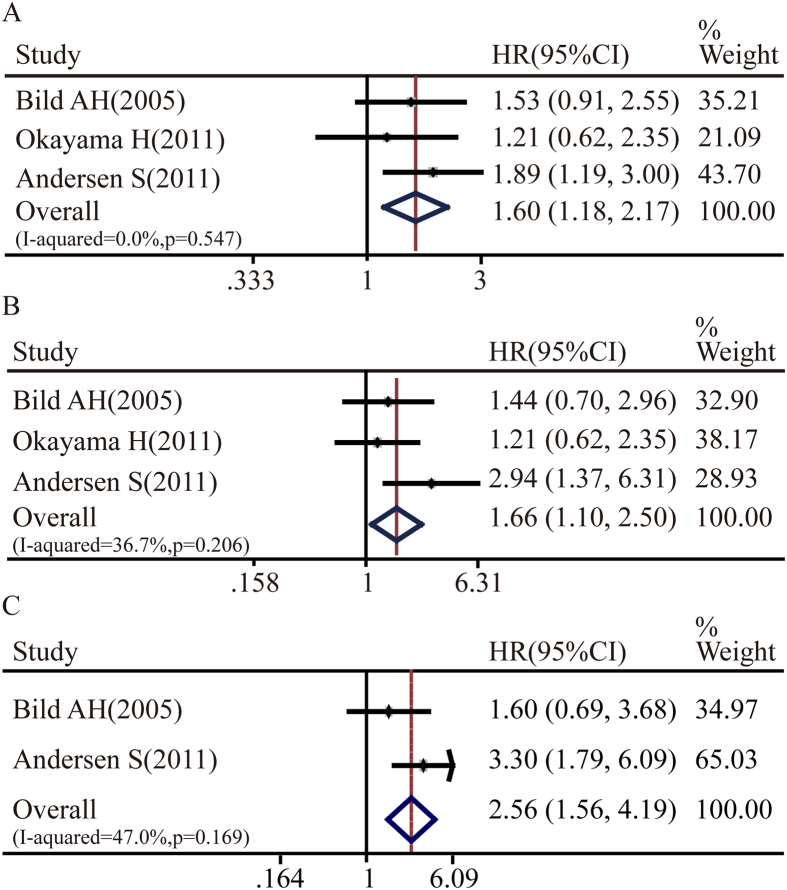
Notch signaling is initiated by ligand binding to Notch receptor; therefore, we further analyzed expressions of Notch ligands and prognosis of NSCLC patients. As shown in Fig. 5, a statistically significant association between DLL4 expression and NSCLC overall survival rate was found (NSCLC: pooled HR = 1.60, 95%CI: 1.18-2.17, p = 0.547 and I2 = 0.0%; lung adenocarcinoma: pooled HR = 1.66, 95%CI: 1.10-2.50, p = 0.206 and I2 = 36.7%; lung squamous cell carcinoma: pooled HR = 2.56, 95%CI: 1.56-4.19, p = 0.169 and I2 = 47%). However, there was no significant relation between the expressions of DLL1 and DLL3 and overall survival of NSCLC patients.
Figure 5.
Forest plot of hazard ratio (HR). CI, confidence interval. A. Association between DLL4 and NSCLC OS. B. Association between DLL4 and ADC OS. C. Association between DLL4 and SCC OS.
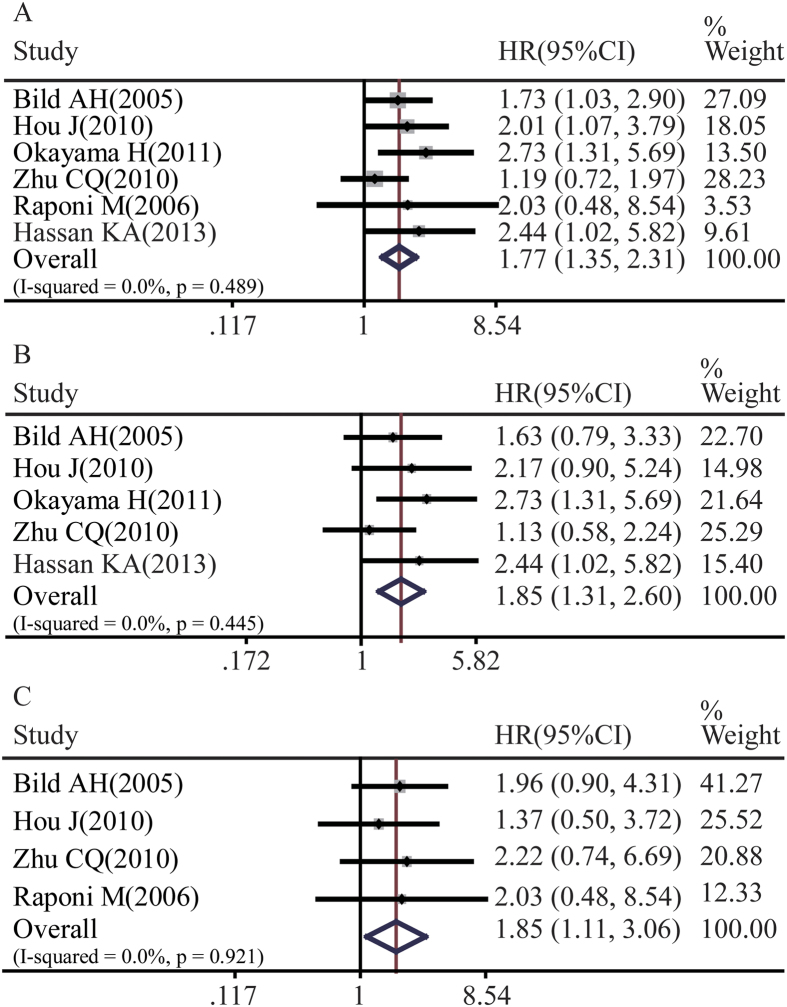
Next, we explored whether key targets of Notch1 and Notch3 could represent Notch signaling activity. The relationship between HES1 expression and NSCLC prognosis is illustrated in Fig. 6. Six studies including a total of 1087 patients demonstrated that HES1 overexpression was statistically associated with the overall survival rate of NSCLC patients (pooled HR = 1.77, 95%CI: 1.35-2.31, p = 0.489 and I2 = 0.0%) (Fig. 6A). In the subgroup analysis stratified by cancer histological types, HES1 expression was also associated with both lung adenocarcinoma overall survival rate (pooled HR = 1.85, 95%CI: 1.31-2.60, p = 0.445 and I2 = 0.0%) (Fig. 6B) and lung squamous cell carcinoma overall survival rate (pooled HR = 1.85, 95%CI: 1.11-3.06, p = 0.921 and I2 = 0.0%) (Fig. 6C).
Figure 6.
Forest plot of hazard ratio (HR). CI, confidence interval. A. Association between HES1 and NSCLC OS. B. Association between HES1 and ADC OS. C. Association between HES1 and SCC OS.
Whole genomic expression profiles provide unbiased quantitative measure of mRNA abundance and have been proved to correlated with histo-pathological characteristics and predict prognosis24. To investigate whether HES1 mRNA abundance correlated with NSCLC survival, we analyzed published gene expression database of NSCLC with survival information. Kaplan –Meier survival curve of GSE31210 comprised of 226 NSCLC patients at stage I-II demonstrated that the overall survival rate was lower in patients with higher expression of HES1 (p = 0.007) (Fig. 7A). To explore whether mutation of EGFR interferes with HES1, NSCLC patients were subdivided into EGFR mutation and wild type (wt) group. The overall survival rate of patients with EGFR mutation was correlated with the abundance of HES1 expression (p = 0.008) (Fig. 7B). However, there is no statistical relation between HES1 expression and OS in EGFR wt patients (p = 0.244) (Fig. 7C). Together, analysis results from HES1 mRNA profile are in consisting with protein abundance, both indicating that higher Notch signaling activity predicts worse prognosis.
Figure 7.
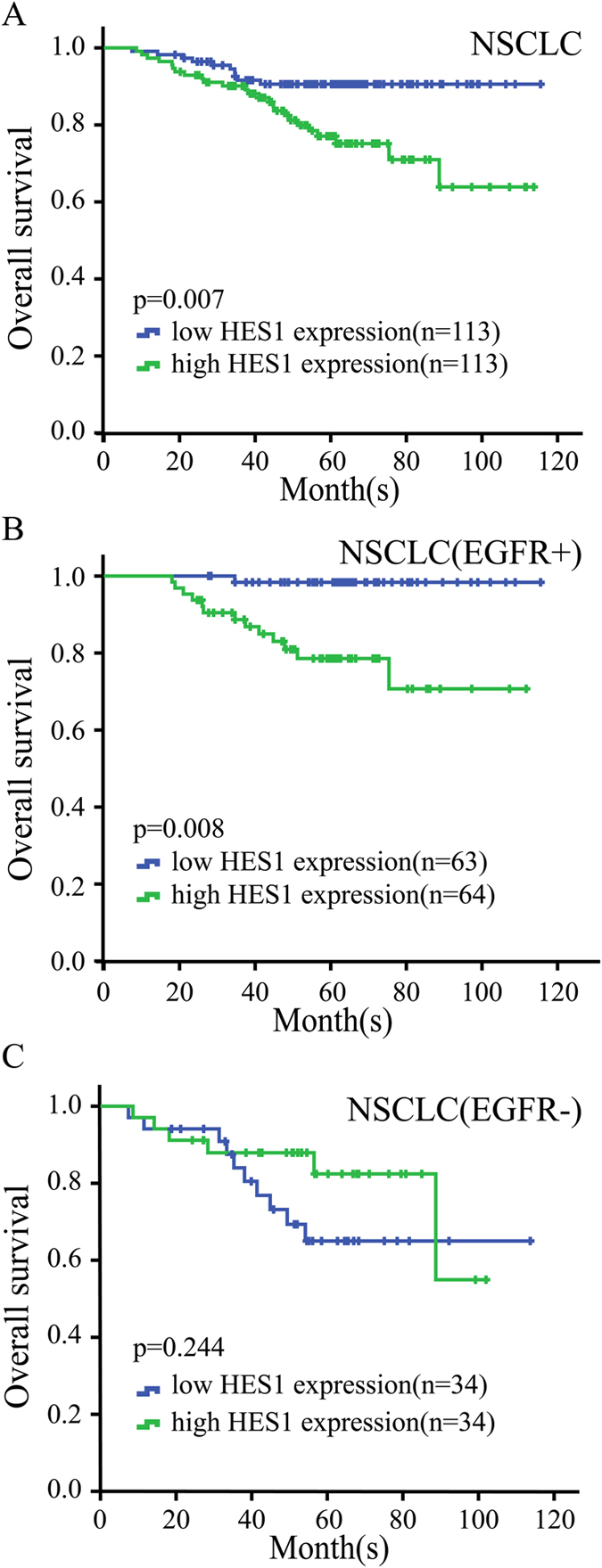
HES1 expression is correlated with overall survival of NSCLC patients(GSE31210). A. Overall survival rate was analyzed in 226 patients with NSCLC cancer, in relation to HES1 expression. B. Overall survival rate was analyzed in 127 patients with EGFR mutation+ NSCLC cancer, in relation to HES1 expression. C. Overall survival rate was analyzed in 68 patients with EGFR mutation- NSCLC cancer, in relation to HES1 expression.
Publication bias
Publication bias statistics were obtained using Begg’s test and Egger’s test, which did not indicate evidence of significant publication bias of Notch1 expression in NSCLC patients: NSCLC: Begg’s test, p = 0.296, Egger’s test, p = 0.591; LNM: Begg’s test, p = 1,Egger’s test, p = 0.553; TNM stages: Begg’s test, p = 1,Egger’s test, p = 0.713; lung adenocarcinoma: Begg’s test, p = 0.876, Egger’s test, p = 0.852; OS of NSCLC: Begg’s test, p = 0.221, Egger’s test, p = 0.123; OS of lung adenocarcinoma: Begg’s test, p = 0.089, Egger’s test, p = 0.148. Similar results were found for OS of Notch3, DLL4 and HES1 expression: Notch3 expression in NSCLC: Begg’s test, p = 0.072, Egger’s test, p = 0.97; Notch3 expression in adenocarcinoma: Begg’s test, p = 1,Egger’s test, p = 0.636; Notch3 expression in squamous cell carcinoma: Begg’s test, p = 0.806, Egger’s test, p = 0.402; DLL4 expression in NSCLC: Begg’s test, p = 0.296, Egger’s test, p = 0.176; DLL4 expression in adenocarcinoma: Begg’s test, p = 0.296,Egger’s test, p = 0.232; HES1 expression in NSCLC: Begg’s test, p = 0.806,Egger’s test, p = 0.291; HES1 expression in adenocarcinoma: Begg’s test, p = 0.462,Egger’s test, p = 0.259; HES1 expression in squamous cell carcinoma: Begg’s test, p = 1, Egger’s test, p = 0.9.
Discussion
It is generally assumed that Notch1 and Notch3 activity were higher in advanced NSCLC and predicted poor prognosis16,19; However, opposite result was also reported in lung squamous carcinoma10. Precisely measuring the prognostic value of Notch1 and Notch3 may help to guide an individual therapy for NSCLC patients24. In our study, a combined analysis of 19 eligible clinical studies revealed a predictive value of Notch1 and Notch3 expression in NSCLC patients. The meta-analysis results suggested that Notch1 expression was significantly higher in lung cancer compared with normal tissues, and correlated with lymph node metastasis and TNM stages. However, Notch 3 was only associated with lymph node metastasis. In addition, our meta-analysis suggested that overexpression of Notch1 and Notch 3 could be a prognostic marker for OS.
Notch1 is commonly expressed in malignant cells from different types of cancer, participating in multiple functions, including motility, cell-cell connections and cell polarity25,26. Thus, Notch1 signaling could affect invasion, lymph node metastasis and TNM stages. Multiple studies have also demonstrated that Notch3 plays a role in the regulation of cellular proliferation, differentiation, apoptosis, as well as tumorigenesis27. Moreover, high Notch3 expression in lung cancer represented a higher possibility of being resistant to chemotherapy28. In support of clinical-pathological observation, experimental study demonstrated that high Notch activity enhanced tumor sphere formation and knockdown of Notch decreased cell proliferation and induced apoptotic cell death29. It has been shown that block Notch activity by GSI reduced xenograft growth of Notch abundant cells and decreased tumor incidence upon re-implantation. Correspondingly, molecular analysis confirmed the reduced expression of downstream effectors of Notch pathway from tumor xenografts of mice treated with GSI23, demonstrating a potent antitumor efficacy of Notch1 inhibitor. These findings suggest that GSI may provide novel therapies to improve the efficacy of conventional therapies by directly targeting the CSC, thus delaying tumor recurrence and improving cancer patient survival.
The identification of biomarkers for micro-metastases, disseminated tumors, and residual disease is critical for early detection and treatment of these diseases to prevent metastases and recurrence. To elucidate the correlation between the Notch signaling and NSCLC prognosis, analysis of Notch signaling cascade is necessary. Thus, the ligands and downstream target genes of Notch signaling were evaluated. In previous study, Dll4 was found to positively correlate with VEGFR1. Moreover, by up-regulation of Notch1 and VEGFR1, DLL4 played important roles in tumor angiogenesis and prognosis of lung cancer30. In addition, the activation of HES1 in NSCLC leaded to tumor cell growth and tumor progression31. Our meta-analysis is in agreement with previous report, showing that DLL4/HES1 were positively associated with poor OS of NSCLC patients.
Currently, new target therapies show a significantly improved progression-free survival (PFS) and relatively less toxicity in comparison with standard chemotherapy2. However, the inevitable resistance to target drugs limits theirs clinical efficiency. Identification of molecules cross-talk with target pathways will be critical for further improvements in NSCLC treatment. In vivo experimental studies observed significant antitumor activity in lung xenograft models accompanied with impaired tumor vasculature by reduced expression of several key angiogenic genes after the treatment of GSI RO4929097. Molecular analysis revealed Notch pathway target gene HES1 strongly corresponded with the therapeutic response. Thus, Notch pathway downstream genes could be used to predict the antitumor activity of RO492909732.
EGFR mutations are one of the most important active genetic changes yet discovered in NSCLC. Both EGFR and Notch signaling are known to be deregulated in many human cancers. However, interactions of those two pathways are not well understood. It is observed that in human lung cancer cell line NCI-H292 cells, inhibition of the Notch pathway decreased both the Notch and EGFR/ERK pathways, associated with the reduction of proliferative cells. These results suggest that Notch signaling pathway has crosstalk with EGFR/ERK in human bronchial epithelial cells33. It can be proposed that GSIs might synergize with other signaling pathway inhibitors to treat NSCLC. For example, Notch 1 contributed to the acquired resistance to TKI and inhibition of Notch-1 resulted in effective response to EGFR target therapy (gefitinib)34.
Heterogeneity test is an essential part in meta-analysis. In this study, evidence of minor heterogeneities was observed; however, there was still some problems worth considering. Firstly, multicenter prospective studies based on large, homogeneous patient populations will be required. Another significant heterogeneity was likely due to the variations in assessing Notch signaling expression. The cutoff value was estimated in 5 studies by using the median Notch1 level measured by RT-PCR, remaining 12 articles measured by IHC. Some studies defined Notch1 expression based upon the percentage of immuno-positive cells or staining intensity, whereas other studies used a scoring system that combing those 2 factors together, causing the cutoff values for judging Notch1 expression diverse.
Lastly, publication bias is worth considering in meta-analysis. In this study, analyses of OS and other clinicopathological parameters did not show big variation. It should be noted that some limitations exist in the present meta-analysis. First, the number of included studies was relatively limited. Second, the applied method for detecting Notch signaling expression and the cutoff values were different in the included studies. Third, publication bias is still a concern. We tried to review all the relevant articles, but in some studies detailed description of raw data were not available.
All together, the meta-analysis provides evidence that Notch signaling is high activated in some NSCLC patients, which is associated with cancer progression and predicts bad prognosis. Preclinical models have proved the anti-tumor effects of GSIs. The therapeutic responses from early phase of clinical trials have observed in several types of cancer, including NSCLC. However, the dose -dependent adverse effects limited its clinical benefit 35. The identification and validation of of biomarkers associated with the hyperactivation of Notch signaling and correlated with treatment efficacy are important for a successful development of Notch pathway inhibitors. Incorporation of the specific biomarkers in future clinical trial may eventually demonstrate significant value of GSIs to subpopulation of NSCLC 36.
Methods
Literature search
We conducted a search of electronic databases PubMed, Embase and Cochrane library published up to October 1, 2014 using the search terms Notch (“Notch Receptors”, ”Notch Proteins”) and NSCLC (“Non-Small-Cell Lung Carcinoma,” “Non-Small Cell Lung Cancer”). The citation lists associated with the retrieved articles were also reviewed to identify additional relevant publications.
Inclusion criteria
This meta-analysis collected data from randomized controlled studies (RCTs) or observational studies (case-control or cohort) that evaluated the relationship between Notch signaling expression and NSCLC parameters such as clinicopathological features and prognostic factors. Studies met the following criteria were included: a) patients recruited in the study were pathologically diagnosed to have NSCLC; b) Notch signaling expression was measured by immunohistochemistry (IHC) or real-time reverse transcription polymerase chain reaction (RT-PCR) in primary NSCLC tissue; c) the hazard ratio (HR) and corresponding 95% confidence interval (CI) were reported or could be statistically extracted from the study. When several articles were from the same patient population, the newest or most complete article was included.
Data extraction
All data were abstracted by using a standardized data collection form, with information recorded as follows: first author’s last name, publication year, country of origin, histological type, tumor stage, number of cases and controls, detection methods, cutoff values for Notch1, Notch3, DLL4 and HES1. For articles without HRs, the statistical variables were calculated from published survival curves using methods described by Tierney et al37. We also reviewed Oncomine and found 5 independent human NSCLC microarray datasets with Notch signaling expression and survival data. Overall survival (OS) was evaluated by Cox proportional HRs and 95% CIs using these numerical data.
For observational studies, the Newcastle-Ottawa Quality Assessment Scale (NOS) was employed for assessing the quality of these studies. This scale is based on the identification of 8 sources of potential study bias that estimate patient selection, study comparability, and outcomes. Literature search, study selection, and data extraction were performed independently by two reviewers and disagreements among reviewers were resolved by discussion.
Statistical analysis
Statistical analysis was conducted by the guidelines proposed by Meta-Analysis of Observational Studies38. The prognosis of NSCLC patients with expressions of Notch1, Notch 3, Notch ligands, Notch downstream target gene HES1 and HEY1was calculated by HRs and 95% CIs, respectively. Clinicopathological parameters included histological type, lymph node metastasis (LNM), TNM stage. Heterogeneity of the odds ratio (OR) and HR was appraised by using the Cochran Q and I2 test. A random-effect model was applied when p < 0.1 or I2 > 50%. When heterogeneity was absent, a fixed-effect model was employed. Publication bias was assessed by Begg’s rank correlation method and Egger’s weighted regression method. All p values were two tailed, and all analyses were carried out using STATA software package (version 12.0) (Stata Corp LP, College Station, TX, USA).
Author Contributions
All authors contributed significantly to this work. K.W. and Q. C. designed and wrote the article. X.Y., H.W., H.X. and N. H. collected the data and performed the research study; S.Y. and Y.C. summarized and discussed the data. All authors reviewed this manuscript and approved the final draft.
Additional Information
How to cite this article: Yuan, X. et al. Meta-analysis reveals the correlation of Notch signaling with non-small cell lung cancer progression and prognosis. Sci. Rep. 5, 10338; doi: 10.1038/srep10338 (2015).
Supplementary Material
Acknowledgments
This work was supported by China NSFC No. 81172422, 81072169, 81261120395 and 81301929, and the Natural Science Foundation of Hubei Province No. 2014CFB218.
References
- Siegel R., Ma J., Zou Z. & Jemal A. Cancer statistics, 2014 . Ca-Cancer J. Clin. 64, 9–29 (2014). [DOI] [PubMed] [Google Scholar]
- Johnson D. H., Schiller J. H. & Bunn P.J. Recent clinical advances in lung cancer management. J. Clin. Oncol. 32, 973–82 (2014). [DOI] [PubMed] [Google Scholar]
- Hori K., Sen A. & Artavanis-Tsakonas S. Notch signaling at a glance. J. Cell. Sci. 126, 2135–40 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabher C., von B. H. & Look A. T. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 6, 347–59 (2006). [DOI] [PubMed] [Google Scholar]
- Dang T. P. et al. Chromosome 19 translocation, overexpression of Notch3, and human lung cancer. J. Natl. Cancer. Inst. 92, 1355–57 (2000). [DOI] [PubMed] [Google Scholar]
- Allen T. D., Rodriguez E. M., Jones K. D. & Bishop J. M. Activated Notch1 induces lung adenomas in mice and cooperates with Myc in the generation of lung adenocarcinoma. Cancer Res. 71, 6010–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. P. et al. Cisplatin selects for multidrug-resistant CD133+ cells in lung adenocarcinoma by activating Notch signaling. Cancer Res. 73, 406–16 (2013). [DOI] [PubMed] [Google Scholar]
- Baumgart A. et al. ADAM17 regulates epidermal growth factor receptor expression through the activation of Notch1 in non-small cell lung cancer. Cancer Res. 70, 5368–78 (2010). [DOI] [PubMed] [Google Scholar]
- Cao H. et al. Down-regulation of Notch receptor signaling pathway induces caspase-dependent and caspase-independent apoptosis in lung squamous cell carcinoma cells. APMIS. 120, 441–50, (2012). [DOI] [PubMed] [Google Scholar]
- Li L., Sheehan C. E. & Ross J. S. Notch signaling in non small cell lung cancers (NSCLC) is associated with squamous differentiation and favorable clinical outcome. Lab Invest. 89, 356A–356A (2009). [Google Scholar]
- Li Y. et al. Distinct expression profiles of Notch-1 protein in human solid tumors: Implications for development of targeted therapeutic monoclonal antibodies. Biologics. 4, 163–71 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Cen H. F., Mao Y., Yuan X. D. & Wu B. Study on expression of Notch signal pathway and VEGF in human non-small cell lung cancer. The Chinese Journal of Clinical Pharmacology. 27, 265–67 (2011). [Google Scholar]
- Pesch B. et al. NOTCH1, HIF1A and other cancer-related proteins in lung tissue from uranium miners--variation by occupational exposure and subtype of lung cancer. PLoS One. 7, e45305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X. et al. Expression and significance of Notch1, Jagged1 and VEGF in human non-small cell lung cancer. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 32, 1031–36 (2007). [PubMed] [Google Scholar]
- Zhang S. et al. Inhibition of CK2alpha down-regulates Notch1 signaling in lung cancer cells. J. Cell. Mol. Med. 17, 854–62 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S. et al. Correlation and coexpression of HIFs and NOTCH markers in NSCLC. Anticancer Res. 31, 1603–06 (2011). [PubMed] [Google Scholar]
- Bild A. H. et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 439, 353–57 (2006). [DOI] [PubMed] [Google Scholar]
- Okayama H. et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 72, 100–11 (2012). [DOI] [PubMed] [Google Scholar]
- Westhoff B. et al. Alterations of the Notch pathway in lung cancer. Proc. Natl. Acad. Sci. USA. 106, 22293–98(2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou J. et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 5, e10312 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C. Q. et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J. Clin. Oncol. 28, 4417–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raponi M. et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 66, 7466–72 (2006). [DOI] [PubMed] [Google Scholar]
- Hassan K. A. et al. Notch pathway activity identifies cells with cancer stem cell-like properties and correlates with worse survival in lung adenocarcinoma. Clin. Cancer Res. 19, 1972–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.M. et al. Expression of Notch 1 and 3 is related to inhibition of lymph node metastasis and progression in non-small cell lung carcinomas. Basic Appl. Pathol. 1, 93–7 (2008). [Google Scholar]
- Andersson E. R. & Lendahl U. Therapeutic modulation of Notch signaling-are we there yet? Nat. Rev. Drug Discov. 13, 357–78 (2014). [DOI] [PubMed] [Google Scholar]
- Yuan X. et al. Notch signaling and EMT in non-small cell lung cancer: biological significance and therapeutic application. J. Hematol. Oncol. 7, 87 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye YZ. et al. Notch3 overexpression associates with poor prognosis in human non-small-cell lung cancer. Med. Oncol. 30, 595 (2013). [DOI] [PubMed] [Google Scholar]
- Shi C. et al. Notch 3 protein, not its gene polymorphism, is associated with the chemotherapy response and prognosis of advanced NSCLC patients. Cell Physiol Biochem. 34, 743–52 (2014) [DOI] [PubMed] [Google Scholar]
- Wael H. et al. Notch1 signaling controls cell proliferation, apoptosis and differentiation in lung carcinoma. Lung Cancer. 85, 131–40 (2014). [DOI] [PubMed] [Google Scholar]
- Yu S. et al. Aberrant expression and association of VEGF and Dll4/Notch pathway molecules under hypoxia in patients with lung cancer. Histol Histopathol. 28, 277–84 (2013). [DOI] [PubMed] [Google Scholar]
- Tang Y. et al. Rnd3 Regulates Lung Cancer Cell Proliferation through Notch Signaling. PLoS One. 9, e111897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luistro L. et al. Preclinical profile of a potent gamma-secretase inhibitor targeting notch signaling with in vivo efficacy and pharmacodynamic properties. Cancer Res. 69, 7672–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. H., Lee E. H., Park S. W. & Chung I. Y. MUC5AC expression through bidirectional communication of Notch and epidermal growth factor receptor pathways. J. Immunol. 187, 222–29 (2011). [DOI] [PubMed] [Google Scholar]
- Xie M., He C.S., Wei S.H. & Zhang L. Notch-1 contributes to epidermal growth factor receptor tyrosine kinase inhibitor acquired resistance in non-small cell lung cancer in vitro and in vivo. Eur. J. Cancer. 49, 3559–72 (2013). [DOI] [PubMed] [Google Scholar]
- Takebe N., Nguyen D. & Yang SX. Targeting notch signaling pathway in cancer: clinical development advances and challenges. Pharmacol Ther. 141 2140–9(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza I. & Miele L. Notch inhibitors for cancer treatment. Pharmacol Ther. 139, 95–110(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney J. F., Stewart L. A., Ghersi D., Burdett S. & Sydes M. R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 8, 16 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroup D. F. et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 283, 2008–12 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.