Abstract
Streptococcus pyogenes (Group A Streptococcus, GAS) remains a major public health burden worldwide, infecting over 750 million people leading to over 500,000 deaths annually. GAS pathogenesis is complex, involving genetically distinct GAS strains and multiple infection sites. To overcome fastidious genetic manipulations and accelerate pathogenesis investigations in GAS, we developed a mariner-based system (Krmit) for en masse monitoring of complex mutant pools by transposon sequencing (Tn-seq). Highly saturated transposant libraries (Krmit insertions in ca. every 25 nucleotides) were generated in two distinct GAS clinical isolates, a serotype M1T1 invasive strain 5448 and a nephritogenic serotype M49 strain NZ131, and analyzed using a Bayesian statistical model to predict GAS essential genes, identifying sets of 227 and 241 of those genes in 5448 and NZ131, respectively. A large proportion of GAS essential genes corresponded to key cellular processes and metabolic pathways, and 177 were found conserved within the GAS core genome established from 20 available GAS genomes. Selected essential genes were validated using conditional-expression mutants. Finally, comparison to previous essentiality analyses in S. sanguinis and S. pneumoniae revealed significant overlaps, providing valuable insights for the development of new antimicrobials to treat infections by GAS and other pathogenic streptococci.
In 2002, Streptococcus pyogenes (Group A Streptococcus, GAS) was reported by the World Health Organization among the top 10 causes of morbidity and mortality due to bacterial infections worldwide; and it currently remains a major public health problem1. GAS is responsible for a wide range of diseases, most commonly presenting as self-limiting infections of the skin (impetigo) and throat (pharyngitis)2,3,4,5. GAS can also enter normally sterile sites and cause severe life-threatening invasive infections (e.g. necrotizing fasciitis and toxic shock syndrome), leading to over 150,000 deaths worldwide each year2,4. Invasive GAS infections typically spread rapidly causing extended tissue damage that require drastic surgical intervention6. Moreover, immune-mediated complications following GAS infections (e.g., glomerulonephritis and acute rheumatic fever, ARF), affect children and young adults in developing countries, resulting in over 350,000 deaths each year2,4. Although GAS is generally treatable with antibiotics and remains one of the few pathogens susceptible to beta-lactams (e.g., penicillin), treatment failures can occur and antibiotic treatment does not guarantee prevention of immune sequelae7,8,9. Low availability of antibiotics in developing countries and the emergence of antibiotic-resistant GAS strains emphasizes the need for novel interventions to address GAS diseases4. To help guide the development of new treatment strategies, a better understanding of GAS physiology and pathogenesis during human infection is key.
Essential genes, defined as those necessary for growth and survival under a given condition, represent attractive targets for the discovery of new therapeutics against bacterial pathogens10. Identification of these genes on a genomic scale has been of particular interest as it provides candidates for guiding subsequent drug development11,12. A gene is considered essential when one is unable to generate a viable knockout mutation in that gene under the tested conditions. Gene-by-gene knockout strategies have been successfully implemented to screen the genomes of a few bacteria for essential genes, such as Escherichia coli13, Bacillus subtilis14 and Streptococcus sanguinis15; while, in S.aureus, Forsyth et al.16 successfully used a shotgun antisense RNA method towards this goal. However, these approaches require organisms for which extensive and efficient genetic tools are available (e.g., natural competence). To circumvent the labor and technical difficulties inherent to these approaches, transposon mutagenesis has become a method of choice for genome-scale gene essentiality studies17,18. Genome-wide identification of mutable genes, and therefore of the non-mutable essential genes, has been carried out using conventional DNA sequencing19,20, genetic footprinting21, and microarray hybridization (transposon site hybridization or TraSH)22. More recently, transposon sequencing (Tn-seq) using parallel sequencing of transposon-adjacent chromosome has allowed for the identification of insertions at nucleotide resolution in complex mutant libraries23,24,25,26.
Essentiality screens have been successfully carried out in two pathogenic streptococci, the oral pathogen S. sanguinis using directed mutagenesis15 and the respiratory pathogen S. pneumoniae both by TraSH27 and Tn-seq26; however, these analyses have not yet been undertaken for GAS. Towards this goal, we recently developed a mariner transposon (Oskar) for stable random mutagenesis in GAS28 and used a complex mutant library for TraSH to identify genes that are important for GAS fitness in an ex vivo model of human blood infection29. Though successful, our study revealed technical bottlenecks of TraSH such as the incomplete representation of the GAS genome on the microarray and the inability to accurately map transposon mutations29.
Here, we present the development and application of a modified mariner-based transposon (Krmit) for the application of Tn-seq in GAS to allow significantly increased mutant monitoring through next-generation sequencing. High saturation mutagenesis was achieved in two divergent GAS strains that are representative of strains with tropism for the throat alone (serotype M1T1 strain 5448) and both the throat and skin (serotype M49 strain NZ131), respectively. Tn-seq was combined with Bayesian analyses integrated over multiple in vitro passages to identify highly conserved genes in the GAS core genome that are essential for growth in rich media under optimal conditions. This work establishes a baseline for future Tn-seq studies of GAS growing in disease-relevant environments and provides invaluable information for the discovery of new targets for antimicrobial drugs as well as the study of GAS pathogenesis on a genome-scale.
Results
Development of Krmit: a transposon for Tn-seq in GAS
Identification of essential genes by Tn-seq requires the production of saturated mutant libraries in a given bacterial genome by transposition30. To accomplish this, we modified the mariner transposon (Oskar) developed for GAS by our group28,29 for use in Tn-seq. The pKRMIT in vivo mariner delivery plasmid was produced through PCR-mediated modification of the pOSKAR plasmid to include MmeI restriction sites in Oskar mini-transposon ITR sequences, creating the new transposon Krmit (for Kanamycin-resistant transposon for massive identification of transposants) (Fig. S1AB). The presence of MmeI sites allows for digestion of genomic DNA containing Krmit transposon insertion sites (TIS), producing uniform 20-nt regions of adjacent chromosomal DNA (insertion tags) for next-generation sequencing and Tn-seq screens31. Initial tests for in vivo transposition using pKRMIT in a GAS serotype M1T1 strain revealed that Krmit transposed comparably to Oskar28,29, exhibiting an average transposition frequency of 4 × 10−3 with insertions occurring exclusively within the dinucleotide TA (Fig. S1C). Krmit transposition in different GAS serotypes (e.g. M1T1, M3, M6, M49) showed similar results (data not shown), demonstrating that introduction of the two MmeI sites did not affect in vivo transposon delivery in GAS. Since pKRMIT relies on the pWV01 replicon, it has the potential to be used in other closely related Gram-positive genera (e.g., Streptococcus, Enterococcus, and Lactococcus).
Selection of M1T1 GAS Krmit libraries under in vitro growth
Krmit transposition was performed in the GAS strain 5448, a representative of the invasive M1T1 serotype circulating worldwide that is associated with a throat tropism (Pattern A, Class I emm gene)32. For in vivo Krmit transposition in M1T1 5448, the protocol developed for Oskar was used28,29 as detailed in Materials and Methods. Following transformation with pKRMIT, a set of individual colonies (ca. 120) was subjected to phenotype screening to verify the presence of the intact plasmid and to select appropriate mutant libraries (ca. 24) for further genetic analyses. A low percentage of spectinomycin-resistance (Spr) among Krmit transposants (< 5%) was preferred to limit the frequency of the pKRMIT plasmid within the mutant pools; an unwanted phenomenon previously observed29. A total of 15 libraries with a plasmid integration frequency ranging from 0.05% to 5% (av. 2.6%) were selected for arbitrary-primed PCR (AP-PCR) analyses to identify the Krmit transposon insertion site (TIS) in at least 20 randomly picked mutants per library. Transposon insertion randomness was found to vary widely between the tested mutant pools (35% to 95%).
To determine the set of genes essential for M1T1 5448 growth in vitro in "rich" Todd-Hewitt Yeast (THY) medium under typical laboratory conditions (37°C, 5% CO2), four independent Krmit mutant libraries with optimal randomness were selected and subjected to three additional 24-hour passages under these conditions. This approach allowed for en masse mutant selection over continued in vitro growth in THY (T0 to T3), with mutants in essential genes predicted to be lost from the library in the early passages (T0, T1) and those with reduced fitness being lost only during the final passages (T2 to T3). The screen resulted in a total of 16 different passaged libraries (4 libraries at 4 time points) for the Tn-seq analyses.
Tn-seq analyses of in vitro-grown M1T1 5448
The complexity of the Krmit mutant libraries in M1T1 5448 was examined by Tn-seq26,31 with modifications for its use in GAS with Krmit. Briefly, genomic DNA was isolated from the different mutant pools (T0 to T3), subjected to complete digestion by MmeI followed by ligation to MmeI adapters, and PCR amplification to produce DNA-seq libraries consisting of 176-bp Krmit-specific insertion tags containing adjacent genomic sequence, Illumina-specific sequences and one of 8 distinct barcodes for multiplexing. DNA-seq reads containing both the barcode sequence (specific for Krmit mutant libraries) and the Krmit ITR sequence were assigned into 16 distinct Tn-seq datasets, each containing, on average, a total of ca. 13.7 millions reads (Table S1).
For genome alignment of the reads, the 20-nt sequence corresponding to the Krmit TIS was retained and the dinucleotide TA used as a means to orient the transposon on the chromosome. The complete annotated genome of M1T1 5448 is not publically available; however, the draft genome sequence was found to be almost identical to the reference genome of M1T1 MGAS500533,34 and the latter was used for mapping. Sequencing reads that aligned to more than one position on the MGAS5005 chromosome (ca. 5% of the total read counts, Table S1) were kept in the datasets and randomly mapped to one candidate position on the genome. The orientation of Krmit insertions showed an equal distribution of TIS on the forward and reverse strand of the chromosome (data not shown).
Over 90% of the reads matched perfectly to the reference GAS genome in the different datasets, with the exception of the T0 datasets where this proportion varied from 69% to 6% (Table S1). The reads that did not align to the GAS genome were found to correspond to sequences on pKRMIT, with the vast majority aligning to the region adjacent to Krmit on the plasmid (data not shown). This indicated that pKRMIT was still present during the initial passage of the mutant libraries (T0); however, it was lost during further passages in THY (T1, T2 and T3) (Table S1).
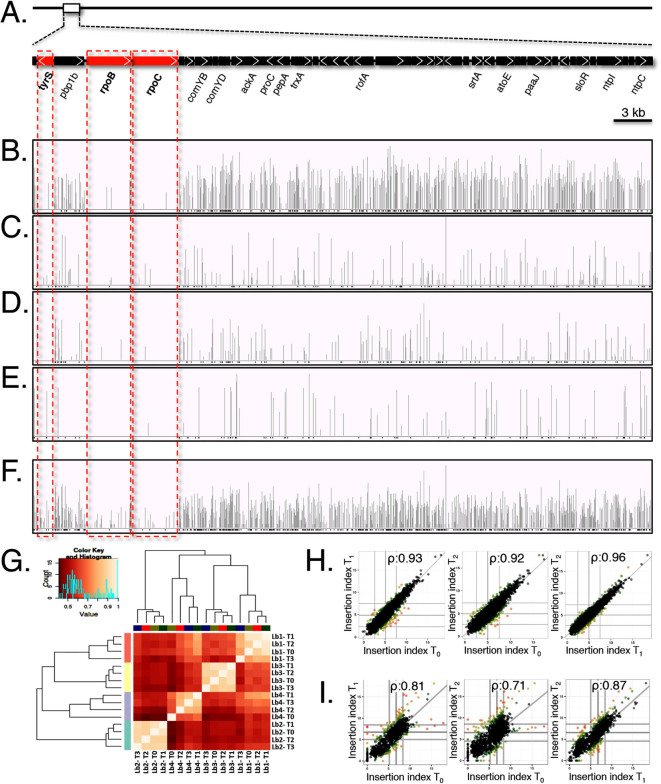
Visualization of the read alignments using IGV35,36 revealed that the initial libraries (T0) contained distinct Krmit insertion patterns as shown for the genomic region from the tyrS to ntpC genes (Fig. 1A) with TIS density being highest in library 1 (Fig. 1B) compared to libraries 2 to 4 (Fig. 1C-E). Determination of the unique Krmit TIS and the percentage of TAs containing a transposon correlated with this result (Table S1). Library 1 TIS density (ca. 45% of TAs targeted) was significantly higher than the TIS density in the other libraries (20% or less). Our results also showed that TIS density typically increased between T0 and T1 (Table S1), likely due to continued transposition in T0 libraries that still contained pKRMIT.
Figure 1. Comprehensive Krmit mutant libraries in GAS M1T1 5448 and M49 NZ131.
(A) Schematic of GAS M1T1 5448 chromosomal region from tyrS to ntpC. Genes that possess limited Krmit transposon insertion sites (TIS) determined by Tn-seq and Bayesian analysis are shown in red. Scale bar is indicated at right. (B to E) Location (horizontal axis) and depth (vertical axis) of all Krmit TIS identified within the 5448 genomic region in four independent mutant libraries as determined by Tn-seq at time point T0 presented using IGV. (F) Location and density of Krmit TIS in the “Master” M1T1 5448 library generated by merging the 4 independent libraries B-E. (G) Heat map summarizing the pairwise comparison of the four independent M1T1 5448 Krmit mutant libraries as determined by Tn-seq at each time point (T0, T1, T2 and T3; 16 total libraries) using Spearman’s rank correlation coefficients. Scatter plot summarizing the pairwise comparison of the Krmit TIS in the M1T1 5448 “Master” library (H) and the M49 NZ131 library (I) at different time points using Spearman’s rank correlation coefficients.
Pairwise comparison analyses, including Principal Component Analysis (Fig. S2A), Euclidean distance (Fig. S2B), and Spearman’s rank correlation (Fig. 1G), were performed to determine the degree of similarity between the 16 different mutant pools. The results revealed that the members of each Krmit mutant library (i.e., same library at different time points) were much more similar to one another than to the other libraries. Thus, although the individual libraries were random, they were not saturated enough individually to permit optimal library comparison and gene essentiality predictions. Therefore, data from the four independent Krmit libraries (Lb1, Lb2, Lb3 and Lb4) were combined to generate a single high-density Krmit insertion dataset for each of the four passages (T0 to T3). This “Master” library in M1T1 5448 was found to contain on average 45.8 million aligned reads corresponding to an average of over 85,000 unique TIS in all four passages (Table S1), representing Krmit integration in over 64% of the available TAs on the M1T1 5448 genome or one Krmit TIS for every 22 nucleotides. Read alignments revealed the increased complexity of the library (Fig. 1F, data not shown), showing high numbers of Krmit insertions in most genes. Spearman’s correlation rank analyses found that the master libraries at the 4 time points were highly comparable with ρ values over 0.92 (Fig. 1H).
Bayesian prediction of GAS 5448 essential genes
A recently developed Bayesian statistical model37 was used to analyze the Tn-seq data generated from M1T1 5448 to rigorously predict the essentiality of individual genes. Essential genes are typically defined as those that do not tolerate disruption and should be identified in our datasets as loci that lack Krmit insertions. However, recent Tn-seq analyses in other bacteria have shown that in addition to genes completely lacking transposon insertions, essential genes may also have insertions in the extreme 5’ and 3’ ends of the open reading frame or in sequences encoding non-essential domains (Fig. 1F; tyrS, rpoB and rpoC)17,38. To account for this, the DeJesus Bayesian method37 makes rigorous predictions of statistically significant stretches of TA sites within a given gene lacking transposon (i.e., Krmit) insertions regardless of the presence of insertions in other regions in the ORF. For stringency, TA sites with just one insertion were discarded from our datasets as these could represent sequencing errors. This filter resulted in a significant coverage reduction for the Tn-seq dataset generated at time point T3 (Table S2) and consequently this dataset was excluded from all further analyses.
The Bayesian analysis generated a posterior probability of essentiality Zi score for every M1T1 5448 gene in each remaining dataset (T0, T1 and T2), assigning each gene into 4 distinct fitness categories: non-essential gene (NEBay) (0.0 < Zi < 0.05), essential gene (EBay) (Zi> 0.995), non-conclusive data (scarcity of insertions) (0.05 < Zi < 0.995), and gene too small for analysis (Zi = −1)37. The resulting Bayesian analysis for M1T1 5448 is illustrated in Fig. 2A and detailed in Table S3.
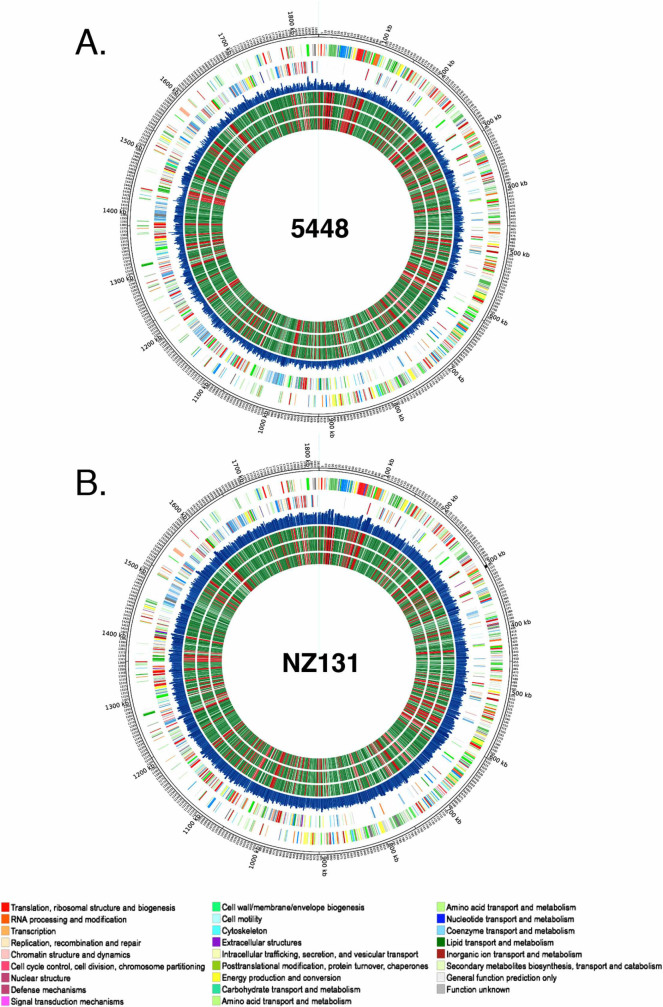
Figure 2. Bayesian prediction of gene essentiality in GAS M1T1 5448 and M49 NZ131 genomes at multiple time points.
Circos atlas representation of M1T1 5448 (A) and M49 NZ131 (B) genomes are shown with base pair (bp) ruler on outer ring. Next two outer circles represent GAS open reading frames on the (+) and (−) strands, respectively, with colors depicting COG categories (legend at bottom). The next circle (blue) indicates the frequency of Krmit TIS in each genome at T0. The inner three circles present the results of Bayesian analysis of GAS gene essentiality at time points T0, T1 and T2 in order towards center; with essential genes (EBay) in red, non-essential genes (NEBay) in green, and excluded genes in black.
To integrate the results of the Bayesian analysis at all three time points (T0, T1 and T2), we established the following four penultimate categories: a gene was “Essential” when found to be EBay at all 3 time points (T0, T1 and T2) or the latter 2 time points (T1 and T2); a gene was “Critical” for fitness when found to be EBay only at the final T2 time point; and a gene was “Non Essential” if found NEBay at all 3 time points. Those genes that did not meet any of these three criteria were categorized as “Non-conclusive”. Using those stringent criteria on the 1841 annotated genes in the M1T1 5448 genome, our analyses found that 227 (~12%) were “Essential”, 71 (~4%) were “Critical”, 1337 genes (~73%) were “Non-Essential”, and 206 genes (~11%) were “Non-Conclusive” for in vitro growth (Table S4).
Genes “Essential” for in vitro growth of GAS M49 strain NZ131
To compare essential genes between two divergent GAS strains, Krmit mutant libraries were produced in the GAS serotype M49 isolate NZ131, a nephritogenic representative of “generalist” GAS strains associated with both throat and skin infections (Pattern E, Class II emm gene)32. NZ131 has a fully annotated genome sequence that is publically available39. Krmit transposition proved more efficient in the highly transformable NZ131, facilitating the selection of complex Krmit mutant libraries (data not shown). Tn-seq analysis was performed as described above on a selected mutant library passaged 4 times in THY broth. Even though read alignments showed that the M49 NZ131 Krmit mutant library did not have the depth (18 millions aligned reads on average) of the master library in M1T1 5448 (45.8 millions aligned reads on average), the TIS saturation of NZ131 was reasonably similar to that observed for 5448 with a Krmit insertion every 27 nucleotides compared to every 22 nucleotides, respectively (Table S1). Furthermore, a Spearman’s correlation rank analyses showed that the passaged libraries were very similar (Fig. 1I).
The Tn-seq data from the M49 NZ131 Krmit libraries at each time point was subjected to Bayesian analysis as described for M1T1 5448 (Fig. 2B and Table S5), followed by integration of time points into our defined gene categories (Table S6). As with 5448, the T3 time point was removed from the analysis due to low Krmit coverage. Out of the 1698 annotated genes in the NZ131 genome, 241 genes (~14%) were “Essential”, 45 genes (~3%) were “Critical”, 1177 genes (~69%) were considered “Non-Essential”, and 235 genes (~14%) were found “Non-Conclusive” (Table S6).
Conserved "Essential" genes in the core GAS genome
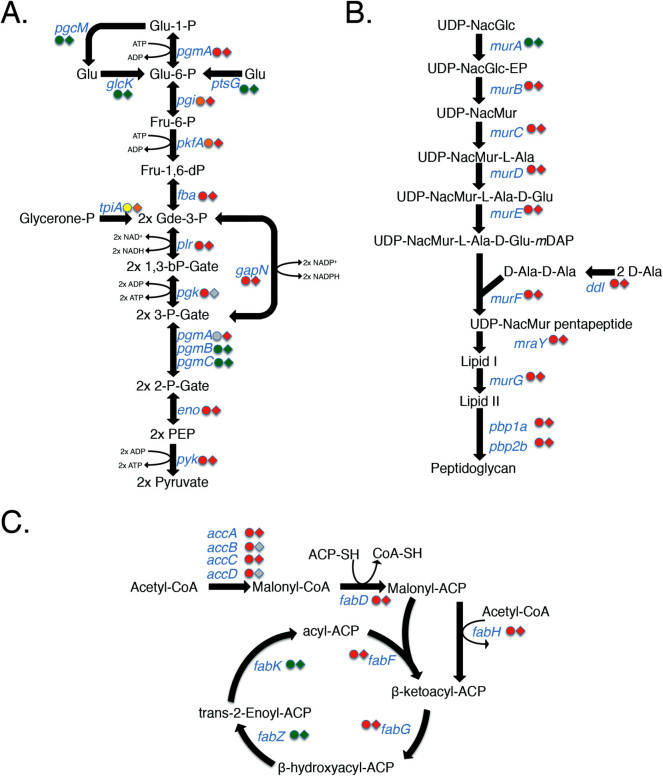
The integrated essentiality results from M1T1 5448 and M49 NZ131 were compared (Fig. 3 and Table S7). Of the genes found "Essential" in 5448 (227) and NZ131 (241), 187 were found to overlap in both strains and most likely represent essential genes shared by many GAS strains (Fig. 3A and Table S7). Comparison of the GAS 5448 and NZ131 datasets revealed the importance of conserved essential genes in key metabolic pathways such as glycolysis (Fig. 4A), peptidoglycan biosynthesis (Fig. 4B) and fatty acid synthesis (Fig. 4C).
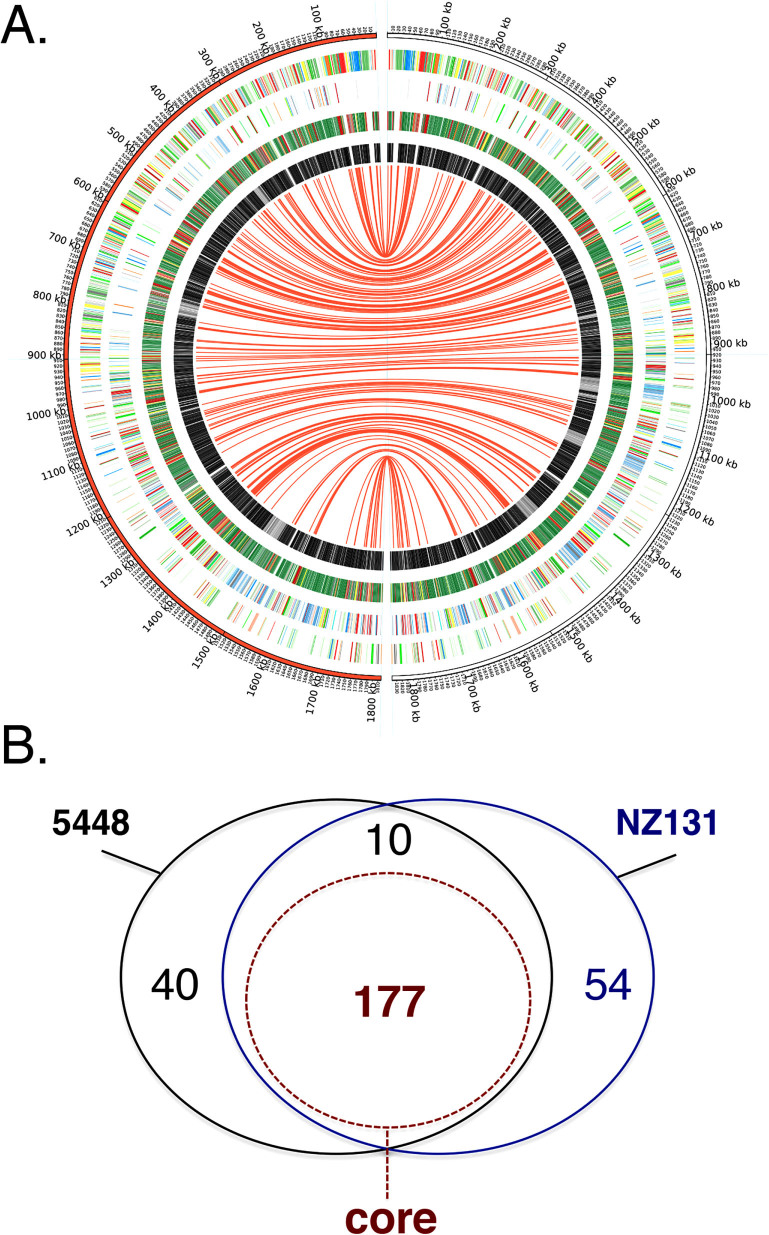
Figure 3. Conserved GAS gene essentiality based on integration of Bayesian analysis at all time points.
Bayesian prediction datasets at the time points T0, T1 and T2 were integrated for both M1T1 5448 and M49 NZ131 into the categories "essential", "critical", "non-essential", or "non-conclusive". (A) Circos atlas representation of M1T1 5448 (left, red semicircle) and M49 NZ131 (right, white semicircle) genomes are shown with base pair (bp) ruler on outer ring. Next two outer circles represent GAS open reading frames on the (+) and (−) strands, respectively, with colors depicting COG categories (see Fig. 2). The next circle presents the integrated analysis of GAS gene essentiality at all time points; with "essential" (red bars), "critical" (yellow bars), "non-essential" (green bars), and "non-conclusive" (black bars) genes indicated. Inner circle represents a genomic comparison between the M1T1 5448 and M49 NZ131 with homology (black) and non-homology (grey) shown. Red centerlines connect conserved "essential" genes shared between the two GAS genomes. (B) Venn diagram representing a comparison between the integrated "essential" genes found in GAS M1T1 5448 (227 genes, black) and M49 NZ131 (234 genes, blue) genomes. Shared "essential" genes also found within the GAS "core" genome (177 genes, see Fig. 4) are shown (dashed red line). See Table S7 for detailed list of genes.
Figure 4. GAS essential genes correlate to key metabolic pathways and cellular functions.
Schematics of the pathways for (A) Glycolysis, (B) Peptidoglycan biosynthesis, and (C) Fatty acid biosynthesis are shown, including reactants/products (black font) and genes encoding key enzymes (blue font). Integrated Bayesian analysis results for key enzymes are shown for GAS 5448 (circles) and NZ131 (diamonds) with "essential" genes in red; "critical" genes in yellow; "non-essential" genes in green; and "non conclusive" genes in grey.
Of the non-overlapping "Essential" genes (i.e. 40 genes for 5448; 54 genes for NZ131), only a few (i.e., 1 in 5448 and 2 in NZ131) were located in prophages found in one GAS strain, but not the other (Fig. 3B). The remaining genes were found in both strains, but classified in different categories: "Essential" in 1 strain, but either "Critical", "Non-essential", or "Non-conclusive" in the other. Overall, the comparison revealed a set of 187 conserved "essential" genes found in both 5448 and NZ131 (Fig. 3B and Table S7).
A "core genome" is defined as the genes shared by all strains of a given species as determined by comparative genomics of multiple sequenced genomes40. There are currently 20 annotated GAS genome sequences publically available representing emm serotypes correlated with tissue tropism32, including pattern A-C associated with throat infections (M1, M3, M5, M6, M12, M14, and M18), pattern D associated with skin infections (M53), and pattern E (generalists) that can colonize both sites (M2, M4, M28, M49, and M59). The GAS core genome was determined using these sequenced genomes (see Materials and Methods), identifying a total of 1224 orthologous genes common to all of the GAS strains (Fig. S3 and Table S7). The vast majority of the genes categorized as either "Essential" or "Critical" in M1T1 5448 (271/298 or 91%) and M49 NZ131 (267/286 or 93%) were part of the GAS core genome (Table S7). Furthermore, of the 187 "Essential" genes shared by both 5448 and NZ131, 177 were present in the GAS core genome (Fig. 3B, Fig. S4 and Table S7) and likely to be essential for all GAS strains.
Validation of selected GAS "Essential" genes
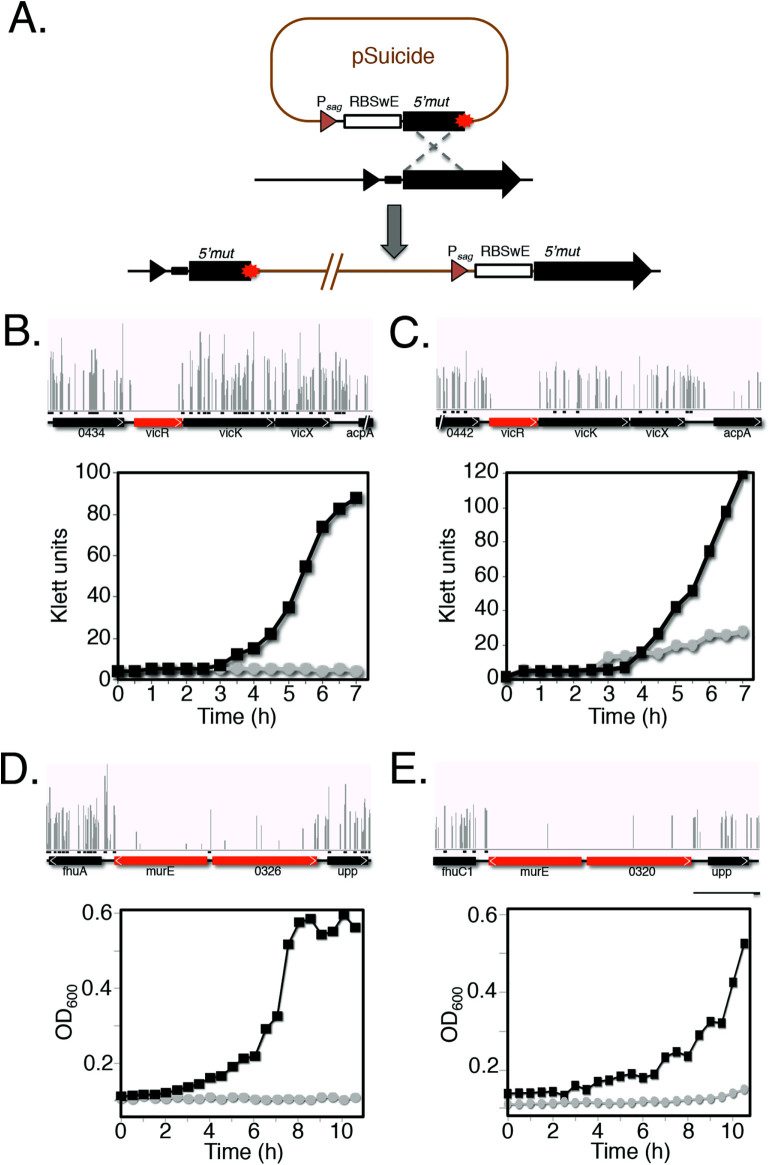
To confirm that conserved genes identified in our screen are required for in vitro growth of GAS, we utilized a conditionally lethal approach that took advantage of a theophylline-sensitive synthetic riboswitch functional in GAS41. A suicide/helper system pSin-pHlp was created for GAS to allow for stable insertional gene inactivation (see Materials and Methods). Stable plasmid integration was generated in the GAS chromosome resulting in a merodiploid strain containing the heterologous Psag promoter followed by the theophylline-inducible riboswitch E controlling expression of the full length targeted gene, while the wild type promoter of the targeted gene transcribed a truncated non-functional allele (Fig. 5A).
Figure 5. Experimental validation of selected essential genes.
(A) Schematic showing construction of GAS conditional expression mutants controlled by riboswitch E. The 5’-end of the selected gene was cloned along with the Psag promoter and a theophylline-inducible riboswitch into the new pSinS vector for stable chromosomal integration in GAS. Merodiploid integrants have target gene expression under the control of the riboswitch, while the wild-type promoter controls the expression of a truncated target gene. (B to C) Validation of vicR (B to C) and murE (D to E) as an essential gene in GAS 5448 (B and D) and NZ131 (C and D). The top section of each panel represents the distribution of Krmit TIS in the each locus. The bottom section shows the growth of an inducible mutant in the presence (black squares) or absence (grey circles) of theophylline.
Two conserved essential genes were selected to validate our essentiality findings in both serotypes; murE, encoding a UDP-N-acetylmuramoylalanyl-D-glutamate-L-lysine ligase involved in peptidoglycan synthesis, and vicR, encoding a two-component system response regulator shown to be essential in various Gram-positive bacteria, including B. subtilis42, E. faecalis43, S. aureus44, S. pneumoniae26 and S. sanguinis15. Both 5448 and NZ131 vicR-inducible strains were viable when theophylline was provided into the growth medium, yet showed significantly compromised growth in the absence of theophylline (Fig. 5BC). Similarly, the murE-inducible mutants were not viable in the absence of theophylline yet grew normally in the presence of the inducer (Fig. 5DE). Together, these data validate that murE and vicR do represent genes essential for in vitro growth of both M1T1 5448 and M49 NZ131 as indicated by our analysis.
Essential genes conserved amongst pathogenic streptococci
In order to identify essential genes conserved across the genus Streptococcus, we compared the conserved 177 genes found "Essential" in the GAS core genome to the results obtained from published essentiality studies performed in two other pathogenic streptococci. Xu et al.15 identified 218 genes essential for S. sanguinis growth in vitro in THY using a gene-by-gene inactivation strategy, while van Opijnen et al.26 used Tn-seq to report 397 genes in S. pneumoniae that were either essential or likely essential for growth under comparable in vitro conditions. Orthologs of the GAS core "Essential" genes were determined within the S. sanguinis and S. pneumoniae genomes using the COG database and then compared to the published essential gene lists. Of the 177 genes identified as essential in GAS, 129 and 148 genes were shared in the essential gene sets of S. sanguinis and S. pneumoniae, respectively (Fig. 6); and a total of 120 genes (Table S7) were found to be "Essential" in all three Streptococcus species. We also identified a total of 8 genes "Essential" for GAS, but unambiguously dispensable for S. sanguinis and S. pneumoniae, including those encoding the heat shock protein gene grpE, the cobalt ABC transporter gene cbiO, group A carbohydrate biosynthesis genes (gac), and 3 genes encoding poorly characterized proteins. Overall, the 120 conserved streptococcal "Essential" genes identified here represent attractive targets for the potential development of antimicrobials against multiple streptococcal human pathogens.
Figure 6. Essential gene set conserved between pathogenic streptococci.
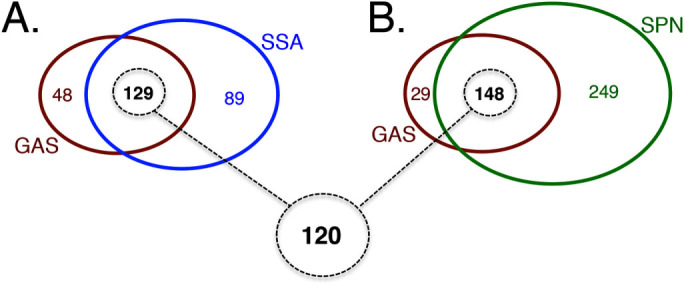
Venn diagram showing the comparison of the 177 essential genes identified in the GAS core genome to found in (A) Streptococcus sanguinis (blue circle) and (B) Streptococcus pneumoniae (green circle). Overlap of essential genes conserved between all three streptococcal pathogens is shown at bottom (dashed circle).
Discussion
GAS remains a considerable disease burden worldwide, resulting in over half a million deaths each year1. Despite considerable efforts, progress in designing new vaccines against this significant pathogen has been slow in comparison to other infectious diseases45. GAS pathogenesis is complex as it involves multiple infection sites and genetically divergent GAS clinical isolates. Systematic genome-wide forward-genetic approaches should provide invaluable resources for a better, large-scale understanding of GAS physiology and virulence29,46,47.
In this work, we modified our recently developed mariner system for whole-genome TraSH screens in GAS28,29 to create Krmit, a transposon system for Tn-seq analyses. This work significantly improves high-resolution whole-genome mutant screens in GAS, as we are now able to track complex mutant pools at nucleotide resolution. As a first application of the new Krmit system, we identified a set of 177 genes essential for growth in rich medium in vitro and conserved in the GAS core genome, as well as identified those essential genes conserved in the genomes of multiple pathogenic streptococci and representing valuable targets for vaccine and drug development.
Krmit and the production of Krmit libraries for Tn-seq in GAS
Tn-seq26 allows for en masse identification of mutants in highly complex pools through the production of specific TIS tags. To allow for both qualitative (TIS tag location) and quantitative (gene fitness index) analyses of the mutant pool composition, a mariner transposon with modified MmeI-containing ITRs is used to produce TIS tags of identical size. However, the ITR modification must have no deleterious effect on the mariner transposition. We were able to show that the ITR modification of Krmit did not significantly impact in vivo mariner delivery in different GAS genetic backgrounds (Fig. S1) compared to the published parental Oskar mariner system.
The M1T1 strain 5448 represents a model for the study of GAS invasive pathogenesis as it produces strong virulence phenotypes in different in vivo and ex vivo infections models. However, genetic manipulation of 5448 has proven fastidious and impacts mariner transposition efficiency28,29. Thus, phenotype screens and AP-PCR analyses were necessary on 120 independent 5448 Krmit libraries in order to select for the most desirable libraries prior to the production and sequencing of Krmit TIS tags. Tn-seq analyses of four of these 5448 Krmit libraries emphasized the heterogeneity in the mariner mutagenesis randomness and the necessity to pool together different libraries to achieve near-saturation mutagenesis in GAS 5448 (Fig. 1). In contrast, Krmit library production in the highly transformable GAS NZ131 was found to be more straightforward, needing analysis of far fewer independent libraries and not necessarily requiring pooling (Fig. 1). Thus, as previously observed28, ease of in vivo Krmit mariner delivery in GAS appears to strongly correlate with the transformation efficiency of the strain and this fact should be kept in mind during future Tn-seq studies using Krmit in GAS.
Tn-seq revealed the presence of the pKRMIT delivery plasmid in the first passage of Krmit libraries (T0) (Table S1). Originally seen as an inconvenient contamination of the mutant libraries that could affect library stability28,29, our Tn-seq data showed that the plasmid presence in T0 leads to significantly increased library TIS in the subsequent passage (T1) combined with plasmid dilution and disappearance. This observation shows that maximum coverage and near saturation mutagenesis is achieved in poorly transformable GAS strains only after two consecutive overnight passages in vitro. Furthermore, these T1 libraries should represent the "input" libraries for comparison to different ex vivo and in vivo GAS growth environments.
Tn-seq also revealed high mutagenesis saturation at T1; with Krmit transposons found ca. every 25 nucleotides in both the GAS 5448 and NZ131 genomes (Fig. 2). To our knowledge, this work provides the most comprehensive mutant libraries yet produced in GAS and represents an invaluable resource for the study of GAS physiology and pathogenesis. As Tn-seq provides the means to track the libraries’ complexity at a nucleotide scale, this is a significant improvement over our recent TraSH screens where, due to the limitations of microarrays, we could only assess ca. 60% of the GAS genome29. Moving forward, Tn-seq’s resolution has the potential to monitor qualitatively (saturation index) and quantitatively (fitness index) libraries’ composition. We are currently using Tn-seq with the 5448 and NZ131 Krmit libraries to investigate GAS physiology and pathogenesis. The level of Krmit saturation achieved in GAS 5448 and NZ131 chromosomes should provide the means to determine the contribution of both protein-coding genes and non-coding regions.
Prediction of essential genes for GAS 5448 and NZ131
As a first application of our new resources, we chose to identify GAS essential genes as these provide valuable targets for the development of novel therapeutic interventions and identify genes to be removed from future Tn-seq analyses of GAS fitness under ex vivo and in vivo growth conditions. Tn-seq data analyses present some challenges for gene essentiality prediction and different methods are emerging for computational analyses. One approach is to determine the frequency of a given mutant in the population (read depth) and average the results to generate a fitness value (fitness index) for each gene23,24,25,26,31,48,49,50,51. Initial attempts revealed that this approach was not suited for essential gene prediction using our datasets (data not shown), likely due to the protocol used to produce mutant libraries. Since transposition events were not synchronized during our in vivo mariner delivery procedure, mutants that appeared early during the procedure had the potential to become overrepresented within the mutant pool, thus affecting the read depth for the corresponding location. Read mapping analyses confirmed this hypothesis showing overrepresented clones (data not shown).
An alternative approach for gene essentiality prediction consists of searching for genes with significant gaps of transposon insertions (coverage) regardless of read depth37,52,53,54,55. Here we used the computational pipeline developed by DeJesus et al.37 to make prediction of essential genes by means of a Bayesian statistical model that scanned the GAS genome to determine the distance between Krmit insertions within the GAS genomes and identify abnormal insertion gaps as a call for gene essentiality (Fig. 2; Tables S3 and S5). We then integrated the analyses obtained from 3 consecutive in vitro passages of the Krmit libraries (T0, T1 and T2) into 4 categories: essential, critical, non-essential and non-conclusive (Tables S4 and S6). Using this approach, we identified a set of 227 and 241 essential genes in GAS 5448 and NZ131, respectively. Perhaps not surprisingly, a large proportion of the genes found essential in both GAS strains corresponded to genes involved in basic cell functions such as DNA replication, RNA transcription, aminoacyl-tRNA synthesis, ribosomal proteins, central carbon metabolism, generation of proton motive force, peptidoglycan biosynthesis, and fatty acid synthesis. The paucity of known virulence factors56 in the list of genes essential for GAS growth in vitro likely reflects the host-specific requirement for these genes in vivo. There was a striking overlap with our data and a "minimal" essential gene set57 developed through the integration of data collected from different Gram-negative and Gram-positive bacterial species using various experimental approaches (mutagenesis, bioinformatics) (data not shown). This provides a strong validation of the integrated Bayesian approach used to define GAS essential genes.
When directly comparing the results obtained in 5448 and NZ131, we found a total of 187 essential genes common to the two diverse GAS strains (Fig. 3). Therefore, our analysis also identified 94 genes essential exclusively in one GAS strain, but not in the other. There are several explanations that could account for these findings.
First, some of these essential genes were identified in strain-specific phages, where they typically encode for phage repressor proteins from the cI/Cro-family, including the 5448 PhiRamid phage58 (M5005_Spy_1464) or the ΦNZ131.1 phage39 (Spy49_0369). Additional phage cI/Cro repressor-encoding genes were also found to be critical (Table S4, S6 and S7). The suggestion would be that loss of the repressor leads to induction of the lytic phage and loss of the mutant from the population.
Second, the conservative nature of our essentiality call could exclude a gene that was deemed non-conclusive in the other strain leading to an underestimation of the shared GAS essential genes. We found that analysis of the data within metabolic pathways using the KEGG database (e.g., central carbon metabolism, peptidoglycan and fatty acid biosynthesis) (Fig. 4) and/or comparison of visualized read alignments for data in both GAS strains (Fig. 1 and 5) helped to reveal the critical or essential roles of non-conclusive genes in vital cell processes.
Third, GAS strain-specific essential genes may also reflect loci that are more related to fitness (e.g., environment stress response) than being truly essential, where the competitive environment of the en masse mutant selection led to an underrepresentation or disappearance of mutants that were less fit in one of the GAS strains. The distinction between "gene essentiality" and "fitness disadvantage" can be quite subtle in Tn-seq26,59,60,61. Recently, Valentino et al.51 found, as they were attempting to experimentally validate genes identified by Tn-seq as essential in S. aureus, that 50% of the conditional mutants tested presented delayed growth. We found a number of genes linked to stress responses that were called essential in one of the strains but not in the other. For example, codY and ccpA were found essential in NZ131, but critical and non-conclusive, respectively, in 5448. These two genes encode transcriptional regulators involved in stress responses to nutrient limitation that are likely to be experienced by the GAS cells in our experimental setting. Previous analyses have shown that both codY62 and ccpA63,64,65 were mutable in various serotypes of GAS. Similarly, we found covS, encoding the histidine-kinase of the CovRS two-component system in GAS, to be essential in NZ131 but not in 5448. Although covS has been mutated by many groups66,67,68,69, Dalton and Scott showed that a covS mutant was sensitive to physicochemical stresses67.
Another explanation for GAS strain-specific essential genes might be that the mechanisms involved in the environmental stress response during in vitro library selection could be different in the two GAS strains.
Conserved essential genes in the "Core" GAS genome and other streptococci
Molecular epidemiology has identified a GAS population genetic structure with 3 discrete subpopulations based on tissue tropism32, including skin specialists (Pattern D), throat specialists (Pattern A-C) and generalists (Pattern E). Importantly, any effective therapeutic should target all 3 distinct subpopulations. By comparing the available genome sequences of 20 different GAS strains reflecting these subpopulations, we were able to identify a set of 1224 genes common to all GAS strains (GAS core genome). Of these, 177 genes were unambiguously identified as essential in our Tn-seq analysis of GAS 5448 and NZ131. Furthermore, 120 of these core GAS essential genes were also found to be essential in S. sanguinis and S. pneumoniae (Fig. 6). These two sets of essential genes represent interesting candidates for the development of drugs against GAS and multiple streptococcal pathogens, respectively. Currently, the most successful antibiotics exploit a small number of targets, i.e. the ribosome, protein synthesis, cell wall synthesis, folic acid metabolism, RNA polymerase, DNA gyrase and DNA topoisomerase70. Our data identifies additional essential genes and diverse pathways that could be the targets for novel antimicrobial approaches (Table S7).
By identifying genes that are non-essential, essential or critical for in vitro GAS fitness, our data also provides invaluable information for the investigation of GAS pathogenesis as it provides a roadmap for GAS genetic manipulation. Whereas genes shown as non-essential in both GAS 5448 and NZ131 are likely to be easy targets for null mutations, essential or critical genes might potentially require more effort. GAS genetic manipulations of these genes that require extensive strain passaging could give rise to compensatory (suppressor) mutations masking the true phenotype of the gene initially targeted. One example from our data is vicR, which has previously been shown to be mutable albeit with considerable effort71, but here is clearly found to be an essential gene both by Tn-seq and through conditional gene expression (Fig. 6 and Table S7).
Concluding remarks
The development of Krmit and its use for Tn-seq in GAS now allows for unparalleled en masse monitoring of mutants to assess gene essentiality and fitness on a genome-wide scale in this important human pathogen. In addition to the invasive M1T1 5448 and the nephritogenic M49 NZ131 strains, we have established complex libraries in many other GAS serotypes as a resource for the GAS pathogenesis community. Using Tn-seq in conjunction with RNA-seq, we can now explore the functional implications of genetic elements (coding and non-coding genes) at an unprecedented level of resolution in GAS disease-relevant environments.
Methods
Bacterial strains and media
Bacterial strains used in this study are shown in Table S8. 544872 is a GAS clinical isolate representative of the globally disseminated invasive serotype M1T1 clone. NZ13173 is a GAS strain isolated from a patient with Acute Post-Streptococcal Glomerulonephritis (APSGN). GAS strains were routinely cultured in Todd-Hewitt medium (Alpha Biosciences) supplemented with 0.2% yeast extract (THY) as described elsewhere74. Escherichia coli strains DH5α75 and C43[DE3]76 were used as hosts for plasmid construction and preparation and were cultured in Luria-Bertani (LB) medium (EMD Chemicals). Antibiotics (Fischer Scientific; Gold Biotechnology) were used at the following concentrations: Ampicillin (Ap) at 100 μg/ml for E. coli, Spectinomycin (Sp) at 100 μg/ml for both E. coli and GAS, and Kanamycin (Km) at 50 μg/ml for E. coli and 300 μg/ml for GAS.
Molecular genetics
Oligonucleotides used in this study were synthesized by Integrated DNA Technologies, Inc. and are listed in Table S9. Plasmids used in this study are shown in Table S8. Plasmids were isolated using the Wizard Plus SV Minipreps kit (Promega) or the QIAGEN Plasmid Purification Midi Kit (QIAGEN). Restriction enzymes, Antarctic Phosphatase and T4 DNA ligase (New England Biolabs) were used according to the manufacturer’s instructions. PCR was performed using either Taq DNA polymerase (New England Biolabs) or High-Fidelity AccuPrime Pfx DNA polymerase (Life Technologies) with 1 μg of DNA template and 10 pmol of the appropriate primers (Table S9). When necessary, PCR products were purified using Wizard SV Gel and PCR Clean-Up System kit (Promega). Transformations were performed with the Gene Pulser Xcell System apparatus (Bio-Rad) as recommended by the manufacturer, using electrocompetent cells of E. coli or GAS prepared as described by Ausubel et al.77 or Le Breton and McIver28, respectively. Genomic DNA (gDNA) from GAS was purified using the MasterPure Complete DNA Purification kit (Epicentre Biotechnologies). Genewiz, Inc performed the Sanger DNA sequencing.
Generation of the Krmit mariner transposon
The pOSKAR mariner delivery system29 was modified for Tn-seq analyses as follows: an MmeI restriction site was introduced into both inverted terminal repeat (ITR) sequences of Oskar by PCR using pOSKAR (Table S8) and the primer oKrmit1 (Table S9) to generate the Krmit transposon (Fig. S1A). The resulting Krmit PCR product was digested with PstI and used to replace Oskar in PstI-digested pOSKAR and generate the mariner delivery plasmid pKRMIT (Fig. S1B, Table S8). Like pOSKAR28,29, pKRMIT is unstable in E. coli and the use of C43[DE3]76 was required for efficient amplification.
Generation of Krmit mutant libraries in GAS
In vivo transposition of Krmit for random mutagenesis in GAS was accomplished as described for pOSKAR28,29 with some modifications. GAS 5448 and NZ131 were transformed with 300 μg and 50 μg pKRMIT, respectively, and allowed to outgrow in THY broth at 30°C for 4 h. Transformants were plated on THY agar containing Km and Sp, and then incubated at 30°C for 48 h. Naturally occurring kanamycin resistance has not been observed in GAS 5448 or NZ131. The presence of intact pKRMIT was tested as previously described28 and proper GAS transformants were stored at −80°C. For Krmit transposition, an individual GAS (pKRMIT) freezer stock was used to inoculate 250 ml THY containing Km for overnight growth at 37°C (T0). The quality of Krmit transposition was tested as previously described28. The complexity or randomness of Krmit mutant libraries was assessed by amplifying the insertion sites of random mutants by AP-PCR28,29, Sanger DNA sequencing, and mapping to their appropriate GAS genome. Percent randomness was determined by defining a ratio of unique insertions (by AP-PCR sequencing) among a tested population.
Tn-seq analyses of GAS following growth in THY
A 10 ml aliquot of Krmit mutant libraries in either GAS 5448 or NZ131 (T0) was grown overnight in 250 ml THY containing Km at 37°C (T1). 10 ml of the resulting T1 culture was inoculated in 250 ml THY containing Km and incubated overnight at 37°C a second (T2) and third (T3) time. At each time point, 10 ml aliquots were collected to harvest cells by centrifugation for subsequent gDNA isolation.
Tn-seq was performed as originally described by van Opijnen et al.31 with some modifications: GAS gDNA was isolated from Krmit mutant pools and 5 µg was subjected to complete digestion with MmeI, treated with Antarctic Phosphatase (NEB), and purified by phenol/chloroform extraction. The MmeI-digested gDNA fragments were then ligated to an MmeI adapter. Eight different MmeI adapters (Adapter-501 to Adapter-508) were generated by annealing oligonucleotide pairs (Table S9) containing Illumina barcode sequences (501 to 508) to allow sample multiplexing during massively parallel sequencing. The ligation mixture was then used as a template for a 20-cycle PCR with the primers oKrmit-Tnseq2 and oAdapterPCR (Table S9), resulting in the production of 176-bp Krmit insertion tags (Fig. S2) that were purified from a 2% agarose gel. Quality and yield of the resulting tags was assessed using both a NanoDrop 8000 spectrophotometer (Thermo Scientific) and a Bioanalyzer (Agilent).
Libraries of Krmit insertion tags were analyzed by massively parallel sequencing (50-nt single end reads) on an Illumina HiSeq 1500 platform in the Institute for Bioscience and Biotechnology Research (IBBR) Sequencing Facility located at the University of Maryland, College Park. The detailed procedures for analysis of the Tn-seq data are presented in Text S1. Succinctly, the quality of read datasets (Sanger FastQ format) was determined using FastQC78, data filtered and trimmed using Biopieces (biopieces.org) to select for reads containing the Tn-seq barcodes and Krmit ITR end. Reads were then de-multiplexed and count tables generated using SamTools79 and HTseq80. Reads were mapped to the GAS MGAS5005 (for 5448) or NZ131 genome using Bowtie81 and data visualized using the Integrative Genomics Viewer (IGV) browser (broadinstitute.org/igv/home)35,36. Gene essentiality was determined using a Bayesian statistical model based on the Metropolis-Hastings algorithm using the Python script (saclab.tamu.edu/essentiality) developed by DeJesus et al.37.
Determination of the GAS "core" genome
Twenty GAS whole genome sequences publicly available at the time of this study (i.e. MGAS5005, NZ131, SF370, MGAS8232, MGAS315, MGAS10394, MGAS6180, MGAS9429, MGAS10270, MGAS2096, MGAS10750, MGAS15252, MGAS1882, Alab49, A20, M1-476, HSC5, HKU, Manfredo and SSI-1; Table S10) were subjected to whole genome multiple sequence alignment using the Mugsy software82 with default parameters. Clusters of syntenic orthologs were generated using the Mugsy-Annotator software83 based on the annotated genes from each genome and using synteny information derived from the Mugsy alignment. Core clusters were then selected by requiring that each cluster contain at least one gene from each of the 20 genomes analyzed.
Validation of gene essentiality by riboswitch-inducible expression
Mutants in two selected genes identified as essential in the Tn-seq screen (murE, vicR) were constructed in order to have their mRNA translation under control of a synthetic theophylline-dependent riboswitch developed by Topp et al.84 as follows. First, a new mutagenesis system pSin/pHlp was created for stable plasmid integration into the GAS chromosome comprised of: (i) a pSinS suicide plasmid unable to independently propagate in GAS derived by PCR from the pCRS plasmid backbone29 using the primers RepAminus1 and RepAminus2 to delete a portion of the repAts gene, (ii) a pHlpK helper plasmid possessing a thermosensitive pWV01 replicon and constructed by PCR from the pCRK backbone29 using the primers KmR1NotI and OTS2 to delete pCRK’s multiple cloning site and its ColE1 origin of replication. The mutagenic plasmid was obtained by cloning a DNA fragment into the pSinS vector produced by SOE-PCR to fuse the Psag promoter along with the synthetic riboswitch E contained on pEU7742-E to the 5’-end (ca. 600 bp) of the target gene (Fig. 5A, Tables S8 and S9). The construct was then introduced into pHlpK-containing GAS cells and transformants selected in the presence of Km and Sp at permissive temperature (30°C) to allow replication of the two plasmids. Chromosomal integration of the mutagenic plasmid was achieved by culturing the clone at non-permissive temperature (37°C) in the presence of 2 mM theophylline. A merodiploid GAS mutant was produced in which the wild type gene had its expression controlled by the theophylline-inducible riboswitch, while the wild-type promoter of the targeted gene controlled the expression of a truncated allele (Fig. 5A). The growth of the integrative mutant in THY in the presence or absence of theophylline was monitored by measuring the culture turbidity using a Klett-Summerson colorimeter equipped with an A filter (Klett Units) or an automated BMG FLUOstar Omega microplate spectrophotometer (OD600 nm).
Accession number for public deposition of Tn-seq data
Illumnia sequencing reads from the Tn-seq analyses were deposited in the NCBI Sequence Read Archive (SRA) under the accession number (PRJNA280537).
Author Contributions
Y.L.B. and K.S.M. conceived and designed the research plan, supervised the project, analyzed the data and interpreted the results. Y.L.B. performed most of the experiments with technical contributions from K.M.V. (pKRMIT design), E.I. (construction of Krmit libraries, pSin/pHlp system and inducible mutants) and P.C. (AP-PCRs for NZ131 Krmit mutants). A.T.B. and N.M.E. performed the bioinformatics analyses. H.T. determined the GAS core genome. M.E.S. provided intellectual input and guidance. Y.L.B., A.T.B., H.T., N.M.E. and K.S.M. wrote the manuscript. All authors read and approved the final manuscript.
Additional information
Supplementary information accompanies this paper at http://www.nature.com/scientificreports
How to cite this article: Le Breton, Y. et al. Essential Genes in the Core Genome of the Human Pathogen Streptococcus pyogenes. Sci. Rep. 5, 9838; DOI:10.1038/srep09838 (2015).
Supplementary Material
Supplementary Information
Acknowledgments
We thank Ganesh (Surya) Sundar and Luis Vega for critical comments of this manuscript. Helpful suggestions provided by Andrew Camilli, Tim van Opijnen and Gary Port are greatly appreciated. The Tn-seq originates from funding by a 2014–2015 UMB/UMCP Seed Grant (Y.L.B., K.S.M., and M.E.S.). This work was directly supported by grants from the NIH National Institute of Allergy and Infectious Diseases (K.S.M., AI047928; N.M.E.S., AI094773) and in part by a NIH F31 predoctoral fellowship (K.M.V., AI100576).
References
- Carapetis J. R., Steer A. C., Mulholland E. K. & Weber M. The global burden of group A streptococcal diseases. Lancet Infect. Dis. 5, 685–694, doi:10.1016/S1473-3099(05)70267-X (2005). [DOI] [PubMed] [Google Scholar]
- Cunningham M. W. Pathogenesis of group A streptococcal infections and their sequelae. Adv. Exp. Med. Biol. 609, 29–42, doi:10.1007/978-0-387-73960-1_3 (2008). [DOI] [PubMed] [Google Scholar]
- Olsen R. J. & Musser J. M. Molecular pathogenesis of necrotizing fasciitis. Ann. Rev. Pathol. 5, 1–31 (2010). [DOI] [PubMed] [Google Scholar]
- Steer A. C., Lamagni T., Curtis N. & Carapetis J. R. Invasive group a streptococcal disease: epidemiology, pathogenesis and management. Drugs 72, 1213–1227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tart A. H., Walker M. J. & Musser J. M. New understanding of the group A Streptococcus pathogenesis cycle. Trends Microbiol. 15, 318–325, 10.1016/j.tim.2007.05.001 (2007). [DOI] [PubMed] [Google Scholar]
- Cole J. N., Barnett T. C., Nizet V. & Walker M. J. Molecular insight into invasive group A streptococcal disease. Nat. Rev. Microbiol. 9, 724–736, 10.1038/nrmicro2648 (2011). [DOI] [PubMed] [Google Scholar]
- Kaplan E. L. & Johnson D. R. Unexplained reduced microbiological efficacy of intramuscular benzathine penicillin G and of oral penicillin V in eradication of group a streptococci from children with acute pharyngitis. Pediatrics 108, 1180–1186, 10.1542/peds.108.5.1180 (2001). [DOI] [PubMed] [Google Scholar]
- Pichichero M. E. Group A beta-hemolytic streptococcal infections. Pediatr. Rev. 19, 291–302 (1998). [DOI] [PubMed] [Google Scholar]
- Pichichero M. E. & Casey J. R. Systematic review of factors contributing to penicillin treatment failure in Streptococcus pyogenes pharyngitis. Otolaryngol. Head Neck Surg. 137, 851–857, 10.1016/j.otohns.2007.07.033 (2007). [DOI] [PubMed] [Google Scholar]
- Osterman A. L. & Gerdes S. Y. Comparative approach to analysis of gene essentiality. Methods Mol. Biol. 416, 459–466, doi:10.1007/978-1-59745-321-9_31 (2008). [DOI] [PubMed] [Google Scholar]
- Shaw K. J. Overview of Whole-Genome Essentiality Analysis. Microbial Gene Essentiality: Protocols and Bioinformatics. Osterman, A.L.; Gerdes, S.Y. (eds.), 3–8, doi:10.1007/978-1-59745-321-9_1 (2008). [DOI] [PubMed] [Google Scholar]
- Mobegi F. M. et al. From microbial gene essentiality to novel antimicrobial drug targets. BMC Genomics 15, 958, 10.1186/1471-2164-15-958. (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T. et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biology 2 10.1038/msb4100050 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K. et al. Essential Bacillus subtilis genes. Proc. Natl. Acad. Sci. U.S.A. 100, 4678–4683, 10.1073/pnas.0730515100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P. et al. Genome-wide essential gene identification in Streptococcus sanguinis. Sci. Rep. 1, 125, 10.1038/srep00125 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsyth R. A. et al. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43, 1387–1400, doi:10.1046/j.1365-2958.2002.02832.x (2002). [DOI] [PubMed] [Google Scholar]
- Akerley B. J. et al. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl. Acad. Sci. U.S A. 95, 8927–8932 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson N. & Mekalanos J. J. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 18, 740–745, 10.1038/77305 (2000). [DOI] [PubMed] [Google Scholar]
- Hutchison C. A. et al. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286, 2165–2169, 10.1126/science.286.5447.2165 (1999). [DOI] [PubMed] [Google Scholar]
- Glass J. I. et al. Essential genes of a minimal bacterium. Proc. Natl. Acad. Sci. U.S.A. 103, 425–430, 10.1073/pnas.0510013103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholle M. D. & Gerdes S. Y. Whole-genome detection of conditionally essential and dispensable genes in Escherichia coli via genetic footprinting. Methods Mol. Biol. 416, 83–102, doi:10.1007/978-1-59745-321-9_6. (2008). [DOI] [PubMed] [Google Scholar]
- Sassetti C. M., Boyd D. H. & Rubin E. J. Comprehensive identification of conditionally essential genes in mycobacteria. Proc. Natl. Acad. Sci. U.S.A. 98, 12712–12717, 10.1073/pnas.231275498 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawronski J. D., Wong S. M., Giannoukos G., Ward D. V. & Akerley B. J. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc. Natl. Acad. Sci. U.S.A. 106, 16422–16427, 10.1073/pnas.0906627106 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman A. L. et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe 6, 279–289, 10.1016/j.chom.2009.08.003 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langridge G. C. et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 19, 2308–2316, 10.1101/gr.097097.109 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T., Bodi K. L. & Camilli A. Tn-seq: high-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat. Meth. 6, 767–772, 10.1038/nmeth.1377 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijlsma J. J. et al. Development of genomic array footprinting for identification of conditionally essential genes in Streptococcus pneumoniae. Appl. Environ. Microbiol. 73, 1514–1524, 10.1128/AEM.01900-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton Y. & McIver K. S. Genetic Manipulation of Streptococcus pyogenes (The Group A Streptococcus, GAS). Curr. Protoc. Microbiol. 30, 9D.3.1–9D.3.29, 10.1002/9780471729259.mc09d03s30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Breton Y. et al. Genome-wide identification of genes required for fitness of group a streptococcus in human blood. Infect. Immun. 81, 862–875, 10.1128/IAI.00837-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T. & Camilli A. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat. Rev. Microbiol. 11, 435–442, 10.1038/nrmicro3033 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T., Lazinski D. W. & Camilli A. Genome-Wide Fitness and Genetic Interactions Determined by Tn-seq, a High-Throughput Massively Parallel Sequencing Method for Microorganisms. Curr. Protoc. Mol. Biol. 106, 7.16.11–17.16.24, 10.1002/0471142727.mb0716s106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessen D. E. Population biology of the human restricted pathogen, Streptococcus pyogenes. Infect. Genet. Evol. 9, 581–593, 10.1016/j.meegid.2009.03.002 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumby P. et al. Evolutionary origin and emergence of a highly successful clone of serotype M1 group A streptococcus involved multiple horizontal gene transfer events. J. Infect. Dis. 192, 771–782, 10.1086/432514 (2005). [DOI] [PubMed] [Google Scholar]
- Maamary P. G. et al. Tracing the evolutionary history of the pandemic group A streptococcal M1T1 clone. FASEB J. 26, 4675–4684, 10.1096/fj.12-212142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26, 10.1038/nbt.1754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J. T. & Mesirov J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192, 10.1093/bib/bbs017 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus M. A. et al. Bayesian analysis of gene essentiality based on sequencing of transposon insertion libraries. Bioinform. 29, 695–703, 10.1093/bioinformatics/btt043 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B. et al. The essential genome of a bacterium. Mol. Syst. Biol. 7 10.1038/msb.2011.58 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShan W. M. et al. Genome sequence of a nephritogenic and highly transformable M49 strain of Streptococcus pyogenes. J. Bacteriol. 190, 7773–7785, 10.1128/JB.00672-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H., Riley D., Cattuto C. & Medini D. Comparative genomics: the bacterial pan-genome. Curr. Opin. Microbiol. 11, 472–477 (2008). [DOI] [PubMed] [Google Scholar]
- Bugrysheva J. V., Froehlich B. J., Freiberg J. A. & Scott J. R. The histone-like protein Hlp is essential for growth of Streptococcus pyogenes: comparison of genetic approaches to study essential genes. Appl. Environ. Microbiol. 77, 4422–4428, 10.1128/AEM.00554-11 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrac S., Bisicchia P., Devine K. M. & Msadek T. A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. Mol. Microbiol. 70, 1307–1322, doi:10.1111/j.1365-2958.2008.06483.x (2008). [DOI] [PubMed] [Google Scholar]
- Le Breton Y. et al. Molecular characterization of Enterococcus faecalis two-component signal transduction pathways related to environmental stresses. Environ. Microbiol. 5, 329–337, doi:10.1046/j.1462-2920.2003.00405.x (2003). [DOI] [PubMed] [Google Scholar]
- Dubrac S., Boneca I. G., Poupel O. & Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J. Bacteriol. 189, 8257–8269, 10.1128/JB.00645-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steer A. C., Dale J. B. & Carapetis J. R. Progress toward a global group a streptococcal vaccine. Ped. Infect. Dis. J. 32, 180–182 (2013). [DOI] [PubMed] [Google Scholar]
- Kizy A. E. & Neely M. N. First Streptococcus pyogenes signature-tagged mutagenesis screen identifies novel virulence determinants. Infect. Immun. 77, 1854–1865, 10.1128/IAI.01306-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson-Smith M. et al. A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J. Infect. Dis. 210, 1325–1338, 10.1093/infdis/jiu260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A. et al. A comprehensive transposon mutant library of Francisella novicida, a bioweapon surrogate. Proc. Natl. Acad. Sci. U.S.A. 104, 1009–1014, 10.1097/INF.0b013e318281da11 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moule M. G. et al. Genome-wide saturation mutagenesis of Burkholderia pseudomallei K96243 predicts essential genes and novel targets for antimicrobial development. mBio 5, e00926–00913, 10.1128/mBio.00926-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmele C. W. et al. Transcriptional landscape and essential genes of Neisseria gonorrhoeae. Nuc. Acids Res. 42, 10579–10595, 10.1093/nar/gku762 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentino M. D. et al. Genes contributing to Staphylococcus aureus fitness in abscess- and infection-related ecologies. mBio 5, e01729–01714, 10.1128/mBio.01729-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao M. C. et al. High-resolution definition of the Vibrio cholerae essential gene set with hidden Markov model-based analyses of transposon-insertion sequencing data. Nuc. Acids Res. 41, 9033–9048, 10.1093/nar/gkt654 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus M. A. & Ioerger T. R. A Hidden Markov Model for identifying essential and growth-defect regions in bacterial genomes from transposon insertion sequencing data. BMC Bioinform. 14 10.1186/1471-2105-1114-1303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin J. E. et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Path. 7, e1002251, 10.1371/journal.ppat.1002251 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane G. et al. A postgenomic method for predicting essential genes at subsaturation levels of mutagenesis: application to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S A. 100, 7213–7218, 10.1073/pnas.1231432100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker M. J. et al. Disease manifestations and pathogenic mechanisms of group a Streptococcus. Clin. Microbiol. Rev. 27, 264–301, 10.1128/CMR.00101-13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil R., Silva F. J., Peretó J. & Moya A. Determination of the core of a minimal bacterial gene set. Microbiol. Mol. Biol. Rev. 68, 518–537, 10.1128/MMBR.68.3.518-537.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K. et al. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J. Bacteriol. 187, 3311–3318, 10.1128/JB.187.10.3311-3318.2005 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Elia M. A., Pereira M. P. & Brown E. D. Are essential genes really essential? Trends Microbiol. 17, 433–438, 10.1016/j.tim.2009.08.005 (2009). [DOI] [PubMed] [Google Scholar]
- Gallagher L. A., Shendure J. & Manoil C. Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. mBio 2, e00315–00310, 10.1128/mBio.00315-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatiwara A. et al. Genome scanning for conditionally essential genes in Salmonella enterica Serotype Typhimurium. Appl. Environ. Microbiol. 78, 3098–3107, 10.1128/AEM.06865-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malke H., Steiner K., McShan W. M. & Ferretti J. J. Linking the nutritional status of Streptococcus pyogenes to alteration of transcriptional gene expression: the action of CodY and RelA. Int. J. Med. Microbiol. 296, 259–275, 10.1016/j.ijmm.2005.11.008 (2006). [DOI] [PubMed] [Google Scholar]
- Almengor A. C., Kinkel T. L., Day S. J. & McIver K. S. The catabolite control protein CcpA binds to Pmga and influences expression of the virulence regulator Mga in the group A streptococcus. J. Bacteriol. 189, 8405–8416, 10.1128/JB.01038-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kietzman C. C. & Caparon M. G. CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect. Immun. 78, 241–252, 10.1128/IAI.00746-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne S. A., Davenport M. T., Keith D. B. & Musser J. M. The role of complex carbohydrate catabolism in the pathogenesis of invasive streptococci. Trends Microbiol. 16, 318–325, 10.1016/j.tim.2008.04.002 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton T. L., Hobb R. I. & Scott J. R. Analysis of the role of CovR and CovS in the dissemination of Streptococcus pyogenes in invasive skin disease. Microb. Pathog. 40, 221–227, 10.1016/j.micpath.2006.01.005 (2006). [DOI] [PubMed] [Google Scholar]
- Dalton T. L. & Scott J. R. CovS inactivates CovR and is required for growth under conditions of general stress in Streptococcus pyogenes. J. Bacteriol. 186, 3928–3937, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollands A. et al. Genetic switch to hypervirulence reduces colonization phenotypes of the globally disseminated group A Streptococcus M1T1 clone. J. Infect. Dis. 202, 11–19, 10.1086/653124 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevino J. et al. CovS simultaneously activates and inhibits the CovR-mediated repression of distinct subsets of group A Streptococcus virulence factor-encoding genes. Infect. Immun. 77, 3141–3149, 10.1128/IAI.01560-08 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K. Platforms for antibiotic discovery. Nature Reviews in Drug Discovery 12, 371–387, 10.1038/nrd3975 (2013). [DOI] [PubMed] [Google Scholar]
- Liu M. et al. Defects in ex vivo and in vivo growth and sensitivity to osmotic stress of group A Streptococcus caused by interruption of response regulator gene vicR. Microbiol. 152, 967–978, 10.1099/mic.0.28706-0 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatellier S. et al. Genetic relatedness and superantigen expression in group A streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect. Immun. 68, 3523–3534, 10.1128/IAI.68.6.3523-3534.2000 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon D. & Ferretti J. J. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol. Lett. 66, 219–224 (1991). [DOI] [PubMed] [Google Scholar]
- Gera K. & McIver K. S. Laboratory growth and maintenance of Streptococcus pyogenes (the Group A Streptococcus, GAS). Curr. Protoc. Microbiol. 30, Unit 9D.2 10.1002/9780471729259.mc09d02s30 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. & Meselson M. Plasmid screening at high colony density. Meth. Enzymol. 100, 333–342 (1983). [DOI] [PubMed] [Google Scholar]
- Miroux B. & Walker J. E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298, 10.1006/jmbi.1996.0399 (1996). [DOI] [PubMed] [Google Scholar]
- Ausubel F. et al. Short Protocols in Molecular Biology, 5th edition (John Wiley & Sons, Inc., New York, 2002), ISBN 0-471-25092-9. [Google Scholar]
- Patel R. K. & Jain M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. PloS One 7, e30619, 10.1371/journal.pone.0030619 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinform. 25, 2078–2079, 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S. & Huber W. Differential expression analysis for sequence count data. Genome Biol. 11, R106, 10.1186/gb-2010-11-10-r106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M. & Salzberg S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiuoli S. V. & Salzberg S. L. Mugsy: fast multiple alignment of closely related whole genomes. Bioinform. 27, 334–342, 10.1093/bioinformatics/btq665 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiuoli S. V., Dunning Hotopp J. C., Salzberg S. L. & Tettelin H. Improving pan-genome annotation using whole genome multiple alignment. BMC Bioinform. 12 10.1186/1471-2105-12-272 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp S. et al. Synthetic riboswitches that induce gene expression in diverse bacterial species. Appl. Environ. Microbiol. 76, 7881–7884, 10.1128/AEM.01537-10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information