Abstract
Microbiota assembly is perturbed in children with undernutrition, resulting in persistent microbiota immaturity that is not rescued by current nutritional interventions. Evidence is accumulating that this immaturity is causally related to the pathogenesis of undernutrition and its lingering sequelae. Preclinical models in which human gut communities are replicated in gnotobiotic mice have provided an opportunity to identify and predict the effects of different dietary ingredients on microbiota structure, expressed functions, and host biology. This capacity sets the stage for proof-of-concept tests designed to deliberately shape the developmental trajectory and configurations of microbiota in children representing different geographies, cultural traditions, and states of health. Developing these capabilities for microbial stewardship is timely given the global health burden of childhood undernutrition, the effects of changing eating practices brought about by globalization, and the realization that affordable nutritious foods need to be developed to enhance our capacity to cultivate healthier microbiota in populations at risk for poor nutrition.
Introduction
Understanding the determinants of the nutritional value of different foods has never been more important, with population stabilization being unlikely this century (Gerland et al., 2014) and growing challenges related to sustainable agriculture. An integral part of understanding how best to deliver nutritious food to a burgeoning population is understanding how the microbial denizens in our gut, the gut microbiota, are shaped by what we eat and how that community in turn shapes our development and health. Nowhere will this kind of insight be more crucial than in raising the world’s children.
Current obstacles to achieving healthy and productive lives and societies are reflected in the United Nations’ millennium development goals that include reductions in child mortality and hunger and improvements in maternal health (http://www.un.org/millenniumgoals/). The scope of the problem of childhood undernutrition is described by parameters such as the International Food Policy Research Institute’s Global Hunger Index (http://www.ifpri.org/publication/2014-global-hunger-index), which is an aggregate measure of calorie intake plus the rates of children being underweight and childhood mortality within a given region and/or country.
Much has been said about how changing patterns of food preferences brought about by economic development, globalization, and changes in food technology and food distribution systems are producing dramatic changes in how, what, and when we eat. These changes, combined with rapid population expansion and issues related to sustainable agriculture, create the need and the opportunity to drive innovation in the area of identifying new, affordable, and nutritious foods. Here, we focus on the importance of understanding the postnatal developmental biology of our gut microbial community—a highly adaptable microbial “organ” that is critically involved in the biotransformation of foods to products that can shape many aspects of human biology. In our view, studies of human gut microbial communities will markedly revise current thinking about many aspects of human nutrition. The knowledge gained could and should catalyze efforts to integrate agricultural policies, food production, and nutritional recommendations for consumers representing different ages, cultural traditions, and geographies. Preclinical research platforms are now available to evaluate the effects of foods that we currently consume and those that we envision creating in the future on the gut microbial community and host biology in ways that can inform clinical studies. Furthermore, studies of children with undernutrition are highlighting the importance of postnatal development of the gut microbiota for achieving healthy growth and providing us with a new set of metrics to define the efficacy of nutritional recommendations and interventions directed at infants, the maternal-infant dyad, and children. Finally, we emphasize the importance of addressing ethical, social, and regulatory issues related to research in this area now rather than later.
Defining Human Postnatal Development from a Microbial Perspective
The human gut microbial community, microbiota, is composed of all three domains of life; Bacteria, which predominate, Archaea, and Eukarya, plus viruses. The gut microbiota is composed of relatively few bacterial phyla compared to other body habitats and is notable for its strain-level diversity. Application of low-error sequencing methods to PCR amplicons generated from the bacterial phylogenetic marker gene encoding the principal RNA in the small subunit of ribosomes (16S rRNA) has indicated that, once acquired, the majority of bacterial strains in a healthy adult are retained for long periods of time (Faith et al., 2013). Thus, early colonizers, once established in the gut ecosystem, have the potential to exert their effects on our biological features and health status for most and perhaps all of our adult lives. This latter finding emphasizes the importance of understanding whether there is a definable program of community assembly in healthy infants/children and whether such a program is shared or varies considerably across populations with distinct dietary habits and traditions residing in different geographic locations. If such a developmental program were definable and a significant contributor to healthy growth, fostering its proper and full execution could represent the basis of an arm of preventive medicine designed to ensure long-term health through informed microbial stewardship.
Food is a major factor that shapes the proportional representation of microorganisms present in the gut microbiota and the relative abundance of its genes (microbiome). Reciprocally, the configuration of the microbiota/microbiome influences the nutritional value of food. One illustration of this interrelationship comes from a culture-independent metagenomic analysis of the gut microbiomes of infants, children, and adults belonging to 150 families living in three countries located on three different continents (metropolitan areas of the USA plus rural villages in southern Malawi and the Amazonas state of Venezuela). The results revealed that the relative abundances of genes in the microbiome that are related to vitamin biosynthesis (e.g., folate, cobalamin, thiamine, and biotin), amino acid metabolism, and processing of complex polysaccharides change in an identifiable sequence during the postnatal period (Yatsunenko et al., 2012). In addition, differences between Westernized (USA) and non-Westernized populations were evident, with breastfed Malawian and Amerindian babies having higher relative abundances of microbial genes encoding enzymes involved in carbohydrate metabolism, vitamin biosynthesis (e.g., components of the biosynthetic pathway for riboflavin, a component of breast milk, dairy products, and meat), and urease (Yatsunenko et al., 2012). Urea represents up to 15% of breast milk nitrogen; its degradation to ammonia can be used for microbial biosynthesis of essential amino acids, potentially benefiting both the microbiota and host when diets are deficient in protein. Significant differences in microbiome configuration were also observed between breast-fed and formula-fed infants, with the latter showing increased representation of genes involved in various aspects of carbohydrate and amino acid metabolism and cobalamin (vitamin B12) biosynthesis (Yatsunenko et al., 2012). Cobalamin is not only important for the host—the ability to transport cobalamin and other substituted corrins is an important determinant of survival for members of the microbiota (Degnan et al., 2014).
Together, these findings suggested that the gut community should be considered when assessing the nutritional requirements at different stages of the human life cycle and in different geographic/cultural settings. They also raised the question of whether perturbations in the functional development of the microbiota/microbiome were related to childhood undernutrition, the major cause of childhood deaths worldwide and a manifestation of a complex set of still poorly understood intra- and intergenerational factors, rather than food insecurity alone (Lazzerini et al., 2013; Caulfield et al., 2014; Richard et al., 2014).
Undernutrition and Gut Microbiota Immaturity
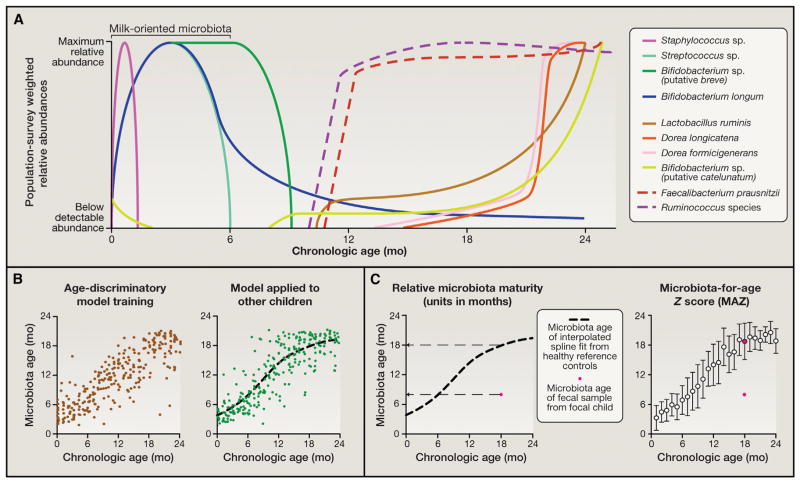
The World Health Organization’s (WHO) Multi-Center Growth Reference Study (http://www.who.int/childgrowth/mgrs/en/) defined three anthropometric (physical) parameters (weight-for-age, height-for-age, and weight-for-height Z scores) to describe normal early childhood growth and nutritional status by evaluating 8,440 infants and children living in six distinct sites around the world (USA, Oman, Norway, Brazil, Ghana, and India). A recent study provided another definition of healthy growth but from a microbial perspective. It did so by examining gut microbiota assembly in 50 children residing in Dhaka, Bangladesh whose anthropometry during their first 2 years of life indicated healthy growth. Fecal samples were collected monthly from birth through the end of the second postnatal year, and the relative abundances of bacterial strains were analyzed by 16S rRNA amplicon sequencing. The results revealed that interpersonal variation in the bacterial component of their gut communities was significantly smaller than the variation associated with age. Applying Random Forests, a machine-learning method, to regress relative abundances of bacterial taxa across these children revealed age-discriminatory bacterial strains. Separating these 50 children into training and validation cohorts, the regression approach was subsequently pruned to include the most informative taxa for accurate prediction of microbiota “age.” The results were formally validated to prevent over-fitting and over-estimation of generalizability and produced a sparse model composed of 24 strains that could be used in aggregate as a microbial signature for describing a shared program of microbiota development in healthy individuals and two derived metrics for defining deviations from that normal program: “relative microbiota maturity” and “microbiota-for-age”’ Z (MAZ) score (Figure 1).
Figure 1. Developing Metrics for Describing Gut Microbial Community Development.
(A) Bacterial taxa that discriminate different stages of development were identified by a machine learning-based (Random Forests) regression of 16S rRNA data sets produced from monthly fecal samples collected from anthropometrically healthy infants and children living in an urban slum in Dhaka, Bangladesh during their first 2 years of postnatal life to their respective chronologic ages at the time of sample collection (Subramanian et. al, 2014). Shown are depictions of the typical distributions of these age-discriminatory taxa across the population. Taxa were selected based on their relative importance to the accuracy of the Random Forests model using a permutation-based “feature importance.”
(B) The most discriminatory taxa, as defined by their feature importance, were used as inputs into a sparse 24 taxon model whose output (“microbiota age”) is a microbiota-based prediction of the chronologic age of a healthy child. The plot on the left of the panel shows microbiota age against chronologic age of healthy children used as a training set to fit the regression (each dot is a fecal sample from an individual child). The plot on the right of the panel shows application of the sparse model to a validation set composed of a different group of children living in the same location that were not used to train the model. Applying the model to a separate validation set controls for over-fitting of the model to the training set and ensures its wider usability.
(C) Two metrics of microbiota maturation based on application of the model to two separate validation sets of singletons and a separate study of Bangladeshi twins/triplets. “Relative microbiota maturity” is the deviation, in months, from a smooth-spline fit of microbiota age values with respect to chronologic age, fitted using the validation data sets (see black dashed curve). The red dot represents a fecal sample collected from a focus child that is 11 months below the spline fit, indicating negative relative microbiota maturity (i.e., an immature microbiota). A MAZ is computed by dividing the difference between the focal child’s microbiota age and the median microbiota age of healthy controls in the same monthly chronologic age bin over the SD within the same age bin. The median and SD of each bin are computed using the validation data sets. The distribution of microbiota maturity and MAZ scores in birth-cohort studies have been studied using linear mixed models that take into account random variation specific to each serially sampled child and family while estimating the fixed variation attributable to a factor observed across different children (e.g., diarrheal episodes) (Subramanian et al., 2014).
Note that using Random Forests to study microbiota maturation is advantageous because of its non-parametric assumptions and utility in the context of high dimensional data sets (large numbers of predictors). Nonetheless, it is one of several methods that can be useful. For example, the rank-order Spearman correlation metric has been applied to infant microbiome data sets to detect monotonic relationships between microbiome-encoded functions/bacterial taxa and postnatal age (Yatsunenko et. al, 2012).
Severe acute malnutrition (SAM) is defined by weight-for-height Z (WHZ) scores more than 3 SDs below those of children with healthy growth phenotypes in the WHO reference cohort. Application of this sparse model to 64 Bangladeshi children with SAM (WHZ −4.2 ± 0.72 [SD]) revealed they had gut microbiota that appeared significantly “younger” than their chronological age (relative microbiota maturity of −6 ± 0.7 months and MAZ scores of −1.7 ± 0.2). Moreover, this immaturity was incompletely and only transiently rescued following a customary period of administration of either one of two types of ready-to-use therapeutic foods (RUTFs; typically given for 2 weeks until a 15% increase in weight gain is achieved; http://www.ClinicalTrials.gov, number NCT01331044). Bangladeshi children with moderate acute malnutrition (WHZ between −3 and −2) also exhibited significant microbiota immaturity, although less severe than children with SAM (Subramanian et al., 2014). These results indicate that children with SAM have a persistent developmental abnormality affecting their gut microbial “organ” that is not durably repaired with existing therapy.
These observations raise a critical question: is microbiota immaturity a cause or an effect of childhood undernutrition? Many studies have shown that, although current protocols for treating children with (acute) undernutrition reduce mortality, they do not rescue its long-term morbidities, including stunting, immune dysfunction, and neurodevelopmental abnormalities (Victora et al., 2008, Gaayeb et al., 2014, Kosek et al., 2013, Galler et al., 2012). For example, given the remarkable metabolic requirements of the neonatal brain, alterations in the normal postnatal development of the gut microbiota may trigger marked impairments in brain development and lead to persistent disorders of cognition.
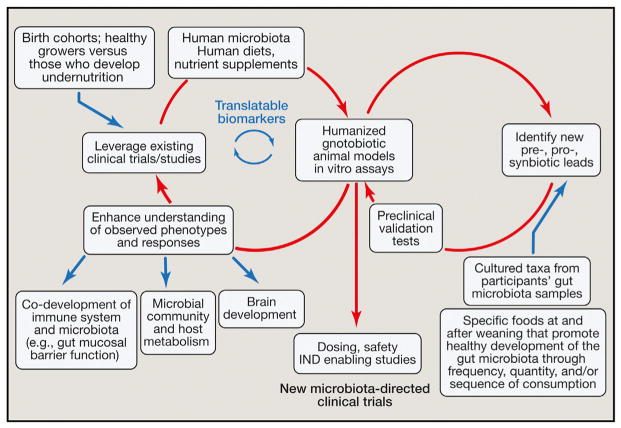
Support for a causal role for the gut microbiota in SAM comes from studies of gnotobiotic mice. In recent years, methods have been developed for transplanting previously frozen fecal samples from human donors into groups of germ-free mice at a selected stage of their lives (e.g., young, rapidly growing animals that have been recently weaned or older animals) and with a designated genetic background. If the human microbiota sample is frozen shortly after it is produced and maintained at −80°C, the bacterial strains represented in the donor’s community can be transmitted efficiently and reproducibly to recipient mice (e.g., Turnbaugh et al., 2009a; Smith et al., 2013; Ridaura et al., 2013; Palm et al., 2014; Kau et al., 2015). The recipient mice can then be fed diets that contain ingredients used in foods consumed by the microbiota donor. Moreover, the ingredients and methods for preparing (cooking) such diets can be varied systematically. This approach allows myriad types of models to be constructed for studying the interaction of foods and the human gut microbiota in vivo. For example, diets can be given that are representative of those consumed by populations other than those of the donor to anticipate the effects of changes in food consumption patterns associated with Westernization or composed of ingredients that represent new potential sources of affordable, nutritious foods such as landraces and waste streams from current food manufacturing processes. Critically, these preclinical gnotobiotic animal models allow proof-of-concept tests of whether a donor phenotype is transmissible via his/her gut microbiota, the extent to which phenotypic transmission generalizes across different donor microbiota, and the sensitivity or robustness of phenotypic transmission to diet type. These preclinical models also permit simulations of existing or anticipated therapeutic interventions, including the opportunity to “randomize” a given individual to not just one but multiple treatment arms in order to directly compare the effect (and effect size) of the treatments on both the microbiota and host, to characterize underlying mechanisms, and to identify surrogate- or mechanism-based biomarkers that could be translatable to the microbiota donor or donor population (Figure 2).
Figure 2. Integration of Existing Clinical Observational and Interventional Studies into Gnotobiotic Mouse Models to Identify Interactions between the Gut Microbiota, Food, and Host Biology.
The discovery process depicted by the left circle illustrates how gnotobiotic animal models colonized with human donor microbiota and fed human diets can lead to a greater understanding of how diet-by-microbiota interactions are causally related to healthy growth and to phenotypes associated with undernutrition: e.g., immune system development, brain development, and host and microbial community metabolism. New surrogate- or mechanism-based biomarkers of nutritional state emanating from these gnotobiotic models can be validated using biospecimens collected from the donors used to construct these gnotobiotic models, as well as from other members of the study population. The discovery/development process depicted on the right illustrates how dietary and microbial “leads” can be tested in the context of humanized gnotobiotic animals to assess how they modulate biological processes already known, discovered, or postulated to be involved in healthy growth and/or the pathogenesis of undernutrition. The downward-pointing arrow in the middle of the figure points to next steps in clinical translation. See the main text for a discussion of the regulatory, ethical, societal, and commercial implications of these efforts. Abbreviation: IND, investigational new drug.
Transplanting fecal microbiota from same-gender Malawian twins discordant for kwashiorkor, a form of SAM, into separate groups of adult germ-free mice and feeding the recipient animals a representative micro- and macronutrient-deficient Malawian diet disclosed that the healthy and kwashiorkor co-twins’ microbiota transmitted discordant weight loss and metabolic phenotypes (as well as an enteropathy characterized by disruption of the small intestinal and colonic epithelial barrier in animals harboring kwashiorkor but not healthy microbiota) (Smith et al., 2013; Kau et al., 2015). Unlike the transplanted healthy co-twins’ microbiota, the kwashiorkor microbiota was structurally and metabolically labile, reconfiguring itself upon exposure to a peanut-based RUTF, but not in a sustained way when animals were returned to the Malawian diet. The combination of a nutrient-deficient Malawian diet and a kwashiorkor microbiota was required to produce pathology in the recipient “humanized” mice, including inhibition of steps within the tricarboxylic acid cycle in host cells (Smith et al., 2013). These findings not only provided evidence for a causal relationship between the gut microbiota and SAM but also highlighted the importance of diet-by-microbiota interactions in disease pathogenesis.
If we consider children with persistent microbiota immaturity from the perspective of developmental biology, we can pose a number of basic and applied scientific questions. One question is whether the developmental program defined in Bangladeshi infants and children is generalizable to other populations representing different geographic and cultural settings. If so, it would reveal a fundamental shared aspect of postnatal human development and raise mechanistic questions about the factors that specify a healthy microbial community “fate.” Initial support for generalizability comes from an analysis of concordant healthy Malawian twin pairs, which showed that a number of the age-discriminatory bacterial strains with the highest feature importance scores in the Bangladeshi Random Forests model are also represented in the Malawian population (Subramanian et al., 2014; Yatsunenko et al., 2012). The designation “same strain” was based on the same 16S rRNA sequence; whole-genome sequencing of a given age-discriminatory strain identified by its 16S rRNA sequence will be needed to determine its degree of gene conservation across different Bangladeshi and Malawian hosts. Bacterial 16S rRNA analyses of fecal samples obtained at monthly intervals from infants and children with healthy growth phenotypes enrolled in birth cohorts living at multiple low-income countries allow country/community site-specific, sparse Random-Forests-based models of microbiota maturation to be constructed, as well as an aggregate model representing data pooled from all sites. “Generalizability” can be established through reciprocal tests of the accuracy of the site-specific models (and aggregate model) for healthy individuals living at the different sites and whether these models reveal similar relationships between anthropometry and relative microbiota maturity/MAZ scores for undernourished children living at each of these sites.
A second question has to do with the relationship between microbiota development, enteropathogen load, and environmental enteric dysfunction (EED, also known as environmental enteropathy), an enigmatic and as-yet-incompletely defined disorder of gut barrier function (Keusch et al., 2014; Kosek et al., 2014). Does a primary failure to execute normal maturation of the microbiota directly influence risk for enteropathogen invasion, perturbations in development of mucosal immune system, and abnormalities in nutrient processing and absorption that ultimately results in growth faltering? Alternatively, is a holistic view required that considers each these features of enteric biology as intimately and integrally related to one another? Large birth cohort studies such as MAL-ED and GEMS have provided an opportunity to measure the contributions of enteropathogen load/carriage and diarrheal incidence to growth faltering (MALED Network Investigators, 2014; Platts-Mills et al., 2014; Kotloff et al., 2013). Evidence is emerging that some of the age-discriminatory taxa that define normal microbiota maturation also protect the host from enteropathogen infection. Intriguingly, studies of Bangladeshi adults with acute cholera have shown that recovery from the diarrheal phase involves recapitulation of the sequence of appearance of the same age-discriminatory bacterial strains that define the normal pattern of assembly of the microbiota in healthy Bangladeshi infants/children, suggesting that an essential set of rules governs this assembly process (Hsiao et al., 2014). For example, Ruminococcus obeum, a bacterium that directly correlates with recovery from Vibrio cholerae infection in adult Bangladeshi subjects and defines later stages of normal gut microbiota maturation in healthy Bangladeshi children, restricts V. cholerae colonization of gnotobiotic mice harboring a representative human gut microbiota. Its mechanism involves production of an autoinducer-2 (AI-2) that causes quorum-sensing mediated repression of V. cholerae colonization and virulence factor expression (Hsiao et al., 2014).
A third related question is the manner in which the mucosal immune system and the microbiota co-develop. How do these complex organs talk and educate each other? The answers could help identify factors that legislate a normal developmental trajectory for a gut community and how developmental arrest of the microbiota could be become fixed and difficult to overcome/ advance. Immaturity of the microbiota may be associated with relative immaturity of mucosal immunity in ways that impede responsiveness to vaccines or enteropathogens. If so, can we use members of the microbiota as next-generation adjuvants to prime the immune system in the context of a defined antigen (Yilmaz et al., 2014)? One way to characterize maturation of the mucosal immune system is to use fluorescence-activated cell sorting (FACS) to identify microbial taxa targeted by its IgA responses as a function of chronologic age in hosts with healthy growth phenotypes and in those with undernutrition (critically, IgA targeting is not simply a reflection of the abundances of organisms in the gut community; Kau et al., 2015). This method, known as BugFACS, has identified bacterial targets of gut mucosal IgA responses using fecal samples from children with healthy growth phenotypes or those with varying degrees of undernutrition, as well as fecal samples harvested from gnotobiotic mice harboring transplanted microbiota from healthy and undernourished donors fed diets representative of those that these children consume. BugFACS-purified viable IgA-targeted bacterial taxa were subsequently introduced into germ-free animals fed nutrient-deficient or -sufficient diets to characterize their functional properties. The results disclosed that IgA responses to members of the microbiota can be used as biomarkers of growth faltering, that they are influenced by enteropathogen load, and that they mediate a diet-dependent enteropathy characterized by small intestinal and colonic epithelial barrier disruption. Moreover, IgA-targeted bacterial strains purified from healthy donor microbiota can prevent development of gut mucosal barrier disruption in humanized gnotobiotic mice (Kau et al., 2015), indicating that this method may have utility that extends beyond diagnostics to therapeutic lead discovery and defining mechanisms underlying EED pathogenesis.
A fourth and critical question is whether age-discriminatory taxa are not only just biomarkers but also effectors of growth? If so, they become potential therapeutic agents and targets for manipulation, including food-based manipulations that allow for their establishment in an individual or population at the time of presentation with manifest disease or prior to that time. One way we are currently determining whether age-indicative taxa are also growth indicative is by transplanting microbial communities from children exhibiting varying degrees of growth faltering (defined by anthropometry), representing a particular geographic region, into young, actively growing germ-free animals fed diets representative of the donor population and then defining the effects of the different transplanted communities on the growth and metabolic and immunologic phenotypes of recipient gnotobiotic mice (Figure 2). 16S rRNA data sets generated from the animals’ fecal samples can be used to correlate strain abundances to these phenotypes. These strains can then be cultured from the microbiota of different donor populations. Determining the effects of subsequently introducing these strains—singly or as components of defined consortia—into young gnotobiotic mice harboring microbiota from different undernourished donors represents a way to address several challenges that would be faced when designing and interpreting a clinical study. For example, these preclinical studies could help to (1) define criteria used to select strains beyond their feature importance scores in the Random Forests models and cultivability (e.g., the extent of representation of virulence determinants in their genomes); (2) assess how to encapsulate these organisms, including anaerobes, in ways that permit their long-term storage and viability; (3) determine the extent to which consortia can invade and establish themselves in different microbiota representing individuals from a given population or different populations; (4) assess the nature of their effects on growth (e.g., gain of lean body mass), metabolism, and gut barrier function as a function of the degree of donor undernutrition and microbiota immaturity; and (5) ascertain the degree to which invasion and establishment of these strains in the targeted microbiota and their host effects are impacted by diet. Determining whether these strains are interchangeable between countries will influence the generalizability of microbial interventions or whether there would have to be local sourcing of these biological resources by or for the communities who are themselves afflicted by undernutrition.
Establishing Microbiota and the Maternal Influence
The origins of the microbes that colonize an infant’s gastrointestinal tract are complex, given that infants are exposed to different environmental sources. A major source is the mother and includes microbes from her vagina, skin, gut, and as some have reported, breast milk and possibly the placenta (Dominguez-Bello et al., 2010; Hunt et al., 2011; Grönlund et al., 2011; Cabrera-Rubio et al., 2012; Aagaard et al., 2014).
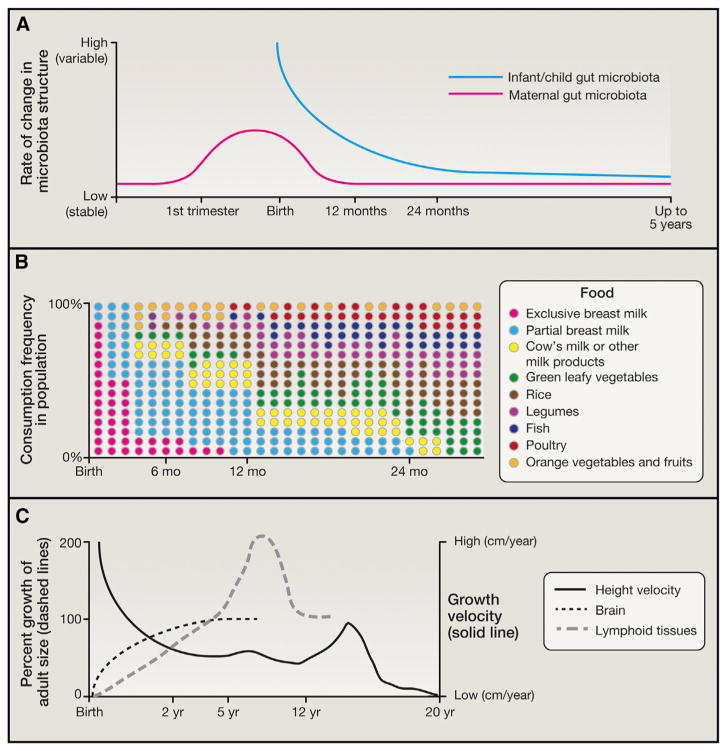
A key knowledge gap relates to the “anthropology of microbes”: knowing how practices associated with pregnancy, including micronutrient supplementation, as well as traditional (and changing) societal “prescriptions” for dietary practices, impact a mother’s microbial ecology prior to parturition and how this may impact transmission of her microbes to her infant. A study of 91 pregnant Finnish women showed that the maternal microbiota changes between the first and third trimester (Koren et al., 2012) (Figure 3). Another analysis of Bangladeshi mothers revealed marked changes in their gut microbiota in the first month post-partum, followed by less substantial changes in the ensuing 9 months (Subramanian et al., 2014). One testable hypothesis is that the maternal microbiota, much like the infant microbiota, undergoes stereotypical alterations during normal pregnancy designed to enhance maternal health and to promote transfer of strains to the infant. Testing this hypothesis will require detailed time series sampling of maternal microbiota throughout pregnancy and of the maternal-infant dyad, plus other environmental sources, including other family members and caregivers. If a program of pregnancy-associated changes in the maternal gut microbiota can be identified using approaches analogous to those described above to describe maturation of the infant microbiota, it could provide an opportunity to use the most indicative or transmissible taxa as biomarkers of nutritional status and as reporters of the effects of different dietary practices or the efficacy of prescribed prenatal nutritional interventions.
Figure 3. Co-variation in Gut Microbiota Assembly/Maturation, Dietary Patterns, and Other Facets of Human Postnatal Development.
(A) Illustration of the rate of change occurring in gut microbiota structure of both mother and child. Note that infant variation curves are known from both longitudinal and cross-sectional study designs (Yatsunenko et al., 2012; Subramanian et al., 2014). In the case of mothers, the curve is interpolated based on studies of pregnant Finnish mothers prior to delivery (Koren et al., 2012) and Bangladeshi mothers following parturition (Subramanian et al., 2014).
(B) The food consumption pattern shown is at a population level and does not depict the great deal of temporal variation observed in food consumption patterns within a given child. Depicting the fractional contribution of each food to the consumption patterns of children in Bangladesh underscores how dietary changes occur simultaneously (lowering of breast milk and increase in legumes and cow’s milk) and not in an orderly fashion (small fluctuations from month to month; re-entry and dropout of certain foods). It also underscores the challenge encountered in ascertaining how food and the microbiota interact to effect maturation of the community.
(C) Major processes related to growth and how they vary in rate and magnitude over time. Curves are adapted from Bogin (1999). Note that the newborn brain represents 12% of body weight (a value 6 times greater than in adults). By the end of the first decade, the brain represents 6% of body weight and consumes twice the amount of glucose and 1.5 times the amount of oxygen as the adult brain. Approximately 30% of the glucose consumed by the infant brain is accounted for by aerobic glycolysis (versus 12% in adults) (Goyal et al., 2014). The dramatic changes in brain metabolism that occur over the first two decades of life coincide with the initial proliferation and then pruning of synapses to adult levels. Central questions that need to be addressed in this area include the biological effects of the gut microbial community on neurogenesis, synaptic connectivity, gliogenesis and glial-neuron interactions, neural circuit function and higher cognitive processes in the context of healthy growth versus undernutrition, and whether/how the gut-brain axis operates to influence/regulate other aspects of host physiology, metabolism, and immunity in the infant/child. Moreover, if persistent immaturity of the gut microbiota is causally related to undernutrition and its long-term sequelae, including neurodevelopmental abnormalities, does durable repair of this immaturity require that nutritional interventions be administered earlier before disease becomes fully manifest (and the microbial ecosystem is so perturbed that restoration becomes very difficult)? Do nutritional interventions need to be applied for more sustained periods of time? Do new types of therapeutic foods need to be developed, or is a microbial intervention also needed?
Pregnancy is also a time of increased susceptibility to infection. Rowe et al. (2011) demonstrated that pregnant mice show increased bacterial burden in models of Listeria monocytogenes and Salmonella typhimurium infection, mediated by active immune suppression by a population of FoxP3+ regulatory T cells (Tregs). Moreover, ablation of the Treg compartment resulted in near-complete resorption of fetuses, indicating a delicate balance between immunological tolerance of the fetus and defense against enteropathogens (Rowe et al., 2011). It is not known how this period of deliberate immune suppression impacts the maternal microbiota and, in turn, vertical transfer of pathogens (and other microbial community members) to the infant.
The Impact of First Foods
Breast Milk
The association between healthy postnatal growth and exclusive breastfeeding has led to the WHO’s recommendation for a minimum of 6 months of exclusive breastfeeding (Kramer and Kakuma, 2002). Human milk is composed of lipids (tri-, di-, and monoglycerides, phospholipids, glycolipids, and free fatty acids), protein components (including immunoglobulins, lacto-ferrin, lysozyme, and cytokines), and a large repertoire of human milk oligosaccharides (HMOs). Over time, this composition changes from colostrum, which is HMO rich, to mature milk, which contains fewer HMOs and protein while the fat content remains relatively stable (Coppa et al., 1993; Lemons et al., 1982).
HMOs and other milk glycoconjugates pass undigested through the proximal gut (Engfer et al., 2000) and serve as nutrient substrates for saccharolytic microbiota in the colon. The microbiota of healthy exclusively breastfed infants is dominated by members of the genus Bifidobacterium (Figure 1; Yatsunenko et al., 2012; Subramanian et al., 2014). These infant-associated bifidobacteria, notably Bifidobacterium longum subsp. infantis, possess a suite of genes involved in importing complex fucosylated and sialylated milk glycans, their further degradation, and subsequent utilization (Sela et al., 2008). The functions encoded by this suite of genes allow them to outcompete other saccharolytic taxa (Marcobal et al., 2010). Bifidobacteria also actively reshape milk composition. For example, they release N-linked glycans conjugated to milk glycoproteins for use as a growth substrate. However, the effect of deglycosylation on milk protein digestibility and function is as-yet unknown (Garrido et al., 2012, 2013).
Colonization by Bifidobacterium species during nursing is associated with a range of benefits, including improved vaccine responses (Huda et al., 2014) and enhanced gut barrier function (Ewaschuk et al., 2008; Weng et al., 2014), including stabilized epithelial tight junctions noted in both animal models (Bergmann et al., 2013) and human cell lines (Chichlowski et al., 2012). Recent work has shown that infants with high Bifidobacterium population densities exhibit a corresponding decrease in fecal milk glycans (De Leoz et al., 2015; Wang et al., 2015), a relationship that could serve as the basis for developing inexpensive diagnostics to predict a healthy gut microbiota in nursing infants.
Development of a healthy infant gut microbiota can be threatened by maternal undernutrition and premature birth. Maternal undernutrition during pregnancy increases risk for underweight and preterm births (Kramer et al., 1992). Children of undernourished mothers receive substantially less than the recommended intake of priority micronutrients during lactation (Allen, 2005). Fortified milk obtained from donors who have had a full-term pregnancy likely does not provide sufficient protein to preterm infants (Arslanoglu et al., 2009). Even when mothers of preterm infants can produce sufficient milk, alterations in milk fat, protein, oligosaccharide content (Weber et al., 2001; De Leoz et al., 2012), and the repertoire of immunoactive components (Castellote et al., 2011) are observed, leading to a call for identifying additional elements for infant nutritional support (Gabrielli et al., 2011; De Leoz et al., 2012).
A vicious cycle of maternal undernutrition and poor infant nutritional status can reflect alterations in the immune, HMO, and/or other components of mother’s milk. This has critical implications for infant health. Poor maternal health is associated with variations in breast milk immunoglobulins and glycoprotein structures during lactation (Smilowitz et al., 2013) and with decreased lactoferrin, a protein with antimicrobial activities (Hennart et al., 1991). Parasite-specific breast milk IgA titers to Entamoeba histolytica and Cryptosporidium spp. correlate with nutritional status in a Bangladeshi infant population in which the burden of infection with these enteropathogens is very high (Korpe et al., 2013). Preterm delivery is associated with atypical variations in milk glycan structures (De Leoz et al., 2012), which poses additional risks. As HMOs have structural similarities to epithelial cell surface and mucus glycans, they can have anti-adhesive effects on enteropathogens. Sialic acid or fucose moieties are key determinants of this activity. Thus, variations in fucosylated HMOs associated with preterm birth may reduce the efficacy of milk oligosaccharides as anti-adhesive decoy molecules for pathogens (Ruiz-Palacios et al., 2003; Jantscher-Krenn et al., 2012).
Understanding how breast milk glycan repertoires correlate with normal microbiota assembly and with impaired microbiota maturation and undernutrition provides an opportunity to identify new glycan streams that could be used to treat undernourished infants. Commercial prebiotics are commonly added to infant formula, where they increase bifidobacteria titers in infant feces (Haarman and Knol, 2005; Knol et al., 2005; Boehm et al., 2002) and lower the incidence of pathogens (Knol et al., 2005). However, current prebiotics, namely fructooligosaccharides and galactooligosaccharides, do not represent the constellation of complex glycan structures delivered in human milk. Moreover, their consumption is not restricted to the population of microbes that define normal gut microbiota maturation (Everard et al., 2014; Dewulf et al., 2013). Numerous efforts to recreate the glycan landscape present in human milk are underway. The technology for chemical and chemoenzymatic construction of complex “milk” oligosaccharides has advanced tremendously, enabling wholesale construction of a limited number of HMO-like structures present in milk (Muthana et al., 2009). Alternatively, purification from animal milks presents another opportunity for rapid and large-scale acquisition of milk oligosaccharides and glycoconjugates. At present, a number of enriched or purified bovine milk glycoproteins, including immunoglobins, lactoferrin, and glycomacropeptide, and glycolipids are commercially available or could be readily produced at scale for use in preclinical and clinical studies. Bovine milk contains a relatively low concentration of free oligosaccharides, but the distribution of structures observed roughly matches the most abundant species present in HMOs (Aldredge et al., 2013). Importantly, bovine milk oligosaccharides (BMOs) can be sourced from numerous points in dairy processing, including cheese whey, suggesting an opportunity for large-scale production of fractions enriched for given (or similar) structures (Zivkovic and Barile, 2011).
Serial Introduction of Complementary Foods in Ways that Promote Maturation of the Gut Microbiota
A recent study compared the microbiota and immune system in bottle-fed versus breastfed macaques. The results showed that breastfed infant macaques develop more robust TH17 cells in the memory pool, suggesting that the timing and trajectory of dietary exposures during early life may have lasting functional consequences beyond that period (Ardeshir et al., 2014). In breastfed humans, the transition to formula feeding and family foods (complementary feeding practices) varies considerably in terms of which food types are consumed, the order of their presentation, and the duration of their consumption. Documenting which foods growing infants consume and in what quantities has required innovative approaches, particularly in low-income countries in which undernutrition is prevalent (Caulfield et al., 2014) (Figure 3). This makes it difficult to determine how the sequence and representation of various first foods affect maturation of the microbiota. Efforts are underway to overcome this challenge. For example, data collection protocols across eight different countries have been harmonized to enable quantification of variations in child feeding practices in the MAL-ED consortium (Caulfield et al., 2014).
The co-linearity between the introduction of various types of solid foods, reduction in breast milk consumption, and maturation of the gut microbiota makes it challenging to identify causal relationships between specific ingredients and the representation of specific microbes through human studies. However, studies in gnotobiotic mice colonized with defined collections of cultured (and sequenced) human gut-derived bacteria have been successful in interrogating specific food-microbe associations (Faith et. al., 2011). These relationships were identified using an experimental design in which a given gnotobiotic animal harboring a defined microbial consortium received a sequence of diets, composed of several different combinations of foods, whose concentrations are intentionally varied between diets. The order of presentation of the different diets was also varied between different mice in order to limit confounding from hysteresis effects. This approach has identified associations between various commercially available foods given in the USA during the complementary feeding period and specific microbes independent of their order of presentation, which would be virtually impossible to identify in clinical studies of developing human infants (Faith et al., 2011). This approach can be applied to young mice colonized with the age- and healthy growth-associated bacterial strains identified using the methods described above to determine which complementary foods promote their representation and expressed functional features. The results could lead to a recommended sequence of complementary feeding that reflects local food availability, affordability, and cultural practices and that sponsors healthy microbiota maturation. This information would advance current recommendations, which are not microbiota based and quite general (Kleinman, 2000).
Additional Considerations Regarding the Developmental Biology of the Gut Microbiota
Microbiota and Metabolic Syndrome
Although we have emphasized the global challenge of undernutrition in children, another vexing global health problem is the growing burden of obesity and associated metabolic dysfunction in children. Increasing attention is being paid to delineating differences in the gut microbiota of children who become obese in the hopes that early recognition of perturbed microbiota development may permit early interventions in at risk populations. For example, a recent culture-independent study of a Singaporean birth cohort disclosed that precocious maturation of the microbiota during the first 6 months of postnatal life was associated with significantly increased adiposity at 18 months (Dogra et al., 2015). Specifically, an unsupervised clustering approach based on bacterial 16S rRNA sequence data sets revealed three clusters of fecal microbiota configurations. The number of samples that binned into one of these clusters (cluster 3), which is characterized by high levels of Bifidobacteria and Collinsella and low levels of Streptococcus and Enterobacteriaceae, increased with age. A faster time to achieving a cluster 3 configuration was associated with significantly greater adiposity measured at age 18 months. Given the rapid rate of change in eating practices and incidence of childhood obesity, longitudinal studies of this type are timely. They should be strategically applied to populations representing different manifestations of these economic, anthropologic, and epidemiologic transitions and accompanied by comprehensive, quantitative assessments of food consumption during the pre-weaning, weaning, and postweaning periods.
Obesity is associated with reduced organismal and genetic diversity in the gut microbiota/microbiome of adults (Turnbaugh et al., 2009b; Le Chatelier et al., 2013). Transplantation of intact fecal microbiota samples, or derived culture collections, from adult twins stably discordant for obesity into germ-free mice transmitted the donors’ discordant adiposity phenotypes, as well as obesity-associated metabolic dysfunction (Ridaura et al., 2013). Co-housing mice just after they received the obese donor’s (Ob) microbiota with mice just after they received the lean co-twin’s (Ln) microbiota, before their discordant adiposity/metabolic phenotypes became evident, prevented development of obesity and metabolic abnormalities in the Ob cagemate. This prevention was associated with unidirectional invasion of bacteria from the Ln cagemate’s gut community to the Ob cagemate’s microbiota. Invasion was diet dependent, occurring in mice fed a human diet formulated to reflect the lower third of saturated fat and upper third of fruit and vegetable consumption in the USA, but not when animals received an unhealthy diet representing the upper third of saturated fat and lower third of fruit and vegetable consumption (Ridaura et al., 2013). These results illustrate how niches can be filled in the Ob microbiota by Ln-derived bacterial taxa to prevent disease and how important diet is to the installation of these health-promoting strains. The results raise important questions about the origins of the reduced bacterial diversity observed in Ob microbiota.
Impact of Antibiotics
One active area of investigation is the role of frequent consumption of broad-spectrum antibiotics in determining the diversity and functional features of the developing microbiota. Studies in conventionally raised mice treated with low-dose penicillin from birth to 4, 8, or 28 weeks of age revealed that early and brief exposure was sufficient to produce durable changes in body composition (Cox et al., 2014). Practical issues (in many parts of the world, antibiotic consumption in children is pervasive and poorly documented), ethical considerations, and the identification of suitable controls all confound the design of human studies that would seek to determine the effects of antibiotic administration on the developmental biology of the human infant gut microbiota and growth. In principle, pre-clinical tests that administer various classes of antibiotics in varying doses—together with representative human diets to gnotobiotic mice harboring transplanted microbiota from infants and children living in various parts of the world—followed by transplantation of their antibiotic-treated microbiota to a next generation of (antibiotic-free) gnotobiotic recipients, would provide one way to explore these questions.
Affordable Nutritious Foods: Societal Implications and Challenges
An imbalance of carbohydrate, fat, and protein consumption, food insecurity, and changing diets in low-income countries brought about by globalization, increases in food prices at the point of retail, and a global protein supply that needs to double by 2050 are some of the drivers for developing new types of affordable nutritious foods that are culturally acceptable, suitable for storage, and distributable given current and envisioned future infrastructure. A sustainable economic model in which local economies benefit from producing and/or distributing foods is also required to ensure long-term supplies. Moreover, there is a paucity of generally accepted metrics for defining foods that provide optimal nutrition at affordable cost (e.g., see the nutrient-rich foods index developed based on FDA recommendations; Drewnowski, 2010).
We propose that the gut microbiota provides a parameter that needs to be considered when developing nutrition options and that the type of preclinical gnotobiotic models described above will be vital for testing and ultimately defining dietary parameters. Studies with mice and other species provide means for defining interactions between food ingredients (at different levels of ingredient resolution and including culturally relevant spices and sweeteners), their methods of preparation and preservation, the gut microbiota of various consumer populations, and human metabolic, immunologic, and other physiologic features. These research platforms offer the promise of yielding next-generation foods designed to be satiating, delicious, nutritious, and able to manipulate microbiota and host properties in ways that promote healthy growth and wellness. However, fulfilling this promise demands a holistic view of the nexus of human gut microbial ecology research, agricultural practices, food production, evolving consumer tastes in an era of rapid globalization, envisioned commercialization strategies, current regulatory structures/practices, ethical issues, and public education. For example, there is a need to more thoroughly and rapidly characterize, through readily searchable, accessible, well-annotated databases, emerging food consumption patterns in countries representing different cultural traditions, stages of economic development, and land/water resources. At the commercial level, there is an opportunity to define and differentiate foods based on their effects on different consumer populations with distinct biological phenotypes and with different gut microbial community configurations. There is an accompanying need to frame intellectual property laws in ways that provide appropriate incentives for private investment while protecting the public good.
To effectively and responsibly apply this knowledge in ways that benefit society, there is a need to work with government agencies to provide efficient and sensible regulatory schemes. These regulatory frameworks vary between nations and are evolving. Currently, the US Food and Drug Administration (FDA) defines “medical foods” as foods that make medical claims. A “dietary supplement” is a product intended for ingestion that contains a dietary ingredient designed to add further nutritional value to a diet. Dietary supplements can only contain ingredients that are “generally regarded as safe” (GRAS) or approved as food additives by the FDA after filing a “new dietary ingredient” (NDI) notification with full description of the ingredient and product in which it will be marketed, the basis for the manufacturer’s conclusion that it is an NDI, recommended use and proposed labeling, plus a history of its use and evidence of its safety to support the proposed use. Probiotics have been defined in various ways, including “live microorganisms that, when administered in adequate amounts, confer a health benefit on the host” (Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics, 2001), whereas prebiotics have been considered to be “a selectively fermented ingredient that allows specific changes both in the composition and/or activity of the gastrointestinal microbiota that confer benefits upon host well-being and health” (Roberfroid, 2007). Synbiotics are combinations of prebiotics and probiotics. Regulation of prebiotics, probiotics, and synbiotics remains a work in progress, although any health claims they make will likely require a clinical development pathway that is the same as that employed for biologics.
Opening the Public Discussion
For public acceptance and societal benefit, a thoughtful proactive, science-based, educational outreach is needed with an understandable vocabulary tailored to targeted consumer populations and respectful of their cultural traditions. The goal would be to objectively describe the extent to which the nutritional value of food is related to a consumer’s microbiota and how food ingredients, food choices, and the microbiota are connected to health benefits.
We suggest that one way of framing a public discussion regarding the impact of human gut microbiome research on the nexus of food, agriculture, and nutrition is to divide it into three “sectors”: science and technology, ethics, and policy and governance.
Science and Technology
Ongoing and new studies will help to define (1) methods for selection and production of new food sources, (2) design of new foods/diets, (3) definitions of nutritional value and benefit and metrics for differentiation of foods, and (4) the role of the gut microbiota in determining nutritional status in pregnant women, infants and children, and adults throughout the course of their lives.
Ethics
The impact of gut microbiota research extends beyond conceptions of health to human rights. Key issues include (1) concepts of self and ownership of microbes and the shaping of these views by cultural, religious, socioeconomic, educational, and political factors; (2) use of a person’s microbes to improve nutritional status within and beyond family, community, and country; (3) strategies for responsible stewardship of our (human) microbial resources; and (4) personal, familial, and societal impact (and shared benefit) of methods envisioned to promote intergenerational transmission of beneficial microbes and to effect durable repair of defective gut microbial community development early in life or functional restoration later in life.
Policy and Governance
Advances in gut microbiota research will have long-term impact on regulatory and other governmental policies and agencies as they relate to agriculture, food, and nutritional health. These effects include (1) definitions of food safety, including the products of microbial biotransformation of food ingredients; (2) definitions of nutritional benefit within and outside of the context of specific human health claims; (3) laws concerning ownership of microbial strains and their distribution within and across national borders (for example, in October 2014, the Convention on Biological Diversity/Nagoya Protocol on Access to Genetic Resources and the Fair and Equitable Sharing of Benefits from their Utilization entered into international force “stringent requirements for prior informed consent and benefit sharing for research and commercial activities involving genetic resources from plants, animals, and microorganisms” [http://www.cbd.int/abs/]); (4) laws concerning intellectual property related to microbes, microbial consortia, and the products of microbial interactions with food ingredients, including diagnostics and therapeutics; (5) policies related to standards of manufacture, purity, and composition of probiotics and synbiotics; and (6) incentives for linking plans for food production and distribution with gut microbiota health. A key challenge is how to construe (1)–(6) in the context of a reference set of “representative” countries.
Closing Thoughts
Given the intricate links between first foods and long-term human health, ensuring availability of appropriate food sources is of high priority. Because undernutrition is such a widespread affliction, it is critical to consider how to categorize the targeted populations, the cost and economic sustainability, the efficacy (effect size and durability), and the cultural acceptability of various therapeutic or preventative approaches, as well as the generalizability of both food-based and microbial interventions to large populations within and across national/societal boundaries. One way of conceptualizing this complex set of challenges for treatment and prevention is to place, on one end of the spectrum of undernutrition, children with already manifest SAM and significant microbiota immaturity who could be treated with locally produced, readily and reproducibly manufactured, affordable and safe, culturally acceptable next-generation RUTFs, with or without microbial interventions of the type described above. Moving along this continuum, another group would consist of individuals who manifest growth faltering (stunting) in the first 1,000 days after conception, where the envisioned targets for interventions are pregnant and lactating women and their infants. At the other end of the continuum is a third group that are the targets of locally produced, consumer-focused, affordable nutrition products designed to improve dietary quality and increase the diversity of food choices.
Looking back over 800 million years of metazoan evolution, we appreciate more now than ever before the splendid innovation of having a gut that assembles microbial resources that enable efficient utilization of available nutrients (McFall-Ngai et al., 2013). We, humans, are now in a position to not only understand but to deliberately influence this process of microbial community acquisition in order to ensure its optimal execution. The challenges we face in designing and improving food systems and nutritional health are great and pressing. Hopefully, our gut instinct will be to honor and harness the intimate interrelationship between foods and “our” microbes in an attempt to address this challenge now and throughout the course of this defining century for our species and planet.
Acknowledgments
Work cited from the authors’ labs was supported in part by grants from the Bill & Melinda Gates Foundation (BMGF) and the NIH (DK078669, DK30292, and DK70977). We thank our colleagues in the BMGF-sponsored Breast Milk, Gut Microbiome, and Immunity Project; Tahmeed Ahmed and other members of our collaboration with the International Center for Diarrheal Disease Research, Bangladesh (ICDDR,B); Michael Barratt, Jeanette Gehrig, Siddarth Venkatesh and other members of our laboratories; and William Petri and Andrew Serazin for their insights and inspiration. J.I.G. is a co-founder of Matatu, Inc., a company characterizing the role of diet-by-microbiota interactions in animal health. D.A.M. is a co-founder and President of Evolve Biosystems, Inc., a company focused on diet-based manipulation of the gut microbiota.
References
- Aagaard K, Ma J, Antony KM, Ganu R, Petrosino J, Versalovic J. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aldredge DL, Geronimo MR, Hua S, Nwosu CC, Lebrilla CB, Barile D. Annotation and structural elucidation of bovine milk oligosaccharides and determination of novel fucosylated structures. Glycobiology. 2013;23:664–676. doi: 10.1093/glycob/cwt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen LH. Multiple micronutrients in pregnancy and lactation: an overview. Am J Clin Nutr. 2005;81:1206S–1212S. doi: 10.1093/ajcn/81.5.1206. [DOI] [PubMed] [Google Scholar]
- Ardeshir A, Narayan NR, Méndez-Lagares G, Lu D, Rauch M, Huang Y, Rompay KKAV, Lynch SV, Hartigan-O’Connor DJ. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med. 2014;6:252ra120. doi: 10.1126/scitranslmed.3008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslanoglu S, Moro GE, Ziegler EE. Preterm infants fed fortified human milk receive less protein than they need. J Perinatol. 2009;29:489–492. doi: 10.1038/jp.2009.50. [DOI] [PubMed] [Google Scholar]
- Bergmann KR, Liu SXL, Tian R, Kushnir A, Turner JR, Li HL, Chou PM, Weber CR, De Plaen IG. Bifidobacteria stabilize claudins at tight junctions and prevent intestinal barrier dysfunction in mouse necrotizing enterocolitis. Am J Pathol. 2013;182:1595–1606. doi: 10.1016/j.ajpath.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm G, Lidestri M, Casetta P, Jelinek J, Negretti F, Stahl B, Marini A. Supplementation of a bovine milk formula with an oligosaccharide mixture increases counts of faecal bifidobacteria in preterm infants. Arch Dis Child Fetal Neonatal. 2002;86:F178–F181. doi: 10.1136/fn.86.3.F178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogin B. Patterns of Human Growth. Cambridge University Press; 1999. [Google Scholar]
- Cabrera-Rubio R, Collado MC, Laitinen K, Salminen S, Isolauri E, Mira A. The human milk microbiome changes over lactation and is shaped by maternal weight and mode of delivery. Am J Clin Nutr. 2012;96:544–551. doi: 10.3945/ajcn.112.037382. [DOI] [PubMed] [Google Scholar]
- Castellote C, Casillas R, Ramírez-Santana C, Pérez-Cano FJ, Castell M, Moretones MG, López-Sabater MC, Franch A. Premature delivery influences the immunological composition of colostrum and transitional and mature human milk. J Nutr. 2011;141:1181–1187. doi: 10.3945/jn.110.133652. [DOI] [PubMed] [Google Scholar]
- Caulfield LE, Bose A, Chandyo RK, Nesamvuni C, de Moraes ML, Turab A, Patil C, Mahfuz M, Ambikapathi R, Ahmed T MAL-ED Network Investigators . Infant feeding practices, dietary adequacy, and micronutrient status measures in the MAL-ED study. Clin Infect Dis. 2014;59 (4):S248–S254. doi: 10.1093/cid/ciu421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichlowski M, De Lartigue G, German JB, Raybould HE, Mills DA. Bifidobacteria isolated from infants and cultured on human milk oligosaccharides affect intestinal epithelial function. J Pediatr Gastroenterol Nutr. 2012;55:321–327. doi: 10.1097/MPG.0b013e31824fb899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppa GV, Gabrielli O, Pierani P, Catassi C, Carlucci A, Giorgi PL. Changes in carbohydrate composition in human milk over 4 months of lactation. Pediatrics. 1993;91:637–641. [PubMed] [Google Scholar]
- Cox LM, Yamanishi S, Sohn J, Alekseyenko AV, Leung JM, Cho I, Kim SG, Li H, Gao Z, Mahana D, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leoz MLA, Gaerlan SC, Strum JS, Dimapasoc LM, Mirmiran M, Tancredi DJ, Smilowitz JT, Kalanetra KM, Mills DA, German JB, et al. Lacto-N-tetraose, fucosylation, and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J Proteome Res. 2012;11:4662–4672. doi: 10.1021/pr3004979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leoz MLA, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, German JB, Mills DA, Lebrilla CB. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: a proof-of-concept study. J Proteome Res. 2015;14:491–502. doi: 10.1021/pr500759e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Barry NA, Mok KC, Taga ME, Goodman AL. Human gut microbes use multiple transporters to distinguish vitamin B12 analogs and compete in the gut. Cell Host Microbe. 2014;15:47–57. doi: 10.1016/j.chom.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewulf EM, Cani PD, Claus SP, Fuentes S, Puylaert PG, Neyrinck AM, Bindels LB, de Vos WM, Gibson GR, Thissen JP, Delzenne NM. Insight into the prebiotic concept: lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut. 2013;62:1112–1121. doi: 10.1136/gutjnl-2012-303304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogra S, Sakwinska O, Soh S-E, Ngom-Bru C, Brück WM, Berger B, Brüssow H, Lee YS, Yap F, Chong Y-S, et al. Dynamics of infant gut microbiota are influenced by delivery mode and gestational duration and are associated with subsequent adiposity. MBio. 2015;6:e02419–14. doi: 10.1128/mBio.02419-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello MG, Costello EK, Contreras M, Magris M, Hidalgo G, Fierer N, Knight R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewnowski A. The Nutrient Rich Foods Index helps to identify healthy, affordable foods. Am J Clin Nutr. 2010;91:1095S–1101S. doi: 10.3945/ajcn.2010.28450D. [DOI] [PubMed] [Google Scholar]
- Engfer MB, Stahl B, Finke B, Sawatzki G, Daniel H. Human milk oligosaccharides are resistant to enzymatic hydrolysis in the upper gastrointestinal tract. Am J Clin Nutr. 2000;71:1589–1596. doi: 10.1093/ajcn/71.6.1589. [DOI] [PubMed] [Google Scholar]
- Everard A, Lazarevic V, Gaïa N, Johansson M, Ståhlman M, Backhed F, Delzenne NM, Schrenzel J, François P, Cani PD. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014;8:2116–2130. doi: 10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewaschuk JB, Diaz H, Meddings L, Diederichs B, Dmytrash A, Backer J, Looijervan Langen M, Madsen KL. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1025–G1034. doi: 10.1152/ajpgi.90227.2008. [DOI] [PubMed] [Google Scholar]
- Faith JJ, McNulty NP, Rey FE, Gordon JI. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science. 2011;333:101–104. doi: 10.1126/science.1206025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. http://dx.doi.org/10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaayeb L, Sarr JB, Cames C, Pinçon C, Hanon J-B, Ndiath MO, Seck M, Herbert F, Sagna AB, Schacht A-M, et al. Effects of malnutrition on children’s immunity to bacterial antigens in Northern Senegal. Am J Trop Med Hyg. 2014;90:566–573. doi: 10.4269/ajtmh.12-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrielli O, Zampini L, Galeazzi T, Padella L, Santoro L, Peila C, Giuliani F, Bertino E, Fabris C, Coppa GV. Preterm milk oligosaccharides during the first month of lactation. Pediatrics. 2011;128:e1520–e1531. doi: 10.1542/peds.2011-1206. [DOI] [PubMed] [Google Scholar]
- Galler JR, Bryce C, Waber DP, Zichlin ML, Fitzmaurice GM, Eaglesfield D. Socioeconomic outcomes in adults malnourished in the first year of life: a 40-year study. Pediatrics. 2012;130:e1–e7. doi: 10.1542/peds.2012-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Nwosu C, Ruiz-Moyano S, Aldredge D, German JB, Lebrilla CB, Mills DA. Endo-β-N-acetylglucosaminidases from infant gut-associated bifidobacteria release complex N-glycans from human milk glycoproteins. Mol Cell Proteomics. 2012;11:775–785. doi: 10.1074/mcp.M112.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido D, Dallas DC, Mills DA. Consumption of human milk glycoconjugates by infant-associated bifidobacteria: mechanisms and implications. Microbiology. 2013;159:649–664. doi: 10.1099/mic.0.064113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerland P, Raftery AE, Sevčíková H, Li N, Gu D, Spoorenberg T, Alkema L, Fosdick BK, Chunn J, Lalic N, et al. World population stabilization unlikely this century. Science. 2014;346:234–237. doi: 10.1126/science.1257469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal MS, Hawrylycz M, Miller JA, Snyder AZ, Raichle ME. Aerobic glycolysis in the human brain is associated with development and neotenous gene expression. Cell Metab. 2014;19:49–57. doi: 10.1016/j.cmet.2013.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grönlund MM, Grześkowiak Ł, Isolauri E, Salminen S. Influence of mother’s intestinal microbiota on gut colonization in the infant. Gut Microbes. 2011;2:227–233. doi: 10.4161/gmic.2.4.16799. [DOI] [PubMed] [Google Scholar]
- Haarman M, Knol J. Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl Environ Microbiol. 2005;71:2318–2324. doi: 10.1128/AEM.71.5.2318-2324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennart PF, Brasseur DJ, Delogne-Desnoeck JB, Dramaix MM, Robyn CE. Lysozyme, lactoferrin, and secretory immunoglobulin A content in breast milk: influence of duration of lactation, nutrition status, prolactin status, and parity of mother. Am J Clin Nutr. 1991;53:32–39. doi: 10.1093/ajcn/53.1.32. [DOI] [PubMed] [Google Scholar]
- Hsiao A, Ahmed AM, Subramanian S, Griffin NW, Drewry LL, Petri WA, Jr, Haque R, Ahmed T, Gordon JI. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huda MN, Lewis Z, Kalanetra KM, Rashid M, Ahmad SM, Raqib R, Qadri F, Underwood MA, Mills DA, Stephensen CB. Stool microbiota and vaccine responses of infants. Pediatrics. 2014;134:e362–e372. doi: 10.1542/peds.2013-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt KM, Foster JA, Forney LJ, Schütte UME, Beck DL, Abdo Z, Fox LK, Williams JE, McGuire MK, McGuire MA. Characterization of the diversity and temporal stability of bacterial communities in human milk. PLoS ONE. 2011;6:e21313. doi: 10.1371/journal.pone.0021313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jantscher-Krenn E, Lauwaet T, Bliss LA, Reed SL, Gillin FD, Bode L. Human milk oligosaccharides reduce Entamoeba histolytica attachment and cytotoxicity in vitro. Br J Nutr. 2012;108:1839–1846. doi: 10.1017/S0007114511007392. [DOI] [PubMed] [Google Scholar]
- Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics. Probiotics in Food: Health and Nutritional Properties and Guidelines for Evaluation (FAO and WHO) 2001. Health and nutrition properties of probiotics in food including powder milk with live lactic acid bacteria; pp. 1–30. [Google Scholar]
- Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu T-C, Stappenbeck TS, Maleta KM, et al. Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med. 2015;7:276ra24. doi: 10.1126/scitranslmed.aaa4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keusch GT, Denno DM, Black RE, Duggan C, Guerrant RL, Lavery JV, Nataro JP, Rosenberg IH, Ryan ET, Tarr PI, et al. Environmental enteric dysfunction: pathogenesis, diagnosis, and clinical consequences. Clin Infect Dis. 2014;59(4):S207–S212. doi: 10.1093/cid/ciu485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman RE. American Academy of Pediatrics recommendations for complementary feeding. Pediatrics. 2000;106:1274. [PubMed] [Google Scholar]
- Knol J, Boehm G, Lidestri M, Negretti F, Jelinek J, Agosti M, Stahl B, Marini A, Mosca F. Increase of faecal bifidobacteria due to dietary oligosaccharides induces a reduction of clinically relevant pathogen germs in the faeces of formula-fed preterm infants. Acta Paediatr Suppl. 2005;94:31–33. doi: 10.1111/j.1651-2227.2005.tb02152.x. [DOI] [PubMed] [Google Scholar]
- Koren O, Goodrich JK, Cullender TC, Spor A, Laitinen K, Bäckhed HK, Gonzalez A, Werner JJ, Angenent LT, Knight R, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150:470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korpe PS, Liu Y, Siddique A, Kabir M, Ralston K, Ma JZ, Haque R, Petri WA. Breast milk parasite-specific antibodies and protection from amebiasis and cryptosporidiosis in Bangladeshi infants: a prospective cohort study. Clin Infect Dis. 2013;56:988–992. doi: 10.1093/cid/cis1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosek M, Haque R, Lima A, Babji S, Shrestha S, Qureshi S, Amidou S, Mduma E, Lee G, Yori PP, et al. MAL-ED network . Fecal markers of intestinal inflammation and permeability associated with the subsequent acquisition of linear growth deficits in infants. Am J Trop Med Hyg. 2013;88:390–396. doi: 10.4269/ajtmh.2012.12-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosek M, Guerrant RL, Kang G, Bhutta Z, Yori PP, Gratz J, Gottlieb M, Lang D, Lee G, Haque R, et al. MAL-ED Network Investigators . Assessment of environmental enteropathy in the MAL-ED cohort study: theoretical and analytic framework. Clin Infect Dis. 2014;59(4):S239–S247. doi: 10.1093/cid/ciu457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet. 2013;382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- Kramer MS, Kakuma R. Optimal duration of exclusive breast-feeding. Cochrane Database Syst Rev. 2002:CD003517. doi: 10.1002/14651858.CD003517. [DOI] [PubMed] [Google Scholar]
- Kramer MS, McLean FH, Eason EL, Usher RH. Maternal nutrition and spontaneous preterm birth. Am J Epidemiol. 1992;136:574–583. doi: 10.1093/oxfordjournals.aje.a116535. [DOI] [PubMed] [Google Scholar]
- Lazzerini M, Rubert L, Pani P. Specially formulated foods for treating children with moderate acute malnutrition in low- and middle-income countries. Cochrane Database Syst Rev. 2013;6:CD009584. doi: 10.1002/14651858.CD009584.pub2. [DOI] [PubMed] [Google Scholar]
- Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, et al. Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500:541–546. doi: 10.1038/nature12506. [DOI] [PubMed] [Google Scholar]
- Lemons JA, Moye L, Hall D, Simmons M. Differences in the composition of preterm and term human milk during early lactation. Pediatr Res. 1982;16:113–117. doi: 10.1203/00006450-198202000-00007. [DOI] [PubMed] [Google Scholar]
- MAL-ED Network Investigators . The MAL-ED study: a multinational and multidisciplinary approach to understand the relationship between enteric pathogens, malnutrition, gut physiology, physical growth, cognitive development, and immune responses in infants and children up to 2 years of age in resource-poor environments. Clin Infect Dis. 2014;59(4):S193–S206. doi: 10.1093/cid/ciu653. [DOI] [PubMed] [Google Scholar]
- Marcobal A, Barboza M, Froehlich JW, Block DE, German JB, Lebrilla CB, Mills DA. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–5340. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFall-Ngai M, Hadfield MG, Bosch TCG, Carey HV, Domazet-Lošo T, Douglas AE, Dubilier N, Eberl G, Fukami T, Gilbert SF, et al. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci USA. 2013;110:3229–3236. doi: 10.1073/pnas.1218525110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthana S, Cao H, Chen X. Recent progress in chemical and chemoenzymatic synthesis of carbohydrates. Curr Opin Chem Biol. 2009;13:573–581. doi: 10.1016/j.cbpa.2009.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platts-Mills JA, McCormick BJJ, Kosek M, Pan WK, Checkley W, Houpt ER MAL-ED Network Investigators . Methods of analysis of enteropathogen infection in the MAL-ED Cohort Study. Clin Infect Dis. 2014;59(4):S233–S238. doi: 10.1093/cid/ciu408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard SA, McCormick BJJ, Miller MA, Caulfield LE, Checkley W MAL-ED Network Investigators . Modeling environmental influences on child growth in the MAL-ED cohort study: opportunities and challenges. Clin Infect Dis. 2014;59(4):S255–S260. doi: 10.1093/cid/ciu436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberfroid M. Prebiotics: the concept revisited. J Nutr. 2007;137(2):830S–837S. doi: 10.1093/jn/137.3.830S. [DOI] [PubMed] [Google Scholar]
- Rowe JH, Ertelt JM, Aguilera MN, Farrar MA, Way SS. Foxp3(+) regulatory T cell expansion required for sustaining pregnancy compromises host defense against prenatal bacterial pathogens. Cell Host Microbe. 2011;10:54–64. doi: 10.1016/j.chom.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278:14112–14120. doi: 10.1074/jbc.M207744200. [DOI] [PubMed] [Google Scholar]
- Sela DA, Chapman J, Adeuya A, Kim JH, Chen F, Whitehead TR, Lapidus A, Rokhsar DS, Lebrilla CB, German JB, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smilowitz JT, Totten SM, Huang J, Grapov D, Durham HA, Lammi-Keefe CJ, Lebrilla C, German JB. Human milk secretory immunoglobulin a and lactoferrin N-glycans are altered in women with gestational diabetes mellitus. J Nutr. 2013;143:1906–1912. doi: 10.3945/jn.113.180695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, Kau AL, Rich SS, Concannon P, Mychaleckyj JC, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339:548–554. doi: 10.1126/science.1229000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, Huq S, Yatsunenko T, Haque R, Mahfuz M, Alam MA, Benezra A, DeStefano J, Meier MF, Muegge BD, et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009a;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009b;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victora CG, Adair L, Fall C, Hallal PC, Martorell R, Richter L, Sachdev HS Maternal and Child Undernutrition Study Group. Maternal and child undernutrition: consequences for adult health and human capital. Lancet. 2008;371:340–357. doi: 10.1016/S0140-6736(07)61692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Li M, Wu S, Lebrilla CB, Chapkin RS, Ivanov I, Donovan SM. Fecal microbiota composition of breast-fed infants is correlated with human milk oligosaccharides consumed. J Pediatr Gastroenterol Nutr. 2015 doi: 10.1097/MPG.0000000000000752. Published online February 2, 2015 http://dx.doi.org/10.1097/MPG.0000000000000752. [DOI] [PMC free article] [PubMed]
- Weber A, Loui A, Jochum F, Bührer C, Obladen M. Breast milk from mothers of very low birthweight infants: variability in fat and protein content. Acta Paediatr. 2001;90:772–775. [PubMed] [Google Scholar]
- Weng M, Ganguli K, Zhu W, Shi HN, Walker WA. Conditioned medium from Bifidobacteria infantis protects against Cronobacter sakazakii-induced intestinal inflammation in newborn mice. Am J Physiol. 2014;306:G779–G787. doi: 10.1152/ajpgi.00183.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz B, Portugal S, Tran TM, Gozzelino R, Ramos S, Gomes J, Regalado A, Cowan PJ, d’Apice AJF, Chong AS, et al. Gut microbiota elicits a protective immune response against malaria transmission. Cell. 2014;159:1277–1289. doi: 10.1016/j.cell.2014.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic AM, Barile D. Bovine milk as a source of functional oligosaccharides for improving human health. Adv Nutr. 2011;2:284–289. doi: 10.3945/an.111.000455. [DOI] [PMC free article] [PubMed] [Google Scholar]