Abstract
The Ras/mitogen-activated protein kinase (MAPK) signalling cascade regulates various biological functions, including cell growth, proliferation and survival. As such, this pathway is often deregulated in cancer, including melanomas, which frequently harbour activating mutations in the NRAS and BRAF oncogenes. Hyperactive MAPK signalling is known to promote protein synthesis, but the mechanisms by which this occurs remain poorly understood. Here, we show that expression of oncogenic forms of Ras and Raf promotes the constitutive activation of the mammalian target of rapamycin (mTOR). Using pharmacological inhibitors and RNA interference we find that the MAPK-activated protein kinase RSK (p90 ribosomal S6 kinase) is partly required for these effects. Using melanoma cell lines carrying activating BRAF mutations we show that ERK/RSK signalling regulates assembly of the translation initiation complex and polysome formation, as well as the translation of growth-related mRNAs containing a 5’ terminal oligopyrimidine (TOP) motif. Accordingly, we find that RSK inhibition abrogates tumour growth in mice. Our findings indicate that RSK may be a valuable therapeutic target for the treatment of tumours characterized by deregulated MAPK signalling, such as melanoma.
Keywords: Melanoma, BRAF, MAPK, RSK, mTOR
Introduction
The Ras/mitogen-activated protein kinase (MAPK) signalling cascade regulates a wide range of cellular functions, including cell growth, proliferation, and survival (1, 2). In this pathway, ligand binding to growth factor receptors leads to activation of the small GTPase Ras, which allows membrane recruitment and activation of the Raf protein kinases. Once activated, Raf phosphorylates and activates MEK1/2, which are dual-specificity kinases capable of phosphorylating and activating the MAPKs ERK1/2. Hyperactivation of the MAPK pathway is a highly prevalent event in human cancer and usually results from aberrant activation of receptor tyrosine kinases or by gain-of-function mutations in Ras or Raf isoforms (3). One prominent example is melanoma, where activating mutations in BRAF or NRAS (in ~50% and ~15% of cases, respectively) are the earliest genotypic changes observed (3-5). More than 90% of BRAF mutations encode a protein harbouring the V600E mutation, which constitutively activates ERK1/2 signalling (6). Malignant melanoma is highly resistant to conventional chemotherapy (7), but recently developed therapies that target components of the MAPK pathway have demonstrated survival advantage in patients with BRAF-mutated melanoma (8).
The Ras/MAPK pathway promotes cell growth and proliferation by controlling the activity of various transcription factors and cell cycle regulators (9). In addition, this pathway was shown to stimulate protein synthesis (10, 11), but the molecular mechanisms involved in this process remain poorly understood. The MAPK-activated protein kinases MNK1/2 were found to phosphorylate the translation initiation factor eIF4E (12, 13), providing a potential mechanism by which MAPK signalling regulates mRNA translation (reviewed in 14, 15). However, the MNKs have been shown to be dispensable for global protein synthesis (16), and may only regulate the translation of a specific subset of mRNAs (17). The p90 ribosomal S6 kinase (RSK) family comprises four protein kinases that are directly phosphorylated and activated by ERK1/2 (18). RSK regulates various substrates involved in cell proliferation and survival (19-21), and was found to be hyperactivated in several types of cancer (22), including melanoma (23). Increasing evidence indicates that ERK/RSK signalling participates in the regulation of the mammalian target of rapamycin (mTOR)(24, 25), providing an additional mechanism by which MAPK signaling regulates the translational machinery (26).
mTOR is an evolutionarily conserved Ser/Thr kinase that plays essential roles in the regulation of cell growth (27, 28). mTOR associates with different protein partners to form two functionally distinct complexes, the rapamycin-sensitive and -insensitive mTOR complex (mTORC) 1 and 2, respectively (3, 29, 30). mTORC1 positively regulates mRNA translation by stimulating two main translational regulators, the eukaryotic translation initiation factor 4E (eIF4E) and the p70 ribosomal S6 kinases (S6K1 and S6K2) (26, 31). While eIF4E promotes recruitment of the ribosome at the 5’ cap of mRNAs (32), activated S6Ks phosphorylate proteins involved in pre-mRNA processing and ribosome biogenesis, including the 40S ribosomal protein (rp) S6 (33). mTORC1 is frequently deregulated in human cancer and its activation is often linked with mutations in components of the phosphoinositide-3 kinase (PI3K) pathway (34). In melanoma, increased mTORC1 activity has been observed in ~70% of tumours (35), but it is currently unknown whether ERK/RSK signalling contributes to this deregulation.
In this study, we demonstrate that oncogenic activation of the MAPK pathway promotes constitutive mTORC1 activity and function. More specifically, we identify RSK as a key MAPK substrate involved in the regulation of mTORC1 in melanoma. Inhibition of RSK expression or activity results in decreased mTORC1 activity and function, as well as reduced melanoma cell and tumour growth. Together, these data suggest that RSK may be a valuable therapeutic target for the treatment of metastatic melanoma.
Results
Oncogenic Ras and Raf promote mTORC1 signalling in an ERK/RSK-dependent manner
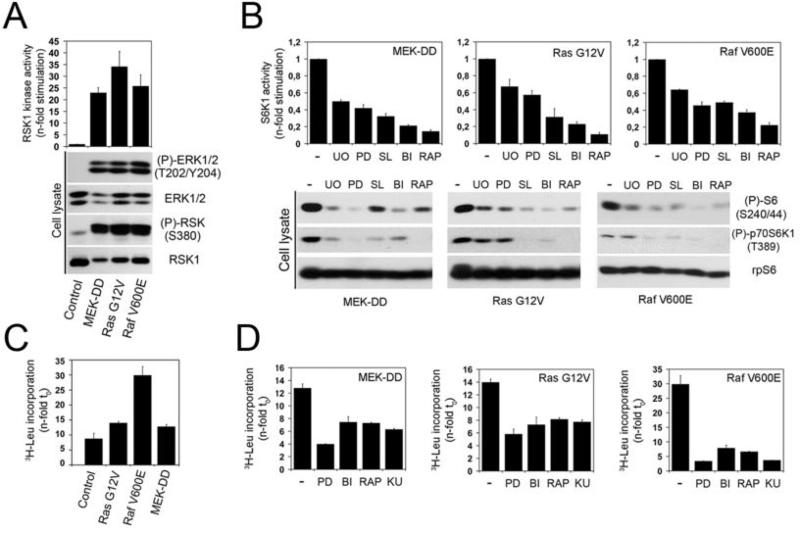
To examine the contribution of oncogenic MAPK signalling in the regulation of mTORC1, we generated stable HEK293 cell lines expressing activated alleles of MEK1 (MEK-DD), H-Ras (G12V) or B-Raf (V600E) (Fig. 1A). These cells were starved of serum overnight to reduce basal mTORC1 activity resulting from growth factors present in serum. To monitor mTORC1 activity, we have quantified several established mTORC1-dependent events, such as S6K1 phosphorylation at its mTORC1-regulated site (Thr389), rpS6 phosphorylation at S6K1-specific residues (Ser240/244), as well as the kinase activity of immunoprecipitated endogenous S6K1. Using these three approaches, we found that all three cell lines displayed constitutively high levels of mTORC1 activity (Fig. 1B, lane 1 of each panel), which were sensitive to rapamycin treatment (Fig. 1B, lane 6 of each panel). To characterize the involvement of the MAPK pathway, cells were pre-treated with inhibitors of MEK1/2 (U0126 or PD184352) or RSK (BI-D1870 or SL0101) for one hour prior to being harvested. These inhibitors resulted in a robust decrease in mTORC1 activity (Fig. 1B), suggesting that oncogenic ERK/RSK signalling contributes significantly to the regulation of mTORC1 activity. To measure mTORC1-dependent functions, we tested the effect of MEK1/2 and RSK inhibitors on global protein synthesis. As expected, we found that oncogenic activation of the MAPK pathway increased tritiated leucine incorporation compared to control cells (Fig. 1C). Interestingly, we found that inhibition of ERK/RSK signalling significantly suppressed global protein synthesis in all three cell lines (Fig. 1D). These reductions were similar to those obtained with two mTOR inhibitors (rapamycin and KU-0063794), suggesting an important role for ERK/RSK signalling in global protein synthesis.
Fig. 1.
Oncogenic RAS and RAF promote mTORC1 signalling in a RSK-dependent manner. (A) HEK293 cells stably expressing constitutively-activated MEK1 (MEK-DD), Ras (G12V) or Raf (V600E) were serum-starved overnight, and analyzed for ERK and RSK phosphorylation by immunoblotting. Immunoprecipitated RSK1 kinase activity was assayed using GST-rpS6 as substrate, in the presence of γ[32P]ATP. Samples were subjected to SDS-PAGE and the dried coomassie-stained gel autoradiographed. (B) HEK293 cells stably expressing constitutively-activated MEK1, Ras or Raf were serum-starved overnight, pre-treated with U0126 (U0), PD184352 (PD), SL0101 (SL), BI-D1870 (BI) or rapamycin (RAP) for 1h, prior to being harvested. Immunoprecipitated S6K1 kinase activity was assayed using GST-rpS6 as substrate, in the presence of γ[32P]ATP. The histogram shows quantifications of phosphorylated rpS6 from three independent experiments. Phosphorylation of endogenous rpS6 and S6K1, and total rpS6 protein level were monitored by immunoblotting. (C) Control and constitutively-activated cells were maintained in low-serum (2%) for 18 hr. Global protein synthesis was measured by adding 0.5 μCi/ml [3H]leucine to the medium for 6 hours. Histograms show radioactivity incorporation normalized to time 0. (D) As in (C), except that cells were treated with indicated inhibitors for 60 minutes prior to addition of 0.5 μCi/ml [3H]leucine to the medium.
ERK/RSK signalling contributes to the constitutive activation of mTORC1 in melanoma
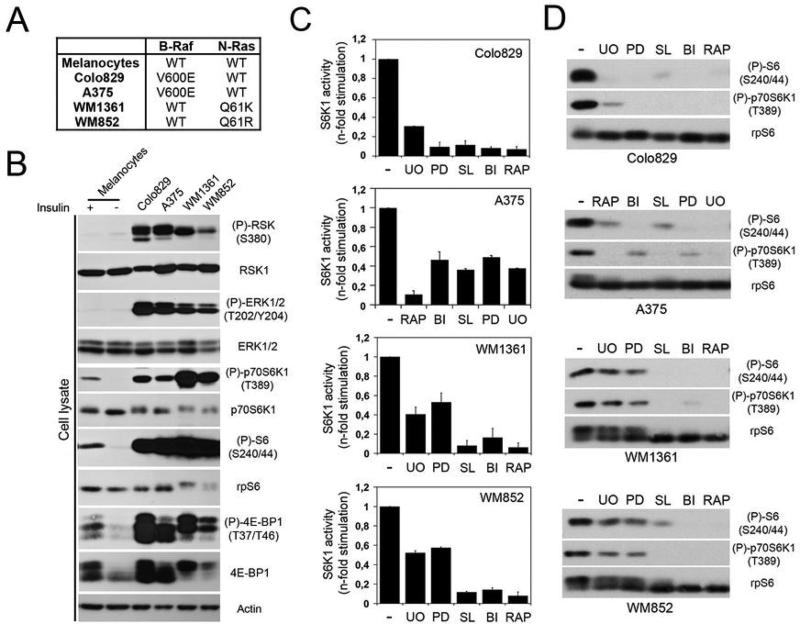
To evaluate the role of RSK in melanoma, we used four human melanoma cell lines harbouring gain-of-function mutations in NRAS (WM852 and WM1361) or BRAF (Colo829 and A375)(Fig. 2A). These cells have constitutively high ERK/RSK activity compared to normal human melanocytes (Fig. 2B), and are highly sensitive to MEK1/2 inhibitors irrespective of their genotypes (Fig. S1A). Interestingly, these cells also display constitutively high mTORC1 activity compared to serum-starved or insulin-treated normal human melanocytes (Fig. 2B), suggesting the involvement of the MAPK pathway. Consistent with this, we found that mTORC1 activity in melanoma cells was highly sensitive to inhibitors of ERK/RSK signalling (Fig. 2C and 2D). This effect was not due to a modulation of the PI3K/Akt pathway, as we did not detect variations in Akt phosphorylation in serum-starved A375 cells treated with inhibitors at the same concentration (Fig. S1B). While cells harbouring activating mutations in BRAF and NRAS were all sensitive to the RSK inhibitors, it is noteworthy that MEK1/2 inhibition resulted in a weaker inhibitory effect in NRAS cells.
Fig. 2.
Inhibition of ERK/RSK signalling decreases constitutive activation of mTORC1 in melanoma. (A) Four melanoma cell lines were used in this study. While A375 and Colo829 cells harbour a B-Raf V600E mutation, WM852 and WM1361 cells carry an N-Ras mutation at Q61 (R or K). (B) Phosphorylation of endogenous RSK, ERK1/2, rpS6, S6K1 and 4E-BP1 was monitored in total extracts from serum-starved melanoma cell lines and normal human melanocytes treated or not with insulin (100 nM) for 30 min. Cell lysates were also immunoblotted for total protein levels (RSK1, ERK1, rpS6, S6K1, 4E-BP1 and β-actin). (C) Serum-starved melanoma cells were treated with the indicated inhibitors for 60 minutes. Immunoprecipitated S6K1 kinase activity was assayed as in Fig. 1. (D) Phosphorylation of endogenous rpS6 and S6K1, and total rpS6 protein level were monitored by immunoblotting.
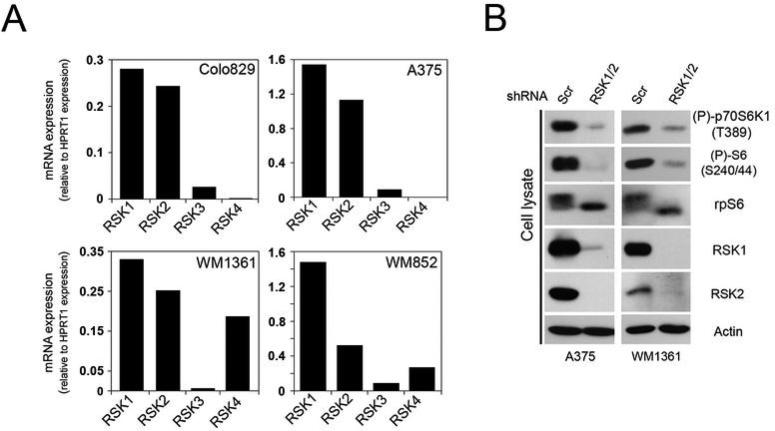
To validate the requirement for RSK activity, we used RNA interference (RNAi) to specifically silence RSK expression in melanoma cells. First, we determined the expression levels of all RSK isoforms and found that RSK1 and RSK2 were the most abundant isoforms at the mRNA level (Fig. 3A). To reduce RSK expression, we used two complementary approaches using small interfering RNA (siRNA)- or short hairpin (shRNA)-mediated RNAi. Using these approaches, we found that transient knockdown of RSK1/2 expression in Colo829 cells resulted in a significant inhibition of mTORC1 activity (Fig. S2). The role of the MAPK pathway was also verified using siRNA duplexes targeting ERK1/2 expression, which resulted in a similar inhibitory effect (Fig. S2). We also generated A375 and WM1361 stable cell lines expressing shRNAs targeting RSK1 and RSK2. When these cells were serum-starved overnight, we found that RSK knockdown significantly decreased mTORC1 activity (Fig. 3B). Similar results were obtained when depleting RSK in other melanoma cell lines, such as WM852 and SK-MEL-2 (data not shown). Together, these results demonstrate that endogenous RSK contributes to the regulation of mTORC1 signalling in melanoma cells.
Fig. 3.
RSK contributes to the constitutive activation of mTORC1 in melanoma. (A) Relative mRNA expression of the RSK isoforms in Colo829, A375, WM1361 and WM852 melanoma cell lines. Histograms show relative abundance of all RSK mRNAs monitored by quantitative real-time PCR and normalized to control HPRT1 mRNA abundance. (B) A375 and WM1361 cells stably expressing a control shRNA or shRNAs against RSK1/2 were serum-starved for 18 hours. Phosphorylation of endogenous rpS6 and S6K1, and total rpS6, RSK1, RSK2 and β-actin protein level were monitored by immunoblotting.
ERK/RSK signalling promotes mTORC1-mediated eIF4F assembly and mRNA translation in melanoma
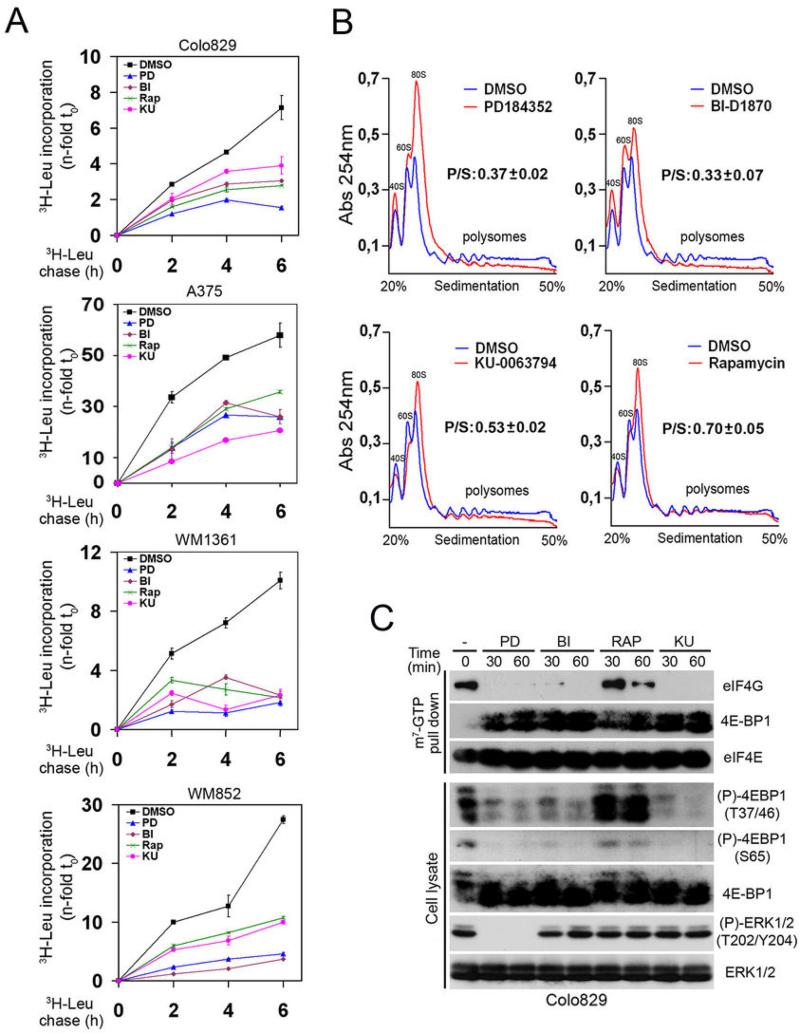
Next, we investigated whether ERK/RSK signalling regulates mTORC1-dependent protein synthesis in melanoma. First, cells were serum-starved and treated with MEK1/2 or RSK inhibitors prior to measuring tritiated leucine incorporation. Consistent with results from HEK293 cells, we found that ERK/RSK signalling was required for global protein synthesis in the four melanoma cell lines tested (Fig. 4A). To determine whether ERK/RSK signalling regulates mRNA translation, we examined polysome assembly using sucrose gradient sedimentation. Notably, we found that treatment of Colo829 cells with MEK1/2 or RSK inhibitors significantly decreased polysome assembly and proportionally increased the number of free ribosomes (Fig. 4B). Indeed, we found that inhibitor treatment reduced the polysome/subpolysome (P/S) ratio by ~65% compared to vehicle-treated cells (Fig. 4B and S3), suggesting that ERK/RSK signalling plays a major role in regulating mRNA translation in melanoma.
Fig. 4.
ERK/RSK signalling promotes mTORC1-mediated eIF4F assembly and translation in melanoma. (A) Serum-starved melanoma cells were treated with PD184352, BI-D1870, KU-0063794 or rapamycin (50 nM) for 60 minutes. Time course analysis of [3H]leucine incorporation was performed as in Fig. 1, except that cells were collected at 0, 2, 4 and 6 h after addition of [3H]leucine. (B) Serum-starved Colo829 cells were treated with indicated inhibitors as in (A). Cell extracts were size-fractionated by centrifugation through sucrose gradients (20-50%). The absorbance of polysomes (P) and subpolysomal (S) particles was continuously monitored at 260 nm. Representative A260 nm traces are shown (n = 3). The area under the curves was calculated and the P/S ratio refers to the percentage of ribosomes engaged in translation. The data are normalized to P/S ratio of control condition (DMSO) and presented as a mean ± S.E. (n = 3). (C) Association of 4E-BP1 and eIF4G to the 7-methylguanosine cap complex was monitored by immunoblotting 7-methylguanosine precipitates from serum-starved Colo829 cells, pre-treated with the indicated inhibitors for 60 minutes. Equal levels between precipitates were monitored through analysis of eIF4E. Phosphorylation and total protein level of endogenous 4E-BP1 and ERK1/2 were monitored by immunoblotting.
mTORC1 promotes mRNA translation by phosphorylating and inactivating the eIF4E-binding proteins (4E-BPs), resulting in their dissociation from eIF4E and assembly of the eIF4F complex (eIF4E, eIF4G and eIF4A). As with S6K1, we found that melanoma cells display high levels of 4E-BP1 phosphorylation (Fig. 2B and 4C), which reflects the constitutively high mTORC1 activity found in these cells (Fig. 2). To determine the role of ERK/RSK signalling in eIF4F assembly, we used 7-methylguanosine-conjugated sepharose beads to purify eIF4E and associated proteins from Colo829 cells. Interestingly, we found that treatment of cells with MEK1/2 or RSK inhibitors significantly increased the association of hypophosphorylated 4E-BP1 to eIF4E (Fig. 4C), suggesting that oncogenic ERK/RSK signalling promotes mTORC1-dependent phosphorylation of 4E-BP1 in melanoma cells. We observed that MEK1/2 and RSK inhibition dramatically decreased 4E-BP1 phosphorylation on three sites (Thr37/Thr46/Ser65) known to be regulated by mTORC1 (Fig. 4C), suggesting that RSK promotes eIF4F formation through the control of mTORC1-dependent 4E-BP1 phosphorylation. Consistent with this, we found that MEK1/2 and RSK inhibition robustly reduced eIF4G recruitment to eIF4E. As expected, inhibition of mTOR using KU-0063794 also reduced eIF4F complex formation, and as shown by others (36), we found that rapamycin treatment was less efficient than KU-0063794 to inhibit eIF4F formation (Fig. 4C). Together, these data demonstrate that ERK/RSK signalling plays an important role in mTORC1-dependent translation initiation in melanoma cells.
ERK/RSK signalling promotes translation of growth-related mRNAs in melanoma
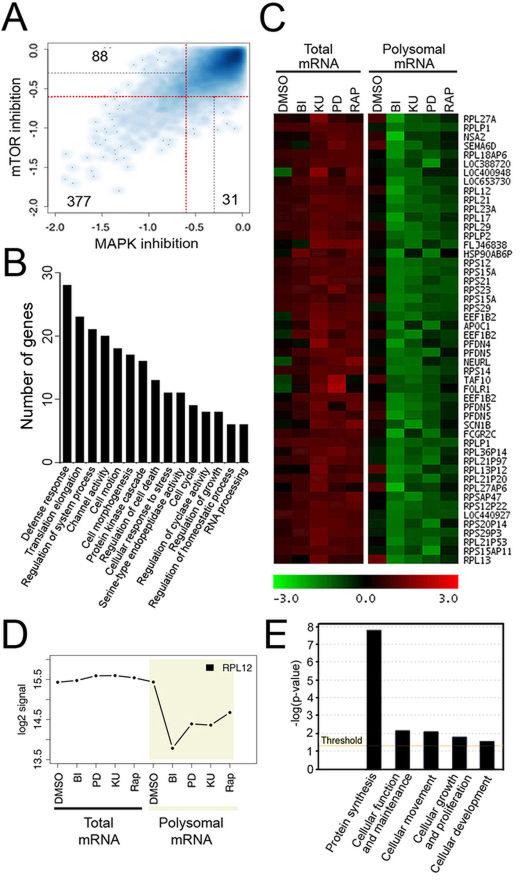
Next, we investigated the type of transcripts that were regulated by ERK/RSK signalling in melanoma cells using a global microarray-based approach. We compared total and polysome-bound RNAs to globally identify mRNAs that are actively being translated (polysomal mRNA) and normalize their levels to their overall abundance in the cell (total mRNA). Total and polysomal RNAs were extracted from Colo829 cells treated with MEK1/2 (PD184352), RSK (BI-D1870) or mTORC1 (KU-0063794 and rapamycin) inhibitors, and hybridized to human NimbleGen 60-mer oligonucleotides arrays. Cellular treatments were limited to one hour to minimize potential transcriptional effects of these drugs. Resulting data were analyzed and compared using several bioinformatics approaches (as described in Materials and Methods), and a statistically significant (p < 0.05) cut-off value of 1.5-fold (log2-coefficient=−0.58) decrease in the relative abundance of total or polysomal mRNA upon inhibitor treatment was used to designate specific regulation. Overall, the number of downregulated genes upon inhibition of the ERK/RSK pathway was greater in the polysomal RNA fraction compared with total RNAs (Fig. S4A), consistent with the idea that ERK/RSK signalling regulates mRNA translation (Fig. 4). As expected, we observed similar effects when blocking mTORC1 activity for one hour (Fig. S4A). Overall, we identified 577 genes that displayed at least a 1.5-fold decrease in the relative amount of polysomal mRNA as a function of MAPK or mTORC1 inhibition (Dataset S1). The vast majority of these genes (377) were regulated by both pathways, while 31 and 88 genes were specifically regulated by the MAPK and mTOR pathways, respectively (Fig. 5A). These results strongly support the notion that ERK/RSK signalling controls the translation of mTORC1-dependent mRNAs, and gene function enrichment analysis revealed that these mRNAs belong to growth-related biological functions, such as translation initiation, elongation, cellular response to stress or cell cycle (Fig. 5B).
Fig. 5.
ERK/RSK signalling promotes translation of growth-related mRNAs in melanoma. (A) Scatter plot of mRNAs log2-coefficients in polysomal mRNA from serum-starved Colo829 cells treated with mTOR (rapamycin or KU-0063794) or MAPK (BI-D1870 or PD184352) pathway inhibitors. Red and grey lines represent cutoffs of 1.5-fold (log2-coefficient=−0.58) and 1.25-fold (log2-coefficient=−0.32) decrease, respectively. mRNAs regulated by both types of inhibitors are found in the bottom-left quadrant delimited by two red lines whereas mRNAs regulated by only one inhibitor class are found at the top-left (MAPK) or bottom-right of the plot (mTOR). (B) Gene categories found as enriched by DAVID analysis in the list of downregulated mRNAs identified as translationally repressed by both mTOR and MAPK inhibitors (treatment effect<1.5-fold decrease and pvalue <0.05). (C) K-Means clustering of downregulated mRNAs in the presence of both mTOR and MAPK inhibitors led to the identification of a cluster whose mRNAs are translationally repressed. mRNAs and sample class are presented vertically and horizontally, respectively. Green and red represent respectively a relative decrease and a relative increase in mRNA abundance compared to the mRNA mean abundance. (D) Profile of a selected mRNA identified by clustering with log2 signal on the y-axis and the different sample classes on the x-axis. (E) Enrichment analysis using Ingenuity Pathways Analysis of mRNAs from identified clusters.
A search for genes having similar response patterns to MAPK and mTORC1 signalling was performed using K-Means clustering. Among identified clusters, two groups of mRNAs whose translation are repressed upon inhibitor treatment were identified (Fig. 5C and S4B). Enrichment analysis identified one predominant biological group in the cluster, namely, protein synthesis (Fig. 5E). Fifteen different ribosomal proteins, one translation initiation and two elongation factors were present within these groups (Fig. 5C, 5D and S4C), suggesting that many ERK/RSK-regulated mRNAs are involved in ribosome biogenesis or encode components of the translational machinery. A common feature of RNAs encoding components of the translational machinery is the presence in their 5’ untranslated region (UTR) of a terminal oligopyrimidine (TOP) motif that is known to be associated with mTORC1-dependent translation regulation (37). Strikingly, one cluster was found to be significantly enriched in mRNAs that have been previously shown to contain a 5' TOP motif (22 out of 50 mRNAs, Fisher exact test p-value=1.555e-12)(38). Many of these mRNAs have previously been shown to be regulated by mTOR and/or rapamycin treatment, such as eEF1b and Rpl12 (39). Together, these data highly suggest that ERK/RSK signalling converges on mTORC1 to regulate the translation of mRNAs involved in melanoma cell growth.
RSK is required for efficient melanoma cell proliferation and tumour development in mice
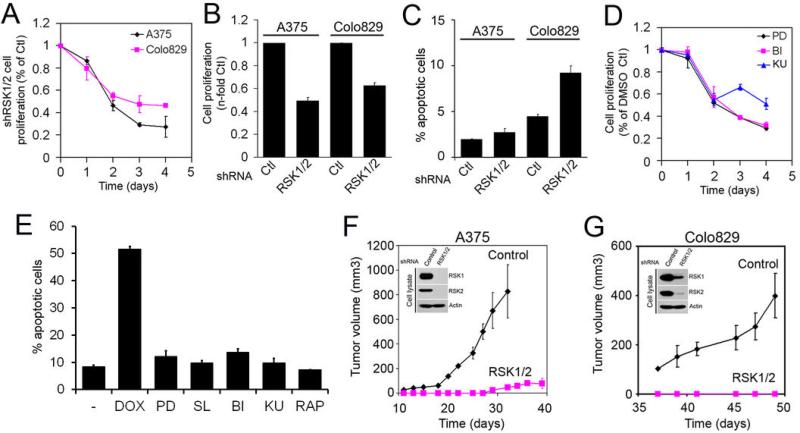
To determine the biological function of ERK/RSK signalling in melanoma, we next evaluated the impact of knocking down RSK1/2 on melanoma cell proliferation. Depletion of RSK1/2 by shRNA-mediated RNAi in serum-growing Colo829 and A375 cells resulted in a significant decrease in proliferation rates, as shown by measuring global metabolic activity (Fig. 6A) or using cell counts (Fig. 6B). Because RSK was previously shown to inhibit apoptotic cell death (21, 40), we determined whether RSK1/2 knockdown increased Annexin V binding. Our results indicate that the number of apoptotic cells was significantly increased by RSK knockdown (Fig. 6C), but that these changes could not entirely explain the large reduction in cell proliferation (Fig. 6A and 6B). Using pharmacological inhibitors, we also found that inhibition of ERK/RSK signalling reduced melanoma cell proliferation to levels similar to cells treated with mTOR inhibitors (Fig. 6D), and again, these treatments did not significantly increased apoptosis of melanoma cells (Fig. 6E).
Fig. 6.
RSK is essential for melanoma cell proliferation and tumour development in mice. (A) A375 and Colo829 cells stably expressing control vector or shRNA against RSK1/2 were grown in culture medium containing 5% FBS. The relative number of viable cells was measured during four days using an MTS assay. The data are normalized to control vector (Ctl). (B) As in (A), except that after 72 h of culture, the relative number of viable cells was measured by cell counts. (C) Graph showing the relative number of apoptotic cells after 48 h of culture in serum-growing conditions, as measured by FACS analysis of Annexin V binding. (D) As in (A), except that A375 cells were grown in the presence of indicated inhibitors. (E) As in (C), except that A375 cells were treated with the indicated inhibitor for 24 h prior to FACS analysis. As positive control, cells were treated with 0.5 μM doxorubicin (DOX). (F) and (G) A375 and Colo829 cells stably expressing a control shRNA or shRNAs against RSK1/2 were injected subcutaneously into the flanks of athymic mice. Mice were monitored for tumour development and graphs represent the growth rate of subcutaneous tumours. Values represent the average volume +/− SEM of 3 to 5 tumours (3 to 5 mice). Insets: Endogenous RSK1, RSK2 and β-actin protein levels in injected cells were monitored by immunoblotting.
Finally, we investigated the role of RSK in melanoma tumour growth using a xenotransplantation assay in nude mice. For this, A375 or Colo829 cells were infected with lentiviruses expressing RSK1/2 shRNAs and populations of transduced cells (2 × 106 cells) were injected subcutaneously in the flanks of nude mice. As shown in Fig. 6F and 6G, A375 and Colo829 cells expressing a control shRNA rapidly formed tumours in mice, reaching 0.5 cm3 within ~30-50 days of injection. In sharp contrast, depletion of RSK1/2 almost completely inhibited tumour formation from A375 and Colo829 cells, indicating that RSK is essential for melanoma tumour growth in vivo. We monitored RSK1/2 expression in xenografts from A375 cells obtained two months post-injection (Fig. 6F), and found that these tumours recovered RSK expression to almost wild-type levels. Together, our data indicate that ERK/RSK signalling contributes to the constitutively high mTORC1 activity in melanoma cells, and that RSK inhibition abrogates tumour development in mice.
Discussion
The data presented here demonstrate that oncogenic MAPK signalling promotes constitutive mTORC1 activation and downstream signalling to its main targets, S6K1 and 4E-BP1 (Fig. 1-4). Further, our results indicate that the ERK1/2-activated protein kinase RSK plays an important role in this process, as RSK inhibitors and RNAi strongly reduced mTORC1 activity in melanoma (Fig. 2, 3 and 4). Consistently, we found that ERK/RSK signalling regulates the translation of growth-related mRNAs also controlled by the mTOR pathway (Fig. 5), supporting the notion that oncogenic ERK/RSK signalling promotes mTORC1-dependent functions. RSK depletion was found to inhibit melanoma cell proliferation and tumour growth (Fig. 6), indicating that melanoma cells are highly dependent on ERK/RSK signalling for their growth and proliferation.
Translational regulation plays a pivotal role in tumour formation and metastasis (41, 42). Accordingly, our data indicate that RSK controls melanoma cell growth by modulating the phosphorylation status of important components of the translational machinery. Particularly, the inhibitory activity of 4E-BP1 was found to be highly dependent on ERK/RSK signalling, as inhibition of MEK1/2 and RSK dramatically decreased eIF4F complex formation and polysome formation. These results suggest that 4E-BP1 plays an important role in mediating MAPK-dependent melanoma cell growth. This is supported by observations that depletion of Raptor in melanoma cells is lethal (data not shown), and by studies showing that tumours in which 4E-BP1 expression is reduced are less addicted to oncogenic MAPK signalling (43). Our findings are in a good agreement with recent studies demonstrating that 4E-BP1 is a major integrator of oncogenic signalling pathways (44) and that mTORC1-dependent cell proliferation is mediated by the 4E-BPs (45).
Although we have shown that ERK/RSK signalling regulates cell growth via mTORC1, we cannot exclude additional modes of translational regulation. Indeed, rapamycin was found to be less effective than RSK inhibitors in reducing global protein synthesis (Fig. 4 and 6), suggesting that ERK/RSK signalling regulates additional proteins involved in mRNA translation. Consistent with this, RSK was previously shown to phosphorylate the translation initiation factor eIF4B and thereby stimulate its recruitment to the translation initiation complex (46). The ribosomal protein S6 has also been shown to be directly phosphorylated by RSK at Ser235/236 (47), but the exact outcome of this phosphorylation event remains elusive. Therefore, ERK/RSK signalling appears to participate at several levels of translational control, which may explain the acute dependency of melanoma cells to ERK/RSK activity. Consistent with these findings, ERK and RSK were recently shown to regulate LKB1 (48), a tumour suppressor protein that is involved in the activation of the AMP-activated protein kinase (AMPK). ERK- and RSK-mediated phosphorylation of LKB1 was shown to be important for cell proliferation (48), suggesting that several ERK/RSK-dependent mechanisms may contribute to the regulation of melanoma growth.
Our data demonstrate a molecular link between oncogenic MAPK signalling and mTOR-dependent growth in melanoma, and indicate that RSK may be a valuable therapeutic target for the treatment of metastatic melanomas, as well as other cancer types characterized by the hyperactivation of the MAPK pathway. RSK inhibitors could also be used in combination with drugs that target the PI3K pathway, as was done with MEK1/2 inhibitors (49), which may inhibit mTORC1 more efficiently in tumours that also display activating mutations in PI3K pathway components.
Material and Methods
Cell Culture and treatments
The cell lines used in this study were purchased from ATCC (A375, Colo829 and SK-MEL-2) or Coriell (WM1361 and WM852). The human epidermal melanocytic (105-25N) cell line was purchased from Cell Applications (San Diego, CA) and grown in Melanocyte Growth Medium (135-500, Cell Applications). HEK293, A375 and SK-MEL-2 cells were grown in DMEM supplemented with 10% foetal bovine serum (FBS) and antibiotics. Colo829 cells were grown in RPMI medium supplemented with 10% FBS and antibiotics. WM1361 and WM852 were maintained in (4:1) MCDB 153:Leibovitz L-15 media supplemented with 2% FBS, 1.69 mM CaCl2, 5 μg/ml insulin and antibiotics. HEK293 stable cell lines were generated using pBabe-puro-derived retroviral particles, and expressing cells were selected using puromycin (2 μg/ml). Cells were serum-starved overnight and treated with agonists/inhibitors as indicated. Starved cells were pre-treated with U0126 (20 μM), PD184352 (10 μM), SL0101 (50 μM), BI-D1870 (10 μM), KU-0063794 (5 μM) or rapamycin (25 nM, except when indicated) (Biomol, Plymouth Meeting, PA).
Antibodies
Antibodies targeted against phospho-rpS6 (Ser240/244 and Ser235/236), phospho-Akt (Ser473), ERK1/2, phospho-4EBP1 (Thr37/46), S6K1 and phospho-S6K1 (Thr389) were purchased from Cell Signaling Technologies (Beverly, MA). Anti-phospho-ERK1/2 monoclonal antibodies were purchased from Sigma-Aldrich (Oakville, ON). Anti-RSK1 and RSK2 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Invitrogen (Carlsbad, CA), respectively. The phospho-RSK (Ser380) antibody was purchased from R&D Systems (Minneapolis, MN). The phospho-4EBP1 Ser65 antibody was kindly provided by Dr. Nahum Sonenberg (McGill University). All secondary horseradish peroxidase (HRP)-conjugated antibodies used for immunoblotting were purchased from Chemicon (Temecula, CA).
Immunoprecipitations, Cap Binding Assays and Immunoblotting
Cell lysates were prepared as described elsewhere (50). Briefly, cells were washed with ice-cold PBS and lysed in 10 mM K3PO4, 1 mM EDTA, 5 mM EGTA, 10 mM MgCl2, 50 mM β-glycerophosphate, 0.5% NP-40, 0.1% Brij 35, 0.1% deoxycholic acid, 1 mM sodium orthovanadate (Na3VO4), 1 mM phenylmethylsulfonyl fluoride, and a complete protease inhibitor cocktail tablet (Roche). For immunoprecipitations, cell lysates were incubated with the indicated antibody for 2 h, followed by 1 h of incubation with protein A-Sepharose CL-4B beads (GE Healthcare). For cap binding assays, lysates were incubated with 7-methyl-GTP Sepharose (GE Healthcare) for 2 h. Immunoprecipitates were washed thrice in lysis buffer and eluted in reducing sample buffer. Eluates and total cell lysates were subjected to SDS-PAGE, and resolved proteins were transferred onto polyvinylidene difluoride (PVDF) membranes for immunoblotting.
Protein Phosphotransferase Assays
For S6K1 and RSK1 assays, beads from immunoprecipitations were washed twice in lysis buffer and twice in kinase buffer (25 mM Tris-HCl [pH 7.4], 10 mM MgCl2, 5 mM β-glycerophosphate). Kinase assays were performed with bacterially purified recombinant GST-S6 fusion proteins (3 μg per assay) as substrate, under linear assay conditions. Assays were performed for 10 min at 30°C in kinase buffer supplemented with 5 μCi [γ-32P]ATP. All samples were subjected to SDS-PAGE, and incorporation of radioactive [32P]phosphate was determined by autoradiography. The data presented are representative of at least three independent experiments.
RNA Interference and Viral Infections
For small interfering RNA (siRNA)-mediated knockdown of RSK1, RSK2, ERK1 and ERK2, validated 21-nucleotide cRNAs with symmetrical two nucleotide overhangs were obtained from Qiagen (Valencia, CA). Colo829 cells were transfected using Oligofectamine (Invitrogen) and 50 nM siRNA per 35-mm dish. At 24 hours following transfection, cells were serum starved overnight before being harvested. For short hairpin RNA (shRNA)-mediated knockdown, lentiviruses were produced using vectors from the Mission TRC shRNA library. Cells were infected in the presence of 4 μg/ml polybrene, and 2 days after viral infection, cells were treated and selected with 2 μg/ml puromycin. shRNA constructs were obtained from Sigma-Aldrich (shRSK1, TRCN470; shRSK2, TRCN537). Retroviruses were produced using the pBabe-puro vector to overexpress ectopic RasG12V, MEK1-DD or B-Raf-V600E mutants. Two days after infection, positive pools were selected in 2 μg/ml puromycin. Plasmids encoding HA-tagged RasG12V, B-Raf-V600E and flag-tagged MEK1-DD were previously described (50).
Protein Synthesis Measurements
Cells were seeded in 6-well plates for 24h then serum starved or maintained in low-serum (2%) medium for 16 to 18 h. After treatment with indicated inhibitors for 1 h, 0.5 μCi/ml [3H]leucine was added to the medium. At indicated times of labeling, the medium was aspirated and the cells were incubated 30 min in cold 5% TCA. The wells were then washed once with 5% TCA and three times with tap water. The radioactivity incorporated into TCA-precipitable material was measured by liquid scintillation counting after solubilization in 0.1 M NaOH.
Polysomal Fractionation and RNA Extraction
Sucrose density gradient centrifugation was employed to separate the subpolysomal from the polysomal ribosome fractions. Fifteen minutes before harvest, 100 μg/ml cycloheximide was added to the culture medium. Cells were washed in ice-cold PBS supplemented with 100 μg/ml cycloheximide, and harvested in polysome lysis buffer (PLB; 50 mm HEPES [pH 7.4], 250 mm KCl, 5 mm MgCl2, 250 mm sucrose, 1% Triton X-100, 1.3% sodium deoxycholate, 100 μg/ml cycloheximide, and protease inhibitors). Samples were incubated on ice for 15 min and then centrifuged at 10,000 × g for 10 min at 4 °C. The resulting supernatant was layered on a 20–50% linear sucrose gradient (in PLB) and centrifuged in a SW40 rotor at 145,000 × g for 165 min at 4 °C. Following centrifugation, the A260 was continuously monitored and recorded using a Gradient Station IP (Biocomp, Fredericton, NB) attached to a UV-MII (GE Healthcare) spectrophotometer. Polysomal fractions were collected and RNA was extracted using RNeasy Mini Kit (Qiagen). For RT-qPCR experiments, total RNA was extracted using the same procedure.
Quantitative Real-Time PCR and Microarray Experiments
Total RNA was reverse transcribed using the cDNA Reverse Transcription Kit (Applied Biosystems, Carlsbad, CA) as described by the manufacturer. Gene expression levels of endogenous controls GAPDH and ACTB were determined using pre-validated Taqman Gene Expression Assays (Applied Biosystems), and gene expression level for RSK isoforms was determined using assays designed with the Universal Probe Library (UPL) from Roche. Microarray experiments were performed in triplicate using Nimblegen Human Gene Expression 4x72K Arrays according to the manufacturer's instructions (Roche NimbleGen, Madison, WI). For each condition, 6.5 μg of polysomal or total RNA was reverse transcribed and labeled using Cy3 (Trilink Biotechnology). The labeled material was then hybridized to the microarray overnight at 42°C, washed and scanned at 5 μm resolution.
Microarray Data Analysis
Data was quantified and Robust Multichip Average (RMA) normalized using NimbleScan software. Normalized data was log2-transformed and analyzed using Bioconductor packages and R statistical language. Using the R package limma, we fit linear models to identify genes regulated between different conditions. A batch effect term was added to the model to account for experimental bias and an interaction term allowed the identification of translational regulation independent of cytosolic mRNA level change. Genes having log2-coefficients smaller than −0.58 (1.5 fold decrease) and p-value smaller than 0.05 were considered as significantly regulated. The replicate average log2- signals of genes downregulated in polysomal mRNA in the presence of mTOR and MAPK inhibitors were used as input for K-Means clustering using MeV Software. Enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) and through the use of Ingenuity Pathways Analysis (Ingenuity® Systems).
Proliferation and Apoptosis Assay
For proliferation assays, 2×103 cells were seeded in 96-well plates. Cells were grown in the presence of inhibitors when indicated. The relative number of viable cells was measured every 24 h during four consecutive days using a nonradioactive cell proliferation assay from Promega (Madison, WI). For cell counting, 5×104 cells were seeded in 6-well plates and grown as described above. After 48h, cells were stained with trypan blue and counted using a hemacytometer. Results displayed represent the mean of triplicates ± standard error (SE). For analysis of apoptotic cells, 3 × 105 A375 cells were seeded in 35-mm dishes and grown in 10% FBS for 48 h. After trypsination, cells were washed twice with ice cold PBS containing 5 mM CaCl2 and incubated with PE labeled annexin V (BD Pharmingen). Cells were washed twice and processed on a BD FACS Canto II cytometer.
Mouse Xenotransplantation Studies
All animals were housed under pathogen-free conditions, and experiments were performed in accordance with CCAC guidelines. Female Balb/c athymic nude mice (nu/nu) were purchased from Harlan and used at 6–8 weeks of age. For subcutaneous tumor model studies, A375 or Colo829 cells stably expressing shRSK1 and shRSK2 were harvested from sub-confluent cultures by brief exposure to 0.25% trypsin and 0.02% EDTA. The cells were washed once in PBS, and 2 × 106 cells in a final volume of 200 μl were injected subcutaneously in the flanks of the mouse. The mice were monitored regularly and the tumors were measured every 2–3 days using a caliper.
Supplementary Material
Fig. S1. RSK is constitutively activated in melanoma cell lines. (A) Phosphorylation of endogenous RSK at S380 and ERK1/2 at T202 and Y204 were monitored in total extracts from melanoma cell lines pre-treated with U0126 (20 μM) or PD184352 (20 μM) for 60 minutes. The cell lysates were also immunoblotted for total protein levels (RSK1, ERK1/2). (B) Phosphorylation of Akt was monitored in serum-starved A375 cells treated with U0126 (U0), PD184352 (PD), SL0101 (SL), BI-D1870 (BI) or rapamycin (RAP). Following inhibitor treatment for 1h, cell lysates were immunoblotted as in (A).
Fig. S2. Colo829 cells were transiently transfected with a control siRNA or siRNAs targeting RSK1/2 or ERK1/2. After 48 h, cells were serum-starved overnight and phosphorylation of endogenous rpS6 was monitored. Cell lysates were also immunoblotted for total protein levels (rpS6, RSK1, RSK2, Actin).
Fig. S3. RSK regulates polysome assembly in melanoma. Colo829 cells were serum-starved for 18 hours and treated with the indicated inhibitors for 60 minutes. Cell extracts were size-fractionated and the absorbance of polysomes (P) and subpolysomal (S) particles was monitored at 260 nm. The area under the curves was calculated and the P/S ratio refers to the percentage of ribosomes engaged in translation. The data are normalized to P/S ratio of control condition (DMSO) and presented as a mean ± S.E. (n = 3). * p < 0.05, ** p < 0.01, relative to control.
Fig. S4. RSK inhibition impairs the translation of specific mRNAs. (A) The histogram shows the number of genes that displayed 1.5-fold decrease (log2-coefficient=−0.6) in transcription (Total mRNA) or in translation (Polysomal mRNA) upon inhibitor treatment compared to the control condition. (B) K-Means clustering of downregulated mRNAs in the presence of both mTOR and MAPK inhibitors led to the identification of a second cluster whose mRNAs are translationally repressed. mRNAs and sample class are presented vertically and horizontally, respectively. Green and red represent respectively a relative decrease and a relative increase in mRNA abundance compared to the mRNA mean abundance. (C) Profile of selected mRNAs identified by clustering with log2 signal on the y-axis and the different sample classes on the x-axis.
Acknowledgments
Grant support
This work was supported by grants from the Canadian Cancer Society Research Institute (P.P.R.), the Cancer Research Society (P.P.R and S.M.), and the Natural Sciences and Engineering Research Council of Canada (P.P.R.). We thank Dr. Nahum Sonenberg for the phospho-4E-BP1 antibody. P.P. Roux holds a Canada Research Chair in Signal Transduction and Proteomics and a Career Development Award from the Human Frontier Science Program (HFSP). K. Borden and S. Meloche hold Canada Research Chairs in Molecular Biology of the Cell Nucleus, and in Cellular Signalling, respectively. IRIC core facilities are supported by the FRSQ.
Footnotes
Disclosure of potential conflict of interest
The authors declare no conflict of interest.
References
- 1.Murphy LO, Blenis J. MAPK signal specificity: the right place at the right time. Trends Biochem Sci. 2006 May;31(5):268–75. doi: 10.1016/j.tibs.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Ballif BA, Blenis J. Molecular mechanisms mediating mammalian mitogen-activated protein kinase (MAPK) kinase (MEK)-MAPK cell survival signals. Cell Growth Differ. 2001 Aug;12(8):397–408. [PubMed] [Google Scholar]
- 3.Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007 Apr;7(4):295–308. doi: 10.1038/nrc2109. [DOI] [PubMed] [Google Scholar]
- 4.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, et al. Mutations of the BRAF gene in human cancer. Nature. 2002 Jun 27;417(6892):949–54. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 5.Fecher LA, Amaravadi RK, Flaherty KT. The MAPK pathway in melanoma. Curr Opin Oncol. 2008 Mar;20(2):183–9. doi: 10.1097/CCO.0b013e3282f5271c. [DOI] [PubMed] [Google Scholar]
- 6.Gray-Schopfer VC, da Rocha Dias S, Marais R. The role of B-RAF in melanoma. Cancer Metastasis Rev. 2005 Jan;24(1):165–83. doi: 10.1007/s10555-005-5865-1. [DOI] [PubMed] [Google Scholar]
- 7.Miller AJ, Mihm MC., Jr. Melanoma. N Engl J Med. 2006 Jul 6;355(1):51–65. doi: 10.1056/NEJMra052166. [DOI] [PubMed] [Google Scholar]
- 8.Scheier B, Amaria R, Lewis K, Gonzalez R. Novel therapies in melanoma. Immunotherapy. 2011 Dec;3(12):1461–9. doi: 10.2217/imt.11.136. [DOI] [PubMed] [Google Scholar]
- 9.Meloche S, Pouyssegur J. The ERK1/2 mitogen-activated protein kinase pathway as a master regulator of the G1- to S-phase transition. Oncogene. 2007 May 14;26(22):3227–39. doi: 10.1038/sj.onc.1210414. [DOI] [PubMed] [Google Scholar]
- 10.Servant MJ, Giasson E, Meloche S. Inhibition of growth factor-induced protein synthesis by a selective MEK inhibitor in aortic smooth muscle cells. J Biol Chem. 1996 Jul 5;271(27):16047–52. doi: 10.1074/jbc.271.27.16047. [DOI] [PubMed] [Google Scholar]
- 11.Rajalingam K, Schreck R, Rapp UR, Albert S. Ras oncogenes and their downstream targets. Biochim Biophys Acta. 2007 Aug;1773(8):1177–95. doi: 10.1016/j.bbamcr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Waskiewicz AJ, Flynn A, Proud CG, Cooper JA. Mitogen-activated protein kinases activate the serine/threonine kinases Mnk1 and Mnk2. Embo J. 1997 Apr 15;16(8):1909–20. doi: 10.1093/emboj/16.8.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Waskiewicz AJ, Johnson JC, Penn B, Mahalingam M, Kimball SR, Cooper JA. Phosphorylation of the cap-binding protein eukaryotic translation initiation factor 4E by protein kinase Mnk1 in vivo. Mol Cell Biol. 1999 Mar;19(3):1871–80. doi: 10.1128/mcb.19.3.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cargnello M, Roux PP. Activation and Function of the MAPKs and Their Substrates, the MAPK-Activated Protein Kinases. Microbiol Mol Biol Rev. 2011 Mar;75(1):50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buxade M, Parra-Palau JL, Proud CG. The Mnks: MAP kinase-interacting kinases (MAP kinase signal-integrating kinases). Front Biosci. 2008;13:5359–73. doi: 10.2741/3086. [DOI] [PubMed] [Google Scholar]
- 16.Ueda T, Watanabe-Fukunaga R, Fukuyama H, Nagata S, Fukunaga R. Mnk2 and Mnk1 are essential for constitutive and inducible phosphorylation of eukaryotic initiation factor 4E but not for cell growth or development. Mol Cell Biol. 2004 Aug;24(15):6539–49. doi: 10.1128/MCB.24.15.6539-6549.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furic L, Rong L, Larsson O, Koumakpayi IH, Yoshida K, Brueschke A, et al. eIF4E phosphorylation promotes tumorigenesis and is associated with prostate cancer progression. Proc Natl Acad Sci U S A. 2010 Aug 10;107(32):14134–9. doi: 10.1073/pnas.1005320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaestel M. Specificity of signaling from MAPKs to MAPKAPKs: kinases' tango nuevo. Front Biosci. 2008;13:6050–9. doi: 10.2741/3136. [DOI] [PubMed] [Google Scholar]
- 19.Romeo Y, Zhang X, Roux PP. Regulation and function of the RSK family of protein kinases. Biochem J. 2012 Jan 15;441(2):553–69. doi: 10.1042/BJ20110289. [DOI] [PubMed] [Google Scholar]
- 20.Carriere A, Ray H, Blenis J, Roux PP. The RSK factors of activating the Ras/MAPK signaling cascade. Front Biosci. 2008;13:4258–75. doi: 10.2741/3003. [DOI] [PubMed] [Google Scholar]
- 21.Anjum R, Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat Rev Mol Cell Biol. 2008 Oct;9(10):747–58. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 22.Romeo Y, Roux PP. Paving the way for targeting RSK in cancer. Expert Opin Ther Targets. 2011 Jan;15(1):5–9. doi: 10.1517/14728222.2010.531014. [DOI] [PubMed] [Google Scholar]
- 23.Old WM, Shabb JB, Houel S, Wang H, Couts KL, Yen CY, et al. Functional proteomics identifies targets of phosphorylation by B-Raf signaling in melanoma. Mol Cell. 2009 Apr 10;34(1):115–31. doi: 10.1016/j.molcel.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005 Apr 22;121(2):179–93. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 25.Roux PP, Ballif BA, Anjum R, Gygi SP, Blenis J. Tumor-promoting phorbol esters and activated Ras inactivate the tuberous sclerosis tumor suppressor complex via p90 ribosomal S6 kinase. Proc Natl Acad Sci U S A. 2004 Sep 14;101(37):13489–94. doi: 10.1073/pnas.0405659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010 Oct 22;40(2):310–22. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caron E, Ghosh S, Matsuoka Y, Ashton-Beaucage D, Therrien M, Lemieux S, et al. A comprehensive map of the mTOR signaling network. Mol Syst Biol. 2011 Dec 21;6:453. doi: 10.1038/msb.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006 Feb 10;124(3):471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Jacinto E. What controls TOR? IUBMB Life. 2008 Aug;60(8):483–96. doi: 10.1002/iub.56. [DOI] [PubMed] [Google Scholar]
- 30.Foster KG, Fingar DC. Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J Biol Chem. 2010 May 7;285(19):14071–7. doi: 10.1074/jbc.R109.094003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009 May;10(5):307–18. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 32.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009 Feb 20;136(4):731–45. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma XM, Yoon SO, Richardson CJ, Julich K, Blenis J. SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell. 2008 Apr 18;133(2):303–13. doi: 10.1016/j.cell.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 34.Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2008 Dec;27(Suppl 2):S43–51. doi: 10.1038/onc.2009.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karbowniczek M, Spittle CS, Morrison T, Wu H, Henske EP. mTOR is activated in the majority of malignant melanomas. J Invest Dermatol. 2008 Apr;128(4):980–7. doi: 10.1038/sj.jid.5701074. [DOI] [PubMed] [Google Scholar]
- 36.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2(67):pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 37.Meyuhas O, Dreazen A. Chapter 3 Ribosomal Protein S6 Kinase From TOP mRNAs to Cell Size. Prog Mol Biol Transl Sci. 2009;90:109–53. doi: 10.1016/S1877-1173(09)90003-5. [DOI] [PubMed] [Google Scholar]
- 38.Yamashita R, Suzuki Y, Takeuchi N, Wakaguri H, Ueda T, Sugano S, et al. Comprehensive detection of human terminal oligo-pyrimidine (TOP) genes and analysis of their characteristics. Nucleic Acids Res. 2008 Jun;36(11):3707–15. doi: 10.1093/nar/gkn248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bilanges B, Argonza-Barrett R, Kolesnichenko M, Skinner C, Nair M, Chen M, et al. Tuberous sclerosis complex proteins 1 and 2 control serum-dependent translation in a TOP-dependent and -independent manner. Mol Cell Biol. 2007 Aug;27(16):5746–64. doi: 10.1128/MCB.02136-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisenmann KM, VanBrocklin MW, Staffend NA, Kitchen SM, Koo HM. Mitogen-activated protein kinase pathway-dependent tumor-specific survival signaling in melanoma cells through inactivation of the proapoptotic protein bad. Cancer Res. 2003 Dec 1;63(23):8330–7. [PubMed] [Google Scholar]
- 41.Bilanges B, Stokoe D. Mechanisms of translational deregulation in human tumors and therapeutic intervention strategies. Oncogene. 2007 Sep 6;26(41):5973–90. doi: 10.1038/sj.onc.1210431. [DOI] [PubMed] [Google Scholar]
- 42.Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006 Oct 16;25(48):6416–22. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- 43.O'Reilly KE, Warycha M, Davies MA, Rodrik V, Zhou XK, Yee H, et al. Phosphorylated 4E-BP1 is associated with poor survival in melanoma. Clin Cancer Res. 2009 Apr 15;15(8):2872–8. doi: 10.1158/1078-0432.CCR-08-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, et al. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010 Jul 13;18(1):39–51. doi: 10.1016/j.ccr.2010.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010 May 28;328(5982):1172–6. doi: 10.1126/science.1187532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shahbazian D, Roux PP, Mieulet V, Cohen MS, Raught B, Taunton J, et al. The mTOR/PI3K and MAPK pathways converge on eIF4B to control its phosphorylation and activity. Embo J. 2006 Jun 21;25(12):2781–91. doi: 10.1038/sj.emboj.7601166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007 May 11;282(19):14056–64. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng B, Jeong JH, Asara JM, Yuan YY, Granter SR, Chin L, et al. Oncogenic B-RAF negatively regulates the tumor suppressor LKB1 to promote melanoma cell proliferation. Mol Cell. 2009 Jan 30;33(2):237–47. doi: 10.1016/j.molcel.2008.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay R, et al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med. 2008 Dec;14(12):1351–6. doi: 10.1038/nm.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carriere A, Cargnello M, Julien LA, Gao H, Bonneil E, Thibault P, et al. Oncogenic MAPK signaling stimulates mTORC1 activity by promoting RSK-mediated raptor phosphorylation. Curr Biol. 2008 Sep 9;18(17):1269–77. doi: 10.1016/j.cub.2008.07.078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. RSK is constitutively activated in melanoma cell lines. (A) Phosphorylation of endogenous RSK at S380 and ERK1/2 at T202 and Y204 were monitored in total extracts from melanoma cell lines pre-treated with U0126 (20 μM) or PD184352 (20 μM) for 60 minutes. The cell lysates were also immunoblotted for total protein levels (RSK1, ERK1/2). (B) Phosphorylation of Akt was monitored in serum-starved A375 cells treated with U0126 (U0), PD184352 (PD), SL0101 (SL), BI-D1870 (BI) or rapamycin (RAP). Following inhibitor treatment for 1h, cell lysates were immunoblotted as in (A).
Fig. S2. Colo829 cells were transiently transfected with a control siRNA or siRNAs targeting RSK1/2 or ERK1/2. After 48 h, cells were serum-starved overnight and phosphorylation of endogenous rpS6 was monitored. Cell lysates were also immunoblotted for total protein levels (rpS6, RSK1, RSK2, Actin).
Fig. S3. RSK regulates polysome assembly in melanoma. Colo829 cells were serum-starved for 18 hours and treated with the indicated inhibitors for 60 minutes. Cell extracts were size-fractionated and the absorbance of polysomes (P) and subpolysomal (S) particles was monitored at 260 nm. The area under the curves was calculated and the P/S ratio refers to the percentage of ribosomes engaged in translation. The data are normalized to P/S ratio of control condition (DMSO) and presented as a mean ± S.E. (n = 3). * p < 0.05, ** p < 0.01, relative to control.
Fig. S4. RSK inhibition impairs the translation of specific mRNAs. (A) The histogram shows the number of genes that displayed 1.5-fold decrease (log2-coefficient=−0.6) in transcription (Total mRNA) or in translation (Polysomal mRNA) upon inhibitor treatment compared to the control condition. (B) K-Means clustering of downregulated mRNAs in the presence of both mTOR and MAPK inhibitors led to the identification of a second cluster whose mRNAs are translationally repressed. mRNAs and sample class are presented vertically and horizontally, respectively. Green and red represent respectively a relative decrease and a relative increase in mRNA abundance compared to the mRNA mean abundance. (C) Profile of selected mRNAs identified by clustering with log2 signal on the y-axis and the different sample classes on the x-axis.