Abstract
Antibodies to DNA (anti-DNA) are the serological hallmark of systemic lupus erythematosus and markers of underlying immune system disturbances. These antibodies bind to both single-stranded and double-stranded DNA, mediating pathogenesis by forming immune complexes. As shown recently, DNA in blood exists in both free and particulate forms, with DNA representing an important component of microparticles. Microparticles are membrane-bound vesicles containing nuclear molecules, released by membrane blebbing during cell death and activation. A panel of monoclonal NZB/NZW F1 anti-DNA antibodies was tested for binding to microparticles generated from apoptotic THP-1 and Jurkat cells. These studies showed that only certain anti-DNA antibodies in the panel, specific for double-stranded DNA, bound to microparticles. Binding to particles was reduced by soluble DNA or DNase treatment. Together, these results indicate that particle binding is a feature of only certain anti-DNA antibodies, reflecting immunochemical properties of the antibodies and the nature of the exposed DNA antigens.
Keywords: Microvesicle, anti-DNA, lupus, autoimmunity, apoptosis
1. Introduction
Antibodies to DNA (anti-DNA) are the serological hallmark of systemic lupus erythematosus (SLE), a prototypic autoimmune disease characterized by generalized immune system disturbances in association with tissue inflammation and injury [1, 2]. As shown with antibodies from patients as well as murine models, anti-DNA antibodies can bind to antigenic sites on both double stranded (ds) and single stranded (ss) DNA although antibodies to dsDNA are most closely related to disease pathogenesis [3–5]. In addition to their role of serological markers, anti-DNA antibodies represent important effector elements in disease and can mediate tissue inflammation, especially nephritis, by the formation of immune complexes; these complexes can also stimulate cytokine production by plasmacytoid dendritic cells via activation of internal nucleic acid sensors, including toll-like receptor 9 (TLR9) [6–9]. Because of the close association of anti-DNA antibodies with disease manifestations in many patients, defining mechanisms for the formation of immune complexes addresses fundamental issues in the etiology of lupus.
While the molecular properties of anti-DNA antibodies have been intensely investigated, much less is known about the actual form of DNA that contacts the immune system in vivo during normal or aberrant immunity. Importantly, to stimulate autoantibody responses, form immune complexes or promote immunological danger in innate immunity, DNA must leave the cell. Current evidence indicates that this translocation event is a prominent feature of cell death which can occur by a variety of mechanisms characterized by the role of different enzyme cascades which can affect the integrity of DNA as well as lead to post-translation modification of histones and other binding molecules [10, 11]. In lupus, defects in the clearance of dead cell debris may lead to both increased levels of DNA in the extracellular space as well as its persistence [12].
Whatever the mechanisms for extracellular DNA release, levels of DNA are significantly elevated in the blood of patients or experimental animal models in a wide range of conditions marked by cell injury or death such as shock and malignancy. These conditions often show elevations in the levels of histones and nucleosomes [13–15]. These findings suggest that much of the extracellular DNA exists in the form of nucleosomes in which a length of DNA of approximately 147 bases is wrapped around a core octamer of two molecules each of histones H2A, H2B, H3, and H4; the nucleosome represents the main structural element of chromatin and allows dynamic interaction with proteins to mediate processes such as replication, transcription and repair [16, 17]. DNA, histones and nucleosomes all show immunological activity and drive immune responses via pattern recognition receptors that include toll-like receptors (TLRs) as well as internal nucleic acid sensors that can trigger the inflammasome [18–20].
The presence of DNA in the blood does not imply its existence in a soluble form (whether or not associated with proteins on the nucleosome) since, during cell death, nuclear as well as cytoplasmic molecules can transit into the extracellular space in the form of microparticles (MPs). MPs are small membrane-bound vesicles that range in size from 0.1 to 1.0 μm and originate from a blebbing process during cell death; MPs release can also occur during platelet activation [21, 22]. During apoptosis, nuclear molecules, including DNA, most likely in the form of nucleosomes or chromatin, can translocate to the blebs which can encapsulate a wide variety of cellular components [23–28]. Depending on the cell type, MPs can also be a source of cytokines [23]. In view of their composition, MPs can serve numerous physiological functions including thrombosis, hemostasis and inflammation and are elevated in many of the same diseases as is circulating DNA.
As shown recently, DNA and other nuclear molecules on MPs are antigenically active and can be bound by monoclonal antibodies, plasma of patients as well as plasmas of murine models of lupus [28–30]. The binding occurs because DNA and other nuclear molecules reside on the particle surface or in an otherwise accessible form inside the particle itself. The relevance of particle binding to immune complex formation is demonstrated by the presence of IgG on particles in the blood of lupus patients. While the full range of autoantibodies that bind to particles is not known, studies on patients indicate a correlation between the presence of IgG on particles and anti-DNA levels, suggesting that anti-DNA bind particles in vivo. Particles in the blood of patients also have bound complement, further establishing their importance as circulating immune complexes [29].
Previous studies from our laboratory have demonstrated that the binding of autoantibodies may reflect fine specificity of antibodies to DNA as well as other nucleosomal components. Among a panel of murine monoclonal autoantibodies, we showed that certain specificities bound to particles generated in vitro from cell lines undergoing apoptosis [30]. Furthermore, we showed that MRL-lpr/lpr and NZB/NZW F1 mice differ in the content of MPs with bound IgG in the blood as well as the ability of plasma IgG to bind MPs generated in vitro. Since both strains express anti-DNA, these findings could suggest differences in the expressed antibodies in their recognition of DNA epitopes displayed by particles. In the current studies, we have further investigated the relationship between anti-DNA specificity and particle binding with monoclonal anti-DNA antibodies from NZB/NZW F1 mice and MPs generated by cultured cells undergoing apoptosis. Results presented herein indicate that MP binding is a property of only certain anti-DNA antibodies, a property related to differential specificity for ds and ssDNA. As such, these findings suggest the utility of particle binding as a measure of anti-DNA specificity potentially related to pathogenicity.
2. Materials and Methods
2.1 Anti-DNA ELISA
96-well microtiter plates (Thermo, Milford, MA) were coated overnight at 4°C with DNA from calf thymus (Sigma, St. Louis, MO) at a final concentration of 5 μg/ml in sodium citrate buffer (SSC) pH 8. The ds DNA was denatured by boiling for 10 minutes to prepare ssDNA to coat microtiter wells similarly. The coating solution was aspirated and the plates washed three times in phosphate buffered saline (PBS). Five-fold serial dilutions (10 μg/ml to 3.25 ng/ml) of a panel of monoclonal antibodies (described below) were prepared in phosphate buffered saline containing 0.5% BSA, 0.05% Tween 20 (Sigma) (PBS-BSA-T) and transferred to the plates. Plates were incubated for 1 h at room temperature (RT) followed by washing in PBS. Peroxidase-labeled goat anti- mouse IgG γ-chain specific (Sigma) diluted in PBS-BSA-T was added to the wells and plates were incubated for a further 1 h at RT. Plates were washed three times and developed following a 30 minute incubation at RT with substrate solution of 3,3’,5,5’-tetramethylbenzidine and hydrogen peroxide (Sigma) in 0.1M citrate buffer, pH 4.0. The reaction was terminated by adding 10% sulfuric acid solution, and the OD 450 was determined with a microplate reader.
2.2 Preparation of microparticles from in vitro cell cultures
Jurkat and THP-1 human cell lines obtained from the Duke University Comprehensive Cancer Center Cell Culture Facility were cultured in RPMI 1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT) and 20 μg/ml gentamicin (Invitrogen). Cells were cultured at 37°C and 5% CO2, plated at a concentration of 2.5 × 106 cells/ml and induced to undergo apoptosis by treatment with 1 μM staurosporine (STS) or 10 μM etoposide (ETO) (Sigma) for 18 h. The cells in the treated culture medium were collected and washed by centrifuging at 500 × g for 5 minutes. The cell free supernatant was centrifuged further at 16,000 × g for 45 minutes to pellet MPs. The resulting MP pellet was resuspended in Ca++ and Mg++ free PBS (Invitrogen) and adjusted to a concentration of 5000 MPs/μl after counting by flow cytometry (described below, section 2.3). MPs were also isolated from untreated cell cultures in normal growth cycle following 24 h culture.
2.3 Binding of monoclonal antibodies to MPs
MPs were incubated with a panel of monoclonal antibodies (mAbs) to DNA prepared from hybridomas from NZB/NZW F1 mice previously described [31–33]. The anti-DNA mAbs were serially diluted to concentrations ranging from 10 μg/ml to 3.25 ng/ml in 100 μl of PBS containing 5000 MPs/μl. Monoclonal antibody binding to MPs was detected using R-phycoerythrin labeled F(ab’)2 fragment of anti-mouse IgG (whole molecule) prepared in sheep (Sigma) by flow cytometry. MPs were evaluated using a FACScan flow cytometer (BD, San Jose, CA) with all detectors set to logarithmic mode. FlowJo Collector’s Edition (Tree Star, Ashland, OR) was used for data acquisition and analysis.
To test binding specificity of the mAbs, 100 μl of MPs (5000 MPs/μl) were treated with 100 U/ml of RNase-free DNase I (Invitrogen) and incubated at 37°C for 2 h. Nuclease digested MPs were incubated with mAbs and stained with labeled anti-mouse IgG to detect binding by flow cytometry. The mAbs were also incubated in five-fold serial dilutions (10 μg/ml to 3.25 ng/ml) of ssDNA or dsDNA before incubating with MPs to test competitive inhibition of anti-DNA binding to MPs.
MPs from apoptotic Jurkat and THP-1 cells were also pre-incubated with DNA to assess effects on monoclonal antibody binding to particles. For this purpose, 106 THP-1 and Jurkat MPs were incubated with various concentrations of ssDNA or dsDNA for 1 h at RT. The MPs were washed by centrifuging the suspension at 16,000 × g for 30 min to pellet the treated MPs. The MP pellet was resuspended in Ca++ and Mg++ free PBS at a concentration of 5000 MPs/μl for staining with mAbs as described above.
3. Results
3.1 Binding specificity of monoclonal antibodies to ssDNA and dsDNA
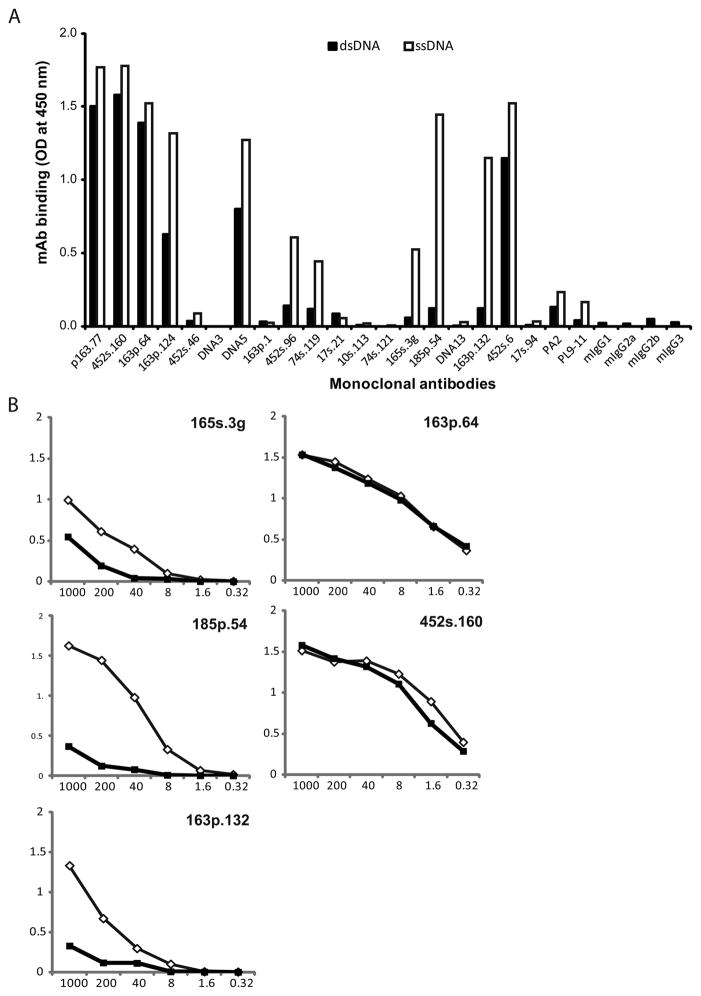
The specificity of the monoclonal anti-DNA in the panel was first assessed using a direct ELISA to test the capacity of the monoclonal antibodies (mAbs) to bind ssDNA and dsDNA [32]. These antibodies were isolated from NZB/NZW F1 mice and previously characterized with respect to binding properties and ability to induce nephritis. As results in Figure 1A indicate, the antibodies varied significantly in their relative binding to the two antigenic forms of DNA. Of the 19 antibodies, six demonstrated strong binding to both ssDNA and dsDNA (p163.77, 452s.160, 163p.64, 163p.124, DNA5 and 452s.6). Five other mAbs showed preferential binding to ssDNA and limited activity against dsDNA (452s.96, 74s.119, 165s.3g, 185p.54 and 163p.132), while the remaining monoclonal antibodies showed limited activity against both forms of DNA under the conditions of this assay.
Figure 1.
DNA specificity of monoclonal antibodies. A panel of 19 monoclonals (mAb) previously prepared from NZB/NZW F1 lupus mice was assayed by ELISA for binding specificity to ds or ssDNA. Microtiter plates were coated with 5 μg/ml of native (dsDNA) or denatured (ssDNA) calf-thymus DNA, treated with 250 ng of mAb (panel A) or five-fold serial dilutions (1000 – 0.32 ng) (panel B) in 100 μl of reaction volume and incubated for 1 h at 37°C. Peroxidase-labeled sheep anti-mouse IgG was used to measure bound antibodies. Black bars (A) or squares (B) represent mAb binding to dsDNA and white bars (A) or diamonds (B) show binding to ssDNA. PA2 and PL9-11 are control mAbs. Results are representative of three replicate experiments.
To assess further the specificity of these antibodies, selected members of the panel were measured over a broader range of concentration. As these results indicated, the relative binding of antibodies varied significantly. Whereas two of the mAbs in this selected group bound similarly to ds and ssDNA (163p.64, 452s.160), the others showed much greater binding to ssDNA compared to dsDNA at the concentrations of 10 – 0.4 μg/ml, (Figure 1B).
3.2 The binding of anti-DNA antibodies to MPs isolated from cells undergoing apoptosis
Previous studies showed that plasmas from patients with SLE as well as lupus mice show significant binding for MPs from apoptotic cell cultures [28, 34]. To determine the relationship between antibody specificity and particle binding, we therefore, tested the ability of the anti-DNA mAbs to bind to MPs generated from cells induced to undergo apoptosis in vitro. For these experiments, cells were treated with either staurosporine, a broad-spectrum kinase inhibitor, or etoposide, a topoisomerase inhibitor; supernatants were then collected and MPs isolated by differential centrifugation.
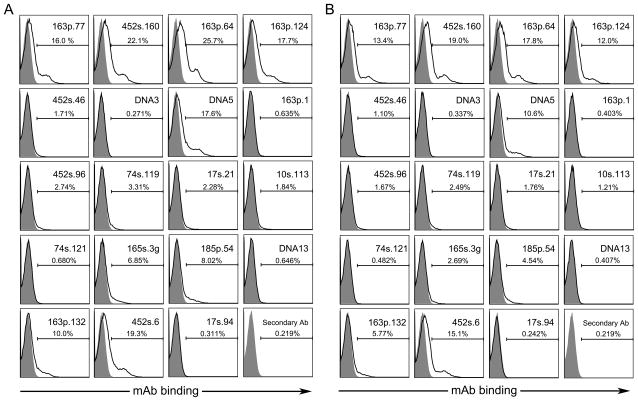
Figure 2 presents results of experiments with particles from cultures treated with staurosporine (Panel A) or etoposide (Panel B). As these findings indicate, among the panel of monoclonal antibodies, only some bound to the MPs from in vitro cell cultures although the extent of binding was similar with MPs from cultures with either inducer. These results confirm previous findings on the similarity in particles from different cell lines in terms of antibody binding [25, 28, 34]. Importantly, reference to the specificity analysis in Figure 1 indicates that the antibodies with high binding to dsDNA bound to MPs, whereas mAbs that preferentially bound ssDNA did not bind to MPs significantly.
Figure 2.
Binding of mAbs to MPs generated in vitro. Jurkat cells were treated with 1 μM staurosporine (A) or 10 μM etoposide (B) for 18 h to induce apoptosis. MPs were isolated from culture media by centrifugation and diluted in PBS to a concentration of 5000 MPs/μl for treatment with 500 ng of mAb for 1 h at 37°C. Binding of mAbs to MPs (black line) was detected by flow cytometry using R-phycoerythrin labeled goat anti-mouse IgG and compared to isotype controls (filled grey peaks). Presented data are representative of three replicate experiments.
3.3 Effects of DNase digestion on antibody binding
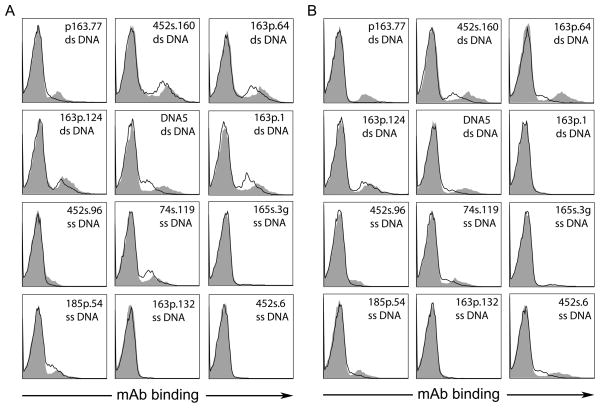
To characterize further the basis of antibody binding to the MPs, we tested the antigenicity of MPs from two different cell types that had been treated with DNase. For this purpose, MPs from THP-1 and Jurkat cells cultured with staurosporine were treated with DNase before staining with the mAbs. As these results indicate, treatment with DNase led to some reduction of mAb binding (Figure 3) although the extent was variable among antibodies. These results indicate that some of the DNA antigen can be removed by specific nuclease treatment although some may be in a resistant or protected form.
Figure 3.
The effects of nuclease treatment on the binding of mAbs to MPs. MPs prepared from Jurkat (A) and THP-1 (B) cells were treated with 100 U/ml of DNase (black line) or left untreated (filled peak). MPs were incubated with the mAbs (5 μg/ml) for 1 h at RT followed by staining with R-PE labeled goat anti-mouse IgG. Stained MPs were assayed by flow cytometry. Results are representative of four replicate experiments.
3.4 The effect of soluble DNA on antibody binding
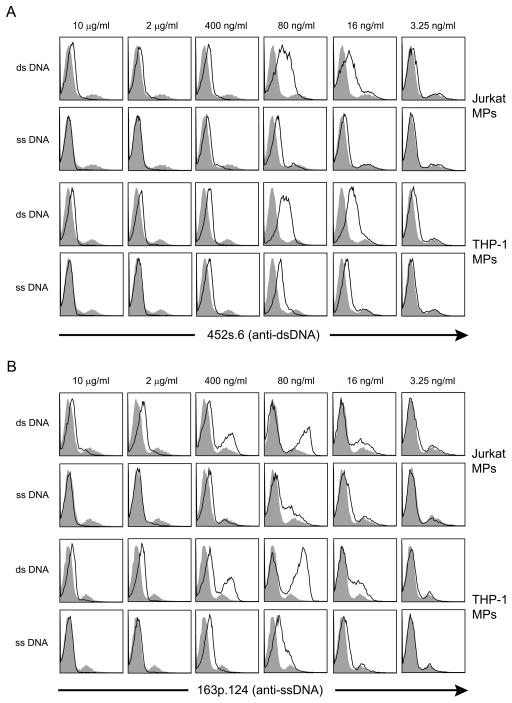
To determine whether antibody binding to particles could be affected by free DNA, we assessed the effect of pre-incubating two representative antibodies (452s.6 and 163p.124) with ssDNA or dsDNA prior to incubation with particles. For this experiment, the antibodies were first incubated in five-fold serial dilutions (10 μg/ml – 3.25 ng/ml) of ssDNA or dsDNA and then added to the THP-1 and Jurkat MP suspension for further incubation. As results of these experiments show, the effects of added DNA were complex. Thus, at higher DNA concentrations, the binding of the MPs was inhibited by pre-incubation with either ds or ssDNA. As the concentration of DNA was decreased to 80 and 16 ng/ml, however, the binding of mAbs to the MPs was enhanced, with a greater effect of ds than ssDNA (Figure 4). The lowest concentration of exogenous DNA (3.25 ng/ml) did not affect mAb binding significantly. These findings suggest that soluble DNA can influence antibody binding to MPs although the outcome depends on the concentration.
Figure 4.
The effects of DNA pre-incubation on the binding of mAbs to MPs. At a concentration of 5μg/ml, two anti-DNA mAbs, 452s.6 (A) and 163p.124 (B), were incubated in five-fold serial dilutions of ssDNA or dsDNA (10 μg/ml – 3.25 ng/ml). The mAb was then added to a suspension of 500,000 MPs from THP-1 or Jurkat cells treated with staurosporine. MAb binding to MPs (black line) was detected by flow cytometry and compared to the binding of mAb with control pre-incubation (filled peak). Results are representative of experiments performed three times.
3.5 The effect of added DNA on the antigenicity of MPs
One explanation for enhanced antibody interactions with MPs at lower DNA concentrations relates to an increase in the antigen content of MPs from the added DNA that could be expected to act as an inhibitor. This increase could occur at low DNA concentrations because of a preferential interaction of antibodies to particles (whose antigenic content is increased by binding of the free DNA) rather than free DNA. In contrast, at higher concentrations, the free DNA would compete effectively with DNA on the particles and inhibit antibody binding. We therefore performed experiments to determine whether DNA can bind to MPs and thereby increase their antigenicity. In these experiments, we also assessed MPs from untreated cells to determine whether induction of apoptosis can affect either the DNA content of MPs or their capacity to bind added DNA.
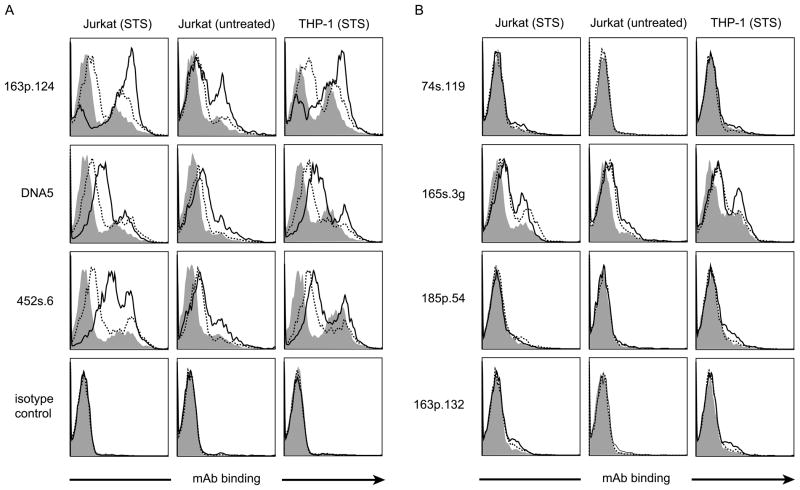
In these experiments, MPs were first incubated with dsDNA or ssDNA, washed and then incubated with the monoclonal antibodies. We assessed the antibodies that either bound to dsDNA (163p.124, DNA5, 452s.6) or ssDNA (74s.119, 165s.3g, 185p.54 and 163p.132). As these experiments indicate (Figure 5), incubation of particles with either ds or ssDNA increased the binding of antibodies that bound well for both antigenic forms (Panel A). With antibodies that bound to ssDNA (Panel B), the effects of the pre-incubation with ssDNA differed from those of antibodies binding dsDNA. Thus, only one of the four antibodies tested (165s3.g) showed an augmentation of binding to particles following pre-incubation with either form of DNA. Together, these results indicate that particles can interact with DNA to increase antigenicity and possibly affect relative specificity for an antibody for ss and dsDNA. Thus, in addition to their display of DNA arising during apoptosis, MPs can provide a matrix to bind DNA to which it may be exposed following release from the cell.
Figure 5.
The effect of incubation of particles with DNA on the binding of mAb. Jurkat and THP-1 MPs from cultures treated with staurosporine were incubated in 10 μg/ml solutions of ssDNA (broken line), dsDNA (solid line) or left untreated (filled peak). Treated MPs were washed by centrifugation and resuspended in fresh PBS. MPs were incubated with 5 μg/ml of anti-dsDNA mAb (163p.124, DNA5 and 452s.6) (panel A) and anti-ssDNA mAb (74s.119, 165s.3g, 185p.54 and 163p.132) (panel B). Bound anti-DNA was detected by flow cytometry using R-PE labeled goat anti-mouse IgG. Results are representative of 3 experiments.
4. Discussion
Results presented herein provide new insight into the binding of anti-DNA antibodies to MPs and indicate that the specificity for dsDNA can influence these interactions. These studies extend prior observations on the binding of anti-DNA antibodies in blood from patients and animal models as well as a limited panel of monoclonal anti-DNA and anti-nucleosomal antibodies that were derived from various mouse models [25, 28, 34]. That panel contained two anti-DNA antibodies, one of which bound to MPs generated in vitro and one of which didn’t; these findings suggested the antigenicity of particle DNA and free DNA differ. Similarly, among antibodies to histones and DNA-histones complexes, only some bound particles. By characterizing a much larger panel of antibodies, the current studies fully support the heterogeneity of anti-DNA interactions with MPs and identify dsDNA as an important epitope for antibody interaction. These findings support conclusions from studies on the binding of monoclonal antibodies to apoptotic blebs and the display of various nucleosomal antigens on particles [35, 36].
While antibody binding to MPs could suggest interaction with surface antigen, particles are permeable and antibodies may penetrate the membrane to contact DNA on the particle interior. Little is known about the internal structure of particles and the relationship of the interior to nucleoplasm, for example. The current study, in concert with others, nevertheless suggests a greater availability of ds DNA for antibody binding. This display could relate to preferential retention of dsDNA compared to ssDNA following nuclease degradation during apoptotic death; alternatively, single stranded regions may have greater interaction with particle proteins that limits or prevents the binding of anti-DNA antibodies. While the basis of this difference is not known, the greater binding of anti-dsDNA supports the idea that antibodies with this specificity may have a more significant role in disease pathogenesis than antibodies to ssDNA including the presentation of dsDNA to induce immune responses.
The current findings may also be relevant to the mechanisms by which self-antigens become available in vivo to form immune complexes in lupus, especially in the kidney. Such complexes may form from nuclear antigen that circulates in the blood or forms in situ in the kidney [37]. As particle release appears be a common, if not an invariable, feature of cell death, particles may be a relevant source of DNA antigen for complex formation whatever the location. As such, particle binding may be related to pathogenicity. It is of interest therefore that some of the antibodies with particle interaction also showed glomerular localization and binding to mesangial matrix in in vivo transfer models. In the in vivo studies, the antibodies with nephritogenic properties showed robust binding to dsDNA whereas antibodies with preferential binding to ssDNA did not show glomerular localization [38]. Since at least some of the nephritogenic antibodies also bound particles, a relationship of MP interaction and disease manifestations is possible. In this regard, nucleases in the blood may degrade the DNA on particles to modify antigenicity; on the other hand, a loss of DNase in the kidney may allow more persistent expression of DNA on the microparticles which lead to increased antibody binding and complex formation [37, 39–41].
These studies also suggest that comparisons between the binding of antibodies and monoclonal antibodies from lupus mice may provide a valuable perspective to assess pathogenicity and elucidate the mechanisms of immune complex formation. This perspective derives from our previous studies which suggested an apparently different role of MPs in the process of immune complex formation in MRL-lpr/lpr and NZB/NZW F1 mice. Thus, using flow cytometry, we showed that plasma from MRL-lpr/lpr mice showed higher levels of MPs with bound IgG than particles from NZB/NZW F1 mice. Furthermore, we showed that purified IgG from MRL-lpr/lpr mice had much more robust interaction with particles than IgG from NZB/NZW F1 mice. In view of these prior results, the significant binding of certain NZB/NZW F1 monoclonal anti-DNA antibodies to particles is perhaps surprising. We would have predicted that, similar to the expressed antibodies in plasma, the monoclonal antibodies would show limited binding to particles generated in vitro. Nevertheless, we found that some of the antibodies bound well to particles in a DNA dependent manner.
One explanation for these differences relates to sampling issues. It is possible that the monoclonal antibodies in our panel are not representative of the expressed repertoire of the strain. This situation could arise if B cells that can form a hydridoma are less likely to be expressed in vivo perhaps related to their differentiation or maturation stage or quantitative representation. In this regard, in our original study, we observed some differences among individual NZB/NZW mice in the levels of IgG on particles [34]. It is thus possible that the mice used as a source of spleen cells for the creation of the hybridoma lines expressed during disease IgG anti-DNA that can bind to particles. In this regard, an environmental influence is also possible as the original studies involved mice purchased from the Jackson Laboratory and raised in North Carolina whereas the source of the spleens for hybridoma lines came from mice raised in Tennessee. Unfortunately, we do not have blood from these mice to assess either the presence of IgG positive particles or the ability of serum IgG to bind to particles. We are performing further experiments to define the various influences on particle binding both in vitro and in vivo.
In conclusion, these studies demonstrate that the ability of an antibody to bind microparticles relates to its fine specificity and preference for single and double stranded DNA. Furthermore, these studies suggest that particle binding represents a useful serological assay to characterize autoantibody specificity as particles may represent an important antigenic form of DNA that is exposed to the immune system in vivo. This DNA may have unique associations with other nuclear and cellular components on particles which may affect antigenicity as well as immunogenicity. Indeed, particles can display cytokines and serve as adjuvants [21, 22]. Future studies will clarify the relationship between particle binding and pathogenicity, in terms of the formation of immune complexes that deposit in the kidney or stimulate cytokine production.
Acknowledgments
These studies were funded by a VA Merit Review grant, the Kirkland Scholar Program from the Hospital for Special Surgery in New York City (Mary Kirkland Center for Lupus Research), National Institutes of Health (5U19-AI-056363) and the Alliance for Lupus Research.
Abbreviations
- dsDNA
double-stranded DNA
- MPs
microparticles
- mAbs
monoclonal antibodies
- SLE
systemic lupus erythematosus
- ssDNA
single-stranded DNA
References
- 1.Hahn BH. Antibodies to DNA. N Engl J Med. 1998;338:1359–1368. doi: 10.1056/NEJM199805073381906. [DOI] [PubMed] [Google Scholar]
- 2.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 3.Jang YJ, Stollar BD. Anti-DNA antibodies: aspects of structure and pathogenicity. Cell Mol Life Sci. 2003;60:309–320. doi: 10.1007/s000180300026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mortensen ES, Rekvig OP. Nephritogenic potential of anti-DNA antibodies against necrotic nucleosomes. J Am Soc Nephrol. 2009;20:696–704. doi: 10.1681/ASN.2008010112. [DOI] [PubMed] [Google Scholar]
- 5.van der Vlag J, Berden JH. Lupus nephritis: role of antinucleosome autoantibodies. Semin Nephrol. 2011;31:376–389. doi: 10.1016/j.semnephrol.2011.06.009. [DOI] [PubMed] [Google Scholar]
- 6.Koffler D, Agnello V, Thoburn R, Kunkel HG. Systemic lupus erythematosus: prototype of immune complex nephritis in man. J Exp Med. 1971;134:169–179. [PMC free article] [PubMed] [Google Scholar]
- 7.Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, Hua J, An LL, Audoly L, La Rosa G, Bierhaus A, Naworth P, Marshak-Rothstein A, Crow MK, Fitzgerald KA, Latz E, Kiener PA, Coyle AJ. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–496. doi: 10.1038/ni1457. [DOI] [PubMed] [Google Scholar]
- 8.Vallin H, Perers A, Alm GV, Ronnblom L. Anti-double-stranded DNA antibodies and immunostimulatory plasmid DNA in combination mimic the endogenous IFN-alpha inducer in systemic lupus erythematosus. J Immunol. 1999;163:6306–6313. [PubMed] [Google Scholar]
- 9.Shrivastav M, Niewold TB. Nucleic Acid Sensors and Type I Interferon Production in Systemic Lupus Erythematosus. Front Immunol. 2013;4:319. doi: 10.3389/fimmu.2013.00319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galluzzi L, Vitale I, Abrams JM, Alnemri ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry WS, Fulda S, Gottlieb E, Green DR, Hengartner MO, Kepp O, Knight RA, Kumar S, Lipton SA, Lu X, Madeo F, Malorni W, Mehlen P, Nunez G, Peter ME, Piacentini M, Rubinsztein DC, Shi Y, Simon HU, Vandenabeele P, White E, Yuan J, Zhivotovsky B, Melino G, Kroemer G. Molecular definitions of cell death subroutines: recommendations of the Nomenclature Committee on Cell Death 2012. Cell Death Differ. 2012;19:107–120. doi: 10.1038/cdd.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pisetsky DS. The translocation of nuclear molecules during inflammation and cell death. Antioxid Redox Signal. 2014;20:1117–1125. doi: 10.1089/ars.2012.5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shao WH, Cohen PL. Disturbances of apoptotic cell clearance in systemic lupus erythematosus. Arthritis Res Ther. 2011;13:202. doi: 10.1186/ar3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holdenrieder S, Stieber P. Clinical use of circulating nucleosomes. Crit Rev Clin Lab Sci. 2009;46:1–24. doi: 10.1080/10408360802485875. [DOI] [PubMed] [Google Scholar]
- 14.Johansson PI, Windelov NA, Rasmussen LS, Sorensen AM, Ostrowski SR. Blood levels of histone-complexed DNA fragments are associated with coagulopathy, inflammation and endothelial damage early after trauma. J Emerg Trauma Shock. 2013;6:171–175. doi: 10.4103/0974-2700.115327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeerleder S, Zwart B, Wuillemin WA, Aarden LA, Groeneveld AB, Caliezi C, van Nieuwenhuijze AE, van Mierlo GJ, Eerenberg AJ, Lammle B, Hack CE. Elevated nucleosome levels in systemic inflammation and sepsis. Crit Care Med. 2003;31:1947–1951. doi: 10.1097/01.CCM.0000074719.40109.95. [DOI] [PubMed] [Google Scholar]
- 16.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259–266. doi: 10.1038/nsmb.2470. [DOI] [PubMed] [Google Scholar]
- 18.Huang H, Chen HW, Evankovich J, Yan W, Rosborough BR, Nace GW, Ding Q, Loughran P, Beer-Stolz D, Billiar TR, Esmon CT, Tsung A. Histones activate the NLRP3 inflammasome in Kupffer cells during sterile inflammatory liver injury. J Immunol. 2013;191:2665–2679. doi: 10.4049/jimmunol.1202733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H, Evankovich J, Yan W, Nace G, Zhang L, Ross M, Liao X, Billiar T, Xu J, Esmon CT, Tsung A. Endogenous histones function as alarmins in sterile inflammatory liver injury through Toll-like receptor 9 in mice. Hepatology. 2011;54:999–1008. doi: 10.1002/hep.24501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindau D, Ronnefarth V, Erbacher A, Rammensee HG, Decker P. Nucleosome-induced neutrophil activation occurs independently of TLR9 and endosomal acidification: implications for systemic lupus erythematosus. Eur J Immunol. 2011;41:669–681. doi: 10.1002/eji.201040593. [DOI] [PubMed] [Google Scholar]
- 21.Buzas EI, Gyorgy B, Nagy G, Falus A, Gay S. Emerging role of extracellular vesicles in inflammatory diseases. Nat Rev Rheumatol. 2014 doi: 10.1038/nrrheum.2014.19. [DOI] [PubMed] [Google Scholar]
- 22.Pisetsky DS, Ullal AJ, Gauley J, Ning TC. Microparticles as mediators and biomarkers of rheumatic disease. Rheumatology (Oxford) 2012;51:1737–1746. doi: 10.1093/rheumatology/kes028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berda-Haddad Y, Robert S, Salers P, Zekraoui L, Farnarier C, Dinarello CA, Dignat-George F, Kaplanski G. Sterile inflammation of endothelial cell-derived apoptotic bodies is mediated by interleukin-1alpha. Proc Natl Acad Sci U S A. 2011;108:20684–20689. doi: 10.1073/pnas.1116848108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane JD, Allan VJ, Woodman PG. Active relocation of chromatin and endoplasmic reticulum into blebs in late apoptotic cells. J Cell Sci. 2005;118:4059–4071. doi: 10.1242/jcs.02529. [DOI] [PubMed] [Google Scholar]
- 25.Reich CF, 3rd, Pisetsky DS. The content of DNA and RNA in microparticles released by Jurkat and HL-60 cells undergoing in vitro apoptosis. Exp Cell Res. 2009;315:760–768. doi: 10.1016/j.yexcr.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 26.Schiller M, Bekeredjian-Ding I, Heyder P, Blank N, Ho AD, Lorenz HM. Autoantigens are translocated into small apoptotic bodies during early stages of apoptosis. Cell Death Differ. 2008;15:183–191. doi: 10.1038/sj.cdd.4402239. [DOI] [PubMed] [Google Scholar]
- 27.Suber T, Rosen A. Apoptotic cell blebs: repositories of autoantigens and contributors to immune context. Arthritis Rheum. 2009;60:2216–2219. doi: 10.1002/art.24715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ullal AJ, Reich CF, 3rd, Clowse M, Criscione-Schreiber LG, Tochacek M, Monestier M, Pisetsky DS. Microparticles as antigenic targets of antibodies to DNA and nucleosomes in systemic lupus erythematosus. J Autoimmun. 2011;36:173–180. doi: 10.1016/j.jaut.2011.02.001. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen CT, Ostergaard O, Stener L, Iversen LV, Truedsson L, Gullstrand B, Jacobsen S, Heegaard NH. Increased IgG on cell-derived plasma microparticles in systemic lupus erythematosus is associated with autoantibodies and complement activation. Arthritis Rheum. 2012;64:1227–1236. doi: 10.1002/art.34381. [DOI] [PubMed] [Google Scholar]
- 30.Pisetsky DS. Microparticles as autoantigens: making immune complexes big. Arthritis Rheum. 2012;64:958–961. doi: 10.1002/art.34377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marion TN, Lawton AR, 3rd, Kearney JF, Briles DE. Anti-DNA autoantibodies in (NZB X NZW)F1 mice are clonally heterogeneous, but the majority share a common idiotype. J Immunol. 1982;128:668–674. [PubMed] [Google Scholar]
- 32.Marion TN, Tillman DM, Jou NT, Hill RJ. Selection of immunoglobulin variable regions in autoimmunity to DNA. Immunol Rev. 1992;128:123–149. doi: 10.1111/j.1600-065x.1992.tb00835.x. [DOI] [PubMed] [Google Scholar]
- 33.Tillman DM, Jou NT, Hill RJ, Marion TN. Both IgM and IgG anti-DNA antibodies are the products of clonally selective B cell stimulation in (NZB x NZW)F1 mice. J Exp Med. 1992;176:761–779. doi: 10.1084/jem.176.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ullal AJ, Pisetsky DS. The role of microparticles in the generation of immune complexes in murine lupus. Clin Immunol. 2013;146:1–9. doi: 10.1016/j.clim.2012.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neeli I, Richardson MM, Khan SN, Nicolo D, Monestier M, Radic MZ. Divergent members of a single autoreactive B cell clone retain specificity for apoptotic blebs. Mol Immunol. 2007;44:1914–1921. doi: 10.1016/j.molimm.2006.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radic M, Marion T, Monestier M. Nucleosomes are exposed at the cell surface in apoptosis. J Immunol. 2004;172:6692–6700. doi: 10.4049/jimmunol.172.11.6692. [DOI] [PubMed] [Google Scholar]
- 37.Seredkina N, Van Der Vlag J, Berden J, Mortensen E, Rekvig OP. Lupus nephritis: enigmas, conflicting models and an emerging concept. Mol Med. 2013;19:161–169. doi: 10.2119/molmed.2013.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krishnan MR, Wang C, Marion TN. Anti-DNA autoantibodies initiate experimental lupus nephritis by binding directly to the glomerular basement membrane in mice. Kidney Int. 2012;82:184–192. doi: 10.1038/ki.2011.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martinez Valle F, Balada E, Ordi-Ros J, Vilardell-Tarres M. DNase 1 and systemic lupus erythematosus. Autoimmun Rev. 2008;7:359–363. doi: 10.1016/j.autrev.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 40.Napirei M, Ludwig S, Mezrhab J, Klockl T, Mannherz HG. Murine serum nucleases--contrasting effects of plasmin and heparin on the activities of DNase1 and DNase1-like 3 (DNase1l3) FEBS J. 2009;276:1059–1073. doi: 10.1111/j.1742-4658.2008.06849.x. [DOI] [PubMed] [Google Scholar]
- 41.Thiyagarajan D, Fismen S, Seredkina N, Jacobsen S, Elung-Jensen T, Kamper AL, Fenton CG, Rekvig OP, Mortensen ES. Silencing of renal DNaseI in murine lupus nephritis imposes exposure of large chromatin fragments and activation of Toll like receptors and the Clec4e. PLoS One. 2012;7:e34080. doi: 10.1371/journal.pone.0034080. [DOI] [PMC free article] [PubMed] [Google Scholar]