Abstract
Isoform-specific signaling by Class IA PI 3-kinases depends in part on the interactions between distinct catalytic subunits and upstream regulatory proteins. From among the class IA catalytic subunits (p110α, p110β, and p110δ), p110β has unique properties. Unlike the other family members, p110β directly binds to Gβγ subunits, downstream from activated G-protein coupled receptors, and to activated Rab5. Furthermore, the Ras-binding domain (RBD) of p110β binds to Rac and Cdc42 but not to Ras. Defining mutations that specifically disrupt these regulatory interactions is critical for defining their role in p110β signaling. This chapter describes the approach that was used to identify the Rab5 binding site in p110β, and discusses methods for the analysis of p110β-Rab5 interactions.
Keywords: PIK3CB, Class IA PI 3-kinase, p110beta, Rab5, Lipid kinases, Phosphoinositide 3-kinases, Small GTPases
1 Introduction
The Class I Phosphoinositide 3-kinases (PI 3-kinases) are activated by signals from receptor tyrosine kinases and G-protein-coupled receptors, and they produce phosphatidylinositol [3,4,5]-trisphosphate (PIP3) in metazoan cells. Of the four catalytic isoforms of PI 3-kinase, the PIK3CB gene product p110β is unique in that it couples to both RTKs and GPCRs [1, 2], has a so-called Ras-binding domain that instead binds to activated Rac and Cdc42 [3], and binds directly to the endosomal GTPase Rab5 in its activated, GTP-bound state [4].
Rab5 plays crucial roles in the sorting of internalized endocytic vesicles. Through its interactions with its effectors EEA1 and the Class III PI3 Kinase hVps34, Rab5 regulates docking and fusion of early endosomes, as well as their attachment to and movement along microtubules [5]. GTP-bound Rab5 has been shown to interact with a number of proteins involved in endocytic sorting, including the Rabaptin-5/Rabex-5 complex [6], the endosomal tethering protein EEA1 [7], and the hVps45-associated Rabenosyn-5 [8], as well as signaling proteins like APPL1/2 [9] and p110β [4].
Previous studies on p110β binding to Rab5 suggested that the binding site lay within residues 136–270, containing portions of the Adaptor-binding domain (ABD) and the Ras-binding domain (RBD), and residues 658–759, containing portions of the helical and kinase domains [10]. We have previously described a fully active p110α/p110β chimera containing the ABD and RBD domains of p110α linked to the C2, helical, and kinase domains of p110β [11]. This chimera showed specific GTP-dependent binding to GTP-Rab5 (Fig. 1). Taken together, these data and the published work suggested that the Rab5 binding site was within the helical and kinase domains of p110β.
Fig. 1.
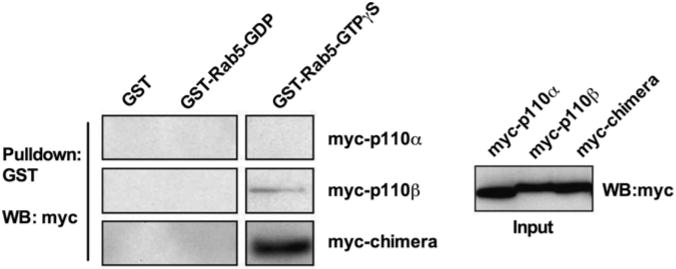
Binding of p110β/p110β chimera to Rab5GTP Chimera of p110α ABD-RBD and p110β C2-helical-kinase domains binds Rab5. HEK293T cells were transfected with HA-p85 and myc-tagged p110α, p110β, or a p110α/p110β chimera. Cleared lysates were subjected to a GST-Rab5 pulldown, and samples were run on a western blot probing for myc
In order to identify point mutations that would disrupt p110β binding to Rab5, we mutated candidate residues within the helical and kinase domains based on the following criteria.
The residues should be poorly conserved between human p110β and p110δ, which is the isoform most homologous to p110β, but which does not bind to Rab5.
We eliminated residues that were predicted to be poorly surface accessible, based on the crystal structure of p110β.
We eliminated residues that were poorly conserved in p110β orthologs from other species.
This analysis (see Note 1) defined 22 residues as potential binding sites for Rab5 (Fig. 2). The binding assay described below was then used to screen for mutants that disrupted p85/p110β binding to GST-Rab5-GTP beads (see Note 2). Two consecutive residues were found to be required for Rab5 binding: Q596C and I597S (Fig. 3a). Although p85 has been reported to directly bind to Rab5 [12], we detected no binding to either p85 or dimers of p85 and p110βI597S in this assay (Fig. 3b).
Fig. 2.
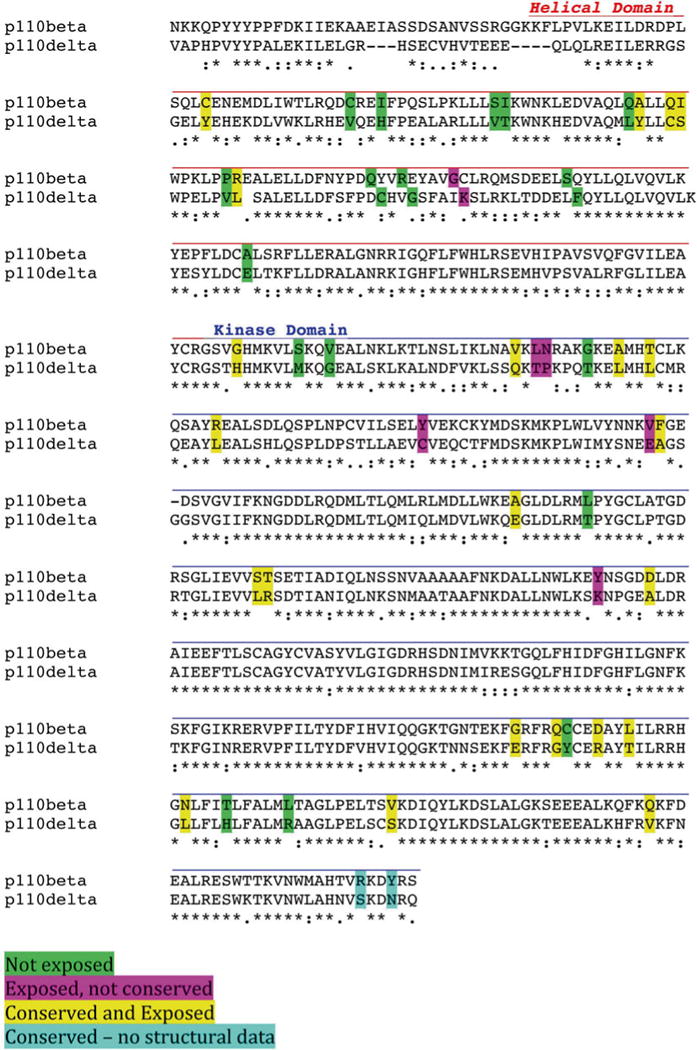
Sequence analysis of p110β. The helical and kinase domains of p110β and p110δ were aligned using T-Coffee, and surface exposure was analyzed using NACCESS [17]. Residues that were not similar between p110β and p110δ are color coded as follows: Green—not surface exposed; Magenta—exposed but not conserved in p110β orthologs; Yellow —exposed and conserved; Blue—conserved but not visible in existing structures
Fig. 3.
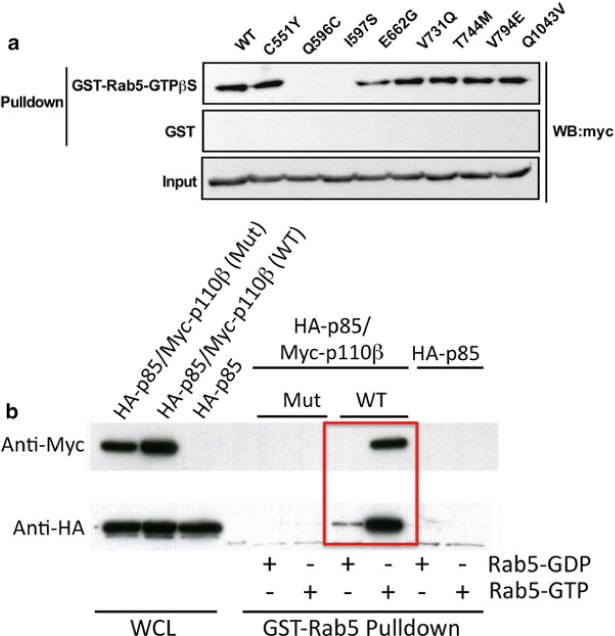
(a) Identification of Rab5-binding deficient p110β mutants. HEK293T cells were transfected with HA-p85 and myc-p110β WT or one of eight p110β point mutants. Cleared lysates were subjected to a GST-Rab5-GTPγS pulldown, and samples analyzed by western blot. (b) HEK 293T cells were transfrected with HA-p85, HA-p85 plus wild type myc-p110β, or HA-p85 plus myc-p110βI597S (Mut). Cell lysates were incubated with immobilized GST-Rab loaded with GDP or GTP-γS, and the beads were washed and analyzed by western blot for p110β (myc) or p85 (HA)
The p110β residues required for Rab5 binding lie immediately behind the Gβγ loop in p110β (Fig. 4). Subsequent analysis showed that these mutants had no effect on in vitro activation of p85/p110β by tyrosine phosphorylated peptides or by recombinant Gβγ (data not shown). The Rab5-uncoupled p110β mutants defined by these methods were subsequently used to implicate Rab5-p110β interactions in macroautophagy [13].
Fig. 4.
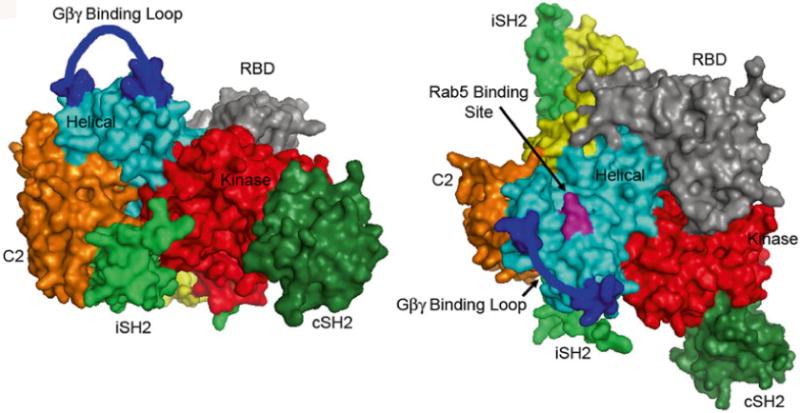
Location of p110β mutants that disrupt binding to Rab GTP The crystal structure of p85(iSH2-cSH2)/ p110β [18] is shown with the C2 and kinase domains of p110β facing frontward (left panel), and rotated 90 to show the Rab5 binding site, which lies between the Gβγ binding loop and the RBD (right panel)
In this chapter, we describe the assay for Rab5-p110β binding that was used to identify the Rab5 binding site in p110β. The protocol has four major parts: purification of GST-Rab5 from bacteria, preparation of recombinant p85/p110β in HEK293T cells, loading of immobilized GST-Rab5 with GDP or GTP-γS, and GST- Rab5 pulldowns of recombinant p85/p110β.
2 Materials
2.1 Plasmids
2.2 Buffers for Bacterial Expression of GST-Rab5
Resuspension Buffer: 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 2 mM EDTA, 2 mM DTT, 10 % glycerol, 1 % CHAPS, Roche Protease Inhibitor tablet (1 tablet per 10 ml buffer), 0.35 mg/ml PMSF (1:100 dilution from freshly made 35 mg/ml PMSF in ethanol) (see Note 3).
Wash Buffer 1: Phosphate buffered saline containing 0.5 % NP-40 and 2 mM DTT.
Wash buffer 2: 50 mM Tris–HCl pH 8.0, 100 mM NaCl, 2 mM EDTA, 2 mM DTT, 10 % glycerol, 1 % CHAPS.
Elution Buffer: Wash Buffer 2 containing 15 mM reduced glutathione.
2.3 Buffers for Mammalian Expression of p85/ p110β
HEK lysis buffer: 20 mM Tris–HCl pH 7.5, 137 mM NaCl, 5 mM MgCl2, 1 mM CaCl2, 2 mM DTT, 10 % glycerol, 1.0 % NP-40, Roche protease inhibitor tablet (1 tablet/10 ml lysis buffer), PSMF (1:100 dilution from freshly made 35 mg/ml PMSF in ethanol), CalBiochem phosphatase inhibitor Cocktail 1 (1:100 dilution), Sigma phosphatase inhibitor Cocktail 2 (1:100 dilution).
2.4 Buffers for Rab5 Nucleotide Loading
Nucleotide Loading Buffer: 25 mM Tris–HCl pH 7.5, 50 mM NaCl, 10 mM EDTA, 5 mM MgCl2, 2 mM DTT, 0.06 % CHAPS.
Nucleotide Stabilization Buffer: 25 mM Tris–HCl pH 7.5, 50 mM NaCl, 10 mM MgCl2, 2 mM DTT, 0.06 % CHAPS.
3 Methods
3.1 Production of Recombinant GST-Rab5 in Bacteria
Day 1: Transform BL21 bacteria with GST-Rab plasmid and plate onto LB-Agar plates (100μg/ml ampicillin).
Day 2: Take a single colony from the plate and inoculate 5 ml of LB containing 100μg/ml ampicillin. Shake overnight at 37°C.
Day 3. Empty the 5 ml culture into 500 ml LB-amp and shake in a 21 flask at 37°C until the OD600 measures 0.6–0.8. Remove 50μl sample for analysis, boil in Laemmli Sample Buffer, and store at −20°C for subsequent analysis (see Note 4).
Add IPTG to the bacterial culture, to a final concentration of 0.2 mM. Shake overnight (16–18 h) at 18°C. Remove 50μl sample for analysis at the end of the induction, and process and store as above.
3.2 Purification of GST-Rab5 (Adapted from Ref. 14)
Centrifuge bacterial culture in refrigerated GSA rotor at 9,000×g for 10 min. Discard the supernatant. Supernatant should be clear and the pellet tightly packed.
Resuspend pellet in Resuspension Buffer (10 ml for a 500 ml culture) and transfer to 50 cc conical tube. Keep on ice at all times.
At this point, resuspended pellet can be flash frozen in dry ice/ethanol bath and stored at −80°C. Allow room in tube for expansion during freezing. To resume the purification, thaw resuspended pellets in ice water. Add fresh PMSF (1:100 dilution of 35 mg/ml in ethanol) once thawed.
Lyse the resuspended bacteria by sonicating for 20 s in ice water, followed by 40 s recovery on ice, 4 times (total = 80 s sonication). Typical sonication uses a Branson Sonicator with a microprobe tip at output level 5. Keep sample tubes in a beaker with ice water during sonication.
Add Triton X-100 to a final concentration of 1 % v/v. Incubate at 4°C on rotating wheel in cold room for 20 min.
Centrifuge at 15,000×g in a Sorvall SS-34 or equivalent rotor for 30 min to remove the insoluble material.
When spin is finished, filter the supernatant using a 0.45μm filter. Remove 50μl sample for analysis, and process and store as above.
Prepare a glutathione Sepharose column. For a 0.5 L culture, transfer 4 ml of 50 % GST bead slurry to a plastic column. Let the storage buffer drain out and then wash with 10 bed volumes of Wash Buffer 2.
Apply the filtered lysate to the glutathione Sepharose column, adjusting the outlet tube so that sample takes 30–60 min to run through. Save the flow through. Alternatively, incubate beads with filtered lysate in a 15 cc conical tube, rotating slowly at 4°C for 2 h, then pour into plastic column. Save the flow through. (In either case, remove 50μl sample of flow through for analysis; process and store as above.)
Wash column with 30–50 column volumes of ice cold Wash Buffer 1.
Wash column with 10 volumes ice cold Wash Buffer 2.
The GST-Rab5 beads can be used in pulldown assays at this point. The beads can be stored by diluting into 10 column volumes of Wash Buffer 2 made up to 50 % glycerol. After mixing on a wheel at 4°C for 10 min, the beads can be stored for several weeks at −20°C. Alternatively, GST-Rab5 can be eluted, dialyzed, and stored at −80°C as described below.
To determine the amount of bound GST-Rab5, resuspend the beads 1:1 with Wash Buffer 2. Remove 30μl of slurry (cut the pipette tip to avoid clogging), and spin the beads briefly at 13,000×g Remove the supernatant, and add 30μl of Laemmli Sample Buffer containing 100 mM DTT. Boil for 3 min, spin at 13,000×g for 2 min, and analyze by reducing SDS-PAGE.
3.3 Elution of GST-Rab5
While Rab5 pulldown experiments can be performed using the beads as described above, eluting and dialyzing the protein have several advantages. First, the protein can be stored at −80°C, enhancing its stability as compared to storage on beads at −20°C in glycerol. Second, when comparing GST-Rab5 to other proteins (e.g., other Rabs, or GST as a control), one can easily prepare sets of glutathione beads containing identical amounts of bound GST fusion protein.
Elute washed beads (from step 12, above) with 20 column volumes Elution Buffer. Collect 1 ml fractions.
Measure OD 280 of each fraction, blanked against Elution Buffer. Yield for a 500 ml bacterial prep is approximately 5–10 mg of GST-Rab5.
Pool peak fractions, and dialyze 2 times for at least 8 h against Wash Buffer 2, with at least a 1000-fold excess of buffer over sample. Alternatively, dialyze 3 times with a 100-fold excess of buffer over sample.
Analyze protein purity by reducing SDS-PAGE.
Freeze and store in aliquots at −80°C.
3.4 Analysis of Protein Concentration
If the eluted GST-Rab5 (or Rab of interest) appears as a single band on SDS-PAGE, then conventional protein assays (such as Biorad DC) can be used to determine protein concentration. If contaminating proteins are present in the preparation, or for analysis of GST-Rab5 bound to glutathione beads, then protein concentration of the Rab5 can be estimated by comparison to a Coomassie stained standard curve. Varying amounts of eluted protein or bead- bound protein (e.g., 10–40μl of protein or 1:1 bead slurry) are analyzed by reducing SDS-PAGE in parallel with a standard curve of a known protein (BSA, or ideally a recombinant purified Rab). After fixing and Coomassie staining, the bands can be quantitated using a LI-COR Odyssey scanner, reading at 700 nm. The slope of the standard curve and the sample curve are determined, and the estimated protein concentration is:
3.5 Preparation of Recombinant p85/p110 β
Recombinant p85/p110β can be readily prepared in HEK293T cells as epitope tagged proteins. Class IA PI 3-kinase catalytic subunits are unstable as monomers and must be co-expressed with p85 regulatory subunits [15]. For HEK293T cells, we obtain high level expression using the pSG5 vector from Stratagene. We typically use myc-p110β and HA-p85.
Plate HEK293T cells on 10 cm tissue cultures dishes in DMEM/10 % fetal bovine serum. One dish yields enough protein for three pulldown assays.
When cells reach 70 % confluence, transfect with 3μg each of plasmids coding for human p85α and p110β. While we have used Fugene HD, any commercial transfection reagent will work.
2 days after transfection, lyse the cells in 1 ml/dish HEK Lysis Buffer. Pool the samples, rotate for 15 min at 4°C, and then spin at 13,000×g for 5 min. The supernatant can be used directly for analysis of p110β-Rab5 binding (Subheading 4 below).
3.6 Nucleotide Loading of GST- Rab5A (Adapted from Ref. 14)
This section assumes the use of eluted and dialyzed GST-Rab5, which is then bound to glutathione sepharose beads. If using GST-Rab5 beads stored in 50 % glycerol, proceed as indicated but start at step 2 Use enough beads to provide 30μg of GST-Rab5. Use at least 15μl of beads per assay so the pellet will be visible during the washes.
Incubate glutathione beads with GST-Rab5 for 2 h in Wash Buffer 2 at 4°C on a rotating wheel. Use 30μg GST- Rab5 per 15μl of packed beads. To achieve effective mixing during rotation, the total volume of Wash Buffer should be at least 500μl per 1.5 ml tube.
Wash beads four times with Wash Buffer.
Wash beads 2 times with Nucleotide Loading Buffer.
Add GDP or GTP-γS to a final concentration of 1 mM. Mix and incubate at 30°C for 15 min.
Add MgCl2 to a final concentration of 20 mM.
Mix and incubate at 30°C for 3 min.
Hold on ice.
3.7 Analysis of Rab5-p85/p110 β Binding
All samples are kept on ice. Use of a refrigerated centrifuge is recommended.
Pool lysates from transfected HEK293T, then dilute and divide into the number of aliquots needed. We typically use the lysate from one 10 cm dish for three GST-Rab5 pulldowns, and dilute so as to have 0.5 ml of lysate per pulldown. Remove 50μl for analysis of total protein expression by western blot.
Add GDP or GTP-γS (10μM final) to each tube.
Add 15μl of GDP or GTP-γS-loaded GST-Rab beads per tube.
Incubate at 4°C for 2 h on wheel.
Spin down samples for 10 s at 13,000×g
Wash the pellets three times in Nucleotide Stabilization Buffer. For each wash, resuspend the pellet in buffer, invert several times to mix, and then centrifuge (see Note 5).
Remove final wash, and remove last residues of wash buffer with 25G insulin syringe.
Add 40μl Laemmli Sample Buffer containing 100 mM DTT.
Boil samples for 3 min.
Separate proteins by reducing SDS-PAGE (7.5 % resolving).
Analyze Rab5 pulldown by western blot (anti-myc for p110β, anti-HA for p85), using standard methods.
Footnotes
The combination of structural analysis (to define surface residues) and sequence analysis (to define residues conserved in orthologs) can provide a powerful approach for identifying putative binding sites that mediate protein–protein interactions. In the case of PI 3-kinase, this approach identified mutations that specifically disrupted p110β binding to Gβγ and Rab5, and p110γ binding to Gβγ. Since mutations can have conformational effects that act at a distance, this analysis should be verified using an empirical approach such as deuterium exchange/mass spectrometry [2, 16].
The pulldown experiment can be readily adapted for testing small molecule inhibitors or Rab5-p110β binding by including the inhibitors in the binding and wash steps.
The use of CHAPS in the GST-Rab5 preparation is critical to maintain solubility of the enzyme. Preparations without CHAPS precipitate during dialysis.
The samples taken for analysis at various stages (before and after induction, after lysis and clearing by centrifugation, etc.) are needed for trouble shooting in case of a poor yield. Similarly, the whole cell lysate sample from the HEK293T cells is required to insure that the substrate for the binding assay (p85/p110β) was successfully produced, and to determine whether the mutants being analyzed show altered expression.
Use of GTP-γS in the GST-Rab5 pulldown prevents hydrolysis of GTP by the intrinsic GTPase activity of Rab5. Nonetheless, the assay should be performed quickly, so that the entire washing procedure takes approximately 15 min.
References
- 1.Maier U, Babich A, Nürnberg B. Roles of non-catalytic subunits in Gβγ-induced activation of class I phosphoinositide 3-kinase isoforms beta and gamma. J Biol Chem. 1999;274(41):29311–29317. doi: 10.1074/jbc.274.41.29311. [DOI] [PubMed] [Google Scholar]
- 2.Dbouk HA, Vadas O, Shymanets A, Burke JE, Salamon RS, Khalil BD, Barrett MO, Waldo GL, Surve C, Hsueh C, Perisic O, Harteneck C, Shepherd PR, Harden TK, Smrcka AV, Taussig R, Bresnick AR, Nurnberg B, Williams RL, Backer JM. G protein-coupled receptor-mediated activation of p110beta by Gbetagamma is required for cellular transformation and invasiveness. Sci Signal. 2012;5(253):ra89. doi: 10.1126/scisignal.2003264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fritsch R, de Krijger I, Fritsch K, George R, Reason B, Kumar MS, Diefenbacher M, Stamp G, Downward J. RAS and RHO families of GTPases directly regulate distinct phosphoinositide 3-kinase isoforms. Cell. 2013;153(5):1050–1063. doi: 10.1016/j.cell.2013.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christoforidis S, Miaczynska M, Ashman K, Wilm M, Zhao L, Yip SC, Waterfield MD, Backer JM, Zerial M. Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat Cell Biol. 1999;1(4):249–252. doi: 10.1038/12075. [DOI] [PubMed] [Google Scholar]
- 5.Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 6.Horiuchi H, Lippe R, McBride HM, Rubino M, Woodman P, Stenmark H, Rybin V, Wilm M, Ashman K, Mann M, Zerial M. A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell. 1997;90(6):1149–1159. doi: 10.1016/s0092-8674(00)80380-3. [DOI] [PubMed] [Google Scholar]
- 7.Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394(6692):494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen E, Christoforidis S, Uttenweiler-Joseph S, Miaczynska M, Dewitte F, Wilm M, Hoflack B, Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol. 2000;151(3):601–612. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miaczynska M, Christoforidis S, Giner A, Shevchenko A, Uttenweiler-Joseph S, Habermann B, Wilm M, Parton RG, Zerial M. APPL proteins link Rab5 to nuclear signal transduction via an endosomal compartment. Cell. 2004;116(3):445–456. doi: 10.1016/s0092-8674(04)00117-5. [DOI] [PubMed] [Google Scholar]
- 10.Kurosu H, Katada T. Association of phosphatidylinositol 3-kinase composed of p110β-catalytic and p85-regulatory subunits with the small GTPase Rab5. J Biochem. 2001;130(1):73–78. doi: 10.1093/oxfordjournals.jbchem.a002964. [DOI] [PubMed] [Google Scholar]
- 11.Dbouk HA, Pang H, Fiser A, Backer JM. A biochemical mechanism for the oncogenic potential of the p110beta catalytic subunit of phosphoinositide 3-kinase. Proc Natl Acad Sci U S A. 2010;107(46):19897–19902. doi: 10.1073/pnas.1008739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chamberlain MD, Anderson DH. Measurement of the interaction of the p85alpha subunit of phosphatidylinositol 3-kinase with Rab5. Methods Enzymol. 2005;403:541–552. doi: 10.1016/S0076-6879(05)03047-8. [DOI] [PubMed] [Google Scholar]
- 13.Dou Z, Pan JA, Dbouk HA, Ballou LM, DeLeon JL, Fan Y, Chen JS, Liang Z, Li G, Backer JM, Lin RZ, Zong WX. Class IA PI3K p110beta subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell. 2013;50(1):29–42. doi: 10.1016/j.molcel.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadano S, Ikeda JE. Purification and functional analyses of ALS2 and its homologue. Methods Enzymol. 2005;403:310–321. doi: 10.1016/S0076-6879(05)03026-0. [DOI] [PubMed] [Google Scholar]
- 15.Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110-alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol. 1998;18:1379–1387. doi: 10.1128/mcb.18.3.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vadas O, Dbouk HA, Shymanets A, Perisic O, Burke JE, Abi Saab WF, Khalil BD, Harteneck C, Bresnick AR, Nurnberg B, Backer JM, Williams RL. Molecular determinants of PI3Kgamma-mediated activation downstream of G-protein-coupled receptors (GPCRs) Proc Natl Acad Sci U S A. 2013;110(47):18862–18867. doi: 10.1073/pnas.1304801110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubbard SJ, Thornton JM. NACCESS. Dept. of Biochemistry and Molecular Biology. University College London; London: 1993. [Google Scholar]
- 18.Zhang X, Vadas O, Perisic O, Anderson KE, Clark J, Hawkins PT, Stephens LR, Williams RL. Structure of lipid kinase p110 beta/p85beta elucidates an unusual SH2-domain- mediated inhibitory mechanism. Mol Cell. 2011;41(5):567–578. doi: 10.1016/j.molcel.2011.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]


