Abstract
Antimicrobial peptides (AMPs) are promising candidates to help circumvent antibiotic resistance, which is an increasing clinical problem. Amino-terminal copper and nickel (ATCUN) binding motifs are known to actively form reactive oxygen species (ROS) upon metal binding. The combination of these two peptidic constructs could lead to a novel class of dual-acting antimicrobial agents. To test this hypothesis, a set of ATCUN binding motifs were screened for their ability to induce ROS formation, and the most potent were then used to modify AMPs with different modes of action. ATCUN binding motif-containing derivatives of anoplin (GLLKRIKTLL-NH2), pro-apoptotic peptide (PAP; KLAKLAKKLAKLAK-NH2), and sh-buforin (RAGLQFPVGRVHRLLRK-NH2) were synthesized and found to be more active than the parent AMPs against a panel of clinically relevant bacteria. The lower minimum inhibitory concentration (MIC) values for the ATCUN-anoplin peptides are attributed to the higher pore-forming activity along with their ability to cause ROS-induced membrane damage. The addition of the ATCUN motifs to PAP also increases its ability to disrupt membranes. DNA damage is the major contributor to the activity of the ATCUN-sh-buforin peptides. Our findings indicate that the addition of ATCUN motifs to AMPs is a simple strategy that leads to AMPs with higher antibacterial activity and possibly to more potent, usable antibacterial agents.
Keywords: antimicrobial peptides, amino-terminal copper & nickel (ATCUN) binding motifs, bioinorganic chemistry, cytotoxicity
Introduction
The emergence of bacterial strains resistant to small-molecule antibiotics drives the need to develop agents with a unique mode of action. Due to their broad-spectrum antimicrobial potency and low propensity to induce drug resistance, antimicrobial peptides (AMPs) constitute an important model for the design of novel antibiotics.[1,2] AMPs differ in sequence but are usually short, amphipathic, and contain an overall positive charge.[1–3] They serve as natural antibiotics to certain multicellular organisms and exert their action against pathogens either by immune modulation or by direct interaction with pathogens, via membrane solubilization or by binding to internal microbial targets.[1,3] Because of their diverse modes of action, a large effort has been made to develop new classes of anti-infective agents based on naturally occurring AMPs.[4–8]
The formation of reactive oxygen species (ROS) has been argued to play a role in the action of the naturally occurring AMPs histatins and indolicidin.[9–11] The ROS hypothesis proposes that along with the classical antimicrobial action, histatins and indolicidin kill bacteria by inducing ROS formation.[10,11] The ROS formed can result in an increase in the susceptibility to antimicrobial agents.[10–13] Specifically, the antimicrobial action of histatins, histidine-rich peptides present in the saliva at micromolar concentrations, has been related to their ability to generate ROS after binding copper and iron ions.[14] In the case of indolicidin, it has been recently reported that electron donors, such as NADH from the tricarboxylic acid cycle play an important role in the generation of superoxides. Thus, it is clear that the synthesis of bactericidal agents with dual action could lead to the development of clinically useful molecules.[11,15] Notably, peptides have been combined with ROS production to achieve antimicrobial defense. Conjugation of photosensitizers to AMPs have led to improved antimicrobial action due to the production of ROS upon irradiation with visible light.[16–18]
The amino-terminal copper and nickel (ATCUN) binding unit,[19] H2N-AA1-AA2-His, is a naturally occurring structural feature present in peptides and proteins that binds certain metal ions through a free NH2 terminus, a histidine residue in the third position, and two backbone amide groups from residue AA2 and histidine (Figure 1).[19–21] It is commonly found in proteins that interact with metal ions, such as albumins and protamins.[19] Interestingly, the ATCUN motif is also found in AMPs histatin-3 and histatin-5.[14] Decades of investigations have shown that ATCUN sequences can interact with copper ions to form metal complexes with nuclease and protein cleavage activity.[22–27] This lytic activity is the result of oxidative damage caused by ROS formed from the CuII/CuIII cycling that arises from interaction of the bound copper ions to O2 or H2O2.[26,27] The ROS-forming ability of the ATCUN motif has been exploited to design catalytic metallodrugs.[24, 25, 28]
Figure 1.
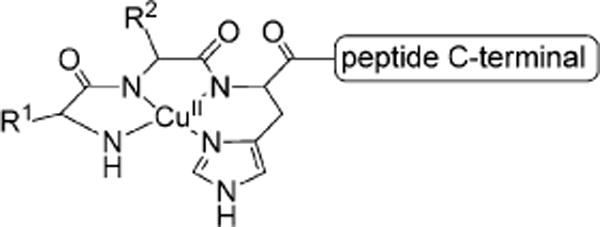
Schematic representation of the ATCUN sequence bound to a Cu2+ ion.
In the present work, several ATCUN sequences were assayed for their ability to form ROS when bound to Cu2+ ions. Three ATCUN binding motifs were selected, the two most active sequences and a weakly active ATCUN sequence as a control, and these were added to the amino terminus of three AMPs with different modes of action: 1) the membrane-disrupting peptides, anoplin (GLLKRIKTLL-NH2)[29–31] and PAP (KLAKLAKKLAKLAK-NH2),[32–34] as well as 2) the non-membrane-active peptide, sh-buforin (RAGLQFPVGRVHRLLRK-NH2).[35–37] The goal was to develop dual-activity compounds that combine the selectivity and potency of AMPs with the oxidative stress brought about by the reactivity of the ATCUN motif to yield a new class of metal binding AMPs with improved efficacies.
Results
Design and preparation of ATCUN–AMPs
A 13-membered ATCUN library was prepared by established methods using solid-phase peptide synthesis (SPPS) and standard fluorenylmethyloxycarbonyl (Fmoc) protocols. Most of the ATCUN sequences occur naturally in a variety of proteins (Table 1). We first assessed the ability of these ATCUN sequences to produce ROS after binding copper ions by using an assay that measures the rate at which the Cu–ATCUN complexes undergo multiple turnover redox process (Table 1).[24] The rate at which reduced ascorbic acid is consumed in the presence and absence of hydrogen peroxide was monitored to measure the turnover rate of the Cu–ATCUN complexes using either dioxygen or hydrogen peroxide as a co-reactant. Initial rates in the presence of hydrogen peroxide are higher than in its absence—a behavior that has been observed in other copper peptide complexes.[24] Cu–GGH and Cu–VIH gave the highest rates for ascorbic acid consumption under both conditions, indicating rapid production of ROS.
Table 1.
Summary of initial rates for ascorbic acid consumption promoted by CuII–ATCUN complexes.
| Complex | Source protein | Initial rate [μm min−1]a | |
|---|---|---|---|
| with H2O2 | without H2O2 | ||
| Cu–DAH | Human serum albumin | 1.45 ± 0.07 | 0.20 ± 0.02 |
| Cu–DMH | Neuromedin K | 1.36 ± 0.02 | 0.16 ± 0.01 |
| Cu–DSH | Histatin 5 | 1.31 ± 0.03 | 0.13 ± 0.01 |
| Cu–DTH | Bovine serum albumin | 2.89 ± 0.07 | 1.28 ± 0.06 |
| Cu–EAH | Rat serum albumin | 1.14 ± 0.04 | 0.12 ± 0.02 |
| Cu–GGH | Simplest ATCUN | 52.54 ± 0.76 | 19.35 ± 0.43 |
| Cu–GKH | – | 1.21 ± 0.09 | 0.13 ± 0.01 |
| Cu–GNH | Neuromedin C | 1.15 ± 0.04 | 0.096 ± 0.002 |
| Cu–LKH | Xnf7 | 10.8 ± 0.27 | 4.52 ± 0.17 |
| Cu–NGH | Met Lyase | 1.29 ± 0.09 | 0.18 ± 0.01 |
| Cu–RTH | Human protamine 2 | 13.12 ± 0.27 | 5.67 ± 0.20 |
| Cu–SMH | Human TBX3 | 1.39 ± 0.04 | 0.11 ± 0.01 |
| Cu–VIH | – | 39.21 ± 1.08 | 15.5 ± 0.07 |
| Cu2+ only | – | 57.21 ± 0.81 | 20.03 ± 0.37 |
| No Cu–XXH | – | 1.13 ± 0.03 | 0.19 ± 0.02 |
Data represent the mean ± standard deviation of three independent experiments.
Since the most efficient catalysts for ROS formation were the copper complexes of GGH and VIH, both sequences were selected for addition to the AMPs. DAH showed slower rates of ROS formation when compared to GGH and VIH, thus, it was selected to serve as a low ROS-producing control sequence. The ATCUN sequences were incorporated at the N terminus of the AMPs. For instance, GGH–anoplin corresponds to the amino acid sequence: GGHGLLKRIKTLL-NH2. All of the ATCUN–AMPs were synthesized by SPPS using Fmoc protocols.
Antimicrobial activity of ATCUN–AMPs
The efficacy of the ATCUN–AMPs was assessed by determining their minimum inhibitory concentration (MIC) against four different bacterial strains: Gram-positive Bacillus subtilis and Staphylococcus epidermidis and Gram-negative Escherichia coli and Enterobacter aerogenes. Data obtained using standard antimicrobial susceptibility testing are shown in Table 2.[38]
Table 2.
Minimum inhibitory concentration (MIC) values of synthesized peptides. The MIC is defined as the lowest concentration required to inhibit visual growth of bacteria.
| Peptide | MIC [μm]a | |||
|---|---|---|---|---|
| B. subtilis | S. epidermidis | E. coli | E. aerogenes | |
| Anoplin | 4 | 8 | 16 | >32 |
| DAH–anoplin | 2 | 16 | 8 | >32b |
| GGH–anoplin | 2 | 8 | 4 | >32b |
| VIH–anoplin | 0.5 | 2 | 2 | 32[b] |
| PAP | 0.5 | 4 | 1 | >32 |
| DAH–PAP | 0.25 | 4 | 0.25 | >32b |
| GGH–PAP | 0.125 | 1 | 0.125 | >32b |
| VIH–PAP | 0.125 | 0.25 | 0.25 | 32[b] |
| sh-Buforin | 8 | 8 | 32 | >32 |
| DAH–sh-buforin | 2 | 4 | 16 | >32b |
| GGH–sh-buforin | 4 | 16 | 16 | >32b |
| VIH–sh-buforin | 2 | 8 | 8 | >32b |
Data are the mode of three independent experiments. Mann–Whitney test was used to compared these results with those of the parental AMP; numbers in bold correspond to P≤0.05; all other results are not statistically significant.
Chi-square (Χ2) test was performed due to presence of ties.
The microbiological characterization showed that the ATCUN–AMPs were generally more effective against the Grampositive bacteria used in this study. Incorporation of the ATCUN motif to anoplin, PAP, and sh-buforin largely resulted in an increase in their activity, particularly when the ATCUN sequence was VIH. For instance, against E. coli, VIH–anoplin was eight-times more active than the parent peptide, whereas VIH–sh-buforin as well as VIH–PAP were four-times more active than sh-buforin and PAP, respectively. GGH was the second most active ATCUN sequence when bound to the membrane-active peptides anoplin and PAP. In the case of sh-buforin, DAH was the second most active sequence.
Since the ATCUN–AMPs bind metal ions, we tested whether the addition of copper ions could affect the antimicrobial activity. When medium was supplemented with Cu2+ ions, no decrease in the activity of the peptides against B. subtilis and E. coli was observed (Table 3). These results suggest that this family of peptides do not induce starvation for copper ions.
Table 3.
Minimum inhibitory concentration (MIC) values of ATCUN-AMPs in the presence of CuII ions.
| Cu–Peptide | MIC [μm]a | |
|---|---|---|
| B. subtilis | E. coli | |
| Cu–Anoplin | 4 | 16 |
| Cu–DAH–Anoplin | 2 | 8 |
| Cu–GGH–Anoplin | 2 | 4 |
| Cu–VIH–Anoplin | 0.5 | 2 |
| Cu–PAP | 0.25 | 1 |
| Cu–DAH–PAP | 0.5 | 0.25 |
| Cu–GGH–PAP | 0.06 | 0.06 |
| Cu–VIH–PAP | 0.03 | 0.125 |
| Cu–sh-Buforin | 8 | 16 |
| Cu–DAH–sh-Buforin | 2 | 16 |
| Cu–GGH–sh-Buforin | 4 | 8 |
| Cu–VIH–sh-Buforin | 1 | 4 |
Cu2+ ions (32 μm) were added to the culture media; data are the mode of three independent experiments.
ATCUN motif increases the membrane-permeabilizing activity of an AMP
Since the addition of the ATCUN motif could affect the ability of the AMPs to turn the bacterial membranes permeable and allow the cellular contents to leak out,[39] we assayed membrane permeabilization in E. coli by measuring the amount of β-galactosidase leakage from the bacterial cytoplasm after exposure to the ATCUN–AMPs. After isolating the leaked β-galactosidase, its activity was monitored by adding 2-nitrophenyl-β-d-galactopyranoside and measuring the increase in absorbance at 405 nm. Upon addition of the DAH motif to anoplin, we found that the membrane lytic activity of the AMP was improved (Figure 2). The addition of GGH and VIH to anoplin did not result in an enhancement of the membrane-permeabilizing property of the peptide. In contrast to the ATCUN–anoplin peptides, the addition of the GGH and VIH motifs to PAP increased its membrane-permeabilizing activity, while DAH conjugation to PAP did not result in increased lytic activity. A four-and fivefold increase in activity was observed for GGH–sh-buforin and VIH–sh-buforin, respectively.
Figure 2.
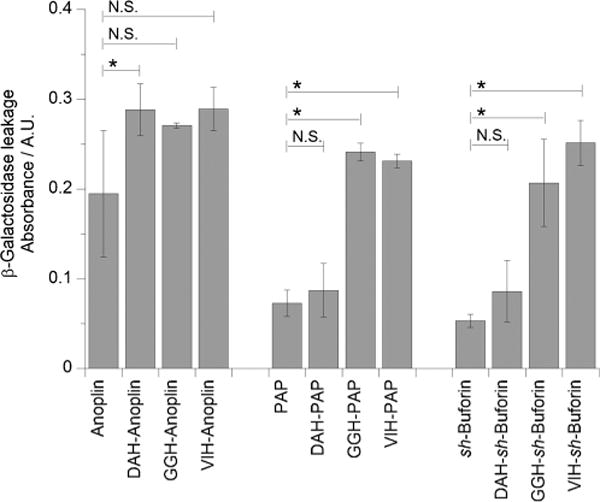
ATCUN motif increases the membrane-permeabilizing activity of anoplin, PAP, and sh-buforin. Absorbance at 405 nm obtained from leakage of β-galactosidase induced by ATCUN–AMPs. E. coli cells were incubated with 16 μm anoplin and ATCUN-anoplin; 1 μm PAP and ATCUN–PAP; and 32 μm sh-buforin and ATCUN-sh-buforin. Background leakage was subtracted from absorbance of the experimental runs. The mean ± standard deviation of three independent experiments is shown (*, P ≤ 0.05; N.S. = not significant).
ATCUN–AMPs bound to copper ions can peroxidate lipids
Since ROS formation can induce damage to bacterial membranes,[16, 40] we then evaluated whether the diffusible and short-lived ROS generated by the ATCUN–AMPs bound to copper ions (Cu–ATCUN–AMP) could potentially destabilize the bacterial membrane through oxidative stress. The oxidative damage brought about by ROS production was assessed by measuring lipid peroxidation in small unilamellar vesicles (SUVs) that mimic the E. coli outer membrane. The Cu–ATCUN–AMP complexes were prepared by mixing 1.5 equivalents of peptide with 1 equivalent of Cu2+ and pre-incubating at room temperature for 30–45 min. The slight excess of ATCUN–AMP is used to ensure no free copper ions are present, as suggested by Cowan et al.[25] Cu–ATCUN–AMPs were incubated with 80:20 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE)/1,2-dioleoyl-sn-glycero-3-phospho-1′-rac-glycerol sodium salt (DOPG) SUVs in the presence of hydrogen peroxide and sodium ascorbate. The malonyldialdehyde formed was tested via a standard thiobarbituric acid test.
As can be seen in Figure 3, the Cu–ATCUN–PAP and –sh-buforin complexes showed no differences among themselves in the amount of peroxidized lipid formed. Interestingly, the Cu–ATCUN–anoplin peptides show a trend that trails the observed MIC values of the ATCUN–AMPs; that is, Cu–VIH–anoplin and Cu–GGH–anoplin being more active than Cu–DAH–anoplin. Together, these results suggest that the increased antimicrobial action of the ATCUN derivatives of the membrane-disrupting peptide anoplin is largely influenced by the oxidative stress on the microbial membrane caused by the peptides.
Figure 3.
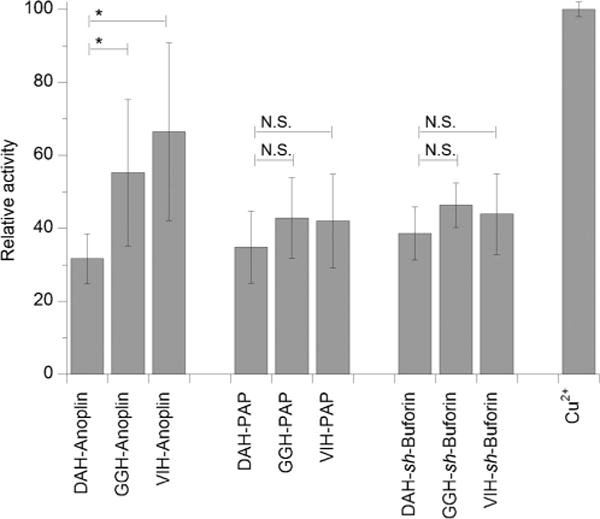
Lipid peroxidation promoted by copper complexes of the ATCUN–AMPs. Percent lipid peroxidized were normalized against the activity of free Cu2+, which was set to 100%. The mean ± standard deviation of three independent experiments is shown (*, P ≤0.05; N.S. = not significant).
Cellular localization of ATCUN–AMPs
Since modifications in the amino acid sequence of the AMPs could result in cellular localization changes, we then evaluated the localization of the AMPs and ATCUN–AMPs. We utilized laser confocal microscopy to visualize the fluorescently labeled AMPs in E. coli (Figure 4). 5(6)-Carboxyfluorescein-labeled peptides were synthesized by attaching the fluorophore (Fluo) to the ε-amino group of an additional lysine residue. The lysine residue was added to the N-terminal residue of the parent peptides anoplin (Fluo-anoplin), PAP (Fluo-PAP), and sh-buforin (Fluo-sh-buforin). The fluorescently labeled VIH–AMPs were synthesized by inserting a lysine residue after the VIH fragment.
Figure 4.
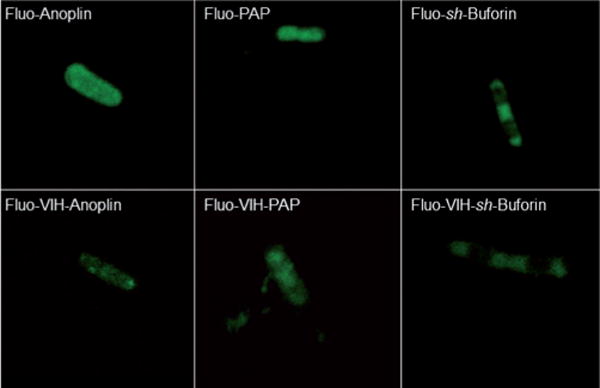
Laser confocal microscopy fluorescence images of live E. coli cells exposed to 5(6)-carboxyfluorescein-labeled anoplin (4 μm), VIH–anoplin (0.5 μm), PAP (0.25 μm), VIH–PAP (0.06 μm), sh-buforin (8 μm) and VIH–sh-buforin (2 μm) for 60 min.
Fluo-Anoplin, Fluo-PAP and their VIH derivatives localized at the bacterial outer membrane. PAP and VIH–PAP were also observed bound to the flagellum (see movie available as Supporting Information). No differences were observed in the localization of the Fluo-AMPs when compared to the VIH–Fluo-AMPs. Fluo-sh-Buforin and VIH–Fluo-sh-buforin also localize in the cytoplasmic space and bind to bacterial DNA. The affinity of Fluo-sh-buforin and its VIH derivative for DNA is nicely reflected in the images as the area where the nucleoid is localized showed decreased fluorescence intensity. This phenomenon originates from the fluorescence quenching of carboxyfluorescein by adenine and guanine bases through a photoinduced electron-transfer process.[41]
ATCUN–sh-buforin peptides bound to copper ions generate ROS inside E. coli
Considering the ability of the fluorescently labeled VIH–sh-buforin to penetrate into bacteria, we next examined the potential ability of the ATCUN–sh-buforin derivatives to generate intracellular ROS. The intracellular probe 2′,7′-dichlorofluorescein diacetate (DCFDA) was used to measure the ability of the ATCUN–sh-buforin peptides to produce ROS when combined with copper ions in E. coli (Figure 5). DCFDA diffuses through the bacterial cell membranes and is enzymatically deacetylated by intracellular esterases to the nonfluorescent 2′,7′-dihydrodichlorofluorescein.[42, 43] This highly polar molecule cannot cross the membrane and is effectively trapped inside the cell. Upon reaction with ROS, it generates the fluorescent molecule dichlorofluorescein. VIH– and GGH–sh-buforin peptides produced the largest amounts of intracellular ROS as compared to the DAH derivative. The intracellular ROS formed by VIH–sh-buforin is similar to the amount of ROS produced by 32 mm Cu2+ ions, although considerably less than the amount formed after exposure to 10 μm hydrogen peroxide.
Figure 5.
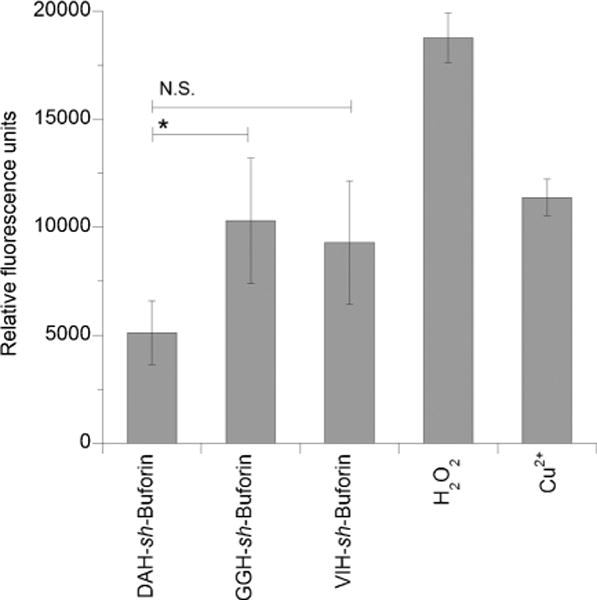
Cu-ATCUN-sh-buforin complexes promote formation of intracellular ROS in E. coli. Relative fluorescence units were used as a measure of generalized oxidative stress caused by exposing the cells to Cu-ATCUN-sh-buforin complexes ([Cu-ATCUN-sh-buforin] = MIC/4), 10 μm H2O2 or 32 μm Cu2+ ions. The mean ± standard deviation of three independent experiments is shown (*, P ≤0.05; N.S. = not significant).
ATCUN–sh-buforin peptides bound to copper ions can cleave DNA
Given that our data shows internalization of the ATCUN–sh-buforin AMPs, and it is well known that the parent buforin II peptide targets nucleic acids,[36, 37] we next investigated the effect of the ROS generated by the copper complexes of the ATCUN–sh-buforin peptides on DNA. DNA cleavage promoted by the Cu–ATCUN–sh-buforin was measured electrophoretically in vitro using pUC19 plasmid as a target. Damage to the DNA plasmid pUC19 by ROS was monitored after 30 min and 2 h of incubation with the Cu–ATCUN–sh-buforin complexes. As observed in Figure 6, a time-dependent cleavage of pUC19 was observed when incubated with all of the Cu–ATCUN–sh-buforin complexes. For the ATCUN–sh-buforin peptides, the percentage of supercoiled plasmid decreased during the 2 h incubation time, and inversely correlated with an increase in the linearized form. Altogether, this evidence points to a capability of Cu–ATCUN–sh-buforin to induce generalized oxidative stress within the bacteria, generating DNA nicks that could be responsible for the antibacterial action of ATCUN–sh-buforin.
Figure 6.
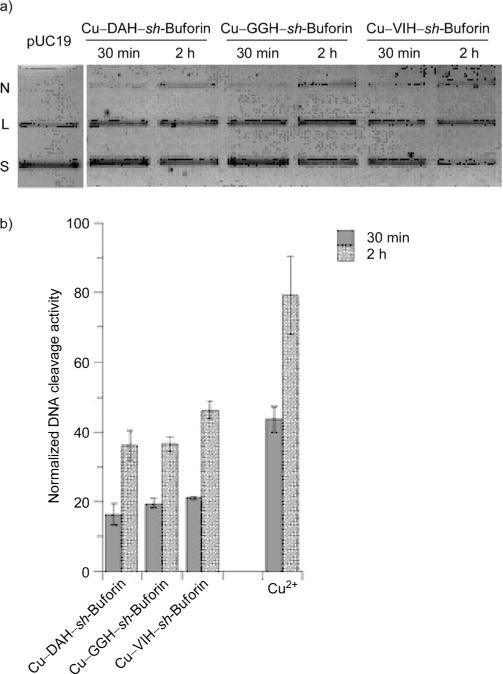
ATCUN–sh-buforin peptides bound to copper ions cleave DNA in a time-dependent fashion: a) agarose gel electrophoresis experiments showing time-dependent conversion of pUC19 from supercoiled (S) form to nicked (N) and linearized (L) forms; b) normalized DNA cleavage activity promoted by Cu–ATCUN–sh-buforin complexes.
ATCUN–AMP peptides exhibited low cytotoxicity against mammalian cells
To assess the selectivity of the tested compounds towards bacteria over mammalian cells, their hemolytic activity was investigated. Release of hemoglobin from packed human red blood cells (RBCs) was measured spectrophotometrically, and hemolysis of the peptides was normalized against the hemolytic activity of Triton X-100. As observed in Table 4, all of the synthesized peptides had low hemolytic activity at the observed MIC value for E. coli as all of the peptides had less than 10% hemolysis (with the exception of VIH–PAP with 13%), suggesting a selectivity of these peptides to lyse bacterial cells instead of RBCs. The data also showed that, upon addition of the ATCUN motif to anoplin and PAP, general membrane lytic activity increased.
Table 4.
Hemolytic activity and therapeutic index of the synthesized peptides.
| Peptide | Hemolysis [%]a | HD10 [μm]b | TIc |
|---|---|---|---|
| Anoplin | 6 ± 1 | >64 | >4 |
| DAH–anoplin | 7 ± 3 | 64 | 8 |
| GGH–anoplin | 6 ± 2 | 16 | 4 |
| VIH–anoplin | 7 ± 1 | 4 | 2 |
| PAP | 3 ± 1 | 64 | 64 |
| DAH–PAP | 4 ± 1 | 8 | 32 |
| GGH–PAP | 4 ± 1 | 4 | 32 |
| VIH–PAP | 13 ± 5 | <1 | <1 |
| sh-Buforin | 8 ± 1 | 64 | 2 |
| DAH–sh-buforin | 8 ± 2 | 32 | 2 |
| GGH–sh-buforin | 7 ± 2 | 64 | 4 |
| VIH–sh-buforin | 7 ± 2 | 64 | 8 |
% Hemolysis observed when treated with test compound at the MIC value against E.coli; data represent the mean ± standard deviation of four independent experiments.
Peptide concentration that results in lysis of 10% red blood cells (HD10); data are representative of 4 independent determinations.
The therapeutic index (TI) was calculated as HD10/MIC(E. coli).
Discussion
This study presents the development of bifunctional antimicrobial agents that combine the bactericidal/bacteriostatic action of AMPs with the ROS-forming ability of a copper(II)–ATCUN complex. We hypothesize that the new ATCUN–AMPs will take advantage of the presence of the existing pool of labile copper ions in bacteria that flux in response to environmental stimuli.[44, 45] The large affinity of the ATCUN motif for copper ions (log K~14–15) would allow successful competition for this endogenous pool of copper.[19] For this purpose, the ability of certain ATCUN sequences to form ROS was initially assessed by monitoring consumption of reduced ascorbic acid. In the presence of the oxidant hydrogen peroxide and reductant ascorbic acid, hydroxyl radicals (HO·) and other ROS are produced via the Fenton reaction,[22–26, 46] as the bound copper cycles back and forth between its +2 and +3 oxidation states. Our experiments showed that both Cu–GGH and Cu–VIH produced ROS at the highest rates, and as such were chosen to be added at the N terminus of anoplin, PAP, and sh-buforin. DAH only exhibited weak ROS forming ability, and as such, the corresponding DAH–AMPs were used as negative controls.
When the ATCUN–AMPs were tested for their potency against a panel of four different bacteria, the ATCUN motif was found to increase the activity of anoplin, PAP and sh-buforin up to a maximum of 16-fold (in the case of VIH–PAP against S. Epidermis). Some AMPs use metal chelation as their mechanism of action and do not involve the production of ROS.[47] In those cases, the addition of metal ions such as Cu2+ to the medium negatively affects their antimicrobial activity.[47] This is not the case for the ATCUN–AMPs presented in this study, since when 32 μm of Cu2+ ions were deliberately added to the growth media, the antibacterial activity of the ATCUN derivatives of anoplin, PAP, and sh-Buforin was retained.[48] This clearly indicates that metal depletion due to the chelating ability of the ATCUN–AMPs synthesized in this work is not the mechanism of action used by these peptides. The ATCUN–AMPs have a low toxicity towards mammalian cells, as tested by an assay that measures their hemolytic activity against RBCs. Although the ATCUN derivatives are slightly more hemolytic than their parent peptides, none of them cause more than 33% hemolysis at a concentration as high as 32 μm.
ATCUN–anoplin and ATCUN–PAP peptides
Anoplin is a membrane-acting AMP, isolated from the venom of the wasp Anoplius samariensis, with broad-spectrum antimicrobial activity.[29–31] Fluorescently labeled anoplin and VIH–anoplin localize within E. coli at the bacterial membrane, indicating that the addition of the ATCUN motif does not change to a major extent the affinity of the anoplin sequence for its target. The highest antibacterial activity against the four bacterial strains tested was observed for VIH–anoplin. The poreforming ability of the ATCUN derivatives seems to be larger than that of anoplin; however, no correlation between the membrane-permeabilizing activity and the MIC values is found. Importantly, ROS-mediated damage of lipids by the ATCUN–anoplin peptides in combination with copper ions correlates with the antimicrobial activity. Thus, it is reasonable to suggest that the increased antimicrobial activity of the anoplin derivatives has as the origin the nonenzymatic oxidative damage of membrane phospholipids.[49] The process is likely initiated by hydroxyl radicals since peroxyl radicals are unable to react with fatty acids with rates that are biochemically important.[40] The hydroxyl radicals produced via the ATCUN–CuIII/ATCUN–CuII cycling can initiate membrane peroxidation through a hydrogen atom abstraction from a CH2 group alpha to a carbon–carbon double bond of an unsaturated fatty acid [reaction (1)]. Next, a lipid peroxyl radical can be formed from the reaction of dioxygen with the lipid radical [reaction (2)]. The radical can then be propagated via reaction (3).
| (1) |
| (2) |
| (3) |
The de novo peptide PAP was designed to be amphipathic in its helical conformation and strongly interact with bacterial membranes.[32–34, 50] Initial studies have established that PAP is a highly active AMP with low toxicity towards mammalian cells.[32, 51] Overall, the addition of the ATCUN motif increased the antimicrobial activity of PAP. The confocal microscopy images corroborate the strong interaction of PAP towards the microbial membrane. The addition of both GGH and VIH doubled the membrane-permeabilizing activity of PAP, whereas no significant difference was observed with the DAH derivative. The increased ability to disrupt the membrane structure by the VIH and GGH derivatives can be explained by analyzing the helical wheel diagram of the three peptides (Figure 7). If a peptide adopts an α-helical conformation, as expected for PAP, the helical wheel diagram provides a simple tool for the analysis of amphipathicity.[52] As observed in Figure 7, VIH–PAP can potentially have the highest amphipathicity, followed by GGH–PAP. In DAH–PAP, the Asp residue is detrimental to the amphipaticity of the peptide, since it would appear surrounded by hydrophobic residues. The close correlation between the membrane-permeabilizing activity and antimicrobial activity is an indication that the main mechanism of action of the ATCUN–PAP peptides is the disruption of the prokaryotic cell membrane.
Figure 7.
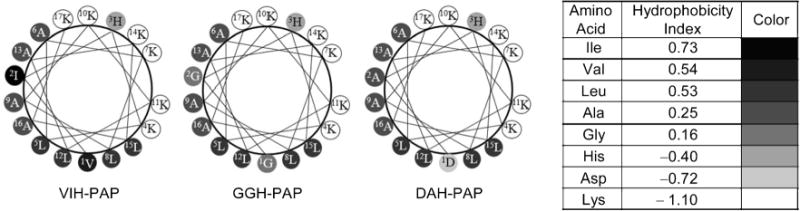
Helical wheel diagrams of ATCUN–PAP peptides. Eisenberg consensus hydrophobicity scale was used.[52]
ATCUN–sh-buforin peptides
sh-Buforin is a truncated analogue of buforin II (TRSSRAGLQFPVGRVHRLLRK), a potent AMP that kills bacteria without cell lysis and has a strong affinity for oligonucleotides.[35–37] The N-terminal random structure (residues 1–4: TRSS) does not appear to be important to the antibacterial activity of buforin II,[37] thus, the addition of the ATCUN motif to the N-terminal region of sh-buforin was not expected to perturb its mode of action. The presence of the ATCUN sequences increases the membrane-permeabilizing activity of sh-buforin in E. coli. We can evaluate the amphipathicity of the sh-buforin derivatives using a helical wheel diagram. However, in this family of peptides, we only need to analyze up to the proline residue, since this residue would act as a hinge if the α-helix were to extend throughout all the residues.[55] As observed in Figure 8, the VIH and GGH derivatives have five hydrophobic residues on one side of the helix, whereas the DAH derivative has only four. The parent peptide, sh-buforin, has only three hydrophobic residues (not shown). This indicates that the VIH– and GGH–sh-buforin peptides are more amphipathic than DAH–sh-buforin and the parent sh-buforin, resulting in an increase in the membrane-disrupting activity.
Figure 8.
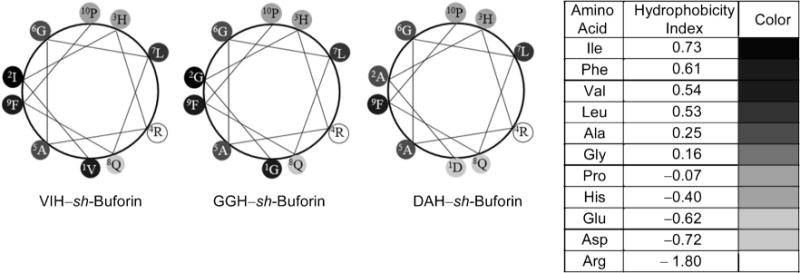
Helical wheel diagrams of ATCUN–sh-buforin peptides up to the Pro residue. Eisenberg consensus hydrophobicity scale was used.[52]
Despite the increase in membrane-permeabilization activity, it is unlikely that this property is the sole contributor to the mode of action since the increase of the antibacterial activity does not trace the lytic properties of the sh-buforin derivatives. The microscopy results reveal that both sh-buforin and VIH-sh-buforin are present in the cytoplasmic space of E. coli and bind to DNA after incubation for 1 h. In addition, the GGH and VIH derivatives produced the largest amounts of intracellular ROS as expected from the initial assay of the ATCUN motifs; however, the intracellular ROS production in E. coli does not correlate with the antimicrobial activity against this microorganism. Against E. coli, VIH-sh-buforin was the most active peptide, while DAH-sh-buforin was as active as GGH-sh-buforin. This activity correlates with the observed DNA cleavage results, in which the activity follows the order: VIH-sh-buforin > DAH-sh-buforin ≈ GGH-sh-buforin. Altogether, the results indicate that ROS reacting with the bacterial DNA is more important than the total production of intracellular ROS. Interestingly, similar nuclease activity has been recently observed and suggested as the main contributor to the increase in antibacterial activity in a metallopeptide containing the GGH motif bound to the DNA-targeting antibacterial sequence WRWYCR.[53,54]
Conclusions
The present study reveals that the addition of the ATCUN motifs DAH, GGH, and VIH leads to an increase in antimicrobial activity of the parental AMPs anoplin, PAP, and sh-buforin. Although the ATCUN–AMPs can induce the formation of ROS, the mechanistic mode of action differs among peptides and depends largely on the parental peptide. The addition of the ATCUN motif to anoplin and PAP results in an increase of their membrane-permeabilizing activity. In the case of anoplin, the membrane-permeabilization is caused by ROS-mediated damage to the lipids; whereas the activity of the PAP derivatives is related to the increase in amphipathicity. On the other hand, the ATCUN–sh-buforin peptides damage bacterial DNA due to the production of intracytoplasmic ROS and the known affinity of the sh-buforin sequence for DNA. In the field of drug design, it is well accepted that the ability to attack microbes with multiple mechanisms of activity may have better chances of generating new antibiotics with broader-spectrum and enhanced antimicrobial activity. Therefore, agents such as the ATCUN–AMPs described in this work are promising lead compounds that deserve further studies to fully describe their mechanism of action and their potential therapeutic applications.
Experimental Section
Peptide synthesis, purification, and quantification
Peptides in their C-terminal amidated forms were synthesized manually on a Rink amide resin (ChemPep Inc, Wellington, FL, USA) using standard fluorenylmethyloxycarbonyl (Fmoc)-protected L-amino acids (Matrix Innovation, Quebec, Canada). Coupling reactions were done with 4 equiv of amino acids and 4 equiv of 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU), deprotection using 20% piperidine in DMF and cleavage using 95:2.5:2.5 trifluoroacetic acid (TFA)/H2O/triisopropylsilane (TIS). Free peptides were purified via reverse phase-HPLC (Schimadzu LC-20AD) on a C18 semi-prep 250×10 mm column (Grace Davison, Deerfield, IL, USA) using 0.1 % TFA in H2O (Buffer A) and 0.1 % TFA in CH3CN (Buffer B). Tripeptides were subjected to a linear gradient of 5–95% Buffer B over 20 min, and ATCUN-AMPs were purified using a linear gradient of 30–60% Buffer B over 30 min.
5(6)-Carboxyfluorescein was purchased from Sigma–Aldrich, and labeled peptides were synthesized by attaching the fluorophore to the ε-amino group of an additional Lysine residue and purified in the same way as the ATCUN–AMPs. Pure fractions were collected, and the identities confirmed by electrospray ionization-mass spectrometry (ESI-MS) run in the positive ionization mode. All purified peptides were subsequently re-injected on a C18 analytical 250× 4.6 mm column (Grace Davison, Deerfield, IL, USA) and were found to be ≥95% pure.
All peptides were quantified using a method suggested by Gruppen.[56] Briefly, lyophilized pure peptides were dissolved in nanopure H2O, and a small aliquot (10 μL) was diluted 1:40 in 80:20:0.1 H2O/CH3CN/formic acid, and the absorbance of this solution was read using a Varian Cary 50Scan UV–Vis spectrometer at 214 nm. Molar extinction coefficients for each peptide were calculated on a sequence-specific manner based on the reported values for each amino acid and the peptide bond.
Peptide handling and preparation
All water used in this study was obtained from a Barnstead NANOpure Diamond filtration system with a 0.2 μm pore size filter. Purified peptides dissolved in nanopure water were stored at 4°C, and were diluted to the required concentration on the day of use. Concentrations were regularly checked via UV-Vis spectrophotometry. To form CuII–peptide complexes, 1.5 equiv of peptide was mixed with 1 equiv of Cu2+ (excess peptide used to ensure no free metal ions present), and the solutions were pre-incubated at room temperature for 30–45 min.[25]
Determination of ascorbic acid consumption
The rate at which ascorbic acid was consumed by the CuII–ATCUN complex was measured by the decrease in absorbance of reduced ascorbic acid. Reaction mixture contained 10 μm CuII–XXH complex with 1 mm ascorbic acid with and without 1 mm H2O2 in 20 mm 4-(2-hydroxyethyl)piperazine-1-ethanesulfonic acid (HEPES), 100 mm NaCl at pH 7.40. Absorbance at 300 nm was measured on a clear 96-well plate using a Molecular Devices FlexStation 3 plate reader. Absorbance was plotted versus time, and the slope of the linear portion of the curve was used to determine the rate constant of reaction. Ascorbic acid molar absorptivity was used to convert units of rate to μm ascorbic acid/min. Average values were taken from three trials and presented as the mean ± standard deviation.
Antimicrobial assay
Antimicrobial susceptibility testing were done using the broth microdilution method as suggested by Hancock.[38] Gram-positive bacteria Bacillus subtilis (PS832), Staphylococcus epidermidis (ATCC 12228) and Gram-negative bacteria Escherichia coli (DL7), and Enterobacter aerogenes (ATCC 13048) were grown in Mueller–Hinton broth (MHB; Difco) for 3–5 h until midlog phase was reached. Peptide stock solutions were diluted in phosphate-buffered saline (PBS; Gibco), pH 7.40, and 50 μL aliquots of two-fold serial dilutions (starting from 32 μm) were placed on a sterile 96-well poplypropylene plate (Greiner). To each well, 50 μL of a bacterial suspension was added, in a final inoculum of 5× 105 CFUmL−1 per well. Ampicillin (Sigma–Aldrich) was used as a positive control, and PBS as a negative control. Plates were incubated at 37°C (30°C for E. aerogenes) for 18–20 h. For experiments with the Cu–ATCUN–AMP complexes, the same dilutions and bacterial manipulations were done using MHB containing Cu2+. A small aliquot (62.5 μL) of a 20.5 mm Cu2+ solution was added to 40 mL of MHB to bring the final Cu2+ concentration to 32 μm (well below the toxic level of Cu2+ in E. coli, which is 3.5 mm).[57] The minimum inhibitory concentration (MIC) was defined as the concentration that prevented visual growth of bacteria, and results were confirmed by matching to a plate reader OD600nm measurement. MIC values reported here are the average of three independent trials.
β-Galactosidase leakage assay
The membrane disruption caused by the peptides was assessed by measuring the amount of leaked β-galactosidase. Overnight cultures of E. coli were inoculated in fresh Luria-Bertani (LB) broth and were grown to OD600nm≈0.6. Overexpression of β-galactosidase was induced for 1 h by addition of isopropyl-β-d-thiogalactopyranoside (IPTG; Fisher) at 1 mm final concentration. The cells were washed three times with PBS and resuspended in fresh LB broth. A 75 μL aliquot of the bacterial suspension was mixed with 75 μL of two-fold serial dilutions of the peptides (starting from 32 μm) in sterile microcentrifuge tubes. The mixture was incubated at 37°C for 1 h. After incubation, tubes were spun down at 4400 rpm at 4°C for 10 min, then 100 μL of the supernatant was transferred to a clear 96-well plate. A 50 μL aliquot of 2-nitrophenyl-β-d-galactopyranoside (ONPG; Thermo Scientific) in PBS was added at 0.8mgmL−1 final concentration. The β-galactosidase activity was monitored by measuring the increase in absorbance at 405 nm every 5 min for a period of 1 h. Data shown are the final absorbance readings for three independent trials and are presented as the mean ± standard deviation.
Lipid peroxidation assay
The extent of oxidative damage to the surface of the cell was measured by quantifying the peroxidation products of unsaturated phospholipids. 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) and 1,2-dioleoyl-sn-glycero-3-phospho-1′-rac-glycerol sodium salt (DOPG) were purchased from Avanti Polar Lipids (Alabaster, AL, USA), and an 80:20 mole percent mixture of DOPE and DOPG in CHCl3 was used to make a model E. coli membrane. The solvent was evaporated under a steady stream of N2, and the lipid cake was dried further overnight on a high vacuum line. The lipid cake was rehydrated with 20 mm HEPES, 100 mm NaCl, at pH 7.40 for 1 h, vortexed five times and sonicated for 20 min to generate small unilamellar vesicles (SUVs). A 100 μm total lipid solution of SUVs was incubated with 10 μm of CuII–ATCUN–AMP complexes with 1 mm H2O2 and 1 mm sodium ascorbate in rehydration buffer for 1 h. Then, 50 μL of butylated hydroxytoluene was added followed by 1.5 mL of 0.44 m H3PO4. The mildly acidic mixture was incubated for 10 min prior to addition of 500 μL of 2-thiobarbituric acid (TBA). The mixture was then heated on a dry block heater set at 90°C for 30 min. After cooling to room temperature, the amount of malonyldialdehyde–TBA adduct was quantified by injecting 50 μL of the final reaction mixture in a C18 analytical RP-HPLC column. Elution was done using 35% MeOH and 65% 50 mm KH2PO4/KOH buffer at pH 7.00, with monitoring at 532 nm. The peak at ~4.6 min corresponds to the pink MDA-TBA adduct, and the area under the curve was used to quantify the amount of adduct present. The average value obtained from three independent trials are reported and presented as the mean ± standard deviation.
Measurement of intracellular oxidative damage
To measure the extent of generalized intracellular oxidative stress brought about by the Cu–ATCUN–AMP complexes, the fluorescence of dichlorofluorescein was quantified. E. coli cells in mid-logarithmic phase were washed with fresh MHB and resuspended in M9 + glucose minimal media containing 10 μm of the profluorescent compound 2′,7′-dichlorofluorescein diacetate. The dye was loaded into the cells for 1 h before washing unloaded dye away. The loaded cells were resuspended in fresh MHB and allowed to grow for an additional 30 min, after which the cells were incubated with the Cu–ATCUN–AMP complexes at one-quarter the MIC value for 1 h. The resulting fluorescence was measured using a Molecular Devices FlexStation 3 plate reader set to a top read mode. H2O2 (10 μm) and Cu2+ (32 μm) was used as a positive control, while loaded cells without any peptide was used as a negative control to account for oxidation of the dye by other cellular components. Media containing 10 μm of 2′,7′-dichlorofluorecein diacetate was used as background to account for autooxidation of extracellular dye by components of the media. The average value obtained from three independent trials are shown as mean ± standard deviation.
Confocal fluorescence microscopy imaging
Images were acquired on Zeiss LSM-510 confocal microscope using a 63 × (1.4 N.A.) oil immersion objective at a pixel resolution of 512 × 512, and then processed utilizing ImageJ (US National Institutes of Health).
DNA cleavage studies
In vitro DNA damage brought about by the CuII–peptide complexes were assessed by agarose gel electrophoresis. Reaction mixtures contained 10 μm base pair pUC19, 100 nm CuII–ATCUN–AMP with 1 mm H2O2 and 1 mm sodium ascorbate in 20 mm HEPES, 100 mm NaCl, at pH 7.40. Reaction was incubated at room temperature and quenched by adding 3X loading dye containing 1 mm EDTA at two time points—30 min and 2 h. Then, a 15 μL aliquot was loaded on a 1 % agarose gel containing ethidium bromide (EtBr) and run at 80 V for 90 min. Gels were imaged using a Bio-Rad GelDoc XR+ Imager, and bands were quantified using the accompanying Image Lab 5.0 software. A correction factor or 1.47 was applied to the intensity of the supercoiled form to account for its decreased ability to intercalate EtBr.[25] Normalized DNA cleavage activity was calculated using the initial and final amounts of supercoiled DNA according to the formula: [(initial–final)/initial] × 100. Quantified supercoiled DNS is shown as mean ± standard deviation of three independent measurements.
Hemolytic assay
To assess the selectivity of the peptides towards bacterial membrane disruption, extent of hemolysis was measured in human red blood cells (RBCs). Packed human erythrocytes (ZenBio Inc, Research Triangle, NC, USA) with anticoagulant citrate dextrose were washed three times with sterile PBS. A small aliquot of washed cells were resuspended in fresh PBS to make a 0.8% (v/v) solution of RBCs. Then, a 75 μL aliquot of RBCs was mixed with a 75 μL aliquot of a two-fold serial dilution series of the peptides, and then incubated at 37°C for 1 h. Triton X-100 and PBS were used as positive and negative controls, respectively. The tubes were then spun down at 4400 rpm at 4°C for 10 min, and 100 μL of the supernatant was transferred to a clear 96-well plate. The absorbance at 414 nm was measured and normalized against the absorbance of the positive and negative controls. Data were obtained from four independent trials and presented as the mean ± standard deviation.
Statistical analysis
Data were analyzed for statistical differences using GraphPad Prism software 5.0. The Mann–Whitney asymptotic U-test was used for comparison because a Gaussian distribution could not be assumed. Whenever ties were present, comparison was done using a Chi-square test. Statistical significance for all tests was set at P < 0.05.
Supplementary Material
Acknowledgments
A.M.A.-B. acknowledges the University of Connecticut (USA) for start-up funds. The authors are grateful to Prof. Peter Setlow and Prof. Ashis Basu for their generous gift of B. subtilis and E. coli, respectively.
Footnotes
Supporting Information
Synthesis and characterization of peptides, and plots of ascorbic acid consumption are provided in the Supporting Information available via http://dx.doi.org/10.1002/cmdc.201402033.
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cmdc.201402033.
References
- 1.Hancock REW, Sahl HG. Nat Biotechnol. 2006;24:1551–1557. doi: 10.1038/nbt1267. [DOI] [PubMed] [Google Scholar]
- 2.Zasloff M. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 3.Epand RM, Vogel HJ. Biochim Biophys Acta Biomembr. 1999;1462:11–28. doi: 10.1016/s0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 4.Slootweg JC, van Schaik TB, Quarles HC, Breukink E, Liskamp RMJ, Rijkers DTS. Bioorg Med Chem Lett. 2013;23:3749–3752. doi: 10.1016/j.bmcl.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 5.Meinike K, Hansen PR. Protein Pept Lett. 2009;16:1006–1011. doi: 10.2174/092986609789055421. [DOI] [PubMed] [Google Scholar]
- 6.Bagheri M, Beyermann M, Dathe M. Bioconjugate Chem. 2012;23:66–74. doi: 10.1021/bc200367f. [DOI] [PubMed] [Google Scholar]
- 7.Anantharaman A, Sahal D. J Med Chem. 2010;53:6079–6088. doi: 10.1021/jm100483y. [DOI] [PubMed] [Google Scholar]
- 8.Chu-Kung AF, Bozzelli KN, Lockwood NA, Haseman JR, Mayo KH, Tirrell MV. Bioconjugate Chem. 2004;15:530–535. doi: 10.1021/bc0341573. [DOI] [PubMed] [Google Scholar]
- 9.Veerman EC, Nazmi K, Van’t Hof W, Bolscher JG, Den Hertog AL, Nieuw Amerongen AV. Biochem J. 2004;381:447–452. doi: 10.1042/BJ20040208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helmerhorst EJ, Troxler RF, Oppenheim FG. Proc Natl Acad Sci USA. 2001;98:14637–14642. doi: 10.1073/pnas.141366998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z, Cai Y, Young AW, Totsingan F, Jiwrajka N, Shi Z, Kallenbach NR. Med Chem Commun. 2012;3:1548–1554. [Google Scholar]
- 12.Fang FC. Nat Rev Microbiol. 2004;2:820–832. doi: 10.1038/nrmicro1004. [DOI] [PubMed] [Google Scholar]
- 13.Jiang N, Tan NS, Ho B, Ding JL. Nat Immunol. 2007;8:1114–1122. doi: 10.1038/ni1501. [DOI] [PubMed] [Google Scholar]
- 14.Melino S, Santone C, Di Nardo P, Sarkar B. FEBS J. 2014;281:657–672. doi: 10.1111/febs.12612. [DOI] [PubMed] [Google Scholar]
- 15.Pokrovskaya V, Baasov T. Expert Opin Drug Discovery. 2010;5:883–902. doi: 10.1517/17460441.2010.508069. [DOI] [PubMed] [Google Scholar]
- 16.Johnson GA, Muthukrishnan N, Pellois JP. Bioconjugate Chem. 2013;24:114–123. doi: 10.1021/bc3005254. [DOI] [PubMed] [Google Scholar]
- 17.Liu F, Ni ASY, Lim Y, Mohanram H, Bhattacharjya S, Xing B. Bioconjugate Chem. 2012;23:1639–1647. doi: 10.1021/bc300203d. [DOI] [PubMed] [Google Scholar]
- 18.Dosselli R, Gobbo M, Bolognini E, Campestrini S, Reddi E. ACS Med Chem Lett. 2010;1:35–38. doi: 10.1021/ml900021y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harford C, Sarkar B. Acc Chem Res. 1997;30:123–130. [Google Scholar]
- 20.Du X, Wang X, Liu Q, Ni J, Sun H. Chem Commun. 2013;49:9134–9136. doi: 10.1039/c3cc45360j. [DOI] [PubMed] [Google Scholar]
- 21.Donaldson LW, Skrynnikov NR, Choy WY, Muhandiram DR, Sarkar B, Forman-Kay JD, Kay LE. J Am Chem Soc. 2001;123:9843–9847. doi: 10.1021/ja011241p. [DOI] [PubMed] [Google Scholar]
- 22.Long EC, Fang Y, Lewis MA. ACS Symp Ser. 2009;1012:219–241. [Google Scholar]
- 23.Cowan JA. Pure Appl Chem. 2008;80:1799–1810. [Google Scholar]
- 24.Jin Y, Lewis MA, Gokhale NH, Long EC, Cowan JA. J Am Chem Soc. 2007;129:8353–8361. doi: 10.1021/ja0705083. [DOI] [PubMed] [Google Scholar]
- 25.Joyner JC, Reichfield J, Cowan JA. J Am Chem Soc. 2011;133:15613–15626. doi: 10.1021/ja2052599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Margerum DW. Pure Appl Chem. 1983;55:23–34. [Google Scholar]
- 27.Neupane KP, Aldous AR, Kritzer JA. Inorg Chem. 2013;52:2729–2735. doi: 10.1021/ic302820z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joyner JC, Cowan JA. Braz J Med Biol Res. 2013;46:465–485. doi: 10.1590/1414-431X20133086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Konno K, Hisada M, Fontana R, Lorenzi CCB, Naoki H, Itagaki Y, Miwa A, Kawai N, Nakata Y, Yasuhara T, Ruggiero NJ, de Azevedo WF, Palma MS, Nakajima T. Biochim Biophys Acta Protein Struct Mol Enzymol. 2001;1550:70–80. doi: 10.1016/s0167-4838(01)00271-0. [DOI] [PubMed] [Google Scholar]
- 30.Ifrah D, Doisy X, Ryge T, Hansen PR. J Pept Sci. 2005;11:113–121. doi: 10.1002/psc.598. [DOI] [PubMed] [Google Scholar]
- 31.Dos Santos Cabrera MP, Arcisio-Miranda M, Costa STB, Konno K, Ruggiero JR, Procopio J, Ruggiero NJ. J Pept Sci. 2008;14:661–669. doi: 10.1002/psc.960. [DOI] [PubMed] [Google Scholar]
- 32.Javadpour MM, Juban MM, Lo WCJ, Bishop SM, Alberty JB, Cowell SM, Becker CL, McLaughlin ML. J Med Chem. 1996;39:3107–3113. doi: 10.1021/jm9509410. [DOI] [PubMed] [Google Scholar]
- 33.McGrath DM, Barbu EM, Driessen WHP, Lasco TM, Tarrand JJ, Okhuysen PC, Kontoyiannis DP, Sidman RL, Pasqualini R, Arap W. Proc Natl Acad Sci USA. 2013;110:3477–3482. doi: 10.1073/pnas.1221924110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim HY, Kim S, Youn H, Chung JK, Shin DH, Lee K. Biomaterials. 2011;32:5262–5268. doi: 10.1016/j.biomaterials.2011.03.074. [DOI] [PubMed] [Google Scholar]
- 35.Park CB, Kim MS, Kim SC. Biochem Biophys Res Commun. 1996;218:408–413. doi: 10.1006/bbrc.1996.0071. [DOI] [PubMed] [Google Scholar]
- 36.Yi G, Park CB, Kim SC, Cheong C. FEBS Lett. 1996;398:87–90. doi: 10.1016/s0014-5793(96)01193-3. [DOI] [PubMed] [Google Scholar]
- 37.Park CB, Yi KS, Matsuzaki K, Kim MS, Kim SC. Proc Natl Acad Sci USA. 2000;97:8245–8250. doi: 10.1073/pnas.150518097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wiegand I, Hilper K, Hancock REW. Nat Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 39.Brogden KA. Nat Rev Microbiol. 2005;3:238–250. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 40.Bielski BHJ, Arudi RL, Sutherland MW. J Biol Chem. 1983;258:4759–4761. [PubMed] [Google Scholar]
- 41.Torimura M, Kurata S, Yamada K, Yokomaku T, Kamagata Y, Kanagawa T, Kurane R. Anal Sci. 2001;17:155–160. doi: 10.2116/analsci.17.155. [DOI] [PubMed] [Google Scholar]
- 42.Zhu H, Bannenberg GL, Moldeus P, Shertzer HG. Arch Toxicol. 1994;68:582–587. doi: 10.1007/s002040050118. [DOI] [PubMed] [Google Scholar]
- 43.Jakubowski W, Bartosz G. Cell Biol Int. 2000;24:757–760. doi: 10.1006/cbir.2000.0556. [DOI] [PubMed] [Google Scholar]
- 44.Rubino JT, Franz KJ. J Inorg Biochem. 2012;107:129–143. doi: 10.1016/j.jinorgbio.2011.11.024. [DOI] [PubMed] [Google Scholar]
- 45.a Fung DKC, Lau WY, Chan WT, Yan A. J Bacteriol. 2013;195:4556–4568. doi: 10.1128/JB.00543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Outten FW, Munson GP. J Bacteriol. 2013;195:4553–4555. doi: 10.1128/JB.00891-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Retsky KL, Freeman MW, Frei B. J Biol Chem. 1993;268:1304–1309. [PubMed] [Google Scholar]
- 47.Silva FD, Rezende CA, Rossi DCP, Esteves E, Dyszy FH, Schreier S, Gueiros-Filho F, Campos CB, Pires JR, Daffre S. J Biol Chem. 2009;284:34735–34746. doi: 10.1074/jbc.M109.016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Copper MIC value for E. coli was found to be 60 mm, a concentration well above the supplemented amount. For antibacterial activity data of ATCUN–AMPs in combination with 32 μm of Cu2+ ions, see Supporting Information.
- 49.Girotti AW. J Lipid Res. 1998;39:1529–1542. [PubMed] [Google Scholar]
- 50.Dathe M, Wieprecht T, Nikolenko H, Handel L, Maloy WL, MacDonald DL, Beyermann M, Bienert M. FEBS Lett. 1997;403:208–212. doi: 10.1016/s0014-5793(97)00055-0. [DOI] [PubMed] [Google Scholar]
- 51.Angeles-Boza AM, Erazo-Oliveras A, Lee Y-J, Pellois J-P. Bioconjugate Chem. 2010;21:2164–2167. doi: 10.1021/bc100130r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eisenberg D, Weiss RM, Terwilliger TC, Wilcox W. Faraday Symp Chem Soc. 1982;17:109–120. [Google Scholar]
- 53.Joyner JC, Hodnick WF, Cowan AS, Tamuly D, Boyd R, Cowan JA. Chem Commun. 2013;49:2118–2120. doi: 10.1039/c3cc38977d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kepple KV, Boldt JL, Segall AM. Proc Natl Acad Sci USA. 2005;102:6867–6872. doi: 10.1073/pnas.0409496102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee JK, Gopal R, Park SC, Ko HS, Kim Y, Hahm KS, Park Y. PLoS One. 2013;8:e67597. doi: 10.1371/journal.pone.0067597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kuipers BJH, Gruppen H. J Agric Food Chem. 2007;55:5445–5451. doi: 10.1021/jf070337l. [DOI] [PubMed] [Google Scholar]
- 57.Grass G, Rensing C. J Bacteriol. 2001;183:2145–2147. doi: 10.1128/JB.183.6.2145-2147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


