Abstract
Small molecule based therapeutic intervention of amyloids has been limited by their low solubility and poor pharmacokinetic characteristics. We report here, the use of water soluble poly lactic-co-glycolic acid (PLGA)-encapsulated curcumin and emetine nanoparticles (Cm-NPs and Em-NPs, respectively), as potential modulators of gelsolin amyloidogenesis. Using the amyloid-specific dye Thioflavin T (ThT) as an indicator along with electron microscopic imaging we show that the presence of Cm-NPs augmented amyloid formation in gelsolin by skipping the pre-fibrillar assemblies, while Em-NPs induced non-fibrillar aggregates. These two types of aggregates differed in their morphologies, surface hydrophobicity and secondary structural signatures, confirming that they followed distinct pathways. In spite of differences, both these aggregates displayed reduced toxicity against SH-SY5Y human neuroblastoma cells as compared to control gelsolin amyloids. We conclude that the cytotoxicity of gelsolin amyloids can be reduced by either stalling or accelerating its fibrillation process. In addition, Cm-NPs increased the fibrillar bulk while Em-NPs defibrillated the pre-formed gelsolin amyloids. Moreover, amyloid modulation happened at a much lower concentration and at a faster rate by the PLGA encapsulated compounds as compared to their free forms. Thus, besides improving pharmacokinetic and biocompatible properties of curcumin and emetine, PLGA conjugation elevates the therapeutic potential of both small molecules against amyloid fibrillation and toxicity.
Introduction
Pathogenic manifestations in several neurodegenerative and systemic diseases including Alzheimer’s, Parkinson’s, Huntington’s, type II diabetes and prion diseases have been mainly attributed to fibrillar protein aggregates called amyloids [1–4]. Amyloid formation is the resultant of perturbed protein folding due to mutations, weakened ubiquitin-proteasome machinery, oxidative stress and metal imbalance [5, 6]. The current pharmacological intervention of amyloid diseases is limited owing to lack of information on the mechanism and the actual toxic species involved. Several non-specific chaperones and small molecules have been shown to alter amyloid pathway in a variety of amyloidogenic proteins but their pharmacokinetic relevance is yet to be tested [7–11]. Nevertheless, the ability of small molecules to induce conformational changes in proteins at low concentrations, negligible immunogenicity and ease of in vitro and in vivo testing makes them a preferred choice as amyloid modulators [12–14].
Gelsolin is an actin-binding protein that remodels the actin cytoskeleton as well as scavenges actin fibrils released from injured tissues [15]. However, a mutation (D187N/Y) in gelsolin causes Familial Amyloidosis of Finnish Type (FAF), a disease characterized by amyloid deposits of an 8 kDa protein fragment (fAGel, residues 173–243, Fig 1). FAF is a systemic amyloidosis disease characterized by cranial lattice dystrophy which progresses with age and is associated with severe neuropathy [16]. Previously, we reported curcumin and emetine as potent binders of amyloidogenic stretch in gelsolin and modulate its amyloidogenic pathway [17]. However, a major drawback in the applicability of both curcumin and emetine in physiological systems is their poor solubility, high hydrophobicity and instability [18].
Fig 1. The crystal structure of human plasma gelsolin (PDB: 3FFN) showing the presence of 8 kDa amyloidogenic fragment (fAGel; highlighted in red).
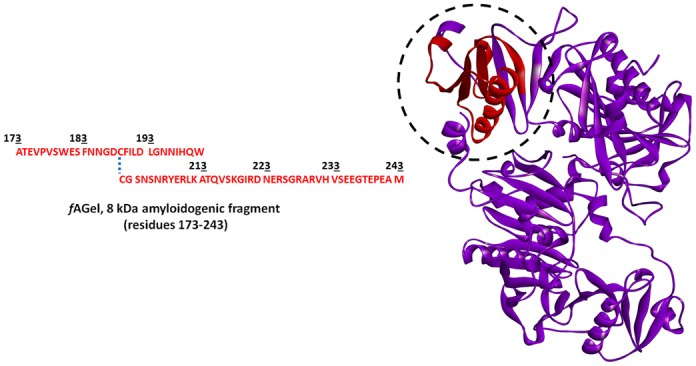
The amino acid sequence of fAGel is also shown.
A practical way to overcome this drawback is the preparation of nanoparticles (NPs) that entail higher solubility and surface availability, along with improved stability [19–22]. Generally, an increase in solubility enhances bioavailability that delays rapid metabolism and systemic elimination [23]. Moreover, NPs can pass through the blood-brain barrier and due to high levels of surface free energies it shows increased adsorption [24–26]. PLGA (Poly (lactic-co-glycolic acid)) is formed by copolymerization of the cyclic dimers of glycolic acid and lactic acid. The ester linkage of PLGA undergoes hydrolysis in aqueous systems, releasing the monomers which are easily metabolized and removed from the body [27]. Moreover, PLGA nanoparticles with some surface modification having sizes below 300 nm have been shown to pass through the blood brain-barrier [28, 29].
Thus, owing to the biocompatible and biodegradable properties, PLGA has been approved by the Food and Drug Administration for usage in therapeutics [30]. Here we report that PLGA-loaded curcumin and emetine nanoparticles modulate the fibrillation of a disease-associated gelsolin amyloidogenic fragment (fAGel) in a much efficient way as compared to the non-encapsulated forms of the compounds.
Materials and Methods
Chemicals and reagents
Curcumin, Poly (D,L-lactide-co-glycolide) (PLGA), MW (40,000–75,000) co-polymer ratio (50:50), Poly (vinyl alcohol) (PVA), average MW (30,000–70,000), viscosity 4–6 cP (4% aqueous solution, 20°C), Dimethyl sulfoxide (DMSO) and Dichloro-methane (DCM) were purchased from Sigma-Aldrich, USA. Emetine was purchased from Calbiochem, India. The cell culture medium (RPMI 1640), penicillin-streptomycin and fetal bovine serum (FBS) were obtained from Gibco BRL (Life technology, Paisley, Scotland). All other chemicals of analytical grade were purchased from Merck (India).
Cloning, expression and purification of Protein
The 213 base pair DNA sequence coding for the 8 kDa (Ala 173-Met 243) amyloidogenic fragment (fAGel) was amplified by polymerase chain reaction (PCR) using human plasma gelsolin cDNA template (Gene bank accession number AK295572.1). The following primer pairs CCGTCCATGGGCCACCGAGGTACCTGTG and CCGTCTCGAGCATCGCCTCGGGCTCAGTG were used. The amplified cDNA sequence was inserted in the pGEX-6P2 vector (GE Healthcare, USA) downstream of a GST coding sequence (NcoI and XhoI). This GST- fAGel fusion clone was transformed into E. coli BL21 (DE3) cells (Promega, USA) for protein expression using the standard IPTG induction protocol. The cells were grown in Luria broth (LB), containing 100 μM ampicillin at 37°C shaking, until an OD600nm of 0.6 was reached. At this stage, the cells were induced by 1 mM IPTG and further grown for 4–5 h before harvesting by centrifugation (3000 rcf). The cell pellet obtained was re-suspended in buffer A (50 mM Tris, 100 mM NaCl; pH 8.0) containing 1X protease inhibitor cocktail (Sigma Inc., USA) and incubated for 30 minutes on ice before sonicating for 15 cycles (2 min ON/1 min OFF) at 40 W (output 6; 50% duty cycle) in ice in a Q700 sonicator (QSonica, USA). The sonicated cell suspension was centrifuged at 12000 rcf for 1 h to remove cell debris. The resulting cell lysate was mixed with glutathione sepharose resin (GST resin, GE Healthcare, USA) pre-equilibrated with buffer A and left for incubation at 4°C for 1 h to allow the protein to bind. After the incubation period, the mixture was loaded onto a column (column volume 5 ml) in batch mode and the flow through was collected. After washing the column with buffer A (5 column volumes), protein was eluted with pre-chilled elution buffer (50 mM Tris, 100 mM NaCl and 10 mM reduced glutathione; pH 8.0). Following this, the eluted fractions were collected and dialyzed extensively against buffer A at 4°C to remove glutathione. The purified GST-tagged protein was incubated overnight with precision protease (0.2 U per mg protein) at 4°C to remove the GST-tag. The cleavage was confirmed by SDS-PAGE (15%). The residual GST-tags were removed by loading the protein solution onto a GST column, pre-equilibrated with buffer A. A second step of purification involved 10 kDa centrifugal filters to remove any residual GST impurity. The obtained 8 kDa fAGel fragment was confirmed on 20% SDS PAGE, was concentrated, lyophilized and stored at -80°C.
Preparation of Nanoparticles
Emetine and curcumin were encapsulated in PLGA by water/oil/water and oil/water method, respectively [31, 32]. A 10 mM stock solution of both curcumin and emetine were made in DMSO. Initially, 90 mg of PLGA was added to 2 ml of DCM along with 1 ml stock solution of curcumin and vortexed to prepare the primary emulsion. In the case of emetine, stock solution of emetine was added in PLGA-DCM solution step by step (100 μl at a time) and vortexed, followed by addition into the 2% PVA solution (12 ml) and mixed. Both kinds of suspensions (~15 ml each) were sonicated (output 6; 40% duty cycle) for 3 minutes for secondary emulsion and were left on stirrer overnight to evaporate the DCM by desiccation. Following three washing steps by centrifugation (45,000 rcf at 4°C, 20 minutes), pellets were resuspended in miliQ water. Larger aggregates were removed by centrifugation and supernatant was lyophilized.
Particle size and zeta potential of Nanoparticles
Zeta potential and size of the curcumin and emetine nanoparticles was determined using Zetasizer Nano ZS 3000 (Malvern Instruments, UK) as described earlier [33]. For particle size, scans were taken in duplicates with a run of 10 seconds correlation time. Intensity of distribution (%) of particles was plotted against hydrodynamic radius (nm). For zeta potential, measurements of blank and NPs in milliQ water were taken using default parameters of dielectric constant, refractive index, and viscosity. All measurements were done in triplicates at 25°C. Total counts of particles were plotted against Zeta potential (mV). The built in Zetasizer software 6.01 was used for all analysis.
Determination of entrapment efficiency (EE)
Curcumin and emetine content in the NPs were determined using UV-spectrophotometric analysis. A standard curve was made by taking absorbance of the compounds with a range of known concentrations. The absorbance was taken at 425 nm and 236 nm for curcumin and emetine, respectively [34, 35]. The test samples were prepared by dissolving the NPs in 0.1 N NaOH and heating at 50°C for an hour to ensure complete release of the compounds. The absorbance of resulting supernatant was taken at respective wavelengths and concentration was calculated from the standard curve. All absorbance values were recorded in triplicates and the entrapment efficiency was calculated using the following equation, where the fraction is defined as actual loading (μg/mg) of compounds [36]
FTIR spectroscopy
FTIR analysis of NPs was performed to confirm the functional groups of the compounds. For this, dried PLGA-NPs, Cm-NPs and Em-NPs were mixed with spectroscopic grade KBr (Sigma-Aldrich), pressed into a pellet and the spectrum taken immediately. All spectra were recorded using a Nicolet 6700 FTIR spectrometer (Thermo Scientific, USA) with an inbuilt (MCT A) photo detector purged with dry air. For characterizing the secondary structural signatures of the aggregates, ATR-FTIR was used. For this, 24 h incubated aggregates were centrifuged and resuspended in 50 μl of 0.1 M acetate buffer, pH 5.0. A drop of aggregate solution was applied on the germanium crystal of the horizontal ATR sampling accessory. In all cases data was acquired at a resolution of 4 cm-1 and 128 scans were averaged per sample. The background (buffer/KBr) was subtracted. The amide I region between 1700 cm-1 to 1600 cm-1 was analysed for characteristic changes in the polypeptide chain of aggregated species based on previously described peak assignments [37, 38]. The OMNIC 32 software was used to record and process the spectra.
Aggregation assay
For aggregation reactions, samples containing fAGel alone (25 μM, 2 mg/ml) or with varying concentrations of NPs (0.01, 0.05, 0.1 mg/ml) were made in 50 mM acetate buffer (pH 4.0) containing 150 mM NaCl, 0.02% NaN3 and incubated at 37°C, without agitation [39]. For defibrillation assay, fAGel fibrils were grown up to 4 days and then NPs were added (0.05, 0.1 mg/ml) into the solution and incubated at 37°C, without agitation.
Dye binding assays
Aliquots withdrawn at different time points and were diluted into buffer (50 mM acetate buffer, pH 5.0) containing 10 μM ThT and adjusted to a final volume of 100 μl. Fluorescence spectra of these samples were recorded by using LS 55 fluorescence spectrometer (Perkin Elmer, MA, USA), keeping the excitation and emission wavelengths at 440 nm and 485 nm, respectively. The averaged fluorescence data points from triplicate reactions for each concentration were fitted to sigmoidal curve using the following equation in Origin 8.0.
Here, A1 and A2 represent initial and final fluorescence intensities respectively, t0.5 is the time at half-maximum fluorescence and k is the rate constant. The lag phase was calculated as t0.5-2k.
For Nile red binding assay, aliquots were diluted with 50 mM acetate buffer, pH 5.0 containing 5 μM Nile red and adjusted to a final volume of 100 ul. Fluorescence emission spectra of triplicate samples were recorded from 550–700 nm after excitation at 530 nm. In all cases, the excitation and emission slit width were kept at 5 nm and 10 nm respectively.
Transmission Electron Microscopy (TEM)
TEM images of aggregating samples were acquired using Tecnai transmission electron microscope (FEI, Eindhoven, The Netherlands) equipped with a bright field/dark field detector operating at 120 kV. The fAGel aggregates formed in presence or absence of Cm-NPs or Em-NPs obtained after one week of incubation were imaged. 2–3 μl of 5-fold diluted samples were placed on copper grid for 2 minutes. Following this, the samples were negatively stained using a 2% (w/v) uranyl acetate solution and washed with milliQ water, air dried and imaged. The imaging was done at a magnification of 86,000X with an electron dose of 20–30 e/Å2.
Atomic Force Microscopy (AFM)
Aqueous suspension of NPs or three fold diluted aggregating samples (~5 ul) were applied on freshly peeled mica surface. After 5–10 minutes, samples were washed and dried under nitrogen flush. Imaging was done using Bioscope Catalyst AFM (Bruker, USA) in tapping mode. Images were processed using nanoscope analysis v.1.4.
Scanning Electron Microscopy (SEM)
Aqueous suspensions of NPs were spread evenly over a slab, dried and gold-coated. Similarly, fAGel aggregating samples incubated for 2 weeks, with or without NPs were also prepared. Each sample was visualized using scanning electron microscope (Evo50, Zeiss, UK) operating at 5 keV.
Cell culture and toxicity assessment
The human SH-SY5Y cell line (ATCC, USA) was maintained in DMEM supplemented with 10% fetal bovine serum (Gibco, UK) and incubated at 37°C under 5% CO2. About 6x104 viable cells were seeded in 96 well plates with overnight media replacement. The fAGel aggregates formed after 24 hour of incubation either alone or in the presence of different concentrations of NPs were pelleted and resuspended in PBS (NaCl 137 mM, KCl 2.7 mM, Na2HPO4 10 mM, KH2PO4 1.8 mM). These aggregates (25 μl) were added to the cells and incubated for 24 hours. Following this, the medium was replaced with the MTT containing medium (1 mg/ml) and the cells were further incubated for 4 hours at 37°C. After incubation, medium was removed and cells were diluted in 200 μl of DMSO. The relative formazan formation in each well was measured by determination of absorbance at 570 nm in a microplate reader (MultiSkan, Thermo Scientific, USA). The resulting absorbance values were converted to percentage viability with respect to the untreated control cells. The differences between the sampling groups were checked for statistical significance using student's t-test.
Results
Preparation and characterization of nanoparticles
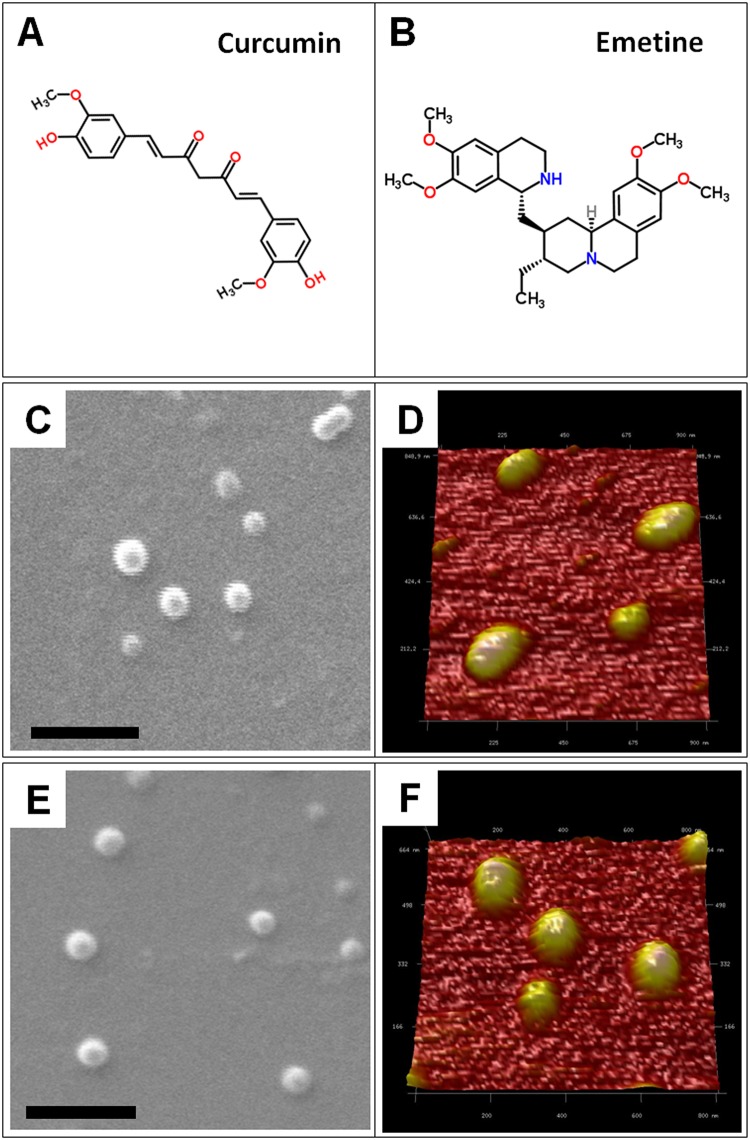
The biological activity of both the compounds depend on their specific functional groups viz. phenol rings in curcumin and isoquinoline ring in emetine (Fig 2A and 2B). PLGA polymer was used for encapsulation of curcumin or emetine by oil in water and water in oil in water solvent evaporation method, respectively. The corresponding SEM and AFM analysis showed roughly spherical morphology of both the NPs (Fig 2C & 2D; 2E & 2F). The physico-chemical properties of the NPs are presented in Table 1. The nanoparticles showed narrow size distribution with an average diameter of 261.5±8 nm. To assess the surface charge, their average zeta potential was determined and was found as -30.8 mV, -37.6 mV and -35.9 mV for the Em-NPs, Cm-NPs the control PLGA NPs, respectively (S1 Fig). The FTIR spectra of NPs are shown in Fig 3. A characteristic C = O absorption band at 1750 cm-1 of PLGA was observed in all the three cases. In the Cm-NPs, additional peak of curcumin at 3379 cm-1 characteristic of O-H stretching was observed. For the Em-NPs, very sharp peak of isoquinoline at 3395 cm-1 characteristic of N-H stretching was observed. The aromatic signatures of C-C were found overlapping in all the samples. Based on actual loading of compounds, entrapment efficiency (EE) of Cm-NPs and Em-NPs was found ~19% and ~27%, respectively (Table 1).
Fig 2. Chemical and morphological characteristics of nanoparticles.
The chemical structure of curcumin (A) and emetine (B), SEM (C) and AFM (D) images of Cm-NPs, SEM (E) and AFM (F) images of Em-NPs. The scale bar in SEM images represents 1 μm.
Table 1. Physicochemical characteristics of nanoparticles.
| Formation | Size (nm) | Polydispersity | Zeta Potential (mV) | Entrapment efficiency (%) | Actual Loading (μg/mg NP) |
|---|---|---|---|---|---|
| NPs | 263.5±7.8 | 0.071±0.06 | -35.9±1.5 | - | - |
| Cm-NPs | 261±2.4 | 0.039±0.08 | -37.6±1.1 | 19 | 190 |
| Em-NPs | 260.1±8.1 | 0.085±0.05 | -30.8±1.3 | 27 | 270 |
Fig 3. Fourier transformed infrared spectra of nanoparticles.
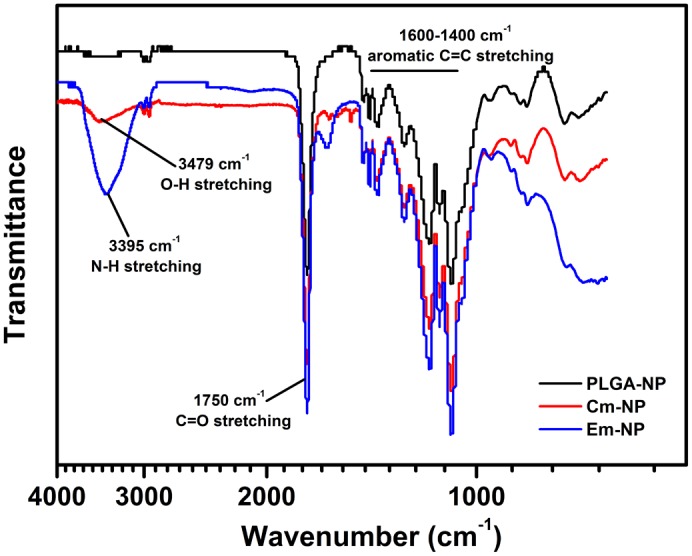
FTIR spectra of Cm-NPs (red), Em-NPs (blue) and free PLGA-NPs (black) showing overlapping peaks of C = O stretching, corresponding to PLGA. Distinct—OH and—NH stretching peaks characteristic of curcumin and emetine, respectively are also shown.
fAGel exhibits nucleation-dependent aggregation
The aggregation kinetics of fAGel at pH 4.0, 37°C was followed by monitoring the changes in Thioflavin T (ThT) fluorescence, a widely used fluorescent dye used for characterizing amyloid kinetics [40]. Evidently, the fAGel aggregation showed a time-dependent increase (k = 7.9 h-1) in the ThT fluorescence, followed a sigmoidal trace before saturating around 60 h (Fig 4A). A distinct lag phase (~18 h) was observed, suggesting that the polypeptide undergoes nucleation-dependent polymerization under these conditions, a phenomenon generally reported for other amyloidogenic polypeptides [41–43]. The TEM images taken after 24 h showed the presence of protofilaments and early fibrillar aggregates (Fig 4B). The corresponding AFM images taken after 60 h incubation showed the presence of mature fibrils of 8.6 nm average width and 5.2 nm average height having smooth morphologies (Fig 4C, S2 Fig).
Fig 4. Kinetics and morphology of fAGel aggregation.
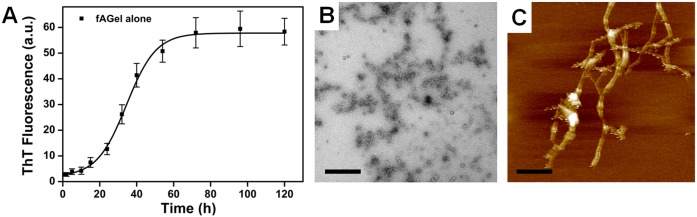
(A) Amyloid formation kinetics of fAGel alone monitored by ThT fluorescence showing sigmoidal trend with 18 h lag phase. (B) TEM image of amyloids formed after 24 h incubation. Scale bar 500 nm. (C) AFM image taken after 60 h showing fibrillar aggregates of fAGel. Scale bar 200 nm. The fAGel concentration was 2 mg/ml.
Testing the effect of NPs on fAGel aggregation
Initially, the kinetics of amyloid aggregation in fAGel was characterized by ThT fluorescence assay along with TEM and AFM imaging. The presence of protofilaments and mature fibrils at 24 h and 60 h respectively, during fAGel aggregation led us to choose these time points as benchmarks for structural characterization of fAGel aggregates formed in presence of NPs. For characterizing the effect of different concentrations of NPs on aggregation kinetics of fAGel, ThT assay of aliquots withdrawn at different time points was done. Following this, samples were characterized for morphological differences (24 h and 60 h) using TEM and AFM imaging. The surface hydrophobicity and secondary structural changes were compared only for the 24 h samples since they exhibited highest degree of morphological differences. Since we were also interested in investigating the effect of NPs on amyloid toxicity, pre-fibrillar aggregates of fAGel observed at the end of 24 h were chosen for relative assessment of toxicity of aggregates formed in presence of NPs at the same time point. In addition, for checking the effect of NPs on pre-formed amyloids, fAGel was allowed to aggregate for an extended period of time (4 days) to ascertain that the sample only contains mature fibrils. Following this, the effect of NPs was then substantiated using ThT assay in a time-wise manner and TEM imaging. The details are described in the methods section.
Cm-NPs augment and Em-NPs arrest fAGel fibrillation
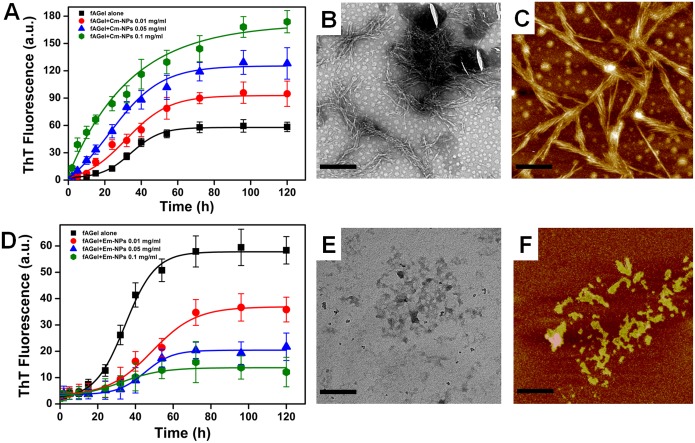
We tested the effect of Cm-NPs and Em-NPs at varying concentrations (0.01, 0.05, 0.1 mg/ml) on fAGel aggregation kinetics using ThT fluorescence and compared it with control. In contrast to the nucleation polymerization kinetics seen in fAGel alone, the presence of Cm-NPs augmented the fibrillation process. Both the ThT fluorescence maxima (Fmax) and the apparent rate of polymerization increased by more than 150% (Fmax = 92.8; k = 11.8 h-1) in presence of minimum Cm-NP concentration (0.01 mg/ml) (Fig 5A). On increasing the concentration of Cm-NPs, the aggregation kinetics increased exponentially with no apparent lag phase. A strikingly high ThT fluorescence maxima (>300% at 0.1 mg/ml Cm-NP concentration) indicated the presence of large amount of fibrillar aggregates. This was confirmed by TEM micrographs of Cm-NP samples that showed dense fibrillation with individual fibrils sticking and interweaving together (Fig 5B). Further, on analysing the AFM images of 60 h incubated samples, laterally stacked fibrils in bunches of 8–10 individual fibers were observed (Fig 5C). The fibrillar bunches were having an average width and height of 33.3 nm and 27.2 nm respectively indicating relatively high fibrillar bulk (S3 Fig). One possible explanation could be the availability of larger number of soluble curcumin moieties in the form of NPs, resulting into higher occurrence of stacking interactions. This corroborates well with our recent report of curcumin binding to the core amyloidic stretch of gelsolin (AGel, 182–192 residue) through tri-planar π-π stacking interactions with PHE 183 and PHE 189 residues [17]. This also supports previous reports where aromatic interactions were shown to affect amyloid aggregation [44–46].
Fig 5. Kinetics and morphologies of NP- incubated fAGel aggregates.
fAGel aggregation kinetics monitored by ThT fluorescence in the absence (black trace) or in the presence (coloured traces) of different concentrations of NPs. (A) Acceleration of aggregation in the presence of increasing concentrations of Cm-NPs. The corresponding TEM (B) and AFM (C) images are shown. (D) Suppression of aggregation in the presence of increasing concentrations of Em-NPs with corresponding TEM (E) and AFM (F) images. In each case the TEM and the AFM images were taken after 24 h and 60 h of fAGel incubation with 0.1 mg/ml of respective NPs. Scale bars represent 500 nm for TEM and 200 nm for AFM images.
Contrary to this, in the presence of Em-NPs, the fAGel aggregation kinetics de-escalated in a dose dependent manner. At increasing concentrations of Em-NPs, the ThT fluorescence subdued significantly, with less than 25% fluorescence observed at the highest NP concentration of 0.1 mg/ml (Fig 5D). With a gradual increase in the lag phase with increasing concentrations (~26 h at 0.01 mg/ml and ~35 h at 0.05 mg/ml), the sigmoidal kinetics of fAGel aggregation was lost and subsequently arrested at the highest Em-NP concentration. In these samples, irregular shaped aggregates were observed under TEM, suggesting the inhibition of amyloid fibrillation in the presence of Em-NPs (Fig 5E). In this case, nano-encapsulation plausibly increased the number of emetine molecules accessing the fAGel polypeptide, in turn accelerating the steric blockage on fAGel aggregation by the bulky isoquinoline rings [17]. These observations were also ascertained by the AFM images of 60 h incubated samples, showing randomly shaped structures devoid of any distinct morphology (Fig 5F). The width and height of these unstructured aggregates were unevenly distributed, indicating low structural content (S6 Fig).
Cm-NPs and Em-NPs alter hydrophobic exposure and secondary structure
The amyloid assembly progresses through interaction of hydrophobic surfaces often associated with secondary structural changes. Since, the extent of hydrophobic exposure is associated with the toxicity of the amyloid aggregates, we were interested in evaluating the same under the influence of NPs [47, 48]. For this, we employed Nile Red (NR) fluorescence assay (previously used for assessing hydrophobicity) on the 24 h incubated fAGel samples (both with and without NPs), since they exhibited highest degree of morphological differences in TEM and AFM images [49]. Evidently, Cm-NP treatment resulted in reduced NR binding that indicates reduction in surface/volume ratio in Cm-NP samples containing mature fibrils as compared to control that correspond to early/premature aggregates (Fig 6A). On the other hand, Em-NPs caused dramatically reduced NR fluorescence as compared to control fAGel aggregates confirming the unstructured and random aggregate species seen in TEM and AFM images. To further associate these alterations in the hydrophobic surfaces with any secondary structural changes under the influence of NPs, we performed ATR-FTIR analysis of these aggregates.
Fig 6. Hydrophobicity and secondary structure assessment of the aggregates.
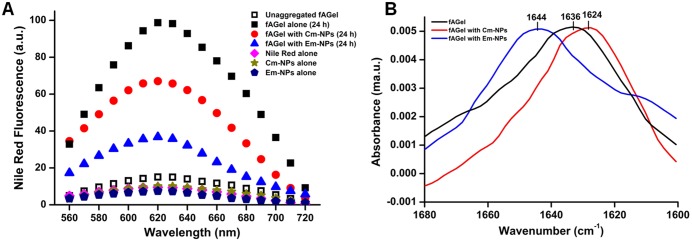
(A) Nile Red fluorescence of fAGel alone (black squares), with Cm-NPs (red circles) and Em-NPs (blue triangles). The fluorescence spectra of controls are also shown in colour. (B) ATR-FTIR spectra of aggregating samples of fAGel alone (black), with Cm-NPs (red) and with Em-NPs (blue) are compared. In each case the samples were taken after 24 h of incubation with NPs at 0.1 mg/ml concentration. A characteristic 1644 cm-1 peak for Em-NP sample corresponds to majorly unstructured aggregates.
ATR-FTIR is a sensitive spectroscopic probe to evaluate secondary structural contents in proteins. The FTIR spectrum of fAGel showed a major peak at 1636 cm-1 which is attributed to the presence of β-sheets rich aggregates (Fig 6B). On the other hand, the Cm-NP influenced aggregates showed a major shift in peak position at 1624 cm-1, indicating the presence of fibrillar axis composed of extensive inter-molecular cross-β sheets [37, 50]. However, the presence of major peak at 1644 cm-1 in Em-NP samples was suggestive of random and unstructured aggregate species that validates the observed reduction in hydrophobic surface.
Cm-NPs and Em-NPs reduce toxic assembly of fAGel amyloids
The fibrillar and non-fibrillar aggregates formed in the presence of Cm-NPs and Em-NPs were tested for their toxic effects on the human neuroblastoma cells (SH-SY5Y). Since early proto- or pre-fibrillar aggregates are shown as the toxic amyloid species, we chose the 24 h incubated samples showing significant morphological differences [51]. The fAGel aggregates alone displayed high cellular toxicity reducing the cell viability to less than 40% after 24 h of incubation (Fig 7). Interestingly, while both the Cm- and Em-NP-induced aggregates displayed reduced cellular toxicity, the viability showed an upward trend in the case of Cm-NPs in a dose dependent manner. In the case of Em-NPs however, there was a loss of cell viability at higher concentrations which is probably due to the inhibitory effect of emetine on protein synthesis [52].
Fig 7. Cytotoxicity profile of aggregates.
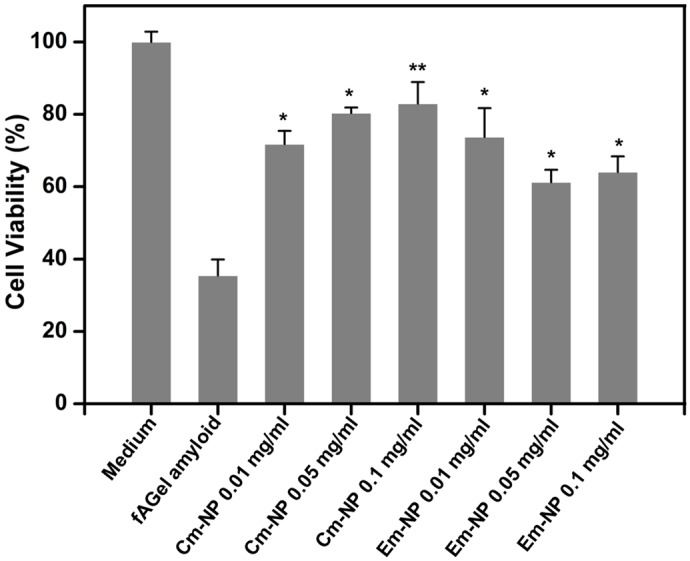
Toxicity assessment of fAGel amyloids and aggregates formed at different concentrations of Cm-NPs or Em-NPs on SH-SY5Y cells, represented as percentage of untreated control. In each case the 24 h incubated aggregates were used.
Cm-NPs increase fibrillar bulk and Em-NPs defibrillate amyloid fibers
We further checked whether both the NPs have any effect on pre-formed amyloids of fAGel. For this, we incubated fAGel amyloid fibrils with Cm-NPs or Em-NPs at 0.05 mg/ml and 0.1 mg/ml, and monitored the ThT response in a time-wise manner. We observed that at both the concentrations of Cm-NPs, a time dependent increase in ThT fluorescence was noted (Fig 8A). To visualize the associated morphological changes, we obtained TEM micrographs of the 7 day incubated samples which showed dense fibrillation and increase in fibril bulk (Fig 8B) [17]. The resulting higher order aggregates were thicker (~25–30 nm), rich in β-sheets and exhibited enhanced ThT response (>80% increase). In contrast, fAGel amyloids incubated with Em-NPs exhibited a decline in ThT response with time (~65% decline) and showed smaller, detached fragments with reminiscent of fibrillar axis (Fig 8C).
Fig 8. Effect of nanoparticles on pre-formed amyloids.
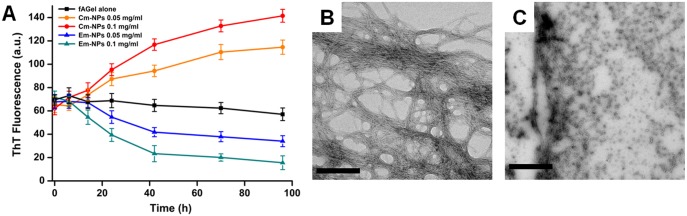
(A) Time-dependent ThT fluorescence showing effect of Cm-NPs (solid circles) and Em-NPs (solid triangles) on pre-formed amyloids of fAGel (control, black solid squares). The electron micrographs of aggregates after incubation at 37°C for a week in the presence of 0.1 mg/ml of Cm-NPs (B) and Em-NPs (C). Scale bars represent 500 nm in each case.
Discussion
The present study describes the synthesis of PLGA nanoparticles encapsulating curcumin and emetine molecules, and their differential effects on gelsolin amyloids. Although small molecule inhibitors have been promoted as potential anti-amyloidogenic agents, their applicability falls way below the standard guidelines defined for drug candidates [53]. We had previously reported the interaction of curcumin and emetine, on the amyloidogenic propensity of a gelsolin-derived core amyloidogenic peptide AGel [17]. However, the poor solubility of the compounds and insufficient information about the actual disease-associated gelsolin fragment (fAGel), led us to initiate this investigation.
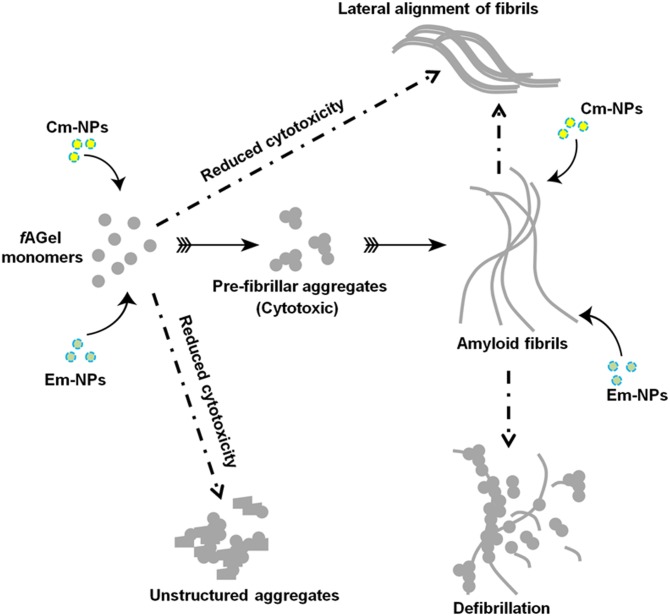
We report diametrically opposite effects of curcumin and emetine loaded PLGA NPs on the amyloid aggregation of disease-associated gelsolin fragment (fAGel). While Cm-NPs accelerate fibrillar aggregation in fAGel, Em-NPs divert the aggregation into an alternate non-amyloid state. Clearly, these categorically distinct aggregates indicate discrete set of molecular interactions between each compound-loaded NPs and fAGel. A complete disappearance of lag phase at higher Cm-NP concentrations suggested a nucleation-independent polymerization event, culminating into thick mature fibrils at latter stages of incubation (S5a Fig, and Fig 5C). This effect was similar to other reports on amyloid acceleration by heparin, Benzalkonium Chloride and TiO2 nanoparticles [54–56]. We reasoned that this is due to lateral alignment of fibrils in the presence of highly hydrophobic curcumin moieties. In the case of emetine, the irregular shaped assemblies resulted from steric blockage by the bulky isoquinolines of emetine as reported earlier, and as seen in the corresponding SEM images (S5 Fig). In each one of these cases, the amount of compound required to bring about the same effect as that of free compound is drastically reduced (Table 2). This is because PLGA nano-encapsulation enhanced the solubility of the compounds, causing several fold increase in their accessibility to fAGel. This results in effective modulation of the aggregation pathway even at 1000-times lower concentrations as compared to free compounds (Table 2, Fig 9). The SEM images taken after extended periods of incubation (2 weeks) displayed sustained and extended amyloid modulation induced by the two compounds (S5 Fig). In the case of PLGA-curcumin, our study clearly indicates that the amyloid-modulating effect persisted long after the in vivo absorption and elimination half lives (0.3 h and 1.7 h, respectively) of free curcumin [57]; a desirable quality for any potential drug candidate. This property is particularly useful for the effectiveness of emetine as an amyloid modulator. Emetine affects protein synthesis and thus is detrimental for cell growth at its higher concentrations [52]. PLGA encapsulation however, bypasses this drawback and enables controlled and prolonged release at drastically reduced sub-nanomolar concentrations. Another interesting aspect is that the resulting aggregates in both the cases showed reduced cytotoxicity, re-enforcing the concept that cytotoxic species in amyloid pathways are intermediates, rather than the end products [58]. In our case, both the NPs altered or bypassed the formation of these toxic intermediates and resulted in significant reduction in amyloid toxicity (Fig 9).
Table 2. Comparison of parameters between nano-encapsulated versus free compounds.
| Parameter | Curcumin (free) | Curcumin Nanoparticle (Cm-NPs) | Emetine (free) | Emetine Nanoparticle (Em-NPs) |
|---|---|---|---|---|
| Solubility | Low | high | Moderate | high |
| Concentration* | 25 μM | 25.7x10-3μM | 25 μM | 28x10-3μM |
| Time # | 10.9 h | NIL | 21.4 h | 34.6 h |
| Release | Poor | Sustained | low | Sustained |
* Concentration of free curcumin and emetine used in Arya P et al. 2014.
# Reduction of lag phase (curcumin) and increase in lag phase (emetine). Free compounds data taken from Arya P et al. 2014.
Fig 9. Schematic representation of differential effect of NPs on fAGel amyloidogenesis.
Conclusion
Rational design of effective therapeutics against amyloid diseases requires a targeted approach, owing to the complex mechanism of amyloidogenesis. The disease progression arrogates biological defence mechanisms that involve multiple proteins interacting together. Biologically compatible drug delivery systems may selectively interfere at different steps of the complex protein aggregation pathway. Curcumin, a traditionally known multi-faceted Indian herb has been utilized mostly as a food supplement rather than a drug molecule. On the other hand, emetine is a plant alkaloid used as an antiprotozoal drug. In both cases, poor pharmacokinetic properties hinder their sustained usage as multi-targeted therapeutic agents. From our study, we conclude that as compared to free compounds, PLGA encapsulation not only increases the solubility of curcumin and emetine but also reduces their concentration dependence for aiding amyloid modulation (~1000 fold reduction). Although further conclusions on druggability of both these NPs depend on in vivo studies, our preliminary results suggest therapeutic importance of curcumin and emetine as starting molecules for designing anti-amyloidogenic drugs. As a whole, with the advantage of controlled release, the biocompatibility and biodegradability of PLGA makes our conjugate NPs novel drug-like therapeutics against amyloid toxicity.
Supporting Information
(TIF)
(TIF)
(TIF)
(TIF)
The images were captured after two weeks of incubation with 0.1 mg/ml of respective NPs. The scale bars represent 100 μm.
(TIF)
Acknowledgments
The authors thank the Indian Institute of Technology Delhi and the CSIR Institute of Microbial Technology Chandigarh for infrastructural support. The work was supported by funding from ICMR, Govt of India. PA and SG acknowledge CSIR India and AS acknowledges ICMR India for research fellowships.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was supported by funding from The Indian Council of Medical Research, Government of India. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006; 75: 333–66. [DOI] [PubMed] [Google Scholar]
- 2. Murphy MP, LeVine H 3rd. Alzheimer's disease and the amyloid-beta peptide. J Alzheimers Dis. 2010; 19: 311–323. 10.3233/JAD-2010-1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lotharius J, Brundin P. Pathogenesis of Parkinson's disease: dopamine, vesicles and alpha-synuclein. Nat Rev Neurosci. 2002; 3: 932–942. [DOI] [PubMed] [Google Scholar]
- 4. Stewart LA, Rydzewska LH, Keogh GF, Knight RS. Systematic review of therapeutic interventions in human prion disease. Neurology. 2008; 70: 1272–1281. 10.1212/01.wnl.0000308955.25760.c2 [DOI] [PubMed] [Google Scholar]
- 5. Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012; 148: 1188–1203. 10.1016/j.cell.2012.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rana A, Gnaneswari D, Bansal S, Kundu B. Prion Metal Interaction: Is prion pathogenesis a cause or a consequence of metal imbalance? Chemico-Biological Interaction. 2009; 181: 282–91. 10.1016/j.cbi.2009.07.021 [DOI] [PubMed] [Google Scholar]
- 7. Muchowski PJ. Protein misfolding, amyloid formation, and neurodegeneration: a critical role for molecular chaperones? Neuron. 2002; 35: 9–12. [DOI] [PubMed] [Google Scholar]
- 8. Tomar R, Garg DK, Mishra R, Thakur A, Kundu B. N-terminal domain of Pyrococcus furiosus l-asparaginase functions as a non-specific, stable, molecular chaperone. FEBS J. 2013; 280: 2688–99. 10.1111/febs.12271 [DOI] [PubMed] [Google Scholar]
- 9. Khan JM, Chaturvedi SK, Rahman SK, Ishtikhar M, Qadeer A, Ahmad E, et al. Protonation favors aggregation of lysozyme with SDS. Soft Matter. 2014; 10: 2591–2599. 10.1039/c3sm52435c [DOI] [PubMed] [Google Scholar]
- 10. Bhatia NK, Srivastava A, Katyal N, Jain N, Khan MA, Kundu B, et al. Curcumin binds to the pre-fibrillar aggregates of Cu/Zn superoxide dismutase (SOD1) and alters its amyloidogenic pathway resulting in reduced cytotoxicity. Biochim Biophys Acta. 2015; 1854: 426–436. 10.1016/j.bbapap.2015.01.014 [DOI] [PubMed] [Google Scholar]
- 11. Porat Y, Abramowitz A, Gazit E. Inhibition of amyloid fibril formation by polyphenols: structural similarity and aromatic interactions as a common inhibition mechanism. Chem Biol Drug Des. 2006; 67: 27–37. [DOI] [PubMed] [Google Scholar]
- 12. Ahmad E, Rabbani G, Zaidi N, Khan MA, Qadeer A, Ishtikhar M, et al. Revisiting ligand-induced conformational changes in proteins: essence, advancements, implications and future challenges. J Biomol Struct Dyn. 2013; 31: 630–648. 10.1080/07391102.2012.706081 [DOI] [PubMed] [Google Scholar]
- 13. Belluti F, Rampa A, Gobbi S, Bisi A. Small-molecule inhibitors/modulators of amyloid-β peptide aggregation and toxicity for the treatment of Alzheimer's disease: a patent review. Expert Opin Ther Pat. 2013; 23: 581–596. 10.1517/13543776.2013.772983 [DOI] [PubMed] [Google Scholar]
- 14. Mohd I, Rabbani G, Khan S, Khan RH. Biophysical investigation of thymoquinone binding to ‘N’ and ‘B’ isoforms of human serum albumin: exploring the interaction mechanism and radical scavenging activity. RSC Adv. 2015; 5: 18218–18232. [Google Scholar]
- 15. Peddada N, Sagar A, Rathore YS, Choudhary V, Pattnaik UB, et al. Global shapes of F-actin depolymerization-competent minimal gelsolins: insight into the role of g2-g3 linker in pH/Ca2+ insensitivity of the first half. J Biol Chem. 2013; 288: 28266–82. 10.1074/jbc.M113.463224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Solomon JP, Page LJ, Balch WE, Kelly JW. Gelsolin amyloidosis: genetics, biochemistry, pathology and possible strategies for therapeutic intervention. Crit. Rev. Biochem. Mol. Biol. 2012; 47: 282–96. 10.3109/10409238.2012.661401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arya P, Srivastava A, Vasaikar SV, Mukherjee G, Mishra P, Kundu B. Selective interception of gelsolin amyloidogenic stretch results in conformationally distinct aggregates with reduced toxicity. ACS Chem Neurosci. 2014; 5: 982–92. 10.1021/cn500002v [DOI] [PubMed] [Google Scholar]
- 18. Tihanyi K, Vastag M. Solubility, Delivery and ADME Problems of Drugs and Drug-Candidates, first ed., Bentham Science, UAE: 2011. [Google Scholar]
- 19. Wilczewska AZ, Niemirowicz K, Markiewicz KH, Car H. Nanoparticles as drug delivery systems. Pharmacol Rep. 2012; 64: 1020–37. [DOI] [PubMed] [Google Scholar]
- 20. Zaman M, Ahmad E, Qadeer A, Rabbani G, Khan RH. Nanoparticles in relation to peptide and protein aggregation. Int J Nanomedicine. 2014; 9: 899–912. 10.2147/IJN.S54171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bao G, Mitragotri S, Tong S. Multifunctional nanoparticles for drug delivery and molecular imaging. Annu Rev Biomed Eng. 15: 253–82. 10.1146/annurev-bioeng-071812-152409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rabbani G, Khan MJ, Ahmad A, Maskat MY, Khan RH. Effect of copper oxide nanoparticles on the conformation and activity of β-galactosidase. Colloids Surf B Biointerfaces. 2014; 123: 96–105. 10.1016/j.colsurfb.2014.08.035 [DOI] [PubMed] [Google Scholar]
- 23. Zhang L, Gu FX, Chan JM, Wang AZ, Langer RS, Farokhzad OC. Nanoparticles in medicine: therapeutic applications and developments. Clin Pharmacol Ther. 2008; 5: 761–769. [DOI] [PubMed] [Google Scholar]
- 24. Silva GA. Nanotechnology approaches for drug and small molecule delivery across the blood brain barrier. Surg Neurol. 2007; 67: 113–16. [DOI] [PubMed] [Google Scholar]
- 25. Kreuter J. Mechanism of polymeric nanoparticle-based drug transport across the blood-brain barrier (BBB). J Microencapsul. 2013; 30: 49–54. 10.3109/02652048.2012.692491 [DOI] [PubMed] [Google Scholar]
- 26. Wohlfart S, Gelperina S, Kreuter J. Transport of drugs across the blood-brain barrier by nanoparticles. J Control Release. 2012; 161: 264–273. 10.1016/j.jconrel.2011.08.017 [DOI] [PubMed] [Google Scholar]
- 27. Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V. PLGA-based nanoparticles: an overview of biomedical applications. J Control Release. 2012; 161: 505–22. 10.1016/j.jconrel.2012.01.043 [DOI] [PubMed] [Google Scholar]
- 28. Li J, Sabliov C. PLA/PLGA nanoparticles for delivery of drugs across the blood-brain barrier. Nanotechnology Reviews. 2013; 2: 241–257. [Google Scholar]
- 29. Gan CW, Feng SS. Transferrin-conjugated nanoparticles of poly(lactide)-D-alpha-tocopheryl polyethylene glycol succinate diblock copolymer for targeted drug delivery across the blood-brain barrier. Biomaterials, 2010; 31:7748–7757. 10.1016/j.biomaterials.2010.06.053 [DOI] [PubMed] [Google Scholar]
- 30. Jagur-Grodzinski J. Biomedical application of functional polymers. React. Funct. Polym. 1999; 39: 99–138. [Google Scholar]
- 31. Murakami H, Kobayashi M, Takeuchi H, Kawashima Y. Preparation of poly (DL-lactide-co-glycolide) nanoparticles by modified spontaneous emulsification solvent diffusion method. Int J Pharm. 1999; 187: 143–52. [DOI] [PubMed] [Google Scholar]
- 32. Cohen-Sela E, Chorny M, Koroukhov N, Danenberg HD, Golomb G. A new double emulsion solvent diffusion technique for encapsulating hydrophilic molecules in PLGA nanoparticles. J Control Release. 2009; 133: 90–95. 10.1016/j.jconrel.2008.09.073 [DOI] [PubMed] [Google Scholar]
- 33. Rabbani G, Ahmad E, Zaidi N, Fatima S, Khan RH. pH-induced molten globule state of Rhizopus niveus lipase is more resistant against thermal and chemical denaturation than its native state. Cell Biochem Biophys. 2012; 62: 487–499. 10.1007/s12013-011-9335-9 [DOI] [PubMed] [Google Scholar]
- 34. Jayaprabhaa KN, Joy PA. Citrate modified β-cyclodextrin functionalized magnetite nanoparticles: a biocompatible platform for hydrophobic drug delivery. RSC Adv. 2015; 5: 22117–22125. [Google Scholar]
- 35. Sero L, Sanguinet L, Blanchard P, Dang BT, Morel S, Richomme P, et al. Tuning a 96-well microtiter plate fluorescence-based assay to identify AGE inhibitors in crude plant extracts. Molecules. 2013; 18: 14320–14339. 10.3390/molecules181114320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shankarayan R, Kumar S, Mishra P. Differential permeation of piroxicam-loaded PLGA micro/nanoparticles and their in vitro enhancement. J Nanopart Res. 2013; 15: 1–13. [Google Scholar]
- 37. Ridgley DM, Ebanks KC, Barone JR. Peptide mixtures can self-assemble into large amyloid fibers of varying size and morphology. Biomacromolecules. 2011; 12: 3770–79. 10.1021/bm201005k [DOI] [PubMed] [Google Scholar]
- 38. Shivu B, Seshadri S, Li J, Oberg KA, Uversky VN, Fink AL. Distinct β-sheet structure in protein aggregates determined by ATR-FTIR spectroscopy. Biochemistry. 2013; 52: 5176–5183. 10.1021/bi400625v [DOI] [PubMed] [Google Scholar]
- 39. Ratnaswamy G, Koepf E, Bekele H, Yin H, Kelly JW. The amyloidogenicity of gelsolin is controlled by proteolysis and pH. Chem Biol. 1999; 6: 293–304. [DOI] [PubMed] [Google Scholar]
- 40. Biancalana M, Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta. 2010; 1804: 1405–12. 10.1016/j.bbapap.2010.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kundu B, Maiti NR, Jones EM, Surewicz KA, Vanik DL, Surewicz WK. Nucleation-dependent conformational conversion of the Y145Stop variant of human prion protein: structural clues for prion propagation. Proc. Natl. Acad. Sci. U.S.A. 2003; 100: 12069–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Prigent S, Ballesta A, Charles F, Lenuzza N, Gabriel P, Tine LM, et al. An efficient kinetic model for assemblies of amyloid fibrils and its application to polyglutamine aggregation. PLoS One. 2012; 7: e43273 10.1371/journal.pone.0043273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Powers ET, Powers DL. Mechanisms of protein fibril formation: nucleated polymerization with competing off-pathway aggregation. Biophys J. 2008; 94: 379–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tu LH, Raleigh DP. Role of aromatic interactions in amyloid formation by islet amyloid polypeptide. Biochemistry. 2013; 52: 333–342. 10.1021/bi3014278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gazit E. A possible role for pi-stacking in the self-assembly of amyloid fibrils. FASEB J. 2002; 16: 77–83. [DOI] [PubMed] [Google Scholar]
- 46. Ladiwala AR, Dordick JS, Tessier PM. Aromatic small molecules remodel toxic soluble oligomers of amyloid beta through three independent pathways. J Biol Chem. 2011; 286: 3209–3218. 10.1074/jbc.M110.173856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Janson J, Ashley RH, Harrison D, McIntyre S, Butler PC. The mechanism of islet amyloid polypeptide toxicity is membrane disruption by intermediate-sized toxic amyloid particles. Diabetes. 1999; 48: 491–498. [DOI] [PubMed] [Google Scholar]
- 48. Williams TL, Serpell LC. Membrane and surface interactions of Alzheimer's Aβ peptide-insights into the mechanism of cytotoxicity. FEBS J. 2011; 278: 3905–17. 10.1111/j.1742-4658.2011.08228.x [DOI] [PubMed] [Google Scholar]
- 49. Singh PK, Kotia V, Ghosh D, Mohite GM, Kumar A, Maji SK. Curcumin modulates α-synuclein aggregation and toxicity. ACS Chem Neurosci. 2013; 4: 393–407. 10.1021/cn3001203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bouchard M, Zurdo J, Nettleton EJ, Dobson CM, Robinson CV. Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 2000; 9: 1960–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002; 416: 507–511. [DOI] [PubMed] [Google Scholar]
- 52. Grollman AP. Structural basis for inhibition of protein synthesis by emetine and cycloheximide based on an analogy between ipecac alkaloids and glutarimide antibiotics. Proc. Natl. Acad. Sci. U.S.A. 1966; 56: 1867–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gillmore JD, Hawkins PN. Drug Insight: emerging therapies for amyloidosis. Nat Clin Pract Nephrol. 2006; 2: 263–70. [DOI] [PubMed] [Google Scholar]
- 54. Solomon JP, Bourgault S, Powers ET, Kelly JW. Heparin binds 8 kDa gelsolin cross-β-sheet oligomers and accelerates amyloidogenesis by hastening fibril extension. Biochemistry. 2011; 50: 2486–98. 10.1021/bi101905n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kato Y, Yagi H, Kaji Y. Benzalkonium chloride accelerates the formation of the amyloid fibrils of corneal dystrophy-associated peptides. J Biol Chem. 2013; 288: 25109–118. 10.1074/jbc.M113.477695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Wu WH, Sun X, Yu YP, Hu J, Zhao L, Liu Q, et al. TiO2 nanoparticles promote beta-amyloid fibrillation in vitro. Biochem Biophys Res Commun. 2008; 373: 315–18. 10.1016/j.bbrc.2008.06.035 [DOI] [PubMed] [Google Scholar]
- 57. Anand P, Kunnumakkara AB, Newman RA, Aggarwal BB. Bioavailability of curcumin: problems and promises. Mol Pharm. 2007; 4: 807–18. [DOI] [PubMed] [Google Scholar]
- 58. Fändrich M.. Oligomeric intermediates in amyloid formation: structure determination and mechanisms of toxicity. J Mol Biol. 2012; 421: 427–40. 10.1016/j.jmb.2012.01.006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(TIF)
(TIF)
(TIF)
The images were captured after two weeks of incubation with 0.1 mg/ml of respective NPs. The scale bars represent 100 μm.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.