Summary
Bone morphogenetic proteins (BMPs) belong to the transforming growth factor-beta (TGFβ) superfamily. BMPs mediate a highly conserved signal transduction cascade through the type I and type II serine/threonine kinase receptors and intracellular Smad proteins. The BMP pathway regulates multiple developmental and homeostatic processes. Mutations in this pathway can cause various diseases in humans, such as skeletal disorders, cardiovascular diseases and various cancers. Multiple levels of regulation, including extracellular regulation, help to ensure proper spatiotemporal control of BMP signaling in the right cellular context. The family of repulsive guidance molecules (RGMs) and the type I trans-membrane protein neogenin, a paralog of DCC (Deleted in Colorectal Cancer), have been implicated in modulating the BMP pathway. In this review, we discuss the properties and functions of RGM proteins and neogenin, focusing on their roles in the modulation of BMP signal transduction.
Keywords: RGM, neogenin, BMP, HJV, DRAG-1, TGFβ
The BMP signal transduction pathway
Bone morphogenetic proteins (BMPs) were first discovered in 1965, when Marshall Urist found that dead bone matrix fragments have bone-inducing activity (Urist. 1965). It's not until the late 1980s when the proteins responsible for bone induction were purified and their genes cloned (Wozney et al. 1988) using an ectopic bone induction assay developed in 1981 (Sampath and Reddi. 1981). The identified molecules, called BMP2 through BMP7, belong to the transforming growth factor β (TGFβ) superfamily (Wozney. 1992). The BMP subfamily is known to contain over 20 members (Bragdon et al. 2011). BMPs have been shown to regulate a wide variety of cellular processes including cell fate specification, cell proliferation, cell migration, and cell death during the development of multi-cellular organisms (Wu and Hill. 2009). Malfunction of the pathway causes many somatic and hereditary disorders in humans, including skeletal abnormalities, cardiovascular diseases and various types of cancers (Gordon and Blobe. 2008,Massague. 2012,Cai et al. 2012).
The BMP signal transduction pathway has been well studied. The ligand is generated as a pre-protein containing a N-terminal prodomain and a C-terminal mature domain that is bioactive. The prodomain is then cleaved off by serine endoproteases such as furin in the secretory pathway. The resulting active forms of ligands form dimers that are stabilized by hydrophobic interactions and an inter-subunit disulfide bond (Shi and Massague. 2003). The BMP receptors belong to the receptor serine/threonine kinase family that is divided into two classes: type I and type II (Attisano et al. 1994,Massague. 2012,Xu et al. 2012). Both types of receptors are type I trans-membrane proteins, comprised of an extracellular domain, a trans-membrane region, and a C-terminal serine/threonine kinase domain. The extracellular domains of type I and type II receptors are capable of binding to the ligands. Unlike the TGFβ subfamily of ligands that display a higher affinity for the type II receptors than the type I receptors, BMP ligands, such as BMP-2, exhibit a higher affinity for the type I receptors and a lower affinity for the type II receptors (Groppe et al. 2002,Allendorph et al. 2006). So far, four type I receptors (ALK3/BMPR1A, ALK6/BMPRIB, ALK1/Acvrl1 and ALK2/ActRI,) and three type II receptors (BMPRII, ActRIIA and ActRIIB) have been described to function in the BMP pathway. The type I, but not the type II, receptors contain a characteristic SGSGSG sequence, also known as the GS domain, which is located N-terminal to the kinase domain. Once the ligand binding brings the type I and type II receptors to close proximity, the constitutively active type II receptor phosphorylates type I receptor at its GS domain (Shi and Massague. 2003).
The activated type I receptor is poised to relay the BMP signal from the cytoplasm to the nucleus through the Smad proteins (Feng and Derynck. 2005,Heldin and Moustakas. 2012). Two classes of Smad proteins are required for signal transduction: receptor regulated Smads (R-Smads) and common mediator Smads (co-Smads). R-Smads are directly phosphorylated at the C-terminal Ser-X-Ser (SXS) motif by the type I receptors. The phosphorylated R-Smads then complex with the constitutively phosphorylated Co-Smad (Nakao et al. 1997,Roelen et al. 2003) and enter the nucleus, where the complex incorporates different DNA-binding cofactors to regulate downstream gene expression (Feng and Derynck. 2005, Massague. 2012). A third class of Smads, the inhibitory Smads, can associate with type I receptors and competitively block R-Smad recruitment and phosphorylation (Feng and Derynck. 2005). Among the R-Smads, the BMP subfamily utilizes Smads 1, 5 and 8, while the TGFβ subfamily utilizes Smad2 and Smad3. In addition to the Smad pathway, BMP signals can also activate other “non-canonical” or non-Smad signaling pathways, such as the P38 MAPK pathway, that complement Smad function (Massague. 2012).
Modulation of BMP signaling at the ligand-receptor level
Because of its various roles in development and homeostasis, the BMP pathway is modulated at multiple levels, including receptor activation outside the cell, Smad access to the receptors in the cytoplasm, Smad nucleocytoplasmic shuttling, and Smad-dependent transcription in the nucleus (Balemans and Van Hul. 2002,Umulis et al. 2009,Huang and Chen. 2012,Massague. 2012,Ramel and Hill. 2012).
A number of proteins bind to the ligand, receptors and/or ligand-receptor complexes to regulate BMP signaling at the level of receptor activation. One group of them is the extracellular secreted proteins that can function as ligand binding traps (Balemans and Van Hul. 2002,Shi and Massague. 2003). These include Noggin, Follistatin, Chordin/Short gastrulation (Sog), members of the Gremlin/Cerberus/DAN/Coco family and matrix GLA protein (MGP) (Balemans and Van Hul. 2002,Zakin and De Robertis. 2010,Cai et al. 2012). Noggin and Follistatin are probably the best known BMP antagonists. They sequester the ligands and block the ligands from binding to the type I and II receptors (Groppe et al. 2002). MGP appears to function in a similar way (Zebboudj et al. 2002). Chordin and its Drosophila homolog Sog can form a complex with Tsg (Twisted gastrulation). This complex sequesters BMPs to attenuate BMP signaling locally but enhances long range BMP signaling by transporting BMPs through tissues (O'Connor et al. 2006). Crossveinless 2 (CV2), which is also referred to as BMPER (BMP endothelial cell precursor derived regulator), can activate or inhibit BMP signaling by directly binding to the BMP ligands, Chordin or the Chordin-BMP complex (Ambrosio et al. 2008,Serpe et al. 2008,Zhang et al. 2008). Members of the Gremlin/Cerberus/DAN/Coco family of proteins are multivalent BMP inhibitors that also affect Nodal and Wnt signaling pathways (Hsu et al. 1998,Avsian-Kretchmer and Hsueh. 2004).
A number of membrane or membrane-associated proteins are also important modulators of BMP signaling. The extracellular matrix (ECM) protein, Type IV Collagen, has been shown to bind to the Drosophila BMP homolog Dpp (Decapentaplegic) and affect the formation of the Dpp morphogenetic gradient (Wang et al. 2008). In both Drosophila and C. elegans, Dally and LON-2, members of glycosylphosphatidylinositol (GPI)-anchored proteoglycan family, can bind to the BMP ligands and modulate BMP signaling in vivo. In particular, Dally positively regulates the distribution of the Dpp morphogen in Drosophila (Grisaru et al. 2001,Han et al. 2005,Takeo et al. 2005), while LON-2 functions as a negative regulator of the BMP-like Sma/Mab pathway in C. elegans (Gumienny et al. 2007,Taneja-Bageshwar and Gumienny. 2012). Betaglycan, also known as the TGFβ type III receptor, not only binds to TGFβ ligands and helps present them to the type II receptors, but also binds to BMP molecules and increases BMP-mediated signaling output (Kirkbride et al. 2008,Bilandzic and Stenvers. 2011). Similarly, endoglin, another glycoprotein that acts as a TGFβ co-receptor, can potentiate BMP signaling (Scherner et al. 2007). BMPs can also interact with the decoy receptor BAMBI (BMP and activin membrane-bound inhibitor), which resembles the type I receptors but lacks an active kinase domain. BMP-BAMBI interaction leads to inhibition of BMP signaling by sequestering the BMP ligands from the active receptors (Onichtchouk et al. 1999). More recently, RGM (Repulsive Guidance Molecule) proteins and a RGM interaction protein neogenin have been implicated in the modulation of BMP signaling. RGMs and neogenin will be the focus of the reminder of this article.
The roles of RGM proteins in modulating BMP signaling
Introduction
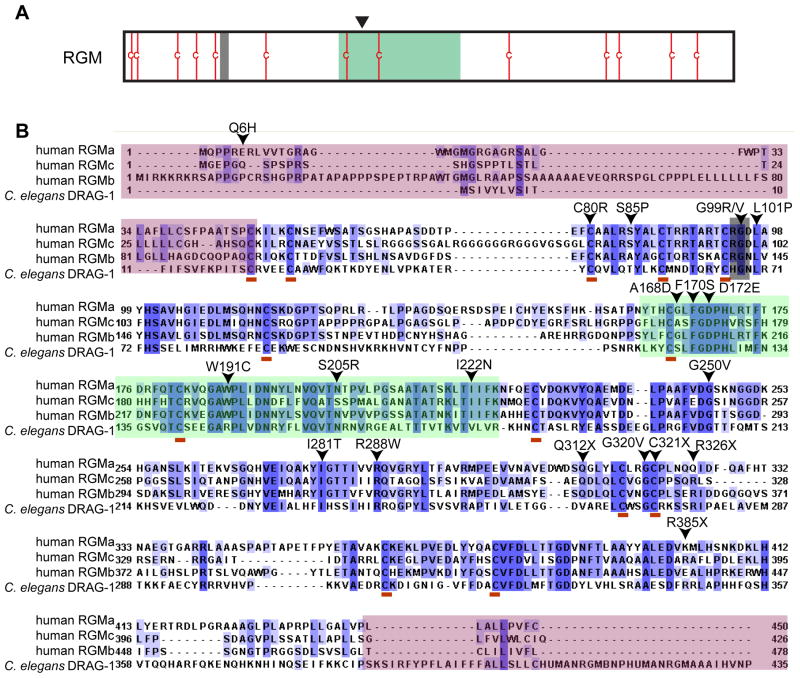
RGM proteins are named after the discovery of RGMa, which is expressed in a gradient in the optic tectum in chick embryos and functions as a repulsive guidance molecule for the temporal retinal axons (Monnier et al. 2002). It is now known that the RGM family members in vertebrates include RGMa, RGMb and RGMc/Hemojuvelin (HJV), while invertebrates, such as C. elegans and sea urchin, only have a single RGM protein encoded in each genome (Camus and Lambert. 2007). The three vertebrate RGMs share approximately 40-50% identity in primary amino acid sequences. They share similarities in predicted protein domains and overall structure, as inferred by ab initio molecular modeling (Severyn et al. 2009). Specifically, all RGMs have an N-terminal signal peptide, a partial von Willebrand type D domain (vWF-type D), which includes a highly conserved autoproteolysis site, and a C-terminal GPI (Glycosylphosphatidylinositol)-anchor. RGMa and RGMc/HJV, but not RGMb, have a RGD motif. RGM proteins also contain a number of highly conserved cysteine residues (Figure 1). RGMa and RGMc have been shown to go through autoproteolytic cleavage at a conserved FGDPH site to generate two fragments that are linked by disulfide bonds (Zhang et al. 2005,Tassew et al. 2012). RGMa and RGMc/HJV also undergo additional processing (more discussion later). Recent studies have shown that RGM proteins function as BMP co-receptors and may allow cells to selectively respond to low levels of BMP ligands (Corradini et al. 2009,Severyn et al. 2009).
Figure 1. Conserved features of RGM proteins.
A. A schematic highlighting features of the mature RGM proteins, including an RGD motif (grey line, only in RGMa and RGMc), a partial vWF type D domain (shaded in green) with an autocleavage site (arrowhead), and conserved cysteine residues (red lines). B. Sequence comparison between human RGMa, RGMb, RGMc, and C. elegans DRAG-1, highlighting conserved residues (blue), the RGD motif (grey), partial vWF type D domain (green), conserved cysteine residues (red underline). The JH (juvenile hemochromatosis) disease-associated hypomorphic mutations in RGMc/HJV are shown (De Gobbi et al. 2002,Lee et al. 2004,Lanzara et al. 2004). Notice that many of them lie in the conserved residues between C. elegans DRAG-1 and human RGM proteins. Sequences shaded in pink are highly divergent between DRAG-1 and its vertebrate counterparts.
RGMa
RGMa was the first discovered RGM family member (Monnier et al. 2002) and it has a broad expression pattern during mouse development (Corradini et al. 2009). RGMa is expressed in the central nervous system in a mostly non-overlapping fashion with RGMb (Oldekamp et al. 2004,Schmidtmer and Engelkamp. 2004,van den Heuvel et al. 2013). It is also expressed in the developing mouse cochlea, lung, limb, and gut (Oldekamp et al. 2004,Metzger et al. 2005). In the adult mouse, RGMa expression is found in the brain, skin, heart, liver, lung, kidney, testis and gut (Babitt et al. 2005,Metzger et al. 2005,Corradini et al. 2009,van den Heuvel et al. 2013). The broad expression pattern of RGMa in the developing mouse embryo suggests that it likely plays important roles in development, especially in central nervous system development. In fact, a large fraction of RGMa knockout mice die due to failure of neural tube closure (Niederkofler et al. 2004). RGMa has also been found to function in various other processes, such as axon guidance in the developing visual system and in axon tract formation in the embryonic brain in vivo (Monnier et al. 2002,Matsunaga et al. 2004,Tassew et al. 2008), in neural tube closure (Kee et al. 2008), neuronal differentiation (Matsunaga et al. 2006), cell survival (Koeberle et al. 2010), as well as cell migration and adhesion (Lah and Key. 2012). Neogenin, as a receptor for RGMa that functions in cells abutting those expressing RGMa, is involved in all the processes described above (see more on neogenin below). Recently, RGMa has also been implicated in colorectal, prostate, and breast cancer (Li et al. 2011,Li et al. 2012,Zhao et al. 2012), and in the immune response (Nohra et al. 2010,Muramatsu et al. 2011,Mirakaj et al. 2011).
RGMa was found to be a BMP co-receptor in vitro only after its homolog RGMb was found to be so (Samad et al. 2005). Both overexpression in LLC-PK1 porcine kidney epithelial cells (Babitt et al. 2005) and siRNA knockdown in mouse myoblast C2C12 cells (Halbrooks et al. 2007) showed that RGMa specifically enhances BMP signaling activity through the canonical BMP signaling pathway, but not the TGFβ signaling pathway. The enhancement depends on BMP ligands, because it was blocked by co-transfection of Noggin, a BMP inhibitor, or BMP2 and BMP4 neutralizing antibodies (Babitt et al. 2005). Besides the requirement of the ligand by RGMa, BMP type I receptors ALK3 and ALK6 as well as Smad1, 5, and 8 are also involved, because dominant negative forms of ALK3 and ALK6 blocked the RGMa-induced enhancement of both BMP-responsive reporter expression and Smad1/5/8 phosphorylation (Babitt et al. 2005).
Purified RGMa.Fc (the extracellular domain of RGMa fused to the Fc portion of human IgG) protein binds directly and selectively to 125I-BMP2 and 125I-BMP4 with a KD of 2.4 nM and 1.4nM, respectively, but not to BMP7 or TGFβ1 ligands (Babitt et al. 2005,Halbrooks et al. 2007,Xia et al. 2007). Results from two Surface Plasmon Resonance Biacore assays also supported direct binding between RGMa and different BMP molecules, the binding of RGMa.Fc to BMP2 has a KD ∼2.46nM (Babitt et al. 2005,Halbrooks et al. 2007) and the binding between RGMa and BMP2 has a KD ∼22nM (Wu et al. 2012). The different results obtained from the two Biocore assays are likely due to the fact that RGMa.Fc fusion proteins form dimers, while the free RGMa proteins act as monomers (Wu et al. 2012). In addition to the BMP ligands, RGMa has also been found to bind to the BMP type I receptor ALK6 (Babitt et al. 2005). RGMa.Fc binds to ALK6.Fc with or without the presence of BMP2, and the binding between RGMa and ALK6 increased the binding of 125I-BMP2 to ALK6 (Babitt et al. 2005). However, there is no evidence that RGMa binds to any of the type II receptors in BMP signaling.
RGMa may function to allow cells to respond to low levels of BMP ligands by changing the utilization of type II receptors from BMPRII alone to both BMPRII and ActRIIA (Xia et al. 2007). In both human ovarian granulosa KGN cells and mouse pulmonary artery smooth muscle cells, where BMP2 and BMP4 signaling is primarily transduced by BMPRII but not ActRIIA or ActRIIB, both BMPRII and ActRIIA are being used to transduce BMP2 and BMP4 signaling when these cells are transfected with RGMa. Furthermore, the addition of RGMa not only increased the binding of 125I-BMP2 to the BMP type I receptor ALK3.Fc, but also increased the binding of 125I-BMP2 to the type II receptor ActRIIA.Fc in the presence of ALK3.Fc (Xia et al. 2007).
Despite the above in vitro studies, there is no in vivo evidence that RGMa functions as a BMP co-receptor. As described earlier, RGMa has been implicated in a diverse array of functions during development. However, it is not know which, if any, of these RGMa-mediated processes is due to the potentiation of BMP signaling by RGMa. For example, BMP signaling occurs in neurons of the mouse adult spinal cord that expresses RGMa (Babitt et al. 2005), however, it is not known if RGMa functions by mediating BMP signaling there. Many of the processes that RGMa is involved in require the function of the RGM receptor neogenin (discussed more later). However, it is not known whether both proteins function together via BMP signaling to regulate these processes in vivo.
RGMb
RGMb is currently less well characterized than RGMa. RGMb, also known as DRAGON, was first identified by using a genomic DNA-binding array to identify downstream genes controlled by DRG11, a homobox transcription factor expressed in embryonic dorsal root ganglion (DRG) (Samad et al. 2004). In the developing brain, RGMa and RGMb exhibit non-overlapping expression patterns. Unlike RGMa, RGMb does not have any detectable repulsive guidance role in embryonic and neonatal DRG neuritis (Samad et al. 2004). RGMb expression is also found in the brain, bone, heart, lung, liver, kidney, testis, ovary, uterus, epididymis and pituitary in adult mouse (Corradini et al. 2009). Because of this wide expression of RGMb, it is not surprising that RGMb knockout mice die 2-3 weeks after birth (Xia et al. 2011). Even though the exact cause of this lethality has not been determined, several studies have provided in vitro and in vivo evidence supporting the functions of RGMb in the nervous system and the immune system: RGMb controls aggregation and migration of neogenin-positive dentate precursor cells (Conrad et al. 2010), promotes neurite outgrowth and peripheral nerve regeneration (Liu et al. 2009,Ma et al. 2011), and negatively regulates IL-6 expression in macrophage cells (Xia et al. 2011). Furthermore, RGMb appears to be a negative regulator of breast cancer proliferation, adhesion, and migration in vitro (Li et al. 2012).
RGMb was the first RGM family member shown to be a BMP co-receptor. The reason to test its involvement in BMP signaling is due to the shared expression pattern between RGMb and the BMP type I and type II receptors in the developing mouse and Xenopus embryos (Samad et al. 2005,Babitt et al. 2005). Similar to RGMa, RGMb enhances BMP signaling but not TGFβ signaling in multiple cell lines, including kidney epithelial LLC-PK1 cells, 10 T1/2 cells, and human liver-derived HepG2 cells, and in Xenopus embryos (Samad et al. 2005), while siRNA treatment of RGMb inhibits BMP signaling in C2C12 cells (Halbrooks et al. 2007). Like RGMa, RGMb enhancement of BMP signaling, as shown in kidney epithelial LLC-PK1 cells, is ligand-dependent and through the canonical BMP signaling pathway (Samad et al. 2005). Also like RGMa, RGMb.Fc binds directly to 125I-BMP2 (KD ∼ 1.5nM) and this binding can be competed by the addition of unlabeled BMP2 and BMP4, but not BMP7 or TGFβ (Samad et al. 2005). Biacore assays confirmed that both RGMb.Fc and RGMb bind directly to BMP2 and BMP4 with similar binding affinities (KD ∼ 5.5 nM for BMP2 and KD ∼2.6nM for BMP4 (Wu et al. 2012). RGMb also physically associates with BMP type I receptors (ALK2, ALK3 and ALK6) and BMP type II receptors (ActRII and ActRIIB) as indicated by co-immunoprecipitation (co-IP) experiments when these components are expressed in HEK293T cells (Samad et al. 2005). The function of RGMb to promote BMP signaling requires its membrane association, as RGMb with the C-terminal GPI anchor deleted, failed to increase BMP signaling (Samad et al. 2005).
While most of the studies have shown that RGMb positively promotes BMP signaling, one study showed that RGMb inhibits constitutively active BMP receptor-induced, or constitutively activated Smad1-induced, BMP signaling in C2C12 myoblasts (Kanomata et al. 2009). This inhibitory effect of RGMb on BMP signaling seems to be via a novel mechanism. How RGMb mediates this inhibition and the functional significance of this inhibition need to be further investigated.
The in vivo relevance of RGMb as a BMP co-receptor has just started to be discovered. As described earlier, RGMb negatively regulates the expression of IL-6 expression specifically in macrophage cells in vitro and in vivo. RGMb knockout mice have increased IL-6 expression in the lungs and liver (Xia et al. 2011). Interestingly, this BMP-dependent regulation of IL-6 expression by RGMb appears to be through the non-canonical P38 MAPK and Erk1/2 pathway, rather than the canonical Smad1/5/8 pathway (Xia et al. 2011). Further studies are needed to determine the mechanistic basis of this regulation. RGMb also promotes neurite outgrowth and peripheral nerve regeneration by modulating BMP signaling (Ma et al. 2011). DRG (dorsal root ganglion) explants from RGMb knockout mice exhibit reduced neurite outgrowth, which can be rescued by BMP2. In addition, the BMP inhibitor Noggin inhibits neurite outgrowth in wild-type DRG explants. Reduced BMP signaling and axonal regeneration have also been observed in vivo in RGMb knockout mice or by Noggin treatment in wild-type mice (Ma et al. 2011). Whether this function is mediated via the canonical Smad1/5/8 pathway or the non-canonical P38 MAPK pathway is not clear. Like RGMa, RGMb also physically associates with neogenin (Conrad et al. 2010). It is not known if neogenin functions together with RGMb in these processes to mediate BMP signaling.
RGMc/HJV
RGMc was identified by positional cloning of the locus that is associated with juvenile hemochromatosis (JH) (Papanikolaou et al. 2004). JH is a rare autosomal recessive disease characterized by high penetrance, early-onset systemic iron overload that affects young patients and leads to severe clinical complications typically in the first and second decade of life (De Gobbi et al. 2002,Camaschella et al. 2002,De Domenico et al. 2008a). JH is also caused by the lack of functional hepcidin (Roetto et al. 2003). Hepcidin is a defensin-like small peptide secreted predominantly from hepatocytes that is essential for iron homeostasis (Park et al. 2001,Pigeon et al. 2001). It acts by binding to and degrading Ferroportin, a rate-limiting sole iron exporter (De Domenico et al. 2008b). Hepcidin expression is up-regulated by high iron level in the body; therefore it is a feedback regulator of iron absorption (Pigeon et al. 2001,Kautz et al. 2008).
HJV is both required and sufficient for hepcidin expression. JH patients with HJV mutations exhibit a reduction of hepatic hepcidin expression, which results in severe iron accumulation in liver, heart and pancreas (Papanikolaou et al. 2004). Hjv-/- mice exhibit a similarly low hepcidin mRNA expression and iron deposition in liver, heart and pancreas (Huang et al. 2005). Transfection of hepatoma-derived Hep3B cells with HJV cDNA increases hepcidin mRNA expression and hepcidin promoter activity in a luciferase assay (Babitt et al. 2006). Introducing HJV into the hepatocytes of Hjv-/- mice using adeno-associated virus 2/8 as a vector is sufficient to completely restore the decreased hepatic hepcidin expression and to lower the high serum iron level to the wild type level (Zhang et al. 2010). Consistent with HJV regulating hepcidin expression in the liver, HJV is expressed predominantly in the liver, skeletal muscle and heart (Niederkofler et al. 2004,Papanikolaou et al. 2004,Samad et al. 2004,Rodriguez et al. 2007). The functional significance for skeletal muscle and heart-expressed HJV is currently not fully understood. Skeletal muscle-expressed HJV has been suggested to be a reservoir for soluble HJV (Zhang et al. 2010; see section on HJV processing). However, skeletal muscle specific knockout of HJV led to no change in hepcidin expression or systemic iron homeostasis (Gkouvatsos et al. 2011,Chen et al. 2011), suggesting that muscle HJV is dispensable for iron metabolism.
Like other RGM family members, HJV functions as a co-receptor in BMP signaling. Transfection of HJV into hepatoma-derived HepG2 and Hep3B cells enhances BMP, but not TGFβ, signaling activity (Babitt et al. 2005,Halbrooks et al. 2007). siRNA knockdown of HJV in C2C12 myoblast reduces BMP2 signaling (Babitt et al. 2005,Halbrooks et al. 2007). Similar to RGMa and RGMb, HJV's role in potentiating BMP signaling is dependent on the BMP ligand, because HJV-induced BMP signaling is abolished by the BMP inhibitor Noggin, and by BMP2 and BMP4 neutralizing antibodies (Babitt et al. 2006). Like RGMa and RGMb, purified HJV.Fc directly binds to 125I-BMP2 and 125I-BMP4, and this binding can be competed away by excess of unlabeled BMP2 and BMP4, but not by BMP7 or TGFβ1 (Babitt et al. 2006). HJV.Fc also directly binds to BMP6 (Andriopoulos et al. 2009). Several Surface Plasmon Resonance Biocore assays have shown that HJV can bind to multiple BMP ligands with high but differential affinity and that the dimeric form of HJV appears to have a stronger affinity to BMP than the monomeric form (Halbrooks et al. 2007,Yang et al. 2008,Wu et al. 2012). In particular, HJV exhibits the highest affinity for BMP6 (KD ∼ 8.1nM), and this binding affinity is much higher than those of RGMb-BMP6 binding (KD ∼ 28nM) and of RGMa-BMP6 binding (KD ∼ 55nM) (Wu et al. 2012). HJV also binds BMP5 (KD ∼ 17nM) and BMP7 (KD ∼ 20nM) with higher affinities higher than those of RGMb and RGMa (KD ∼ 33nM for RGMb-BMP5, ∼83nM for RGMa-BMP5, 166nM for RGMb-BMP7, and 63nM for RGMa-BMP7) (Wu et al. 2012). In addition to binding to the ligand, HJV can form a complex with the type I receptor ALK6 in the presence of BMP2, as indicated by a co-IP assay in HEK293 cells (Babitt et al. 2006). By using siRNA knockdown of different BMP ligands, type I or type II receptors in various cell lines, including Hep3B, Huh-7 and HepG2 cells, and examining the expression of these different ligands and receptors in human liver, Xia and colleagues suggested that HJV likely uses the ligands BMP2, BMP4 and BMP6, the type I receptors ALK2 and ALK3, and the type II receptor ActRIIA in vivo (Xia et al. 2008).
Results from multiple studies have convincingly shown that the underlying mechanism of HJV's role in regulating hepcidin expression is through BMP signaling. Multiple BMP ligands are able to increase hepcidin expression through phosphorylated Smad1/5/8 signaling in hepatocytes in vitro (Babitt et al. 2006,Truksa et al. 2006,Andriopoulos et al. 2009). Furthermore, BMP2-induced hepcidin expression is enhanced by HJV (Babitt et al. 2006). Babitt and colleagues also showed that while wild-type HJV can increase hepcidin expression, HJV carrying JH disease-causing mutations failed to increase hepcidin expression, in liver cells (Babitt et al. 2006). Consistent with these in vitro data, Hjv-/- mice have reduced hepatic hepcidin expression and deceased level of liver pSmad, an indication of reduced BMP signaling activity. Hepatic expression of HJV in the liver of Hjv-/- mice leads to increased hepcidin expression and pSmad1/5/8 level (Zhang et al. 2010). The critical role of BMP signaling in regulating hepcidin expression and iron homeostasis is further supported by the reports that mutations in the genes encoding the ligand BMP6, the type I BMP receptors ALK2 and ALK3, and the co-Smad SMAD4, all result in hepcidin deficiency and iron overload (Wang et al. 2005,Andriopoulos et al. 2009,Meynard et al. 2009,Steinbicker et al. 2011).
In addition to binding to the BMP ligands and receptors, RGMc/HJV also binds to neogenin. The relationship between RGM, neogenin and BMP signaling will be discussed in more detail in the “neogenin” section of this article.
HJV post-translational processing and significance
Because of the role of HJV in regulating iron homeostasis and the fact that mutations in HJV cause JH, HJV has been intensively studied since its discovery. Much of the analysis has been based on HJV proteins over produced in different mammalian cell lines. These studies showed that the HJV protein undergoes complex post-translational modification and processing. It has three predicted N-glycosylation sites. The muscle-expressed and liver-expressed HJV proteins are differentially glycosylated (Fujikura et al. 2011). However, the functional significance for this differential glycosylation is unknown.
Under reducing conditions, cell membrane localized HJV proteins that are overexpressed in HEK293 cells have three major species, with apparent molecular weight of 50, 35, and 20kD respectively. Under non-reducing conditions, HJV only runs as one 50kD band, which corresponds to the full-length form. The 35kD and 20kD species are from an intracellular autoproteolytic cleavage event that happens in a mildly acidic environment. The cleavage site is located in a conserved FGDPH sequence motif in the vWF type D domain of HJV between amino acid 172D (aspartic acid) and 173P (proline), and the resulting two polypeptides are joined together by disulfide bonds to form a two-chain form (Zhang et al. 2005,Kuninger et al. 2006). The membrane-bound two-chain species of HJV is the predominant isoform (Kuninger et al. 2006,Silvestri et al. 2007). Several JH disease-causing mutations appear to affect this autocleavage event. These mutations include those that lie in or near the cleavage site (D172E, F170C and W191C), as well as one predominant disease causing mutation G320V (Silvestri et al. 2007,Pagani et al. 2008). It has been found that these mutant forms of HJV are retained in the endoplasmic reticulum and have low signaling activities, while other disease causing mutations, such as G99V and C119F, that have no effect on HJV autocleavage but have reduced signaling activities do reach the plasma membrane (Silvestri et al. 2007). These studies suggest that HJV cleavage is essential for their export to the plasma membrane.
HJV also undergoes active cleavage by the furin proprotein convertase, resulting in a soluble form of cleaved HJV (s-HJV) that has a molecular mass of ∼40kD (Kuninger et al. 2008,Lin et al. 2008,Silvestri et al. 2008). The cleavage site has been mapped to a RNRR sequence that is not conserved in RGMa and RGMb (Lin et al. 2008,Silvestri et al. 2008). Furin is predominantly localized in the trans-Golgi network (TGN) compartment, with only a small portion of it being on the plasma membrane (Thomas. 2002). Consistent with this, it has been shown that HJV cleavage occurs in the TGN after full-length HJV is being endocytosed from the plasma membrane (Maxson et al. 2009). Furthermore, the binding of neogenin to HJV on the plasma membrane triggers the retrograde trafficking and subsequent release of soluble HJV, s-HJV (Zhang et al. 2008). s-HJV is present not only in transfected cell lines and cell lines that endogenously express HJV, but also in human and rat serum (Kuninger et al. 2004,Lin et al. 2005,Silvestri et al. 2007,Zhang et al. 2007). However, concerns have been raised regarding a commercial assay used to measure s-HJV in humans (Gutiérrez et al. 2012).
s-HJV is an inhibitor of BMP signaling by binding to BMP2, BMP4, and BMP6 to suppress hepcidin expression in vitro (Lin et al. 2007,Kuns-Hashimoto et al. 2008,Andriopoulos et al. 2009,Nili et al. 2010). Injection of s-HJV decreases hepatic hepcidin mRNA level in vivo (Babitt et al. 2007). s-HJV is hypothesized to compete with hepatocyte membrane-bound HJV for BMP ligands, acting as a negative regulator of the BMP-mediated hepcidin expression. Because hepcidin expression is positively regulated by body iron load, it is reasonable to hypothesize that body iron load may negatively regulate the amount of s-HJV. In fact, s-HJV release is negatively regulated by iron level in vitro (Lin et al. 2005,Silvestri et al. 2007,Zhang et al. 2007). In acutely iron-deficient rats, decreased serum Transferrin and Ferritin saturation is associated with decreased hepcidin expression and increased serum s-HJV level (Zhang et al. 2007). Therefore, s-HJV is a negative regulator of hepcidin expression in response to body iron status.
A type II transmembrane serine protease matriptase-2 (MT2), encoded by the gene TMPRSS6, is also involved in regulating body iron load (Silvestri et al. 2008). In mice and zebrafish, MT2 mutations cause hepatic hepcidin overexpression and iron-deficient anemia (Du et al. 2008,Folgueras et al. 2008,Silvestri et al. 2008,Ramsay et al. 2009). In humans, mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA), a familial anemia disorder (Finberg et al. 2008). MT2 and HJV are co-expressed in the liver (Velasco et al. 2002). Co-IP experiments in HeLa cells suggest that they physically interact with each other (Silvestri et al. 2008). This interaction results in the cleavage of cellular HJV into many fragments that are released to the media (Silvestri et al. 2008). Analysis of double mutants between Hjv-/- and Tmprss6-/- or a catalytic mutation Tmprss6mask showed that the double mutants exhibit the Hjv-/- single mutant phenotypes, suggesting that MT2 acts through HJV in regulating hepcidin expression and iron homeostasis (Truksa et al. 2009,Finberg et al. 2010). Consistent with the genetic analysis, over-expression of MT2 can inhibit HJV- and BMP-induced hepcidin expression (Du et al. 2008,Silvestri et al. 2008). However, the Hjv-/-; Tmprss6-/- double mutant phenotype is also consistent with the hypothesis that Tmprss6 functions in a separate pathway from, but is also dependent upon, the HJV pathway for full induction of hepcidin. Consistent with this hypothesis, Tmprss6 mutations that have lost their protease activity can still down regulate hepcidin expression when expressed in HeLa cells and the hepatocyte cell line Huh7, suggesting that the serine protease activity of MT2 may not be important for the regulation of Tmprss6 on hepcidin (Guillem et al. 2012). Moreover, liver membrane HJV level is decreased rather than increased in Tmprss6-/- mutant mice, suggesting that the relationship between HJV and MT2 is more complex in vivo (Krijt et al. 2011). Tmprss6 mRNA expression is stimulated by iron loading treatment or BMP6 protein injection and is blocked by injection of neutralizing antibody against BMP6 in mice (Meynard et al. 2011). Since iron induces hepcidin expression, the up-regulation of MT2, a negative regulator of hepcidin, by iron indicates that MT2 likely represents a negative feedback regulating mechanism to balance hepcidin overexpression induced by iron to maintain systemic iron homeostasis. Further experiments are needed to determine whether MT2 is required for cleaving HJV in vivo, and what the precise relationships among MT2, HJV, BMP signaling and hepcidin expression are.
In addition to RGMc/HJV, processing of RGMa has also been observed (Tassew et al. 2012). Tassew and colleagues showed that RGMa is N-glycosylated, processed by autocleavage and by proprotein convertases SKI-1 (Subtilisin Kexon Isozyme-1) and Furin. These cleavages together with the formation of a disulfide bridge result in the production of 4 membrane-bound and 3 soluble RGMa species, which are all capable of inhibiting neurite outgrowth via neogenin in vitro. The in vivo significance of these cleavages and their relationship with BMP signaling are not known. It's also unclear whether RGMb undergoes similar complicated post-translational processing.
RGM protein function in invertebrates: the sole C. elegans RGM protein is a positive modulator of a BMP-like signaling pathway
RGM proteins are not only found in vertebrates, but also found in invertebrates, such as mollusks, echinoderms and nematodes, which contain one RGM homolog in each corresponding genome (Camus and Lambert. 2007). Surprisingly, no RGM sequence is found in the Drosophila genome (Camus and Lambert. 2007). C. elegans RGM protein DRAG-1 share high degree of amino acid sequence conservation with human RGMs, including conserved vWF type D domain and cysteine residues (Figure 1A,B). Moreover, most of the JH-disease-associated hypomorphic mutations lie in residues that are conserved in DRAG-1 (Figure 1B). We have shown that the sole C. elegans RGM protein DRAG-1 positively modulates a BMP-like signaling pathway, called the Sma/Mab pathway (Tian et al. 2010).
There are two TGFβ related signaling pathways in C. elegans: the Sma/Mab pathway that regulates body size, mail tail patterning (Patterson and Padgett. 2000,Savage-Dunn. 2005), and postembryonic mesoderm development (Foehr et al. 2006); and the dauer pathway that controls formation of dauer larvae in response to environmental stresses (Ren et al. 1996). The Sma/Mab pathway utilizes the BMP-like molecule DBL-1 as the ligand, SMA-6 as the type I receptor, DAF-4 as the type II receptor (shared with the dauer pathway), SMA-2 and SMA-3 as the R-Smads, and SMA-4 as the co-Smad. Loss-of-function mutations in each of the above components cause a small body size and male tail patterning defects (Suzuki et al. 1999,Suzuki et al. 1999,Savage-Dunn et al. 2003). They also suppress the postembryonic mesoderm dorsoventral patterning defect of sma-9 mutants (Foehr et al. 2006). sma-9 encodes a zinc finger transcription factor that is the C. elegans homolog of the Drosophila and vertebrate Schnurri (SHN) proteins (Liang et al. 2003).
drag-1 was identified in a sma-9 suppressor screen (Tian et al. 2010). In addition to suppressing the sma-9 mesoderm patterning defects, drag-1(0) mutants are also small. However, they do not exhibit dauer formation defects. These mutant phenotypes and results from genetic epistasis analysis between drag-1(0) and null mutants of the Sma/Mab pathway demonstrate that drag-1 functions exclusively in the Sma/Mab pathway at the ligand/receptor level to positively modulate Sma/Mab signaling. DRAG-1 appears to function in a cell type-specific manner because drag-1 is not expressed in the male tail and drag-1(0) mutants do not have male tail patterning defects. Consistent with the vertebrate RGM proteins acting as a BMP co-receptor, DRAG-1 is membrane-associated, and is expressed and functions in the signal-receiving cells to modulate Sma/Mab signaling. Despite the lack of biochemical evidence of interactions between DRAG-1 and the Sma/Mab pathway members, this study establishes a direct link between RGM proteins and BMP signaling in vivo, and provides a simple genetic system for further mechanistic studies on RGM protein regulation of BMP signaling in vivo.
The roles of neogenin in modulating BMP signaling
Introduction
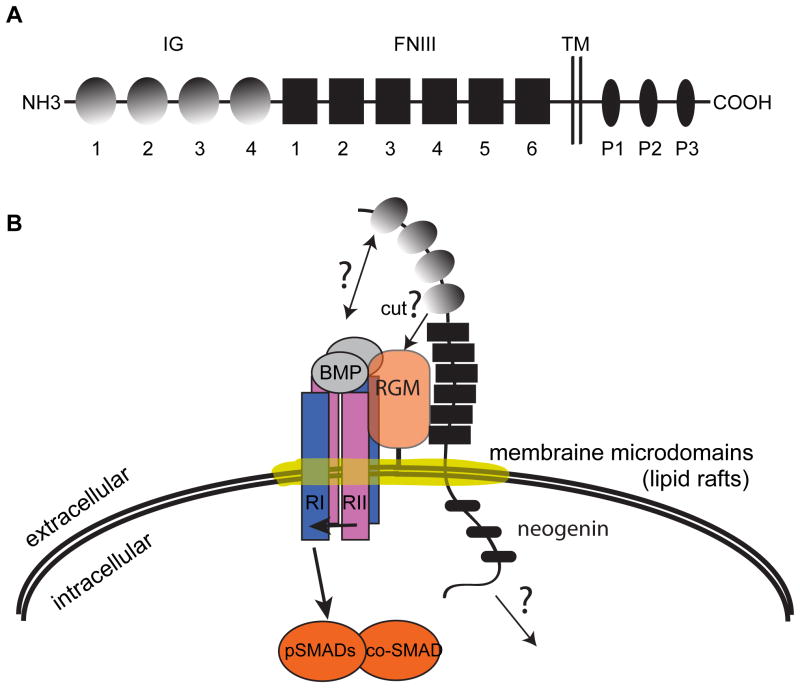
Neogenin encodes a type I transmembrane protein that is homologous to the tumor suppressor gene DCC (Deleted in Colorectal Cancer) (Fearon et al. 1990,Vielmetter et al. 1994). Neogenin and DCC proteins share about 50% sequence identity and similar secondary structures. Neogenin, like DCC, has an extracellular domain, a transmembrane domain and an intracellular domain (Figure 2A). The extracellular domains of neogenin and DCC are closely related and both contain four immunoglobuin (IG) domains and six fibronectin type III (FNIII) domains. The intracellular regions of the two proteins share little homology except for three conserved motifs: P1, P2, and P3. Vertebrates such as zebrafish, Xenopus and mammals contain both neogenin and DCC (Cole et al. 2007,Wilson and Key. 2007,Yamashita et al. 2007). However, in invertebrates such as Drosophila and C. elegans, only a single protein that resembles both DCC and neogenin is present in each organism, namely, Frazzled in Drosophila and UNC-40 in C. elegans (Chan et al. 1996,Kolodziej et al. 1996).
Figure 2. A model on RGM and neogenin functioning in modulating BMP signaling.
A. A schematic of the neogenin protein, which has four immunoglobulin (IG) repeats and six fibronectin type III (FNIII) repeats located in the extracellular domain, a transmembrane (TM) domain, and three conserved P1, P2 and P3 motifs located in the intracellular domain. B. A model based on one proposed by Zhou et al. (2010) on how RGM and neogenin modulate BMP signaling. RGM and neogenin may form a super complex with the ligand and receptors of the BMP pathway in lipid raft membrane microdomains to mediate signaling. However, many questions remain as to how neogenin may modulate BMP signaling, which include, but are not limited to those marked by the question marks. For example, does neogenin play a role in the processing of RGM? What other proteins interact with neogenin? What is the role of the intracellular domain of neogenin in BMP signaling? See text for more discussions.
Unlike DCC, whose expression in vertebrates is primarily restricted to the developing nervous system (Gad et al. 1997), neogenin has a much broader expression pattern. Neogenin is found in the dividing neurogenic and gliogenic progenitors as well as maturing neurons throughout the embryonic and adult central nervous system (Vielmetter et al. 1994,Keeling et al. 1997,Fitzgerald et al. 2006a,Fitzgerald et al. 2006b,van den Heuvel et al. 2013,). Outside of the nervous system, neogenin is found in many developing and adult tissues, including gut, heart, kidney, lung, liver, skeletal muscle and bone cells (Gad et al. 1997,Rodriguez et al. 2007). The broad presence of neogenin outside of the nervous system suggests that it may have non-neuronal functions. Recent research is beginning to shed light on these functions of neogenin outside of the nervous system.
Neogenin as a netrin receptor
DCC has been well established to be a receptor for netrin in axon guidance and migration (Kennedy et al. 1994,Colavita and Culotti. 1998,Hong et al. 1999,Wang et al. 1999,Qin et al. 2007). Because of the sequence similarity shared by DCC and neogenin, earlier work on neogenin focused on its interaction with netrins. Although less well studied compared to DCC, neogenin has a netrin-dependent role in mediating chemoattractant responses to netrin in supraoptic axons in the Xenopus forebrain (Wilson and Key. 2006). Neogenin also regulates cell-cell adhesion and tissue organization through interacting with netrins (Srinivasan et al. 2003,Kang et al. 2004,Park et al. 2004,Jarjour et al. 2008,Lejmi et al. 2008). The netrin-binding site in neogenin has not been mapped. But because DCC binds to netrin via the fourth and fifth FNIII domains (FNIII-4, FNIII-5) (Geisbrecht et al. 2003,Kruger et al. 2004), and DCC and neogenin share high degree of sequence similarity, neogenin is expected to bind netrin via similar domains. Consistently, the binding affinity between netrin and neogenin appears similar to the DCC-netrin binding affinity, which is around 2 nM (Keino-Masu et al. 1996,Wang et al. 1999).
The signal transduction cascades initiated upon neogenin-netrin interaction are not well understood. While DCC-netrin interaction has been found to activate signal transduction cascades that include intracellular tyrosine kinases, second messengers, and Rho GTPases (Gomez and Zheng. 2006,Lin and Holt. 2007,Moore et al. 2007,Round and Stein. 2007,Rajasekharan and Kennedy. 2009,Lai Wing Sun et al. 2011), limited evidence suggests neogenin may have differential downstream effects and interaction partners than DCC (Li et al. 2004,Liu et al. 2004,Xie et al. 2005,Ren et al. 2004, 2008). For example, while both DCC and neogenin become tyrosine phosphorylated in response to netrin-1 in cortical neurons, tyrosine phosphorylated DCC and neogenin exhibit differential binding to effector proteins, such as SHIP-1 (Src Homology 2-containing Inositol 5-Phosphatase 1, Ren et al. 2008). Similarly, in mouse migratory GnRH neuron-derived NLT cells, DCC and neogenin both interact with myosin X (Myo X), an unconventional actin-based motor protein important for filopodium formation, but they exert differential regulatory roles on myosin X activity (Zhu et al. 2007,Liu et al. 2012). Much research is needed to further illustrate the downstream signal transduction events of neogenin-netrin interaction.
Neogenin interaction with RGM proteins
In addition to netrins, neogenin also binds to RGM proteins. Neogenin-RGMa interaction is first identified to be required for the chemorepulsion of temporal retinal axons in the anterior tectum of the chick (Rajagopalan et al. 2004). Later on, neogenin-RGMa interaction has been recognized in many of the processes that RGMa regulates, including axon tract formation in the embryonic brain (Monnier et al. 2002,Matsunaga et al. 2004,Tassew et al. 2008), neural tube closure (Kee et al. 2008), neuronal differentiation (Matsunaga et al. 2006), cell survival (Koeberle et al. 2010), cell migration, and cell-cell adhesion (Lah and Key. 2012). The expression patterns of neogenin and RGMa have been documented in some of these studies, mostly by using mRNA in situ hybridization. Baring the resolution of this technique, neogenin and RGMa appear to exhibit complementary or abutting expression domains and appear to function in trans as a ligand-receptor interaction (Monnier et al. 2002,Rajagopalan et al. 2004,Kee et al. 2008,Tassew et al. 2012).
The interaction between RGMa and neogenin has been dissected in more detail. By performing AP (Alkaline phosphatase) binding assays in monkey COS-7 transfected cells and co-immunoprecipitation experiments in HEK293 cells, Rajagopalan and colleagues (Rajagopalan et al. 2004) showed that the binding affinity of chick RGMa to mouse neogenin is about 10-fold higher than that of netrin to neogenin, suggesting that RGMa may be the primary ligand for neogenin in the CNS (Rajagopalan et al. 2004). They further showed that the binding site for chick RGMa resides within the FNIII domains of mouse neogenin; and that the binding of RGMa to neogenin is unaffected by netrin-1 binding, suggesting that neogenin likely binds to these two ligands via distinct binding sites (Rajagopalan et al. 2004). Surprisingly, both the N- and C-terminal chick RGMa fragments, which are the protease cleavage products of full length RGMa and share no sequence similarity to each other, bind to the same FNIII-(3-4) region of neogenin (Tassew et al. 2012). Whether these regions mediate neogenin-RGMa interaction in vivo will need to be further determined.
Besides RGMa, neogenin also directly binds to RGMb and RGMc/HJV. RGMb-neogenin interaction controls aggregation and migration of a population of dentate precursor cells in vitro and in mice (Conrad et al. 2010). But the interaction between neogenin and HJV has been investigated in much more depth because of the involvement of HJV in JH. Neogenin was first shown to interact with HJV via immunoprecipitation in human embryonic kidney 293 (HEK293) cells transfected with both HJV and neogenin (Zhang et al. 2005). Zhang and colleagues further showed that co-expression of HJV and neogenin in HEK293 cells increases iron accumulation, whereas HJV carrying a disease-causing G320V mutation fails to interact with neogenin (Zhang et al. 2005). Yang and colleagues (Yang et al. 2008,Yang et al. 2011) subsequently purified the ectodomain of human neogenin and showed that the HJV-binding site is localized to FNIII-5, FNIII-6 and the proximal juxtamembrane region of neogenin, with FNIII-6 mediating most of neogenin-HJV interaction. They further solved the crystal structure of FNIII-5 and FNIII-6 of neogenin, and proposed, by comparing neogenin FNIII-6 domain (that binds to HJV) to neogenin FNIII-5 domain and DCC FNIII-6 domain (that do not bind to HJV), that two loops (C-C′ loop and E-F loop) and the C′ strand in the FNIII-6 domain of neogenin may be involved in mediating HJV-neogenin interaction (Yang et al. 2011). However, these regions have not been directly tested for roles in mediating HJV-neogenin interaction. Notably, the HJV binding site (FNIII-5, 6) in neogenin differs from the RGMa binding site (FNIII-3, 4) in neogenin (Rajagopalan et al. 2004,Tassew et al. 2012). Further studies are needed to determine whether this difference is due to intrinsic differences between the RGMa and HJV/RGMc proteins, or differences between the specific assay conditions used. It is intriguing though that HJV/RGMc and neogenin are co-expressed in hepatocytes (Zhang et al. 2009), thus likely function in cis in regulating iron homeostasis, while RGMa and neogenin are expressed in adjacent cells, and thus likely function in trans, in regulating axon growth (Rajagopalan et al. 2004,Tassew et al. 2012). These different modes of action may be another reason underlying the differences between the RGM binding domains in neogenin described above.
Discrepancy also exists regarding the neogenin-binding site in RGM proteins. As described above, Tassew and colleagues showed that both the N- and C-terminal chick RGMa fragments, which are the protease cleavage products of full length chick RGMa and share no sequence similarity to each other, bind to the same FNIII-(3-4) region of mouse neogenin (Tassew et al. 2012). In another study, Itokazu and colleagues (Itokazu et al. 2012) showed, via co-immunoprecipitation assays in HEK293T cells, that amino acids 259-295, which are located in between the vWF type D motif and the hydrophobic region of human RGMa, directly bind to the ectodomain of neogenin. While the discrepancy between the two studies could be due to the specific cell lines and assay conditions used, an additional variable may be the sources of the RGMa and neogenin sequences: chick, mouse vs. human cDNAs being used. Future work should also address in vivo the functional importance of RGM-neogenin interaction and the interaction motifs in each protein.
Modulation of BMP signaling by Neogenin
Earlier work using tissue culture cells has suggested conflicting roles of neogenin in BMP signaling. Zhang and colleagues showed that HJV and neogenin are co-expressed in hepatocytes and that knocking down of neogenin in human hepatoma-derived HepG2 cells leads to decreased BMP signaling and reduced hepcidin expression, suggesting a positive role of neogenin in modulating BMP signaling (Zhang et al. 2009). In a separate study using Hep3B cells, also hepatoma-derived, Xia and colleagues showed that HJV-mediated BMP signaling and hepcidin regulation could occur independently of neogenin (Xia et al. 2008). The use of different cell lines in these studies may have contributed to the conflicting results. More recently, the analysis of the neogenin knockout mice provides convincing evidence supporting a role of neogenin in promoting BMP signaling. Homozygous mice of a hypomorphic gene-trap line of neogenin, which exhibit ∼90% reduction of the neogenin protein level, die within a month of birth and share many defects resembling those caused by reduced BMP signaling (Bae et al. 2009,Lee et al. 2010). In particular, these neogenin-hypomorphic mice show impaired digit/limb development and endochondral ossification, apparently due to reduced Smad1/5/8 phosphorylation by BMP2 (Zhou et al. 2010). Importantly, defective BMP signaling and chondrogenesis in the neogenin-hypomorphic chondrocytes can be rescued by high dose of BMP2, supporting a role of neogenin in modulating BMP signaling and function (Zhou et al. 2010). Similarly, neogenin- hypomorphic mice have reduced hepatic hepcidin expression, severe iron overload, and reduced BMP signaling (Lee et al. 2010), which are similar, although not identical, phenotypes to those seen in Hjv-/- mice (Niederkofler et al. 2005,Huang et al. 2005). Major differences include the level of reduction of hepcidin expression and the severity and onset of iron overload (Lee et al. 2010,Niederkofler et al. 2005,Huang et al. 2005). These differences may partly be due to the hypomorphic nature of the neogenin mutant mice. Alternatively, neogenin and HJV may have overlapping as well as distinct functions in regulating iron homeostasis. It is important to note that neogenin and HJV share similar expression patterns in the liver, while neogenin and RGMa, RGMb and RGMc are all expressed in chondrocytes, suggesting that neogenin and RGM proteins may function in the same cells in cis to modulate BMP signaling (Lee et al. 2010, Zhou et al. 2010).
The mechanism of how neogenin modulates BMP signaling is controversial and poorly understood. Neogenin has been implicated in the processing and secretion of HJV. As described earlier, cellular HJV can be cleaved by furin, matriptase-2 (MT2) and via auto-cleavage to release soluble HJV (s-HJV), which can bind BMP molecules and suppress BMP-induced hepcidin expression (Zhang et al. 2010). On the one hand, neogenin appears to promote the secretion of s-HJV: neogenin can bind to both MT2 and HJV to facilitate the cleavage of HJV by MT2 in HepG2 and HEK293 cells; and knockdown of neogenin in HepG2 cells abolished HJV secretion (Zhang et al. 2005,Zhang et al. 2007,Enns et al. 2012). On the other hand, Lee and colleagues reported that neogenin inhibits HJV secretion in HEK293 cells, which is consistent with their in vivo observations that neogenin positively modulates hepcidin expression and BMP signaling (Lee et al. 2010). The discrepancy might be due to different cell lines being used. It is also worth noting that little is known about whether neogenin-MT2 interaction is also involved in regulating the cleavage of RGMa and RGMb proteins and how neogenin-MT2 interaction intersects with the BMP pathway in vivo.
Zhou and colleagues (Zhou et al. 2010) showed that in mouse chondrocytes upon BMP stimulation, both neogenin and BMP receptors become associated with the lipid raft microdomains and the association of BMP receptors to these membrane microdomains requires neogenin. Interestingly, RGM proteins appear to bridge neogenin with BMP receptors in these lipid raft microdomains as neogenin does not directly associate with the BMP receptors. Based on these findings, the authors proposed a model in which neogenin positively modulates BMP signaling by promoting the formation of BMP-induced super receptor complexes, including RGMs, neogenin and BMP receptors, in membrane microdomains (Zhou et al. 2010). However, results from a different study showed that neogenin negatively regulates BMP2-induced osteoblastic differentiation of C2C12 cells and phosphorylation of Smads1/5/8, apparently by activating RhoA independently of the BMP receptors (Hagihara et al. 2011). Because of these results and the finding that neogenin directly associates with recombinant human BMP1, BMP4, BMP6 and BMP7, the authors proposed that neogenin acts as a receptor for BMPs to negatively regulate BMP function (Hagihara et al. 2011). This negative role of neogenin in BMP signaling may be context dependent and requires further support from in vivo analysis.
Conclusions and perspective
In summary, RGM proteins clearly play important roles in modulating the BMP signaling pathway. Neogenin, the RGM receptor, also appears to be required for BMP signaling. However, much work is still needed, especially in the case of neogenin, to dissect the mechanistic basis on how RGM and neogenin proteins function in modulating BMP signaling. In the case of RGM proteins, further studies are needed to provide in vivo evidence for the involvement of RGMa in BMP signaling. RGM proteins, in particular, RGMc/HJV, appear to undergo complex post-translational modifications. However, except for the processing of HJV by matriptase-2, the in vivo significance of these cleavages in BMP signal transduction is not known. Nor is it known about how the activities of the different processing enzymes are regulated to ensure tight control of proper processing and function of the substrates. All three RGM proteins have been found to bind to the ligand and BMP receptors in vitro, the specific motifs in each of the proteins that are involved in each pair-wise interaction have not been identified, let alone the in vivo significance of these interactions. Finally, the impact of various JH-disease causing mutations on BMP signal transduction should also be further investigated. These natural mutations affect many highly conserved residues among RGM proteins and may provide powerful entry points to uncovering new molecular interactions or functions for RGMs in general.
Compared to RGM proteins, our understanding of the function of neogenin in BMP signal transduction is much more fragmented, and in some cases contradictory. It will be worth investigating whether the reported conflicting roles of neogenin in BMP signaling are due to differences of the various cell culture systems used, or reflect the in vivo diversity of cell type-specific functions of neogenin in modulating BMP signaling. It will also be important to determine the functional significance of RGM-neogenin interaction in modulating BMP signaling in vivo. For example, the neogenin hypomorphic mutant mice exhibit BMP loss of function phenotypes in bone development and iron homeostasis (Zhou et al. 2010). Do any of the RGM protein knockout models exhibit similar phenotypes? Does disruption of RGM-neogenin interaction affect BMP signaling in vivo? Zhou and colleagues proposed a model (Figure 2B) in which RGM and neogenin form a super complex with the ligand and receptors of the BMP pathway in lipid raft membrane microdomains. Is RGM-neogenin interaction required for the formation of this complex in vivo? How does the complex get recruited to these membrane microdomains? What other components are also located and function in these membrane microdomains in mediating BMP signaling? How do we reconcile this model with the model in which neogenin is involved in matriptase-2-mediated processing of HJV?
Neogenin is involved in both netrin-mediated signaling and RGM-mediated signaling. Are these two independent processes or does neogenin represent a point of crosstalk between the two signaling pathways? Unlike RGM proteins, which are GPI-anchored proteins, neogenin has a large cytoplasmic domain (CTD) containing three conserved motifs. While the CTD is clearly required for neogenin to mediate netrin signaling, it is not known whether CTD is also required for neogenin's role in mediating BMP signaling, and if it is required, what the downstream effectors are in BMP signal transduction. Okamura and colleagues reported that neogenin can be cleaved by the tumor necrosis factor-alpha converting enzyme TACE, which in turn desensitizes cortical neurons to RGMa-induced repulsive behavior (Okamura et al. 2011). While the importance of this cleavage in RGMa function in vivo has not been determined, in light of the roles of neogenin in modulating the BMP pathway, one wonders whether the cleavage of neogenin also plays a role in BMP signaling.
In addition to the research on these proteins in vertebrate systems, studies of RGM proteins and neogenin in invertebrate model systems such as C. elegans and Drosophila may also help shed light on whether and how these proteins function in modulating BMP signaling in vivo. Drosophila does not contain a RGM homology but encodes a single protein Frazzled, which is a homolog of both neogenin and DCC (Kolodziej et al. 1996). In C. elegans, we have previously shown that the single RGM protein DRAG-1 positively modulates BMP signaling (Tian et al. 2010). C. elegans also contains one protein UNC-40, which is homologous to both DCC and neogenin (Chan et al. 1996). The functions of UNC-40 in axon guidance and cell migration have been well established (Quinn and Wadsworth. 2008). In light of the interaction between vertebrate RGM proteins and neogenin, it will be interesting to determine whether UNC-40 also plays a role in RGM mediated BMP signaling. If it does, the power of genetics in C. elegans will allow for functional dissection of the relevance of RGM-neogenin interaction in modulating BMP signaling in vivo, and allow us to determine at single cell resolution whether RGM and neogenin function in cis or in trans to modulate BMP signaling. From an evolutionary perspective, it will also be interesting to determine whether the Drosophila DCC/neogenin protein Frazzled can function in modulating BMP signaling, and if yes, how it functions independent of a RGM protein.
Acknowledgments
We thank Ken Kemphues, Mariana Wolfner and two anonymous reviewers for their helpful comments and suggestions and apologize to colleagues whose work was not cited due to space limitation. This work was supported by NIH R01 GM066953 (to J.L.).
Grant support: NIH R01 GM066953 to J.L.
Abbreviations
- BMP
bone morphogenetic protein
- TGFβ
transforming growth factor-beta
- RGM
repulsive guidance molecule
- DCC
deleted in colorectal cancer
- R-Smad
receptor regulated Smad
- co-Smad
common mediator Smad
- MAPK
mitogen activated protein kinase
- Sog
short gastrulation
- MGP
matrix GLA protein
- Tsg
twisted gastrulation
- CV2
crossveinless 2
- ECM
extracellular matrix
- GPI
glycosylphosphatidylinositol
- Sma/Mab
small/male tail abnormal
- BAMBI
BMP and activin membrane-bound inhibitor
- HJV
hemojuvelin
- vWF
von Willebrand factor
- DRG
dorsal root ganglion
- co-IP
co-immunoprecipitation
- JH
juvenile hemochromatosis
- s-HJV
soluble HJV
- TGN
trans-Golgi network
- MT2
matriptase-2
- IRIDA
iron-refractory iron deficiency anemia
- IG
immunoglobulin
- FNIII
fibronectin type III
- CTD
cytoplasmic domain
- TACE
tumor necrosis factor-alpha converting enzyme
References
- Allendorph GP, Vale WW, Choe S. Structure of the ternary signaling complex of a TGF-beta superfamily member. Proc Natl Acad Sci U S A. 2006;103:7643–7648. doi: 10.1073/pnas.0602558103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio AL, Taelman VF, Lee HX, Metzinger CA, Coffinier C, De Robertis EM. Crossveinless-2 Is a BMP feedback inhibitor that binds Chordin/BMP to regulate Xenopus embryonic patterning. Dev Cell. 2008;15:248–260. doi: 10.1016/j.devcel.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriopoulos B, Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, Knutson MD, Pietrangelo A, Vukicevic S, Lin HY, Babitt JL. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet. 2009;41:482–487. doi: 10.1038/ng.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attisano L, Wrana JL, Lopez-Casillas F, Massague J. TGF-beta receptors and actions. Biochim Biophys Acta. 1994;1222:71–80. doi: 10.1016/0167-4889(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Avsian-Kretchmer O, Hsueh AJ. Comparative genomic analysis of the eight-membered ring cystine knot-containing bone morphogenetic protein antagonists. Mol Endocrinol. 2004;18:1–12. doi: 10.1210/me.2003-0227. [DOI] [PubMed] [Google Scholar]
- Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–539. doi: 10.1038/ng1777. [DOI] [PubMed] [Google Scholar]
- Babitt JL, Huang FW, Xia Y, Sidis Y, Andrews NC, Lin HY. Modulation of bone morphogenetic protein signaling in vivo regulates systemic iron balance. J Clin Invest. 2007;117:1933–1939. doi: 10.1172/JCI31342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babitt JL, Zhang Y, Samad TA, Xia Y, Tang J, Campagna JA, Schneyer AL, Woolf CJ, Lin HY. Repulsive guidance molecule (RGMa), a DRAGON homologue, is a bone morphogenetic protein co-receptor. J Biol Chem. 2005;280:29820–29827. doi: 10.1074/jbc.M503511200. [DOI] [PubMed] [Google Scholar]
- Bae GU, Yang YJ, Jiang G, Hong M, Lee HJ, Tessier-Lavigne M, Kang JS, Krauss RS. Neogenin regulates skeletal myofiber size and focal adhesion kinase and extracellular signal-regulated kinase activities in vivo and in vitro. Mol Biol Cell. 2009;20:4920–4931. doi: 10.1091/mbc.E09-06-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balemans W, Van Hul W. Extracellular regulation of BMP signaling in vertebrates: a cocktail of modulators. Dev Biol. 2002;250:231–250. [PubMed] [Google Scholar]
- Bilandzic M, Stenvers KL. Betaglycan: a multifunctional accessory. Mol Cell Endocrinol. 2011;339:180–189. doi: 10.1016/j.mce.2011.04.014. [DOI] [PubMed] [Google Scholar]
- Bragdon B, Moseychuk O, Saldanha S, King D, Julian J, Nohe A. Bone morphogenetic proteins: a critical review. Cell Signal. 2011;23:609–620. doi: 10.1016/j.cellsig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- Cai J, Pardali E, Sanchez-Duffhues G, ten Dijke P. BMP signaling in vascular diseases. FEBS Lett. 2012;586:1993–2002. doi: 10.1016/j.febslet.2012.04.030. [DOI] [PubMed] [Google Scholar]
- Camaschella C, Roetto A, De Gobbi M. Juvenile hemochromatosis. Semin Hematol. 2002;39:242–248. doi: 10.1053/shem.2002.35635. [DOI] [PubMed] [Google Scholar]
- Camus LM, Lambert LA. Molecular evolution of hemojuvelin and the repulsive guidance molecule family. J Mol Evol. 2007;65:68–81. doi: 10.1007/s00239-006-0241-5. [DOI] [PubMed] [Google Scholar]
- Chan SS, Zheng H, Su MW, Wilk R, Killeen MT, Hedgecock EM, Culotti JG. UNC-40, a C. elegans homolog of DCC (Deleted in Colorectal Cancer), is required in motile cells responding to UNC-6 netrin cues. Cell. 1996;87:187–195. doi: 10.1016/s0092-8674(00)81337-9. [DOI] [PubMed] [Google Scholar]
- Chen W, Huang FW, de Renshaw TB, Andrews NC. Skeletal muscle hemojuvelin is dispensable for systemic iron homeostasis. Blood. 2011;117:6319–6325. doi: 10.1182/blood-2010-12-327957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colavita A, Culotti JG. Suppressors of ectopic UNC-5 growth cone steering identify eight genes involved in axon guidance in Caenorhabditis elegans. Dev Biol. 1998;194:72–85. doi: 10.1006/dbio.1997.8790. [DOI] [PubMed] [Google Scholar]
- Cole SJ, Bradford D, Cooper HM. Neogenin: A multi-functional receptor regulating diverse developmental processes. Int J Biochem Cell Biol. 2007;39:1569–1575. doi: 10.1016/j.biocel.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Conrad S, Stimpfle F, Montazeri S, Oldekamp J, Seid K, Alvarez-Bolado G, Skutella T. RGMb controls aggregation and migration of Neogenin-positive cells in vitro and in vivo. Mol Cell Neurosci. 2010;43:222–231. doi: 10.1016/j.mcn.2009.11.003. [DOI] [PubMed] [Google Scholar]
- Corradini E, Babitt JL, Lin HY. The RGM/DRAGON family of BMP co-receptors. Cytokine Growth Factor Rev. 2009;20:389–398. doi: 10.1016/j.cytogfr.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Domenico I, McVey Ward D, Kaplan J. Regulation of iron acquisition and storage: consequences for iron-linked disorders. Nat Rev Mol Cell Biol. 2008a;9:72–81. doi: 10.1038/nrm2295. [DOI] [PubMed] [Google Scholar]
- De Domenico I, Nemeth E, Nelson JM, Phillips JD, Ajioka RS, Kay MS, Kushner JP, Ganz T, Ward DM, Kaplan J. The hepcidin-binding site on ferroportin is evolutionarily conserved. Cell Metab. 2008b;8:146–156. doi: 10.1016/j.cmet.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- De Gobbi M, Roetto A, Piperno A, Mariani R, Alberti F, Papanikolaou G, Politou M, Lockitch G, Girelli D, Fargion S, Cox TM, Gasparini P, Cazzola M, Camaschella C. Natural history of juvenile haemochromatosis. Br J Haematol. 2002;117:973–979. doi: 10.1046/j.1365-2141.2002.03509.x. [DOI] [PubMed] [Google Scholar]
- Du X, She E, Gelbart T, Truksa J, Lee P, Xia Y, Khovananth K, Mudd S, Mann N, Moresco EM, Beutler E, Beutler B. The serine protease TMPRSS6 is required to sense iron deficiency. Science. 2008;320:1088–1092. doi: 10.1126/science.1157121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enns CA, Ahmed R, Zhang AS. Neogenin interacts with matriptase-2 to facilitate hemojuvelin cleavage. J Biol Chem. 2012;287:35104–35117. doi: 10.1074/jbc.M112.363937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Cho KR, Nigro JM, Kern SE, Simons JW, Ruppert JM, Hamilton SR, Preisinger AC, Thomas G, Kinzler KW. Identification of a chromosome 18q gene that is altered in colorectal cancers. Science. 1990;247:49–56. doi: 10.1126/science.2294591. [DOI] [PubMed] [Google Scholar]
- Feng XH, Derynck R. Specificity and versatility in tgf-beta signaling through Smads. Annu Rev Cell Dev Biol. 2005;21:659–693. doi: 10.1146/annurev.cellbio.21.022404.142018. [DOI] [PubMed] [Google Scholar]
- Finberg KE, Heeney MM, Campagna DR, Aydinok Y, Pearson HA, Hartman KR, Mayo MM, Samuel SM, Strouse JJ, Markianos K, Andrews NC, Fleming MD. Mutations in TMPRSS6 cause iron-refractory iron deficiency anemia (IRIDA) Nat Genet. 2008;40:569–571. doi: 10.1038/ng.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finberg KE, Whittlesey RL, Fleming MD, Andrews NC. Down-regulation of Bmp/Smad signaling by Tmprss6 is required for maintenance of systemic iron homeostasis. Blood. 2010;115:3817–3826. doi: 10.1182/blood-2009-05-224808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald DP, Cole SJ, Hammond A, Seaman C, Cooper HM. Characterization of neogenin-expressing neural progenitor populations and migrating neuroblasts in the embryonic mouse forebrain. Neuroscience. 2006a;142:703–716. doi: 10.1016/j.neuroscience.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Fitzgerald DP, Seaman C, Cooper HM. Localization of Neogenin protein during morphogenesis in the mouse embryo. Dev Dyn. 2006b;235:1720–1725. doi: 10.1002/dvdy.20744. [DOI] [PubMed] [Google Scholar]
- Foehr ML, Lindy AS, Fairbank RC, Amin NM, Xu M, Yanowitz J, Fire AZ, Liu J. An antagonistic role for the C. elegans Schnurri homolog SMA-9 in modulating TGFbeta signaling during mesodermal patterning. Development. 2006;133:2887–2896. doi: 10.1242/dev.02476. [DOI] [PubMed] [Google Scholar]
- Folgueras AR, de Lara FM, Pendas AM, Garabaya C, Rodriguez F, Astudillo A, Bernal T, Cabanillas R, Lopez-Otin C, Velasco G. Membrane-bound serine protease matriptase-2 (Tmprss6) is an essential regulator of iron homeostasis. Blood. 2008;112:2539–2545. doi: 10.1182/blood-2008-04-149773. [DOI] [PubMed] [Google Scholar]
- Fujikura Y, Krijt J, Necas E. Liver and muscle hemojuvelin are differently glycosylated. BMC Biochem. 2011;12:52. doi: 10.1186/1471-2091-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad JM, Keeling SL, Wilks AF, Tan SS, Cooper HM. The expression patterns of guidance receptors, DCC and Neogenin, are spatially and temporally distinct throughout mouse embryogenesis. Dev Biol. 1997;192:258–273. doi: 10.1006/dbio.1997.8756. [DOI] [PubMed] [Google Scholar]
- Geisbrecht BV, Dowd KA, Barfield RW, Longo PA, Leahy DJ. Netrin binds discrete subdomains of DCC and UNC5 and mediates interactions between DCC and heparin. J Biol Chem. 2003;278:32561–32568. doi: 10.1074/jbc.M302943200. [DOI] [PubMed] [Google Scholar]
- Gkouvatsos K, Wagner J, Papanikolaou G, Sebastiani G, Pantopoulos K. Conditional disruption of mouse HFE2 gene: maintenance of systemic iron homeostasis requires hepatic but not skeletal muscle hemojuvelin. Hepatology. 2011;54:1800–1807. doi: 10.1002/hep.24547. [DOI] [PubMed] [Google Scholar]
- Gomez TM, Zheng JQ. The molecular basis for calcium-dependent axon pathfinding. Nat Rev Neurosci. 2006;7:115–125. doi: 10.1038/nrn1844. [DOI] [PubMed] [Google Scholar]
- Gordon KJ, Blobe GC. Role of transforming growth factor-beta superfamily signaling pathways in human disease. Biochim Biophys Acta. 2008;1782:197–228. doi: 10.1016/j.bbadis.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Grisaru S, Cano-Gauci D, Tee J, Filmus J, Rosenblum ND. Glypican-3 modulates BMP- and FGF-mediated effects during renal branching morphogenesis. Dev Biol. 2001;231:31–46. doi: 10.1006/dbio.2000.0127. [DOI] [PubMed] [Google Scholar]
- Groppe J, Greenwald J, Wiater E, Rodriguez-Leon J, Economides AN, Kwiatkowski W, Affolter M, Vale WW, Belmonte JC, Choe S. Structural basis of BMP signalling inhibition by the cystine knot protein Noggin. Nature. 2002;420:636–642. doi: 10.1038/nature01245. [DOI] [PubMed] [Google Scholar]
- Guillem F, Kannengiesser C, Oudin C, Lenoir A, Matak P, Donadieu J, Isidor B, Mechinaud F, Aguilar-Martinez P, Beaumont C, Vaulont S, Grandchamp B, Nicolas G. Inactive matriptase-2 mutants found in IRIDA patients still repress hepcidin in a transfection assay despite having lost their serine protease activity. Hum Mutat. 2012;33:1388–1396. doi: 10.1002/humu.22116. [DOI] [PubMed] [Google Scholar]
- Gumienny TL, MacNeil LT, Wang H, de Bono M, Wrana JL, Padgett RW. Glypican LON-2 is a conserved negative regulator of BMP-like signaling in Caenorhabditis elegans. Curr Biol. 2007;17:159–164. doi: 10.1016/j.cub.2006.11.065. [DOI] [PubMed] [Google Scholar]
- Gutierrez OM, Sun CC, Chen W, Babitt JL, Lin HY. Statement of concern about a commercial assay used to measure soluble hemojuvelin in humans. Am J Nephrol. 2012;36:332–3. doi: 10.1159/000342519. author reply 333. [DOI] [PubMed] [Google Scholar]
- Hagihara M, Endo M, Hata K, Higuchi C, Takaoka K, Yoshikawa H, Yamashita T. Neogenin, a receptor for bone morphogenetic proteins. J Biol Chem. 2011;286:5157–5165. doi: 10.1074/jbc.M110.180919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbrooks PJ, Ding R, Wozney JM, Bain G. Role of RGM coreceptors in bone morphogenetic protein signaling. J Mol Signal. 2007;2:4. doi: 10.1186/1750-2187-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Yan D, Belenkaya TY, Lin X. Drosophila glypicans Dally and Dally-like shape the extracellular Wingless morphogen gradient in the wing disc. Development. 2005;132:667–679. doi: 10.1242/dev.01636. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Moustakas A. Role of Smads in TGFbeta signaling. Cell Tissue Res. 2012;347:21–36. doi: 10.1007/s00441-011-1190-x. [DOI] [PubMed] [Google Scholar]
- Hong K, Hinck L, Nishiyama M, Poo MM, Tessier-Lavigne M, Stein E. A ligand-gated association between cytoplasmic domains of UNC5 and DCC family receptors converts netrin-induced growth cone attraction to repulsion. Cell. 1999;97:927–941. doi: 10.1016/s0092-8674(00)80804-1. [DOI] [PubMed] [Google Scholar]
- Hsu DR, Economides AN, Wang X, Eimon PM, Harland RM. The Xenopus dorsalizing factor Gremlin identifies a novel family of secreted proteins that antagonize BMP activities. Mol Cell. 1998;1:673–683. doi: 10.1016/s1097-2765(00)80067-2. [DOI] [PubMed] [Google Scholar]
- Huang F, Chen YG. Regulation of TGF-beta receptor activity. Cell Biosci. 2012;2:9. doi: 10.1186/2045-3701-2-9. 3701-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FW, Pinkus JL, Pinkus GS, Fleming MD, Andrews NC. A mouse model of juvenile hemochromatosis. J Clin Invest. 2005;115:2187–2191. doi: 10.1172/JCI25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itokazu T, Fujita Y, Takahashi R, Yamashita T. Identification of the neogenin-binding site on the repulsive guidance molecule a. PLoS One. 2012;7:e32791. doi: 10.1371/journal.pone.0032791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarjour AA, Bull SJ, Almasieh M, Rajasekharan S, Baker KA, Mui J, Antel JP, Di Polo A, Kennedy TE. Maintenance of axo-oligodendroglial paranodal junctions requires DCC and netrin-1. J Neurosci. 2008;28:11003–11014. doi: 10.1523/JNEUROSCI.3285-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JS, Yi MJ, Zhang W, Feinleib JL, Cole F, Krauss RS. Netrins and neogenin promote myotube formation. J Cell Biol. 2004;167:493–504. doi: 10.1083/jcb.200405039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanomata K, Kokabu S, Nojima J, Fukuda T, Katagiri T. DRAGON, a GPI-anchored membrane protein, inhibits BMP signaling in C2C12 myoblasts. Genes Cells. 2009;14:695–702. doi: 10.1111/j.1365-2443.2009.01302.x. [DOI] [PubMed] [Google Scholar]
- Kautz L, Meynard D, Monnier A, Darnaud V, Bouvet R, Wang RH, Deng C, Vaulont S, Mosser J, Coppin H, Roth MP. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood. 2008;112:1503–1509. doi: 10.1182/blood-2008-03-143354. [DOI] [PubMed] [Google Scholar]
- Kee N, Wilson N, De Vries M, Bradford D, Key B, Cooper HM. Neogenin and RGMa control neural tube closure and neuroepithelial morphology by regulating cell polarity. J Neurosci. 2008;28:12643–12653. doi: 10.1523/JNEUROSCI.4265-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling SL, Gad JM, Cooper HM. Mouse Neogenin, a DCC-like molecule, has four splice variants and is expressed widely in the adult mouse and during embryogenesis. Oncogene. 1997;15:691–700. doi: 10.1038/sj.onc.1201225. [DOI] [PubMed] [Google Scholar]
- Keino-Masu K, Masu M, Hinck L, Leonardo ED, Chan SS, Culotti JG, Tessier-Lavigne M. Deleted in Colorectal Cancer (DCC) encodes a netrin receptor. Cell. 1996;87:175–185. doi: 10.1016/s0092-8674(00)81336-7. [DOI] [PubMed] [Google Scholar]
- Kennedy TE, Serafini T, de la Torre JR, Tessier-Lavigne M. Netrins are diffusible chemotropic factors for commissural axons in the embryonic spinal cord. Cell. 1994;78:425–435. doi: 10.1016/0092-8674(94)90421-9. [DOI] [PubMed] [Google Scholar]
- Kirkbride KC, Townsend TA, Bruinsma MW, Barnett JV, Blobe GC. Bone morphogenetic proteins signal through the transforming growth factor-beta type III receptor. J Biol Chem. 2008;283:7628–7637. doi: 10.1074/jbc.M704883200. [DOI] [PubMed] [Google Scholar]
- Koeberle PD, Tura A, Tassew NG, Schlichter LC, Monnier PP. The repulsive guidance molecule, RGMa, promotes retinal ganglion cell survival in vitro and in vivo. Neuroscience. 2010;169:495–504. doi: 10.1016/j.neuroscience.2010.04.079. [DOI] [PubMed] [Google Scholar]
- Kolodziej PA, Timpe LC, Mitchell KJ, Fried SR, Goodman CS, Jan LY, Jan YN. frazzled encodes a Drosophila member of the DCC immunoglobulin subfamily and is required for CNS and motor axon guidance. Cell. 1996;87:197–204. doi: 10.1016/s0092-8674(00)81338-0. [DOI] [PubMed] [Google Scholar]
- Krijt J, Fujikura Y, Ramsay AJ, Velasco G, Necas E. Liver hemojuvelin protein levels in mice deficient in matriptase-2 (Tmprss6) Blood Cells Mol Dis. 2011;47:133–137. doi: 10.1016/j.bcmd.2011.04.009. [DOI] [PubMed] [Google Scholar]
- Kruger RP, Lee J, Li W, Guan KL. Mapping netrin receptor binding reveals domains of Unc5 regulating its tyrosine phosphorylation. J Neurosci. 2004;24:10826–10834. doi: 10.1523/JNEUROSCI.3715-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuninger D, Kuns-Hashimoto R, Nili M, Rotwein P. Pro-protein convertases control the maturation and processing of the iron-regulatory protein, RGMc/hemojuvelin. BMC Biochem. 2008;9:9. doi: 10.1186/1471-2091-9-9. 2091-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuninger D, Kuns-Hashimoto R, Kuzmickas R, Rotwein P. Complex biosynthesis of the muscle-enriched iron regulator RGMc. J Cell Sci. 2006;119:3273–3283. doi: 10.1242/jcs.03074. [DOI] [PubMed] [Google Scholar]
- Kuninger D, Kuzmickas R, Peng B, Pintar JE, Rotwein P. Gene discovery by microarray: identification of novel genes induced during growth factor-mediated muscle cell survival and differentiation. Genomics. 2004;84:876–889. doi: 10.1016/j.ygeno.2004.07.013. [DOI] [PubMed] [Google Scholar]
- Kuns-Hashimoto R, Kuninger D, Nili M, Rotwein P. Selective binding of RGMc/hemojuvelin, a key protein in systemic iron metabolism, to BMP-2 and neogenin. Am J Physiol Cell Physiol. 2008;294:C994–C1003. doi: 10.1152/ajpcell.00563.2007. [DOI] [PubMed] [Google Scholar]
- Lah GJ, Key B. Novel roles of the chemorepellent axon guidance molecule RGMa in cell migration and adhesion. Mol Cell Biol. 2012;32:968–980. doi: 10.1128/MCB.06128-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Wing Sun K, Correia JP, Kennedy TE. Netrins: versatile extracellular cues with diverse functions. Development. 2011;138:2153–2169. doi: 10.1242/dev.044529. [DOI] [PubMed] [Google Scholar]
- Lanzara C, Roetto A, Daraio F, Rivard S, Ficarella R, Simard H, Cox TM, Cazzola M, Piperno A, Gimenez-Roqueplo AP, Grammatico P, Volinia S, Gasparini P, Camaschella C. Spectrum of hemojuvelin gene mutations in 1q-linked juvenile hemochromatosis. Blood. 2004;103:4317–4321. doi: 10.1182/blood-2004-01-0192. [DOI] [PubMed] [Google Scholar]
- Lee DH, Zhou LJ, Zhou Z, Xie JX, Jung JU, Liu Y, Xi CX, Mei L, Xiong WC. Neogenin inhibits HJV secretion and regulates BMP-induced hepcidin expression and iron homeostasis. Blood. 2010;115:3136–3145. doi: 10.1182/blood-2009-11-251199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PL, Beutler E, Rao SV, Barton JC. Genetic abnormalities and juvenile hemochromatosis: mutations of the HJV gene encoding hemojuvelin. Blood. 2004;103:4669–4671. doi: 10.1182/blood-2004-01-0072. [DOI] [PubMed] [Google Scholar]
- Lejmi E, Leconte L, Pedron-Mazoyer S, Ropert S, Raoul W, Lavalette S, Bouras I, Feron JG, Maitre-Boube M, Assayag F, Feumi C, Alemany M, Jie TX, Merkulova T, Poupon MF, Ruchoux MM, Tobelem G, Sennlaub F, Plouet J. Netrin-4 inhibits angiogenesis via binding to neogenin and recruitment of Unc5B. Proc Natl Acad Sci U S A. 2008;105:12491–12496. doi: 10.1073/pnas.0804008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Ye L, Kynaston HG, Jiang WG. Repulsive guidance molecules, novel bone morphogenetic protein co-receptors, are key regulators of the growth and aggressiveness of prostate cancer cells. Int J Oncol. 2012;40:544–550. doi: 10.3892/ijo.2011.1251. [DOI] [PubMed] [Google Scholar]
- Li J, Ye L, Mansel RE, Jiang WG. Potential prognostic value of repulsive guidance molecules in breast cancer. Anticancer Res. 2011;31:1703–1711. [PubMed] [Google Scholar]
- Li J, Ye L, Sanders AJ, Jiang WG. Repulsive guidance molecule B (RGMB) plays negative roles in breast cancer by coordinating BMP signaling. J Cell Biochem. 2012 doi: 10.1002/jcb.24128. [DOI] [PubMed] [Google Scholar]
- Li W, Lee J, Vikis HG, Lee SH, Liu G, Aurandt J, Shen TL, Fearon ER, Guan JL, Han M, Rao Y, Hong K, Guan KL. Activation of FAK and Src are receptor-proximal events required for netrin signaling. Nat Neurosci. 2004;7:1213–1221. doi: 10.1038/nn1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Lints R, Foehr ML, Tokarz R, Yu L, Emmons SW, Liu J, Savage-Dunn C. The Caenorhabditis elegans schnurri homolog sma-9 mediates stage- and cell type-specific responses to DBL-1 BMP-related signaling. Development. 2003;130:6453–6464. doi: 10.1242/dev.00863. [DOI] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Local translation and directional steering in axons. EMBO J. 2007;26:3729–3736. doi: 10.1038/sj.emboj.7601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Goldberg YP, Ganz T. Competitive regulation of hepcidin mRNA by soluble and cell-associated hemojuvelin. Blood. 2005;106:2884–2889. doi: 10.1182/blood-2005-05-1845. [DOI] [PubMed] [Google Scholar]
- Lin L, Nemeth E, Goodnough JB, Thapa DR, Gabayan V, Ganz T. Soluble hemojuvelin is released by proprotein convertase-mediated cleavage at a conserved polybasic RNRR site. Blood Cells Mol Dis. 2008;40:122–131. doi: 10.1016/j.bcmd.2007.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Valore EV, Nemeth E, Goodnough JB, Gabayan V, Ganz T. Iron transferrin regulates hepcidin synthesis in primary hepatocyte culture through hemojuvelin and BMP2/4. Blood. 2007;110:2182–2189. doi: 10.1182/blood-2007-04-087593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Beggs H, Jurgensen C, Park HT, Tang H, Gorski J, Jones KR, Reichardt LF, Wu J, Rao Y. Netrin requires focal adhesion kinase and Src family kinases for axon outgrowth and attraction. Nat Neurosci. 2004;7:1222–1232. doi: 10.1038/nn1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Hashimoto M, Horii H, Yamaguchi A, Naito K, Yamashita T. Repulsive guidance molecule b inhibits neurite growth and is increased after spinal cord injury. Biochem Biophys Res Commun. 2009;382:795–800. doi: 10.1016/j.bbrc.2009.03.115. [DOI] [PubMed] [Google Scholar]
- Liu Y, Peng Y, Dai PG, Du QS, Mei L, Xiong WC. Differential regulation of myosin X movements by its cargos, DCC and neogenin. J Cell Sci. 2012;125:751–762. doi: 10.1242/jcs.094946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CH, Brenner GJ, Omura T, Samad OA, Costigan M, Inquimbert P, Niederkofler V, Salie R, Sun CC, Lin HY, Arber S, Coppola G, Woolf CJ, Samad TA. The BMP coreceptor RGMb promotes while the endogenous BMP antagonist noggin reduces neurite outgrowth and peripheral nerve regeneration by modulating BMP signaling. J Neurosci. 2011;31:18391–18400. doi: 10.1523/JNEUROSCI.4550-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J. TGFbeta signalling in context. Nat Rev Mol Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga E, Nakamura H, Chedotal A. Repulsive guidance molecule plays multiple roles in neuronal differentiation and axon guidance. J Neurosci. 2006;26:6082–6088. doi: 10.1523/JNEUROSCI.4556-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga E, Tauszig-Delamasure S, Monnier PP, Mueller BK, Strittmatter SM, Mehlen P, Chedotal A. RGM and its receptor neogenin regulate neuronal survival. Nat Cell Biol. 2004;6:749–755. doi: 10.1038/ncb1157. [DOI] [PubMed] [Google Scholar]
- Maxson JE, Enns CA, Zhang AS. Processing of hemojuvelin requires retrograde trafficking to the Golgi in HepG2 cells. Blood. 2009;113:1786–1793. doi: 10.1182/blood-2008-08-174565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger M, Conrad S, Alvarez-Bolado G, Skutella T, Just L. Gene expression of the repulsive guidance molecules during development of the mouse intestine. Dev Dyn. 2005;234:169–175. doi: 10.1002/dvdy.20506. [DOI] [PubMed] [Google Scholar]
- Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet. 2009;41:478–481. doi: 10.1038/ng.320. [DOI] [PubMed] [Google Scholar]
- Meynard D, Vaja V, Sun CC, Corradini E, Chen S, Lopez-Otin C, Grgurevic L, Hong CC, Stirnberg M, Gutschow M, Vukicevic S, Babitt JL, Lin HY. Regulation of TMPRSS6 by BMP6 and iron in human cells and mice. Blood. 2011;118:747–756. doi: 10.1182/blood-2011-04-348698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirakaj V, Brown S, Laucher S, Steinl C, Klein G, Kohler D, Skutella T, Meisel C, Brommer B, Rosenberger P, Schwab JM. Repulsive guidance molecule-A (RGM-A) inhibits leukocyte migration and mitigates inflammation. Proc Natl Acad Sci U S A. 2011;108:6555–6560. doi: 10.1073/pnas.1015605108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnier PP, Sierra A, Macchi P, Deitinghoff L, Andersen JS, Mann M, Flad M, Hornberger MR, Stahl B, Bonhoeffer F, Mueller BK. RGM is a repulsive guidance molecule for retinal axons. Nature. 2002;419:392–395. doi: 10.1038/nature01041. [DOI] [PubMed] [Google Scholar]
- Moore SW, Tessier-Lavigne M, Kennedy TE. Netrins and their receptors. Adv Exp Med Biol. 2007;621:17–31. doi: 10.1007/978-0-387-76715-4_2. [DOI] [PubMed] [Google Scholar]
- Muramatsu R, Kubo T, Mori M, Nakamura Y, Fujita Y, Akutsu T, Okuno T, Taniguchi J, Kumanogoh A, Yoshida M, Mochizuki H, Kuwabara S, Yamashita T. RGMa modulates T cell responses and is involved in autoimmune encephalomyelitis. Nat Med. 2011;17:488–494. doi: 10.1038/nm.2321. [DOI] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, ten Dijke P. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- Niederkofler V, Salie R, Arber S. Hemojuvelin is essential for dietary iron sensing, and its mutation leads to severe iron overload. J Clin Invest. 2005;115:2180–2186. doi: 10.1172/JCI25683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederkofler V, Salie R, Sigrist M, Arber S. Repulsive guidance molecule (RGM) gene function is required for neural tube closure but not retinal topography in the mouse visual system. J Neurosci. 2004;24:808–818. doi: 10.1523/JNEUROSCI.4610-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nili M, Shinde U, Rotwein P. Soluble repulsive guidance molecule c/hemojuvelin is a broad spectrum bone morphogenetic protein (BMP) antagonist and inhibits both BMP2- and BMP6-mediated signaling and gene expression. J Biol Chem. 2010;285:24783–24792. doi: 10.1074/jbc.M110.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohra R, Beyeen AD, Guo JP, Khademi M, Sundqvist E, Hedreul MT, Sellebjerg F, Smestad C, Oturai AB, Harbo HF, Wallstrom E, Hillert J, Alfredsson L, Kockum I, Jagodic M, Lorentzen J, Olsson T. RGMA and IL21R show association with experimental inflammation and multiple sclerosis. Genes Immun. 2010;11:279–293. doi: 10.1038/gene.2009.111. [DOI] [PubMed] [Google Scholar]
- O'Connor MB, Umulis D, Othmer HG, Blair SS. Shaping BMP morphogen gradients in the Drosophila embryo and pupal wing. Development. 2006;133:183–193. doi: 10.1242/dev.02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y, Kohmura E, Yamashita T. TACE cleaves neogenin to desensitize cortical neurons to the repulsive guidance molecule. Neurosci Res. 2011;71:63–70. doi: 10.1016/j.neures.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Oldekamp J, Kramer N, Alvarez-Bolado G, Skutella T. Expression pattern of the repulsive guidance molecules RGM A, B and C during mouse development. Gene Expr Patterns. 2004;4:283–288. doi: 10.1016/j.modgep.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Onichtchouk D, Chen YG, Dosch R, Gawantka V, Delius H, Massague J, Niehrs C. Silencing of TGF-beta signalling by the pseudoreceptor BAMBI. Nature. 1999;401:480–485. doi: 10.1038/46794. [DOI] [PubMed] [Google Scholar]
- Pagani A, Silvestri L, Nai A, Camaschella C. Hemojuvelin N-terminal mutants reach the plasma membrane but do not activate the hepcidin response. Haematologica. 2008;93:1466–1472. doi: 10.3324/haematol.12508. [DOI] [PubMed] [Google Scholar]
- Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, Nemeth E, Thompson J, Risler JK, Zaborowska C, Babakaiff R, Radomski CC, Pape TD, Davidas O, Christakis J, Brissot P, Lockitch G, Ganz T, Hayden MR, Goldberg YP. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. doi: 10.1038/ng1274. [DOI] [PubMed] [Google Scholar]
- Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806–7810. doi: 10.1074/jbc.M008922200. [DOI] [PubMed] [Google Scholar]
- Park KW, Crouse D, Lee M, Karnik SK, Sorensen LK, Murphy KJ, Kuo CJ, Li DY. The axonal attractant Netrin-1 is an angiogenic factor. Proc Natl Acad Sci U S A. 2004;101:16210–16215. doi: 10.1073/pnas.0405984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson GI, Padgett RW. TGF beta-related pathways. Roles in Caenorhabditis elegans development. Trends Genet. 2000;16:27–33. doi: 10.1016/s0168-9525(99)01916-2. [DOI] [PubMed] [Google Scholar]
- Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem. 2001;276:7811–7819. doi: 10.1074/jbc.M008923200. [DOI] [PubMed] [Google Scholar]
- Qin S, Yu L, Gao Y, Zhou R, Zhang C. Characterization of the receptors for axon guidance factor netrin-4 and identification of the binding domains. Mol Cell Neurosci. 2007;34:243–250. doi: 10.1016/j.mcn.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Quinn CC, Wadsworth WG. Axon guidance: asymmetric signaling orients polarized outgrowth. Trends Cell Biol. 2008;18:597–603. doi: 10.1016/j.tcb.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan S, Deitinghoff L, Davis D, Conrad S, Skutella T, Chedotal A, Mueller BK, Strittmatter SM. Neogenin mediates the action of repulsive guidance molecule. Nat Cell Biol. 2004;6:756–762. doi: 10.1038/ncb1156. [DOI] [PubMed] [Google Scholar]
- Rajasekharan S, Kennedy TE. The netrin protein family. Genome Biol. 2009;10:239. doi: 10.1186/gb-2009-10-9-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramel MC, Hill CS. Spatial regulation of BMP activity. FEBS Lett. 2012;586:1929–1941. doi: 10.1016/j.febslet.2012.02.035. [DOI] [PubMed] [Google Scholar]
- Ramsay AJ, Hooper JD, Folgueras AR, Velasco G, Lopez-Otin C. Matriptase-2 (TMPRSS6): a proteolytic regulator of iron homeostasis. Haematologica. 2009;94:840–849. doi: 10.3324/haematol.2008.001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- Ren XR, Hong Y, Feng Z, Yang HM, Mei L, Xiong WC. Tyrosine phosphorylation of netrin receptors in netrin-1 signaling. Neurosignals. 2008;16:235–245. doi: 10.1159/000111566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XR, Ming GL, Xie Y, Hong Y, Sun DM, Zhao ZQ, Feng Z, Wang Q, Shim S, Chen ZF, Song HJ, Mei L, Xiong WC. Focal adhesion kinase in netrin-1 signaling. Nat Neurosci. 2004;7:1204–1212. doi: 10.1038/nn1330. [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Pan P, Parkkila S. Expression studies of neogenin and its ligand hemojuvelin in mouse tissues. J Histochem Cytochem. 2007;55:85–96. doi: 10.1369/jhc.6A7031.2006. [DOI] [PubMed] [Google Scholar]
- Roelen BA, Cohen OS, Raychowdhury MK, Chadee DN, Zhang Y, Kyriakis JM, Alessandrini AA, Lin HY. Phosphorylation of threonine 276 in Smad4 is involved in transforming growth factor-beta-induced nuclear accumulation. Am J Physiol Cell Physiol. 2003;285:C823–30. doi: 10.1152/ajpcell.00053.2003. [DOI] [PubMed] [Google Scholar]
- Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, Loukopoulos D, Camaschella C. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21–22. doi: 10.1038/ng1053. [DOI] [PubMed] [Google Scholar]
- Round J, Stein E. Netrin signaling leading to directed growth cone steering. Curr Opin Neurobiol. 2007;17:15–21. doi: 10.1016/j.conb.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Samad TA, Rebbapragada A, Bell E, Zhang Y, Sidis Y, Jeong SJ, Campagna JA, Perusini S, Fabrizio DA, Schneyer AL, Lin HY, Brivanlou AH, Attisano L, Woolf CJ. DRAGON, a bone morphogenetic protein co-receptor. J Biol Chem. 2005;280:14122–14129. doi: 10.1074/jbc.M410034200. [DOI] [PubMed] [Google Scholar]
- Samad TA, Srinivasan A, Karchewski LA, Jeong SJ, Campagna JA, Ji RR, Fabrizio DA, Zhang Y, Lin HY, Bell E, Woolf CJ. DRAGON: a member of the repulsive guidance molecule-related family of neuronal- and muscle-expressed membrane proteins is regulated by DRG11 and has neuronal adhesive properties. J Neurosci. 2004;24:2027–2036. doi: 10.1523/JNEUROSCI.4115-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath TK, Reddi AH. Dissociative extraction and reconstitution of extracellular matrix components involved in local bone differentiation. Proc Natl Acad Sci U S A. 1981;78:7599–7603. doi: 10.1073/pnas.78.12.7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage-Dunn C. TGF-beta signaling. WormBook. 2005:1–12. doi: 10.1895/wormbook.1.22.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage-Dunn C, Maduzia LL, Zimmerman CM, Roberts AF, Cohen S, Tokarz R, Padgett RW. Genetic screen for small body size mutants in C. elegans reveals many TGFbeta pathway components. Genesis. 2003;35:239–247. doi: 10.1002/gene.10184. [DOI] [PubMed] [Google Scholar]
- Scherner O, Meurer SK, Tihaa L, Gressner AM, Weiskirchen R. Endoglin differentially modulates antagonistic transforming growth factor-beta1 and BMP-7 signaling. J Biol Chem. 2007;282:13934–13943. doi: 10.1074/jbc.M611062200. [DOI] [PubMed] [Google Scholar]
- Schmidtmer J, Engelkamp D. Isolation and expression pattern of three mouse homologues of chick Rgm. Gene Expr Patterns. 2004;4:105–110. doi: 10.1016/s1567-133x(03)00144-3. [DOI] [PubMed] [Google Scholar]
- Serpe M, Umulis D, Ralston A, Chen J, Olson DJ, Avanesov A, Othmer H, O'Connor MB, Blair SS. The BMP-binding protein Crossveinless 2 is a short-range, concentration-dependent, biphasic modulator of BMP signaling in Drosophila. Dev Cell. 2008;14:940–953. doi: 10.1016/j.devcel.2008.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severyn CJ, Shinde U, Rotwein P. Molecular biology, genetics and biochemistry of the repulsive guidance molecule family. Biochem J. 2009;422:393–403. doi: 10.1042/BJ20090978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- Silvestri L, Pagani A, Camaschella C. Furin-mediated release of soluble hemojuvelin: a new link between hypoxia and iron homeostasis. Blood. 2008;111:924–931. doi: 10.1182/blood-2007-07-100677. [DOI] [PubMed] [Google Scholar]
- Silvestri L, Pagani A, Fazi C, Gerardi G, Levi S, Arosio P, Camaschella C. Defective targeting of hemojuvelin to plasma membrane is a common pathogenetic mechanism in juvenile hemochromatosis. Blood. 2007;109:4503–4510. doi: 10.1182/blood-2006-08-041004. [DOI] [PubMed] [Google Scholar]
- Silvestri L, Pagani A, Nai A, De Domenico I, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab. 2008;8:502–511. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K, Strickland P, Valdes A, Shin GC, Hinck L. Netrin-1/neogenin interaction stabilizes multipotent progenitor cap cells during mammary gland morphogenesis. Dev Cell. 2003;4:371–382. doi: 10.1016/s1534-5807(03)00054-6. [DOI] [PubMed] [Google Scholar]
- Steinbicker AU, Bartnikas TB, Lohmeyer LK, Leyton P, Mayeur C, Kao SM, Pappas AE, Peterson RT, Bloch DB, Yu PB, Fleming MD, Bloch KD. Perturbation of hepcidin expression by BMP type I receptor deletion induces iron overload in mice. Blood. 2011;118:4224–4230. doi: 10.1182/blood-2011-03-339952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Yandell MD, Roy PJ, Krishna S, Savage-Dunn C, Ross RM, Padgett RW, Wood WB. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development. 1999;126:241–250. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- Takeo S, Akiyama T, Firkus C, Aigaki T, Nakato H. Expression of a secreted form of Dally, a Drosophila glypican, induces overgrowth phenotype by affecting action range of Hedgehog. Dev Biol. 2005;284:204–218. doi: 10.1016/j.ydbio.2005.05.014. [DOI] [PubMed] [Google Scholar]
- Taneja-Bageshwar S, Gumienny TL. Two functional domains in C. elegans glypican LON-2 can independently inhibit BMP-like signaling. Dev Biol. 2012;371:66–76. doi: 10.1016/j.ydbio.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassew NG, Charish J, Seidah NG, Monnier PP. SKI-1 and Furin generate multiple RGMa fragments that regulate axonal growth. Dev Cell. 2012;22:391–402. doi: 10.1016/j.devcel.2011.11.022. [DOI] [PubMed] [Google Scholar]
- Tassew NG, Chestopolava L, Beecroft R, Matsunaga E, Teng H, Chedotal A, Monnier PP. Intraretinal RGMa is involved in retino-tectal mapping. Mol Cell Neurosci. 2008;37:761–769. doi: 10.1016/j.mcn.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753–766. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Sen D, Shi H, Foehr ML, Plavskin Y, Vatamaniuk OK, Liu J. The RGM protein DRAG-1 positively regulates a BMP-like signaling pathway in Caenorhabditis elegans. Development. 2010;137:2375–2384. doi: 10.1242/dev.051615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truksa J, Gelbart T, Peng H, Beutler E, Beutler B, Lee P. Suppression of the hepcidin-encoding gene Hamp permits iron overload in mice lacking both hemojuvelin and matriptase-2/TMPRSS6. Br J Haematol. 2009;147:571–581. doi: 10.1111/j.1365-2141.2009.07873.x. [DOI] [PubMed] [Google Scholar]
- Truksa J, Peng H, Lee P, Beutler E. Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1 expression independently of Hfe, transferrin receptor 2 (Tfr2), and IL-6. Proc Natl Acad Sci U S A. 2006;103:10289–10293. doi: 10.1073/pnas.0603124103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umulis D, O'Connor MB, Blair SS. The extracellular regulation of bone morphogenetic protein signaling. Development. 2009;136:3715–3728. doi: 10.1242/dev.031534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urist MR. Bone: formation by autoinduction. Science. 1965;150:893–899. doi: 10.1126/science.150.3698.893. [DOI] [PubMed] [Google Scholar]
- van den Heuvel DM, Hellemons AJ, Pasterkamp RJ. Spatiotemporal Expression of Repulsive Guidance Molecules (RGMs) and Their Receptor Neogenin in the Mouse Brain. PLoS One. 2013;8:e55828. doi: 10.1371/journal.pone.0055828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco G, Cal S, Quesada V, Sanchez LM, Lopez-Otin C. Matriptase-2, a membrane-bound mosaic serine proteinase predominantly expressed in human liver and showing degrading activity against extracellular matrix proteins. J Biol Chem. 2002;277:37637–37646. doi: 10.1074/jbc.M203007200. [DOI] [PubMed] [Google Scholar]
- Vielmetter J, Kayyem JF, Roman JM, Dreyer WJ. Neogenin, an avian cell surface protein expressed during terminal neuronal differentiation, is closely related to the human tumor suppressor molecule deleted in colorectal cancer. J Cell Biol. 1994;127:2009–2020. doi: 10.1083/jcb.127.6.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Copeland NG, Gilbert DJ, Jenkins NA, Tessier-Lavigne M. Netrin-3, a mouse homolog of human NTN2L, is highly expressed in sensory ganglia and shows differential binding to netrin receptors. J Neurosci. 1999;19:4938–4947. doi: 10.1523/JNEUROSCI.19-12-04938.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L, Deng CX. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. doi: 10.1016/j.cmet.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Wang X, Harris RE, Bayston LJ, Ashe HL. Type IV collagens regulate BMP signalling in Drosophila. Nature. 2008;455:72–77. doi: 10.1038/nature07214. [DOI] [PubMed] [Google Scholar]
- Wilson NH, Key B. Neogenin: one receptor, many functions. Int J Biochem Cell Biol. 2007;39:874–878. doi: 10.1016/j.biocel.2006.10.023. [DOI] [PubMed] [Google Scholar]
- Wilson NH, Key B. Neogenin interacts with RGMa and netrin-1 to guide axons within the embryonic vertebrate forebrain. Dev Biol. 2006;296:485–498. doi: 10.1016/j.ydbio.2006.06.018. [DOI] [PubMed] [Google Scholar]
- Wozney JM. The bone morphogenetic protein family and osteogenesis. Mol Reprod Dev. 1992;32:160–167. doi: 10.1002/mrd.1080320212. [DOI] [PubMed] [Google Scholar]
- Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA. Novel regulators of bone formation: molecular clones and activities. Science. 1988;242:1528–1534. doi: 10.1126/science.3201241. [DOI] [PubMed] [Google Scholar]
- Wu MY, Hill CS. Tgf-beta superfamily signaling in embryonic development and homeostasis. Dev Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Wu Q, Sun CC, Lin HY, Babitt JL. Repulsive guidance molecule (RGM) family proteins exhibit differential binding kinetics for bone morphogenetic proteins (BMPs) PLoS One. 2012;7:e46307. doi: 10.1371/journal.pone.0046307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Babitt JL, Sidis Y, Chung RT, Lin HY. Hemojuvelin regulates hepcidin expression via a selective subset of BMP ligands and receptors independently of neogenin. Blood. 2008;111:5195–5204. doi: 10.1182/blood-2007-09-111567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Cortez-Retamozo V, Niederkofler V, Salie R, Chen S, Samad TA, Hong CC, Arber S, Vyas JM, Weissleder R, Pittet MJ, Lin HY. Dragon (repulsive guidance molecule b) inhibits IL-6 expression in macrophages. J Immunol. 2011;186:1369–1376. doi: 10.4049/jimmunol.1002047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Yu PB, Sidis Y, Beppu H, Bloch KD, Schneyer AL, Lin HY. Repulsive guidance molecule RGMa alters utilization of bone morphogenetic protein (BMP) type II receptors by BMP2 and BMP4. J Biol Chem. 2007;282:18129–18140. doi: 10.1074/jbc.M701679200. [DOI] [PubMed] [Google Scholar]
- Xie Y, Ding YQ, Hong Y, Feng Z, Navarre S, Xi CX, Zhu XJ, Wang CL, Ackerman SL, Kozlowski D, Mei L, Xiong WC. Phosphatidylinositol transfer protein-alpha in netrin-1-induced PLC signalling and neurite outgrowth. Nat Cell Biol. 2005;7:1124–1132. doi: 10.1038/ncb1321. [DOI] [PubMed] [Google Scholar]
- Xu P, Liu J, Derynck R. Post-translational regulation of TGF-beta receptor and Smad signaling. FEBS Lett. 2012;586:1871–1884. doi: 10.1016/j.febslet.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Mueller BK, Hata K. Neogenin and repulsive guidance molecule signaling in the central nervous system. Curr Opin Neurobiol. 2007;17:29–34. doi: 10.1016/j.conb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- Yang F, West AP, Jr, Allendorph GP, Choe S, Bjorkman PJ. Neogenin interacts with hemojuvelin through its two membrane-proximal fibronectin type III domains. Biochemistry. 2008;47:4237–4245. doi: 10.1021/bi800036h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, West AP, Jr, Bjorkman PJ. Crystal structure of a hemojuvelin-binding fragment of neogenin at 1.8A. J Struct Biol. 2011;174:239–244. doi: 10.1016/j.jsb.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakin L, De Robertis EM. Extracellular regulation of BMP signaling. Curr Biol. 2010;20:R89–92. doi: 10.1016/j.cub.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebboudj AF, Imura M, Bostrom K. Matrix GLA protein, a regulatory protein for bone morphogenetic protein-2. J Biol Chem. 2002;277:4388–4394. doi: 10.1074/jbc.M109683200. [DOI] [PubMed] [Google Scholar]
- Zhang AS, Anderson SA, Meyers KR, Hernandez C, Eisenstein RS, Enns CA. Evidence that inhibition of hemojuvelin shedding in response to iron is mediated through neogenin. J Biol Chem. 2007;282:12547–12556. doi: 10.1074/jbc.M608788200. [DOI] [PubMed] [Google Scholar]
- Zhang AS, Gao J, Koeberl DD, Enns CA. The role of hepatocyte hemojuvelin in the regulation of bone morphogenic protein-6 and hepcidin expression in vivo. J Biol Chem. 2010;285:16416–16423. doi: 10.1074/jbc.M110.109488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AS, West AP, Jr, Wyman AE, Bjorkman PJ, Enns CA. Interaction of hemojuvelin with neogenin results in iron accumulation in human embryonic kidney 293 cells. J Biol Chem. 2005;280:33885–33894. doi: 10.1074/jbc.M506207200. [DOI] [PubMed] [Google Scholar]
- Zhang AS, Yang F, Meyer K, Hernandez C, Chapman-Arvedson T, Bjorkman PJ, Enns CA. Neogenin-mediated hemojuvelin shedding occurs after hemojuvelin traffics to the plasma membrane. J Biol Chem. 2008;283:17494–17502. doi: 10.1074/jbc.M710527200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang AS, Yang F, Wang J, Tsukamoto H, Enns CA. Hemojuvelin-neogenin interaction is required for bone morphogenic protein-4-induced hepcidin expression. J Biol Chem. 2009;284:22580–22589. doi: 10.1074/jbc.M109.027318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JL, Qiu LY, Kotzsch A, Weidauer S, Patterson L, Hammerschmidt M, Sebald W, Mueller TD. Crystal structure analysis reveals how the Chordin family member crossveinless 2 blocks BMP-2 receptor binding. Dev Cell. 2008;14:739–750. doi: 10.1016/j.devcel.2008.02.017. [DOI] [PubMed] [Google Scholar]
- Zhao ZW, Lian WJ, Chen GQ, Zhou HY, Wang GM, Cao X, Yang HJ, Hou YP. Decreased expression of repulsive guidance molecule member A by DNA methylation in colorectal cancer is related to tumor progression. Oncol Rep. 2012;27:1653–1659. doi: 10.3892/or.2012.1693. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Xie J, Lee D, Liu Y, Jung J, Zhou L, Xiong S, Mei L, Xiong WC. Neogenin regulation of BMP-induced canonical Smad signaling and endochondral bone formation. Dev Cell. 2010;19:90–102. doi: 10.1016/j.devcel.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu XJ, Wang CZ, Dai PG, Xie Y, Song NN, Liu Y, Du QS, Mei L, Ding YQ, Xiong WC. Myosin X regulates netrin receptors and functions in axonal path-finding. Nat Cell Biol. 2007;9:184–192. doi: 10.1038/ncb1535. [DOI] [PubMed] [Google Scholar]