Abstract
Little is known regarding how the synthesis and degradation of individual proteins changes during the life of an organism. Such knowledge is vital to understanding the aging process. To fill this knowledge gap, we monitored newly synthesized proteins on a proteome scale in Caenorhabditis elegans over time during adulthood using a SILAC-based label-chase approach. For most proteins, the rate of appearance of newly synthesized protein was high during the first 5 days of adulthood, slowed down between the fifth and the 11th days, and then increased again after the 11th day. However, the magnitude of appearance rate differed significantly from protein to protein. For example, the appearance of newly synthesized protein was fast for proteins involved in embryonic development, transcription regulation, and lipid binding/transport, with >70% of these proteins newly synthesized by day 5 of adulthood, whereas it was slow for proteins involved in cellular assembly and motility, such as actin and myosin, with <70% of these proteins newly synthesized even on day 16. The late-life increase of newly synthesized protein was especially high for ribosomal proteins and ATP synthases. We also investigated the effect of RNAi-mediated knockdown of the rpl-9 (ribosomal protein), atp-3 (ATP synthase), and ril-1 (RNAi-induced longevity-1 ) genes and found that inhibiting the expression of atp-3 and ril-1 beginning in late adulthood is still effective to extend the life span of C. elegans.
Keywords: protein turnover, Caenorhabditis elegans, aging, SILAC, life span, ATP synthase, ribosomal protein
INTRODUCTION
The regulation of proteostasis (i.e., the synthesis, folding, trafficking, and degradation of proteins) plays an essential role in maintaining the health of cells and the entire organism. As organisms age, the balance between protein synthesis and degradation, commonly referred to as protein turnover, gradually becomes disrupted.1-3 An association between protein turnover and aging has been postulated based on the observations that reducing protein synthesis or promoting protein degradation significantly extends the life span of Caenorhabditis elegans.4 Regulated and selective synthesis and degradation of proteins control many cellular processes, such as activation/inhibition of signaling pathways.5, 6 Thus, monitoring the synthesis or degradation of individual proteins on a global scale over the life span of an organism will likely yield novel insights into the underlying physiologic processes that control aging.
Historically, studies of protein turnover in living organisms have involved measuring the incorporation of an isotopically labeled amino acid(s) or radioactive element(s) such as 35S into a protein(s) of interest and then estimating the synthesis rate by monitoring the labeled protein(s).7 Because modern mass spectrometry–based proteomics technologies enable the analysis of individual proteins in complex mixtures, we now have an unprecedented opportunity to monitor appearance of newly synthesized proteins on a proteomic scale. Three different strategies have been employed for this purpose.8 The first strategy involves the uniform labeling of nitrogen or carbon atoms in proteins with either 15N or 13C, respectively.9 The second strategy involves the labeling of proteins with deuterium (2H) or heavy oxygen (18O) by giving living organisms heavy water (2H2O or H 182O).10 The third strategy involves the incorporation of pre-labeled amino acid(s) into proteins.11-16 This latter strategy is often referred to as “stable-isotope labeling by amino acids in cell culture” (SILAC)17 because it was originally developed for in vitro use. From an analytical standpoint, SILAC is superior to other two strategies because the difference in mass between labeled and unlabeled peptide species can be predicted for all peptides; thus, the analysis of mass spectrometry data for protein identification and quantification is straightforward when using SILAC.
We chose C. elegans as a model organism because it has a short life span (2–3 weeks), its genome has been completely sequenced, and the SILAC technique to monitor newly synthesized proteins in this organism was available in our laboratory.18 In this study, newly synthesized proteins were monitored throughout the life of adult worms using a SILAC-based label-chase approach. For most proteins, the rate of appearance of newly synthesized protein was high during the first 5 days of adulthood, slowed down between the fifth and 11th days, and then increased again after the 11th day. This late-life increase of newly synthesized protein was particularly high for ribosomal proteins and ATP synthases. Our RNAi knockdown experiment showed that inhibiting the expression of the atp-3 and ril-1 genes beginning late in adulthood (day 9) extends the life span of C. elegans. To the best of our knowledge, this is the first proteome-scale study of this kind in C. elegans.
MATERIALS AND METHODS
Materials
12C6 - and 13C6-Lys were purchased from Sigma-Aldrich (St. Louis, MO) and Cambridge Isotope Laboratories (Tewksbury, MA), respectively. The endoproteinase Lys-C was purchased from Wako USA (Richmond, VA). All other chemicals were either reagent grade or were of the highest quality that was commercially available.
C. elegans Strain, Maintenance, and Age Synchronization
Wild-type (WT) Bristol N2 strain C. elegans was used in this study. For the incorporation of 12C6- and 13C6-Lys, nematodes were maintained according to standard methods that included culture on peptone-free NGM plates (51 mM NaCl, 25 mm K3PO4, 5 μg/mL cholesterol, 1 mM CaCl2, 1 mM MgSO4) seeded with Escherichia coli strain AT713. The composition of the media and solutions and detailed protocols for their use were described previously.19 To synchronize their age, gravid nematodes were bleached according to a published protocol,19 and the surviving eggs were hatched as age-synchronized nematodes. In all experiments, the pre-fertile period of adulthood was noted as t = 0, with day 1 as the first day of adulthood.
Labeling Bacteria with Light (12C6) and Heavy (13C6) Lysine
Arginine- and lysine-auxotrophic E. coli strain AT713 was obtained from the E. coli Genetic Stock Center at Yale University. Bacteria were first streaked on a lysogeny broth agar plate and cultured overnight at 37°C. A single bacterial colony was then inoculated into 10 mL of lysogeny broth and cultured overnight in an incubator shaker (37°C, 180 rpm). Next, 100 μL of bacterial culture was inoculated into 50 mL of M9 minimal medium (50 mM Na2HPO4, 20 mM KH2PO4, 10 mM NaCl, 20 mM NH4Cl, 2 mM MgSO4, 0.1 mM CaCl2, and 0.2% glucose) supplemented with arginine (100 μg/mL), cysteine (100 μg/mL), and lysine (100 μg/mL, either 12C6- or 13C6-labeled) and continuously cultured in an incubator shaker (37°C, 200 rpm) until the absorbance of the culture at 600 nm (A600) reached 1.0. Next, 10 mL of the labeled bacterial culture was inoculated into 1,000 mL of M9 basal medium supplemented with amino acids and cultured in an incubator shaker (37°C, 200 rpm) until the A600 reached 2.0. The bacteria were then pelleted by a brief centrifugation (8,000 × g, 10 min) and resuspended in 15 mL of sterile water. Bacteria were then spread onto a peptone-free NGM plate (500 μL of culture for each 100-mm plate and 200 μL of culture for each 60-mm plate) and exposed to ultraviolet light at 1,000 mJ/cm2 using a SpectroLinker XL-1500 UV crosslinker (Spectronics Corp., Westbury, NY). The plates containing bacteria in which the proteins were labeled with light or heavy Lys were stored at 4°C until use.
Life Span Assay
Lifespan analysis was conducted at 20°C as described previously.20 Nematodes were grown at 20°C on peptone-free NGM plates containing 12C6 - or 13C6-Lys-labeled E. coli AT713 for at least two generations before the experiments were initiated. Eggs of the 12C6 - and 13C6-Lys-labeled nematodes were prepared as described above and placed on the plates seeded with either 12C6- or 13C6-Lys-labeled bacteria, respectively. Once the eggs were hatched, the L4 worms were moved to new plates and then moved to additional new plates every 2 days during the reproductive phase and then every 7 days for the remainder of the life span analysis. Nematode viability was scored every 2 days. Survival was scored based on touch-provoked movement and pumping of the pharynx. All survival plots refer to life span beginning at adulthood. Nematodes that crawled off the plate, burrowed into the agar, or died from internally hatching progeny were excluded from the analysis. OASIS software was used for statistical analysis of the data.21 In all cases, the log-rank test was used to test the hypothesis that the survival functions among groups were equal.
Label-chase Experiment
Several WT Bristol N2 nematodes were transferred onto a peptone-free NGM plate previously seeded with heavy-Lys-labeled E. coli. Gravid nematodes from the next generation were bleached to collect their live eggs according to a previously described protocol.19 The eggs were transferred onto peptone-free NGM plates seeded with heavy-Lys-labeled E. coli. Bleaching, age synchronization, and plating onto NGM plates seeded with labeled bacteria were then repeated. After hatching, age-synchronized nematodes were cultured to L4 larval stage (day 0) and then transferred to peptone-free NGM plates seeded with heavy-Lys-labeled E. coli and containing 25 mg/L 5-fluoro-2’ -deoxyuridine (Acros Organics, Pittsburgh, PA). On the following day (day 1), the nematodes were transferred to plates seeded with light-Lys-labeled E. coli and containing 25 mg/L 5-fluoro-2’-deoxyuridine. Worms were harvested from the plates at different time points until the 16th day of adulthood. Worms were transferred to fresh plates containing light-Lys-labeled E. coli after days 5, 8, 11, and 15. Note that the worms were harvested using a protocol for separating dead and live worms as described below.
Separation of Dead and Live Worms
Live worms were separated from dead worms using sucrose density centrifugation. Worms were collected from the 10-cm diameter plate with 40 mL of distilled water and placed in a 50-mL Falcon tube and centrifuged at 2,000 × g for 2 min. The collected worms were carefully overlaid onto chilled 30% sucrose and centrifuged at 2,000 × g for 5 min. The upper layer of worms (i.e., those that were alive) were immediately collected in a 15-mL Falcon tube, washed several times with purified water, centrifuged at 2,000 × g for 2 min, then weighed and stored at −80°C until used.
Preparation of Samples for Proteomic Analyses
Worms were suspended in 250 μL of 100 mM ammonium bicarbonate buffer containing 4% perfluorooctanoic acid (w/v),22 protease inhibitor mixture (Sigma-Aldrich), and phosphatase inhibitor mixture 3 (Sigma-Aldrich). Proteins were then extracted by ultrasonication (4.5 kHz 3 times for 9 s with a 3-min pause on ice between the pulses) using a Virsonic 100 ultrasonic cell disrupter (SP Scientific, Warminster, PA), as described previously.18 The resulting protein extract was centrifuged at 15,000 × g for 10 min and the supernatant was collected. The proteins within the supernatant were reduced by addition of 10 mM dithiothreitol (DTT) and incubation at 37°C for 30 min and then S-alkylated by addition of 25 mM iodoacetamide and incubation at 25°C for 45 min. A 9-fold excess volume of ice-cold acetone was then added and the sample was allowed to stand for 2 h at −20°C to precipitate the proteins. The precipitated proteins were then centrifuged at 2,400 × g for 10 min at 4°C, and the pellet was washed with ice-cold 90% acetone. The pellet was air-dried for 5 min and then redissolved in 50 μL of 100 mM ammonium bicarbonate, containing 8 M urea by sonication for 1 min in a Bransonic Ultrasonic 2510R-MT water bath (Bransonic, Danbury, CT). The resulting solution was diluted with 450 μL of 100 mM ammonium bicarbonate to reduce the urea concentration to 0.8 M, and the amount of dissolved protein was determined with a DC protein assay kit (Bio-Rad, Hercules, CA). A total of 25 μg of protein was digested with Lys-C (1:25 Lys-C to protein ratio [w/w]) at 37°C for 18 h. The peptide mixture was desalted using a Vydac C18 UltraMicro Tip Column (The Nest Group, Southborough, MA), resuspended in 0.1% formic acid, and analyzed using LC-MS/MS.
LC-MS/MS Analysis
LC-MS/MS analyses were conducted using an UltiMate 3000 LC system (Dionex Inc., Sunnyvale, California) interfaced with a Velos Pro Ion Trap/Orbitrap Elite hybrid mass spectrometer (Thermo Scientific, Bremen, Germany). The platform was operated in the nano-LC mode, using the standard nano-ESI source. The spray voltage was set to 1.2 kV, and the temperature of the heated capillary was set to 200°C. The rate of solvent flow through the column was maintained at 300 nL/min. Lys-C peptide digests (typically 2 μL) were injected onto a reversed-phase C18 PepMap trapping column (0.3 × 5 mm with a 5-μm particle size; Dionex Inc.) equilibrated with 0.1% formic acid/1% acetonitrile (v/v). The column was washed for 5 min with the equilibration solution at a flow rate of 25 μL/min using an isocratic loading pump operated through an autosampler. The trapping column was then switched in-line with a reversed-phase C18 Acclaim PepMap 100 column (0.075 × 150 mm, Dionex Inc.), and peptides were eluted using a linear gradient of 2 to 37% acetonitrile in aqueous 0.1% formic acid over 200 min at a flow rate of 300 nL/min. The eluent was directly introduced into the mass spectrometer. The mass spectrometer was operated in data-dependent MS to MS/MS switching mode, with the 25 most intense ions in each MS scan subjected to MS/MS. Full MS scanning was performed at a resolution of 120,000 in the Orbitrap detector, and MS/MS was performed in the ion trap detector in collision induced dissociation mode. The threshold intensity for the MS/MS trigger was always set at 3,000. Fragmentation was carried out in the collision induced dissociation mode with a normalized collision energy of 35. Data were collected entirely in the profile mode for the full MS scan and the centroid mode for MS/MS scans. The dynamic exclusion function for previously selected precursor ions applied the following parameters: repeat count of 1, repeat duration of 40 s, exclusion duration of 90 s, and exclusion size list of 500. Xcalibur software (version 2.2, SP1 build 48, Thermo-Finnigan Inc., San Jose, CA) was used for instrument control, data acquisition, and data processing. No technical replicate was used for the LCMS/MS of Lys-C digests.
Identification and Quantification of Light- and Heavy-Lys-Labeled Peptides and Proteins
Proteins were identified by comparing all of the experimental peptide MS/MS spectra against the Wormpep database containing 25391 sequences (November 2012) using Mascot database search software (Version 2.2.0, Matrix Science, London, UK). Carbamidomethylation of cysteine was set as a fixed modification, whereas variable modifications included oxidation of methionine to methionine sulfoxide, acetylation of N-terminal amino groups, and replacement of C-terminal Lys with heavy Lys. The mass tolerance was set at 10 ppm for precursor ions and 0.8 Da for product ions. Strict Lys-C specificity was applied, and missed cleavages were not allowed. BuildSummary23 was used to generate a confident protein list with a peptide false discovery rate ≤ 0.01. Other criteria for significant peptide identifications included the following: peptides must be composed of at least six amino acid residues and have a minimum Mascot score of 20, which corresponds to Mascot absolute probability of 0.01.
SILAC Quantification Suite in ProteomicsTools software,18, 24 version 3.4.3, was used to determine the abundances of light- and heavy-labeled proteins, from which the fraction of light protein (light protein/[light protein + heavy protein]) was calculated. The SILAC Quantification Suite extracts the precursor ion of a peptide to be quantified from a raw LC-MS/MS file based on the information of a MS/MS scan that identified the peptide. If the peptide was identified by multiple spectra, the scans between identified MS/MS spectra were included in quantification scan window. Otherwise, five scans around the identified MS/MS spectrum were included in quantification scan window. The standard deviations (SD) of both light and heavy precursor m/z were calculated based on those scans. Then, the quantification scan window was extended at both ends until the m/z offset of either light or heavy precursor becomes larger than 3-fold of their corresponding SD. Once the quantification scan window is determined, the light/heavy precursor intensities were extracted and the ratio was calculated by non-negative least squares. Proteins identified with multiple peptides were also quantified by non-negative least squares using the light/heavy precursor intensities of all peptides identified from the same protein. R square values of non-negative least square at both peptide and protein level were used to indicate the quantification quality. All the quantified proteins were manually validated with user-friendly graphic interface. Peptides with R square greater than 0.8 were accepted as quantified peptides.
Analysis of Protein Expression
WT Bristol N2 nematodes were cultured on a peptone-free NGM plate seeded with light-Lys-labeled E. coli AT713. Gravid animals were bleached to collect their live eggs according to the protocol described above, and the eggs were grown on peptone-free NGM plates seeded with light-Lys-labeled E. coli. After hatching, age-synchronized animals were grown on light-Lys-labeled bacteria and harvested on adult age days 1, 5, 8, 11, and 16. For reference, 9-day-old heavy-Lys-labeled worms were used in the quantitative analysis of protein expression. For all samples, proteins were extracted from the same wet weight of worms (30 mg), and 50 μg of protein from each sample was digested with Lys-C. A fixed amount of Lys-C digest from the reference sample was spiked into all five experimental Lys-C digests prior to LC-MS/MS analysis. Protein identification and quantification were carried out as described above.
RNA Interference (RNAi) and Life Span Analysis
The identity of all RNAi clones was verified by sequencing the inserts using the M13-forward primer. All clones were obtained from Julie Ahringer's RNAi library. HT115 bacteria transformed with RNAi vectors expressing dsRNAs of genes of interest were grown at 37°C in lysogeny broth with 10 [.proportional]g/mL tetracycline and 50 [.proportional]g/mL carbenicillin and then seeded onto NG-carbenicillin plates supplemented with 100 μL of 0.1 M IPTG. For life span analysis on peptone-free NGM plates, RNAi bacteria were concentrated 10-fold before seeding.
Life span analysis was conducted at 20°C as described previously, with minor modifications.18, 25 Briefly, worms were grown at 20°C for at least two generations before the experiments were initiated. Between 60 and 90 worms were tested in each experiment. RNAi was carried out by adding synchronized eggs (unless otherwise noted) to peptone-free plates seeded with specific RNAi bacteria. Worms were moved to a fresh RNAi plate every 2 days until reproduction ceased. Worms were then moved to a new plate every 5–7 days for the remainder of the life span analysis. Viability was scored every 2 days. In all life span analyses, the pre-fertile period of adulthood was set as t = 0. Stata 12 software was used for statistical analysis to determine means and percentiles. In all cases, P values were calculated using the log-rank (Mantel-Cox) method.
RESULTS
Life Span of C. elegans Fed Light and Heavy Lys
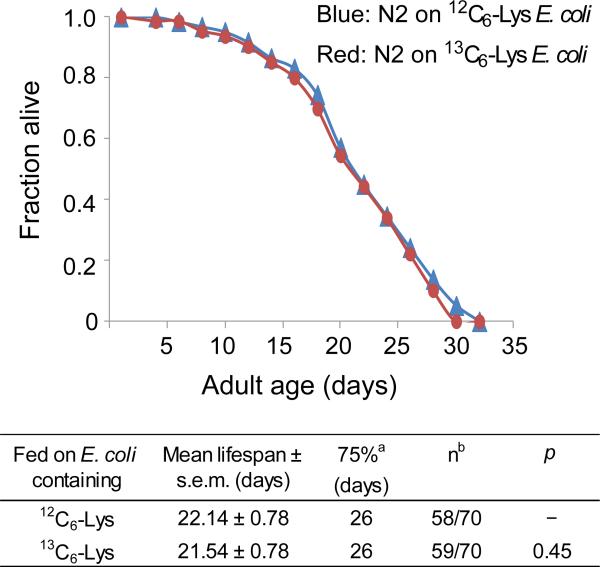
To test whether incorporation of heavy Lys affects the life span of C. elegans, WT N2 Bristol nematodes were fed light Lys (12C6-Lys)- or heavy Lys (13C6-Lys)-labeled E. coli AT713 as their only food source. As shown in Figure 1, the survival curves for nematodes fed light and heavy Lys were essentially indistinguishable, demonstrating that the isotope labels per se did not differentially affect the life span of C. elegans. This result justified the use of isotope-labeled worms for monitoring newly synthesized proteins.
Figure 1.
Effect of heavy and light Lys on the life span of WT N2 C. elegans. Survival curves are shown for WT worms fed E. coli AT713 containing light Lys (blue) or heavy Lys (red). All worms were grown at 20°C on NGM plates without peptone. Statistical details are shown in the table at bottom. aAge at which the fraction of dead worms reached 0.75. bNumber of observed deaths relative to the total starting number of worms. The difference represents the number of worms excluded from analysis during the experiment.
Age-dependent Appearance of Newly Synthesized Proteins in C. elegans
The SILAC label-chase experiment, which had one biological replicate at each time point, was carried out twice to monitor age-dependent appearance of newly synthesized proteins in C. elegans. The experimental workflow is shown in Figure 2. Tables S1 and S2 list all of the peptides that were identified and quantified in the first and second label-chase experiments, respectively. A total of 58979 peptides were identified in the study. Most of these peptides provided quantitative information, except about 2.5% of the peptides (1461 peptides), which were C-terminal peptides of proteins and did not contain a lysine residue, therefore no quantitative information was obtained from those 1461 peptides. The study resulted in quantifying a total of 1,135 proteins in the first experiment (Table S3) and 1,275 proteins in the second experiment (Table S4). We chose 318 proteins from the first experiment (Table S5) and 383 proteins from the second experiment (Table S6), of which 243 were common to both experiments, for subsequent data analyses. The criterion was that proteins must have at least six of the seven time points (day 1, 5, 7, 9, 11, 13, 16).
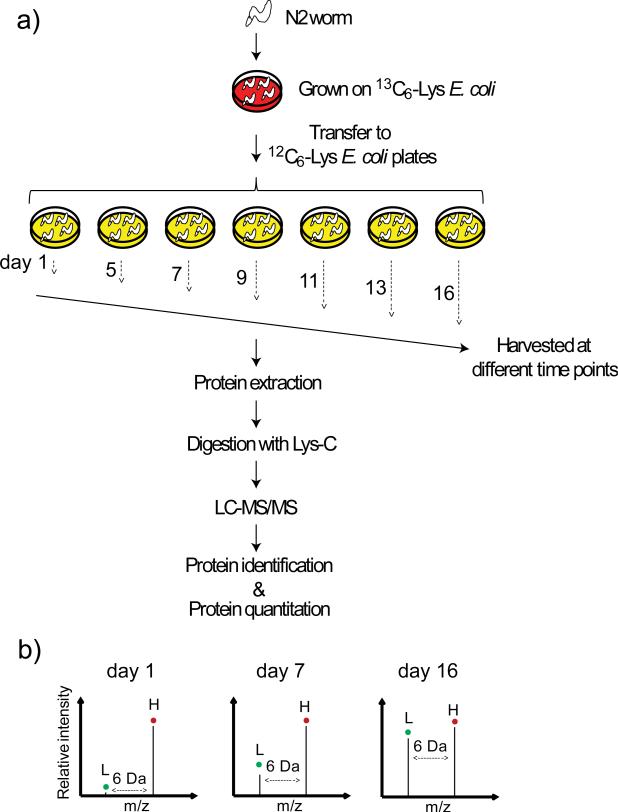
Figure 2.
Strategy for monitoring the appearance of newly synthesized proteins in a proteome scale. a) A single nematode was propagated on heavy Lys (13C6-Lys)-labeled E. coli for two generations, and then age-synchronized adult worms were transferred to light Lys (12C6-Lys)- labeled E. coli plates on day 1 and harvested at various time points. Proteins extracted from these samples were digested with Lys-C and analyzed by LC-MS/MS. The resulting data were then analyzed to identify and quantify the proteins present. b) Hypothetical mass spectra of a peptide from worms at different ages. The 12C6-Lys labeled ‘light’ peak (L) increases relative to the 13C6-Lys labeled ‘heavy’ (H) peak as the worm ages, thus indicating increase of the newly synthesized protein from which the peptide originated.
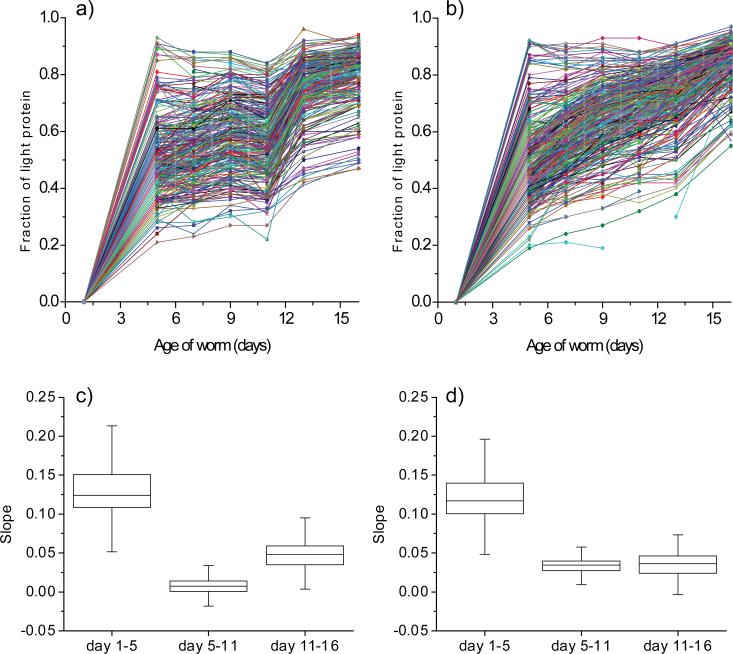
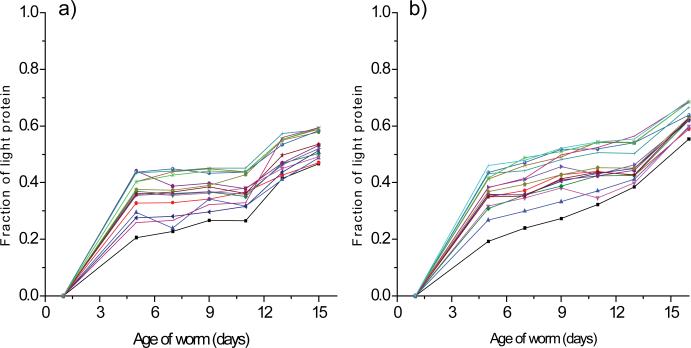
The fraction of light protein (newly synthesized protein) for the 318 and 383 proteins quantified in the first and second experiments is plotted as a function of worm age in Figure 3a and b. Although the age-dependent appearance of newly synthesized protein for the first and second experiments were not identical, they appeared to show a common trend, in which the appearance rate of newly synthesized protein was robust in the first 5 days, slowed between the fifth and 11th days, and then increased again after the 11th day. In order to analyze the data statistically, we calculated the slopes for the changes of the fraction of newly synthesized protein between day 1-5, day 5-11 and day 11-16 for all the proteins observed and performed box plot analysis on them (Figure 3c and d). The analysis clearly showed that the slopes between day 1-5 are statistically higher than those between day 5-11 in both the experiments, indicating that the appearance rates of newly synthesized protein for most proteins were robust in the first 5 days, and then slow down between day 5-11. The analysis also showed that the slopes between day 11-16 from the first experiment are statistically higher than those between day 5-11, while no statistical significance was observed on the data from the second experiment. Even though we did not observed statistically significant difference between the slopes on the data from the second experiment, we found that for majority of proteins (215 proteins out of 279 in the first experiment and 161 proteins out of 276 in the second experiment) the slopes between day 11-16 were higher than those between day 5-11. Thus, our data show that the appearance rates of newly synthesized protein increased for majority of proteins after the 11th day.
Figure 3.
Age-dependent appearance of newly synthesized proteins. The fraction of light protein (newly synthesized protein) is plotted as a function of worm age for 318 proteins from the first experiment (a) and 383 proteins from the second experiment (b). Data for at least six of the seven time points were available for all these proteins. Box plots that display the variability of the slopes between day 1-5, day 5-11 and day 11-16 calculated for all the proteins observed in the first (c) and second experiment (d) are shown. The horizontal bar in the box structures represents the median value, the bottom line of the box represents the first quartile and the upper line of the box represents the third quartile. The 95% confidence intervals for the median value on the plots in (c) were as follows: day 1-5 (0.110 – 0.138, n=279), day 5-11 (0.006 – 0.009, n=279) and day 11-16 (0.043 – 0.053, n=318), and those on the plots in (d) were as follows: day 1-5 (0.105 – 0.129, n=347), day 5-11 (0.031 – 0.038, n=340) and day 11-16 (0.032 – 0.040, n=312).
It should be noted that, in principle, the fraction of newly synthesized protein should only increase as worms age. However, there are discrepant data points whose values were lower than the earlier data points. This discrepancy could be related to variation of culturing condition and instrument performance.
Proteins for Which Appearance of Newly Synthesized Protein is Rapid or Slow
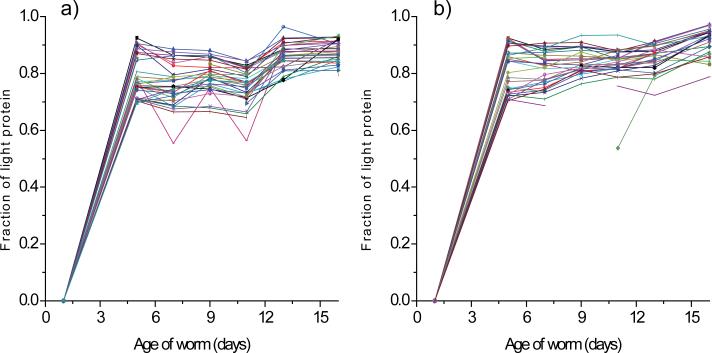
Although most proteins exhibited a common trend in terms of the age-dependent appearance of newly synthesized protein, the magnitude of appearance rate differed significantly from protein to protein. The complements of 35 proteins in the first experiment and 32 proteins in the second experiment were already comprised of >70% light protein (newly synthesized protein) by day 5, and 22 of these proteins were common to both experiments, indicating a rapid synthesis of these proteins in young age (Figure 4a and b). These proteins were found to be involved in embryonic development, transcription regulation, and lipid binding/transport (Table 1). It is worth to note that we identified a total of 14 lipid binding/transport proteins in the study, 9 of which are included in Table 1. The table also includes a number of proteins of unidentified function. We also investigated how the overall levels of the proteins listed in Table 1 changed between days 1 and 5 in a separate experiment using the quantitative mass spectrometry method described in the “Materials and Methods” section. More than 1,000 proteins were quantified in this experiment, including 40 of the 45 proteins listed in Table 1. The expression of 34 of the 40 quantified proteins increased between days 1 and 5, as indicated by the day 5 to day 1 ratios (Table 1). The full results of the protein expression analysis will be reported elsewhere.
Figure 4.
Profiles of proteins for which appearance of newly synthesized protein is rapid during the first five days of adulthood. The fraction of light protein (newly synthesized protein) is plotted as a function of worm age for 35 proteins from the first experiment (a) and 32 proteins from the second experiment (b). These proteins were >70% comprised of newly synthesized protein by day 5. Data for at least six of the seven time points were available for all these proteins.
Table 1.
Proteins for Which Appearance of Newly Synthesized Protein is Rapid
| Wormbase Protein ID |
Gene Name | Protein Name | Function | Fraction of Light-Protein at Day 5 |
Protein Expression (Day 5 / Day 1) |
|
|---|---|---|---|---|---|---|
| First Experiment |
Second Experiment |
|||||
| CE29924 | acp-6 | Unknown | Acid phosphatase activity | ND | 0.80 | 0.68 |
| CE23090 | brp-1 | Unknown | Unknown | ND | 0.85 | ND |
| CE21413 | car-1 | Cytokinesis, Apoptosis, RNA-associated-1 | Unknown | 0.87 | 0.87 | 0.76 |
| CE10598 | cey-2 | C. Elegans Y-box-2 | Regulation of DNA-dependent transcription | 0.91 | 0.90 | 0.62 |
| CE12296 | cey-3 | C. Elegans Y-box-3 | Regulation of DNA-dependent transcription | 0.87 | ND | 0.65 |
| CE00839 | cgh-1 | ATP-dependent RNA helicase | RNA helicase activity | ND | 0.84 | 0.68 |
| CE16333 | cpl-1 | Cathepsin-like protease-1 | Cysteine type peptidase activity | 0.76 | ND | 1.77 |
| CE07251 | cpr-4 | Cathepsin B-like cysteine proteinase-4 | Cysteine-type peptidase activity | 0.76 | ND | 2.57 |
| CE03244 | F21D5.1 | Unknown | Phosphoacetylglucosamine mutase activity | 0.78 | ND | 1.00 |
| CE00134 | far-2 | Fatty Acid/Retinol binding protein-2 | Lipid binding | 0.71 | 0.71 | 1.04 |
| CE21232 | far-6 | Fatty Acid/Retinol binding protein-6 | Lipid binding | 0.75 | 0.74 | 1.09 |
| CE02343 | gpd-1 | Glyceraldehyde 3-phosphate dehydrogenase- 1 | Oxidoreductase activity | 0.78 | 0.73 | 1.06 |
| CE17789 | gst-24 | Glutathione s-transferase | Protein binding | ND | 0.87 | 0.99 |
| CE00503 | iff-1 | Initiation Factor Five (eIF-5A) homolog-1 | Translation elongation factor activity | 0.75 | 0.79 | 1.00 |
| CE04533 | lbp-1 | Fatty acid-binding protein-1 | Lipid binding | 0.70 | ND | 4.33 |
| CE04532 | lbp-2 | Fatty acid-binding protein -2 | Lipid binding | 0.70 | 0.73 | 4.20 |
| CE06531 | mdt-28 | MeDiaTor-28 | Embryonic development | 0.76 | ND | 1.00 |
| CE18083 | mix-1 | Mitotic and X-chromosome associated protein -1 | ATP binding | ND | 0.73 | ND |
| CE08871 | nasp-2 | Histone binding protein-2 | Embryonic development and locomotion | 0.90 | 0.86 | 1.57 |
| CE27133 | nmy-2 | Myosin heavy chain-2 | ATP binding | 0.71 | ND | 1.36 |
| CE27850 | perm-2 | PERMeable eggshell-2 | Embryonic development and reproduction | 0.90 | 0.91 | 3.28 |
| CE00994 | sip-1 | Heat shock hsp20 protein | Unknown | 0.85 | 0.86 | 1.76 |
| CE00475 | ttr-2 | Transthyretin-like family-2 | Unknown | 0.74 | 0.72 | 3.15 |
| CE01921 | ubq-1 | UBiQuitin-1 | Protein binding | 0.76 | 0.74 | 1.34 |
| CE06950 | vit-2 | VITellogenin structural genes-2 | Lipid transporter activity | 0.90 | 0.91 | 3.02 |
| CE20900 | vit-3 | VITellogenin structural genes-3 | Lipid transporter activity | ND | 0.92 | 1.70 |
| CE26817 | vit-4 | VITellogenin structural genes-4 | Lipid transporter activity | ND | 0.92 | 1.75 |
| CE03921 | vit-5 | VITellogenin structural genes-5 | Lipid transporter activity | 0.91 | 0.92 | 1.82 |
| CE18026 | vit-6 | VITellogenin structural genes-6 | Lipid transporter activity | 0.93 | 0.92 | 2.65 |
| CE15560 | B0513.4 | Unknown | Unknown | ND | 0.90 | 3.23 |
| CE02454 | C06A8.3 | Unknown | Unknown | 0.71 | 0.72 | 2.05 |
| CE29195 | C28H8.3 | Unknown | helicase activity | 0.71 | ND | 1.05 |
| CE47099 | C39D10.7 | Unknown | chitin binding | 0.89 | 0.91 | 3.19 |
| CE04189 | C42D4.1 | Unknown | Unknown | 0.76 | 0.74 | 4.22 |
| CE05528 | D1054.10 | Unknown | Unknown | ND | 0.84 | ND |
| CE01220 | E04F6.8 | Unknown | Unknown | 0.77 | 0.77 | 5.62 |
| CE40453 | F13G11.3 | Unknown | Unknown | 0.85 | ND | 2.05 |
| CE27971 | F17E9.4 | Unknown | Unknown | ND | 0.85 | 2.00 |
| CE26681 | H34I24.2 | Unknown | Unknown | 0.81 | ND | 2.31 |
| CE18851 | K06G5.1 | Unknown | Unknown | 0.75 | 0.75 | 2.55 |
| CE33832 | T12D8.5 | Unknown | Unknown | 0.70 | 0.70 | ND |
| CE16638 | Y45F10C.4 | Unknown | Unknown | 0.79 | ND | 0.91 |
| CE29936 | Y92H12BR.3 | Unknown | Unknown | 0.76 | ND | ND |
| CE15209 | ZC410.5 | Unknown | Unknown | 0.70 | 0.71 | 2.20 |
| CE06583 | ZC434.8 | Unknown | kinase activity | 0.71 | ND | 1.14 |
ND: Not determined
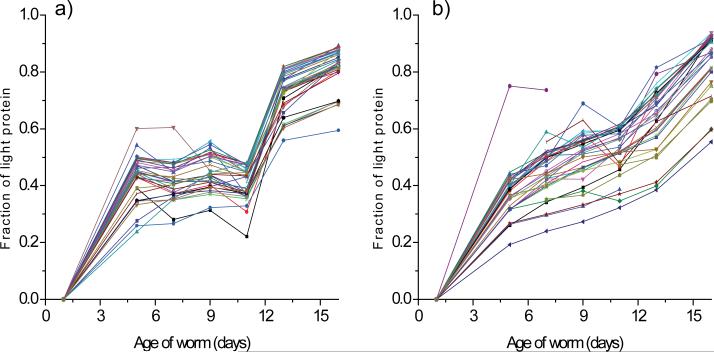
In addition, we identified 15 proteins in the first and second experiments for which the replacement of original protein molecules was very slow (Figure 5a and b). These proteins were comprised of <70% newly synthesized protein even at day 16, indicating that >30% of the original molecules are not replaced during adulthood in C. elegans. These proteins are listed in Table 2, and 12 of 18 proteins were common to both experiments. With the exceptions of ASB-2, MIG-6, and T25F10.6, most of these proteins are involved in cellular assembly and motility and include actin and myosin.
Figure 5.
Profiles of proteins for which appearance of newly synthesized protein is slow. The fraction of light protein (newly synthesized protein) is plotted as a function of worm age for 15 proteins from the first experiment (a) and 15 proteins from the second experiment (b). These proteins were >30% comprised of original molecules even at day 16. Data for at least six of the seven time points were available for all these proteins.
Table 2.
Proteins for Which Appearance of Newly Synthesized Protein is Slow
| Wormbase Protein ID | Gene Name | Protein Name | Function | Fraction of Light-Protein at Day 16 |
|
|---|---|---|---|---|---|
| First Experiment | Second Experiment | ||||
| CE12358 | act-4 | Actin -4 | Structural constituent of muscle | 0.59 | 0.68 |
| CE16463 | act-5 | Actin-5 | Structural constituent of muscle | 0.59 | 0.68 |
| CE07016 | asb-2 | ATP synthase B chain -2 | Mitochondrial ATP synthesis | 0.59 | ND |
| CE04038 | dim-1 | Disorganised muscle protein-1 | Structural constituent of muscle | ND | 0.62 |
| CE02622 | ifb-2 | Intermediate filament protein- 2 | Cytoskeleton structural component | 0.47 | 0.55 |
| CE28782 | lev-11 | LEVamisole resistant -11 | Actin binding structural protein | 0.58 | ND |
| CE17535 | mig-6 | Protease inhibitor -6 | Abnormal cell migration | ND | 0.69 |
| CE20542 | mlc-2 | Myosin light chain -2 | Regulator of myosin ATPase activity | 0.50 | 0.62 |
| CE01236 | mlc-3 | Myosin light chain-3 | Normal development, locomotion, egg laying | 0.52 | 0.62 |
| CE06253 | myo-1 | Myosin heavy chain -1 | Structural constituent of muscle | 0.49 | 0.6 |
| CE31619 | myo-2 | Myosin heavy chain C -2 | ATP binding and motor activity | 0.51 | 0.6 |
| CE34936 | myo-3 | Myosin heavy chain-3 | Structural constituent of muscle | 0.47 | 0.63 |
| CE34313 | tnt-2 | TropoNin T -2 | Locomotion | 0.59 | 0.64 |
| CE09197 | unc-15 | Paramyosin-15 | Cytoskeletal protein binding | 0.49 | 0.59 |
| CE40008 | unc-27 | Troponin I | Locomotion and Muscle constituent | ND | 0.69 |
| CE44668 | unc-22 | UNCoordinated -22 | Regulation of muscle contraction | 0.58 | ND |
| CE09349 | unc-54 | Myosin heavy chain -54 | Structural constituent of muscle | 0.54 | 0.63 |
| CE07537 | T25F10.6 | Calponin-like protein | Unknown | 0.53 | 0.62 |
ND: Not determined
Proteins for Which Appearance of Newly Synthesized Protein are Markedly rapid in Aged Nematodes
In the first label-chase experiment, we identified a group of 36 proteins for which the fraction of newly synthesized protein increased rapidly (>1.8-fold) between days 11 and 16 (Figure 6a), suggesting an increase in protein turnover after day 11. This group of proteins included 5 ATP synthases, 25 ribosomal proteins, 2 proteins that facilitate protein folding (HSP-60 and PDI-2), and 4 proteins with other or unknown functions (DCT-16, GDH-1, H28O16.1, and Y69A2AR.18) (Table 3). We also examined the top 36 proteins for which the fraction of newly synthesized protein increased markedly between days 11 and 16 in the second label-chase experiment (Figure 6b). Consistent with the first experiment, this group included a number of ATP synthases and ribosomal proteins (Table 3). Remarkably, all 6 ATP synthases and 29 of the 36 ribosomal proteins identified in the two experiments were included in Table 3. We also monitored changes in the expression of these proteins between days 11 and 16. The analysis revealed that the overall levels of these proteins decreased after day 11, as indicated by the ratio of protein expression on day 16 to that on day 11 (Table 3).
Figure 6.
Profiles for proteins for which appearance of newly synthesized protein were markedly rapid after day 11 in the first (a) and second (b) label-chase experiments. The fraction of light protein (newly synthesized protein) is plotted as a function of worm age for 36 proteins from the first (a) and the second experiment (b). Data for at least six of the seven time points were available for all these proteins.
Table 3.
Proteins for which the fraction of light protein increased rapidly between days 11 and 16
| Wormbase Protein ID |
Gene Name | Protein Name | Function | Fraction of Light-Protein (Day 16 / Day 11) |
Protein Expression (Day 16 / Day 11) |
#Pathways | |
|---|---|---|---|---|---|---|---|
| First Experiment |
Second Experiment |
||||||
| CE29047 | alh-6 | Aldehyde dehydrogenase-6 | Oxidoreductase activity | ND | 1.48 | ND | a,b |
| CE02183 | alh-8 | Methylmalonate-semialdehyde dehydrogenase-8 | Oxidoreductase activity | ND | 1.95 | 0.65 | c,d,e,f,g,h |
| CE00968 | asb-1 | ATP synthase B chain | ATP synthesis in Mitochondria | 1.92 | ND | 0.68 | g,i |
| CE07016 | asb-2 | ATP synthase B chain -2 | ATP synthesis in Mitochondria | 1.81 | 1.94 | 0.65 | g,i |
| CE29950 | atp-2 | ATP synthase beta chain-2 | ATP synthesis in Mitochondria | 1.93 | 1.57 | 0.65 | g,i |
| CE09719 | atp-3 | ATPase -3 | ATP synthesis in Mitochondria | 1.97 | ND | 0.48 | g,i |
| CE13291 | atp-4 | ATP synthase subunit -4 | ATP synthesis in Mitochondria | ND | 1.50 | 0.66 | Unknown |
| CE15602 | atp-5 | ATP synthase D chain-5 | ATP synthesis in Mitochondria | 1.87 | 1.56 | 0.69 | g,i |
| CE19095 | dct-16 | DAF-16/FOXO Controlled, germline Tumor affecting-16 | Unknown | 1.94 | 2.02 | 0.92 | Unknown |
| CE33098 | F46H5.3 | Unknown | Unknown | ND | 1.55 | 0.57 | b |
| CE06652 | gdh-1 | Glutamate dehydrogenase-1 | Oxidatoreductase activity | 1.83 | 1.51 | 0.68 | a,b,g,j,k |
| CE06092 | gta-1 | 4-aminobutyrate aminotransferase-1 | Enzyme catalysis | ND | 1.55 | 0.67 | a,b,c,d,f,g,m |
| CE18826 | H28O16.1 | Unknown | Unknown | 1.81 | 1.57 | ND | g,i |
| CE27244 | hsp-60 | Heat Shock Protein-60 | Mitochondrion specific chaperonin | 1.83 | ND | 0.61 | n |
| CE31507 | ifa-1 | Intermediate filament protein-1 | Cytoskeleton structural component | ND | 1.60 | 0.61 | Unknown |
| CE02622 | ifb-2 | Intermediate filament protein-2 | Cytoskeleton structural component | ND | 1.72 | ND | Unknown |
| CE37390 | K06G5.1 | Unknown | Unknown | ND | 1.62 | ND | Unknown |
| CE14426 | lbp-6 | Fatty acid-binding protein-6 | Lipid transport | ND | 1.65 | 0.68 | Unknown |
| CE06253 | myo-1 | Myosin heavy chain-1 | Structural constituent of muscle | ND | 1.73 | 0.54 | Unknown |
| CE31619 | myo-2 | Myosin heavy chain C-2 | ATP binding and motor activity | ND | 1.61 | 0.56 | Unknown |
| CE03972 | pdi-2 | Protein disulfide isomerase-2 | Protein folding in ER | 1.81 | ND | 0.66 | o |
| CE08950 | ril-1 | RNAi-Induced Longevity -1 | Unknown | ND | 1.56 | ND | Unknown |
| CE09655 | rla-0 | Ribosomal protein, Large subunit, Acidic (P1) | Protein biosynthesis | 1.89 | ND | 0.62 | p |
| CE25552 | rpl-1 | Ribosomal Protein L1 | Protein biosynthesis | ND | 1.51 | 0.56 | p |
| CE07033 | rpl-11.2 | Ribosomal protein L11.2 | Protein biosynthesis | 2.29 | 1.55 | 0.52 | p |
| CE17986 | rpl-12 | Ribosomal protein L12 | Protein biosynthesis | 2.09 | 1.53 | 0.52 | p |
| CE08526 | rpl-13 | Ribosomal protein L13 | Protein biosynthesis | 2.14 | ND | 0.61 | p |
| CE12148 | rpl-15 | Ribosomal protein L15 | Protein biosynthesis | ND | 1.52 | 0.49 | p |
| CE22195 | rpl-17 | Ribosomal Protein, L17 | Protein biosynthesis | 2.26 | 1.56 | 0.46 | p |
| CE04102 | rpl-22 | Ribosomal protein L22 | Protein biosynthesis | 2.28 | 1.53 | 0.60 | p |
| CE05721 | rpl-25.2 | Ribosomal protein L25.2 | Protein biosynthesis | 2.42 | 1.52 | 0.64 | p |
| CE06313 | rpl-28 | Ribsomal protein L28 | Protein biosynthesis | 2.04 | ND | 0.65 | p |
| CE03709 | rpl-32 | Ribosomal protein L32 | Protein biosynthesis | 2.23 | 1.55 | 0.59 | p |
| CE30781 | rpl-36 | Ribosomal protein L36 | Protein biosynthesis | 3.74 | 1.77 | 0.68 | p |
| CE02255 | rpl-5 | Ribosomal protein L5 | Protein biosynthesis | 2.15 | 1.48 | 0.75 | p |
| CE00744 | rpl-6 | Ribosomal protein L6 | Protein biosynthesis | ND | 1.55 | 0.58 | p |
| CE11024 | rpl-7 | Ribosomal protein L7 | Protein biosynthesis | ND | 1.51 | 0.53 | p |
| CE27398 | rpl-7A | Ribosomal Protein L7A | Protein biosynthesis | 2.14 | 1.49 | 0.53 | p |
| CE01380 | rpl-9 | Ribosomal protein L9 | Protein biosynthesis | 2.62 | ND | 0.61 | p |
| CE00854 | rps-0 | Ribosomal protein S0 | Protein biosynthesis | 1.85 | ND | 0.57 | p |
| CE05860 | rps-11 | Ribosomal protein S11 | Protein biosynthesis | 1.95 | ND | 0.59 | p |
| CE04009 | rps-13 | Ribosomal protein S13 | Protein biosynthesis | 1.85 | ND | 0.60 | p |
| CE12918 | rps-16 | Ribosomal protein S16 | Protein biosynthesis | 1.96 | ND | 0.69 | p |
| CE26948 | rps-17 | Ribosomal protein S17 | Protein biosynthesis | 1.84 | ND | 0.63 | p |
| CE14956 | rps-18 | Ribosomal protein S18 | Protein biosynthesis | 1.91 | ND | 0.65 | p |
| CE04237 | rps-2 | Ribosomal Protein S2 | Protein biosynthesis | 2.01 | ND | 0.58 | p |
| CE40119 | rps-24 | Ribosomal Protein S24 | Protein biosynthesis | 1.84 | ND | 0.64 | p |
| CE04691 | rps-25 | Ribosomal protein S25 | Protein biosynthesis | 1.85 | ND | 0.65 | p |
| CE24278 | rps-4 | Ribosomal Protein S4 | Protein biosynthesis | 1.94 | ND | 0.67 | p |
| CE24592 | rps-6 | Ribosomal protein S6 | Protein biosynthesis | 1.90 | ND | 0.67 | p,q |
| CE06577 | rps-7 | Ribosomal protein S7 | Protein biosynthesis | 1.85 | ND | 0.53 | p |
| CE27514 | Y69A2AR.18 | Unknown | Unknown | 1.85 | ND | 0.65 | g,i |
| CE07373 | spc-1 | Spectrin alpha chain-1 | Calcium binding and muscle formation | ND | 1.49 | 0.59 | Unknown |
| CE03351 | suca-1 | Succinate-CoA ligase-1 | ATP binding | ND | 1.59 | 0.57 | f,g,h,r |
| CE36726 | ttn-1 | TiTiN family -1 | ATP binding | ND | 1.48 | ND | Unknown |
| CE47057 | unc-22 | UNCoordinated -22 | Regulation of muscle contraction | ND | 1.49 | ND | Unknown |
| CE27514 | Y69A2AR.18 | Unknown | Unknown | ND | 1.57 | 0.65 | g,i |
ND: Not determined
Obtained from KEGG database (www.genome.jp/kegg). a: alanine, aspartate, and glutamate metabolism; b: arginine and proline metabolism; c: valine, leucine, and isoleucine degradation; d: beta-alanine metabolism; e: inositol phosphate metabolism; f: propanoate metabolism; g: metabolic pathways; h: carbon metabolism; i: oxidative phosphorylation; j: d-glutamine and d-glutamate metabolism; k: nitrogen metabolism; l: propanoate metabolism; m: butanoate metabolism; n: RNA degradation; o: protein processing in endoplasmic reticulum; p: ribosome; q: mTOR signaling pathway; r: citrate cycle.
Effect of RNAi-mediated Knockdown of the rpl-9, atp-3 , and ril-1 Genes on the Life Span of C. elegans
We also examined the effect on the life span of C. elegans by inhibiting the synthesis of those proteins for which appearance of newly synthesized protein were rapid in aged worms. The effect of RNAi-mediated knockdown of selected genes was examined using life span analysis. One gene encoding a ribosomal protein (rpl-9) and another encoding an ATP synthase (atp-3) were selected, as these genes represented the largest and second largest groups of proteins, respectively, in Table 3. We also examined RNAi-mediated knockdown of ril-1, which encodes a longevity-related protein26 identified in the second label-chase experiment (Table 3) as showing a significant increase in expression after day 11.
RNAi was initiated either at hatching (lifelong) or at day 9 of adulthood (late-life). The results are summarized in Table 4. Consistent with a previous report,26 lifelong RNAi knockdown of atp-3 and ril-1 extended the life span of C. elegans by 42–87%, whereas the life span decreased more than 30% with lifelong RNAi-mediated knockdown of rpl-9. Initiation of RNAi knockdown of atp-3 or ril-3 at day 9 of adulthood also extended the life span of C. elegans (10– 13% and 7–9% extension following knockdown of atp-3 and ril-3, respectively). Considering that knockdown was not initiated until day 9 of the adulhood, the true effect of inhibiting the expression of these genes is even larger (15-23% for atp-3 and 10-16% for ril-1, respectively). Late-life RNAi knockdown of rpl-9 resulted in a small (2–7%) and statistically insignificant life span extension.
Table 4.
Effect of RNAi knockdown of atp-3, ril-1, rpl-9 and dnj-13 on the lifespan of N2 C. elegans
| RNAi | Mean lifespan (days) | 75% (days)a | nb | Extentionc | Extention after D9d | Pe |
|---|---|---|---|---|---|---|
| Trial #1 | ||||||
| Whole-life RNAi knockdown | ||||||
| vector control | 23.97 ± 0.43 | 26 | 67/73 | - | - | - |
| atp-3 | 34.07 ± 0.96 | 38 | 57/82 | 42% | - | <0.0001 |
| ril-1 | 35.02 ± 0.87 | 40 | 58/60 | 46% | - | <0.0001 |
| rpl-9 | 13.98 ± 0.55 | 17 | 58/73 | −42% | - | <0.0001 |
| Late-life RNAi knockdown | ||||||
| vector control | 23.97 ± 0.43 | 26 | 67/73 | - | - | - |
| atp-3 | 26.26 ± 0.61 | 32 | 78/94 | 10% | 15% | <0.0001 |
| ril-1 | 25.53 ± 0.62 | 28 | 78/90 | 7% | 10% | 0.0087 |
| rpl-9 | 24.51 ± 0.56 | 29 | 75/78 | 2% | 4% | 0.0765 |
| Trial #2 | ||||||
| Whole-life RNAi knockdown | ||||||
| vector control | 21.54 ± 0.61 | 24 | 75/88 | - | - | - |
| atp-3 | 40.23 ± 1.47 | 49 | 80/87 | 87% | - | <0.0001 |
| ril-1 | 33.86 ± 0.97 | 40 | 76/76 | 57% | - | <0.0001 |
| rpl-9 | 14.42 ± 0.51 | 17 | 84/90 | −33% | - | <0.0001 |
| Late-life RNAi knockdown | ||||||
| vector control | 21.54 ± 0.61 | 24 | 75/88 | - | - | - |
| atp-3 | 24.39 ± 0.85 | 26 | 73/85 | 13% | 23% | 0.0002 |
| ril-1 | 23.56 ± 0.61 | 28 | 78/90 | 9% | 16% | 0.0028 |
| rpl-9 | 22.99 ± 0.81 | 28 | 79/89 | 7% | 12% | 0.1368 |
The age at which the fraction of animals dead reaches 0.75.
Number of observed deaths relative to total number of animals started. The difference between these numbers represents the number of animals censored during the experiment.
The percentage of total lifespan extension compared to the empty vector control.
The percentage of extension of the lifespan after D9 of adulhood compared to the empty vector control.
Calculated by pair-wise comparisons to the empty vector control, each consisting of control and experimental animals examined at the same time. We used Stata 12 software for statistical analysis and to determine means and percentiles. The logrank (Mantel-Cox) test was used to test the hypothesis that the survival functions among groups were equal.
DISCUSSION
In order to increase our understanding of how the turnover of individual proteins changes with aging, we monitored the appearance of newly synthesized proteins over the adult life span of C. elegans using a SILAC-based label-chase approach. We found that for most C. elegans proteins, the newly synthesized protein increases significantly during the first 5 days of adulthood, slows down between the fifth and 11th days, and then increases again after the 11th day (Figure 3). The observed rapid increase of newly synthesized protein during the first 5 days of adulthood suggests that vigorous protein synthesis occurs during this period. We further found that the expression levels of many proteins concomitantly increased during the same period (Table 1), thus suggesting that the balance between synthesis and degradation is shifted toward synthesis during this period. This finding is consistent with the physiologic state of C. elegans during this same period. During the first 5 days of adulthood, C. elegans grow in terms of both length and volume, which normally plateaus at day 6.27 Moreover, the observed vigorous increase of newly synthesized protein temporally overlaps with the reproductive period (i.e., during which production of yolk proteins occurs). Therefore, it is not surprising that both newly synthesized protein and overall protein levels increased significantly in first 5 days of adulthood.
The observation that increase of newly synthesized protein slowed between the fifth and the 11th days suggests that the metabolic activity of the nematodes declines significantly after day 5. It should be noted that there is a possibility that labeled proteins could have been recycled into newly synthesized proteins. We did not determine the extent to which such recycling of labeled proteins may have occurred in our study. Therefore, the fractions of light protein reported may be underestimates. Also, 5-fluoro-2’-deoxyuridine was used in our experiments to avoid progeny interference. Although it was necessary to use this drug in our experiments, reports indicate that it can alter some aspects of the physiology of C. elegans.28, 29
Our study revealed that newly synthesized protein significantly increases for many C. elegans proteins after the 11th day of adulthood (Table S3). The increase of newly synthesized protein was especially high for ATP synthases, ribosomal proteins, and chaperone proteins (Figure 6 and Table 3); while intriguingly, their expression levels were rather decreased more than 30% during this period (Table 3). A straightforward explanation for this observation would be that the balance between protein synthesis and degradation in aged worms is shifted toward degradation. This explanation, however, assumes that aged worms maintain a healthy protein degradation system. It has been shown that both protein synthesis and degradation decline with aging in a variety of animals, including C. elegans.4, 30, 31 Consistent with these studies, recent studies have demonstrated that the collapse of proteostasis is a major molecular event during aging,32, 33 and this collapse leads to an increase in protein aggregation in C. elegans.34, 35 If we postulate that substantial protein aggregation occurred in the aged worms in our study, the observed increase of newly synthesized protein in aged animals could have been due primarily to removal of old proteins from the system through aggregation. It is worth noting that there is significant overlap between the proteins listed in Table 3 and those previously reported as being aggregation prone.34, 35 Therefore, it appears to be possible that the aged worms are still actively synthesizing proteins to compensate for the proteins lost by aggregation, and yet, they are unable to keep up, as we see with the overall decline of expressions of those proteins. Further work involving simultaneous measurement of the synthesis, degradation, and aggregation rates for individual proteins with and without inhibiting their protein syntheses will be required to clarify whether our observation, increase of newly synthesized proteins in aged nematodes, is associated with the aggregation of proteins.
Some of the genes listed in Table 3 have been shown to extend life span when knocked down in C. elegans, including ril-1, atp-3, and atp-5.26 In addition, inhibition of ribosomal protein expression (e.g., RPL-4, RPL-19, and RPL-30) has been shown to extend life span.36-38 We examined the effect of late-life RNAi-mediated knockdown of three of the genes (rpl-9, atp-3, and ril-1) listed in Table 3. An extension of life span following knockdown of two of these genes (atp-3 and ril-1) was observed even when RNAi was initiated late in adulthood (day 9), although the effect was less pronounced compared with lifelong knockdown of these genes (Table 4). Our results suggest that lowering the expression of aggregation-prone proteins (e.g., ATP-3 and RIL-1) late in life (but before proteostasis is disrupted) may slow down the rate of further aggregation of these proteins and extend the organism's life span. Our data suggest that the worms still actively synthesize these proteins late in life. The fact that inhibition of ATP-3 expression in aged worms extends the life span is somewhat surprising, as mitochondrial respiration genes are known to function only during development to control longevity.39 Thus, ATP-3 may influence the aging process by playing distinct roles during both development and late adulthood.
In conclusion, the results of the present study provide new insights into age-dependent changes in protein turnover in C. elegans, creating a foundation for a more complete understanding of the mechanism of aging in this important model organism.
Supplementary Material
ACKNOWLEDGMENT
We thank Yiyuan Yuan for providing expertise in nematode maintenance, Giridharan Gokulrangan for mass spectrometry analyses, and Kenji Ishida for critically reading the manuscript. We also thank the Invertebrate Animal Core of the University of Michigan's Nathan Shock Center of Excellence for their technical support.
Footnotes
ASSOCIATED CONTENT
Supporting Information
List of peptides quantified in the first pulse-chase labeling experiment (Table S1); list of peptides quantified in the second pulse-chase labeling experiment (Table S2); proteins quantified in at least one time point from the first pulse-chase experiment (Table S3); proteins quantified in at least one time point from the second pulse-chase experiment (Table S4); proteins quantified in at least six time points from the first pulse-chase experiment (Table S5); and proteins quantified in at least six time points from the second pulse-chase experiment (Table S6). This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Lund J, Tedesco P, Duke K, Wang J, Kim SK, Johnson TE. Transcriptional profile of aging in C. elegans. Curr Biol. 2002;12(18):1566–73. doi: 10.1016/s0960-9822(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 2.Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol. 2009;29(4):1095–106. doi: 10.1128/MCB.01227-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275(40):31505–13. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 4.Tavernarakis N. Ageing and the regulation of protein synthesis: a balancing act? Trends Cell Biol. 2008;18(5):228–35. doi: 10.1016/j.tcb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Rao PK, Li Q. Protein turnover in mycobacterial proteomics. Molecules. 2009;14(9):3237–58. doi: 10.3390/molecules14093237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Segref A, Hoppe T. Think locally: control of ubiquitin-dependent protein degradation in neurons. EMBO Rep. 2009;10(1):44–50. doi: 10.1038/embor.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfe RR, Chinkes DL. Isotope Tracers in Metabolic Research: Principles and Practice of Kinetic Analysis. John Wiley & Sons; New Jersey, USA: 2005. [Google Scholar]
- 8.Claydon AJ, Beynon R. Proteome dynamics: revisiting turnover with a global perspective. Mol Cell Proteomics. 2012;11(12):1551–65. doi: 10.1074/mcp.O112.022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, Reckow S, Webhofer C, Boehme M, Gormanns P, Egge-Jacobsen WM, Turck CW. Proteome Scale Turnover Analysis in Live Animals Using Stable Isotope Metabolic Labeling. Anal Chem. 2011:83. doi: 10.1021/ac102755n. [DOI] [PubMed] [Google Scholar]
- 10.Li L, Willard B, Rachdaoui N, Kirwan JP, Sadygov RG, Stanley WC, Previs S, McCullough AJ, Kasumov T. Plasma proteome dynamics: analysis of lipoproteins and acute phase response proteins with 2H2O metabolic labeling. Mol Cell Proteomics. 2012;11(7):M111 014209. doi: 10.1074/mcp.M111.014209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayapal KP, Sui S, Philp RJ, Kok YJ, Yap MG, Griffin TJ, Hu WS. Multitagging proteomic strategy to estimate protein turnover rates in dynamic systems. J Proteome Res. 2010;9(5):2087–97. doi: 10.1021/pr9007738. [DOI] [PubMed] [Google Scholar]
- 12.Michalik S, Bernhardt J, Otto A, Moche M, Becher D, Meyer H, Lalk M, Schurmann C, Schluter R, Kock H, Gerth U, Hecker M. Life and death of proteins: a case study of glucose-starved Staphylococcus aureus. Mol Cell Proteomics. 2012;11(9):558–70. doi: 10.1074/mcp.M112.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Claydon AJ, Beynon R. Proteome dynamics: revisiting turnover with a global perspective. Molecular & cellular proteomics : MCP. 2012;11(12):1551–65. doi: 10.1074/mcp.O112.022186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boisvert FOM, Ahmad Y, Gierlinski M, Charriere F, Lamont D, Scott M, Barton G, Lamond AI. A Quantitative Spatial Proteomics Analysis of Proteome Turnover in Human Cells. Molecular & Cellular Proteomics. 2012;11(3) doi: 10.1074/mcp.M111.011429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westman-Brinkmalm A, Abramsson A, Pannee J, Gang C, Gustavsson MK, von Otter M, Blennow K, Brinkmalm G, Heumann H, Zetterberg H. SILAC zebrafish for quantitative analysis of protein turnover and tissue regeneration. J Proteomics. 2011;75(2):425–434. doi: 10.1016/j.jprot.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Cohen LD, Zuchman R, Sorokina O, Muller A, Dieterich DC, Armstrong JD, Ziv T, Ziv NE. Metabolic turnover of synaptic proteins: kinetics, interdependencies and implications for synaptic maintenance. PLoS One. 2013;8(5):e63191. doi: 10.1371/journal.pone.0063191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 18.Yuan Y, Kadiyala CS, Ching TT, Hakimi P, Saha S, Xu H, Yuan C, Mullangi V, Wang L, Fivenson E, Hanson RW, Ewing R, Hsu AL, Miyagi M, Feng Z. Enhanced Energy Metabolism Contributes to the Extended Life Span of Calorie-restricted Caenorhabditis elegans. J Biol Chem. 2012;287(37):31414–26. doi: 10.1074/jbc.M112.377275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stiernagle T. Maintenance of C. elegans. WormBook. 2006:1–11. doi: 10.1895/wormbook.1.101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300(5622):1142–5. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 21.Yang JS, Nam HJ, Seo M, Han SK, Choi Y, Nam HG, Lee SJ, Kim S. OASIS: online application for the survival analysis of lifespan assays performed in aging research. PLoS One. 2011;6(8):e23525. doi: 10.1371/journal.pone.0023525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kadiyala CS, Tomechko SE, Miyagi M. Perfluorooctanoic Acid for shotgun proteomics. PLoS One. 2010;5(12):e15332. doi: 10.1371/journal.pone.0015332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sheng Q, Dai J, Wu Y, Tang H, Zeng R. BuildSummary: using a group-based approach to improve the sensitivity of peptide/protein identification in shotgun proteomics. J Proteome Res. 2012;11(3):1494–502. doi: 10.1021/pr200194p. [DOI] [PubMed] [Google Scholar]
- 24.Guo Y, Miyagi M, Zeng R, Sheng Q. O18Quant: A Semiautomatic Strategy for Quantitative Analysis of High-Resolution (16)O/(18)O Labeled Data. Biomed Res Int. 2014;2014:971857. doi: 10.1155/2014/971857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ching T-T, Paal AB, Mehta A, Zhong L, Hsu A-L. drr-2 encodes an eIF4H that acts downstream of TOR in diet-restriction-induced longevity of C-elegans. Aging Cell. 2010;9(4):545–557. doi: 10.1111/j.1474-9726.2010.00580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen M, Hsu AL, Dillin A, Kenyon C. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 2005;1(1):119–128. doi: 10.1371/journal.pgen.0010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolanowski MA, Russell RL, Jacobson LA. Quantitative measures of aging in the nematode Caenorhabditis elegans. I. Population and longitudinal studies of two behavioral parameters. Mech Ageing Dev. 1981;15(3):279–95. doi: 10.1016/0047-6374(81)90136-6. [DOI] [PubMed] [Google Scholar]
- 28.Rooney JP, Luz AL, Gonzalez-Hunt CP, Bodhicharla R, Ryde IT, Anbalagan C, Meyer JN. Effects of 5′-fluoro-2-deoxyuridine on mitochondrial biology in Caenorhabditis elegans. Exp Gerontol. 2014;56:69–76. doi: 10.1016/j.exger.2014.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feldman N, Kosolapov L, Ben-Zvi A. Fluorodeoxyuridine improves Caenorhabditis elegans proteostasis independent of reproduction onset. PLoS One. 2014;9(1):e85964. doi: 10.1371/journal.pone.0085964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamer G, Matilainen O, Holmberg CI. A photoconvertible reporter of the ubiquitin proteasome system in vivo. Nat Methods. 2010;7(6):473–8. doi: 10.1038/nmeth.1460. [DOI] [PubMed] [Google Scholar]
- 31.Syntichaki P, Troulinaki K, Tavernarakis N. Protein synthesis is a novel determinant of aging in Caenorhabditis elegans. Ann N Y Acad Sci. 2007;1119:289–95. doi: 10.1196/annals.1404.001. [DOI] [PubMed] [Google Scholar]
- 32.Taylor RC, Dillin A. Aging as an Event of Proteostasis Collapse. Cold Spring Harbor Perspectives in Biology. 2011;3(5) doi: 10.1101/cshperspect.a004440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ben-Zvi A, Miller EA, Morimoto RI. Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc Natl Acad Sci U S A. 2009;106(35):14914–9. doi: 10.1073/pnas.0902882106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reis-Rodrigues P, Czerwieniec G, Peters TW, Evani US, Alavez S, Gaman EA, Vantipalli M, Mooney SD, Gibson BW, Lithgow GJ, Hughes RE. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell. 2012;11(1):120–7. doi: 10.1111/j.1474-9726.2011.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread Protein Aggregation as an Inherent Part of Aging in C. elegans. Plos Biology. 2010;8(8) doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6(1):95–110. doi: 10.1111/j.1474-9726.2006.00267.x. [DOI] [PubMed] [Google Scholar]
- 37.Chen D, Pan KZ, Palter JE, Kapahi P. Longevity determined by developmental arrest genes in Caenorhabditis elegans. Aging Cell. 2007;6(4):525–33. doi: 10.1111/j.1474-9726.2007.00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curran SP, Ruvkun G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 2007;3(4):e56. doi: 10.1371/journal.pgen.0030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Rates of behavior and aging specified by mitochondrial function during development. Science. 2002;298(5602):2398–401. doi: 10.1126/science.1077780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.