Abstract
Hematopoietic stem cell (HSC) research took hold in the 1950s with the demonstration that intravenously injected bone marrow cells can rescue irradiated mice from lethality by reestablishing blood cell production. Attempts to quantify the cells responsible led to the discovery of serially transplantable, donor-derived, macroscopic, multilineage colonies detectable on the spleen surface 1 to 2 weeks posttransplant. The concept of self-renewing multipotent HSCs was born, but accompanied by perplexing evidence of great variability in the outcomes of HSC self-renewal divisions. The next 60 years saw an explosion in the development and use of more refined tools for assessing the behavior of prospectively purified subsets of hematopoietic cells with blood cell–producing capacity. These developments have led to the formulation of increasingly complex hierarchical models of hematopoiesis and a growing list of intrinsic and extrinsic elements that regulate HSC cycling status, viability, self-renewal, and lineage outputs. More recent examination of these properties in individual, highly purified HSCs and analyses of their perpetuation in clonally generated progeny HSCs have now provided definitive evidence of linearly transmitted heterogeneity in HSC states. These results anticipate the need and use of emerging new technologies to establish models that will accommodate such pluralistic features of HSCs and their control mechanisms.
Historical beginnings
We all like stories that have a beginning to capture our interest, a middle to sustain it, and an end that brings closure, but also harks to a future. The story of hematopoietic stem cells (HSCs) fits well into such a framework. The origin of blood cells, first in the developing embryo, and then later throughout life, has intrigued scientists, caregivers, and patients for centuries. As for the many advances, a combination of serendipity and the opportunistic exploitation of new tools have been important determinants of progress. For the HSC field, the development of atomic weaponry in the first half of the 20th century proved to be a game-changing event. It galvanized interest in understanding how ionizing radiation damages normal tissue and whether the effects of a lethal dose could be abrogated by a medically applicable intervention. The microscope helped to reveal the bone marrow to be one of the most radiosensitive of all tissues, but this tool proved inadequate to address the question of rescue.
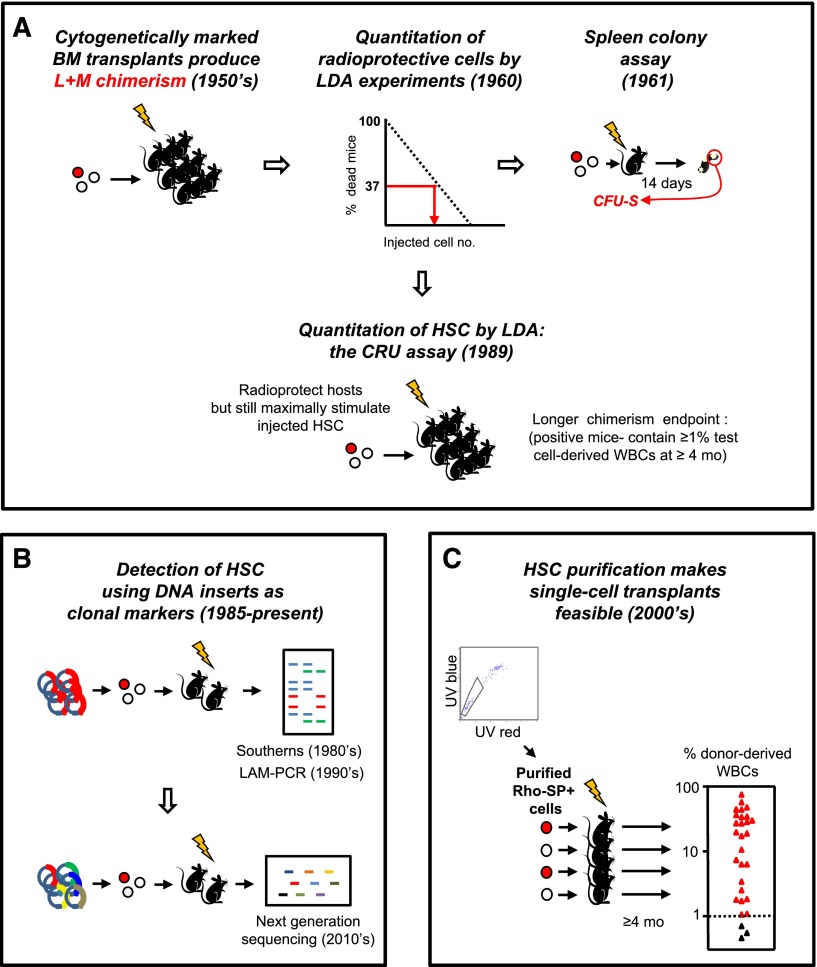
In fact, HSC research as a quantitative science emerged as a by-product of other investigative strategies seeking to determine how the consequences of myeloablation might be overcome. The seminal discovery was the finding of transplantable multipotent adult bone marrow cells with clonally demonstrable hematopoietic activity—a finding that evolved from experiments showing that an intravenous transplant of normal adult mouse bone marrow cells could protect recipients from a lethal dose of radiation1 by replacing the destroyed blood-forming system with a new and sustained source of lymphoid and myeloid cells.2 These observations established the presence in the bone marrow of adult mice of cells with long-term hematopoietic repopulating activity. This finding, in turn, sparked the idea that the original cells with this repopulating activity might then be characterized and even quantified based on the mature cells they could produce in myeloablated recipients (Figure 1A3-5).
Figure 1.
Historical sequence of methods used to detect and quantify mouse HSCs in vivo. (A) Development of LDA approaches to identify the transplantable cells that can rescue mice permanently from radiation-induced lethality by regenerating the inactivated blood-forming system of the host. (B) Genetic approaches to track HSCs by detection of their long-lived clonal outputs in transplanted recipients. Random sites of vector integration into the DNA of the regenerated progeny of transduced transplanted cells were the first unique DNA identifiers used.3,4 More recently, uptake of a single vector encoding a short unique “barcode” sequence from a diverse vector library has been used as a clonal tracking strategy.94-97 (C) Advances in LTRC purification enabling single-cell transplants to reveal the diversity of long-term clonal white blood cell outputs of individual HSCs as previously suggested by limiting dilution transplants and vector-marking experiments. Data shown are for purified Rho−SP+ LTRCs (HSCs) adapted from Uchida et al5 (with permission from Experimental Hematology.) BM, bone marrow; L+M, lymphoid + myeloid; mo, months; LAM-PCR, linear amplification mediated polymerase chain reaction; SP, side population; UV, ultraviolet; WBCs, white blood cells.
Lessons from the first clonal assays for transplantable HSCs
An early experimental design to pursue this idea involved transplanting decreasing numbers of adult mouse bone marrow cells into lethally irradiated recipients. The goal was to determine the minimal number of cells that would then protect the hosts.6 Interpretation of the results of this early type of limiting dilution assay (LDA) experiment (Figure 1A) was based on the assumption that a finite number of repopulating cells would have to be injected to enable the recipients to survive. As is now widely recognized, many subsequent studies have shown that the validity of this assumption is confounded by the presence in many hematopoietic organs of multiple, distinct types of transplantable hematopoietic cells with different, apparently predetermined, regenerative properties.7 Nevertheless, this first experimental attempt to measure the frequency of hematopoietic cells with radioprotective activity is noteworthy in its introduction of an objective biological endpoint to quantify a population of cells that could not be uniquely identified by any directly evident feature.
These early LDA experiments also served as the launch pad for a key derivative observation. This was the detection of distinct “nodules” that become macroscopically visible on the surface of the spleen in irradiated mice injected with low doses of syngeneic bone marrow cells 9 to 14 days previously (Figure 1A).8 Importantly, the finding that the number of these nodules was linearly related to the number of cells injected over a 10-fold range prompted the idea that their formation might offer an alternative—more direct (but still functional)—method to quantify hematopoietic cells with extensive and rapid mature blood cell–producing potential. These initial observations were then followed by a remarkable series of experiments that established the following core principles.
The large spleen nodules generated in such experiments are true clones, each derived from a single parental cell (hence its designation as a colony-forming unit [CFU]8), as shown using injections of bone marrow cells carrying different genetic (chromosomal) markers.9
Many of the spleen colonies thus obtained consist of mixtures of mature cells of the myeloid (erythroid, megakaryopoietic, and granulocyte/macrophage [GM]) lineages.10 Later genetic experiments showed that the cells that produce spleen colonies are derived from cells that can also produce lymphoid progeny, thus proving the maintenance in adult mice of individual cells with all of these differentiation potentialities.11
Some of these spleen colonies also contain daughter cells that generate similar macroscopically visible, multilineage colonies in the spleens of secondary irradiated recipients.12 The revelation of this behavior at the single-cell level was the birth of the concept of “self-renewal”—now widely considered to be a defining “stem cell” property in multiple tissue contexts.
The numbers and types of mature and primitive cells present in individual spleen colonies vary widely and independently.12,13 This diverse behavior led to the concept of stochastic variables underlying the type(s) of progeny generated by individual primitive hematopoietic cells. This idea was captured by the descriptor “hemopoiesis engendered randomly” in contrast to “hemopoietic inductive microenvironment,” suggesting an alternative explanation for the observed variable behavior seen.14,15
Most of the cells in the bone marrow of adult mice that produce these spleen colonies are quiescent (ie, in a dormant or G0 state). Thus, in the absence of any unusual perturbation, they are resistant to drugs or other treatments that specifically target dividing cells.16,17
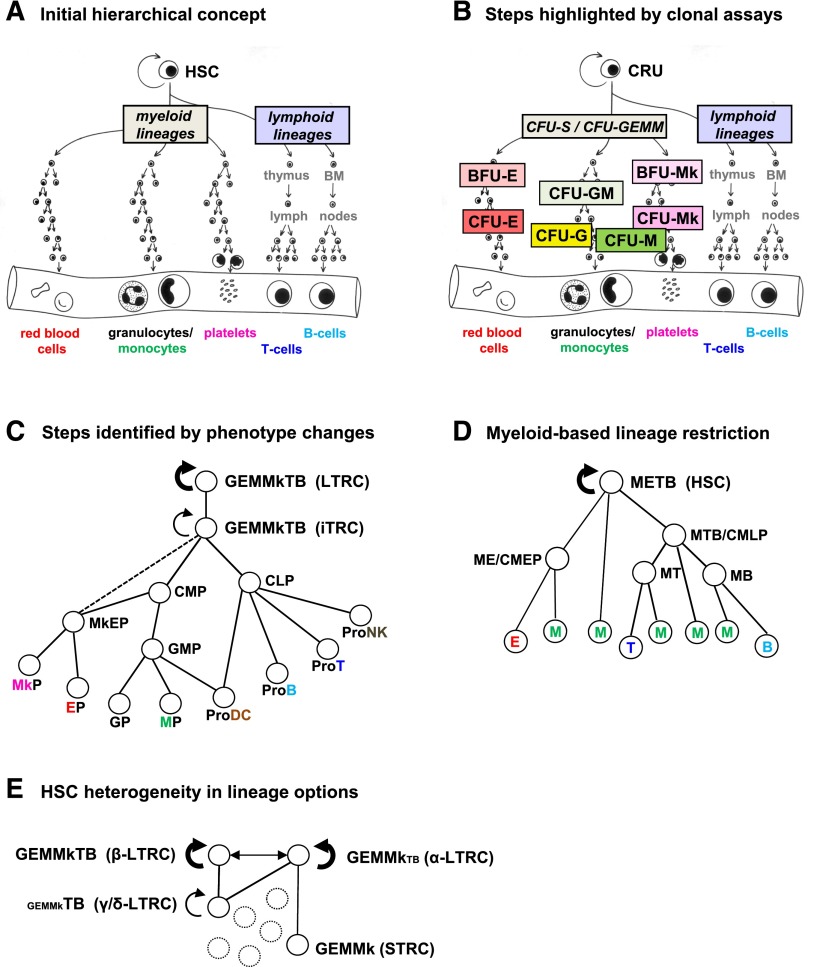
Together, these findings demonstrated the maintenance in the bone marrow of adult mice of rare, mainly quiescent, hematopoietic cells that originate from cells that also have lymphoid potential and can be activated to proliferate and regenerate all known myeloid lineages. The concept that all of the different major types of blood cells are derived throughout normal adult life from a common pool of self-renewing HSCs through a hierarchical differentiation process was thus introduced (Figure 2A). The existence of mechanisms that produce extensive heterogeneity in their self-renewal and differentiation behavior was also recognized. These findings also served to establish the importance of clonogenic assays to detect and quantify cells that are not directly identifiable and the need for new functional names for these cells8—to confer on them the specificity that no morphologically based, or indeed any direct descriptor, could provide. At the same time, this nomenclature side-stepped the unresolved issue of their heterogeneous behavior.
Figure 2.
Hierarchical models of HSC self-renewal and differentiation. Increasingly complex models of HSC differentiation hierarchies reflecting historically changing information about the numbers and types of lineages obtained from single-mouse “HSCs” in various in vitro and in vivo systems. (A) Initial view showing all lymphoid and all myeloid potentialities as the first lineage groupings to be segregated. (B) A more detailed view of the compartmentalization of intermediate, lineage-restricted progenitor subsets based on their behavior in short-term in vitro colony assays and properties allowing their separate isolation. (C) Map of early lineage restriction events based on the predominant functional activities of particular cell phenotypes (redrawn from Figure 1 of Seita and Weissman.38 (D) Concept of multiple origins of GM cells derived from examination of the lymphoid and myeloid activities elicited from different subsets of mouse hematopoietic cells in vitro. The hierarchy shown in this case has been adapted from Figure 1E of Kawamoto et al.107 (E) Differences in lineage potentialities exhibited by LTRCs with durable (serially transplantable) self-renewal activity (HSCs) as reported by Dykstra et al41 and Benz et al.44 BFU-E, burst-forming unit-erythroid; E, erythroid; G, granulocyte; GEMM, granulocyte, erythrocyte, monocyte, megakaryocyte; Mk, megakaryocyte; NK, natural killer; DC, dendritic cell; T, T-lymphoid; B, B-lymphoid; STRC, short-term repopulating cell.
Early in vitro colony assays identify downstream progenitors
Despite the important insights afforded by these early experiments, their restricted application to mouse cells precluded testing whether similar principles would apply to processes governing human hematopoiesis. Over the next 2 decades, however, an expanding capability for growing mammalian cells in vitro and the recognized power of clonal assays enabled the development of protocols that allowed in vitro hematopoietic colony assays to be developed, first for mouse cells and, soon thereafter, for their human counterparts.18-20 The first such colonies obtained were found to consist of GM cells. This finding had 2 possible explanations. One was that the in vitro GM colonies were incomplete reflections of progenitors with a broader differentiation potential; for example, as displayed by the cells that form spleen colonies in vivo (CFU-S). The other possibility was that the in vitro GM colonies originated from a previously unidentified progenitor cell with restricted GM differentiation potential (a CFU-GM). Efforts to distinguish between these alternatives revealed that the in vitro GM colonies are generated from a distinct type of progenitor whose numbers under regenerative conditions (eg, within a spleen colony) make it appear closely related to CFU-S,21 but whose physical and biological properties are largely different.22,23 These findings provided the first evidence for a stage of differentiation intermediate between HSCs (measured as CFU-S at that time) and the terminal stages of blood cell differentiation described by sequential, morphologically defined changes (Figure 2B).
The discovery of in vitro conditions that enable the detection of erythroid, megakaryocyte, and mixed/multilineage colonies soon followed. In each case, measurements of the frequencies, parent–progeny relationships, growth requirements, and biological properties of the initial clonogenic cells reinforced the concept that they represent separate intermediate stages of hematopoietic differentiation (Figure 2B),24,25 analogous to the transit-amplifying populations recognized histologically in other tissues by their more obvious spatial arrangement.
A strong correlation between the multiple changing properties in cells that produce progressively smaller single-lineage colonies in vitro provided further support for a model in which the execution of erythroid, GM, and megakaryocytic differentiation pathways constitute well-coordinated, multistep processes that span many cell divisions (>10 cell cycles to account for the largest sizes of single-lineage colonies typically obtained). At the same time, this coordination had to be sufficiently flexible to generate a progressive loss of correlation within individual spleen colonies in the numbers of progenitors that generate increasingly smaller colonies in vitro.26,27
HSC detection in clonal assays for LTRCs
Subsequent studies revealed that spleen colony formation is obtained from a biologically more heterogeneous population than was originally appreciated, and one that does not overlap extensively with cells with long-term repopulating ability (hereafter referred to as long-term repopulating cells [LTRCs]). Detailed time course analyses showed that both the appearance and disappearance of individual spleen colonies is very rapid, with a precise timing determined by biologically distinct subsets of CFU-S.28,29 Subsequent cell-separation experiments demonstrated that immediate radioprotection and long-term repopulating ability are not necessarily vested in cells with the same physical properties.30 Thus a need to devise an assay specific for LTRCs became apparent.
Two complementary strategies were then pursued. Both adopted endpoints indicative of a more prolonged production of mature cells than is obtained in either the CFU-S or short-term in vitro clonogenic cell assays.31 The in vivo competitive repopulating unit (CRU) assay relies on the ability of the environment of a myelosuppressed or genetically compromised host to support the full spectrum of potential growth and differentiation possibilities of transplanted input cell(s) (Figure 1A). The second approach incorporates features of the in vivo marrow environment, but in an in vitro context, thus offering the possibility that a similar approach might be applicable to human cells.
The mouse CRU assay was designed to enable the sensitive and specific detection within heterogeneous populations of a single cell that is able to regenerate and maintain long-term blood cell production in vivo (Figure 1A). This required identifying conditions that would support the initial as well as the lifelong survival of all recipients while still maintaining a useful sensitivity of detecting the cells of interest. Early experiments showed that this could be achieved by providing another source of cells with transient repopulating activity as well as a minimal source of genetically distinct LTRCs (ie, in numbers just sufficient to keep the recipient alive long-term). The latter maneuver, which determines the sensitivity of the assay, can be met using various ways to deplete or genetically compromise the other LTRCs present in the host. CRU frequencies in the test cell suspension are then quantified using an LDA transplant strategy. To achieve specificity for detecting LTRCs exclusively, a test cell–derived contribution to the white blood cells present 4 to 6 months posttransplant of at least 1% is required.
The term CRU and the assay used to detect CRUs should not be confused with the “competitive repopulation assay” introduced into the field much earlier.32 This older approach quantifies LTRC activity (not numbers) by comparing a similarly long-term contribution of a genetically distinct test population to the total blood cell content of lethally irradiated recipients that have been cotransplanted with a large (and radioprotective) number of HSCs and transient reconstituting cells. This latter experimental design is much more efficient than the CRU assay because the number of recipient mice required is much smaller. This assay is also particularly useful for determining whether a given manipulation has had a gross effect on LTRCs by comparing them with a control cell population. However, it does not discriminate between effects on LTRC numbers and their clonal outputs, which can vary (eg, as with aging33), and are independently measured parameters in the CRU assay.31
The second (in vitro) approach also incorporates an endpoint of prolonged hematopoiesis. In this case, the cell of origin is variably referred to by the operational terms: long-term culture-initiating cell or cobblestone area–forming cell, according to the indicator of continuing hematopoiesis used. The development of this approach was made possible by the initial discovery that prolonged growth of primitive mouse hematopoietic cells can be achieved when adult bone marrow cells are cultured at 33°C in the presence of steroids on a competent (self-formed or provided) stromal feeder layer.34-36 The separate provision of a stromal feeder layer is critical to enable input long-term culture-initiating cell/cobblestone area–forming cell frequencies to be quantified using LDA principles. To confer specificity for very primitive cells, evidence of continuing hematopoietic cell production for at least 4 to 6 (mouse) or 6 to 12 (human) weeks is required. Culture conditions can also be varied to affect the types and lineages of cell(s) produced as well as the duration of their generation. Such modifications can provide specificity for detecting cells with lymphomyeloid differentiation potential or with in vivo assayed LTRCs.31
HSC purification
Methods to prospectively separate viable cells based on their different physical (size and density), biochemical (enzymatic), and surface antigen profiles have enabled distinct subsets of mouse (and human) hematopoietic cells with different repopulating kinetics and activity to be identified, quantified, and further characterized.7 These findings have now led to a widely accepted hierarchical model of hematopoiesis in which the orderly and coordinated loss of multilineage differentiation and durable proliferative potential by an initial compartment of LTRCs can be traced by accompanying changes in their surface and metabolic phenotypes (Figure 2C).37,38
Advances in the development of robust methods for isolating mouse LTRCs at high purities typically combine the use of Rhodamine-123 (Rho) and/or Hoechst 33342 efflux measurements (to isolate Rho−SP+ cells), or antibodies against 2 SLAM markers (CD48 and CD150) in combination with those that isolate Lin−Sca1+Kit+CD34−Flt3− or EPCR+ phenotypes (to give LSK34−FL−48−150+ or ESLAM cells, respectively).39-43 The purities of “durable” LTRCs in these populations (∼20% to 30% using a stringent definition of serial reconstituting activity, not just persisting chimerism at 4 to 6 months) are sufficient to analyze their clonal outputs directly from single-cell transplants (Figure 1C). The results thus obtained indicate that all of these purification strategies isolate the same, or very similar, cell populations.33,41,42,44-46 Interestingly, very few of the markers generally used to purify LTRCs contribute to their functional properties, suggesting that new, more relevant markers may still be awaiting discovery.
In the meantime, the powerful LTRC purification strategies available are already being used in combination with analytical methods that can be applied to small numbers of cells to identify unique molecular features (or unique combinations of molecular features) of LTRCs. These analyses have uncovered numerous types of chromatin regulators, including members of many transcription factor complexes and their downstream targets as well as regulators of the metabolome, protein synthesis, and mediators of signaling responses to external cues.47-52 Indeed, the multiplicity of entities that appear to have nonredundant roles has been disconcerting in suggesting the lack of a singular pathway or state that regulates the maintenance of LTRC potential, or that distinguishes these cells from derivatives that appear irreversibly destined to differentiate in available assay systems. Rather, current findings suggest the involvement of highly complex and possibly alternative molecular networks controlling LTRC self-renewal that may require systems approaches to elucidate.53,54 Indeed, one might advance the somewhat heretical idea that intracellular states responsible for different HSC self-renewal potentialities may not be well-matched to cellular phenotypes that display different transplant-derived endpoints of repopulating activity.
HSC/LTRC heterogeneity and the “new reality”
As noted, broad ranging heterogeneity in the self-renewal and differentiation behavior of individual multipotent hematopoietic cells became evident from early analyses of the cellular composition of individual spleen colonies.10,12,13,27 Later examination of clones generated in vitro from multipotent cells in the presence of soluble factors (and hence more homogeneous environments) confirmed the display of an equivalent degree of heterogeneity in the clonal outputs of multipotent cells, including progeny detectable as CFU-S.55-57 The results of these later studies argue strongly against a deterministic role of external cues as the major regulators of the self-renewal and lineage potentialities of primitive hematopoietic cells. These findings do not, however, exclude the possibility that exposure to external factors can have deterministic influences on LTRC outputs, as has been suggested.58,59 Undoubtedly, a complete understanding of the mechanisms involved in regulating the defining properties of HSCs will likely require a separate analysis of intrinsic and extrinsic parameters that can alter their viability and cycling status as well as their lineage options and self-renewal activity (Figure 3A).
Figure 3.
HSC viability, mitogenesis and self-maintenance can be separately regulated by different external cues. (A) Schema showing the multiple responses that can affect HSC numbers. (B) Demonstration that different extrinsic conditions can separately regulate the survival, proliferation, and self-renewal responses of highly purified, durable LTRCs assessed in single-cell cultures. Shown are examples of different in vitro conditions in which full LTRC activity of the surviving input cells was maintained for 7 days either in the absence of cell death or division, or under different conditions that variably kept the input cells alive but maximally stimulated mitogenesis of the survivors. Also shown are conditions that supported full survival and mitogenesis of the same input cells, but with substantial loss of their original LTRC activity (redrawn from data published in Wohrer et al65 with permission). ESLAM, EPCR+CD48−CD150+; FBS, fetal bovine serum; EP, erythropoietin; SF, Steel factor; CM, UG26 stromal cell conditioned medium; LDA, limiting dilution assay.
LTRC self-renewal control
The development of a protocol to expand LTRCs ex vivo without predisposing the cells to leukemic transformation has long been a driving goal in the field of hematology. A simple view of the needs for such a protocol would be to achieve conditions that promote LTRC survival and proliferation as well as their self-renewal. The ability of external factors to promote the survival and alter the cycling status of primitive hematopoietic cells has also been appreciated for a long time, as exemplified by the pioneering studies of CFU-S cycling control both in vivo and in vitro,16,60 later extended to functionally identified LTRCs.61 An ability of external factors to modulate LTRC self-renewal has been inferred from their ability to enhance LTRC outputs in transplanted mice62 (assuming that such expansions are not due to a decreased rate of LTRC death or to an increased proportion of cycling LTRCs). Assessments of highly purified LTRCs to defined factors in vitro allow requirements for their viability and cycling activity to be directly visualized.63,64 The exploitation of this latter approach has now provided conclusive robust evidence that LTRC self-renewal, mitogenesis, and survival can be independently regulated (Figure 3B).65
Nevertheless, the large (>500-fold) variations in self-renewal of individual LTRCs seen in vivo when these are tracked clonally and measured through at least 2 serial transplants support the operation of an underlying stochastic process.44,66 On the other hand, analysis of clones produced from the first division progeny of stringently defined LTRCs have shown both variable (in vitro)65,67 and more restricted (in vivo)66 patterns of self-renewal behavior in paired daughter cells. The latter findings suggest LTRC self-renewal responses to external stimuli may reflect variable cell–intrinsic elements that can be sustained through multiple cell generations. Such an inference is noteworthy in that it departs from the traditional concept of a singular, shared “ground state” of LTRC self-renewal ability.
Superimposed on these sources of heterogeneity in HSC self-renewal probabilities are those that are incurred during development. Documented differences in key properties of primitive fetal and adult hematopoietic cells date back several decades. These include differences in both the cycling state of these cells in situ16,68 and the self-renewal activity they display posttransplant.69-71 Recent studies have confirmed that these developmental changes also apply to stringently defined mouse LTRCs, and appear to be coordinated by a process that causes an abrupt switch in these same 2 properties (cycling status and self-renewal ability) between 3 and 4 weeks after birth.61,72 These changes are associated with changes in the responsiveness of LTRCs to Steel factor73 and a differential expression of (or dependence on) a panoply of chromatin regulators. The latter include a demonstrated selective dependence of fetal LTRCs on members of the Polycomb2 complex (Eed and Ezh2),74,75 Sox17,76 and Lin28-regulated Hmga2.77,78 Conversely, a selective dependence of adult LTRC self-renewal and/or survival on Bmi179 and Tel/Etv680 has been reported, and a selective dependence of adult LTRC quiescence/dormancy on E47-regulated p21,81-83 Pbx1,84 Gfi-1,85 and C/ebpa.86,87
LTRC differentiation control
Very little is known about the molecular mechanisms that establish the hematopoietic lineage potential of LTRCs. Past models have generally assumed that LTRCs are created in the embryo with an unbiased ability to activate all hematopoietic lineage programs. Indeed, clonal tracking of the progeny of fetal LTRCs indicate this to generally be the case.44 However, it is also well-established that primitive multipotent fetal and adult hematopoietic cells produce lymphoid progeny with different molecular features and biological properties.88-90 Subsequent studies have shown that at least some of these changes are determined at the level of fetal and adult LTRCs,91 and by the same developmentally regulated decline in Lin28b expression and consequent postnatal “switch” effect that alters LTRC self-renewal and proliferation control, albeit via different downstream effectors.77,78
The first definitive indication of specific patterns of lineage outputs sustained through multiple self-renewal divisions of adult LTRCs came from tracking the clonal progeny regenerated from a single LTRC through successive serial transplants.92 These observations have since been confirmed in numerous experiments.41,44-46,93 The stability of these clone-specific lineage output preferences through many LTRC self-renewal divisions argues strongly in favor of some type of preserved epigenetic signature as the responsible mechanism. Clone-specific differentiation patterns associated with LTRCs with durable self-renewal ability are either “balanced” in their output of lymphoid and myeloid progeny, or “lymphoid-deficient.” The latter type of LTRC is also commonly referred to in the literature as “myeloid-biased” or “myeloid-skewed.” However, these latter terms are misleading, because the myeloid outputs characteristic of such clones are not different from those produced by LTRCs that produce a balanced contribution of myeloid and lymphoid cells to the circulating pools of these mature blood cells. Rather, it is the reduced production of lymphoid cells that gives the clone a myeloid-biased appearance.41,44 The other types of LTRCs produce reduced contributions of mature myeloid cells without corresponding increases in lymphoid cell production and thus are best referred to as myeloid-deficient rather than lymphoid-biased for the same reasons. Interestingly, the same spectrum of lineage output patterns has been documented even under conditions in which many clones are generated within a single recipient and tracked individually using powerful vector-mediated DNA barcoding strategies.94-97 Statistical analysis of the various lineage output patterns seen have suggested that these may reflect the existence of a defined repertoire of LTRC subsets with discrete, predetermined lineage options (rather than a continuum),41,98 hence the suggested use of objectively assigned terms for them (ie, α, β, γ, δ; Figure 2E). That myeloid-deficient clones also do not contain sufficient progeny LTRCs to reconstitute secondary recipients41 is notable because it suggests a possible mechanistic link between LTRC retention of myeloid and self-renewal potential.
It has been reported that LTRCs with different outputs can be selectively enriched according to their phenotype (ie, increased or decreased expression of various proteins, such as CD150,42,45 CD41,99 CD229,100 and vWf101). These findings support the operation of cell-intrinsic mechanisms determining the specific differentiation programs that characterize individual LTRCs. However, these markers have proven to be more discriminatory for LTRCs with durable activity than for LTRCs with a particular lineage output profile. It is also important to note that some secondary clones generated from serial transplants of single cells,44 or from paired daughter cells obtained from purified LTRCs stimulated to divide in vitro,65,67,93 have not shown convincing evidence of a preserved differentiation profile. Likewise, the distribution of LTRCs categorized according to the different lineage options they display has been found to change during development and aging33,44,102 and be subject to extrinsic modulation.46 Thus, although the molecular mechanisms involved in predetermining the specific lineage output properties of individual LTRCs remain elusive, they do not appear to overlap entirely with those that control LTRC cycling activity/dormancy or self-renewal potential.
Initial models of HSC differentiation in vivo invoked the principle of a relatively fixed order and timing of sequential lineage restriction events, beginning with a separation of lymphoid and myeloid programs (Figure 2A-C). This concept is also now being eroded by more sensitive analyses of the cell types produced from individual LTRCs and other multipotent cells. For example, different phenotypes with persisting GM differentiation potentialities in combination with other different lineage options have now been reported (Figure 2C-D).103-107 Additional deviations from a unilinear branching model of hematopoietic cell differentiation from a single original compartment include examples of a very rapid loss of LTRC properties with full retention of viability and proliferative responsiveness in vitro65 or a rapid differentiation of their progeny into mature cells in vivo.93,108 Also of note is the suggestion that the unexpectedly accelerated differentiation profiles exhibited by some cells with an LTRC phenotype may be related to their particular “poised” or “primed” gene expression state—as reflected in their expression of certain lineage-specific genes at a low level.101,109,110
Physiological relevance of LTRCs
The number of serially transplantable LTRCs in normal adult mice is maintained at a relatively high level (∼3000 per mouse assuming a total bone marrow content of 2 × 108 cells). These numbers gradually increase with age,102 with a low but measurable and continuous rate of entry into a cycling state.111 The contribution of an individual LTRC to the total output of any mature cell lineage would thus be anticipated to be highly variable given the many cell generations and time likely separating it from most of its ultimately generated mature progeny. A lack of correlation in the clonal representation of cells at these 2 ends of the hematopoietic hierarchy would thus be predicted, as has recently been documented.112 This finding does not, however, imply that LTRCs are superfluous to the long-term integrity of mature blood cell production after adulthood is reached. To address this latter issue requires an inducible LTRC-specific lineage tracing approach and new markers (such as those recently suggested113,114) that might be used to design the required reporters. In addition, there is a compelling need to gain a better understanding of the molecular mechanisms that confer on cells the unique properties of LTRCs detected by transplantation assays so that these might be enhanced for clinical applications.
Of related interest is the growing list of mature myeloid cells produced before birth from Myb-independent embryonic hematopoietic sources that persist throughout life and are not normally reconstituted from adult LTRCs whose generation is Myb-dependent.115 These include the microglia of the brain,116 adult Langerhans cells,117 and the Kupffer cells of the liver.118
Looking forward
The era of predicting biological behavior from knowledge of the molecular mechanisms that define cell types and how they respond to external cues is upon us. An impending challenge in the HSC field is the extent of biological heterogeneity that requires molecular elucidation at the single-cell level. Compounding this challenge are issues related to the essential averaging process inherent in historically used procedures for performing molecular analyses. Fortunately, many new and powerful tools for interrogating individual cells in diverse populations are now becoming available. These include advances in cellular reprogramming and molecular profiling together with new bioinformatic programs to infer cell states and relationships.119-122 We can thus anticipate that current 2-dimensional concepts of how specific primed blood “lineage programs” are activated may soon be replaced by more pluralistic models. These, in turn, will likely require significant revision of present ideas about how molecular mechanisms permit, ignore, or direct changes with biological sequelae—and how these may relate to cell types currently defined by surface phenotypes and growth endpoints. However, the reward may be the generation of models of HSC states and their differentiation that predict how to modify biological outcomes, the molecular trajectories that may be involved, and the speed of their attainment. These issues are not just of academic interest. They may create paradigms relevant to other tissues, in addition to being critical drivers of future methods to detect and treat disease with greater effectiveness.
Acknowledgments
The author acknowledges helpful discussions with A. Eaves and a group of talented past and present trainees whose ideas and experimental results have contributed to many of the ideas presented.
The preparation of this review was aided by a grant from the Terry Fox Run.
Authorship
Contribution: C.E. reviewed the literature and wrote the paper.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: C. J. Eaves, Terry Fox Laboratory, 675 West 10th Ave, Vancouver, BC, Canada V5Z 1L3; e-mail: ceaves@bccrc.ca.
References
- 1.Jacobson LO, Simmons EL, Marks EK, Eldredge JH. Recovery from radiation injury. Science. 1951;113(2940):510–511. doi: 10.1126/science.113.2940.510. [DOI] [PubMed] [Google Scholar]
- 2.Ford CE, Hamerton JL, Barnes DW, Loutit JF. Cytological identification of radiation-chimaeras. Nature. 1956;177(4506):452–454. doi: 10.1038/177452a0. [DOI] [PubMed] [Google Scholar]
- 3.Dick JE, Magli MC, Huszar D, Phillips RA, Bernstein A. Introduction of a selectable gene into primitive stem cells capable of long-term reconstitution of the hemopoietic system of W/Wv mice. Cell. 1985;42(1):71–79. doi: 10.1016/s0092-8674(85)80102-1. [DOI] [PubMed] [Google Scholar]
- 4.Keller G, Paige C, Gilboa E, Wagner EF. Expression of a foreign gene in myeloid and lymphoid cells derived from multipotent haematopoietic precursors. Nature. 1985;318(6042):149–154. doi: 10.1038/318149a0. [DOI] [PubMed] [Google Scholar]
- 5.Uchida N, Dykstra B, Lyons KJ, Leung FY, Eaves CJ. Different in vivo repopulating activities of purified hematopoietic stem cells before and after being stimulated to divide in vitro with the same kinetics. Exp Hematol. 2003;31(12):1338–1347. doi: 10.1016/j.exphem.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 6.McCulloch EA, Till JE. The radiation sensitivity of normal mouse bone marrow cells, determined by quantitative marrow transplantation into irradiated mice. Radiat Res. 1960;13:115–125. [PubMed] [Google Scholar]
- 7.Miller PH, Knapp DJ, Eaves CJ. Heterogeneity in hematopoietic stem cell populations: implications for transplantation. Curr Opin Hematol. 2013;20(4):257–264. doi: 10.1097/MOH.0b013e328360aaf6. [DOI] [PubMed] [Google Scholar]
- 8.Till JE, McCullough EA. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells. Radiat Res. 1961;14:213–222. [PubMed] [Google Scholar]
- 9.Becker AJ, McCullough EA, Till JE. Cytological demonstration of the clonal nature of spleen colonies derived from transplanted mouse marrow cells. Nature. 1963;197:452–454. doi: 10.1038/197452a0. [DOI] [PubMed] [Google Scholar]
- 10.Wu AM, Till JE, Siminovitch L, McCulloch EA. A cytological study of the capacity for differentiation of normal hemopoietic colony-forming cells. J Cell Physiol. 1967;69(2):177–184. doi: 10.1002/jcp.1040690208. [DOI] [PubMed] [Google Scholar]
- 11.Wu AM, Till JE, Siminovitch L, McCulloch EA. Cytological evidence for a relationship between normal hematopoietic colony-forming cells and cells of the lymphoid system. J Exp Med. 1968;127(3):455–464. doi: 10.1084/jem.127.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siminovitch L, McCulloch EA, Till JE. The distribution of colony-forming cells among spleen colonies. J Cell Physiol. 1963;62:327–336. doi: 10.1002/jcp.1030620313. [DOI] [PubMed] [Google Scholar]
- 13.Fowler JH, Wu AM, Till JE, McCulloch EA, Siminovitch L. The cellular composition of hemopoietic spleen colonies. J Cell Physiol. 1967;69(1):65–72. [Google Scholar]
- 14.Till JE, McCulloch EA, Siminovitch L. A stochastic model of stem cell proliferation, based on the growth of spleen colony-forming cells. Proc Natl Acad Sci USA. 1964;51:29–36. doi: 10.1073/pnas.51.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Till JE, McCulloch EA. Hemopoietic stem cell differentiation. Biochim Biophys Acta. 1980;605(4):431–459. doi: 10.1016/0304-419x(80)90009-8. [DOI] [PubMed] [Google Scholar]
- 16.Becker AJ, McCulloch EA, Siminovitch L, Till JE. The effect of differing demands for blood cell production on DNA synthesis by hemopoietic colony-forming cells of mice. Blood. 1965;26:296–308. [PubMed] [Google Scholar]
- 17.Bruce WR, Meeker BE, Valeriote FA. Comparison of the sensitivity of normal hematopoietic and transplanted lymphoma colony-forming cells to chemotherapeutic agents administered in vivo. J Natl Cancer Inst. 1966;37(2):233–245. [PubMed] [Google Scholar]
- 18.Pluznik DH, Sachs L. The cloning of normal “mast” cells in tissue culture. J Cell Physiol. 1965;66(3):319–324. doi: 10.1002/jcp.1030660309. [DOI] [PubMed] [Google Scholar]
- 19.Bradley TR, Metcalf D. The growth of mouse bone marrow cells in vitro. Aust J Exp Biol Med Sci. 1966;44(3):287–299. doi: 10.1038/icb.1966.28. [DOI] [PubMed] [Google Scholar]
- 20.Senn JS, McCulloch EA, Till JE. Comparison of colony-forming ability of normal and leukaemic human marrow in cell culture. Lancet. 1967;2(7516):597–598. doi: 10.1016/s0140-6736(67)90742-8. [DOI] [PubMed] [Google Scholar]
- 21.Wu AM, Siminovitch L, Till JE, McCulloch EA. Evidence for a relationship between mouse hemopoietic stem cells and cells forming colonies in culture. Proc Natl Acad Sci USA. 1968;59(4):1209–1215. doi: 10.1073/pnas.59.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worton RG, McCulloch EA, Till JE. Physical separation of hemopoietic stem cells from cells forming colonies in culture. J Cell Physiol. 1969;74(2):171–182. doi: 10.1002/jcp.1040740209. [DOI] [PubMed] [Google Scholar]
- 23.Iscove NN, Till JE, McCulloch EA. The proliferative states of mouse granulopoietic progenitor cells. Proc Soc Exp Biol Med. 1970;134(1):33–36. doi: 10.3181/00379727-134-34721. [DOI] [PubMed] [Google Scholar]
- 24.Eaves CJ, Eaves AC. Part II. Cell biology and pathobiology. Anatomy and physiology of hematopoiesis. In: Pui C-H, editor. Childhood Leukemias. 2nd ed., Chapter 4. Cambridge: Cambridge University Press; 2006. pp. 69–105. [Google Scholar]
- 25.Kondo M, Wagers AJ, Manz MG, et al. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- 26.Gregory CJ, McCulloch EA, Till JE. Erythropoietic progenitors capable of colony formation in culture: state of differentiation. J Cell Physiol. 1973;81(3):411–420. doi: 10.1002/jcp.1040810313. [DOI] [PubMed] [Google Scholar]
- 27.Gregory CJ. Erythropoietin sensitivity as a differentiation marker in the hemopoietic system: studies of three erythropoietic colony responses in culture. J Cell Physiol. 1976;89(2):289–301. doi: 10.1002/jcp.1040890212. [DOI] [PubMed] [Google Scholar]
- 28.Hodgson GS, Bradley TR. Properties of haematopoietic stem cells surviving 5-fluorouracil treatment: evidence for a pre-CFU-S cell? Nature. 1979;281(5730):381–382. doi: 10.1038/281381a0. [DOI] [PubMed] [Google Scholar]
- 29.Magli MC, Iscove NN, Odartchenko N. Transient nature of early haematopoietic spleen colonies. Nature. 1982;295(5849):527–529. doi: 10.1038/295527a0. [DOI] [PubMed] [Google Scholar]
- 30.Jones RJ, Wagner JE, Celano P, Zicha MS, Sharkis SJ. Separation of pluripotent haematopoietic stem cells from spleen colony-forming cells. Nature. 1990;347(6289):188–189. doi: 10.1038/347188a0. [DOI] [PubMed] [Google Scholar]
- 31.Miller CL, Dykstra B, Eaves CJ. Curr Protoc Immunol. 2008. Characterization of mouse hematopoietic stem and progenitor cells. Chapter 22:Unit 22B.2. [DOI] [PubMed] [Google Scholar]
- 32.Harrison DE. Competitive repopulation: a new assay for long-term stem cell functional capacity. Blood. 1980;55(1):77–81. [PubMed] [Google Scholar]
- 33.Dykstra B, Olthof S, Schreuder J, Ritsema M, de Haan G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J Exp Med. 2011;208(13):2691–2703. doi: 10.1084/jem.20111490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dexter TM, Allen TD, Lajtha LG. Conditions controlling the proliferation of haemopoietic stem cells in vitro. J Cell Physiol. 1977;91(3):335–344. doi: 10.1002/jcp.1040910303. [DOI] [PubMed] [Google Scholar]
- 35.Greenberger JS. Sensitivity of corticosteroid-dependent insulin-resistant lipogenesis in marrow preadipocytes of obese-diabetic (db/db) mice. Nature. 1978;275(5682):752–754. doi: 10.1038/275752a0. [DOI] [PubMed] [Google Scholar]
- 36.Ploemacher RE, van der Sluijs JP, Voerman JS, Brons NH. An in vitro limiting-dilution assay of long-term repopulating hematopoietic stem cells in the mouse. Blood. 1989;74(8):2755–2763. [PubMed] [Google Scholar]
- 37.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10(2):120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Seita J, Weissman IL. Hematopoietic stem cell: self-renewal versus differentiation. Wiley Interdiscip Rev Syst Biol Med. 2010;2(6):640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273(5272):242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 40.Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 41.Dykstra B, Kent D, Bowie M, et al. Long-term propagation of distinct hematopoietic differentiation programs in vivo. Cell Stem Cell. 2007;1(2):218–229. doi: 10.1016/j.stem.2007.05.015. [DOI] [PubMed] [Google Scholar]
- 42.Kent DG, Copley MR, Benz C, et al. Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood. 2009;113(25):6342–6350. doi: 10.1182/blood-2008-12-192054. [DOI] [PubMed] [Google Scholar]
- 43.Rossi L, Challen GA, Sirin O, Lin KK, Goodell MA. Hematopoietic stem cell characterization and isolation. Methods Mol Biol. 2011;750:47–59. doi: 10.1007/978-1-61779-145-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benz C, Copley MR, Kent DG, et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 2012;10(3):273–283. doi: 10.1016/j.stem.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 45.Morita Y, Ema H, Nakauchi H. Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med. 2010;207(6):1173–1182. doi: 10.1084/jem.20091318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Challen GA, Boles NC, Chambers SM, Goodell MA. Distinct hematopoietic stem cell subtypes are differentially regulated by TGF-beta1. Cell Stem Cell. 2010;6(3):265–278. doi: 10.1016/j.stem.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkinson AC, Göttgens B. Transcriptional regulation of haematopoietic stem cells. Adv Exp Med Biol. 2013;786:187–212. doi: 10.1007/978-94-007-6621-1_11. [DOI] [PubMed] [Google Scholar]
- 48.Ito K, Suda T. Metabolic requirements for the maintenance of self-renewing stem cells. Nat Rev Mol Cell Biol. 2014;15(4):243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rossi L, Lin KK, Boles NC, et al. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell. 2012;11(3):302–317. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Signer RA, Magee JA, Salic A, Morrison SJ. Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature. 2014;509(7498):49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hogart A, Lichtenberg J, Ajay SS, Anderson S, Margulies EH, Bodine DM NIH Intramural Sequencing Center. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res. 2012;22(8):1407–1418. doi: 10.1101/gr.132878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cabezas-Wallscheid N, Klimmeck D, Hansson J, et al. Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA Methylome analysis. Cell Stem Cell. 2014;15(4):507–522. doi: 10.1016/j.stem.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 53.Lemischka IR. Stem cell biology: a view toward the future. Ann N Y Acad Sci. 2005;1044:132–138. doi: 10.1196/annals.1349.017. [DOI] [PubMed] [Google Scholar]
- 54.Roeder I, Horn K, Sieburg HB, Cho R, Muller-Sieburg C, Loeffler M. Characterization and quantification of clonal heterogeneity among hematopoietic stem cells: a model-based approach. Blood. 2008;112(13):4874–4883. doi: 10.1182/blood-2008-05-155374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Humphries RK, Jacky PB, Dill FJ, Eaves AC, Eaves CJ. CFU-S in individual erythroid colonies derived in vitro from adult mouse marrow. Nature. 1979;279(5715):718–720. doi: 10.1038/279718a0. [DOI] [PubMed] [Google Scholar]
- 56.Humphries RK, Eaves AC, Eaves CJ. Self-renewal of hemopoietic stem cells during mixed colony formation in vitro. Proc Natl Acad Sci USA. 1981;78(6):3629–3633. doi: 10.1073/pnas.78.6.3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suda T, Suda J, Ogawa M. Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc Natl Acad Sci USA. 1984;81(8):2520–2524. doi: 10.1073/pnas.81.8.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mossadegh-Keller N, Sarrazin S, Kandalla PK, et al. M-CSF instructs myeloid lineage fate in single haematopoietic stem cells. Nature. 2013;497(7448):239–243. doi: 10.1038/nature12026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Florian MC, Nattamai KJ, Dörr K, et al. A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing. Nature. 2013;503(7476):392–396. doi: 10.1038/nature12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Toksöz D, Dexter TM, Lord BI, Wright EG, Lajtha LG. The regulation of hemopoiesis in long-term bone marrow cultures. II. Stimulation and inhibition of stem cell proliferation. Blood. 1980;55(6):931–936. [PubMed] [Google Scholar]
- 61.Bowie MB, McKnight KD, Kent DG, McCaffrey L, Hoodless PA, Eaves CJ. Hematopoietic stem cells proliferate until after birth and show a reversible phase-specific engraftment defect. J Clin Invest. 2006;116(10):2808–2816. doi: 10.1172/JCI28310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Iscove NN, Nawa K. Hematopoietic stem cells expand during serial transplantation in vivo without apparent exhaustion. Curr Biol. 1997;7(10):805–808. doi: 10.1016/s0960-9822(06)00341-1. [DOI] [PubMed] [Google Scholar]
- 63.Audet J, Miller CL, Eaves CJ, Piret JM. Common and distinct features of cytokine effects on hematopoietic stem and progenitor cells revealed by dose-response surface analysis. Biotechnol Bioeng. 2002;80(4):393–404. doi: 10.1002/bit.10399. [DOI] [PubMed] [Google Scholar]
- 64.Lecault V, Vaninsberghe M, Sekulovic S, et al. High-throughput analysis of single hematopoietic stem cell proliferation in microfluidic cell culture arrays. Nat Methods. 2011;8(7):581–586. doi: 10.1038/nmeth.1614. [DOI] [PubMed] [Google Scholar]
- 65.Wohrer S, Knapp DJ, Copley MR, et al. Distinct stromal cell factor combinations can separately control hematopoietic stem cell survival, proliferation, and self-renewal. Cell Reports. 2014;7(6):1956–1967. doi: 10.1016/j.celrep.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sieburg HB, Rezner BD, Muller-Sieburg CE. Predicting clonal self-renewal and extinction of hematopoietic stem cells. Proc Natl Acad Sci USA. 2011;108(11):4370–4375. doi: 10.1073/pnas.1011414108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kent DG, Dykstra BJ, Cheyne J, Ma E, Eaves CJ. Steel factor coordinately regulates the molecular signature and biologic function of hematopoietic stem cells. Blood. 2008;112(3):560–567. doi: 10.1182/blood-2007-10-117820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fleming WH, Alpern EJ, Uchida N, Ikuta K, Spangrude GJ, Weissman IL. Functional heterogeneity is associated with the cell cycle status of murine hematopoietic stem cells. J Cell Biol. 1993;122(4):897–902. doi: 10.1083/jcb.122.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ross EA, Anderson N, Micklem HS. Serial depletion and regeneration of the murine hematopoietic system. Implications for hematopoietic organization and the study of cellular aging. J Exp Med. 1982;155(2):432–444. doi: 10.1084/jem.155.2.432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jordan CT, Astle CM, Zawadzki J, Mackarehtschian K, Lemischka IR, Harrison DE. Long-term repopulating abilities of enriched fetal liver stem cells measured by competitive repopulation. Exp Hematol. 1995;23(9):1011–1015. [PubMed] [Google Scholar]
- 71.Pawliuk R, Eaves C, Humphries RK. Evidence of both ontogeny and transplant dose-regulated expansion of hematopoietic stem cells in vivo. Blood. 1996;88(8):2852–2858. [PubMed] [Google Scholar]
- 72.Bowie MB, Kent DG, Dykstra B, et al. Identification of a new intrinsically timed developmental checkpoint that reprograms key hematopoietic stem cell properties. Proc Natl Acad Sci USA. 2007;104(14):5878–5882. doi: 10.1073/pnas.0700460104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bowie MB, Kent DG, Copley MR, Eaves CJ. Steel factor responsiveness regulates the high self-renewal phenotype of fetal hematopoietic stem cells. Blood. 2007;109(11):5043–5048. doi: 10.1182/blood-2006-08-037770. [DOI] [PubMed] [Google Scholar]
- 74.Mochizuki-Kashio M, Mishima Y, Miyagi S, et al. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 2011;118(25):6553–6561. doi: 10.1182/blood-2011-03-340554. [DOI] [PubMed] [Google Scholar]
- 75.Xie H, Xu J, Hsu JH, et al. Polycomb repressive complex 2 regulates normal hematopoietic stem cell function in a developmental-stage-specific manner. Cell Stem Cell. 2014;14(1):68–80. doi: 10.1016/j.stem.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim I, Saunders TL, Morrison SJ. Sox17 dependence distinguishes the transcriptional regulation of fetal from adult hematopoietic stem cells. Cell. 2007;130(3):470–483. doi: 10.1016/j.cell.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335(6073):1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Copley MR, Babovic S, Benz C, et al. The Lin28b-let-7-Hmga2 axis determines the higher self-renewal potential of fetal haematopoietic stem cells. Nat Cell Biol. 2013;15(8):916–925. doi: 10.1038/ncb2783. [DOI] [PubMed] [Google Scholar]
- 79.Park IK, Qian D, Kiel M, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423(6937):302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 80.Hock H, Meade E, Medeiros S, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004;18(19):2336–2341. doi: 10.1101/gad.1239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cheng T, Rodrigues N, Shen H, et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287(5459):1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 82.van Os R, Kamminga LM, Ausema A, et al. A Limited role for p21Cip1/Waf1 in maintaining normal hematopoietic stem cell functioning. Stem Cells. 2007;25(4):836–843. doi: 10.1634/stemcells.2006-0631. [DOI] [PubMed] [Google Scholar]
- 83.Yang Q, Esplin B, Borghesi L. E47 regulates hematopoietic stem cell proliferation and energetics but not myeloid lineage restriction. Blood. 2011;117(13):3529–3538. doi: 10.1182/blood-2010-07-297689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2(5):484–496. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hock H, Hamblen MJ, Rooke HM, et al. Gfi-1 restricts proliferation and preserves functional integrity of haematopoietic stem cells. Nature. 2004;431(7011):1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 86.Ye M, Zhang H, Amabile G, et al. C/EBPa controls acquisition and maintenance of adult haematopoietic stem cell quiescence. Nat Cell Biol. 2013;15(4):385–394. doi: 10.1038/ncb2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hasemann MS, Lauridsen FK, Waage J, et al. C/EBPα is required for long-term self-renewal and lineage priming of hematopoietic stem cells and for the maintenance of epigenetic configurations in multipotent progenitors. PLoS Genet. 2014;10(1):e1004079. doi: 10.1371/journal.pgen.1004079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ikuta K, Uchida N, Friedman J, Weissman IL. Lymphocyte development from stem cells. Annu Rev Immunol. 1992;10:759–783. doi: 10.1146/annurev.iy.10.040192.003551. [DOI] [PubMed] [Google Scholar]
- 89.Montecino-Rodriguez E, Dorshkind K. B-1 B cell development in the fetus and adult. Immunity. 2012;36(1):13–21. doi: 10.1016/j.immuni.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Palmer DB. The effect of age on thymic function. Front Immunol. 2013;4:316. doi: 10.3389/fimmu.2013.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ghosn EE, Yamamoto R, Hamanaka S, et al. Distinct B-cell lineage commitment distinguishes adult bone marrow hematopoietic stem cells. Proc Natl Acad Sci USA. 2012;109(14):5394–5398. doi: 10.1073/pnas.1121632109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Muller-Sieburg CE, Cho RH, Karlsson L, Huang JF, Sieburg HB. Myeloid-biased hematopoietic stem cells have extensive self-renewal capacity but generate diminished lymphoid progeny with impaired IL-7 responsiveness. Blood. 2004;103(11):4111–4118. doi: 10.1182/blood-2003-10-3448. [DOI] [PubMed] [Google Scholar]
- 93.Yamamoto R, Morita Y, Ooehara J, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154(5):1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 94.Lu R, Neff NF, Quake SR, Weissman IL. Tracking single hematopoietic stem cells in vivo using high-throughput sequencing in conjunction with viral genetic barcoding. Nat Biotechnol. 2011;29(10):928–933. doi: 10.1038/nbt.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Naik SH, Perié L, Swart E, et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496(7444):229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- 96.Verovskaya E, Broekhuis MJ, Zwart E, et al. Heterogeneity of young and aged murine hematopoietic stem cells revealed by quantitative clonal analysis using cellular barcoding. Blood. 2013;122(4):523–532. doi: 10.1182/blood-2013-01-481135. [DOI] [PubMed] [Google Scholar]
- 97.Grosselin J, Sii-Felice K, Payen E, Chretien S, Tronik-Le Roux D, Leboulch P. Arrayed lentiviral barcoding for quantification analysis of hematopoietic dynamics. Stem Cells. 2013;31(10):2162–2171. doi: 10.1002/stem.1383. [DOI] [PubMed] [Google Scholar]
- 98.Muller-Sieburg CE, Sieburg HB. The GOD of hematopoietic stem cells: a clonal diversity model of the stem cell compartment. Cell Cycle. 2006;5(4):394–398. doi: 10.4161/cc.5.4.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gekas C, Graf T. CD41 expression marks myeloid-biased adult hematopoietic stem cells and increases with age. Blood. 2013;121(22):4463–4472. doi: 10.1182/blood-2012-09-457929. [DOI] [PubMed] [Google Scholar]
- 100.Oguro H, Ding L, Morrison SJ. SLAM family markers resolve functionally distinct subpopulations of hematopoietic stem cells and multipotent progenitors. Cell Stem Cell. 2013;13(1):102–116. doi: 10.1016/j.stem.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sanjuan-Pla A, Macaulay IC, Jensen CT, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013;502(7470):232–236. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- 102.Sudo K, Ema H, Morita Y, Nakauchi H. Age-associated characteristics of murine hematopoietic stem cells. J Exp Med. 2000;192(9):1273–1280. doi: 10.1084/jem.192.9.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Adolfsson J, Månsson R, Buza-Vidas N, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121(2):295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 104.Chi AW, Chavez A, Xu L, et al. Identification of Flt3+CD150− myeloid progenitors in adult mouse bone marrow that harbor T lymphoid developmental potential. Blood. 2011;118(10):2723–2732. doi: 10.1182/blood-2010-09-309989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Forsberg EC, Serwold T, Kogan S, Weissman IL, Passegué E. New evidence supporting megakaryocyte-erythrocyte potential of flk2/flt3+ multipotent hematopoietic progenitors. Cell. 2006;126(2):415–426. doi: 10.1016/j.cell.2006.06.037. [DOI] [PubMed] [Google Scholar]
- 106.Luc S, Luis TC, Boukarabila H, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13(4):412–419. doi: 10.1038/ni.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kawamoto H, Ikawa T, Masuda K, Wada H, Katsura Y. A map for lineage restriction of progenitors during hematopoiesis: the essence of the myeloid-based model. Immunol Rev. 2010;238(1):23–36. doi: 10.1111/j.1600-065X.2010.00959.x. [DOI] [PubMed] [Google Scholar]
- 108.Harrison DE, Zhong RK. The same exhaustible multilineage precursor produces both myeloid and lymphoid cells as early as 3-4 weeks after marrow transplantation. Proc Natl Acad Sci USA. 1992;89(21):10134–10138. doi: 10.1073/pnas.89.21.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miyamoto T, Akashi K. Lineage promiscuous expression of transcription factors in normal hematopoiesis. Int J Hematol. 2005;81(5):361–367. doi: 10.1532/ijh97.05003. [DOI] [PubMed] [Google Scholar]
- 110.Månsson R, Hultquist A, Luc S, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26(4):407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 111.Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135(6):1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 112.Sun J, Ramos A, Chapman B, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514(7522):322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kataoka K, Sato T, Yoshimi A, et al. Evi1 is essential for hematopoietic stem cell self-renewal, and its expression marks hematopoietic cells with long-term multilineage repopulating activity. J Exp Med. 2011;208(12):2403–2416. doi: 10.1084/jem.20110447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gazit R, Mandal PK, Ebina W, et al. Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J Exp Med. 2014;211(7):1315–1331. doi: 10.1084/jem.20130428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Schulz C, Gomez Perdiguero E, Chorro L, et al. A lineage of myeloid cells independent of Myb and hematopoietic stem cells. Science. 2012;336(6077):86–90. doi: 10.1126/science.1219179. [DOI] [PubMed] [Google Scholar]
- 116.Ginhoux F, Greter M, Leboeuf M, et al. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010;330(6005):841–845. doi: 10.1126/science.1194637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hoeffel G, Wang Y, Greter M, et al. Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J Exp Med. 2012;209(6):1167–1181. doi: 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Perdiguero EG, Klapproth K, Schulz C, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518(7540):547–551. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bendall SC, Simonds EF, Qiu P, et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science. 2011;332(6030):687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guo G, Luc S, Marco E, et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell. 2013;13(4):492–505. doi: 10.1016/j.stem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bendall SC, Davis KL, Amir el-AD, et al. Single-cell trajectory detection uncovers progression and regulatory coordination in human B cell development. Cell. 2014;157(3):714–725. doi: 10.1016/j.cell.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Riddell J, Gazit R, Garrison BS, et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell. 2014;157(3):549–564. doi: 10.1016/j.cell.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]