Abstract
Autophagy is an evolutionally conserved lysosomal pathway used to degrade and turn over long-lived proteins and cytoplasmic organelles. Since autophagy was discovered, it has been thought to act as a pro-survival response to several stresses, especially starvation, at the cell and organism levels by providing recycled metabolic substrates to maintain energy homeostasis. However, several recent studies suggest that autophagy also plays a pro-death role through an autophagic cell death pathway mostly at the cellular level. The mechanism by which autophagy could perform these seemingly opposite roles as a pro-survival and a pro-death mechanism remained elusive until recently Using C. elegans as a model system, we found that physiological levels of autophagy promote optimal survival of C. elegans during starvation, but either insufficient or excessive levels of autophagy render C. elegans starvation-hypersensitive. Furthermore, we found that muscarinic acetylcholine receptor signaling is important in modulating the level of autophagy during starvation, perhaps through DAP kinase and RGS-2. Our recent study provides in vivo evidence that levels of autophagy are critical in deciding its promotion of either survival or death: Physiological levels of autophagy are pro-survival, whereas insufficient or excessive levels of autophagy are pro-death.
Keywords: autophagy, muscarinic acetylcholine receptor signaling, starvation, Caenorhabditis elegans, MAP kinase, cancer
Dual Roles Of Autophagy as a Prosurvival and a Prodeath Mechanism
Autophagy is a so-called “self-eating” system responsible for degrading long-lived proteins and cytoplasmic organelles, the products of which are recycled to generate macromolecules and ATP so as to maintain cellular homeostasis.1 This ability makes autophagy a good candidate for a survival mechanism in response to several stresses, such as damaged mitochondria, protein aggregation, pathogens, and nutrient starvation.1, 2 The best characterized pro-survival function of autophagy is as a starvation response. When the supply of external nutrients is limited by nutrient deprivation, cells can induce autophagy, thereby generating a source of metabolic substrates to sustain the cellular activity needed for survival.2-4 This autophagic response to starvation has been well-studied in various organisms including yeast, worms, flies, and mice.5-8 These lines of evidence indicate that autophagy is a pro-survival mechanism for both cells and organisms.
However, several recent studies suggest that autophagy also functions as a pro-death mechanism at the cellular level. It has been shown that RNAi knockdown of essential autophagy genes inhibits type II programmed cell death (autophagic cell death) in a variety of cell types under different conditions. 2,3,9 At the organismal level, the fact that excessive autophagy is associated with severe wasting of denervated myofibers in runx1 mutant mice suggests that autophagy could be harmful to an organism,10 however there has been no direct evidence that autophagy could contribute to the actual death of multicellular organisms until recently.
How is it that autophagy performs these seemingly opposite roles with respect to survival and death? In recent studies,11 Levine and colleagues suggested the intriguing possibility that depending on its level, autophagy could act in either a pro-survival or a pro-death role at the cellular level. To test this possibility at the organismal level, we used C. elegans as a model system.12 We found that RNAi of either bec-1 or atg-7 (the C. elegans ortholog of Beclin 1/Atg6 and Atg7, respectively) reduced autophagy in the pharyngeal muscle and decreased survival of wild-type worms after starvation, suggesting that autophagy is required for optimal survival of worms during starvation. The addition of food could reverse the pro-death effect of bec-1 RNAi treatment in wild-type worms during starvation, suggesting that a major defect in bec-1(RNAi) worms was lack of nutrients, rendering worms unable to maintain basal cellular activity. In fact, we found that bec-1 RNAi treatment decreased pharyngeal pumping rates, suggesting that autophagy is required to maintain the basal activity of the pharynx during starvation. Taken together, these data suggest a pro-survival role of autophagy in C. elegans during starvation.
Previously we showed that starvation activates a muscarinic acetylcholine receptor →MAP kinase signaling pathway in pharyngeal muscle and that gpb-2 mutants, in which this starvation signal is overactivated, are hypersensitive to starvation, due in part to malfunction of pharyngeal muscle.13 We hypothesized that overactivated starvation signaling (muscarinic signaling) in gpb-2 mutants induces unrestrained autophagy, which in turn, causes damage to the pharyngeal muscle and eventually contributes to death. We found that autophagy is indeed excessively induced in the pharyngeal muscle of gpb-2 mutants following starvation and that reduction of autophagy by either bec-1 or atg-7 RNAi treatment rescued pharyngeal muscle function and reduced starvation-induced death of gpb-2 mutants, supporting our hypothesis that excessive autophagy plays a pro-death role in C. elegans during starvation. Our results provide in vivo evidence that levels of autophagy are critical at the organismal level in deciding between the pro-survival and pro-death roles.
Bridge Between Starvation and Autophagy
While progress has been made on autophagy-inhibiting signaling pathways at the cellular levels, 14-17 the pathways acting at the organismal level have not been as extensively characterized.18-21 Our results demonstrate that the muscarinic signaling acts as an autophagy-inducing signaling pathway in the multicellular organism C. elegans during starvation. Our results also suggest that muscarinic signaling induced autophagy through DAP kinase and RGS-2.12 These results taken together with the previous study showing that starvation activates muscarinic signaling in C. elegans13 suggest a role of the muscarinic acetylcholine pathway as a bridge between starvation and autophagy (Fig. 1).
Figure 1.
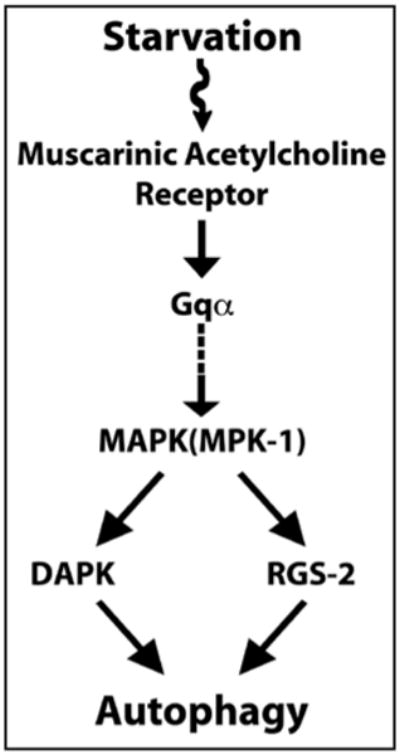
Muscarinic acetylcholine receptor signaling functions as an autophagy-inducing signaling pathway. In the simplified model shown, starvation activates MAPK (MPK-1, C. elegans ortholog of mammalian ERK) through the muscarinic acetylcholine receptor signaling pathway. Activated MAPK positively regulates autophagy, at least in part, through DAP kinase and RGS-2.
Implications for Dual Roles of Autophagy in Tumorigenesis
Based on the ability of autophagy to promote cell survival in response to metabolic stress, it has been suggested that autophagy may contribute to tumor development by providing an energy source to tumor cells located far from the blood supply where nutrients are extremely limited.2,22-24 Recent findings by White and colleagues showing that autophagy promotes cell survival in solid tumors support this hypothesis.25 In this regard, our observation that the overactivation of the Ras signaling pathway induced autophagy, combined with the fact that Ras signaling is frequently overactivated in various cancers, leads to the intriguing hypothesis that autophagy activated by overactivated Ras signaling may provide a survival advantage to cancer cells in the central area of the large tumor masses until vascular support can be established.12
Autophagy is also believed to suppress tumors because monoal-lelic loss of beclin 1 is frequently associated with human cancer, and because mice with heterozygous disruption of beclin 1 are tumor-prone.26 Recent findings support the view that autophagy acts as a tumor suppressor mechanism by limiting genome damage and chromosomal instability. 27,28
These two seemingly contradictory functions of autophagy suggest the possibility that autophagy can act either as cancers friend or foe, depending on the progression of the tumor. Until vascular support is established (and thus nutrient limitation is resolved), autophagy provides a temporary survival advantage to tumor cells where they suffer from metabolic stress. After vascularization, autophagy instead suppresses tumor progression by limiting genome damage and chromosomal instability, and possibly by causing autophagic cell death. At this stage, other selective pressures drive cancer cells to gain additional mutations that impair the autophagy process and further tumor progression. In fact, recent studies 18,29,30 showing that well-known tumor suppressor genes (DAPK1 and p19ARF) can induce autophagy suggest the possibility that mutations in these tumor suppressor genes may decrease the level of autophagy, thereby inhibiting the tumor suppressor activity of autophagy and leading to further tumor progression. With respect to this possibility, it would be interesting to examine the timing of mutations that can affect the autophagy process during tumor progression.
Acknowledgments
We thank B. Levine for helpful discussions and C. Glynn for critical reading of the manuscript. C. Kang thanks M.S. Kim for unfailing support and encouragement. This work was supported by research grant HL46154 from the U.S. Public Heath Service.
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest. 2005;115:2679–88. doi: 10.1172/JCI26390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eskelinen EL. Doctor Jekyll and Mister Hyde: autophagy can promote both cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1468–72. doi: 10.1038/sj.cdd.4401721. [DOI] [PubMed] [Google Scholar]
- 4.Zong WX, Thompson CB. Necrotic death as a cell fate. Genes Dev. 2006;20:1–15. doi: 10.1101/gad.1376506. [DOI] [PubMed] [Google Scholar]
- 5.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 6.Melendez A, Talloczy Z, Seaman M, Eskelinen EL, Hall DH, Levine B. Autophagy genes are essential for dauer development and life-span extension in C. elegans Science. 2003;301:1387–91. doi: 10.1126/science.1087782. [DOI] [PubMed] [Google Scholar]
- 7.Scott RC, Schuldiner O, Neufeld TP. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev Cell. 2004;7:167–78. doi: 10.1016/j.devcel.2004.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–6. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- 9.Nelson DA, White E. Exploiting different ways to die. Genes Dev. 2004;18:1223–6. doi: 10.1101/gad.1212404. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Blagden C, Fan J, Nowak SJ, Taniuchi I, Littman DR, Burden SJ. Runx1 prevents wasting, myofibrillar disorganization, and autophagy of skeletal muscle. Genes Dev. 2005;19:1715–22. doi: 10.1101/gad.1318305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Kang C, You YJ, Avery L. Dual roles of autophagy in the survival of Caenorhabditis elegans during starvation. Genes Dev. 2007;21:2161–71. doi: 10.1101/gad.1573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.You YJ, Kim J, Cobb M, Avery L. Starvation activates MAP kinase through the muscarinic acetylcholine pathway in Caenorhabditis elegans pharynx. Cell Metab. 2006;3:237–45. doi: 10.1016/j.cmet.2006.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gingras AC, Raught B, Sonenberg N. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 2001;15:807–26. doi: 10.1101/gad.887201. [DOI] [PubMed] [Google Scholar]
- 15.Levine AJ, Feng Z, MakTW, You H, Jin S. Coordination and communication between the p53 and IGF-1-AKT-TOR signal transduction pathways. Genes Dev. 2006;20:267–75. doi: 10.1101/gad.1363206. [DOI] [PubMed] [Google Scholar]
- 16.Meijer AJ, Codogno P. Regulation and role of autophagy in mammalian cells. Int J Biochem Cell Biol. 2004;36:2445–62. doi: 10.1016/j.biocel.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Codogno P, Meijer AJ. Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ. 2005;12(Suppl 2):1509–18. doi: 10.1038/sj.cdd.4401751. [DOI] [PubMed] [Google Scholar]
- 18.Inbal B, Bialik S, Sabanay I, Shani G, Kimchi A. DAP kinase and DRP-1 mediate membrane blebbing and the formation of autophagic vesicles during programmed cell death. J Cell Biol. 2002;157:455–68. doi: 10.1083/jcb.200109094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pattingre S, Bauvy C, Codogno P. Amino acids interfere with the ERK1/2-dependent control of macroautophagy by controlling the activation of Raf-1 in human colon cancer HT-29 cells. J Biol Chem. 2003;278:16667–74. doi: 10.1074/jbc.M210998200. [DOI] [PubMed] [Google Scholar]
- 20.Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol. 2007;170:75–86. doi: 10.2353/ajpath.2007.060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talloczy Z, Jiang W, Virgin HW, IV, Leib DA, Scheuner D, Kaufman RJ, Eskelinen EL, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2alpha kinase signaling pathway. Proc Natl Acad Sci USA. 2002;99:190–5. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–34. doi: 10.1038/nrc1692. [DOI] [PubMed] [Google Scholar]
- 23.Levine B. Unraveling the role of autophagy in cancer. Autophagy. 2006;2:65–6. doi: 10.4161/auto.2.2.2457. [DOI] [PubMed] [Google Scholar]
- 24.Botti J, Djavaheri-Mergny M, Pilatte Y, Codogno P. Autophagy signaling and the cogwheels of cancer. Autophagy. 2006;2:67–73. doi: 10.4161/auto.2.2.2458. [DOI] [PubMed] [Google Scholar]
- 25.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, Mukherjee C, Shi Y, Gelinas C, Fan Y, Nelson DA, Jin S, White E. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, Cattoretti G, Levine B. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, White E. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, Chen G, Jin S, White E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gozuacik D, Kimchi A. DAPk protein family and cancer. Autophagy. 2006;2:74–9. doi: 10.4161/auto.2.2.2459. [DOI] [PubMed] [Google Scholar]
- 30.Reef S, Zalckvar E, Shifman O, Bialik S, Sabanay H, Oren M, Kimchi A. A short mitochondrial form of p19ARF induces autophagy and caspase-independent cell death. Mol Cell. 2006;22:463–75. doi: 10.1016/j.molcel.2006.04.014. [DOI] [PubMed] [Google Scholar]


