Abstract
Single-stranded silencing RNAs (ss-siRNAs) are chemically modified single-stranded oligonucleotides that can function through the cellular RNA interference (RNAi) machinery to modulate gene expression. Because their invention is recent, few studies have appeared describing their use and the potential of ss-siRNAs as a platform for controlling gene expression remains largely unknown. Using oligonucleotides to modulate splicing is an important area for therapeutic development and we tested the hypothesis that ss-siRNAs targeting splice sites might also be capable of directing increased production of therapeutically promising protein isoforms. Here we observe that ss-siRNAs alter splicing of dystrophin. Altered splicing requires a seed sequence complementarity to the target and expression of the RNAi factor argonaute 2. These results demonstrate that ss-siRNAs can be used to modulate splicing, providing another option for therapeutic development programs that aim to increase production of key protein isoforms. Splicing is a classical nuclear process and our data showing that it can be modulated through the action of RNA and RNAi factors offers further evidence that RNAi can take place in mammalian cell nuclei.
Introduction
RNA interference (RNAi) in mammalian cells is usually associated with inhibition of gene expression in the cytoplasm by duplex RNAs. This restrictive view of the potential impact of RNAi is slowly changing as evidence accumulates that RNA can also regulate nuclear processes [1–6]. Our laboratory [7] and others [8] have shown that duplex RNAs that are complementary to gene promoters can be used to regulate transcription of target genes. The mechanism of action appears to involve recognition of RNA transcripts that overlap the gene promoters rather than any direct association with DNA. Several reports have suggested that endogenous miRNAs with complementarity to gene promoters may have the capacity to regulate transcription [7,9–11].
Many researchers have assumed that RNAi factors are localized to the cytoplasm but evidence exists suggesting that they are also present and active in mammalian cell nuclei [12–14]. Relative to the cytoplasm, the amounts of most nuclear RNAi factors are substantial, in the range of 20%–40%. The exceptions are RNA loading factors TRAX, Translin, and Hsp90, which are absent from the nucleus [14], suggesting that strand loading is a cytoplasmic event.
The presence of RNAi factors in the nucleus and their involvement in the regulation of transcription suggests the potential for the RNAi machinery to be programmed by small RNAs to control other nuclear processes. Alternative splicing is a classical nuclear process that affects many genes and leads to the production of proteins with different function. In some cases, protein isoforms exist that have the potential to alleviate disease, but to accomplish this goal their production must be enhanced. The potential for increased isoform production to alleviate some diseases has led to testing antisense oligonucleotides that target sequences near splice sites for their ability to shift the processing of pre-mRNA toward mature mRNAs that code for therapeutically valuable proteins. This approach has been successful, leading to several drug candidates and multiple clinical trials [15–17].
Several laboratories have demonstrated the potential for small RNAs to act in concert with the RNAi machinery to direct alternative splicing [18]. Previously, we identified duplex RNAs that efficiently redirected splicing of an engineered luciferase reporter gene, survival motor neuron 2 (SMN2), and dystrophin [19]. The mechanism of action involved argonaute 2 (Ago2), a central factor in RNAi. Other laboratories have implicated Ago1 in controlling splicing through by altering histone modifications and affecting transcription [20–22].
Another widely held assumption in the past has been that efficient RNAi requires duplex RNA. Recently, however, chemically modified single stranded silencing RNAs (ss-siRNAs) have been demonstrated to silence gene expression through the RNA interference (RNAi) pathway [23,24]. ss-siRNAs that target disease-causing trinucleotide repeats have been shown to allele-selectively inhibit expression of the mutant forms of huntingtin [25,26], ataxin-3 [27], and atrophin-1 [28]. An ss-siRNA complementary to an antisense transcript that overlaps the promoter of progesterone receptor has been shown to reduce recruitment of RNA polymerase 2 and block expression of progesterone receptor [29].
Single stranded siRNAs offer a potentially significant new approach to nucleic acid therapeutics because they combine the favorable pharmacology properties of single stranded oligonucleotides with the robust silencing produced by RNAi. We have previously shown that small duplex RNAs that are complementary to sequences near splice junctions can modulate gene expression. We now show that ss-siRNAs can also modulate splicing. This demonstration that single stranded oligonucleotides functioning through the RNAi pathway can control splicing provides a new option for developing nucleic acids to modulate the expression of therapeutic protein isoforms.
Materials and Methods
Cell culture and transfections
Duplex RNAs were synthesized by Integrated DNA Technologies Inc. To silence Ago2 expression, we used a siRNA pool targeting four different regions of Ago2 mRNA. ss-siRNAs were synthesized by Isis Pharmaceuticals Inc. and reconstituted in nuclease-free water.
Patient-derived Duchenne muscular dystrophy (DMD) fibroblast cells (GM03429; Coriell Cell Repositories) were cultured in minimum essential medium Eagle (MEM; Sigma, M4655) supplemented with 15% (v/v) fetal bovine serum and 1% MEM nonessential amino acids (Sigma, M7145). Cells were plated in six-well plates at 100,000 cells per well in supplemented MEM media 2 days prior to transfection. Cells were transfected with siRNAs using RNAiMAX (Invitrogen). Unless indicated otherwise, total RNA was isolated 24 hours after transfection with Trizol (Invitrogen) for reverse transcriptase PCR (RT-PCR). For double-transfection experiments, the first transfection was performed as described above. Media were changed 24 hours later and the second transfection was carried out on the next day. Total RNA was isolated after 24 hours of second transfection for RT-PCR analysis.
RT–PCR
To generate cDNA, total RNA was extracted and treated with DNase I (Worthington Biochemical) at 25 μM for 10 minutes. Reverse transcription was performed using high-capacity reverse transcription kit (Applied Biosystems) according to the manufacturer's protocol. Generally, 2.0 μg of total RNA was used per 20 μL of reaction mixture. Primary PCR was performed on a PCR machine (BioRad, MJ Mini) using iTaq SYBR Green Supermix (BioRad) using the following primers 8 forward primer (FP): 5′- GACAGATCTGTTGAGAAATGGCGGCGTT-3′; 83 reverse primer (RP): 5′-CCGTAATGATTGTTCTAGCCTCTTGATTGC-3′). The primary PCR cycles are as follows: 50°C for 2 minutes; 95°C for 3 minutes; (95°C for 30 seconds; 60°C for 30 seconds; 72°C for 35 seconds)×30 cycles. Then, nested PCR was carried out with1 μL of primary PCR product as template using an inner primer set (61 FP: 5′-TCAGTGGCTAACAGAAGCTGAACAGTTT-3′ and 83 RP) with cycles as follows: 95°C for 3 minutes; (95°C for 30 seconds; 61°C for 30 seconds; 72°C for 40 seconds) for 35 cycles.
Sequence analysis
RT-PCR products were excised from agarose gels and extracted using a Promega Wizard SV Gel and PCR Clean-Up System kit. The McDermott Center Sequencing Core Facility at University of Texas Southwestern carried out direct DNA sequencing.
Splicing correction analysis
All of the PCR products were separated on a 1.2% agarose gel and visualized on an AlphaImager. The bands were quantified using ImageJ software (National Institutes of Health, http://rsb.info.nih.gov/ij). Splicing correction on DMD fibroblast cells was calculated as a percentage of the total amount of spliced mRNA: correct mRNA×100/(correct mRNA+aberrant mRNA).
Purification and western blot analysis of cytoplasmic and nuclear fractions
Purification of DMD fibroblast cytoplasmic and nuclear fractions and western blot analysis was performed as described previously [14]. For comparing nuclear and cytoplasmic fractions by western blot, the same cell equivalents of extract were separated by electrophoresis. Blocked western blot membranes (Hybond-C Extra, GE Healthcare Life Sciences) were incubated with the following primary antibodies for 16 hours at 4°C in PBST (phosphate-buffered saline+0.05% TWEEN-20)+5% milk with rocking: anti-Ago2 at 1:1,000 (Abcam, ab57113), anti-Calreticulin at 1:1,000 (Cell Signalling, 2891S), anti-Histone H3 at 1:10,000 (Abcam, ab1791), anti-Lamin A/C at 1:1,500 (Abcam, ab8984), anti-glyceraldehyde 3-phosphate dehydrogenase at 1:600 (Abcam, ab9484), anti-RNA polymerase 2 at 1:4,000 (Millipore, 05-623).
Results
Structure of ss-siRNAs
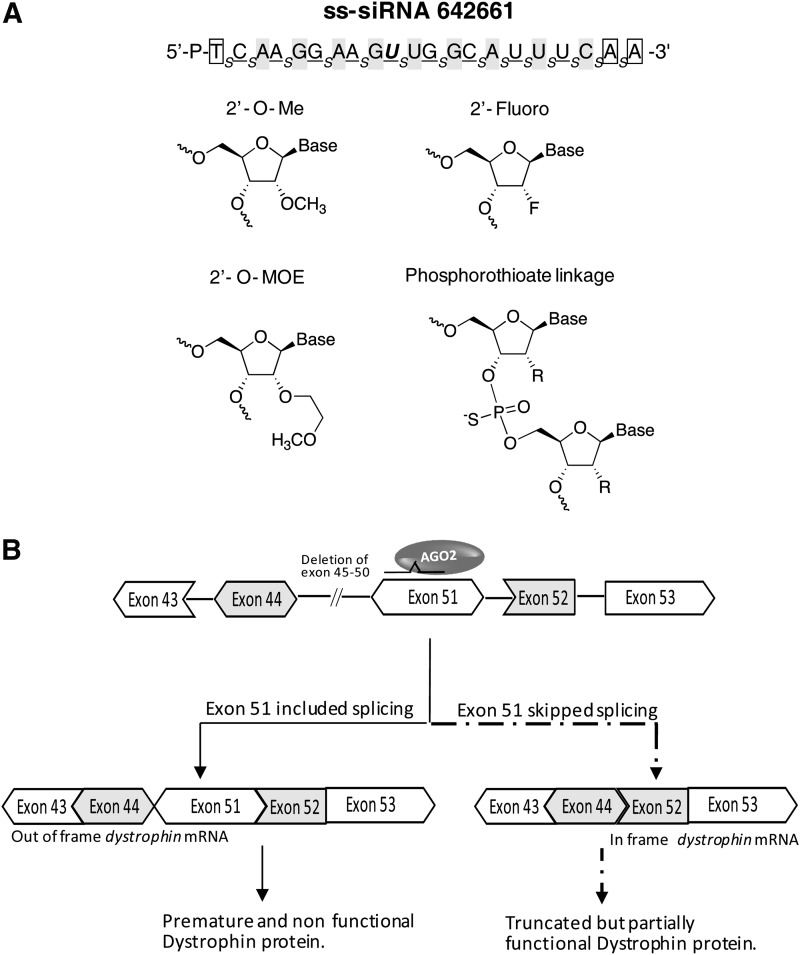
To act through the RNAi pathway inside cells RNA must be chemically modified to be stable to degradation by nucleases. These modifications, however, must be compatible with the ability to bind RNAi proteins and inhibit protein translation through the RNAi-induced silencing complex. Chemical modifications incorporated into the ss-siRNAs include 2′-O-methyl (2′-O-me), 2′-Fluoro, and 2′-O-methoxyethyl nucleosides (Fig. 1A) [23]. Internucleotide linkages are either phosphodiester or phosphorothioate, with the phosphorothioate substitutions contributing to nucleolytic stability. For in vivo applications, ss-siRNAs require a vinyl phosphonate at the 5′ terminus. All studies presented here, however, were done using cultured patient derived cells. ss-siRNAs with 5′-phosphate groups are active in cultured cells and we used 5′-phosphate siRNAs in our experiments because their synthesis is simpler.
FIG. 1.
Design and targeting of single stranded silencing RNAs (ss-siRNAs). (A) ss-siRNA 642661 possess a typical pattern of chemical modifications. Positions of chemical modifications within the sequence are indicated as follows: 2′-O-me (methyl) modified base is background shaded; 2′-O-MOE (methoxyethyl) modified base is boxed; 2′-Fluoro modified base is underlined; mismatched base is bold and italicized; phosphorothioate (PS) linkage is shown as subscript (s). The terminal thymidine has a 5′- phosphate. All other linkages are phosphate. (B) Schematic showing splicing of dystrophin pre-mRNA and how splicing might be modified by a RNA acting through the RNA interference (RNAi) pathway and argonaute 2 (Ago2).
The ss-siRNAs used in our studies target exon 51 of dystrophin (Fig. 1B; Table 1). This target was chosen because antisense oligonucleotides [30,31] and duplex RNAs [19] that are complementary to the exon cause exon 51 to be skipped. Clinical trials have suggested that antisense-mediated exon skipping might be an attractive therapeutic option [32–35], although the refinements to the approach may be needed to increase the likelihood of optimal clinical success [36].
Table 1.
Silencing RNAs Tested in the Experiment to Correct Splicing of Duchenne's Muscular Dystrophy Within Patient-Derived Fibroblast Cells
| Name | Antisense sequence (5′ to 3′) | ss or dsRNA | Target gene | Position of mismatch (if any) |
|---|---|---|---|---|
| CM | 5′-GCUAUACCAGCGUCGUCAUdTdT | ds | Negative control | |
| 66 | 5′-AAGGAAGAUGCCAUUUCUAdTdT | ds | Exon 51, DMD | 11 |
| 68/10 | 5′-UCAAGGAAGUUGGCAUUUCdTdT | ds | Exon 51, DMD | 10 |
| 642661 | 5′-P-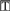 sCsAAsGGsAAsGUsUGsGCsAsUsUsUsCs sCsAAsGGsAAsGUsUGsGCsAsUsUsUsCs s s
|
ss | Exon 51, DMD | 10 |
| 642662 | 5′-P-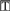 sAsGGsAAsGAsUGsCCsAUsUsUsCsUsAs sAsGGsAAsGAsUGsCCsAUsUsUsCsUsAs s s
|
ss | Exon 51, DMD | 11 |
| 642663 | 5′-P-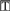 sGsAAsACsCUsCUsUAsCCsUsCsAsGsUs sGsAAsACsCUsCUsUAsCCsUsCsAsGsUs s s
|
ss | Negative control, Luciferase | |
| 642664 | 5′-P-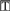 sAsACsUUsCAsGGsGUsCAsGCsUsUsGs sAsACsUUsCAsGGsGUsCAsGCsUsUsGs s s
|
ss | Negative control, EGFP | |
| 522247 | 5′-P-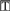 sUsAUsCUsAUsAAsUGsAUsCsAsGsGsUs sUsAUsCUsAUsAAsUGsAUsCsAsGsGsUs s s
|
ss | Negative control, PTEN | |
| 557429 | 5′-P-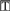 sCsAGsCUsGUsUGsCUsACsUsGsUsUsGs sCsAGsCUsGUsUGsCUsACsUsGsUsUsGs s s
|
ss | Negative control |
Modification positions within the sequence are indicated as follows: boxed, 2′-O-MOE (methoxyethyl); underlined, 2′-Fluoro; background shaded, 2′-O-me (methyl); bold and italicized, mismatched bases; subscripted “s,” phosphorothioate (PS) linkage; d, deoxyribose.
DMD, Duchenne's muscular dystrophy; dsRNA, double stranded RNA; EGFP, enhanced green fluorescent protein; ssRNA, single stranded RNA; PTEN, phosphatase and tensin homolog.
Skipping exon 51 removes a premature stop codon and leads to production of an alternate protein isoform. The alternate protein isoform is naturally produced in a less severe muscular dystrophy, Becker's muscular dystrophy, and is known to possess partial function. By increasing production of the dystrophin variant lacking exon 51 it is hoped that the symptoms of Duchenne muscular dystrophy can be alleviated so that they become more similar to Becker's muscular dystrophy, potentially resulting in a better quality of life and enhanced survival for patients.
Ago2 is a critical RNAi factor that cleaves RNA substrates when guide strands are perfectly complementary [37]. When an ss-siRNA is fully complementary to target it can engage Ago2 leading to cleavage of mRNA. However, structural and biochemical data indicate that RNAs with central mismatches can recognize complementary sequences but cannot cleave them [38]. To avoid potential complications from cleavage of mRNA targets, some ss-siRNAs were designed so that they were mismatched relative to their intended target sequence at positions in the center of molecule (Table 1).
Single stranded siRNAs mediate exon exclusion
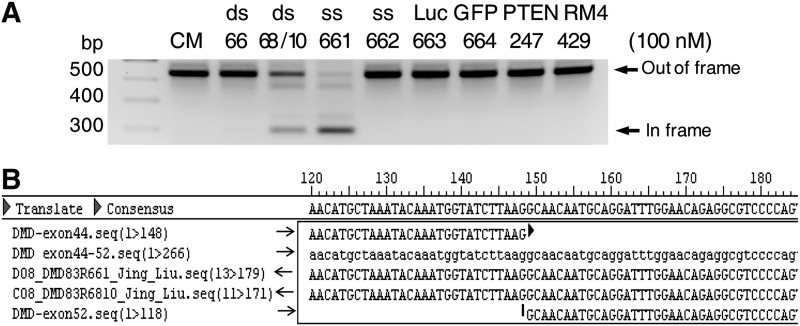
We introduced ss-siRNAs and analogous duplex RNAs (Table 1) into GM03429 patient-derived cells by transfection with cationic lipid. After harvesting, cellular RNA was analyzed by reverse transcriptase PCR to evaluate the effect of RNA addition on the relative production of dystrophin with exon 51 deleted (Fig. 2A). As observed previously [19], duplex RNA 68/10 with a mismatch at position 10 efficiently altered splicing to create product lacking exon 51. A noncomplementary duplex RNA had no effect, nor did duplex RNA 66 containing a mismatch at position 11.
FIG. 2.
An ss-siRNA corrects splicing of Duchenne's muscular dystrophy (DMD) within patient-derived fibroblast cells. (A) Polymerase chain reaction (PCR) amplification followed by gel electrophoresis to separate aberrant and correct splice products upon the addition of small RNAs (50 nM). (B) Sequencing PCR products to confirm that visualize products are due to skipping of exon 51. Patient-derived GM03429 fibroblast cells were used. CM, negative control duplex RNA; GFP, green fluorescent protein; Luc, luciferase; PTEN, phosphatase and tensin homolog; RM, negative control ss-siRNA.
We then tested ss-siRNAs with mismatches at positions 10 (ISIS 642661) or 11 (ISIS 642662). Similar to the duplex RNA, ss-siRNA ISIS 642661 with a position 10 mismatch altered splicing while ss-siRNA ISIS 642661 with a mismatch at position 11 did not. Four different negative control ss-siRNAs that lacked complementarity to dystrophin had no effect on splicing even though they shared the same pattern of chemically modified bases as found in the active compound. Sequencing confirmed that the observed amplified products were derived from RNA lacking exon 51 and had the predicted splice junctions (Fig. 2B).
Sequence specificity
The addition of oligonucleotides into cells has the potential to perturb interactions with splicing factors and possibly alter splicing through indirect off-target effects. During RNA, recognition, complementarity to a “seed sequence” (positions 2–8) within the small RNA is critical for activity. To test the specificity of ss-siRNA mediated production of the deleted exon 51 variant dystrophin we assayed ss-siRNAs with one, two, or three mismatches relative to dystrophin mRNA within the seed sequence in addition to the mismatch at position 10 (Table 2). None of these ss-siRNAs was active (Fig. 3A), supporting the conclusion that the ss-siRNA mediated splicing that we observe is due to an on-target interaction between the ss-siRNA and dystrophin mRNA.
Table 2.
Silencing RNAs Tested in the Experiment to Correct DMD Splicing Using Sequence-Specific siRNA
| Name | Antisense sequence (5′ to 3′) | ss or dsRNA | Target gene | Mismatch position |
|---|---|---|---|---|
| CM | 5′-GCUAUACCAGCGUCGUCAUdTdT | ds | Negative control | |
| 642661 | 5′-P-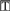 sCsAAsGGsAAsGUsUGsGCsAsUsUUsCs sCsAAsGGsAAsGUsUGsGCsAsUsUUsCs s s
|
ss | Exon 51, DMD | 10 |
| 678870 | 5′-UsCsAsAsGsGsAsAsGsAsUsGsGsCsAsUsUsUsCsU | ss, 2′-OMe | Exon 51, DMD | - |
| 6462662 | 5′-P-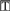 sAsGGsAAsGAsUGsCCsAUsUsUsCsUsAs sAsGGsAAsGAsUGsCCsAUsUsUsCsUsAs s s
|
ss | Exon 51, DMD | 11 |
| 678865 | 5′-P-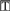 sCAAGGAAsGUsUGsGCsAsUsUsUsCs sCAAGGAAsGUsUGsGCsAsUsUsUsCs s s
|
ss, 3 less PS | Exon 51, DMD | 10 |
| 678866 | 5′-P-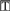 sCAAGGAAGUUGsGCsAsUsUsUsCs sCAAGGAAGUUGsGCsAsUsUsUsCs s s
|
ss, 5 less PS | Exon 51, DMD | 10 |
| 678867 | 5′-P-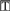 sCsAAsGCsAAsGUsUGsGCsAsUsUsUsCs sCsAAsGCsAAsGUsUGsGCsAsUsUsUsCs s s
|
ss | Exon 51, DMD | 6, 10 |
| 678868 | 5′-P-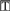 sCsAUsGCsAAsGUsUGsGCsAsUsUsUsCs sCsAUsGCsAAsGUsUGsGCsAsUsUsUsCs s s
|
ss | Exon 51, DMD | 4, 6, 10 |
| 678869 | 5′-P-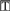 sCsAUsGCsAUsGUsUGsGCsAsUsUsUsCs sCsAUsGCsAUsGUsUGsGCsAsUsUsUsCs s s
|
ss | Exon 51, DMD | 4, 6, 8, 10 |
Modification positions within the sequence are indicated as follows: boxed, 2′-O-MOE (methoxyethyl); underlined, 2′-Fluoro; background shaded, 2′-O-me (methyl); bold and italicized, mismatched bases; subscripted “s,” phosphorothioate (PS) linkage; d,deoxyribose. DMD, Duchenne's muscular dystrophy; dsRNA, double stranded RNA; ssRNA, single-stranded RNA.
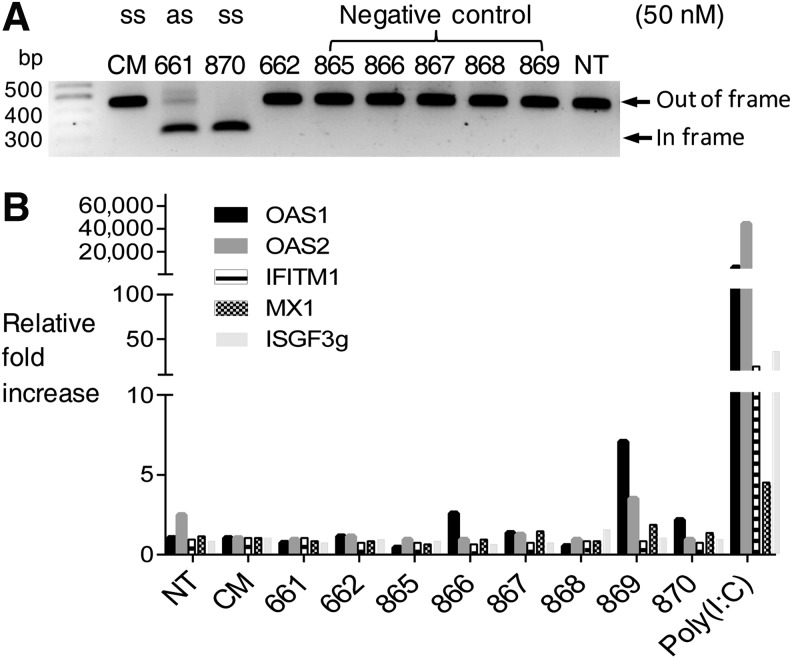
FIG. 3.
Correction of DMD splicing by ss-siRNA is sequence specific. (A) Correction of DMD splicing by a complementary ss-siRNA and ss-siRNAs containing mismatched bases (50 nM). (B) Correction of dystrophin splicing by ss-siRNA (50 nM) does not provoke an interferon response. Five different interferon-responsive genes were monitored by quantitative PCR after treatment with 50 nM ss-siRNA. Poly I:C is a positive control nucleic acid known to stimulate the interferon response. Patient-derived GM03429 fibroblast cells were used. IFITM1, interferon-induced transmembrane protein 1; ISGF3g, interferon-stimulated transcription factor 3 gamma; MX1, interferon-induced GTP-binding protein Mx1; NT, no treatment with RNA; OAS1/2, 2'-5'-oligoadenylate synthetase 1/2.
Another source of off-target effects is induction of the interferon response. To analyze the potential of ISIS 642661 to induce an interferon response that that might lead to off-target splice correction we analyzed the induction of interferon responsive genes upon addition of ss-siRNA. We observed little induction of these genes relative to treatment with known inducer Poly (I:C) (Fig. 3B), further supporting belief that production of the exon 51-deleted product is an on-target effect due to recognition of dystrophin pre-mRNA by ss-siRNA.
To gain another perspective on the susceptibility of splicing to recognition of the target sequence we tested a single stranded antisense 2′-O-methyl oligonucleotide analogous in sequence to ss-siRNA ISIS 642661. This 2′-O-methyl oligomer effectively altered splicing, confirming the susceptibility of the target. We also tested an ss-siRNA that was analogous in sequence to active ss-siRNA ISIS 642661 but possessed fewer phosphorothioate linkages. We found that it was inactive, suggesting that ss-siRNA action is sensitive to the exact nature of chemical modifications introduced during synthesis.
Splice correction is mediated by Ago2
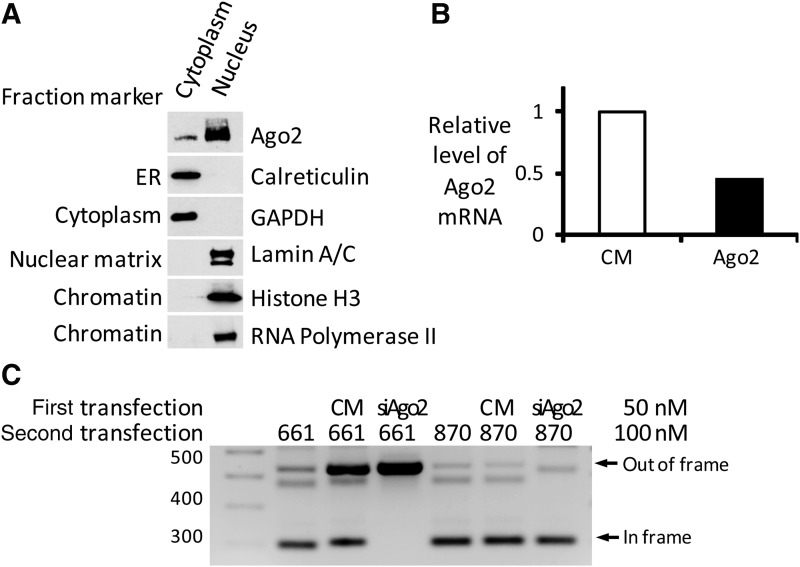
Previous studies had indicated that ss-siRNAs have the potential to act through an RNAi mechanism. ss-siRNAs, however, are chemically modified oligonucleotides that in many ways resemble standard antisense oligonucleotides whose mode of action is independent of RNAi. To determine whether ss-siRNA ISIS 642661 might be functioning through an RNAi pathway to affect splicing we first isolated nuclei from GM03429 patient-derived fibroblast cells and showed that Ago2 can be found in cell nuclei and that our preparations are free of contamination from the endoplasmic reticulum and cytoplasm (Fig. 4A). Interestingly, triplicate purifications all showed a preponderance of Ago2 in cell nuclei relative to cytoplasm, an outcome that differs from other cell lines tested in our laboratory where the amount cytoplasmic Ago2 is equal or greater than nuclear Ago2 [14]. We used an anti-Ago2 siRNA to deplete cells of Ago2 (Fig. 4B) and examined the effect on splice correction of dystrophin mRNA.
FIG. 4.
Ago2 expression affects modulation of splicing by ss-siRNA. (A) Purified cell cytoplasmic or nuclear extract reveals Ago2 in cell nuclei. Calreticulin is a marker for endoplasmic reticulum. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is a marker for cytoplasm. Lamin A/C, RNA polymerase 2, and histone H3 are nuclear markers. ER, endoplasmic reticulum. (B) Knockdown of Ago2 mRNA in DMD patient fibroblast GM03429 using anti-Ago2 siRNA (50 nM) compared with the effect of treating cells with noncomplementary RNA duplex CM. (C) Effect of silencing Ago2 on modulation of splicing by ss-siRNA (100 nm) or an antisense oligonucleotide. Patient-derived GM03429 fibroblast cells were used.
To accomplish this experiment we performed a double transfection. In the first transfection we introduced a noncomplementary control duplex RNA or a siRNA designed to silence Ago2 mRNA. In the second transfection we introduced ISIS 642661 or an analogous 2′-O-methyl oligonucleotide (Table 3). Addition of ISIS 642661 to cells caused formation of the isoform lacking exon 51 when a noncomplementary RNA was added during the first transfection. By contrast, addition of an RNA that reduces Ago2 expression blocked the activity of ISIS 642661 and prevented formation of the Δ51 isoform (Fig. 4C). Interestingly, only ∼50% reduction of AGO2 expression is sufficient to reverse the activity of ISIS 642661. One explanation is that normal cellular processes are relatively resistant to moderate reduction in AGO2 levels while effects from exogenously added ss-siRNAs are more sensitive.
Table 3.
Silencing RNAs Used to Test Effect of Ago2 Expression on Modulation of Splicing by ss-siRNA
| Name | Antisense sequence (5′ to 3′) | ss or dsRNA | Target gene | Position of mismatch (if any) |
|---|---|---|---|---|
| CM | 5′-GCUAUACCAGCGUCGUCAUdTdT | ds | Negative control | |
| 642661 | 5′-P-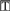 sCsAAsGGsAAsGUsUGsGCsAsUsUsUsCs sCsAAsGGsAAsGUsUGsGCsAsUsUsUsCs s s
|
ss | Exon 51, DMD | 10 |
| 678870 | 5′-UsCsAsAsGsGsAsAsGsAsUsGsGsCsAsUsUsUsCsU | ss, 2′-O-Me | Exon 51, DMD | - |
Modification positions within the sequence are indicated as follows: boxed, 2′-O-MOE (methoxyethyl); underlined, 2′-Fluoro; background shaded, 2′-O-me (methyl); bold and italicized, mismatched bases; subscripted “s,” phosphorothioate (PS) linkage; d, deoxyribose. DMD, Duchenne's muscular dystrophy; dsRNA, double stranded RNA; ssRNA, single-stranded RNA.
We performed a similar experiment using the analogous 2′-O-methyl oligomer. The antisense 2′-O-methyl oligomer also efficiently altered splicing. This alteration, however, was not affected by addition of anti-Ago2 siRNA. These results indicate that the ss-siRNA and the antisense 2′-O-methyl RNA act by different mechanisms and that RNAi drives the observed splice modulation by ss-siRNA.
Discussion
Duchenne muscular dystrophy is an incurable disease and treatments that can slow or halt the progression of symptoms are an urgent unmet need for patients and their families. Clinical trials have shown impressive results in some patients for the 6-minute walk test and elevated dystrophin levels upon biopsy [35]. While promising, small trial size and an absence of definitive data have left many questions unresolved. What is clear is that new approaches to starting points for more potent and efficacious molecules continue to be needed [36].
Our results add ss-siRNAs to the list of nucleic acid chemistries, the most prominent of which are morpholino and phosphorothioate-modified 2′-O-methyl RNA, capable of inducing skipping of exon 51. The skipping that we observe is sequence specific and not due to off-target induction of interferon response. Dependence on Ago2 expression demonstrates involvement of RNAi proteins.
The mechanism of action for splice regulation by ss-siRNAs likely begins with association of the ss-siRNA with AGO2 in the cytoplasm, the known location for strand loading [14]. The AGO2–ss-siRNA complex is then transported into the nucleus, where AGO2 (and probably other RNAi factors like TNRC6A) promote association with the target mRNA. This association of RNA–AGO2 with mRNA disrupts association with the splicing machinery and affects alternative splicing. This mechanism is similar to the mechanism of antisense oligonucleotides that affect splicing, consistent with the fact that our active ss-siRNA was designed to target a dystrophin sequence similar to that targeted by previously known active antisense oligomers [30,31].
Single stranded siRNAs have been studied in a handful of publications to date [23–26,28–9]. One previous paper examined inhibition of transcription by ss-siRNAs targeting a gene promoter [29], suggesting that ss-siRNAs can act within cell nuclei. The results we report here support the conclusion that ss-siRNAs can modulate nuclear processes through an RNAi mechanism by showing that they can affect splicing.
Because ss-siRNAs are single stranded oligomers that act through the RNAi pathway, they have the potential to be a distinct starting point for clinical development relative to antisense oligonucleotides or duplex RNAs. In theory, ss-siRNAs combine the strengths of antisense oligonucleotides (simplified synthesis, better biodistribution upon formulation in saline) with the potential for the RNAi pathway to afford better potency. More research will be necessary to determine whether these theoretical advantages can bring real benefits to drug development but the ability to affect splicing or transcription widens the options for development.
Another significant aspect of this work is that ss-siRNA mediated alteration of splicing in cell nuclei supports the conclusion that RNAi pathways are functional in the nuclei of mammalian cells. While this conclusion may not seem surprising given the existence of nuclear RNAi in many non-mammalian organisms [1], whether or not RNAi acts in mammalian nuclei has met with skepticism in the literature and in general discussions. The consequence of this skepticism and the difficulty of accurately studying RNAi factors in mammalian cell nuclei has slowed progress and clouded vision about potential applications. Our data demonstrate one potential application for using synthetic nucleic acids in combination with RNAi to modulate gene expression.
In summary, this report extends the activity of ss-siRNAs to modulation of splicing within the nuclei of human cells. Specifically, we show that ss-siRNAs can enhance production of dystrophin lacking exon 51 providing a new starting point for the production of a therapeutic isoform of dystrophin protein. Single stranded siRNAs are a relative new and understudied nucleic acid motif. Our data supports the conclusion that they are a promising platform for future development.
Acknowledgments
This work was supported by the National Institutes of Health (GM 106151) and the Robert A. Welch Foundation (I-1244).
Author Disclosure Statement
T.P. Prakash is an employee of ISIS Pharmaceutical.
References
- 1.Gagnon KT. and Corey DR. (2012). Argonaute and the Nuclear RNAs: new pathways for RNA-mediated control of gene expression. Nucleic Acid Ther 22:3–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li LC. (2014). Chromatin remodeling by small RNA machinery in mammalian cells. Epigenetics 9:45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Castel SE. and Martienssen RA. (2013). RNA Interference in the nucleus: roles for small RNAs in transcription, epigenetics, and beyond. Nat Rev Genet 14:100–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schraivogel D. and Meister G. (2014). Import routes and nuclear functions of argonaute and other small RNA-silencing proteins. Trends Biochem Sci 39:420–431 [DOI] [PubMed] [Google Scholar]
- 5.White E, Schacklow M, Kamieniarz-Gdula K, Proudfoot NJ. and Gullerova M. (2014). Human nuclear dicer restricts the deleterious accumulation of endogenous double-stranded RNA. Nat Struc Mol Biol 21:552–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross JP. and Kassir Z. (2014). The varied roles of nuclear argonaute-small RNA complexes and avenues for therapy. Mol Ther Nucl Acids 3:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsui M, Zhang H, Chu Y, Gagnon KT, Manoharan M, Corey DR. and Janowski BA. (2013). A Network of noncoding RNAs links transcription of inflammatory pathway genes COX-2 and PLAG24A. Nucleic Acids Res 41:10086–10109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weinberg MS. and Morris KV. (2013). Long non-coding RNA targeting and transcriptional de-repression. Nucleic Acid Ther 23:9–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Place RF, Li LC, Pookot D, Noonan EJ. and Dahiya R. (2001). MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci (USA) 105:1608–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DH, Saetrom P, Snove O. and Rossi JJ. (2008). MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci 105:16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Younger ST. and Corey DR. (2011). Transcriptional gene silencing in mammalian cells by miRNA mimics that target gene promoters. Nucl Acids Res 39:5682–5691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robb GB, Brown KM, Khurana J. and Rana TM. (2005). Specific and potent RNAi in the nucleus of human cells. Nature Struct Mol Biol 12:133–37 [DOI] [PubMed] [Google Scholar]
- 13.Orht T, Muetze J, Svoboda P. and P Schwille P, (2012). Intracellular localization and routing of miRNA and RNAi pathway components. Curr Top Med Chem 12:79–8 [DOI] [PubMed] [Google Scholar]
- 14.Gagnon KT, Li L, Chu Y, Janowski BA. and Corey DR. (2014). RNAi factors are present and active in mammalian cell nuclei. Cell Reports 6:211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veltrop M. and Aartsma-Rus A. (2014). Antisense-mediated exon skipping: taking advantage of a trick from Mother Nature to treat rare genetic diseases. Exper Cell Res 325:50–55 [DOI] [PubMed] [Google Scholar]
- 16.Jarver P, O'Donovan L. and Gait MJ. (2014). A chemical view of oligonucleotides for exon skipping and related drug applications. Nucleic Acid Ther 24:37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rigo F, Seth PP. and Bennett CF. (2014). Antisense oligonucleotide-based therapies for diseases caused by pre-mRNA processing defects. Adv Exp Med Biol 825:303–352 [DOI] [PubMed] [Google Scholar]
- 18.Battsche E. and Ameyar-Zazoua M. (2014). The influence of argonaute proteins on alternative RNA splicing. Wiley Interdiscip Rev. RNA 6:141–156 [DOI] [PubMed] [Google Scholar]
- 19.Liu J, Hu J, and Corey DR. (2012). Expanding the action of duplex RNAs into the nucleus: redirecting alternative splicing. Nucl Acids Res 40:1240–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allo M, Buggiano V, Fededa JP, Petrillo E, Schor I, de la Mata M, Agirre E, Plass M, Eyras E, et al. (2009). Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struc Mol Biol 16:717–724 [DOI] [PubMed] [Google Scholar]
- 21.Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, et al. (2014). Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat Struc Mol Biol 19:998–1004 [DOI] [PubMed] [Google Scholar]
- 22.Allo M, Agirre E, Bessonov S, Bertucci P, Gomez Acuna L, Buggiano V, Bellora N, Singh B, Petrillo E, et al. (2104). Argonaute-1 binds transcriptional enhances and controls constitutive and alternative splicing in human cells. Proc Natl Acad Sci (USA) 111:15622–15629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lima WF, Prakash TP, Murray HM, Kinberger GA, Li W, Chappell AE, Li CS, Murray SF, Gaus H, et al. (2012). Single-stranded siRNAs activate RNAi in animals. Cell 150: 883–894 [DOI] [PubMed] [Google Scholar]
- 24.Prakash TP, Lima WF, Murray HM, Elbashir S, Cantley W, Foster D, Jayaraman M, Chappell AE, Manoharan M, Swayze EE. and Crooke ST. (2013). Lipid nanoparticles improve activity of single-stranded siRNA and gapmer antisense oligonucleotides in animals. ACS Chem Biol 8:1402–1406 [DOI] [PubMed] [Google Scholar]
- 25.Yu D, Pendergraff H, Liu J, Kordasiewicz HB, Cleveland DW, Swayze E, Lima W, Crooke ST, Prakash T. and Corey DR. (2012). Single-stranded RNAs that function through RNAi are potent and allele-selective inhibitors of huntingtin expression. Cell 150:895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu J, Lui J, Yu D, Aiba Y, Lee S, Pendergraff H, Boubaker J, Arates JW, Lagier-Tourenne C, et al. (2014). Exploring the effect of sequence length and composition on allele-selective inhibition of human huntingtin expression by single-stranded silencing RNAs. Nucleic Acid Ther 24:199–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, Yu D, Aiba Y, Pendergraff H, Swayze EE, Lima WF, Prakash TP. and Corey DR. (2013). ss-siRNAs allele-selectively inhibit ataxin-3 expression: Multiple mechanisms for an alternative gene silencing strategy. Nucl Acids Res 41: 9570–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu J, Liu J, Narayannair KJ, Lackey JG, Kuchimanchi S., Rajeev KG, Manoharan M, Swayze EE, Lima WF, et al. (2014). Allele-selective inhibition of mutant atrophin-1 expression by duplex and single-stranded RNAs. Biochemistry 53,4510–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsui M, Prakash TP. and Corey DR. (2013). Transcriptional silencing by single-stranded RNAs targeting a noncoding RNA that overlaps a gene promoter. ACS Chem Biol 8:122–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aartsma-Rus A, Bremmer-Bout M, Janson AMM, Dunnen JT, van Ommen GTB. and van Deutekom JCT. (2002). Targeted exon skipping as a potential gene correction therapy for Duchenne muscular dystrophy. Neuromuscl Disord 12:S71–S77 [DOI] [PubMed] [Google Scholar]
- 31.Arechavala-Gomeza V, Graham IR, Popplewell LJ, Adams AM, Aartsma-Rus A, Kinali M, Morgan JE, Van Deutekom JC, Wilton SD, Dickson G. and Muntoni F. (2007). Comparative analysis of antisense oligonucleotide sequences for targeting skipping of exon 5 during dystrophin pre-mRNA splicing in human muscle. Hum Mol Gene Ther 18:798–810 [DOI] [PubMed] [Google Scholar]
- 32.van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, et al. (2007). Local dystrophin restoration with antisense oligonucleotide PRO051. New Eng J Med 375:2677–2686 [DOI] [PubMed] [Google Scholar]
- 33.Lu QL, Yokota T, Takeda S, Garcia L, Muntoni F. and Partridge T. (2011). The status of exon skipping as a therapeutic approach to Duchennes muscular dystrophy. Mol Ther 19:9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kole R. and Leppert BJ. (2012). Targeting mRNA splicing as a potential treatment for Duchenne muscular dystrophy. Discov Med 14:59–69 [PubMed] [Google Scholar]
- 35.Voit T, Topaloglu H, Straub V, Muntoni F, Deconinck N, Campion G, de Kimpe SJ, Eagle M, Guglieri M, et al. (2014). Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory randomised, placebo-controlled phase 2 study. Lancet Neurol 13:987–996 [DOI] [PubMed] [Google Scholar]
- 36.Hoffman EP. and Mcnally EM. (2014). Exon-skipping therapy: a roadblock, detour, or a bump in the road. Sci Transl Med 6:230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomsen JM, Song JJ, Hammond SM, Joshua-Tor L. and Hannon GJ. (2004). Argonaute 2 is the catalytic engine of RNAi. Science 305:1437–1441 [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Juranek S, Li H, Sheng G, Tuschl T. and Patel DJ. (2008). Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature 456:921–926 [DOI] [PMC free article] [PubMed] [Google Scholar]