Abstract
Purpose
The Dixon techniques provide uniform water-fat separation but require multiple image sets, which extend the overall acquisition time. Here, an alternative rapid single acquisition method, lipid elimination with an echo-shifting N/2-ghost acquisition (LEENA), was introduced.
Methods
The LEENA method utilized a fast imaging with steady-state free precession sequence to obtain a single k-space dataset in which successive k-space lines are acquired to allow the fat magnetization to precess 180°. The LEENA data were then unghosted using either image-domain (LEENA-S) or k-space domain (LEENA-G) parallel imaging techniques to reconstruct water-only and fat-only images. An off-resonance correction technique was incorporated to improve the uniformity of the water-fat separation.
Results
Uniform water-fat separation was achieved for both the LEENA-S and LEENA-G methods for phantom and human body and leg imaging applications at 1.5T and 3T The resultant water and fat images were qualitatively similar to conventional 2-point Dixon and fat-suppressed images.
Conclusion
The LEENA-S and LEENA-G methods provide uniform water and fat images from a single MRI acquisition. These straightforward methods can be adapted to 1.5T and 3T clinical MRI scanners and provide comparable fat/water separation with conventional 2-point Dixon and fat-suppression techniques.
Keywords: water fat imaging, fat suppression MRI, parallel imaging, SENSE, GRAPPA
Introduction
Fat suppression is an important component on all modern MRI systems because a wide variety of anatomic and pathologic structures can be obscured by the normally bright adipose tissue. The need for effective lipid/off-resonance signal suppression is especially critical in rapid imaging acquisitions, such as echo-planar imaging and non-Cartesian acquisitions (eg, spiral trajectories) where off-resonance spins can result in ghosting artifacts and image blurring, respectively. Of the multiple fat suppression techniques, the Dixon methods provide not only uniform fat suppression but also the potential for lipid quantification (1). These techniques are superior alternatives to the short-tau inversion recovery (2) and spectral excitation fat suppression methods (3), in terms of both uniform fat suppression and quantitative capabilities (4,5).
The original Dixon method, first proposed in 1984, acquires two separate images in which water and fat magnetization vectors have a 0° and a 180° phase difference, respectively, to produce separated water-only and fat-only images (1). This method is called 2-point Dixon (2PD). Since then, many variants of the Dixon techniques have been developed and can be generally sorted into single-point (6,7), two-point (1,8-13), three-point (14-18), and higher-order methods (19-24), depending on how many images are acquired. Fundamentally, these techniques require multiple image sets at different echo times, which can significantly extend the overall acquisition time. This, in turn, could limit image spatial resolution and/or slice coverage in body imaging, which are typically acquired during breath-holding. One straightforward approach to reduce the overall acquisition time is to implement parallel imaging strategies, such as sensitivity encoding (SENSE (25)) or generalized autocalibrating partially parallel acquisitions (GRAPPA (26)) (27) or compressed sensing techniques (28,29) or both (30,31) to decrease the acquisition time for each individual image. An alternative scheme, lipid elimination with an echo-shifting N/2-ghost acquisition (LEENA), uses a specialized trajectory and adapted parallel imaging techniques to reconstruct the water-only and fat-only images. In a preliminary study, LEENA was shown to reliably suppress adipose tissue signal by combining this specialized trajectory with a SENSE-like N/2-ghost reconstruction (32).
In this study, the LEENA acquisition and reconstruction methodology was extended to be used in combination with either image-domain or k-space domain parallel imaging techniques [ie, SENSE (25) and GRAPPA (26)]. These two techniques are referred to as LEENA-S (LEENA acquisition with a SENSE-like reconstruction) and LEENA-G (LEENA acquisition with GRAPPA-like reconstruction), respectively. Furthermore, these methods can be effectively combined with off-resonance correction techniques to ensure uniform water and fat images. In this study, the LEENA trajectory was implemented in a rapid steady-state free precession (FISP) sequence to generate images with ghosting of the fatty tissues. Separate water-only and fat-only images are then generated using the LEENA-S or LEENA-G reconstruction methods. The capabilities of the LEENA-S and LEENA-G techniques are demonstrated in phantom and human imaging studies at 1.5T and 3T.
Methods
LEENA-S and LEENA-G Acquisition Trajectory
The LEENA acquisition trajectory is described using a simple water-fat phantom consisting of tap water and vegetable oil. The LEENA-S and LEENA-G methods were implemented on a Siemens Skyra 3T scanner (Siemens Medical Solutions, Erlangen, Germany) for the phantom experiments. A fully sampled reference image is shown in Figure 1a. At 3T, both the LEENA-S and LEENA-G methodologies utilized a FISP acquisition with a 1.1-ms echo time difference between successively acquired lines in k-space to allow the fat magnetization to precess 180° between adjacent k-space lines (TE1 and TE2, shown as solid lines and dashed lines, respectively, in Figure 1b). Direct Fourier transform of the acquisition data resulted in shifting of the fat component by field of view (FOV)/2 (N/2-ghost) while the on-resonance water component was not shifted (Fig. 1c). Note that this implementation assumes a single-peak lipid model with a 3.5-ppm chemical shift difference from water (ie, methylene peak).
Fig. 1.
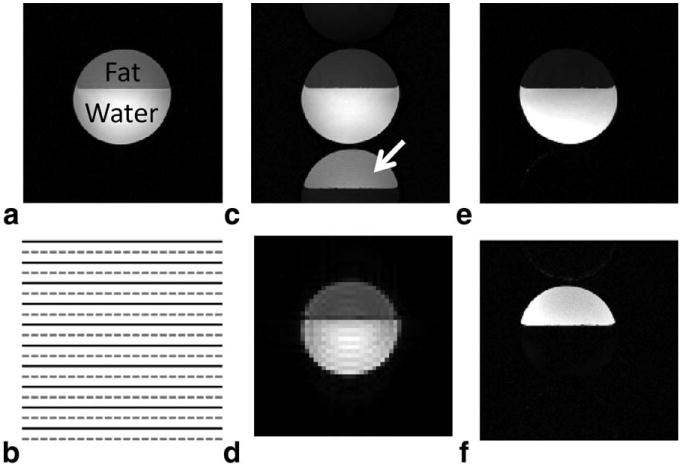
a: A representative fully sampled water-fat phantom image with water and fat components labeled. b: Schematic of the LEENA-S k-space trajectory with solid and dashed lines representing k-space lines with an echo time difference allowing the fat magnetization to precess 180° between adjacent k-space lines (1.1 ms at 3T, 2.2 ms at 1.5T). c: An example “ghosted” image obtained from direct Fourier transform of the LEENA k-space data in which the fat component is shifted by FOV/2 due to echo time variation. d: A low-resolution FISP image used to obtain coil sensitivity maps for the LEENA-S image reconstruction. e,f: Reconstructed (e) LEENA-S water-only image and (f) LEENA-S fat-only image after image separation and off-resonance correction.
LEENA-S Water-Fat Reconstruction Methodology
For the LEENA-S method, a modified version of the well-known SENSE algorithm [ie, PAGE (33,34)] was used to separate the water and fat components in the image domain. As described previously (32), the unghosted water image and the N/2-ghosted fat image were calculated from the individual, ghosted coil images as
| [1] |
where S is the sensitivity matrix obtained from the combination of shifted and unshifted coil sensitivity maps, G is the matrix of ghosted images from the individual coils, and Rn is the noise covariance matrix (assumed to be identity for this initial study). The superscript H represents the transpose of the complex conjugate, and the matrix in brackets is inverted using a least-squares, pseudo-inverse operation. The images resulting from Eq. [1] represent the 0th (water image, f0) and 1st ghosts (fat image, f1), respectively. The unaliasing algorithm was implemented on a pixel-by-pixel basis to generate separate water and fat images with the same resolution as the aliased LEENA image (Fig. 1d and 1e, respectively).
LEENA-G Water-Fat Reconstruction Methodology
For the LEENA-G method, a modified GRAPPA reconstruction algorithm was utilized to unalias the k-space data. A schematic of the LEENA-G methodology is shown in Fig. 2. Similar to the LEENA-S method, the k-space data were acquired with alternating fat magnetization phase, and with spacing of Δky in the phase encoding direction (Fig. 2a). To enable the GRAPPA-like reconstruction, LEENA-G calibration lines were also acquired with alternating fat magnetization phase but with a line spacing of Δky/3 (Fig. 2b). It is important to note that the echo time alternation is required in the calibration lines to ensure that the water and fat components are correctly “unghosted.” The LEENA-G calibration lines were then used to calculate the GRAPPA operators and intermediate k-space lines with a GRAPPA reduction factor of 3 (R=3). Although other reduction factors may be possible, this is the minimum reduction factor to retain the echo time alternation in the calibration lines. Since the reconstructed k-space line spacing is one third of the acquired k-space (ie, 3× FOV, Fig. 2c), the fat (ghosted) component was entirely spatially separated from the water (no ghosting) component after Fourier transform of the GRAPPA-reconstructed k-space data (Fig. 2d). Water and fat phantom images were then directly extracted from this LEENA-G reconstructed image.
Fig. 2.
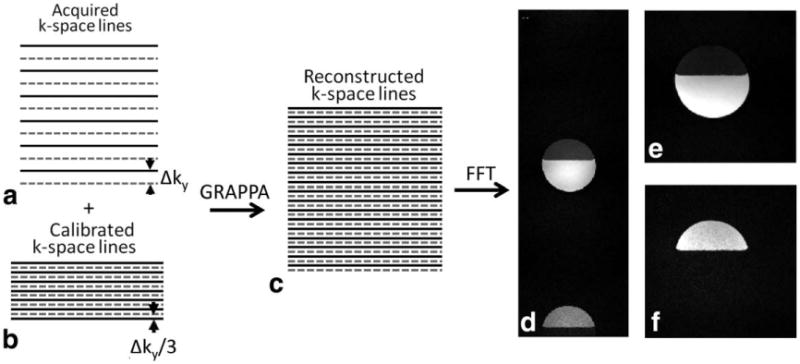
Schematic of the LEENA-G (reduction factor R = 3) acquisition and reconstruction algorithm and phantom imaging results. a: Schematic of LEENA k-space trajectory with solid and dashed lines representing an echo time difference between adjacent k-space lines allowing the fat magnetization to precess 180°. b: Trajectory for LEENA-G calibration lines with Δky/3 line spacing and echo time shifts required for accurate water-fat separation. c: Calculated fully sampled LEENA-G k-space data using a GRAPPA reconstruction (reduction factor R = 3). d: Reconstructed LEENA-G image with spatially distinct water and fat components. e, f: Reconstructed (e) LEENA-G water-only and (f) LEENA-G fat-only images after image separation and off-resonance correction.
Off-Resonance Correction
All image reconstructions were performed offline in MATLAB (MathWorks, Natick, Massachusetts, USA). An off-resonance correction algorithm was applied to the LEENA-G and LEENA-S water and fat images to limit the effects of B0 field inhomogeneity. Following the image reconstructions, the complex water and fat images were algebraically combined to produce complex “Water+Fat” (IW+F) and “Water-Fat” (IW−F) image data sets. A differential phase map (Φi) was then calculated using previously described methods (8,35):
| [2] |
The final off-resonance corrected water and fat images were then calculated as
| [3a] |
| [3b] |
A phase unwrapping algorithm (35) was utilized to correct large field inhomogeneities that would otherwise result in erroneous assignment of water and fat signals.
LEENA-S and LEENA-G Phantom Experiments at 3T
A modified FISP sequence was implemented on a Siemens Skyra 3T scanner to acquire the LEENA-G and LEENA-S phantom data. For this initial implementation, a single echo was acquired for each repetition time (TR) interval. A FISP sequence was selected instead of true FISP or echo-planar imaging acquisitions to limit banding and off-resonance artifacts, respectively (36). The FISP-LEENA imaging parameters for phantom evaluations are listed in Table 1. A separate FISP-LEENA scan was acquired at 3× FOV in the phase direction to acquire the LEENA-G calibration lines. For the LEENA-S reconstruction, a separate FISP scan with low resolution was also acquired to obtain the coil sensitivity maps (25). The coil sensitivities were calculated from established methods (Fig. 1d). All images were obtained for a single 5-mm slice, and the phantom image acquisition parameters are summarized in Table 1.
Table 1. Summary of Imaging Parameters.
| Acquisition | TR (ms) | TE1 (ms) | TE2 (ms) | Flip Angle | Matrix | FOV (mm) | Coil No. | |
|---|---|---|---|---|---|---|---|---|
| Phantom at 3T | ||||||||
| LEENA | FISP | 5.0 | 1.95 | 3.05 | 45° | 256 × 176 | 300 × 225 | 12 |
| LEENA-S (r) | FISP | 3.1 | 1.55 | 45° | 128 × 88 | 300 × 225 | 12 | |
| LEENA-G (c) | FISP | 4.1 | 1.5 | 2.6 | 45° | 64 lines | 300 × 675 | 12 |
| Human liver at 1.5T | ||||||||
| LEENA | FISP | 5.23 | 1.52 | 3.72 | 45° | 256 × 175 | 400 × 275 | 9 |
| LEENA-S (r) | FISP | 2.29 | 1.15 | 45° | 128 × 88 | 400 × 275 | 9 | |
| LEENA-G (c) | FISP | 4.51 | 1.15 | 3.35 | 45° | 64 lines | 400 × 825 | 9 |
| 2PD | GRE | 7 | 2.42 | 4.62 | 25° | 256 × 175 | 400 × 275 | 9 |
| Fat suppression | GRE | 24 | 2.42 | 25° | 256 × 175 | 400 × 275 | 9 | |
| Human liver at 3T | ||||||||
| LEENA | FISP | 4.6 | 1.75 | 2.85 | 45° | 256 × 192 | 400 × 300 | 20 |
| LEENA-S (r) | FISP | 3.52 | 1.76 | 45° | 128 × 96 | 400 × 300 | 20 | |
| LEENA-G (c) | FISP | 3.58 | 1.24 | 2.34 | 45° | 64 lines | 400 × 900 | 20 |
| 2PD | GRE | 8 | 2.2 | 5.5 | 25° | 256 × 192 | 400 × 300 | 20 |
| Fat suppression | GRE | 22 | 2.25 | 25° | 256 × 192 | 400 × 300 | 20 | |
| Human leg at 3T | ||||||||
| LEENA | FISP | 4.6 | 1.75 | 2.85 | 45° | 256 × 192 | 500 × 375 | 20 |
| LEENA-S (r) | FISP | 3.52 | 1.76 | 45° | 128 × 96 | 500 × 375 | 20 | |
| LEENA-G (c) | FISP | 3.58 | 1.24 | 2.34 | 45° | 64 lines | 500 × 1125 | 20 |
| 2PD | GRE | 8 | 2.2 | 5.5 | 25° | 256 × 192 | 500 × 375 | 20 |
| Fat suppression | GRE | 22 | 2.25 | 25° | 256 × 192 | 500 × 375 | 20 |
LEENA-S (r): reference scan for LEENA-S. LEENA-G (c): calibration scan for LEENA-G.
Human LEENA-S and LEENA-G Experiments at 1.5T and 3T
To demonstrate the utility of the LEENA techniques on clinical MRI scanners, axial LEENA-S and LEENA-G liver images were obtained for volunteers on a Siemens Espree 1.5T and a Siemens Skyra 3T MRI scanner. Axial leg images were also obtained for one of the volunteers at 3T. All human imaging studies were conducted in accordance with approved Institutional Review Board, HIPAA-compliant protocols. All volunteers were scanned in a supine position during free breathing. The LEENA-S and LEENA-G imaging parameters for 1.5T liver imaging studies are listed in Table 1. As for the phantom experiments, a separate low-resolution FISP scan was also acquired to calculate coil sensitivity maps for the LEENA-S reconstruction. A low-resolution LEENA-FISP scan was also acquired at 3× FOV in the phase direction to acquire the LEENA-G calibration lines. Fat-suppressed images using conventional spectral excitation/fat saturation scheme and 2PD methods were acquired with a gradient-recalled echo pulse sequence for comparison. The 2PD images were acquired with a GRAPPA reduction factor of 2 for comparison with the LEENA images.
For the 3T liver and leg MRI LEENA scans, an echo time difference of 1.1 ms was implemented as required (Table 1). The human leg experiments used a larger FOV to accommodate both legs in an axial scan for the volunteer. All other imaging parameters for the LEENA, 2PD, and conventional fat suppression gradient-recalled echo acquisitions were similar as applied for the 1.5T human experiments. All of the LEENA-S and LEENA-G image reconstructions were performed off-line in MATLAB. Off-resonance correction was also applied for all human imaging studies as described above.
Results
Figure 1 shows the LEENA-S phantom images obtained with the LEENA-FISP trajectory (Fig. 1b). The N/2-ghost of the fat component is shifted by FOV/2 and is indicated by the white arrow in Figure 1c. A low-resolution FISP image used to obtain coil sensitivity maps for the LEENA-S image reconstruction is shown in Figure 1d. Separate LEENA-S water and fat images after implementation of the LEENA-S unghosting algorithm (Eq. [1]) and off-resonance correction (Eqs. [2] and [3]) are shown in Figures 1e and 1f, respectively. A schematic of the LEENA-G trajectory (Fig. 2a), calibration lines with echo shifting (Fig. 2b), and the calculated LEENA-G k-space (Δky/3, Fig. 2c) are shown in Figure 2 along with the final unghosted water-fat image (Fig. 2d). Note that the resultant 3× increase in FOV from the LEENA-G reconstruction results in complete separation of the water and fat components of the image allowing generation of separate water and fat images (Fig. 2e and 2f).
In vivo LEENA-S and LEENA-G images of the liver and legs of volunteers at 1.5T (liver only) and 3T (legs and liver) are shown in Figures 3, 4, and S1, respectively. The raw, ghosted LEENA images are shown in Figures 3a, 4a, and S1a. Conventional fat-suppressed images obtained from spectral fat suppression are shown in Figures 3b, 4b, and S1b. Separate water and fat images from the LEENA-S (Figs. 3c and 3d, 4c and 4d, and S1c and S1d), LEENA-G (Figs. 3e and 3f, 4e and 4f, and S1e and S1f), and 2PD (Figs. 3g and 3h, 4g and 4h, and S1g and S1h) acquisitions are also shown. The water-only and fat-only images for the LEENA-S and LEENA-G acquisition generated uniform water-fat separation similar to the 2PD acquisition. Increased ghosting artifacts were observed in the LEENA-G and LEENA-S images at 1.5T in comparison with 3T, likely due to fewer coils. The LEENA-S liver images also exhibited visible artifacts, likely due to imperfections in the coil sensitivity maps. In addition, erroneous assignment of water and fat components was observed in the 3T images near the edge of the FOV (arrows in Figures 4 and S1) due to locally increased field inhomogeneities. Otherwise, the LEENA-S and LEENA-G images generated comparable water-fat separation compared with the 2PD method.
Fig. 3.
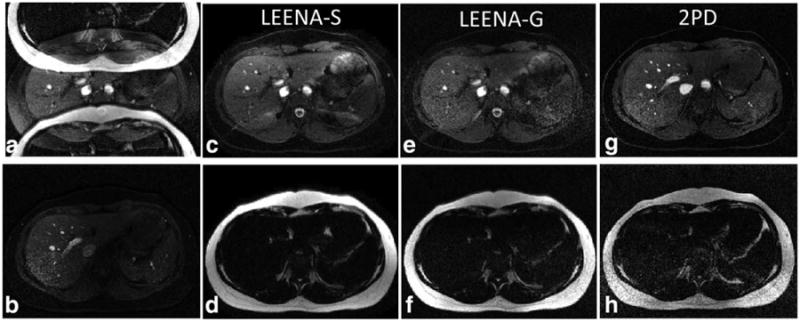
Axial abdominal images from a volunteer acquired on a 1.5T scanner. a: Raw LEENA image with N/2-ghost of the adipose tissue. b: Conventional fat-suppressed image using spectral excitation. c, d: Unghosted and off-resonance corrected LEENA-S water and fat images. e, f: Unghosted and off-resonance corrected LEENA-G water and fat images. g, h: Off-resonance corrected 2PD images as a reference.
Fig. 4.
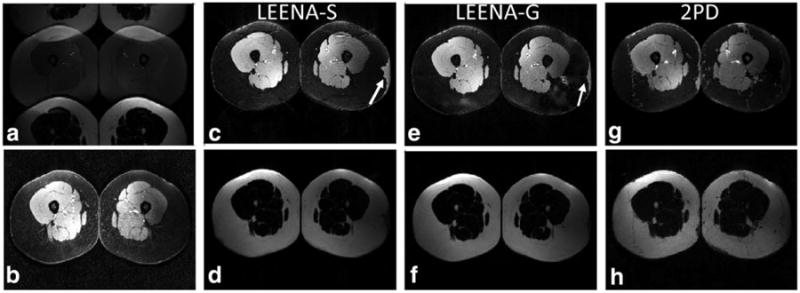
Axial leg images from a volunteer acquired on a 3.0T scanner. a: Raw LEENA image with N/2-ghost of the adipose tissue. b: Conventional fat-suppressed image using spectral excitation. c, d: Unghosted and off-resonance corrected LEENA-S water and fat images. e, f: Unghosted and off-resonance corrected LEENA-G water and fat images. g, h: Off-resonance corrected 2PD images as a reference. Arrows indicate regions of erroneous water-fat assignment due to increased field inhomogeneities near the edges of the FOV.
Discussion
The LEENA algorithm is a rectilinear 1-point Dixon method that combines a rapid echo-shifting trajectory with an unghosting reconstruction using variants of established parallel imaging techniques. According to the applied parallel imaging techniques, the fat/water reconstruction techniques were referred to as LEENA-S and LEENA-G, which use either a SENSE-like reconstruction from image-domain data or a GRAPPA-like reconstruction from k-space data, respectively. Uniform water-fat separation from these two techniques was demonstrated on clinical 1.5T and 3T scanners in both phantom and initial human abdominal and leg imaging experiments. The resulting LEENA-S and LEENA-G water and fat images were qualitatively similar to reference images obtained with conventional 2PD (with a GRAPPA reduction factor of 2) and fat suppression strategies.
In this study, we applied the LEENA techniques with a FISP acquisition to limit image artifacts from other rapid MRI acquisitions such as banding artifacts from true FISP acquisitions and distortion/ghosting artifacts from echo-planar imaging acquisitions (36). The human LEENA imaging results were obtained with free breathing to demonstrate the potential clinical viability of the rapid LEENA techniques. As described above, the LEENA-FISP data were acquired with a single-echo for each TR interval rather than a multiecho approach. Further reductions in acquisition time could be achieved through implementation of a dual-echo approach (ie, 2 k-space lines acquired during each TR interval). However, the dual-echo approach was not implemented in this study to avoid hardware and safety limitations (from peripheral nerve stimulation). In addition, a dual-echo approach at 3T may require bipolar readout gradients, which could result in additional ghosting from both the water and fat components in the images.
The image reconstruction techniques to separate the water and fat signals were similar to the established parallel imaging reconstruction schemes such as SENSE/PAGE or GRAPPA. The LEENA-S reconstruction utilized coil sensitivity maps to enable the SENSE-like reconstruction using the unghosting algorithm shown in Eq. [1]. The LEENA-G acquisition utilized separately acquired GRAPPA calibration lines with the same echo time shift as the LEENA acquisition (Fig. 2b). The echo time shift in the calibration lines was required for accurate separation of the water and fat images. In addition, the GRAPPA calibration lines were acquired with a line spacing of Δky/3, which is the maximum k-space line spacing (minimum FOV) possible to calculate the fully sampled LEENA-G dataset (Fig. 2c). The off-resonance correction method utilized in this study was published previously (8,35) and is typical of off-resonance correction of 2PD water-fat separation strategies. Further optimization of the off-resonance correction method could reduce the erroneous water-fat assignments in the 3T images, but was beyond the scope of this initial implementation.
The LEENA-S and LEENA-G methods described herein were evaluated in abdominal and leg imaging applications at 1.5T and 3T. Multiple other imaging applications such as cardiovascular and neuroimaging applications may be possible. One key advantage of the LEENA techniques is that only the fat components are aliased/ghosted in the raw images, while the water components are not aliased. This is a potentially significant advantage over the 2PD techniques with equal GRAPPA or SENSE reduction factors where both water and fat signal components are aliased. This important difference may provide a significant advantage in specific clinical applications where fatty tissue components are small in size compared with water tissue components, but still clinically relevant (eg, lipoma). In these cases, the LEENA techniques would be expected to outperform the conventional Dixon techniques with equal acquisition time, as minimal aliasing/ghosting would be expected.
One of the limitations of the LEENA techniques as implemented in this study is the assumption for a single lipid peak (i.e., methylene) at a frequency difference of 3.5ppm from water. Multipeak lipid composition strategies have already been shown to achieve accurate water-fat separation and quantification in human MRI studies (37). While not implemented herein, a similar model may be employed with the LEENA techniques and would likely require additional echo time variation similar to the 3-point and 4-point Dixon strategies. The additional echo times would provide the opportunity to incorporate the multipeak lipid models or provide a more comprehensive correction for B0 field inhomogeneities than the off-resonance correction algorithm described above. However, the additional echo time variation would also result in more complex lipid ghosting and therefore increased residual ghosting artifacts in the final LEENA-S and LEENA-G images (ie, increased reduction factors).
In conclusion, the present study presents the LEENA-S and LEENA-G methods to reliably separate water and fat signals from a single MRI acquisition. The uniformity of the LEENA-S and LEENA-G water-fat separation techniques were demonstrated in phantom and human free-breathing abdominal and leg imaging applications at both 1.5T and 3T. The ghosting of fat only is a unique feature of the LEENA trajectory compared with conventional 2PD acquisitions implemented with parallel imaging where both water and fat components are aliased in the undersampled images. This rapid water-fat imaging methodology may offer significant advantages for clinical imaging applications where the extent of lipid distribution is small, as the water tissue components are not aliased.
Supplementary Material
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Dixon WT. Simple proton spectroscopic imaging. Radiology. 1984;153:189–194. doi: 10.1148/radiology.153.1.6089263. [DOI] [PubMed] [Google Scholar]
- 2.Bydder GM, Young IR. MR imaging: clinical use of the inversion recovery sequence. J Comput Assist Tomogr. 1985;9:659–675. [PubMed] [Google Scholar]
- 3.Haase A, Frahm J, Hanicke W, Matthaei D. 1H NMR chemical shift selective (CHESS) imaging. Phys Med Biol. 1985;30:341–344. doi: 10.1088/0031-9155/30/4/008. [DOI] [PubMed] [Google Scholar]
- 4.Hu HH, Kan HE. Quantitative proton MR techniques for measuring fat. NMR Biomed. 2013;26:1609–1629. doi: 10.1002/nbm.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reeder SB, Cruite I, Hamilton G, Sirlin CB. Quantitative assessment of liver fat with magnetic resonance imaging and spectroscopy. J Magn Reson Imaging. 2011;34:729–749. doi: 10.1002/jmri.22580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma J. A single-point Dixon technique for fat-suppressed fast 3D gradient-echo imaging with a flexible echo time. J Magn Reson Imaging. 2008;27:881–890. doi: 10.1002/jmri.21281. [DOI] [PubMed] [Google Scholar]
- 7.Flask CA, Dale B, Lewin JS, Duerk JL. Radial alternating TE sequence for faster fat suppression. Magn Reson Med. 2003;50:1095–1099. doi: 10.1002/mrm.10615. [DOI] [PubMed] [Google Scholar]
- 8.Coombs BD, Szumowski J, Coshow W. Two-point Dixon technique for water-fat signal decomposition with B0 inhomogeneity correction. Magn Reson Med. 1997;38:884–889. doi: 10.1002/mrm.1910380606. [DOI] [PubMed] [Google Scholar]
- 9.Ma J. Breath-hold water and fat imaging using a dual-echo two-point Dixon technique with an efficient and robust phase-correction algorithm. Magn Reson Med. 2004;52:415–419. doi: 10.1002/mrm.20146. [DOI] [PubMed] [Google Scholar]
- 10.Skinner TE, Glover GH. An extended two-point Dixon algorithm for calculating separate water, fat, and B0 images. Magn Reson Med. 1997;37:628–630. doi: 10.1002/mrm.1910370426. [DOI] [PubMed] [Google Scholar]
- 11.Xiang QS. Two-point water-fat imaging with partially-opposed-phase (POP) acquisition: an asymmetric Dixon method. Magn Reson Med. 2006;56:572–584. doi: 10.1002/mrm.20984. [DOI] [PubMed] [Google Scholar]
- 12.Berglund J, Ahlstrom H, Johansson L, Kullberg J. Two-point dixon method with flexible echo times. Magn Reson Med. 2011;65:994–1004. doi: 10.1002/mrm.22679. [DOI] [PubMed] [Google Scholar]
- 13.Eggers H, Brendel B, Duijndam A, Herigault G. Dual-echo Dixon imaging with flexible choice of echo times. Magn Reson Med. 2011;65:96–107. doi: 10.1002/mrm.22578. [DOI] [PubMed] [Google Scholar]
- 14.Berglund J, Johansson L, Ahlstrom H, Kullberg J. Three-point Dixon method enables whole-body water and fat imaging of obese subjects. Magn Reson Med. 2010;63:1659–1668. doi: 10.1002/mrm.22385. [DOI] [PubMed] [Google Scholar]
- 15.Glover GH, Schneider E. Three-point Dixon technique for true water/fat decomposition with B0 inhomogeneity correction. Magn Reson Med. 1991;18:371–383. doi: 10.1002/mrm.1910180211. [DOI] [PubMed] [Google Scholar]
- 16.Lu W, Hargreaves BA. Multiresolution field map estimation using golden section search for water-fat separation. Magn Reson Med. 2008;60:236–244. doi: 10.1002/mrm.21544. [DOI] [PubMed] [Google Scholar]
- 17.Xiang QS, An L. Water-fat imaging with direct phase encoding. J Magn Reson Imaging. 1997;7:1002–1015. doi: 10.1002/jmri.1880070612. [DOI] [PubMed] [Google Scholar]
- 18.Yeung HN, Kormos DW. Separation of true fat and water images by correcting magnetic field inhomogeneity in situ. Radiology. 1986;159:783–786. doi: 10.1148/radiology.159.3.3704157. [DOI] [PubMed] [Google Scholar]
- 19.An L, Xiang QS. Chemical shift imaging with spectrum modeling. Magn Reson Med. 2001;46:126–130. doi: 10.1002/mrm.1167. [DOI] [PubMed] [Google Scholar]
- 20.Hernando D, Haldar JP, Sutton BP, Ma J, Kellman P, Liang ZP. Joint estimation of water/fat images and field inhomogeneity map. Magn Reson Med. 2008;59:571–580. doi: 10.1002/mrm.21522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hernando D, Kellman P, Haldar JP, Liang ZP. Robust water/fat separation in the presence of large field inhomogeneities using a graph cut algorithm. Magn Reson Med. 2010;63:79–90. doi: 10.1002/mrm.22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeder SB, Pineda AR, Wen Z, Shimakawa A, Yu H, Brittain JH, Gold GE, Beaulieu CH, Pelc NJ. Iterative decomposition of water and fat with echo asymmetry and least-squares estimation (IDEAL): application with fast spin-echo imaging. Magn Reson Med. 2005;54:636–644. doi: 10.1002/mrm.20624. [DOI] [PubMed] [Google Scholar]
- 23.Reeder SB, Wen Z, Yu H, Pineda AR, Gold GE, Markl M, Pelc NJ. Multicoil Dixon chemical species separation with an iterative least-squares estimation method. Magn Reson Med. 2004;51:35–45. doi: 10.1002/mrm.10675. [DOI] [PubMed] [Google Scholar]
- 24.Yu H, Shimakawa A, McKenzie CA, Brodsky E, Brittain JH, Reeder SB. Multiecho water-fat separation and simultaneous R2* estimation with multifrequency fat spectrum modeling. Magn Reson Med. 2008;60:1122–1134. doi: 10.1002/mrm.21737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952–962. [PubMed] [Google Scholar]
- 26.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn Reson Med. 2002;47:1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 27.Ma J, Son JB, Bankson JA, Stafford RJ, Choi H, Ragan D. A fast spin echo two-point Dixon technique and its combination with sensitivity encoding for efficient T2-weighted imaging. Magn Reson Imaging. 2005;23:977–982. doi: 10.1016/j.mri.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Doneva M, Bornert P, Eggers H, Mertins A, Pauly J, Lustig M. Compressed sensing for chemical shift-based water-fat separation. Magn Reson Med. 2010;64:1749–1759. doi: 10.1002/mrm.22563. [DOI] [PubMed] [Google Scholar]
- 29.Sharma SD, Hu HH, Nayak KS. Accelerated water-fat imaging using restricted subspace field map estimation and compressed sensing. Magn Reson Med. 2012;67:650–659. doi: 10.1002/mrm.23052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sharma SD, Hu HH, Nayak KS. Chemical shift encoded water-fat separation using parallel imaging and compressed sensing. Magn Reson Med. 2013;69:456–466. doi: 10.1002/mrm.24270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiens CN, McCurdy CM, Willig-Onwuachi JD, McKenzie CA. R2*-corrected water-fat imaging using compressed sensing and parallel imaging. Magn Reson Med. 2014;71:608–616. doi: 10.1002/mrm.24699. [DOI] [PubMed] [Google Scholar]
- 32.Flask CA, Lewin JS, Duerk JL. Lipid elimination with an echo-shifting N/2-ghost acquisition (LEENA). Proceedings of the 12th Annual Meeting of ISMRM; Kyoto, Japan. 2004; p. 269. [Google Scholar]
- 33.Kellman P, Guttman MA, Herzka DA, McVeigh ER. Phased array ghost elimination (PAGE) for segmented SSFP imaging with interrupted steady-state. Magn Reson Med. 2002;48:1076–1080. doi: 10.1002/mrm.10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kellman P, McVeigh ER. Phased array ghost elimination. NMR Biomed. 2006;19:352–361. doi: 10.1002/nbm.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jenkinson M. Fast, automated, N-dimensional phase-unwrapping algorithm. Magn Reson Med. 2003;49:193–197. doi: 10.1002/mrm.10354. [DOI] [PubMed] [Google Scholar]
- 36.Shah T, Lu L, Dell KM, Pagel MD, Griswold MA, Flask CA. CEST-FISP: a novel technique for rapid chemical exchange saturation transfer MRI at 7 T. Magn Reson Med. 2011;65:432–437. doi: 10.1002/mrm.22637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hines CD, Frydrychowicz A, Hamilton G, Tudorascu DL, Vigen KK, Yu H, McKenzie CA, Sirlin CB, Brittain JH, Reeder SB. T independent, T(2) (*) corrected chemical shift based fat-water separation with multi-peak fat spectral modeling is an accurate and precise measure of hepatic steatosis. J Magn Reson Imaging. 2011;33:873–881. doi: 10.1002/jmri.22514. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


