Budding yeast Mph1 helicase and its orthologs drive multiple DNA transactions. Here, Xue et al. show that the conserved histone fold MHF complex promotes Mph1-mediated repair of damaged replication forks but does not influence the outcome of DNA double-strand break repair.
Keywords: homologous recombination, genome replication, replication fork repair, DNA motor proteins, histone fold proteins, structural maintenance of chromosome proteins
Abstract
Budding yeast Mph1 helicase and its orthologs drive multiple DNA transactions. Elucidating the mechanisms that regulate these motor proteins is central to understanding genome maintenance processes. Here, we show that the conserved histone fold MHF complex promotes Mph1-mediated repair of damaged replication forks but does not influence the outcome of DNA double-strand break repair. Mechanistically, scMHF relieves the inhibition imposed by the structural maintenance of chromosome protein Smc5 on Mph1 activities relevant to replication-associated repair through binding to Mph1 but not DNA. Thus, scMHF is a function-specific enhancer of Mph1 that enables flexible response to different genome repair situations.
DNA motor proteins fulfill critical roles in genome maintenance by processing different DNA structures or nucleoprotein complexes in an ATP-dependent manner. The budding yeast motor protein Mph1 and its orthologs in other eukaryotes possess several activities pertinent to genome replication and repair (Kee and D'Andrea 2010; Whitby 2010). When replication is blocked by template lesions, their replication fork regression activity provides an important tool for remodeling the stalled fork to allow lesion bypass (Gari et al. 2008a,b; Sun et al. 2008; Luke-Glaser et al. 2010; Kang et al. 2011; Zheng et al. 2011; Blackford et al. 2012). During DNA double-strand break (DSB) repair, they can dissociate D loops made by the homologous recombination protein machinery, leading to a noncrossover (NCO), as opposed to crossover (CO), outcome (Sun et al. 2008; Prakash et al. 2009; Kang et al. 2011; Crismani et al. 2012; Lorenz et al. 2012; Mazón and Symington 2013). Additional functions have also been demonstrated for some family members. For example, the human FANCM protein, which is linked to the cancer-prone disease Fanconi anemia (FA), facilitates the repair of interstrand cross-linked DNA in conjunction with other FA proteins (e.g., Xue et al. 2008; Wang et al. 2013). Conceptually, differential regulation of the activities of Mph1 family members would be necessary for achieving a flexible response to unique situations occurring during DNA replication versus DNA repair.
The histone fold proteins Mhf1 and Mhf2 form the MHF complex that structurally resembles the histone H3 and H4 heterotetramer and interacts with Mph1 and its orthologs (Singh et al. 2010; Yan et al. 2010; Daee et al. 2012; Tao et al. 2012; Yang et al. 2012; Bhattacharjee et al. 2013; Fox et al. 2014; Zhao et al. 2014). Recent work in fission yeast and human cells suggests that MHFs positively influence Mph1 family proteins. Fission yeast MHF enhances NCO generation and replication fork processing, while human MHF (hMHF) promotes FANCM chromosomal association and DNA motor activities (Singh et al. 2010; Yan et al. 2010; Lorenz et al. 2012; Bhattacharjee et al. 2013; Fox et al. 2014). The mechanisms underlying MHF functions are not completely understood, but the distinct effects of MHF in different organisms suggest that it may be tailored to the specific needs in an organism. Here, we examine how the budding yeast MHF (referred to as scMHF) influences Mph1 functions.
Like its orthologs, Mph1 promotes NCO formation during DSB repair via its D-loop-disruptive activity and supports recombinational repair during replication using its fork regression and branch migration activities (Schurer et al. 2004; Chen et al. 2009; Mankouri et al. 2009; Prakash et al. 2009; Choi et al. 2010; Ede et al. 2011; Kang et al. 2011; Zheng et al. 2011; Xue et al. 2014). The latter functions are selectively inhibited by the Smc5/6 complex, a structural maintenance of chromosomes (SMC) family member important for genome maintenance. Smc5 binds to the Mph1 regulatory domain and selectively inhibits its DNA fork regression and branch migration activities but not its D-loop-disruptive attribute (Chen et al. 2009; Xue et al. 2014). In addition, cellular defects associated with smc5/6 mutants, including sensitivity to replication stress and accumulation of recombination intermediates, are alleviated by mutations or the removal of Mph1 (Chen et al. 2009; Choi et al. 2010; Chavez et al. 2011). These findings indicate that Smc5/6 negatively regulates select Mph1 functions. In order to understand the biological roles of scMHF, we examined how it is related to this negative regulator of Mph1 and whether it also selectively influences Mph1 functions.
We show that scMHF is epistatic with Mph1 in suppressing defects caused by Smc5/6 mutations. In contrast, scMHF, unlike Mph1, does not affect NCO and CO levels during DSB repair. Biochemical evidence reveals that scMHF specifically relieves the negative influence of Smc5 on Mph1-mediated DNA fork regression and branch migration reactions. Our findings establish scMHF as a function-specific regulator that assists Mph1 in DNA replication fork repair and highlight the functional versatility of histone fold protein complexes.
Results and Discussion
Association of scMHF and Smc5 with the Mph1 regulatory domain
We first examined whether the two Mph1 regulators scMHF and Smc5 interact with Mph1 simultaneously. In vivo tests were performed using epitope-tagged Mph1, Mhf1, and Smc5 expressed at endogenous levels from their chromosomal loci. Mhf1 coimmunoprecipitated with Mph1, consistent with their reported interaction (Fig. 1A, top; Daee et al. 2012). In addition, Mhf1 coimmunoprecipitated with Smc5, suggesting that scMHF also associates with Smc5 in vivo (Fig. 1A, bottom). Next, recombinant scMHF, Mph1, and Smc5 were purified from insect cell or Escherichia coli expression systems (Supplemental Fig. S1A,B; see the Materials and Methods) for in vitro pull-down assays. The results showed that Mph1 interacts with scMHF (Supplemental Fig. S1A, lane 3). In addition, Smc5 pulled down Mph1 in complex with scMHF (Fig. 1B, lane 3). As Smc5 directly interacts with Mph1 (Chen et al. 2009; Xue et al. 2014) but not with scMHF (Supplemental Fig. S1B, lane 3), we concluded that Mph1 plays a bridging role in the trimeric complex with Smc5 and scMHF.
Figure 1.
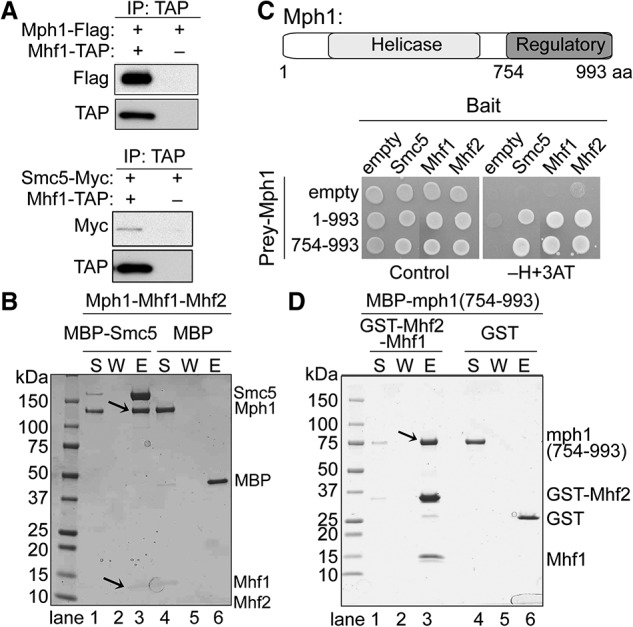
scMHF interacts with Mph1 and Smc5 in vivo and in vitro. (A) Coimmunoprecipitation of Mhf1 with Mph1 and Smc5 from cell extracts. TAP-tagged Mhf1 was immunoprecipitated, and the presence of Flag-tagged Mph1 and Myc-tagged Smc5 in the immunoprecipitates was revealed by Western blot analysis. Extracts containing untagged Mhf1 were included as controls. (B) Smc5 associates with the Mph1–scMHF complex. Mph1–scMHF was retained on amylose resin preloaded with MBP-Smc5 (lanes 1–3) but not MBP (lanes 4–6). Supernatant (S), wash (W), and eluate (E) fractions of the pull-down reactions were analyzed. (C) The Mph1 regulatory region interacts with Smc5 and scMHF. (Top) schematic of Mph1 helicase and regulatory domains. (Bottom) Mph1 (residues 1–993) and its regulatory region (residues 754–993) show two-hybrid interactions with Smc5, Mhf1, and Mhf2, as indicated by growth on the −H+3AT medium. Empty indicates vector-alone situations. (D) The Mph1 regulatory domain binds scMHF. GST-tagged scMHF (lanes 1–3) or GST (lanes 4–6) was incubated with the Mph1 regulatory domain (residues 754–993). Various fractions from the pull-down reactions were examined as in B. The arrow denotes the Mph1 regulatory domain being pulled down by GST-scMHF.
Using yeast two-hybrid (Y2H) and biochemical analyses, we showed previously that Smc5 interacts with the Mph1 C-terminal regulatory domain (residues 754–993) (Fig. 1C; Xue et al. 2014). Here, we found that scMHF also interacts with this Mph1 domain in Y2H analysis (Fig. 1C). In vitro pull-down with purified proteins confirmed this finding, as this domain of Mph1, but not its helicase domain (residues 1–754), binds scMHF (Fig. 1D, lane 3; Supplemental Fig. S1C, lane 9). Taken together, these results show that scMHF and Smc5 can simultaneously interact with Mph1 through its regulatory region.
mhf suppresses smc6 mutant defects in epistasis with mph1
The mph1Δ mutation as well as the mph1 helicase-dead allele suppress the sensitivity of smc6 mutants to replication-blocking agents, such as methyl methanesulfonate (MMS) (Chen et al. 2009; Choi et al. 2010; Chavez et al. 2011). This suppression correlates with a reduction in DNA recombination intermediates that stem from processing of stalled or injured replication forks (Chen et al. 2009; Choi et al. 2010; Chavez et al. 2011). The correlation suggests that suppression is related to the loss of Mph1-mediated replication fork processing. We thus examined smc6 suppression to assess whether scMHF affects this Mph1-mediated function. Removal of either MHF subunit suppressed the MMS sensitivity of the smc6-P4 mutant (Fig. 2A) and the smc6-9 and smc6-56 mutants (Supplemental Fig. S2A). The mhf1Δ mhf2Δ double mutant (mhfΔ) exhibited suppression similar to that of the single mutants (Fig. 2A; Supplemental Fig. S2A), suggesting that the Mhf proteins work together as a functional entity within this context.
Figure 2.
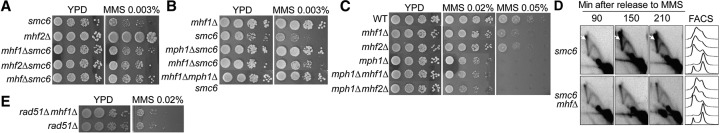
scMHF acts in the same pathway with Mph1 to regulate recombination intermediates generated during replication. (A) mhf1Δ, mhf2Δ, and mhf1Δ mhf2Δ (mhfΔ) suppress the MMS sensitivity of smc6-P4 (marked as smc6) cells. (B) mhf1Δ suppression of smc6-P4 MMS sensitivity shows epistasis with mph1Δ. (C) mhf1Δ and mhf2Δ exhibit mild MMS sensitivity and are epistatic with each other and with mph1Δ. mhf1Δ and mhf2Δ sensitivity is seen at the higher MMS concentration, and their epistatic relationship with mph1Δ is clear at the lower MMS concentration. (D) Two-dimensional gel analyses show reduced X-shaped molecule (X-mol) levels when scMHF was removed in smc6-P4 cells. Log-phase cells were arrested in G2 by nocodazole and released to medium containing 0.03% MMS. Cells were collected at the indicated times with MMS treatment. X-mols are indicated by white arrows. (E) mhf1Δ does not affect the MMS sensitivity of rad51Δ cells.
The strength of suppression of smc6 by mhf1Δ is less than that by mph1Δ, and the suppression conferred by the mhf1Δ mph1Δ double mutant is similar to that of mph1Δ (Fig. 2B). The same results were obtained for mhf2Δ (data not shown). Our interpretation is that scMHF facilitates Mph1-dependent replication-associated repair, but Mph1 retains partial function in its absence. Consistent with this premise, removal of either or both of the Mhf proteins engendered moderate MMS sensitivity but did not sensitize mph1Δ (Fig. 2C; Supplemental Fig. S2B). Collectively, the genetic analyses support the notion that Mhf1 and Mhf2 act together to promote replication-associated repair by Mph1.
scMHF, like Mph1, influences replication-associated recombination structures
To further assess the role of scMHF in replication-associated repair, we examined whether its removal, like mph1Δ, would reduce the level of recombination intermediates in smc6 mutants. As reported previously, smc6-P4 mutant cells accumulated these intermediates, detected as X-shaped molecules (X-mols) in two-dimensional (2D) gel analysis, when cells replicated in the presence of MMS-induced damage (Fig. 2D, arrows). Consistent with its suppression of the MMS sensitivity of smc6-P4 (Fig. 2A), mhfΔ reduced the level of X-mols in smc6-P4 cells (Fig. 2D). These results provide evidence that the smc6-suppressive effect of mph1 and mhf can be partly attributed to a reduction in toxic replication-associated recombination intermediates. Further substantiating this notion, mhf1Δ or mhfΔ did not enhance the MMS sensitivity of cells lacking the recombinase Rad51 (Fig. 2E; data not shown). We note that the suppression of the smc6 mutant sensitivity to MMS through mph1Δ or mhfΔ is not complete, likely because Smc5/6 has additional functions in the replication stress response (Kegel and Sjogren 2010).
scMHF, unlike Mph1, does not affect CO control
Having established that scMHF positively influences Mph1 in replication-associated functions, we tested whether it also affects the ability of Mph1 to regulate the CO:NCO ratio during DNA break repair by homologous recombination (Prakash et al. 2009; Mazón and Symington 2013). We used an established ectopic recombination system to assess whether scMHF also affects the CO:NCO ratio (Prakash et al. 2009). Here, a DSB induced by the HO endonuclease in the MATa sequence on chromosome V is repaired by recombination using the MATa-inc donor sequence on chromosome III (Fig. 3A). The CO and NCO recombination products can be easily discerned by size upon restriction enzyme digestion and Southern blotting (Fig. 3A). Consistent with the published results (Prakash et al. 2009), mph1Δ showed a fivefold greater CO:NCO ratio than wild-type upon DSB induction (Fig. 3B,C). In contrast, mhf1Δ, mhf2Δ, and mhfΔ cells gave the wild-type ratio of CO and NCO recombinants, and introduction of mhf1Δ did not alter the CO:NCO ratio in mph1Δ cells (Fig. 3B,C). DSB repair also proceeds with wild-type efficiency in mhf single and double mutants (Fig. 3B). Thus, unlike Mph1, scMHF has no role in CO versus NCO control. Taking into consideration results presented earlier, these findings reveal that scMHF selectively regulates specific functions of Mph1.
Figure 3.
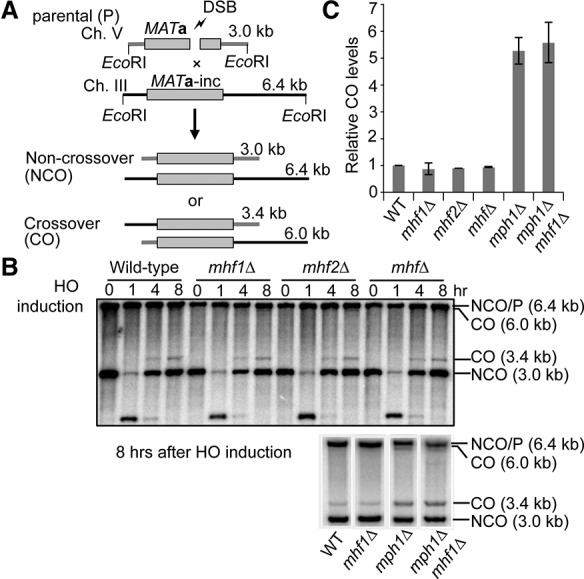
scMHF does not affect CO levels or DSB repair. (A) Schematic of the ectopic recombination assay as described previously (Prakash et al. 2009). (B) Southern blot analysis of gene conversion with and without COs in the indicated strains. (C) Quantification of CO frequency in ectopic recombination. Plotted are the mean values ± SD from at least three independent experiments.
scMHF promotes specific Mph1 functions by relieving Smc5 inhibition
Given that Smc5 exhibits the same selectivity as scMHF in Mph1 regulation and that they both interact with Mph1, we tested the possibility that scMHF may counteract the inhibitory effect of Smc5. As reported previously (Xue et al. 2014), Smc5, when added in an amount stoichiometric to Mph1, reduced Mph1-mediated regression of the mobile replication fork (MRF) (Fig. 4A) and branch migration of the mobile Holliday junction (MHJ) (Fig. 4B). The addition of a similar amount of scMHF restored these activities to an intermediate level, indicative of an ability of scMHF to relieve the inhibitory effect of Smc5 on these Mph1 activities (Fig. 4A,B). Control experiments showed that, in the absence of Smc5, scMHF had no effect on these Mph1 activities (Supplemental Fig. S3A–D). Consistent with a lack of involvement in CO:NCO control, scMHF either alone or in combination with Smc5 did not influence D-loop dissociation by Mph1 (Supplemental Fig. S4A–C). We note that, similar to scMHF, Smc5/6 does not affect CO:NCO control (Lilienthal et al. 2013; S Gangloff, pers. comm.) or D-loop dissociation by Mph1 (Supplemental Fig. S4A–C; Xue et al. 2014). Taken together, the biochemical data indicate that scMHF selectively promotes Mph1 functions by relieving the inhibitory effects of Smc5 in replication fork processing.
Figure 4.
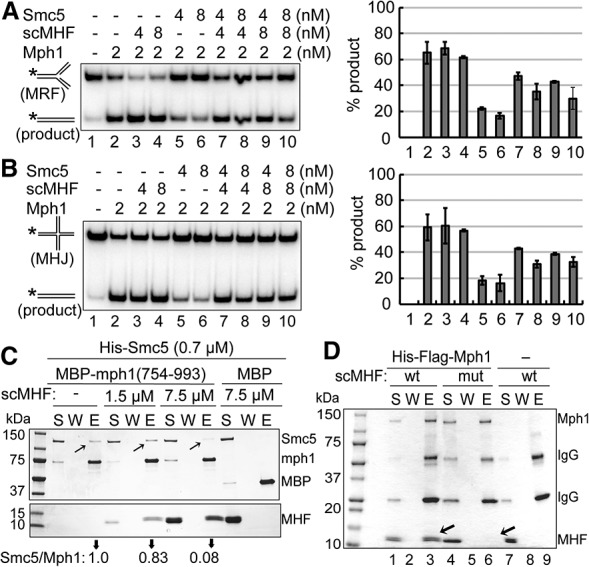
scMHF relieves Smc5 inhibition on Mph1. (A,B) Smc5 inhibits Mph1-mediated regression of a MRF (A) and branch migration of a MHJ (B). The addition of scMHF in the reaction relieves Smc5's inhibitory effect in both reactions. The incubation time was 4 min. The asterisks denote the 32P label. Plotted are the mean values ± SD from three experiments. (C) scMHF competes with Smc5 for Mph1 binding. MBP-tagged mph1 (754–993) prebound to amylose beads was used to pull down Smc5 in the absence or presence of an increasing amount of scMHF. Pull-down reactions were analyzed as in Figure 1C. The relative ratios of Smc5 versus Mph1 in the eluates are indicated. Arrows denote Smc5 bands. (D) mhf-mut does not interact with Mph1 in vitro. Flag-tagged Mph1 was pulled down in combination with scMHF or mhf-mut. Arrows denote scMHF bands or the lack of them.
scMHF attenuates Mph1–Smc5 interaction in vitro
Considering that both scMHF and Smc5 bind to the regulatory domain of Mph1, scMHF may relieve Smc5 inhibition of Mph1 through attenuating Smc5–Mph1 interaction. To test this idea, we examined whether scMHF could affect the interaction of Smc5 with the Mph1 regulatory domain with which both proteins associate. Importantly, in a concentration-dependent manner, the addition of scMHF led to a reduction of Smc5 brought down by the Mph1 regulatory domain (Fig. 4C). This finding provides a plausible mechanism for how scMHF may counteract the Smc5-mediated Mph1 inhibition.
Interaction with Mph1 is indispensable for scMHF function
The above results predicted that scMHF regulation of Mph1 requires its interaction with Mph1. However, as hMHF binds DNA (Singh et al. 2010; Yan et al. 2010), we first clarified whether DNA binding is also involved in scMHF functions. By comparing scMHF and hMHF structures (Yan et al. 2010; Zhao et al. 2014), we found that the former lacks both the C-terminal arm region and positively charged surface residues on hMHF subunits involved in DNA binding (Supplemental Fig. S5A, circled regions). Indeed, DNA mobility shift assays showed that, unlike hMHF, scMHF does not bind DNA fork, HJ DNA, or dsDNA (Supplemental Fig. S5B–D). Moreover, the Mph1–scMHF complex has the same affinity as Mph1 for these DNA substrates (Supplemental Fig. S6A–C). Thus, scMHF does not bind DNA or affect the affinity of Mph1 for DNA.
Next, we tested whether scMHF's selective regulatory effect depends on its interaction with Mph1. Based on the structure of the hMHF–FANCM complex and the sequence alignment of MHF proteins (Singh et al. 2010; Yan et al. 2010; Tao et al. 2012), five residues in scMHF were predicted to be involved in Mph1 interaction (Supplemental Fig. S7A,B). These include three scMhf1 residues (R36, L44, and S54, located in helix 2) and two scMhf2 residues (E70 and L75, located in helix 3). These residues were mutated to create mhf1-R36A/L44A/S54A and mhf2-E70A/L75E mutants (designated as mhf-mut). The mutant proteins expressed well and formed a stable complex that could be readily purified, indicating that the mutations do not affect general protein folding or subunit interactions. In vitro pull-down tests showed that mhf-mut does not bind Mph1 (Fig. 4D, lane 6), suggesting that hMHF–FANCM and scMHF–Mph1 complexes share conserved protein interfaces. Importantly, mhf-mut, unlike the wild-type counterpart, failed to overcome the inhibitory effect of Smc5 on Mph1-mediated catalysis of fork regression and HJ branch migration (Fig. 5A,B), indicating scMHF's function as being reliant on its interaction with Mph1.
Figure 5.
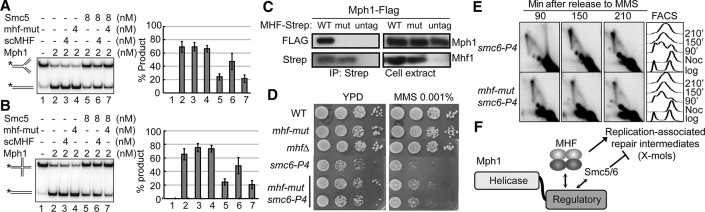
scMHF regulation of Mph1 requires its association with Mph1. (A,B) mhf-mut does not relieve Smc5 inhibition on Mph1. The indicated proteins were incubated with the MRF (A) or MHJ (B) substrate for 4 min. (C) mhf-mut fails to interact with Mph1 in cells. Strains contained Flag-tagged Mph1. (WT) Cells containing strep-tagged Mhf1; (mut) cells containing strep-tagged mhf-mut; (untag) cells containing untagged scMHF. Cell lysates (right) and immunoprecipitation fractions (left) were examined by Western blotting using anti-Flag (top) and anti-Strep (bottom) antibodies. (D) mhf-mut suppresses the MMS sensitivity of smc6-P4 cells. (E) mhf-mut reduces the level of X-mols in smc6-P4 cells, as revealed in 2D gel analysis. (F) Model depicting the role of scMHF in Mph1 regulation. See the text for details.
We then examined mhf-mut in vivo to understand the biological importance of the scMHF and Mph1 interaction. Consistent with the in vitro finding, mhf-mut expressed from the endogenous MHF loci failed to coimmunoprecipitate Mph1 (Fig. 5C). Importantly, mhf-mut, like mhf deletions, suppressed the MMS sensitivity of smc6 mutants (Fig. 5D) and furthermore reduced the levels of X-mols in these mutants in the presence of MMS (Fig. 5E). Taken together, the in vivo and in vitro findings reveal that scMHF promotes specific Mph1 functions during replication-associated repair through its interaction with Mph1.
Mph1 and its orthologs are indispensable for the cellular response to replication stress and DNA break repair, and mutations in FANCM can lead to a predisposition to oncogenesis (Kee and D'Andrea 2010; Whitby 2010). Recent studies identified the histone fold complex MHF as a conserved regulator of these motor proteins (Singh et al. 2010; Yan et al. 2010; Lorenz et al. 2012; Tao et al. 2012; Yang et al. 2012; Bhattacharjee et al. 2013; Sugahara et al. 2013; Wang et al. 2013; Fox et al. 2014; Zhao et al. 2014). Here we provide genetic and biochemical evidence that the budding yeast MHF selectively regulates Mph1 functions to highlight the functional versatility of this family of histone fold complexes.
Several findings suggest that scMHF facilitates Mph1 functions pertaining to replication-associated repair. Like mph1 mutations, mhf mutations suppress the MMS sensitivity of smc6 cells (Fig. 2A,B; Supplemental Fig. S2A); importantly, this correlates with decreased replication-associated recombination intermediates (Fig. 2D). The mhf1 and mhf2 mutations are epistatic with each other and with mph1Δ (Fig. 2A–C; Supplemental Fig. S2), indicating that scMHF collaborates with Mph1 in replication-associated repair. Interestingly, scMHF does not affect the CO:NCO ratio (Fig. 3) even though Mph1 is an important regulator in this regard (Prakash et al. 2009; Mazón and Symington 2013). Thus, scMHF is a selective regulator of Mph1 functions during replication-associated repair but surprisingly has no role in DSB repair. Our study thus provides a new example that distinct functions of multitasking DNA motor proteins can be differentially regulated by histone fold proteins. Our data suggest that scMHF enables versatile and fine-tuned Mph1 responses to genome replication versus DSB repair situations.
Our work also provides mechanistic insights into the function of scMHF. Consistent with our genetic finding, scMHF does not affect D-loop dissociation by Mph1 (Supplemental Fig. S4) but relieves Smc5-mediated inhibition of Mph1 replication fork regression and DNA branch migration activities (Fig. 4A,B). As scMHF alone has no effect on these latter activities of Mph1 (Supplemental Fig. S3), our results indicate that scMHF serves as a specific regulator that relieves the inhibitory function of Smc5/6 on Mph1. Considering the finding that both Smc5 and scMHF interact with the Mph1 regulatory region (Fig. 1C,D; Xue et al. 2014), scMHF may render Mph1 more resistant to Smc5 inhibition through its association with Mph1. This premise is supported by both in vitro binding assays and mutagenesis studies (Figs. 4C,D, 5A–E). Our findings thus provide the first evidence that Mph1-mediated replication fork repair function is subject to regulation by both positive and negative regulators, underscoring the intricate control of this repair pathway (schematized in Fig. 5F). In cells, the role of Smc5 requires the integrity of the Smc5/6 complex. As smc6-P4, smc6-9, and smc6-56 all affect the coiled-coil part of the protein, a region critical for intersubstrate interplay and thus complex function, they likely diminish the ability of Smc5 to bind and/or inhibit Mph1. Testing these ideas would entail biophysical characterization of the Smc5/6 complex and its mutant forms.
Mph1 and its regulators are important for preventing the genome alterations associated with cancer predisposition and other human pathologies. Our work provides mechanistic insights into how antagonistic regulators functionally intersect to affect the engagement of a conserved replication fork repair pathway. These and studies of MHF in other organisms suggest a model that this type of histone fold complex is a situation-dependent regulator of Mph1/FANCM family DNA motors tailored to the specific needs of a given eukaryotic organism. The intellectual and experimental frameworks that we and others have established should facilitate the continuing dissection of functions of these motor proteins in genome maintenance and cancer avoidance.
Materials and methods
Plasmids and yeast strains are listed in Supplemental Table. smc6-P4 contains a K239R mutation (Chen et al. 2009), smc6-9 contains Q903G and S908P mutations, and smc6-56 contains A287V, H379R, and I421T mutations (Sollier et al. 2009). Strain construction and yeast culturing were performed using standard procedures. For damage sensitivity tests, 10-fold serial dilutions of log-phase cultures were spotted on plates with or without MMS. For Y2H assays, the bait plasmids containing GBD fusions (based on the pOBD vector) were cotransformed with the prey plasmids containing GAD fusions (based on the pOAD vector). Methods for Mph1, Smc5, and hMHF expression and purification have been described (Prakash et al. 2009; Zheng et al. 2011; Xue et al. 2014). Expression and purification of scMHF and mutants and assays for DNA mobility shift, coimmunoprecipitation, affinity pull-down, competition pull-down, 2D gel, CO assessment, and DSB repair followed published procedures (Chen et al. 2009; Xue et al. 2014) and/or are detailed in the Supplemental Material.
Supplementary Material
Acknowledgments
We thank Xiao Peng and Prabha Sarangi for their help in editing the manuscript, Xiaofeng Zheng for her help in protein purification, and Bingbing Wan for his help in preparing Supplemental Figure S7. This study was supported by U.S. National Institutes of Health (NIH) grants GM057814, ES007061, ES015632, and ES015252 to P.S.; NIH grant GM080670, American Cancer Society grant RSG-12-013-01-CCG, and a Leukemia and Lymphoma Society Scholar Award to X.Z.; NIH grant GM80600 to G.I.; the Italian Association for Cancer Research (AIRC IG 14171), Fondazione Telethon (GGP12160), and European Research Council (REPSUBREP 242928) grants to D.B.; NIH grant K99/R00 ES021441 to H.N.; and NIH grant T32 GM008539 to J.N.B. as a trainee.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.259143.115.
References
- Bhattacharjee S, Osman F, Feeney L, Lorenz A, Bryer C, Whitby MC. 2013. MHF1–2/CENP-S-X performs distinct roles in centromere metabolism and genetic recombination. Open Biol 3: 130102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Blackford AN, Schwab RA, Nieminuszczy J, Deans AJ, West SC, Niedzwiedz W. 2012. The DNA translocase activity of FANCM protects stalled replication forks. Hum Mol Gen 21: 2005–2016. [DOI] [PubMed] [Google Scholar]
- Chavez A, Agrawal V, Johnson FB. 2011. Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes Smc5/6 complex. J Biol Chem 286: 5119–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y-H, Choi K, Szakal B, Arenz J, Duan X, Ye H, Branzei D, Zhao X. 2009. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc Natl Acad Sci 106: 21252–21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Szakal B, Chen Y-H, Branzei D, Zhao X. 2010. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in S. cerevisiae. Mol Biol Cell 21: 2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R. 2012. FANCM limits meiotic crossovers. Science 336: 1588–1590. [DOI] [PubMed] [Google Scholar]
- Daee DL, Ferrari E, Longerich S, Zheng X-F, Xue X, Branzei D, Sung P, Myung K. 2012. Rad5-dependent DNA repair functions of the S. cerevisiae FANCM homolog Mph1. J Biol Chem 287: 26563–26575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ede C, Rudolph CJ, Lehmann S, Schürer KA, Kramer W. 2011. Budding yeast Mph1 promotes sister chromatid interactions by a mechanism involving strand invasion. DNA Repair (Amst) 10: 45–55. [DOI] [PubMed] [Google Scholar]
- Fox D, Yan Z, Ling C, Zhao Y, Lee D-Y, Fukagawa T, Yang W, Wang W. 2014. The histone-fold complex MHF is remodeled by FANCM to recognize branched DNA and protect genome stability. Cell Res 24: 560–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Décaillet C, Delannoy M, Wu L, Constantinou A. 2008a. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci 105: 16107–16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. 2008b. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell 29: 141–148. [DOI] [PubMed] [Google Scholar]
- Kang Y-H, Munashingha PR, Lee C-H, Nguyen TA, Seo Y-S. 2011. Biochemical studies of the S. cerevisiae Mph1 helicase on junction-containing DNA structures. Nucleic Acids Res 40: 2089–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee Y, D'Andrea AD. 2010. Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev 24: 1680–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kegel A, Sjogren C. 2010. The Smc5/6 complex: more than repair? Cold Spring Harb Symp Quant Biol 75: 179–187. [DOI] [PubMed] [Google Scholar]
- Lilienthal I, Kanno T, Sjögren C. 2013. Inhibition of the Smc5/6 complex during meiosis perturbs joint molecule formation and resolution without significantly changing crossover or non-crossover levels. PLoS Genet 9: e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Osman F, Sun W, Nandi S, Steinacher R, Whitby MC. 2012. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science 336: 1585–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S, Luke B, Grossi S, Constantinou A. 2010. FANCM regulates DNA chain elongation and is stabilized by S-phase checkpoint signalling. EMBO J 29: 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri HW, Ngo H-P, Hickson ID. 2009. Esc2 and Sgs1 act in functionally distinct branches of the homologous recombination repair pathway in S. cerevisiae. Mol Biol Cell 20: 1683–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazón G, Symington LS. 2013. Mph1 and Mus81–Mms4 prevent aberrant processing of mitotic recombination intermediates. Mol Cell 52: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. 2009. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev 23: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurer K, Rudolph C, Ulrich H, Kramer W. 2004. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from homologous recombination, but not from postreplicative repair. Genetics 166: 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Saro D, Ali AM, Zheng X-F, Du C-h, Killen MW, Sachpatzidis A, Wahengbam K, Pierce AJ, Xiong Y, et al. 2010. MHF1–MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Mol Cell 37: 879–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollier J, Driscoll R, Castellucci F, Foiani M, Jackson SP, Branzei D. 2009. The S. cerevisiae Esc2 and Smc5–6 proteins promote sister chromatid junction mediated intra-S repair. Mol Biol Cell 20: 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara R, Mon H, Lee JM, Kusakabe T. 2013. Middle region of FancM interacts with Mhf and Rmi1 in silkworms, a species lacking the Fanconi anaemia (FA) core complex. Insect Mol Biol 23: 185–198. [DOI] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. 2008. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell 32: 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Jin C, Li X, Qi S, Chu L, Niu L, Yao X, Teng M. 2012. The structure of the FANCM–MHF complex reveals physical features for functional assembly. Nat Commun 3: 782–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Leung JW, Jiang Y, Lowery MG, Do H, Vasquez KM, Chen J, Wang W, Li L. 2013. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol Cell 49: 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby MC. 2010. The FANCM family of DNA helicases/translocases. DNA Repair (Amst) 9: 224–236. [DOI] [PubMed] [Google Scholar]
- Xue Y, Li Y, Guo R, Ling C, Wang W. 2008. FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum Mol Gen 17: 1641–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue X, Choi K, Bonner J, Chiba T, Kwon Y, Xu Y, Sanchez H, Wyman C, Niu H, Zhao X, et al. 2014. Restriction of replication fork regression activities by a conserved SMC Complex. Mol Cell 56: 436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z, Delannoy M, Ling C, Daee D, Osman F, Muniandy PA, Shen X, Oostra AB, Du H, Steltenpool J, et al. 2010. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Mol Cell 37: 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Zhang T, Tao Y, Wu L, Li H-t, Zhou J-Q, Zhong C, Ding J. 2012. S. cerevisiae MHF complex structurally resembles the histones (H3–H4)2 heterotetramer and functions as a heterotetramer. Structure 20: 364–370. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Saro D, Sachpatzidis A, Singh TR, Schlingman D, Zheng X-F, Mack A, Tsai M-S, Mochrie S, Regan L, et al. 2014. The MHF complex senses branched DNA by binding a pair of crossover DNA duplexes. Nat Commun 5: 2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X-F, Prakash R, Saro D, Longerich S, Niu H, Sung P. 2011. Processing of DNA structures via DNA unwinding and branch migration by the S. cerevisiae Mph1 protein. DNA Repair (Amst) 10: 1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


