Abstract
Haloarchaeal genomes are generally composed of multiple replicons, and each replicon has a single or multiple replication origin(s). The comparative genomic analysis of replication origins from closely related species can be used to reveal the evolutionary mechanisms that account for the development of multiple origin systems. Multiple replication origins have been in silico and experimentally investigated in Haloarcula hispanica, which raise the possibility for comparisons of multiple replication origins in Haloarcula species. Thus, we performed a comparison of H. hispanica replication origins with those from five additional Haloarcula species. We demonstrated that the multiple replication origins in the chromosome were evolved independently multiple times from the oriC1-dependent ancestral chromosome. Particularly, the two origins oriC1 and oriC2 were conserved in location, and both of them were adjacent to an rRNA operon, suggestive of correlations in replication and expression of surrounding genes that may promote the conservation of these two origins. Some chromosomal variable regions were used as hotspots for origin evolution in which replication origins were continually being acquired, lost, and disrupted. Furthermore, we demonstrated that autonomously replicating sequence plasmids with H. hispanica minichromosomal replication origins were extremely unstable. Because both organization and replication origins of minichromosomes were not conserved, we proposed an association between the evolution of extrachromosomal replicons and origin variation. Taken together, we provided insights into the evolutionary history of multiple replication origins in Haloarcula species, and proposed a general model of association between the dynamics of multiple replication origins and the evolution of multireplicon genome architecture in haloarchaea.
Keywords: multiple replication origins, origin dynamics, evolution, genome architecture, halophilic archaea
Introduction
Similar to bacteria, archaea contain circular chromosomes and initiate chromosome replication at specific sites known as replication origins (Robinson and Bell 2005). However, despite the first description of archaeal replication origins demonstrated that the chromosome of the hyperthermophilic archaeon Pyrococcus abyssi uses a single origin to initiate replication (Myllykallio et al. 2000; Matsunaga et al. 2001, 2003), many archaea characterized to date harbor multiple discrete replication origins (Lundgren et al. 2004; Norais et al. 2007; Robinson and Bell 2007; Coker et al. 2009; Pelve et al. 2012, 2013; Wu et al. 2012, 2014; Hawkins et al. 2013). Among archaea, multiple replication origins have been described in great detail in Sulfolobus species, providing insights into the characterization, utilization, and evolution of the three active replication origins in their single chromosome (Robinson et al. 2004; Dueber et al. 2007; Robinson and Bell 2007; Duggin et al. 2008; Samson et al. 2013). The characterized archaeal origins are normally conserved in structure but vary in sequence among different origins in terms of origin recognition boxes (ORBs) and origin-associated initiator genes (Wu et al. 2012). Recently, the specific recognition of initiator genes to their cognate origins was experimentally established in Sulfolobus islandicus (Samson et al. 2013) and Haloarcula hispanica (Wu et al. 2014). The origins together with their adjacent initiator genes are considered to be distinct replicator-initiator systems, and the integration of extrachromosomal elements has been proposed to account for mosaics of multiple replication origins in specific archaeal chromosomes (Robinson and Bell 2007; Wu et al. 2012). This inferred that the specific linkage between the ORB elements and the corresponding initiator gene is conserved during a long-term evolution, and such a conserved replicator-initiator pairing may translocate frequently among different species.
Haloarchaea are a distinct group of archaea that thrive in hypersaline environments. Haloarchaeal genomes are generally distributed among several replicons, and each replicon has a single or multiple replication origin(s) (Capes et al. 2011), which complicates our understanding of their replication characteristics and evolutionary history. Recently, we performed an in silico study to predict replication origins, and the results demonstrated that the occurrence of multiple replication origins is widespread in haloarchaea and that up to seven putative origins are located on the Haloterrigena turkmenica chromosome (Wu et al. 2012). Furthermore, the active origins have been experimentally studied in three model systems: Halobacterium sp. strain NRC-1 (Berquist and Dassarma 2003; Coker et al. 2009), Haloferax volcanii (Norais et al. 2007; Hawkins et al. 2013), and H. hispanica (Wu et al. 2012, 2014). Remarkably, replication origins are highly diverse in both sequence and utilization in haloarchaea. Unexpectedly, the number of predicted origins was normally greater than that of active origins in each characterized strain, particularly in the extrachromosomal replicons. Thus, it is intriguing to investigate the evolutionary processes that accounted for the development of multiple replication origins in haloarchaea. Insertion, deletion, and genome rearrangement occurred frequently in haloarchaea (Dyall-Smith et al. 2011), and we demonstrated that replication origins were transferred frequently among different haloarchaea (Wu et al. 2012). In addition, a comparative genomic analysis of the replication origins in the chromosomes of H. hispanica and Haloarcula marismortui revealed that strain-specific origins are located in the chromosomal divergent regions (Wu et al. 2012). Thus, there might be correlations between origin diversity and genome variation. Comparative genomic analyses of replication origins have been performed to address the evolution of the replication origins at the structural, locational, and regulatory levels in budding yeasts (Di Rienzi et al. 2012; Muller and Nieduszynski 2012). Thus, a comparison of the replication origins from closely related haloarchaeal species should reveal the evolutionary processes responsible for the development of multiple origins in haloarchaea.
We have previously investigated the utilization of multiple replication origins in H. hispanica. Although both the main chromosome and minichromosome use two active replication origins in vivo, one active replication origin per replicon is sufficient for genome replication (Wu et al. 2012, 2014). The two active replication origins in the chromosome were proposed to originate from integration of the oriC2-cdc6E into an ancestral chromosome that was dependent on oriC1-cdc6A (Wu et al. 2014). In addition, three replication origins, oriC3-cdc6D in the chromosome and oriC4-cdc6G and oriC5-cdc6H in the minichromosome, were proven to be nonfunctional and were considered to be deficient or dormant replication origins (Wu et al. 2012, 2014). To further address how multiple replication origins evolved over evolutionary time, in this study, we performed a comparative genomic analysis of the H. hispanica replication origins with those obtained from five additional Haloarcula species, specifically focusing on the dissection of the three H. hispanica nonfunctional origins. These comparative analyses demonstrated frequent variation of the replication origins in both chromosomal variable regions and unstable extrachromosomal replicons, while the conserved replication origins were maintained in location which may be promoted by their impacts on surrounding genes or on the stabilization of minireplicons. Comparative analyses of the three H. hispanica deficient origins with their homologs in other strains demonstrated that all of them were deficient, which might be due to structural destruction accompanying the frequent insertion and deletion events in the chromosomal variable regions or the frequent rearrangement of variable minireplicons.
Materials and Methods
Strains, Plasmids, and Culture Conditions
Escherichia coli cells were grown in Luria–Bertani medium at 37 °C, and 100 μg/ml ampicillin was added when required. H. hispanica and H. marismortui strains were cultured at 37 °C in nutrient-rich medium AS-168 as previously described (Wu et al. 2012). When required, 3 μg/ml mevinolin was added. The investigation of the autonomous replication ability of replication origins was based on the plasmid pBI101 (Zhou et al. 2007). pBI101 plasmids containing the replication origins pOC1, pOC2, pOC6, pOC7, and pOP were previously constructed (Wu et al. 2014).
Autonomous Replication Ability Assay
The plasmid-based assay of autonomously replicating sequence (ARS) activity was performed as previously described (Wu et al. 2012). In each assay, the origin region together with the cdc6 coding region was amplified from H. hispanica or H. marismortui genomic DNA and was cloned into the nonreplicating plasmid pBI101. After sequencing, the plasmids were then introduced into H. hispanica or the corresponding origin-deletion strains (Wu et al. 2014) using a polyethylene glycol-mediated transformation method (Cline et al. 1989), and the mevinolin-resistant transformants were selected on AS-168 plates with 3 μg/ml mevinolin.
Estimation of ARS Plasmid Stability
The estimated stability of the ARS plasmid with different replication origins was performed as previously described (Norais et al. 2007). The five ARS plasmids were transformed into H. hispanica or the corresponding origin-deletion strains using a polyethylene glycol-mediated transformation method (Cline et al. 1989), and the mevinolin-resistant transformants were selected on AS-168 plates with 3 μg/ml mevinolin. For each transformation, a colony containing the ARS plasmid was selected and inoculated into 10 ml of AS-168 (Mev) broth under selection. The cultures were then propagated twice in AS-168 broth without mevinolin, and at each passage, the cultures were plated on AS168 plates. Finally, the colonies were patched on an AS168 (Mev) plates to determine the fraction of mevinolin-resistant cells.
Genome Resources and Comparative Genomics
Three completed genome sequences, namely those of H. hispanica ATCC 33960 (Liu et al. 2011), H. hispanica N61 (Ding et al. 2014), and H. marismortui ATCC 43049 (Baliga et al. 2004), and contigs from four draft genomes (Lynch et al. 2012), namely those of Haloarcula amylolytica JCM 13557, Haloarcula argentinensis DSM 12282, Haloarcula japonica DSM 6131, and Haloarcula sinaiiensis ATCC 33800, were available through The National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/genome/). Genome visualization and comparative genomics were performed using the CGView Server (Grant and Stothard 2008) with default parameters (http://stothard.afns.ualberta.ca/cgview_server/).
Distribution and Contextual Analysis of Replication Origins
The distribution of the H. hispanica replication origins in Haloarcula species was performed via BLASTP analysis (BLOSUM62 matrix; 1 × 10−6 as an e value cutoff) of origin-associated Cdc6 proteins against a specific Haloarcula genome (http://blast.ncbi.nlm.nih.gov/) (Altschul et al. 1990). Genome context analysis was performed using the NCBI Genome Workbench and scrutinized manually. Pairwise and multiple alignments of replication origins or Cdc6 proteins were generated using the DNAMAM software (for Windows, version 2.6). The identification of ORB elements in the replication origins was performed via motif searches using the MEME software (motif size: 20-40; ZOOPS model) (Bailey et al. 2006).
Phylogenetic Analysis
The RNA polymerase B' subunit (RpoB') was used for phylogenetic analysis as described (Minegishi et al. 2010). The RpoB' amino acid sequences were collected, and multiple alignments were generated using Clustal (substitution matrix = BLOSUM; gap-opening penalty = 10; gap-extension penalty = 0.1). Maximum-likelihood phylogeny was performed with PHYML v3.0 with an LG substitution model and 100 nonparametric bootstrap replicates (Guindon et al. 2010).
Results
Dynamics of Multiple Replication Origins in Haloarcula Species
Recently, multiple replication origins have been in silico predicted and experimentally confirmed in H. hispanica (Wu et al. 2012, 2014). To further understand the evolutionary history of these multiple replication origins, a comparison of replication origins of H. hispanica was performed with those of five additional Haloarcula species that are closely related to H. hispanica. Thus, we first analyzed the distribution of the H. hispanica replication origins in Haloarcula species using BLASTP analyses of the eight Cdc6 proteins associated with putative replication origins against the following five Haloarcula species: H. amylolytica, H. argentinensis, H. japonica, H. marismortui, and H. sinaiiensis (table 1). There were three conserved replication origins in Haloarcula species: oriC1-cdc6A, oriC2-cdc6E, and oriP-cdc6K (table 1). Previous studies (Robinson et al. 2004; Coker et al. 2009; Wu et al. 2012; Raymann et al. 2014) have demonstrated that the oriC1-cdc6A is broadly conserved across the archaeal domain of life, which has been considered to be inherited from the ancestor of archaea. In contrast, the oriC2-cdc6E and oriP-cdc6K were not present in all haloarchaea; thus, the conservation of these two replication origins appeared to be restricted to Haloarcula species, indicating that the acquisition of these two replication origins by Haloarcula occurred before the divergence of individual strains. The remaining five replication origins were either present or absent in an individual strain, that is, the distribution of these origins was variable in Haloarcula species (table 1). It is possible that these variable replication origins can be attributed to strain-specific origin gains and losses, which would account for the diversity of replication origins in Haloarcula species.
Table 1.
Distribution of H. hispanica Putative Replication Origins in Five Additional Haloarcula Species
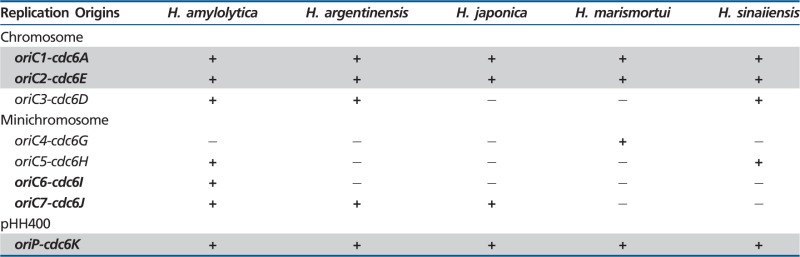 |
Note.—The plus (+) and minus (−) signs indicate the presence and absence, respectively, of the H. hispanica replication origins in the other five Haloarcula genomes. The five functional replication origins of the eight putative origins in H. hispanica are indicated in bold. The conserved replication origins in Haloarcula species are highlighted with a gray background.
Taken together, these findings show that multiple replication origins are dynamic in Haloarcula species, and these origins could be separated into two categories: conserved and variable. The conserved replication origins were present in all of the analyzed strains, and are thus likely inherited from the original archaeal ancestor or a Haloarcula ancestor and maintained stable. The variable origins were not found in all of the strains, and this may be due to strain-specific acquisition or deletion events.
Maintenance of the Conserved Replication Origins via Their Impacts on Surrounding Environments
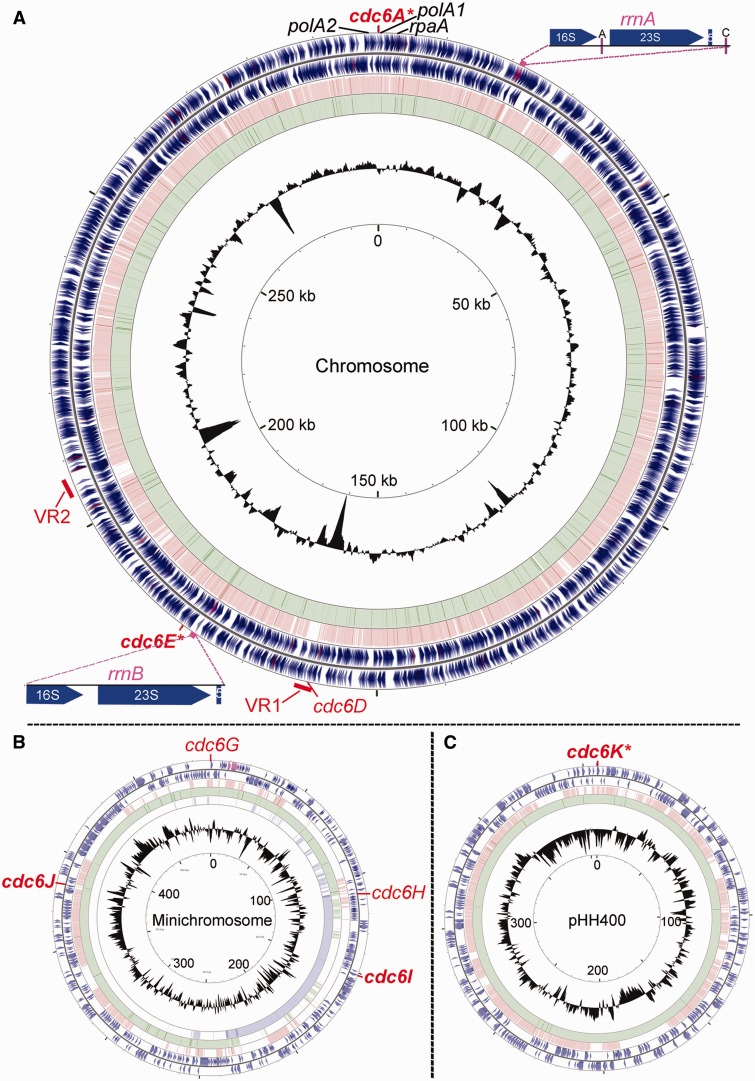
The two conserved replication origins oriC1-cdc6A and oriC2-cdc6E were located in the main chromosome of Haloarcula species (fig. 1A). Our previous marker frequency analysis (MFA) results demonstrated that both of these two replication origins are active in vivo and that each of these replication origins is sufficient for chromosome replication (Wu et al. 2014). Thus, it raises the question of why these origins were conserved over evolutionary time, particularly when we proposed that oriC2-cdc6E appeared to be acquired in the branch leading to Haloarcula. To address this question, we examined the common characteristics between the conserved replication origins, which might play important roles on their evolutionary conservation. We found that oriC1-cdc6A and oriC2-cdc6E are conserved in their chromosomal location (fig. 1A), suggesting that these two conserved origins might be promoted via their surrounding environments. This hypothesis is reinforced by the cluster of replication-associated genes, such as DNA polymerase genes (polA1 and polA2) and replication protein A (rpaA) (fig. 1A), around the oriC1-cdc6A origin. In addition, we found that an rRNA operon (rrn operon) was positioned downstream of each origin, and these two rrn operons are approximately 211 and 20 kb away from oriC1-cdc6A and oriC2-cdc6E, respectively (fig. 1A). Similar genomic organization that rrn operons are close to (and transcribed away from) the two chromosomal replication origins has also been observed in Haloferax volcanii (Norais et al. 2007; Hartman et al. 2010). This organization may ensure that the two rrn operons (and other genes closer to the origins) are able to be replicated earlier compared with the rest of the genome, thereby enhancing the expression of these genes due to a gene dosage effect. In addition, the concurrent rounds of DNA replication has been indicated in Haloferax volcanii (Hawkins et al. 2013), which would be expected to further amplify the dosage. Conversely, high expression of replication-associated genes (such as cdc6 genes) contributes to active utilization of origins that may promote their conservation. Furthermore, both the two rrn operons are directed away from the origins (fig. 1A), which may ensure codirection of replication and transcription that improves the stability of surrounding genes (Paul et al. 2013). We proposed that, similar to oriC1-cdc6A, the effects of the oriC2-cdc6E on surrounding genes (gene expression or gene stability) after its acquisition in the Haloarcula ancestor may promote its conservation in the divergence of individual strains. The other conserved replication origin was oriP-cdc6K from the megaplasmid pHH400, and this replicon was considered to be stable (fig. 1C), suggesting that the conservation of oriP-cdc6K might ensure the stability of pHH400 in Haloarcula species. Based on these findings, we proposed that origin conservation may be promoted by the effects of these origins on their surrounding environments, which may explain why conserved replication origins are also active in vivo (table 1).
Fig. 1.—
Comparative genomic analysis of the replication origins in Haloarcula species. (A) The chromosome. From the outside inward: The first and second circles indicate the annotated protein-coding regions on the plus and minus strands in H. hispanica ATCC 33960. The third and fourth circles represent the BLASTN hits of the chromosome of H. hispanica ATCC 33960 with those of two other Haloarcula genomes: H. marismortui ATCC 43049 and H. hispanica N61, respectively. The innermost circle shows a plot of the GC content along the chromosome of H. hispanica ATCC 33960. The two large variable regions (VR1-2, red) and the positions of two rrn operons are indicated. The two rrn operons are both directed away from the origins. (B and C) Minichromosome and megaplasmid pHH400. The circles in B and C are the same as in A with the exception that the H. hispanica N61 minichromosome was divided into two replicons: The minichromosome and pHH126 (the fourth and fifth circles in B). In all three replicons, the cdc6 genes associated with the putative replication origins in H. hispanica are indicated, and those with active origins are indicated in bold. The conserved ori-cdc6 origins in Haloarcula species, namely oriC1-cdc6A and oriC2-cdc6E on the chromosome and oriP-cdc6K on PHH400, are highlighted with star signs. GC, Guanine-Cytosine.
Replication Origins Evolved Frequently in Genomic Variable Regions
In contrast with conserved replication origins, variable replication origins are normally strain-specific and are located in divergent regions of the genome. We previously proposed that these origins were recently acquired via translocation events (Wu et al. 2012). Thus, we considered that the characterization of these variable origins might contribute to our understanding of the evolutionary history of the development of multiple replication origins in haloarchaeal genomes and of the association between origin diversity and genome variation. To address this hypothesis, analyses of the localization and genome context of the variable origins were performed.
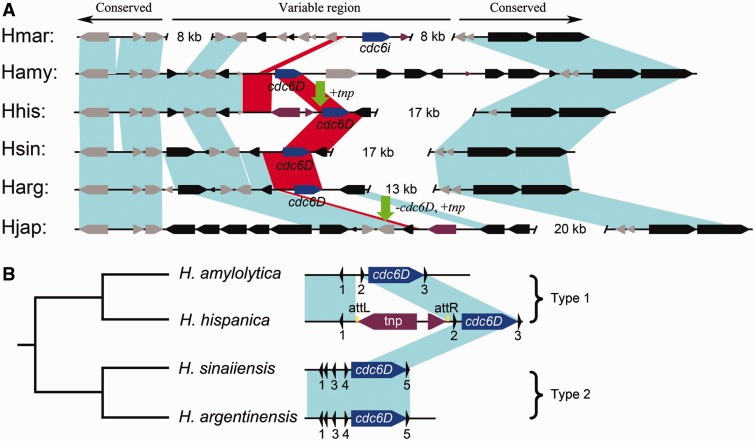
The third replication origin (oriC3-cdc6D) in the H. hispanica chromosome was not conserved in Haloarcula species because its homologs were only observed in three other species, H. amylolytica, H. argentinensis, and H. sinaiiensis (table 1). The chromosomes of H. hispanica and H. marismortui were completely collinear with only two large regions of species-specific variation, and the cdc6D gene was located in VR1 (Wu et al. 2012 and fig. 1A). A detailed comparison with four additional Haloarcula species revealed that this region is highly variable and is located in exactly the same position relative to the conserved regions in each chromosome (fig. 2A). This observation suggests that this region is a hotspot for the integration of foreign sequences, that is, it is a location in which insertion and deletion events occurred frequently, and this conclusion was supported by the distribution of transposases in this region of each chromosome (fig. 2A). Interestingly, similar to the situation in H. hispanica, the cdc6D homolog was also located in this variable region in H. amylolytica, H. argentinensis, and H. sinaiiensis (fig. 2A). It was suggested that the independent gain and loss of oriC3-cdc6D homologs in this frequently variable region (VR1) might account for the variation of this origin among Haloarcula chromosomes.
Fig. 2.—
Replication origins evolved frequently in chromosomal variable regions. (A) Sequence similarity and gene order analyses of the chromosomal variable region VR1. The cdc6 homologs in VR1 are indicated in blue. The variable regions and the edges of the conserved regions were confirmed using BLASTN alignments of the sequences, and a sequence similarity greater than 70% is represented with teal shading; the red-shaded regions indicate homologous regions around the cdc6D gene. The vertical green arrows indicate that the local regions were disrupted by putative transposases (shown in purple). (B) Two types of cdc6D-associated replication origins in VR1. (Left) Phylogenetic relationship of the four Haloarcula species (not to scale). (Right) Physical map of the cdc6D-associated replication origins; the ORB motifs are indicated with small triangles. The shaded teal regions denote a similarity greater than 70%, as determined through BLASTN analyses, and the attachment (att) sites for the insertion of a putative transposase within the oriC3-cdc6 origin of H. hispanica are indicated in yellow.
All four replication origins located in the H. hispanica minichromosome, including the two functional origins oriC6-cdc6I and oriC7-cdc6J, were not conserved in Haloarcula (table 1). The minichromosome appeared to be highly dynamic in Haloarcula species, as well as the two laboratory-derived H. hispanica genomes, H. hispanica ATCC 33960 and H. hispanica N61 (fig. 1B). The 489-kb minichromosome in H. hispanica ATCC 33960 appeared to be divided into two replicons in H. hispanica N61, a 363-kb minichromosome and a 126-kb pHH126. Nevertheless, large and small pairs of orthologs were observed throughout the comparison (fig. 1B). These observations indicate that this replicon was not stable and was reconstructed frequently in the divergence of the species. Thus, we proposed that the frequent reconstruction of the minichromosome might account for the variable replication origins in this replicon because independent gains and losses of replication origins accompanied the reconstruction processes. In addition, oriC4-cdc6G and oriC5-cdc6H were confirmed to be nonfunctional, which might be due to their incomplete acquisition or functional disruption during the reconstruction of the minichromosome.
Taken together, these findings revealed a rapid evolution of replication origins in the variable genome regions during the integration of foreign sequences at chromosomal variable regions or the reconstruction of extrachromosomal replicons, suggesting that variable replication origins might be associated with genomic variable regions.
Addition, Deletion, and Disruption of Replication Origins in Chromosomal Variable Regions
As previously described, the replication origins in the variable regions evolved frequently; thus, these variable origins could serve as models for evolutionary mechanisms responsible for origin diversity. We addressed the evolutionary characteristics of cdc6D-associated replication origins by analyzing the origin region directly upstream of the cdc6D gene in each genome. Interestingly, although the ORB elements showed high sequence conservation, two types of origins with different structures were observed. In particular, the cdc6D-associated origins showed high conservation between H. hispanica and H. amylolytica (type 1), or between H. sinaiiensis and H. argentinensis (type 2); however, these two types of cdc6D-associated origins showed limited conservation both in sequence and in structure (fig. 2B). These results indicated that, although the linkage-specificity of the ORB sequences and Cdc6D protein was conserved over long evolutionary distances, the structure of the origin was only conserved over short evolutionary distances. More importantly, in conjunction with the gene order analysis, we proposed that the cdc6D-associated origin was integrated at this variable region in different chromosomes from two independent origin gains. Integration of extrachromosomal elements that accounts for replicon evolution has been previously proposed in Aeropyrum pernix (Robinson and Bell 2007). The oriC3-cdc6D of H. hispanica was disrupted by a putative transposase in H. hispanica compared with that of H. amylolytica. In addition, the cdc6D-associated origin of H. japonica appeared to be replaced by a putative transposase via coupled insertion and deletion events compared with that of H. argentinensis. Thus, we concluded that frequent strain-specific addition, deletion, and disruption events in the variable region accounted for the diversity of the oriC3-cdc6D origin in different chromosomes. A distinct origin associated with cdc6i was observed in this variable region in the H. marismortui chromosome, which supports the frequent acquisition of replication origins in variable regions. It is also likely the explanation that a cdc6g-associated origin was observed in the other variable region (VR2) of the chromosome in H. marismortui but not in H. hispanica (Wu et al. 2012). Taken together, the results suggest that replication origins evolved multiple times in the variable regions of the chromosome and were continually being created and destroyed.
ARS Plasmids with Minichromosomal Replication Origins Are Significantly Unstable
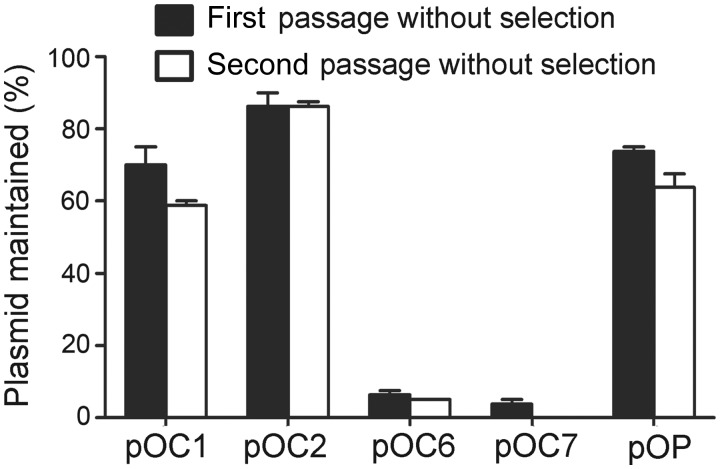
In Haloarcula species, we demonstrated that both the organization and replication origins of the minichromosome are variable, but pHH400 and its origin are stable. Thus, we proposed that there might be correlations between the replication origins and the fate of extrachromosomal replicons. To address this question, we determined the stabilities of ARS plasmids with the five functional replication origins of H. hispanica: oriC1 (pOC1) and oriC2 (pOC2) from the chromosome, oriC6 (pOC6) and oriC7 (pOC7) from the minichromosome, and oriP (pOP) from pHH400. Transformants containing each of these five ARS plasmids were propagated in AS168 broth without mevinolin selection and passaged every 4 days. At each passage, the cultures were diluted and plated on AS168 nonselective medium, and the colonies were subsequently patched on mevinolin-selective plates to determine the fraction of mevinolin-resistant cells. As shown in figure 3, compared with pOC1, pOC2, and pOP, the pOC6 and pOC7 were significantly less stable. After two passages, the oriC6 plasmid pOC6 was maintained in only 5% of the colonies, and the oriC7 plasmid pOC7 was completely lost (fig. 3). Importantly, our results demonstrated that the oriC6- and oriC7-containing ARS plasmids were significantly less stable compared with the oriP-containing plasmid, which suggests that the minichromosomal replication origins are less able to maintain replicon stability than the origin from pHH400. Thus, we proposed that the instability of oriC6- and oriC7-containing ARS plasmids might reflect the unstable nature of the minichromosomes in Haloarcula species, which conversely forced the variation of their bearing replication origins, suggesting an association between the evolution of extrachromosomal replicons and the bearing replication origins.
Fig. 3.—
Instability of ARS plasmids with the two minichromosomal origins oriC6 and oriC7. Transformants with pOC1, pOC2, pOC6, pOC7, and pOP were propagated in nonselective AS168 broth for two passages. At each passage, the cultures were plated on AS168 medium, and the colonies were patched on AS168 (Mev) selective plates to determine the fraction of mevinolin-resistant cells.
Risk Factors for Variable Replication Origins
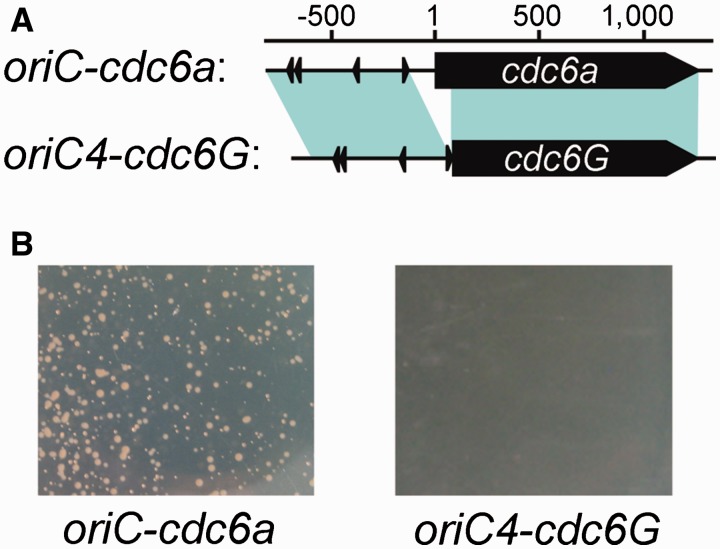
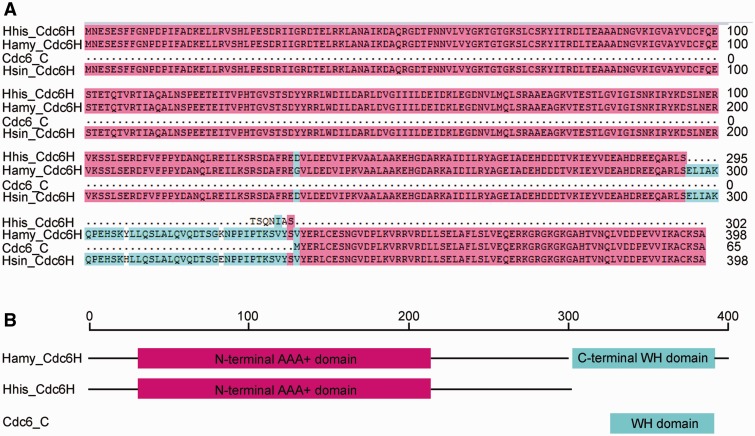
We have revealed the association between origin diversity and genome variation and have confirmed that some of these variable origins are nonfunctional. These findings raise the questions of whether and why frequent evolution of the variable regions affects the function of bearing replication origins. To address this question, we performed comparative analysis to characterize the three H. hispanica nonfunctional replication origins with their homologs from other Haloarcula species. As previously described, we found that the origin region of oriC3-cdc6D was disrupted by an insertion of a putative transposase in H. hispanica (fig. 2B), which might disrupt the function of this origin. In addition, a homolog of oriC4-cdc6G, designated as oriC-cdc6a, was found in the minichromosome of H. marismortui. The alignment analysis of oriC4-cdc6G and oriC-cdc6a revealed high homology extended over the full length of the ori-cdc6 sequence with the exception of a 203-bp sequence loss in oriC4-cdc6G. This sequence included the 5′-terminal 75-bp coding region and the entire promoter of the cdc6 gene (fig. 4A). Indeed, the oriC-cdc6a origin presented ARS activity because it was able to confer replication ability to a nonreplicating plasmid (fig. 4B). Thus, oriC-cdc6a might be the active origin for replication of the H. marismortui minichromosome, which explains why neither of the two functional origins of the H. hispanica minichromosome was observed in the H. marismortui minichromosome (table 1). The comparison analysis of oriC5-cdc6H from different species revealed the lack of the C-terminal winged-helix (WH) domain of Cdc6H in H. hispanica (fig. 5). Thus, the oriC4-cdc6G and oriC5-cdc6H origins in H. hispanica did not contain an intact functional initiator gene, and this absence is a highly likely explanation of why they do not exhibit origin activity. Furthermore, we found a perfect hit to the C-terminal WH domain of Cdc6H in the H. hispanica minichromosome, a small Cdc6 homolog (HAH_4250, designated Cdc6H_C for simplicity) (fig. 5). Remarkably, there was an approximate 123-kb distance between cdc6_c and oriC5-cdc6H. Indicators of translocation processes (integrases or transposases) were observed around both of cdc6_c and oriC5-cdc6H, suggesting that the intact oriC5-cdc6H origin was separated and thus disrupted during the construction of the minichromosome in H. hispanica. Taken together, as these three H. hispanica nonfunctional origins are not conserved and located in variable regions in Haloarcula species, we suggested that their integrity was destroyed during genome variation, either due to incomplete acquisition or functional disruption. In addition, our comparison analyses provided us with the ability to understand the essential elements for a functional replication origin, which would greatly contribute to the dissection of mosaic replication origin systems in haloarchaeal genomes.
Fig. 4.—
Loss of cdc6 promoter broke the ARS activity of the H. hispanica oriC4-cdc6G. (A) Sequence alignment and physical mapping of H. hispanica oriC4-cdc6G and its H. marismortui homolog oriC-cdc6a. The cdc6 genes are indicated, and the start site of H. marismortui cdc6a is numbered one. The ORB elements are indicated with arrowheads. The shaded teal regions denote a similarity greater than 70%, as determined through BLASTN analyses. These two replication origins were highly conserved with the exception of a 203-bp sequence between the cdc6 gene and the origin region, which was lost in H. hispanica oriC4-cdc6G. (B) ARS assay plates for the two origins. Colonies in plates of AS168 (Mev) were observed after 7 days at 37 °C.
Fig. 5.—
Disruption of the integrity of the Cdc6H protein in H. hispanica. (A) Multiple alignments of H. hispanica (Hhis), H. amylolytica (Hamy), and H. sinaiiensis (Hsin) Cdc6H proteins and a small H. hispanica Cdc6_C protein. (B) Functional domain annotation on Cdc6H homologs generated using the CD-search tool in the NCBI website.
Discussion
Multiple replication origins have been previously predicted in most haloarchaeal genomes (Wu et al. 2012), and active origins have been experimentally identified in several model systems (Norais et al. 2007; Coker et al. 2009; Wu et al. 2014). These data indicate the major diversity of replication origins in different strains. Although translocation events have been proposed to account for the mosaics of replication origins in haloarchaeal genomes, the detailed mechanisms of these evolutionary processes are less understood. Recently, multiple replication origins have been in silico and experimentally investigated in H. hispanica (Wu et al. 2014). In this manuscript, based on the replication origins in H. hispanica, we report the first comparative analysis of replication origins from multiple Haloarcula species to understand the evolutionary mechanisms involved in the development of multiple origin systems. Our comparison analyses demonstrated that the dynamics of multiple replication origins is associated with the evolution of multireplicon genome architecture, which indicates that genome evolution forces origin dynamics in variable regions and that maintenance of the conservative replication origins may be promoted by their effects on surrounding genomic environments. This is the first report revealing the dynamics and evolutionary history of multiple replication origins in haloarchaea using comparative genomic analysis of replication origins from closely related species, which not only contributes to understanding of multiple-origin systems in the domain of Archaea but also provides insight into the mechanisms of the more complex replication origins found in eukaryotes.
The comparative analyses revealed that replication origins can be classified into conserved and variable origins in Haloarcula species (table 1, fig. 1 and Wu et al. 2012). The conserved origins are present in all species, and our results demonstrated that these origins were conserved not only in sequence and structure but also in their genomic location. However, the variable origins are either present or absent in a single species and are located in the genomic variable regions. These results suggest that the dynamics of multiple replication origins might be associated with genomic contexts. In particular, genome evolution forced frequent variation of replication origins in haloarchaea with the exception of cases in which the origin was conserved via its effects on surrounding genes.
Because origin variation normally occurs in genomic variable regions, it was unclear to what degree these variable replication origins survive in the face of genomic change and whether origin evolution affects genome replication and genome architecture. The comparative dissection of the H. hispanica nonfunctional replication origins with their homologs in other species revealed that frequent coupled insertion and deletion events were continually creating and destroying replication origins in genomic variable regions. The rapid evolution of replication origins in variable regions might largely account for origin diversity, and our insights into these variable replication origins thus provide us with the ability to understand the evolutionary mechanisms responsible for mosaic origin systems in haloarchaea. In addition, frequent acquisitions and losses of replication origins might fundamentally alter the manner of genome replication, such as the development of a multiple-origin replicon from a single-origin replicon. Interestingly, an Haloferax volcanii strain lacking all active replication origins was constructed, and the recombination-dependent manner of chromosome replication was proposed to be used (Hawkins et al. 2013; Michel and Bernander 2014). The alteration of the manner of replication is very common in reconstructing processes of extrachromosomal replicons, which explains why none of the conserved active origins was observed in the minichromosomes of Haloarcula species (fig. 1B).
It is known that some archaea use multiple origins to initiate chromosome replication, and these multiple-origin chromosomes are considered to originate from the integration of extrachromosomal elements (Robinson and Bell 2007). However, the detailed mechanisms, such as the evolutionary history of the development of multiple replication origins and the factors that govern the conserved or variable origins are unclear. Due to the broad conservation of oriC1 across the archaeal domain of life, this origin was proposed to be present in the archaeal ancestral chromosome (Robinson et al. 2004; Wu et al. 2012). Importantly, replication-associated genes are clustered around oriC1 in the characterized archaeal genomes (Myllykallio et al. 2000; Coker et al. 2009; Pelve et al. 2012). Furthermore, the genes around oriC1 are highly conserved among haloarchaea (Coker et al. 2009). Thus, it is easy to conclude that the oriC1 origin may be preserved from the archaeal ancestor via the conserved genomic environment in which replication-associated genes are abundant. Furthermore, we found that an rRNA operon was located in the oriC1 origin in most haloarchaeal chromosomes, indicating that oriC1 may have an effect on the expression and stability of surrounding genes. Thus, the location of oriC1 may provide a selective advantage for the early replication and high expression of its neighboring replication-associated genes, which might sequentially promote the conservation of oriC1 from the archaeal ancestral chromosome (fig. 6).
Fig. 6.—
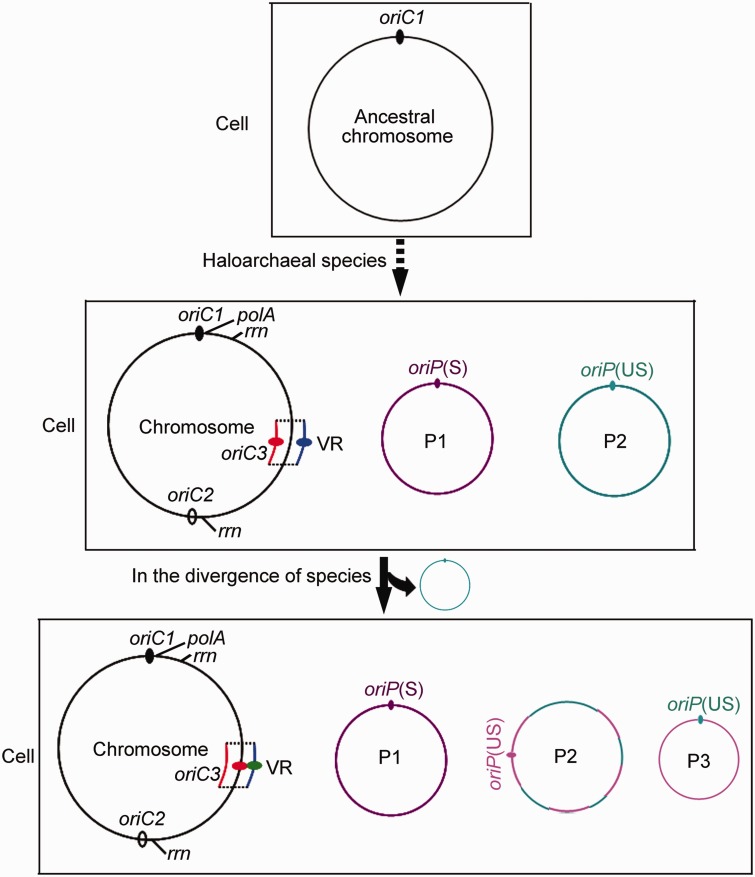
Model of the association between the dynamics of replication origins and genome evolution. The haloarchaeal multireplicon genome evolved in multiple steps (indicated with the dotted arrow) from the oriC1-dependent archaeal ancestral chromosome (up). For example, novel replication origins (oriC2 and oriC3) were independently integrated into the chromosome, and minireplicons were constructed in the branch leading to specific haloarchaeal species, whereas oriC1 was conserved via its effects on surrounding genes (middle). Both the replication origins and the genome architecture (particularly the extrachromosomal replicons) varied frequently in the divergence of species (down). The oriC2 origin was conserved in a similar manner as oriC1 via its effects on surrounding genes, whereas oriC3 was continually being acquired, lost and disrupted in the chromosomal variable regions (in different colors). The fate of extrachromosomal replicons was determined by the hierarchy in the plasmid stabilization of their bearing replication origins. Conversely, unstable extrachromosomal replicons forced variation of their bearing replication origins. For example, the plasmid P1 (in purple) was conserved, as was the stability of the ARS plasmid with its bearing origin oriP(S). In contrast, the plasmid P2 (in dark green) was easily lost because the oriP(US)-based ARS plasmids were unstable. However, parts of the P2 contents might be maintained via the reconstruction of a novel P2 plasmid together with novel genes in the surrounding environments (in mosaic of dark green and pink). In addition, the oriP(US) can be used to construct the novel plasmid (P3) with its surrounding genes (in pink).
There are chromosomal replication origins that are only conserved in specific species, as illustrated by the conservation of oriC2-cdc6E in Haloarcula species. The origin oriC2-cdc6E was proposed to be acquired in the branch leading to the Haloarcula species and was stable via conservation. Unexpectedly, the deletion of oriC2-cdc6E showed no observable growth defects in H. hispanica (Wu et al. 2014), raising the question of what pressure was exerted to maintain this origin in Haloarcula species. Interestingly, the chromosomal location of oriC2 is conserved, and another rRNA operon in the chromosome is located at a distance of only approximately 20 kb from oriC2-cdc6E (fig. 1A), suggesting that, in a similar manner as oriC1, this origin might be conserved via its effects on surrounding genes after its acquisition in the Haloarcula ancestor (fig. 6).
Apart from conserved replication origins, there are some variable origins in the chromosome that are not present in all strains, such as oriC3-cdc6D in the H. hispanica chromosome. Interestingly, these variable origins are normally located in chromosomal variable regions that are integrated at precisely the same position in each chromosome. We speculated that coupled insertion and deletion events occurred frequently in the chromosomal variable regions, resulting in the acquisition, loss, and disruption of replication origins in these regions (fig. 6). It was best to explain that replication origins in chromosomal variable regions are extremely diverse and that some origins are deficient. Taken together, we proposed that multiple-origin haloarchaeal chromosomes were developed in multiple steps, originating from the integration of replication origins into oriC1-dependent ancestral chromosome, and that the surrounding environments determined the fate of these novel origins, that is, conserved or variable (fig. 6).
Haloarchaeal genomes generally harbor extrachromosomal replicon(s), with as many as eight in H. marismortui (Baliga et al. 2004). Compared with the chromosome, extrachromosomal replicons are highly variable within specific species. Consistent with these findings, replication origins in these minireplicons are variable, which indicates that the frequent evolution of extrachromosomal elements might be associated with origin variation. In this study, based on the minichromosome and megaplasmid pHH400, we investigated the evolutionary association between extrachromosomal replicons and their bearing replication origins. We found that oriC6- and oriC7-based ARS plasmids are less stable compared with the oriP plasmid (fig. 3). In conjunction with the dynamics of the minichromosome and conservation of pHH400 in Haloarcula species, we speculated that the hierarchy of origins in plasmid stabilization might reflect the evolution of extrachromosomal replicons. Thus, we proposed a model of the evolutionary association between organization and replication origins of extrachromosomal replicons. In particular, similar to the Haloarcula minichromosome, P2 was unstable and was easily lost in the divergence of the species because the oriP(US)-based ARS plasmids were unstable. However, parts of the genes in P2 might be essential to cell viability; thus, these genes together with genes that promote adaptation to new environments appeared to be clustered in the reconstruction of a novel P2 (fig. 6). Conversely, the frequent reconstruction of P2 resulted in the variation of bearing replication origins. In addition, origins from P2 or from surrounding environments may be employed to construct novel minireplicons with surrounding genes (P3 in fig. 6), which would explain many strain-specific replicons. In contrast, similar to Haloarcula pHH400, P1 was stable in the divergence of species, as was the stability of the ARS plasmid with its bearing origin oriP(S) (fig. 6).
In conclusion, we have addressed the evolutionary association of multiple replication origins and multireplicon genome architecture, which includes evolution of multiple-origin chromosomes and evolution of organization and replication origins of extrachromosomal replicons. More interestingly, we suggested that this mechanism involved in the evolutionary association of multiple replication origins and multireplicon genome architecture is general in haloarchaea: 1) rRNA operon-mediated origin conservation appears to be universal in haloarchaea. Some sequenced haloarchaeal genomes have more than one rRNA operon in the chromosome (Hartman et al. 2010; Capes et al. 2011), and we previously suggested that the rRNA operon might benefit to the preservation of oriCb in Haloferax volcanii, Halogeometricum borinquense, and Halorubrum lacusprofundi from their ancestor (Wu et al. 2012) 2) The mechanism involved in the evolution of architecture and replication origins of extrachromosomal replicons is general in haloarchaea. For example, a comparison of megaplasmids between strain R1 and strain NRC-1 of Halobacterium salinarum revealed that these plasmids can rearrange even in the laboratory (Ng et al. 1998; Pfeiffer et al. 2008). We previously demonstrated that Haloferax mediterranei and Haloferax volcanii employed the same replication origin to construct completely different plasmids (Liu et al. 2013). In addition, it was also demonstrated that ARS plasmids based on replication origin of extrachromosomal replicon pHV1/4 are less stable than oriC1 plasmids (Norais et al. 2007).
Supplementary Material
Acknowledgments
This work was partially supported by grants from the National Natural Science Foundation of China (31100893, 31271334, 31301071).
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga NS, et al. Genome sequence of Haloarcula marismortui: a halophilic archaeon from the Dead Sea. Genome Res. 2004;14:2221–2234. doi: 10.1101/gr.2700304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berquist BR, Dassarma S. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J Bacteriol. 2003;185:5959–5966. doi: 10.1128/JB.185.20.5959-5966.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capes MD, et al. The information transfer system of halophilic archaea. Plasmid. 2011;65:77–101. doi: 10.1016/j.plasmid.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Cline SW, Lam WL, Charlebois RL, Schalkwyk LC, Doolittle WF. Transformation methods for halophilic archaebacteria. Can J Microbiol. 1989;35:148–152. doi: 10.1139/m89-022. [DOI] [PubMed] [Google Scholar]
- Coker JA, et al. Multiple replication origins of Halobacterium sp. strain NRC-1:properties of the conserved orc7-dependent oriC1. J Bacteriol. 2009;191:5253–5261. doi: 10.1128/JB.00210-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Rienzi SC, et al. Maintaining replication origins in the face of genomic change. Genome Res. 2012;22:1940–1952. doi: 10.1101/gr.138248.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JY, Chiang PW, Hong MJ, Dyall-Smith M, Tang SL. Complete genome sequence of the extremely halophilic archaeon Haloarcula hispanica strain N61. Genome Announc. 2014;2:e00178–14. doi: 10.1128/genomeA.00178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–1213. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- Duggin IG, McCallum SA, Bell SD. Chromosome replication dynamics in the archaeon Sulfolobus acidocaldarius. Proc Natl Acad Sci U S A. 2008;105:16737–16742. doi: 10.1073/pnas.0806414105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyall-Smith ML, et al. Haloquadratum walsbyi: limited diversity in a global pond. PLoS One. 2011;6:e20968. doi: 10.1371/journal.pone.0020968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant JR, Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Hartman AL, et al. The complete genome sequence of Haloferax volcanii DS2, a model archaeon. PLoS One. 2010;5:e9605. doi: 10.1371/journal.pone.0009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins M, Malla S, Blythe MJ, Nieduszynski CA, Allers T. Accelerated growth in the absence of DNA replication origins. Nature. 2013;503:544–547. doi: 10.1038/nature12650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, et al. Complete genome sequence of Haloarcula hispanica, a model haloarchaeon for studying genetics, metabolism, and virus-host interaction. J Bacteriol. 2011;193:6086–6087. doi: 10.1128/JB.05953-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, et al. Characterization of the minimal replicon of pHM300 and independent copy number control of major and minor chromosomes of Haloferax mediterranei. FEMS Microbiol Lett. 2013;339:66–74. doi: 10.1111/1574-6968.12052. [DOI] [PubMed] [Google Scholar]
- Lundgren M, Andersson A, Chen L, Nilsson P, Bernander R. Three replication origins in Sulfolobus species: synchronous initiation of chromosome replication and asynchronous termination. Proc Natl Acad Sci U S A. 2004;101:7046–7051. doi: 10.1073/pnas.0400656101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch EA, et al. Sequencing of seven haloarchaeal genomes reveals patterns of genomic flux. PLoS One. 2012;7:e41389. doi: 10.1371/journal.pone.0041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga F, Forterre P, Ishino Y, Myllykallio H. In vivo interactions of archaeal Cdc6/Orc1 and minichromosome maintenance proteins with the replication origin. Proc Natl Acad Sci U S A. 2001;98:11152–11157. doi: 10.1073/pnas.191387498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga F, Norais C, Forterre P, Myllykallio H. Identification of short ‘eukaryotic' Okazaki fragments synthesized from a prokaryotic replication origin. EMBO Rep. 2003;4:154–158. doi: 10.1038/sj.embor.embor732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Bernander R. Chromosome replication origins: do we really need them? Bioessays. 2014;36:585–590. doi: 10.1002/bies.201400003. [DOI] [PubMed] [Google Scholar]
- Minegishi H, et al. Further refinement of the phylogeny of the Halobacteriaceae based on the full-length RNA polymerase subunit B' (rpoB') gene. Int J Syst Evol Microbiol. 2010;60:2398–2408. doi: 10.1099/ijs.0.017160-0. [DOI] [PubMed] [Google Scholar]
- Muller CA, Nieduszynski CA. Conservation of replication timing reveals global and local regulation of replication origin activity. Genome Res. 2012;22:1953–1962. doi: 10.1101/gr.139477.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myllykallio H, et al. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science. 2000;288:2212–2215. doi: 10.1126/science.288.5474.2212. [DOI] [PubMed] [Google Scholar]
- Ng WV, et al. Snapshot of a large dynamic replicon in a halophilic archaeon: megaplasmid or minichromosome? Genome Res. 1998;8:1131–1141. doi: 10.1101/gr.8.11.1131. [DOI] [PubMed] [Google Scholar]
- Norais C, et al. Genetic and physical mapping of DNA replication origins in Haloferax volcanii. PLoS Genet. 2007;3:e77. doi: 10.1371/journal.pgen.0030077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul S, Million-Weaver S, Chattopadhyay S, Sokurenko E, Merrikh H. Accelerated gene evolution through replication-transcription conflicts. Nature. 2013;495:512–515. doi: 10.1038/nature11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelve EA, Lindas AC, Knoppel A, Mira A, Bernander R. Four chromosome replication origins in the archaeon Pyrobaculum calidifontis. Mol Microbiol. 2012;85:986–995. doi: 10.1111/j.1365-2958.2012.08155.x. [DOI] [PubMed] [Google Scholar]
- Pelve EA, Martens-Habbena W, Stahl DA, Bernander R. Mapping of active replication origins in vivo in thaum- and euryarchaeal replicons. Mol Microbiol. 2013;90:538–550. doi: 10.1111/mmi.12382. [DOI] [PubMed] [Google Scholar]
- Pfeiffer F, et al. Evolution in the laboratory: the genome of Halobacterium salinarum strain R1 compared to that of strain NRC-1. Genomics. 2008;91:335–346. doi: 10.1016/j.ygeno.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Raymann K, Forterre P, Brochier-Armanet C, Gribaldo S. Global phylogenomic analysis disentangles the complex evolutionary history of DNA replication in archaea. Genome Biol Evol. 2014;6:192–212. doi: 10.1093/gbe/evu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, Bell SD. Origins of DNA replication in the three domains of life. FEBS J. 2005;272:3757–3766. doi: 10.1111/j.1742-4658.2005.04768.x. [DOI] [PubMed] [Google Scholar]
- Robinson NP, Bell SD. Extrachromosomal element capture and the evolution of multiple replication origins in archaeal chromosomes. Proc Natl Acad Sci U S A. 2007;104:5806–5811. doi: 10.1073/pnas.0700206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NP, et al. Identification of two origins of replication in the single chromosome of the archaeon Sulfolobus solfataricus. Cell. 2004;116:25–38. doi: 10.1016/s0092-8674(03)01034-1. [DOI] [PubMed] [Google Scholar]
- Samson RY, et al. Specificity and function of archaeal DNA replication initiator proteins. Cell Rep. 2013;3:485–496. doi: 10.1016/j.celrep.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Liu H, Liu J, Liu X, Xiang H. Diversity and evolution of multiple orc/cdc6-adjacent replication origins in haloarchaea. BMC Genomics. 2012;13:478. doi: 10.1186/1471-2164-13-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Liu J, Yang H, Liu H, Xiang H. Multiple replication origins with diverse control mechanisms in Haloarcula hispanica. Nucleic Acids Res. 2014;42:2282–2294. doi: 10.1093/nar/gkt1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Zhou M, Sun C, Xiang H, Tan H. Genetic analysis of a novel plasmid pZMX101 from Halorubrum saccharovorum: determination of the minimal replicon and comparison with the related haloarchaeal plasmid pSCM201. FEMS Microbiol Lett. 2007;270:104–108. doi: 10.1111/j.1574-6968.2007.00656.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.