Abstract
Malaria is a mosquito-borne infectious disease caused by parasitic protozoa of the genus Plasmodium. It remains a major problem affecting humans today, especially children. However, the pathogenesis of malaria, especially severe malaria, remains incompletely understood, hindering our ability to treat this disease. Of recent interest is the role that small, non-coding RNAs play in the progression, pathogenesis of, and resistance to, malaria. Independent studies have now revealed the presence of microRNA (miRNA) in the malaria parasite, vector, and host, though these studies are relatively few. Here, we review these studies, focusing on the roles specific miRNA have in the disease, and how they may be harnessed for therapeutic purposes.
Keywords: microRNA, Malaria, Immunopathology, Protective immunity, Inflammation
Introduction
Malaria is a mosquito-borne infectious disease transmitted by the bite of Plasmodium-infected female Anopheles mosquitoes, to vertebrate hosts [1,2]. Malaria remains a major problem affecting humans today, with approximately half of the world's population at risk [3]. Each year, Plasmodium falciparum (Pf) infects over 200 million people worldwide, and is responsible for an estimated 660,000 deaths, mostly of children under five years old. 80% of these deaths occur in sub-Saharan Africa.
The clinical manifestations of malaria are primarily due to schizont rupture, leading to destruction of erythrocytes (RBC), and vary in severity, which depends on host age, immune status, and strain of the parasite. Severe malaria is a multi-system disease caused almost exclusively by untreated Pf infection. Cerebral malaria (CM) and anaemia are the two most common manifestations of severe malaria. CM is a syndrome characterised by unrousable coma and neurological sequel-ae. The fine pathophysiological mechanisms underlying this neurological syndrome are not fully understood, although several hypotheses have been put forward:
-
(i)
The “mechanical” hypothesis, involving cell sequestration, the deformity of RBC and aggregation [1,4,5],
-
(ii)
The “immunological” hypothesis, attributing the development of infection to the stimulation of production and release of pro-inflammatory and anti-inflammatory cytokines [4,6,7], and
-
(iii)
A combination of these [8,9], being the most likely hypothesis, since each alone cannot fully explain all manifestations and clinical signs of CM [10].
Recently, emerging research has focused on miRNA to understand more about their roles in normal and pathophysiological conditions. miRNA are a type of single-stranded, non-coding RNA approximately 22 nucleotides in length, that function as post-transcriptional regulators of targeted gene expression in eukaryotes, thus regulating the translation of mRNAs and proteins. It is now known that miRNA play crucial regulatory roles in numerous biological processes, including apoptosis, cell proliferation, metabolism, development, and differentiation [11,12]. miRNA are also known to be involved in a broad range of infectious diseases and inflammatory pathologies. Recently, our understanding of the cellular and molecular networks that regulate inflammation has improved considerably, including the critical role miRNA play in managing features of the inflammatory process, often associated with a range of pathologies, including chronic inflammation, autoimmunity, and cancer [13]. The dysregulation of miRNA in viral, bacterial, and parasitic diseases has been studied extensively [14-16], though few studies exist examining the role of miRNA in malaria specifically. In this review, we address the ways in which parasite, vector, and host miRNA have been implicated in the pathogenesis of, or protective immunity against, malaria and the dynamic interactions between these three components in malaria transmission. As depicted in Figure 1, miRNA are present in each of the parasite, vector, and host, but a significant sharing of material exists between these entities, and this active sharing process of miRNA has a great impact on the transmission and pathogenesis of, or protective immunity against, malaria.
Figure 1.
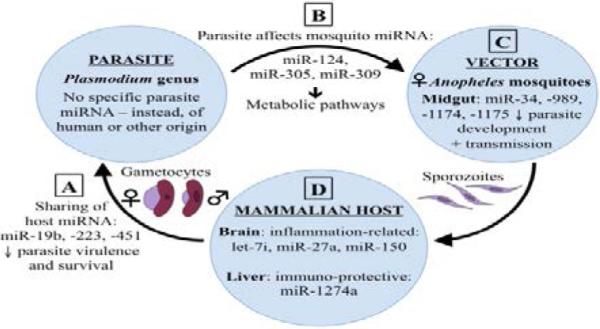
Dynamic interactions between the parasite, vector, and host in malarial infections. Cross-species interactions and regulation affects the transmission and pathogenesis of, and/or resistance to malaria infections. (A) Human miRNA (e.g. miR-19b, −223, and −451) [2] and (C) mosquito miRNA (e.g. miR-34, −989, −1174, and −1175)26 affect malaria parasite biology and survival. (B) Parasite miRNA (e.g. miR-124, −305, and −309)31 regulate metabolic pathways in Anopheles mosquitoes. (D) Host miRNA (e.g. let-7i, miR-27a, −150,51 −1274a42) regulate host gene expression.
miRNA in Host-parasite Interactions
Significant material exchange occurs between the host cell and Pf during the intraerythrocytic developmental cycle [17]. Xue et al. detected the presence of human miRNA within the parasite; however, interestingly, they found no parasite-specific miRNA2, consistent with previous reports by Rathjen et al. [18]. Of the 132 short RNA found by Xue et al., 54 were rRNAs and tRNAs from human blood and Plasmodium, 18 were degraded fragments of human blood and Plasmodium mRNA, 26 were human miRNA, and 24 did not match the human or Plasmodium genome. Notably, among the miRNA found in uninfected normal RBC, human miR-451 was significantly enriched in parasitised RBC (pRBC).
LaMonte et al. investigated how human miRNA are translocated into the parasite, since Pf lacks orthologs of Dicer/Ago [19,20], involved in the conventional mode of RNA interference, which occurs as follows: RNA interference (RNAi) is initiated by the enzyme Dicer, which cleaves dsRNA into shorter molecules that dissociate into ss-RNA. One strand is degraded, and the other is incorporated into the RNA-induced silencing complex. If this strand pairs with a complementary sequence of mRNA, it is degraded by Ago. RNAi is important in defending cells against parasitic nucleotide sequences. Using Illumina deep sequencing, 5’ RACE PCR, and ribonuclease protection assays, LaMonte et al. confirmed that human miRNA transferred into the parasite formed chimeric fusions with Pf mRNA via impaired ribosomal loading, resulting in translational inhibition, eventually impairing parasite biology and survival. It is not yet known what determines the specific enrichment of particular miRNA or their incorporation into specific parasite mRNAs.
Interestingly, La Monte et al. explored the protective immunity and resistance to Plasmodium conferred to RBC by human miRNA and the variant haemoglobin allele (HbS), that causes sickle cell disease [21]. In malaria-endemic regions, sickle cell disease occurs at relatively high frequencies [22,23], and mutations as part of this disease often provide protection against the malaria parasite [24]. This enhanced resistance is thought to be due, at least partly, to more rapid phagocytosis of pRBC by monocytes [25], suggesting that the parasite is more rapidly cleared by the immune system.
The presence of ~100 human miRNA was detected within the parasites, using multiplex real-time PCR, and, in pRBC, found similar enrichment in miR-451 as well as miR-223 and miR-19b [2,21] confirming and expanding previous data [2]. Elevated levels of specific human miRNA were found in RBC with heterozygous (HbAS) and homozygous sickle cell haemoglobin (HbSS), when compared to normal RBC (HbAA), implying a functional role of these miRNA [21]. Specifically, miR-16 was present in abundant levels in HbAA RBC, and miR-181a had no effect upon parasitaemia, whereas transfection with miR-451, miR-223 or let-7i led to a markedly reduced percentage of pRBC in vitro. Combined transfection with miR-451 and miR-223 reduced the infection rate by 46%, with a similar effect seen for additional combination with let-7i. Increased parasite growth was observed in HbAS and HbSS pRBC due to the inhibition of host miRNA, specifically miR-451.
HbAA pRBC showed almost no inhibition of miR-451, and no change in parasite growth. The expression of miR-451 was also examined in Plasmodium berghei-infected mouse RBC by Xue et al. [2], and found to also be independent of erythrocytic stage and parasitaemia, as shown in human erythrocytes by LaMonte et al. [21]. Potential gene targets for these miRNA include PKA-R (cAMP-dependent protein kinase) or are implicated in red cell remodelling. Therefore, miRNA-451 contributes significantly to malaria resistance, by the role played in the differentiation and/or maturation of primary RBCs.
Together, these data confirm that human miRNA regulate protozoan gene expression via cross-species chimeric transcripts, and raise the question of how erythrocyte miRNA modify parasite mRNAs. Examining further these seemingly independent mechanisms of parasite resistance will provide fresh insights into the complex relationship between the host genome and threatening microbial pathogens. In addition to this, further identification of targets genes will shed light on the biological roles of these miRNA of interest. However, what can currently be concluded is that the unidirectional exchange of miRNA from host to parasite protects the host by negatively affecting parasite survival.
miRNA in Vector-parasite Interactions
miRNA in Anopheles gambiae mosquitoes play physiological and protective roles during Plasmodium infection. In the life cycle and transmission of the malaria parasite, the passage Plasmodium takes across the Anopheles midgut constitutes the major bottleneck. Recently, research has focussed on the role of miRNA in regulating the Anopheles defence reaction. Studying interactions between the parasite and insect vector provides an opportunity to identify methods to disrupt or reduce pathogen transmission.
Using a combination of shotgun cloning and bioinformatic analysis, Anopheles mosquito miRNA were analysed, specifically their homology with other species and their localisation within the mosquito [26]. 152 clones were found that could be potential miRNA, however only eighteen met all the criteria to be bona fide miRNA. These eighteen miRNA found in A. gambiae mosquitoes, included three unique to the strain. Twelve were expressed ubiquitously across the body, independent of gender, while the other six exhibited an expression pattern restricted to the digestive system, including the three mosquito-specific miRNA. Interestingly, the expression of four specific miRNA, again including the three unique miRNA, was affected by the presence of Plasmodium. Knocking down Dicer1 and Ago1 mRNAs via gene silencing techniques led to increased susceptibility of the mosquitoes to Plasmodium infection [26-28], therefore providing evidence for miRNA-mediated post-transcriptional regulation of the genes involved in defence reactions, as mosquito genes of the silencing pathway would affect parasite development in the midgut.
Xu et al. too examined the expression profile of miRNA in A. gambiae mosquitoes, and found 200 putative miRNA precursors expressed. Of these precursors, 78 encoded mature miRNA conserved in at least one other animal species [29]. In the Asian malaria mosquito, A. stephensi, 23 conserved and four new miRNA sequences were identified. The expression profile of eight of these miRNA, including four miRNA unique to this mosquito strain, revealed distinct patterns from early embryo to adult stages in the mosquito development. For example, given the expression profile, miR-x-2 was likely involved in female reproduction, and consistent expression of miR-14 suggests its likely importance across all mosquito life stages [30].
During blood feeding, Plasmodium induces several host effector molecules in the mosquito. Blood feeding and parasitised blood feeding revealed regulation of 13 and 16 mosquito miRNA, respectively, for example miR-124, 305, and 309, which are known to target multiple genes in immune pathways. miRNA controlling metabolic, redox homeostasis and protein processing pathways were down-regulated upon parasitised blood feeding. Another set of miRNA showed significant expression changes between 42 h and 5 days p.i. (coinciding with the late phase of midgut invasion by the parasite and initiation of sporozoites release from the oocytes, respectively), indicating a parasite stage-specific role of host miRNA [31]. Biryukova et al., too, showed a shift in miRNA expression following blood feeding and parasitised blood feeding. They found 4 miRNA with a significant expression shift after blood feeding: miR-7, miR-92a, miR-317, and miR-N3; and 6 after parasitised blood feeding, including miR-317, and miR-2940 [32]. Therefore, miRNAs that affect the development and maturation of the parasite within the mosquito are targeted, following a blood meal, indicating some mechanisms of survival and propagation of the parasite.
Understanding miRNA functions contributes to a better under-standing of mosquito reproduction, longevity, and mosquito-pathogen interactions that might provide a revolutionary way to hinder malaria transmission. Further research will enhance this understanding, especially in terms of functional miRNA exchange between the parasite and vector, as well as the little explored topic of miRNA exchange between the vector and the target host.
Mammalian host miRNA
Evidence is accumulating that miRNA are critically implicated in the outcome of diseases, but little information is available regarding their mechanism of action in these cases. To date, Glinksy et al. are the only study to publish research regarding miRNA in human patients. Using PBMCs from malaria patients, they showed, using microarray analysis, that chloroquine therapy reversed the disease-associated phenotypes of nuclear import and inflammasome pathways [33]. Their analysis showed that miRNA and mRNAs controlling the Karyopherin Alpha 1 (KPNA1), and NLR family pyrin domains containing 1 and 3 (NLRP1 and NLRP3) genes (inflammasome-related genes) showed changed levels of expression following chloroquine therapy: decreased expression of KPNA1 and NLRP1 during infection was reversed after chloroquine, and their expression was elevated by 66% and 13%, respectively. Also, infection-induced increased expression of NLRP3 was reduced by 57% after chloroquine.
miRNA have been shown as key components, in the liver and elsewhere, in the immune response following malaria infection, with many miRNA known to control cell growth and proliferation, stress responses, and metabolism, as well as gene expression and susceptibility to infection [34-38]. The liver is a known anti-malaria effector organ, as the site of the pre-erythrocytic development of Plasmodium [39,40], where Kupffer cells and extrathymic lymphocytes are generated following infection to carry out important protective immune functions, such as phagocytosis of pRBC [41].
Delić et al. examined miRNA expression in the liver of malarial-infected mice and found a hepatic miRNA signature in female C57BL/6 mice infected with self-healing P. chabaudi [42]. These mice displayed peak parasitaemia of approximately 50% on day 8 post infection (p.i.) and in 80% of cases these infections were self-healing. Among those mice that survived the initial infection, 100% survived subsequent challenges eight weeks later, with a peak parasitaemia of 1.5%. Liver miRNA profiles were analysed using miRXplore microarrays and quantitative RT-PCR (qRT-PCR). The liver was found to respond to primary infections with an upregulation of three miRNA and a downregulation of sixteen other miRNA, some of which had known roles in apoptosis and cancer. Among them, the high expression of miR-1274a in the liver was confirmed by qRT-PCR. Surprisingly, a similar pattern of miRNA expression was found in immune mice, i.e. mice that survived the first infection, and also upon re-infection of these mice, suggesting that the modulations of expression of these miRNA are specific to the infection status irrespective of their levels. Interestingly, no changes in expression were detected of those miRNA currently known to be critically involved in adaptive immune responses, such as antibody formation, development, B cell maturation, and the formation of memory B cells, CD4+ T cells, and CD8+ T cells. Thus, the development of protective immunity against blood stages of P. chabaudi is associated with a reprogramming of the expression of distinct miRNA species yet to be identified in the mouse liver.
In another study, Al-Quraishy et al. tested testosterone-induced susceptibility to blood-stage malaria of P. chabaudi and measured the coinciding changes in miRNA expression of the anti-malaria effector sites, the spleen and liver [43]. Female C57BL/6 mice were treated with vehicle or testosterone for 3 weeks and, 12 weeks later, were challenged with P. chabaudi. miRNA expression was examined during testosterone treatment, after 12 weeks of testosterone withdrawal, and following this, during infections at peak parasitaemia. Changes could be observed in miRNA expression during testosterone treatment and after its cessation. For example, a specific upregulation during testosterone withdrawal was observed only in the spleen for miR-200a, and only in the liver for miR-142-5p and miR-342-3p. The latter two miRNA have been shown to be upregulated in cancer cells [44], leucocytes [45], in antigen-induced differentiation of T cells [46], and in B cell activation in leukemia [47,48], and the target genes of miR-342-3p are involved in different processes such as cell proliferation [44]. These results imply an influx of cells expressing these two miRNA into the liver. Upon infection, however, these changes in miRNA expression were not maintained. Various changes were observed, notably a downregulated expression of most miRNA species by more than 2-fold in the spleen and liver. The same response to P. chabaudi malaria was observed in miRNA expressions in these two organs in testosterone-pretreated mice and vehicle-treated control mice. This change in expression suggests that testosterone-induced susceptibility to P. chabaudi does not affect miRNA expression and, therefore, gene expression. This susceptibility was shown to be irreversible, i.e. mice pre-treated with testosterone remained susceptible to infection after 12 weeks of withdrawal. This response to infection following testosterone treatment and subsequent withdrawal was also observed by Delic et al. [49]. This study went on to confirm that the testosterone-induced upregulation of specific miRNAs coincided with an at least 2-fold downregulation of some of their known gene targets. Altogether, the reprogramming of miRNA expression in the spleen and liver is essential for the development of protective immunity against blood-stage malaria, and that the responsiveness of this reprogrammed expression is not affected by testosterone-induced susceptibility.
Hentzschel et al. studied the role of miRNA in protection against liver stages of Plasmodium infection. Here, they examined the role specifically of miR-155 in mice infected with genetically attenuated parasites (GAP). Previously, it had been shown that GAP arrested in the liver, induce sterile immunity, but only after several administrations. It was found that GAP-injected mice displayed a broad activation of IFNγ-associated pathways and a significant increase in miR-155 within macrophages in the liver [50]. This upregulation enhanced the protective capacity of GAP substantially, highlighting the crucial role of mammalian miRNAs in Plasmodium liver infection.
The presence of miRNA was also associated with pathogenesis and the development of the neurological syndrome in the murine model of CM. When animals are infected with Plasmodium berghei ANKA (PbA), they develop a neurological syndrome where the brain is the main targeted organ. El Assaad et al. examined the expression of a selection of miRNA, known to be associated with inflammation and infection, by qRT-PCR in the brain and the heart during malaria infection with PbA, and compared it to a non-encephalitogenic strain Plasmodium berghei K173 (PbK)-infected CBA mice, as well as uninfected mice [51]. Significant changes were found in the expression of let7i, miR-27a, and miR-150 in the brain tissue of PbA-infected mice compared to uninfected mice at the time of the neurological syndrome. In contrast, no changes in these miRNA were detected in the heart, an organ with no known pathology, during CM [51]. Distinct expression profiles were observed in the brains of PbA- and PbK-infected and un-infected mice. This difference in expression could not be attributed to parasite load, as mice showed comparable levels of parasitaemia. Using the miRBase database, predicted gene targets of the three miRNA showing significant changes in CM regulate pathways involved in inflammation, innate pathogen recognition, apoptosis, and immune functions [52-54]. Both strains of parasite induced an increase in the expression of let-7i and miR-150 compared with the level in uninfected mice, with PbA inducing a greater response. The let-7 family has previously been shown to control cellular proliferation and innate immune responses [53,54], and miR-150 was highly expressed in monocytes and has been implicated in cell proliferation, development, and differentiation. This change in expression is crucial, as an accumulation of monocytes in the cerebral microvasculature has been associated with fatal murine CM [55]. The sequestration of monocytes within the cerebral microvessels of PbK-infected mice can still be observed, albeit at a lower level than in PbA-infected mice, and this is consistent with the changes in miRNA expression observed. Of the six selected miRNA, miR-27a expression was significantly increased in PbA-infected mice only, suggesting that it may have a more specific role in CM. miR-27a has been shown to induce apoptosis, disrupt mitochondrial membrane potential, increase sensitivity to TNF [56], and control the regulation of T cell proliferation and the NF-KB signalling pathway during inflammation [57]. Thus, overexpression of these three miRNA during PbA infection in mice may be critical for triggering the neurological syndrome. Since these altered expression levels coincided with increased apoptosis, and levels of circulating inflammatory cytokines in the brain, demonstrated by Lackner et al. [58], the regulation of potential targets of these miRNA appears crucial.
Not only does malarial infection modulate particular sets of miRNA, but also the host genetic background may play a role in determining a miRNA profile associated with CM. In these studies, the potential link between alterations in miRNA expression and malaria pathogenesis was explored and these alterations could be reversed following treatment, though evidence of this is only provided in the case of non-cerebral malaria.
Conclusions
The study of miRNA and their importance during malaria infection is at its beginning but is showing promise since miRNA have been shown to play roles in the resistance to and protective immune response against malarial infection, and in the pathogenesis of the infection in some cases. These roles are not strictly limited to host miRNA acting within the host either as evidence of interspecies miRNA transcripts impairing the normal function of Plasmodium in order to confer resistance to the host were demonstrated. miRNA translocation is a dynamic process between parasite, host and vector, allowing for the protective or pathogenic interactions between these elements of malarial transmission. Certain areas, such as the role of miRNA in the interaction between vector and host, have not yet been explored, and more data is needed to support the conclusions regarding the direct role miRNA play in host defence against Pf. Indeed, despite the role miRNA play in infection, whether pathogenic or protective, the mechanisms of action are still incompletely understood. This is partly due to the redundancy in the control of gene expression, where individual miRNA control numerous mRNAs, resulting in innumerable changes in gene expression. Therefore, to attribute one feature of the disease to a single miRNA is practically impossible. While some functional studies have been performed, linking miRNAs with target genes involved in malaria infection or pathogenesis, more research is needed in this area. In addition, unravelling the details of miRNA mechanism of action during infection may give us more hints to understand malarial pathogenesis, serve as biomarkers for disease progression and, most importantly, be useful for developing therapeutic uses for miRNA or their inhibitors in patients with severe malaria.
Acknowledgements
This work was supported by grants from the Australian NHMRC (APP1028241) and NIH (NIH R01NS079873) to Valery Combes and Georges Grau. Amy Cohen is supported by an Australian Postgraduate Award from the Australian Government.
Footnotes
Citation: Cohen A, Combes V, Grau GER (2014) MicroRNAs and Malaria – A Dynamic Interaction Still Incompletely Understood. J Neuroinfect Dis 5: 165. doi: 10.4172/2314-7326.1000165
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Schofield L, Grau GE. Immunological processes in malaria pathogenesis. Nat Rev Immunol. 2005;5:722–735. doi: 10.1038/nri1686. [DOI] [PubMed] [Google Scholar]
- 2.Xue X, Zhang Q, Huang Y, Feng L, Pan W. No miRNA were found in Plasmodium and the ones identified in erythrocytes could not be correlated with infection. Malar J. 2008;7:47. doi: 10.1186/1475-2875-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. World Malaria Report . WHO Press, World Health Organisation; Geneva, Switzerland: 2013. [Google Scholar]
- 4.van der Heyde HC, Nolan J, Combes V, Gramaglia I, et al. A unified hypothesis for the genesis of cerebral malaria: sequestration, inflammation and hemostasis leading to microcirculatory dysfunction. Trends in Parasitology. 2006;22:503–8. doi: 10.1016/j.pt.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 5.Berendt AR, Tumer GD, Newbold CI. Cerebral malaria: the sequestration hypothesis. Parasitol Today. 1994;10:412–414. doi: 10.1016/0169-4758(94)90238-0. [DOI] [PubMed] [Google Scholar]
- 6.Idro R, Jenkins NE, Newton CR. Pathogenesis, clinical features, and neurological outcome of cerebral malaria. Lancet Neurol. 2005;4:827–840. doi: 10.1016/S1474-4422(05)70247-7. [DOI] [PubMed] [Google Scholar]
- 7.Clark IA, Rockett KA. The cytokine theory of human cerebral malaria. Parasitol Today. 1994;10:410–412. doi: 10.1016/0169-4758(94)90237-2. [DOI] [PubMed] [Google Scholar]
- 8.Grau GE, de Kossodo S. Cerebral malaria: mediators, mechanical obstruction or more? Parasitol Today. 1994;10:408–409. doi: 10.1016/0169-4758(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 9.Combes V, El-Assaad F, Faille D, Jambou R, Hunt NH, et al. Microvesiculation and cell interactions at the brain-endothelial interface in cerebral malaria pathogenesis. Prog Neurobiol. 2010;91:140–151. doi: 10.1016/j.pneurobio.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Combes V, Coltel N, Faille D, Wassmer SC, Grau GE. Cerebral malaria: role of microparticles and platelets in alterations of the blood-brain barrier. Int J Parasitol. 2006;36:541–546. doi: 10.1016/j.ijpara.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 11.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 12.Liang H, Zen K, Zhang J, Zhang CY, Chen X. New roles for microRNAs in cross-species communication. RNA Biol. 2013;10:367–370. doi: 10.4161/rna.23663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.O'Connell RM, Rao DS, Baltimore D. microRNA regulation of inflamma-tory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- 14.Eulalio A, Schulte L, Vogel J. The mammalian microRNA response to bacterial infections. RNA Biol. 2012;9:742–750. doi: 10.4161/rna.20018. [DOI] [PubMed] [Google Scholar]
- 15.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hakimi MA, Cannella D. Apicomplexan parasites and subversion of the host cell microRNA pathway. Trends Parasitol. 2011;27:481–486. doi: 10.1016/j.pt.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 17.Deitsch KW, Calderwood MS, Wellems TE. Malaria. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- 18.Rathjen T, Nicol C, McConkey G, Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006;580:5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- 19.Baum J, Papenfuss AT, Mair GR, Janse CJ, Vlachou D, et al. Molecular genetics and comparative genomics reveal RNAi is not functional in malaria parasites. Nucleic Acids Res. 2009;37:3788–3798. doi: 10.1093/nar/gkp239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall N, Karras M, Raine JD, Carlton JM, Kooij TW, et al. A comprehensive survey of the Plasmodium life cycle by genomic, transcriptomic, and proteomic analyses. Science. 2005;307:82–86. doi: 10.1126/science.1103717. [DOI] [PubMed] [Google Scholar]
- 21.LaMonte G, Philip N, Reardon J, Lacsina JR, Majoros W, et al. Translocation of sickle cell erythrocyte microRNAs into Plasmodium falciparum inhibits parasite translation and contributes to malaria resistance. Cell Host Microbe. 2012;12:187–199. doi: 10.1016/j.chom.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livincstone FB. Malaria and human polymorphisms. Annu Rev Genet. 1971;5:33–64. doi: 10.1146/annurev.ge.05.120171.000341. [DOI] [PubMed] [Google Scholar]
- 23.Nagel RL, Roth EF., Jr Malaria and red cell genetic defects. Blood. 1989;74:1213–1221. [PubMed] [Google Scholar]
- 24.Aidoo M, Terlouw DJ, Kolczak MS, McElroy PD, ter Kuile FO, et al. Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet. 2002;359:1311–1312. doi: 10.1016/S0140-6736(02)08273-9. [DOI] [PubMed] [Google Scholar]
- 25.Ayi K, Tumini F, Piga A, Arese P. Enhanced phagocytosis of ring-parasitized mutant erythrocytes: a common mechanism that may explain protection against falciparum malaria in sickle trait and beta-thalassemia trait. Blood. 2004;104:3364–71. doi: 10.1182/blood-2003-11-3820. [DOI] [PubMed] [Google Scholar]
- 26.Winter F, Edaye S, Hüttenhofer A, Brunel C. Anopheles gambiae miRNAs as actors of defence reaction against Plasmodium invasion. Nucleic Acids Res. 2007;35:6953–6962. doi: 10.1093/nar/gkm686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell CL, Black WC, 4th, Hess AM, Foy BD. Comparative genomics of small RNA regulatory pathway components in vector mosquitoes. BMC Genomics. 2008;9:425. doi: 10.1186/1471-2164-9-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mead EA, Li M, Tu Z, Zhu J. Translational regulation of Anopheles gambiae mRNAs in the midgut during Plasmodium falciparum infection. BMC Genomics. 2012;13:366. doi: 10.1186/1471-2164-13-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu Y, Zhou X, Zhang W. MicroRNA prediction with a novel ranking algorithm based on random walks. Bioinformatics. 2008;24:i50–58. doi: 10.1093/bioinformatics/btn175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mead EA, Tu Z. Cloning, characterization, and expression of microRNAs from the Asian malaria mosquito, Anopheles stephensi. BMC Genomics. 2008;9:244. doi: 10.1186/1471-2164-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain S, Rana V, Shrinet J, Sharma A, Tridibes A, et al. Blood feeding and Plasmodium infection alters the miRNome of Anopheles stephensi. PLoS One. 2014;9:e98402. doi: 10.1371/journal.pone.0098402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biryukova I, Ye T, Levashina E. Transcriptome-wide analysis of microRNA expression in the malaria mosquito Anopheles gambiae. BMC Genomics. 2014;15:557. doi: 10.1186/1471-2164-15-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glinsky GV. SNP-guided microRNA maps (MirMaps) of 16 common human disorders identify a clinically accessible therapy reversing transcriptional aberrations of nuclear import and inflammasome pathways. Cell Cycle. 2008;7:3564–76. doi: 10.4161/cc.7.22.7073. [DOI] [PubMed] [Google Scholar]
- 34.Bala S, Marcos M, Szabo G. Emerging role of microRNAs in liver diseases. World J Gastroenterol. 2009;15:5633–5640. doi: 10.3748/wjg.15.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen XM. MicroRNA signatures in liver diseases. World J Gastroenterol. 2009;15:1665–1672. doi: 10.3748/wjg.15.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu LF, Liston A. MicroRNA in the immune system, microRNA as an immune system. Immunology. 2009;127:291–298. doi: 10.1111/j.1365-2567.2009.03092.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci U S A. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kerr TA, Davidson NO. Therapeutic RNA manipulation in liver disease. Hepatology. 2010;51:1055–1061. doi: 10.1002/hep.23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Häussinger D, Kubitz R, Reinehr R, Bode JG, Schliess F. Molecular aspects of medicine: from experimental to clinical hepatology. Mol Aspects Med. 2004;25:221–360. doi: 10.1016/j.mam.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 40.Crispe IN. The liver as a lymphoid organ. Annu Rev Immunol. 2009;27:147–163. doi: 10.1146/annurev.immunol.021908.132629. [DOI] [PubMed] [Google Scholar]
- 41.Krucken J, Delic D, Pauen H, Wotjalla A, El-Khadragy M, et al. Augmented particle trapping and attenuated inflammation in the liver by protective vaccination against Plasmodium chabaudi malaria. Malaria Journal. 2009;8 doi: 10.1186/1475-2875-8-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeliÄ D, Dkhil M, Al-Quraishy S, Wunderlich F. Hepatic miRNA expression reprogrammed by Plasmodium chabaudi malaria. Parasitol Res. 2011;108:1111–1121. doi: 10.1007/s00436-010-2152-z. [DOI] [PubMed] [Google Scholar]
- 43.Al-Quraishy S, Dkhil MA, Delic D, Abdel-Baki AA, Wunderlich F. Organ-specific testosterone-insensitive reponse of miRNA expression of C57BL/6 mice to Plasmodium chabaudi malaria. Parasitology Research. 2012;111:1093–101. doi: 10.1007/s00436-012-2937-3. [DOI] [PubMed] [Google Scholar]
- 44.Van der Auwera I, Limame R, van Dam P, Vermeulen PB, Dirix LY, et al. Integrated miRNA and mRNA expression profiling of the inflammatory breast cancer subtype. Br J Cancer. 2010;103:532–541. doi: 10.1038/sj.bjc.6605787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaefer JS, Montufar-Solis D, Vigneswaran N, Klein JR. Selective up-regulation of microRNA expression in peripheral blood leukocytes in IL-10−/− mice precedes expression in the colon. J Immunol. 2011;187:5834–5841. doi: 10.4049/jimmunol.1100922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, et al. miRNA profiling of naïve, effector and memory CD8 T cells. PLoS One. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ronchetti D, Lionetti M, Mosca L, Agnelli L, Andronache A, et al. An integrative genomic approach reveals coordinated expression of intronic miR-335, miR-34 and miR-561 with deregulated host genes in multiple myeloma. BMC Med Genomics. 2008;1:37. doi: 10.1186/1755-8794-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li S, Moffett HF, Lu J, Werner L, Zhang H, et al. MicroRNA expression profiling identifies activated B cell status in chronic lymphocytic leukemia cells. PLoS One. 2011;6:e16956. doi: 10.1371/journal.pone.0016956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeliÄ D, Grosser C, Dkhil M, Al-Quraishy S, Wunderlich F. Testosterone-induced upregulation of miRNAs in the female mouse liver. Steroids. 2010;75:998–1004. doi: 10.1016/j.steroids.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 50.Hentzschel F, Hammerschmidt-Kamper C, Börner K, Heiss K, Knapp B, et al. AAV8-Mediated In Vivo Overexpression of miR-155 Enhances the Protective Capacity of Genetically Attenuated Malarial Parasites. Molecular Therapy. 2014 doi: 10.1038/mt.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El-Assaad F, Hempel C, Combes V, Mitchell AJ, Ball HJ, et al. Differential microRNA expression in experimental cerebral and noncerebral malaria. Infect Immun. 2011;79:2379–2384. doi: 10.1128/IAI.01136-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bullen DV, Hansen DS, Siomos MA, Schofield L, Alexander WS, et al. The lack of suppressor of cytokine signalling-1 (SOCS1) protects mice from the development of cerebral malaria caused by Plasmodium berghei ANKA. Parasite Immunol. 2003;25:113–118. doi: 10.1046/j.1365-3024.2003.00616.x. [DOI] [PubMed] [Google Scholar]
- 53.Chen XM, Splinter PL, O'Hara SP, LaRusso NF. A cellular micro-RNA, let-7i, regulates Toll-like receptor 4 expression and contributes to cholangiocyte immune responses against Cryptosporidium parvum infection. Journal of Biological Chemistry. 2007;282:28929–38. doi: 10.1074/jbc.M702633200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O'Hara SP, Splinter PL, Gajdos GB, Trussoni CE, Fernandez-Zapico ME, et al. NFkappaB p50-CCAAT/enhancer-binding protein beta (C/EBPbeta)-mediated transcriptional repression of microRNA let-7i following microbial infection. Journal of Biological Chemistry. 2010;285:216–25. doi: 10.1074/jbc.M109.041640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grau GE, Mackenzie CD, Carr RA, Redard M, Pizzolato G, et al. Platelet accumulation in brain microvessels in fatal pediatric cerebral malaria. J Infect Dis. 2003;187:461–466. doi: 10.1086/367960. [DOI] [PubMed] [Google Scholar]
- 56.Chhabra R, Adlakha YK, Hariharan M, Scaria V, Saini N. Upregulation of miR-23a-27a-24-2 cluster induces caspase-dependent and -independent apoptosis in human embryonic kidney cells. PLoS One. 2009;4:e5848. doi: 10.1371/journal.pone.0005848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tourneur L, Chiocchia G. FADD: a regulator of life and death. Trends Immunol. 2010;31:260–269. doi: 10.1016/j.it.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 58.Lackner P, Burger C, Pfaller K, Heussler V, Helbok R, et al. Apoptosis in experimental cerebral malaria: spatial profile of cleaved caspase-3 and ultra-structural alteration in different disease stages. Neuropathology and Application Neurobiology. 2007;33:560–71. doi: 10.1111/j.1365-2990.2007.00833.x. [DOI] [PubMed] [Google Scholar]


