Abstract
CpG island hypermethylation is emerging as one of the main mechanisms for inactivation of cancer related genes in breast tumorigenesis. We examined the changes in methylation patterns during ductal breast cancer progression from atypical ductal hyperplasia to in situ and invasive carcinoma. Paired samples of synchronous pre invasive lesions (Atypical Ductal Hyperplasia and/or Ductal Carcinoma in situ) and invasive ductal breast carcinoma from 31 patients, together with isolated lesions from additional 24 patients were studied. Overall, 95 pathological samples and 20 normal breast tissues were analyzed by Quantitative Methylation Specific PCR (QMSP) on a panel of 9 gene promoters (ESR1, APC, CDH1, CTNNB1, GSTPI, THBS1, MGMT, TMS1 and TIMP3). APC, CDH1, and CTNNB1 promoter regions showed an increase in frequency of methylation and increased methylation levels in pathological samples when compared with normal breast tissues. The analysis of the syncronous paired breast lesions demonstrated also an increase in methylation frequency and level for APC, CDH1, and CTNNB1 genes during progression. By establishing a cutoff value, we were able to distinguish among -invasive and invasive lesions. Synchronous methylation of APC, CDH1, and CTNNB1 was associated only with invasive lesions, whereas simultaneous methylation of APC and CDH1 or APC and CTNNB1 were more frequent in ductal carcinoma in situ and invasive carcinoma. Our data point to direct involvement of APC, CDH1, and CTNNB1 CpG island promoter methylation in the early stages of breast cancer progression, and suggest that these molecular alterations might be involved in the transition to an invasive phenotype.
Keywords: Breast Cancer, DNA methylation/epigenetics, APC, β-catenin, E-cadherin
INTRODUCTION
Breast Cancer is the most common neoplastic disease in women with approximately 1 million new cases diagnosed each year worldwide. Despite of early detection and better management, mammary tumors are still the primary cause of cancer deaths among women (1). The advent of mammography screening has led to an increased detection of pre-invasive mammary lesions, and to a better elucidation of the pathological events that precede the development of invasive breast carcinoma (2). Invasive breast cancer is classified into two main morphological subtypes; ductal carcinoma representing about 80% of breast malignancy, and lobular carcinoma that accounts for 8 to 14% of breast cancer. The remaining malignant tumors are often classified as “special types”. Among the breast lesions classified as pre-invasive, hyperplasia of the usual type is morphologically and phenotypically heterogeneous, whereas atypical ductal hyperplasia and ductal carcinoma in situ are homogenous in cell type and marker expression (3, 4). Retrospective studies indicates that 30 to 50% of the in situ carcinomas evolve into invasive tumors within 6–10 years from the diagnosis, whereas epidemiological and clinical data are less clear about the role of atypical ductal hyperplasia (3). However, genetic studies based on Comparative Genomic Hybridization and Loss of Heterozigosity analysis, demonstrated similar chromosomal abnormalities in both atypical ductal hyperplasia and in situ ductal carcinoma suggesting that they may have a similar clonal origin (2).
Breast cancer is the result of a multistep process characterized by the accumulation of molecular “hits” leading to uncontrolled growth and ultimately to the acquisition of invasive and metastatic potential. There is now a compelling body of evidence supporting the importance of epigenetic mechanisms in the development and progression of cancer (5, 6). In recent years, an increasing number of gene CpG islands aberrantly methylated in cancer were identified. Such DNA methylation mapping has suggested the existence of unique profile of hypermethylated CpG islands that define each neoplasia (5, 6). Changes in the number of methylated genes as well as an increase in methylation density during tumor progression were described in several tumor types, but only a few studies have investigated promoter methylation alterations during breast cancer progression (7).
In a previous study we performed a methylation profile in a series of invasive breast cancer and breast benign lesions (8). A similar pattern of methylation distribution was found for all the genes tested with the exception of CDH1 which was found to be methylated in malignant tumors but not in the benign breast lesions (8). These data suggest that methylation of at least some genes could be directly associated with the malignant phenotype. We sought to characterize frequency and pattern of methylation changes during progression from normal breast to invasive ductal carcinoma. All the genes tested were previously identified as methylated in mammary tumors or breast cancer cell lines (ESR1, CDH1, APC, GSTP1, TIMP3, MGMT THBS1, TMS1) (7). The only exception was CTNNB1 which encodes β-catenin and was described methylated only in metastatic gastric cancer and endometrial tumors (9, 10).
RESULTS
Clinical and Pathological data
We obtained pre-invasive and invasive breast lesions from 55 patients with a median age of 48 years (IQR, Interquartile Range 42–66). Ten cases were staged as Tis (18%), 6 T1a (9%), 10 T1b (16%), 15 T1c (27%), 13 were staged T2 (21%) and the remaining case (4%) was diagnosed with atypical ductal hyperplasia (ADH). Five cases displayed associated ADH, DCIS and IDC, 7 cases ADH and IDC, 13 cases showed associated DCIS and IDC, and 5 cases ADH and DCIS. Among remaining cases seven patients displayed only DCIS, two cases only ADH and seventeen cases only IDC. In total we analyzed 20 ADH from 19 patients (for case BP23 two separate lesion were analyzed), 32 DCIS from 29 patients (for case BP18 and BP22 three and two separate lesions were analyzed respectively) and 43 IDC from 41 patients (for case BP18 two separate lesions were analyzed). Of the 41 cases with invasive ductal carcinoma, 26 were negative for lymph node metastasis (63%) and 15 were positive (37%). Twelve cases showed pathological grade I (29%), 21 were grade II (51%), and 8 were grade III (40%). Six patients were diagnosed with distant metastases. The median values for estrogen receptor, progesterone receptor and ki67/Mib1 measured by immunohistochemical staining were respectively 60% (IQR 18%–90%), 20% (IQR 0%–66%) and 15% (IQR 3%–40%). Median age for the normal breast tissue group was 56 years (IQR 50–63), 19 of the 20 patients were affected by breast cancer (9 pT1cN0, 7 pT1cN1b, 1 pT1cN1a, 1 pT2N0 and 1 pT2N1b) and one patient was diagnosed with fibroadenoma.
Promoter methylation profile of breast pre-invasive and invasive lesions
ADH, DCIS and IDC were analyzed by QMSP for 9 genes including: ESR1, CDH1, APC, TIMP-3, CTNNB1, GSTPI, MGMT, THBS1, and TMS1. The frequency of methylation and the median values are listed on Table 1. Statistically significant differences in methylation frequency and level among NBT, ADH, DCIS and IDC were found for APC, CDH1 and CTNNB1 (P<0.0001, P<0.007, P=0.04). For APC, methylation levels were higher in all three pathological samples as compared to the control group (P<0.0001). CDH1 displayed a statistically significant increase in methylation levels as compared with the normal tissue group in DCIS (P=0.01), and IDC (P<0.001). Whereas CTNNB1 showed a statistically significant increase in methylation levels only in the IDC group (P=0.01). APC, CDH1 and CTNNB1 also showed an increase in methylation frequency in the pathological sample groups as compared with the normal tissue group (P<0.0001, P=0.003 and P=0.04 respectively).
Table 1.
Frequency of positive cases and methylation levels [Median (IQR, Interquartile Range)] in Atypical Ductal Hyperplasia (ADH), Ductal Carcinoma in situ (DCIS) Invasive Ductal carcinoma (IDC) and Normal Breast Tissue (NBT)
| Genes | ADH(n=21) | DCIS(n=32) | IDC(n=43) | NBT(n=20) | P* | ||||
|---|---|---|---|---|---|---|---|---|---|
| n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | n (%) | Median (IQR) | ||
| APC | 13 (65) | 16.66 (0–762.38) | 23 (72) | 167.08 (0–731.54) | 36 (84) | 157.44 (0.29–818.25) | 3 (15) | 0 (0-0) | P<0.0001 |
| CDH1 | 7 (35) | 0 (0–32.21) | 16 (50) | 0.43 (0–8.89) | 28 (65) | 4.70 (0–25.13) | 3 (15) | 0 (0-0) | P<0.004 |
| CTNNB1 | 4 (19) | 0 (0-0) | 11 (34) | 0 (0–2.27) | 19 (44) | 0 (0–3.33) | 2 (10) | 0 (0-0) | P<0.05 |
| ESR1 | 10 (48) | 6.55 (0–11.40) | 11 (34) | 0 (0–14.04) | 21 (51) | 0.41 (0–12.96) | 8 (40) | 0 (0–22.09) | P=NS |
| TIMP3 | 13 (62) | 6.50 (0–47.05) | 14 (44) | 0 (0–11.23) | 24 (56) | 5.31 (0–48.56) | 9 (45) | 0 (0–87.51) | P=NS |
| GSTPI | 2 (10) | 0 (0-0) | 5 (16) | 0 (0–0.75) | 8 (19) | 0 (0-0) | 1 (5) | 0 (0-0) | P=NS |
| MGMT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA |
| THBS1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA |
| TMS1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA |
NA, Not applicable; NS, not significant.
Similar methylation level and frequency were detected in both neoplastic and normal breast tissues for ESR1, TIMP3 and GSTP1. Methylation was not detected in either normal or neoplastic samples for MGMT, THBS1 and TMS1 (Table 1).
In tumor samples no correlation was found between APC, CDH1 and CTNNB1 methylation level and protein levels (ER, PgR status and Mib1/Ki67) or standard clinical/pathological (age, stage, grade, lymph node status) parameters.
Changes in methylation during breast cancer progression
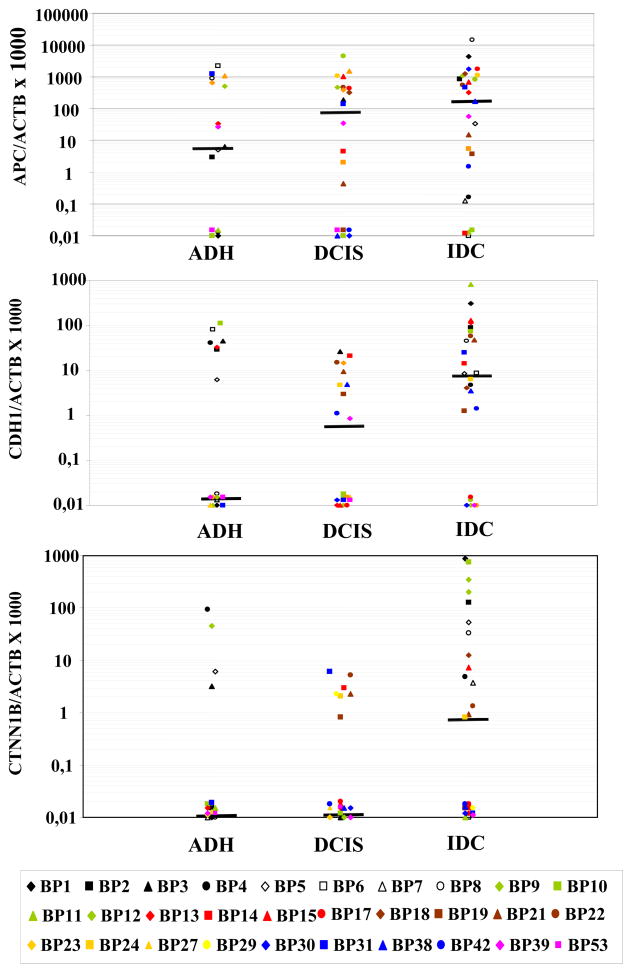
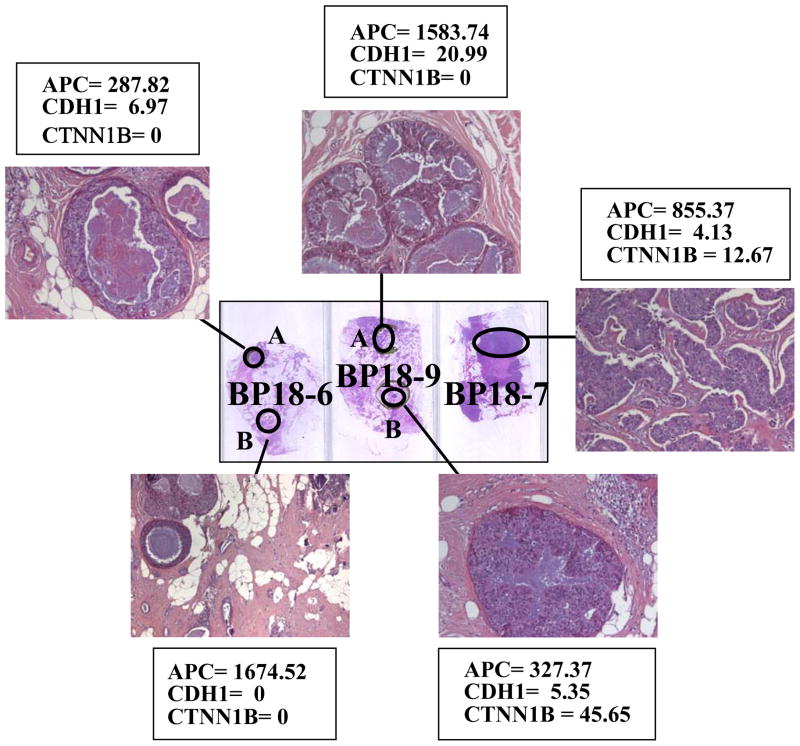
APC, CDH1 and CTNNB1 methylation levels were further analyzed in the paired pre-invasive and invasive lesions (Figure 1). The analysis demonstrated significant differences in methylation frequency for CDH1 and CTNNB1 among pre-invasive and invasive lesions with an increased number of methylated samples in the IDC group (P<0.03 and P<0.04 respectively). APC showed an increase in methylation frequency in IDC as compared with pre-invasive lesions but these differences did not reach statistical significance. The Wilcoxon Matched Pairs test demonstrated significantly higher levels of methylation in IDC as compared with DCIS for APC (P=0.03), and as compared with ADH and DCIS for CTNNB1 (P=0.04). No changes were demonstrated for CDH1 methylation levels. An example of APC, CDH1 and CTNNB1 methylation in multiple lesions from patient BP18 is shown in Figure 2. Although APC methylation was detected in all 5 lesions (two IDC and three DCIS), heterogeneity in the methylation pattern was detected for CDH1 and CTNNB1. Three DCIS and one of the IDC lesions displayed some level of methylation of the CDH1 gene whereas CTNNB1 methylation was detected in only one DCIS and one IDC lesion.
FIGURE 1.
Distribution plots for APC (A), CDH1 (B) and CTNNB1(C) methylation levels in the paired breast lesions: ADH, atypical ductal hyperplasia, DCIS ductal carcinoma in situ, and IDC invasive ductal carcinoma, horizontal bar, median value, * samples with a ratio equal to zero can not be plotted correctly on a log scale and we thus plotted at baseline.
FIGURE 2.
The analysis of multiple breast lesions from patient BP18 displays heterogeneity in APC, CDH1 and CTNNB1. Slide BP18-6 shows a DCIS (A) and an IDC (B), BP18-9 displays two DCIS (A and B) and BP18-7 an IDC. The target gene/ACTB ratio for APC, CDH1 and CTNNB1 are indicated for each of the genes. APC methylation is present in all the lesions with ratios above the cut off of 20.73; CDH1 methylation was above the cut-off of 8.10 only in BP18-9 and CTNNB1 methylation levels were above the cut-off of 2.65 for BP18-9B and BP18-7
APC, CDH1 and CTNNB1 methylation can distinguish pre-invasive from neoplastic breast lesions
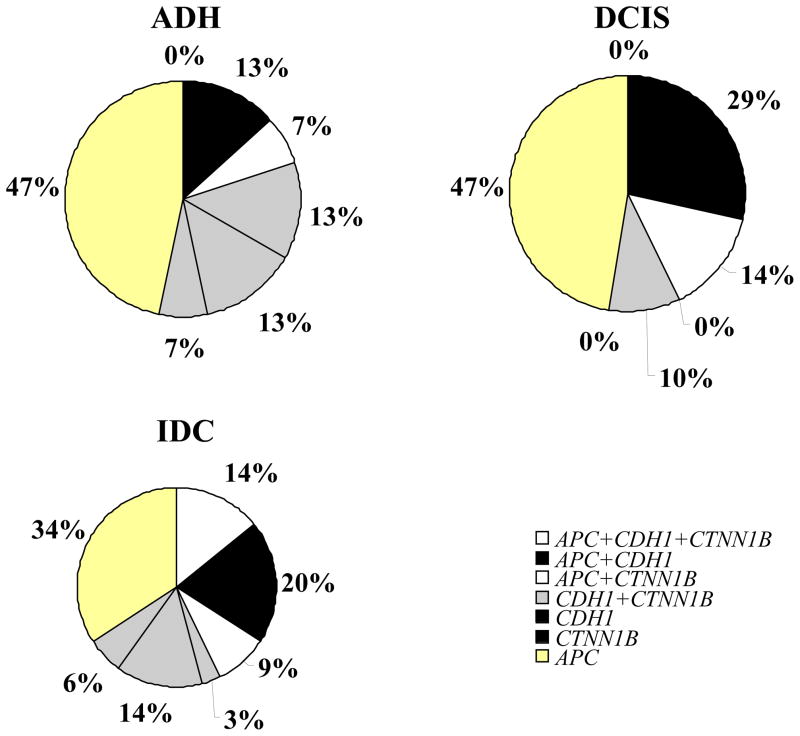
The 90th percentile of the distribution in normal samples was used as cutoff value to distinguish among normal and pathological samples. These values were 20.73 for APC, 8.10 for CDH1 and 2.65 for CTNNB1. Table 2 shows the distribution of the samples methylated above these cutoff values. Methylation in at least one of the genes was detected in 71 out of 96 (74%) preinvasive or neoplastic samples and 3 out of 20 (15%) normal breast tissues (P<0.0001) (Table 2). Methylation at two or three gene loci was found in 5 ADH (25%), 9 DCIS (28%) and 16 IDC (37%) (P=0.03). Synchronous methylation of APC, CDH1, and CTNNB1 promoter regions was found only in IDC (P=0.04), whereas simultaneous methylation of APC and CDH1 or APC and CTNNB1 was more frequent in IDC and DCIS (P=0.01) (Figure 3).
Table 2.
Gene methylation patterns above the optimal cut-off value for individual lesions in breast cancer progression.
| APC* | CDH1* | CTNN1B* | |
|---|---|---|---|
| BP1 IDC | M | M | M |
| BP2 IDC | M | M | M |
| BP5 IDC | M | M | M |
| BP8 IDC | M | M | M |
| BP15 IDC | M | M | M |
| B22A DCIS | M | M | um |
| BP13 IDC | M | M | um |
| BP21 IDC | M | M | um |
| B22 IDC | M | M | um |
| BP31 IDC | M | M | um |
| BP32 IDC | M | M | um |
| BP33 IDC | M | M | um |
| BP34 IDC | M | M | um |
| BP35 DCIS | M | M | um |
| BP13 ADH | M | M | um |
| BP6 ADH | M | M | um |
| BP18-9A DCIS | M | M | um |
| BP3 DCIS | M | M | um |
| B22B DCIS | M | M | um |
| BP23 DCIS | M | M | um |
| BP18-72 IDC | M | um | M |
| BP9 IDC | M | um | M |
| BP12 IDC | M | um | M |
| BP18-9B DCIS | M | um | M |
| B25 DCIS | M | um | M |
| BP31 DCIS | M | um | M |
| BP9 ADH | M | um | M |
| BP10 IDC | um | M | M |
| BP3 ADH | um | M | M |
| BP4 ADH | um | M | M |
| BN10 NBT | um | M | M |
| BP6 IDC | um | M | um |
| BP14 IDC | um | M | um |
| BP36 IDC | um | M | um |
| BP37 IDC | um | M | um |
| BP11 IDC | um | M | um |
| BP14 DCIS | um | M | um |
| BP21 DCIS | um | M | um |
| BP2 ADH | um | M | um |
| BP10 ADH | um | M | um |
| BP4 IDC | um | um | M |
| BP7 IDC | um | um | M |
| BP5 ADH | um | um | M |
| BP38 IDC | M | um | um |
| BP39 IDC | M | um | um |
| BP40 IDC | M | um | um |
| BP41 IDC | M | um | um |
| BP42 IDC | M | um | um |
| BP43 IDC | M | um | um |
| BP44 IDC | M | um | um |
| BP45 IDC | M | um | um |
| BP17 IDC | M | um | um |
| BP18-6 IDC | M | um | um |
| BP29 IDC | M | um | um |
| BP30 IDC | M | um | um |
| BP39 DCIS | M | um | um |
| BP9 DCIS | M | um | um |
| BP12 DCIS | M | um | um |
| BP15 DCIS | M | um | um |
| BP17 DCIS | M | um | um |
| BP18-6 DCIS | M | um | um |
| BP26 DCIS | M | um | um |
| BP27 DCIS | M | um | um |
| BP28 DCIS | M | um | um |
| BP29 DCIS | M | um | um |
| BP39 ADH | M | um | um |
| BP23A ADH | M | um | um |
| BP23B ADH | M | um | um |
| BP27 ADH | M | um | um |
| BP31 ADH | M | um | um |
| BP8 ADH | M | um | um |
| BP16 ADH | M | um | um |
| BN1 NBT | M | um | um |
| BN15 NBT | M | um | um |
| BP19 IDC | um | um | um |
| BP24 IDC | um | um | um |
| BP46 IDC | um | um | um |
| BP47 IDC | um | um | um |
| BP48 IDC | um | um | um |
| BP49 IDC | um | um | um |
| BP50 IDC | um | um | um |
| BP51 IDC | um | um | um |
| BP52 IDC | um | um | um |
| BP38 DCIS | um | um | um |
| BP47 DCIS | um | um | um |
| BP53 DCIS | um | um | um |
| BP54 DCIS | um | um | um |
| BP55 DCIS | um | um | um |
| BP53 ADH | um | um | um |
| BP46 ADH | um | um | um |
| BP10 DCIS | um | um | um |
| BP13 DCIS | um | um | um |
| BP19 DCIS | um | um | um |
| B24 DCIS | um | um | um |
| BP30 DCIS | um | um | um |
| BP20 DCIS | um | um | um |
| BP1 ADH | um | um | um |
| BP7 ADH | um | um | um |
| BP11 ADH | um | um | um |
| BN2 NBT | um | um | um |
| BN3 NBT | um | um | um |
| BN4 NBT | um | um | um |
| BN5 NBT | um | um | um |
| BN6 NBT | um | um | um |
| BN7 NBT | um | um | um |
| BN8 NBT | um | um | um |
| BN9 NBT | um | um | um |
| BN11 NBT | um | um | um |
| BN12 NBT | um | um | um |
| BN13 NBT | um | um | um |
| BN14 NBT | um | um | um |
| BN16 NBT | um | um | um |
| BN17 NBT | um | um | um |
| BN18 NBT | um | um | um |
| BN19 NBT | um | um | um |
| BN20 NBT | um | um | um |
M, methylated sample; um, unmethylated sample.
FIGURE 3.
Methylation distribution in breast tissues samples above the optimal cut-off values of 20.73 for APC, 8.10 for CDH1 and 2.65 for CTNNB1.
DISCUSSION
We have investigated the changes in methylation patterns during breast cancer progression in a series of pre-invasive and invasive breast lesions. The APC promoter was methylated in approximately two thirds of atypical hyperplasia and showed similar methylation levels in in situ and invasive ductal carcinomas. CDH1 and CTNNB1 methylation frequencies and levels increased during progression from pre-invasive to invasive tumors. Atypical ductal hyperplasia and in situ carcinoma showed similar methylation patterns, in agreement with immunophenotipic and genetic data suggesting that atypical hyperplasia should be considered as a well-differentiated or simply small in situ carcinoma (2).
Reduced E-cadherin (CDH1) expression in mammary tumors correlates with loss of differentiation, invasiveness, increased tumor grade, metastases and overall worst prognosis (11). Complete loss of expression is found in 85% of the infiltrative lobular carcinomas, whereas variable levels of expression are found in invasive ductal carcinomas (11–15). Loss of heterozigosity at the E-cadherin gene locus (CDH1) is a frequent event in both ductal and lobular carcinomas, but inactivation of the second allele by mutation was demonstrated only in the lobular subtype (16, 17). Approximately 40% of breast cancers (both lobular and ductal type) harbored methylation at the CDH1 promoter CpG island (17,18). APC mutation or epigenetic inactivation play a key role in colorectal carcinogenesis and in particular in the early stages of disease progression (19). In breast cancer APC is mainly down regulated through promoter hypermethylation which is reported in 30–50% of the cases, and in breast cancer cell lines reduced protein expression correlated with methylation status (8, 17, 20).
Although the role of methylation in CDH1 and APC expression is well known, there are few data on CTNNB1 methylation. In colorectal cancer, β-catenin is activated by oncogenic mutation affecting the phosphorylation site resulting in a constitutively stable protein, or by loss of functional APC (21, 22). However, CTNNB1 methylation (and presumably inactivation) was recently reported in gastric and endometrial cancers (9, 10). In a series of primary metastatic gastric cancer and cell lines derived from metastasis, loss of expression of β-catenin was related to promoter methylation (9). Sequence analysis of the bisulfite treated DNA showed heavy CpG methylation of the CTNNB1 promoter region in a cell line with absent β-catenin expression and treatment with 5-deoxyazacytidine was able to restore β-catenin expression (9). Moreover, Whitcomb et al (10) reported methylation at the CTNNB1 promoter region in 17% of primary endometrial tumors.
To the best of our knowledge, our present study is the first to report CTNNB1 methylation in breast cancer. In mammary tumors, CTNNB1 mutations at the phosphorylation site were not detected, and in the majority of the cases, immunohistochemestry showed loss of β-catenin expression rather than protein accumulation (17, 23, 24). In lobular carcinogenesis, simultaneous loss of expression of E-cadherin and β-catenin seems to be an important step in the formation of in situ lobular carcinoma precursors (23). Moreover, Karayiannakis et al (24) demonstrated that qualitative and quantitative alterations of β-catenin expression occur not only in breast cancer of the lobular type, but also in the ductal type. In a series of 58 ductal carcinomas, they found a heterogeneous protein staining with positive tumor cells interspersed with negative cells in 47 of the tumors (81%) and a complete absence of the staining in 4 cancers (7%) (24). In the same study the staining pattern of synchronous in situ carcinomas was in most of the cases the same as in the invasive carcinoma (24). An analysis of human breast cancer cell lines, also showed reduced or complete loss of β-catenin expression (25). As expected, one cell line with APC truncation displayed β-catenin accumulation (26).
Interestingly, APC, CDH1 and CTNNB1 gene products are functionally linked. Cadherins are a family of transmembrane glycoproteins that play pivotal role in the establishment and maintenance of normal tissue architecture (27). The E-cadherin is mainly expressed in epithelial tissues and in normal breast prevalently in the luminal cells. In developmental animal models, E-cadherin expression is temporarily down regulated when budding lobules invade the stroma (28). Although the cell-to-cell adhesion properties of E-cadherin reside in the extracellular domain, its function depends on its interaction with catenins and in particular β-catenin, which is responsible for the anchorage of E-cadherin to the actin cytoskeleton (27). In a normal cell, β-catenin localize to the cytoplasm, and it is continuously degradated by phosphorylation and subsequent ubiquitination. Phosphorylation occurs in a multiprotein complex, which requires, among other proteins, the presence of APC a scaffolding protein regulated by the Wnt signalling pathway. However, this is not the only function of the APC protein, its terminal C-region can interact with cytoskeleton proteins, contributing to cell migration, proliferation and adhesion (29). Thus, APC, CDH1 and CTNNB1 play an interaction role in the maintenance of cell-to-cell adhesion, and regulation of cell-extracellular matrix interactions (30). The loss of these abilities is necessary, although not sufficient, for the acquisition of invasive properties by cancer cells. A role in “invasion suppression” would be consistent with our results showing changes in promoter methylation in the switch from pre-invasive lesions to invasive breast cancer.
TIMP-3 ESR1, and GSTP1 methylation frequency and levels were similar in normal and neoplastic breast tissues from cancer patients. This finding is puzzling, but might be explained by the occurence of epigenetic changes in apparently normal stromal cells. Hu et al (31) investigated, with a whole genome analysis approach, methylation patterns of epithelial and myoepithelial cells, in stromal fibroblasts from normal breast tissues, as well as in situ and invasive breast carcinoma. The analysis showed distinct epigenetic changes in all three types of cells with some genes differentially methylated in normal and neoplastic samples and others showing the same methylation pattern in cells derived from normal and neoplastic tissues (31). Thus, our results could be explained by the presence of similar levels of methylation of TIMP-3 ESR1, and GSTP1 in stromal cells from normal and cancer specimens. However, these similar epigenetic patterns do not exclude that promoter methylation may play a role in carcinogenesis. Alterations in stromal and myoepithelial cells may establish an abnormal tumor microenviroment and contribute to cancer progression.
We propose a model for the timing of epigenetic modifications in breast cancer development APC methylation would be an early event, correlated with abnormal proliferation of the breast epithelia. CDH1 and CTNNB1 methylation occur later and are likely to play a more direct role in the loss of cell-to-cell adhesion and the acquisition of invasive properties by the cancer cells. The detection of breast cancer in the early stages is a key for a successful treatment of the disease. Our results indicate that methylation not only occurs early during tumor progression, but also that the analysis of specific genes may allow to distinguish between normal and transformed cells and even between pre-invasive and invasive carcinomas, thus representing a promising tool for the identification of tumor cells in clinical specimens. The clinical presentation of breast cancer is often a palpable breast nodule accompanied by cytomorphological analysis of Fine Needle Aspiration (FNA) biopsies for diagnostic evaluation of suspicious breast lesions. However this procedure has false negative rates ranging from 5 to 30%, the accuracy of the analysis also depends on the ability of the operator to collect the sample and the proficiency of the cytopathologist in performing morphological examination (32). DNA methylation has an advantage over other molecular detection methods (e.g. single gene mutation, microsatellite analysis etc.,) because it can be detected with a very high degree of specificity even in the presence of an excess of unmethylated DNA (33). A number of studies have also reported the ability to detect breast cancer cells by DNA methylation analysis in FNAs, Nipple Aspirates or ductal lavages (34–38) with variable sensitivity. Our results suggest that the selection of gene promoter targets is critical based on their frequency and timing in breast cancer development and progression. Thus, it is likely that the combined detection of methylation of APC, CDH1 and CTNNB1 could be more informative than other methylation markers in identifying cancer cells in cytological specimens. This approach might help the pathologist to distinguish definitely malignant from indolent lesions and the clinician to differentiate between lesions with better or worse prognosis.
Materials and Methods
Samples and DNA extraction
Tissue samples from 55 patients were obtained as paraffin embedded specimens from the Department of Pathology “L. Armanni”, University of Naples Italy, the Department of Pathology IRCCS “Casa Sollievo della Sofferenza” San Giovanni Rotondo, Italy and the Department of Pathology IRCCS Oncology Institute, Bari, Italy. Overall 95 pre-invasive and invasive lesions from 55 patients and 20 normal breast tissues from cancer patients were included in the study. Sections, 5-μm-thick, were cut from paraffin blocks and two pathologists (A. Apicella and F. Zito) reviewed each slide to identify the areas of Atypical Ductal Hyperplasia (ADH), Ductal Carcinoma in situ (DCIS) and Invasive Ductal Carcinoma (IDC), and to exclude cancer cell contamination in normal breast tissues (NBT). Sample specimens were then carefully dissected, under a microscope from 12-μm-thick sections to enrich for areas that contained ADH, DCIS or IDC. For each pathological lesion, 6 to 12 sections were dissected depending on the dimension of the lesion. Paraffin-embedded tissues were subsequently placed in xylene to remove the paraffin and genomic DNA was extracted as described previously (39).
Sodium Bisulfite Modification
DNA extracted from tumors was subjected to bisulfite treatment, as described previously with minor modification (40). Briefly, 2 μg of genomic DNA from each sample was denatured with NaOH (final concentration, 0.2 M) in a total volume of 20 μl at 50°C for 20 minutes. The denatured DNA was diluted by adding 500 μL of a freshly prepared solution of 10 mM hydroquinone and 3 M sodium bisulfite, and incubated at 70°C for 3 hours. Bisulfite-modified DNA was purified using a Wizard DNA Clean-Up System (Promega), treated with 0.3 M NaOH at room temperature for 10 minutes, precipitated with ethanol, resuspended in 120 μL of LoTE (2.5 mM EDTA, 10 mM Tris–HCl (PH 8), and stored at −80°C.
Quantitative Methyl Specific PCR (QMSP)
Bisulfite-modified DNA was used as template for fluorescence-based real-time PCR, as previously described (40). Amplification reactions were carried out in triplicate in a volume of 20 μL that contained 3 μL bisulfite-modified DNA, 600 nM forward and reverse primers, 200 nM probe, 5 U of Platinum Taq polymerase (Invitrogen), 200 μM each of dATP, dCTP, dGTP, 200 μM dTTP and 5.5 mM MgCl2. Primers and probes were designed to specifically amplify the promoters of the nine genes of interest and the promoter of a reference gene, ACTB (primer and probe sequences and annealing temperatures are provided in Supplemental Table 1). Amplifications were carried out using the following conditions: one step at 95°C for 3 minutes, 50 cycles at 95°C for 15 seconds, and 60 °C to 62 °C for 1 minute. Supplemental Table 2 lists the nine genes whose promoters were examined, their proposed functions, and the tumors in which these promoters have been shown to be hypermethylated. Amplification reactions were carried out in 384-well plates in a 7900 Sequence detector (Perkin-Elmer Applied Biosystems) and were analyzed by SDS 2.2.1software ( Applied Biosystems). Each plate included patient DNA samples, positive (in vitro methylated leukocyte DNA) and negative (normal leukocyte DNA or DNA from a known unmethylated cell line) controls, and multiple water blanks. Leukocyte DNA from a healthy subject was methylated in vitro with excess SssI methyltransferase (New England Biolabs Inc., Beverly, MA) to generate completely methylated DNA. Serial dilutions (90-.009 ng) of this DNA were used to construct a calibration curve for each plate. All samples were within the assay’s range of sensitivity and reproducibility based on amplification of an internal reference standard (CT value for ACTB of 40 or less). The relative level of methylated DNA for each gene in each sample was determined as a ratio of methylation specific PCR-amplified gene to ACTB (reference gene) and then multiplied by 1000 for easier tabulation (average value of triplicates of gene of interest/average value of triplicates of ACTB × 1000). The samples were categorized as unmethylated or methylated based on the sensitivity of the assay and 90th percentile distribution of methylation levels in known normal (control) breast tissue DNA.
Statistical Analysis
For each gene, the frequency of methylated and unmethylated cases, as well as the median and interquartile range of methylation ratios was determined for the study groups. The Kolmogorov-Smirnov Test allowed for the examination of the appropriateness of a normal distribution assumption for each of the parameters, and then data were analyzed using non-parametric tests. Differences between sample groups were determined using the Kruskal-Wallis test followed by the Mann-Whitney U test when appropriate. For comparing methylation levels between paired samples, the Wilcoxon matched pairs test was performed. The correlation between tumor methylation ratios on the one hand, and age, pathological stage, ER, PgR and Ki67/Mib1 levels, were determined by calculating a Spearman’s correlations coefficient. The χ2 test or Fisher’s Exact test were used for comparison of frequency distribution of the methylated genes among the experimental groups. Statistical analysis was done using SPSS 10.0 software and all tests are two sided with significance at P≤0.05.
Acknowledgments
Grant support: This work was supported by a grant from AIRC (Associazione Italiana Ricerca sul Cancro) and partially supported by a grant from the Italian Ministry of Health (RC 2005 and RC2006). Maria Prencipe was partially supported by an ICRETT grant from the UICC). Under a licensing agreement between Oncomethylome Sciences, SA and the Johns Hopkins University, Dr. Sidransky is entitled to a share of royalty received by the University on sales of products described in this article. Dr. Sidransky owns Oncomethylome Sciences, SA stock, which is subject to certain restrictions under University policy. Dr. Sidransky is a paid consultant to Oncomethylome Sciences, SA and is a paid member of the company’s Scientific Advisory Board.
Non-standard abbreviation
- ADH
Atypical ductal Hyperplasia
- DCIS
Ductal Carcinoma in situ
- IDC
Invasive Ductal carcinoma
- ILC
Invasive Lobular Carcinoma
- NBT
Normal Breast Tissue
Footnotes
The term of this arrangement is being managed by the Johns Hopkins University in accordance with its conflict of interest policies.
References
- 1.Parkin DM, Bray FI, Devesa SS. Cancer burden in the year 2000. The global picture. Eur J Cancer. 2001;37 (Suppl 8):S4–66. doi: 10.1016/s0959-8049(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 2.Reis-Filho JS, Lakhani SR. The diagnosis and management of pre-invasive breast disease: genetic alterations in pre-invasive lesions. Breast Cancer Res. 2003;5:313–319. doi: 10.1186/bcr650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartow SA, Pathak DR, Black WC, Key CR, Teaf SR. Prevalence of benign, atypical, and malignant breast lesions in populations at different risk for breast cancer. A forensic autopsy study. Cancer. 1987;60:2751–2760. doi: 10.1002/1097-0142(19871201)60:11<2751::aid-cncr2820601127>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 4.van Dongen JA, Fentiman IS, Harris JR, Holland R, Peterse JL, Salvadori B, Stewart HJ. In-situ breast cancer: the EORTC consensus meeting. Lancet. 1989;2:25–27. doi: 10.1016/s0140-6736(89)90263-8. [DOI] [PubMed] [Google Scholar]
- 5.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 6.Costello JF, Fruhwald MC, Smiraglia DJ, Rush LJ, Robertson GP, Gao X, Wright FA, Feramisco JD, Peltomaki P, Lang JC, Schuller DE, Yu L, Bloomfield CD, Caligiuri MA, Yates A, Nishikawa R, Su Huang H, Petrelli NJ, Zhang X, O’Dorisio MS, Held WA, Cavenee WK, Plass C. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat Genet. 2000;24:132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 7.Parrella P. CpG island hypermethylation in breast cancer progression and metastasis. In: Esteller M, editor. DNA Methylation Epigenetics and Metastasis. Vol. 7. Dordrecht, The Netherlands: Springer; 2005. pp. 81–132. [Google Scholar]
- 8.Parrella P, Poeta ML, Gallo AP, Prencipe M, Scintu M, Apicella A, Rossiello R, Liguoro G, Seripa D, Gravina C, Rabitti C, Rinaldi M, Nicol T, Tommasi S, Paradiso A, Schittulli F, Altomare V, Fazio VM. Nonrandom distribution of aberrant promoter methylation of cancer-related genes in sporadic breast tumors. Clin Cancer Res. 2004;10:5349–5354. doi: 10.1158/1078-0432.CCR-04-0555. [DOI] [PubMed] [Google Scholar]
- 9.Ebert MP, Yu J, Hoffmann J, Rocco A, Rocken C, Kahmann S, Muller O, Korc M, Sung JJ, Malfertheiner P. Loss of beta-catenin expression in metastatic gastric cancer. J Clin Oncol. 2003;21:1708–1714. doi: 10.1200/JCO.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 10.Whitcomb BP, Mutch DG, Herzog TJ, Rader JS, Gibb RK, Goodfellow PJ. Frequent HOXA11 and THBS2 promoter methylation, and a methylator phenotype in endometrial adenocarcinoma. Clin Cancer Res. 2003;9:2277–2287. [PubMed] [Google Scholar]
- 11.Heimann R, Lan F, McBride R, Hellman S. Separating favorable from unfavorable prognostic markers in breast cancer: the role of E-cadherin. Cancer Res. 2000;60:298–304. [PubMed] [Google Scholar]
- 12.Gamallo C, Palacios J, Suarez A, Pizarro A, Navarro P, Quintanilla M, Cano A. Correlation of E-cadherin expression with differentiation grade and histological type in breast carcinoma. Am J Pathol. 1993;142:987–993. [PMC free article] [PubMed] [Google Scholar]
- 13.Moll R, Mitze M, Frixen UH, Birchmeier W. Differential loss of E-cadherin expression in infiltrating ductal and lobular breast carcinomas. Am J Pathol. 1993;143:1731–1742. [PMC free article] [PubMed] [Google Scholar]
- 14.Vos CB, Cleton-Jansen AM, Berx G, de Leeuw WJ, ter Haar NT, van Roy F, Cornelisse CJ, Peterse JL, van de Vijver MJ. E-cadherin inactivation in lobular carcinoma in situ of the breast: an early event in tumorigenesis. Br J Cancer. 1997;76:1131–1133. doi: 10.1038/bjc.1997.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huiping C, Sigurgeirsdottir JR, Jonasson JG, Eiriksdottir G, Johannsdottir JT, Egilsson V, Ingvarsson S. Chromosome alterations and E-cadherin gene mutations in human lobular breast cancer. Br J Cancer. 1999;81:1103–1110. doi: 10.1038/sj.bjc.6690815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cleton-Jansen AM. E-cadherin and loss of heterozygosity at chromosome 16 in breast carcinogenesis: different genetic pathways in ductal and lobular breast cancer? Breast Cancer Res. 2002;4:5–8. doi: 10.1186/bcr416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarrio D, Moreno-Bueno G, Hardisson D, Sanchez-Estevez C, Guo M, Herman JG, Gamallo C, Esteller M, Palacios J. Epigenetic and genetic alterations of APC and CDH1 genes in lobular breast cancer: relationships with abnormal E-cadherin and catenin expression and microsatellite instability. Int J Cancer. 2003;106:208–215. doi: 10.1002/ijc.11197. [DOI] [PubMed] [Google Scholar]
- 18.Graff JR, Herman JG, Lapidus RG, Chopra H, Xu R, Jarrard DF, Isaacs WB, Pitha PM, Davidson NE, Baylin SB. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–5199. [PubMed] [Google Scholar]
- 19.Fearnhead NS, Wilding JL, Bodmer WF. Genetics of colorectal cancer: hereditary aspects and overview of colorectal tumorigenesis. Br Med Bull. 2002;64:27–43. doi: 10.1093/bmb/64.1.27. [DOI] [PubMed] [Google Scholar]
- 20.Virmani AK, Rathi A, Sathyanarayana UG, Padar A, Huang CX, Cunnigham HT, Farinas AJ, Milchgrub S, Euhus DM, Gilcrease M, Herman J, Minna JD, Gazdar AF. Aberrant methylation of the adenomatous polyposis coli (APC) gene promoter 1A in breast and lung carcinomas. Clin Cancer Res. 2001;7:1998–2004. [PubMed] [Google Scholar]
- 21.Rubinfeld B, Albert I, Porfiri E, Fiol C, Munemitsu S, Polakis P. Binding of GSK3beta to the APC-beta-catenin complex and regulation of complex assembly. Science. 1996;272:1023–1026. doi: 10.1126/science.272.5264.1023. [DOI] [PubMed] [Google Scholar]
- 22.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 23.De Leeuw WJ, Berx G, Vos CB, Peterse JL, Van de Vijver MJ, Litvinov S, Van Roy F, Cornelisse CJ, Cleton-Jansen AM. Simultaneous loss of E-cadherin and catenins in invasive lobular breast cancer and lobular carcinoma in situ. J Pathol. 1997;183:404–411. doi: 10.1002/(SICI)1096-9896(199712)183:4<404::AID-PATH1148>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 24.Karayiannakis AJ, Nakopoulou L, Gakiopoulou H, Keramopoulos A, Davaris PS, Pignatelli M. Expression patterns of beta-catenin in in situ and invasive breast cancer. Eur J Surg Oncol. 2001;27:31–36. doi: 10.1053/ejso.1999.1017. [DOI] [PubMed] [Google Scholar]
- 25.Pierceall WE, Woodard AS, Morrow JS, Rimm D, Fearon ER. Frequent alterations in E-cadherin and alpha- and beta-catenin expression in human breast cancer cell lines. Oncogene. 1995;11:1319–1326. [PubMed] [Google Scholar]
- 26.Schlosshauer PW, Brown SA, Eisinger K, Yan Q, Guglielminetti ER, Parsons R, Ellenson LH, Kitajewski J. APC truncation and increased beta-catenin levels in a human breast cancer cell line. Carcinogenesis. 2000;21:1453–1456. [PubMed] [Google Scholar]
- 27.Behrens J. Cadherins and catenins: role in signal transduction and tumor progression. Cancer Metastasis Rev. 1999;18:15–30. doi: 10.1023/a:1006200102166. [DOI] [PubMed] [Google Scholar]
- 28.Daniel CW, Strickland P, Friedmann Y. Expression and functional role of E- and P-cadherins in mouse mammary ductal morphogenesis and growth. Dev Biol. 1995;169:511–519. doi: 10.1006/dbio.1995.1165. [DOI] [PubMed] [Google Scholar]
- 29.Nathke I. APC at a glance. J Cell Sci. 2004;117:4873–4875. doi: 10.1242/jcs.01313. [DOI] [PubMed] [Google Scholar]
- 30.Welch DR, Steeg PS, Rinker-Schaeffer CW. Molecular biology of breast cancer metastasis. Genetic regulation of human breast carcinoma metastasis. Breast Cancer Res. 2000;2:408–416. doi: 10.1186/bcr87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu M, Yao J, Cai L, Bachman KE, van den Brule F, Velculescu V, Polyak K. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 32.Barrows GH, Anderson TJ, Lamb JL, Dixon JM. Fine-needle aspiration of breast cancer. Relationship of clinical factors to cytology results in 689 primary malignancies. Cancer. 1986;58:1493–1498. doi: 10.1002/1097-0142(19861001)58:7<1493::aid-cncr2820580720>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 33.Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- 34.Pu RT, Laitala LE, Alli PM, Fackler MJ, Sukumar S, Clark DP. Methylation profiling of benign and malignant breast lesions and its application to cytopathology. Mod Pathol. 2003;16:1095–1101. doi: 10.1097/01.MP.0000095782.79895.E2. [DOI] [PubMed] [Google Scholar]
- 35.Jeronimo C, Costa I, Martins MC, Monteiro P, Lisboa S, Palmeira C, Henrique R, Teixeira MR, Lopes C. Detection of gene promoter hypermethylation in fine needle washings from breast lesions. Clin Cancer Res. 2003;9:3413–3417. [PubMed] [Google Scholar]
- 36.Evron E, Dooley WC, Umbricht CB, Rosenthal D, Sacchi N, Gabrielson E, Soito AB, Hung DT, Ljung B, Davidson NE, Sukumar S. Detection of breast cancer cells in ductal lavage fluid by methylation-specific PCR. Lancet. 2001;357:1335–1336. doi: 10.1016/s0140-6736(00)04501-3. [DOI] [PubMed] [Google Scholar]
- 37.Krassenstein R, Sauter E, Dulaimi E, Battagli C, Ehya H, Klein-Szanto A, Cairns P. Detection of breast cancer in nipple aspirate fluid by CpG island hypermethylation. Clin Cancer Res. 2004;10:28–32. doi: 10.1158/1078-0432.ccr-0410-3. [DOI] [PubMed] [Google Scholar]
- 38.Dulaimi E, Hillinck J, Ibanez de Caceres I, Al-Saleem T, Cairns P. Tumor suppressor gene promoter hypermethylation in serum of breast cancer patients. Clin Cancer Res. 2004;10:6189–6193. doi: 10.1158/1078-0432.CCR-04-0597. [DOI] [PubMed] [Google Scholar]
- 39.Parrella P, Sidransky D, Merbs SL. Allelotype of posterior uveal melanoma: implications for a bifurcated tumor progression pathway. Cancer Res. 1999;59:3032–3037. [PubMed] [Google Scholar]
- 40.Jeronimo C, Usadel H, Henrique R, Oliveira J, Lopes C, Nelson WG, Sidransky D. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J Natl Cancer Inst. 2001;93:1747–1752. doi: 10.1093/jnci/93.22.1747. [DOI] [PubMed] [Google Scholar]