Abstract
Ovarian cancer is the leading cause of death among gynecological cancers. It is now recognized that in addition to genetic alterations, epigenetic mechanisms, such as DNA methylation, histone modifications and nucleosome remodeling, play an important role in the development and progression of ovarian cancer by modulating chromatin structure, and gene and miRNA expression. Furthermore, epigenetic alterations have been recognized as useful tools for the development of novel biomarkers for diagnosis, prognosis, therapeutic prediction and monitoring of diseases. Moreover, new epigenetic therapies, such as DNA methyltransferase inhibitors and histone deacetylase inhibitors, have been found to be a potential therapeutic option, especially when used in combination with other agents. Here we discuss current developments in ovarian carcinoma epigenome research, the importance of the ovarian carcinoma epigenome for development of diagnostic and prognostic biomarkers, and the current epigenetic therapies used in ovarian cancer.
Keywords: DNA methylation, DNA methyltransferase, epigenetics, histone deacetylase, histone modification, ovarian cancer
Ovarian cancer is the second most common gynecological cancer and is the leading cause of death among gynecological cancers world-wide [1]. It is expected that in 2010 there will be 21,880 new cases of ovarian cancer in the USA [2] and 13,850 deaths [1]. One of the reasons for the high fatality rate is that more than 70% of cases are diagnosed at an advanced stage. The 5-year survival rates for women with advanced disease are only 20–30%; however, for women who are diagnosed when disease is confined to the ovary, event-free survival rates are approximately 70–90% [3]. The late diagnosis of ovarian cancer is related to the absence of symptoms in the majority of cases during the early stages of the disease, and the lack of truly sensitive and specific screening techniques for early detection of the disease.
Cancer has been considered as a disease driven by progressive genetic alterations, such as mutations involving oncogenes and/or tumor suppressor genes (TSGs), as well as chromosomal abnormalities [3,4]. However, more recently, it has been demonstrated that cancer is also driven by epigenetic alterations, which, unlike genetic alterations, do not alter the primary DNA sequence and occur at the chromosomal level in transformed cells [4]. These epigenetic alterations can influence the transcriptional process, leading to changes in the expression patterns of several genes implicated in diverse cellular processes such as proliferation, differentiation and survival [5,6]. The epigenetic modifications described so far involve [5–9]:
■ DNA methylation
■Histone modifications
■Dysregulations of nucleosomes
Moreover, the recent identification of promoter methylation of miRNAs has become an important epigenetic mechanism that has been described to participate in tumorigenesis. Among these three types of modifications, DNA methylation has been the best studied and the pattern seen in cancer cells is a global hypomethylation with a focal hypermethylation pattern. Global hypomethylation, found to be increased with age, is linked to increased karyotypic instability and activation of tumor-promoting genes by cis or trans effects, which might include altered heterochromatin–euchromatin interactions [10–13]. On the other hand, gene-locus-specific hypermethylation can lead to the transcriptional silencing of TSGs [5–9]. Post-translational epigenetic alterations of histones can modify chromatin structure and, thus, regulate gene expression [5–8,14], and current work on identifying and studying regulators that control nucleosomal remodeling has revealed that some of them are also involved in regulation of DNA methylation and histone modifications [5,7–9,14,15]. Therefore, three epigenetic events – DNA methylation, histone modifications and nucleosomal remodeling – mutually interact with each other to regulate gene expression [6,8,15,16]. A great effort has been made to discern the molecular events that lead to these epigenetic alterations and their consequences, in order to have a better understanding of how cancers initiate, progress and/or relapse, and their clinical consequences.
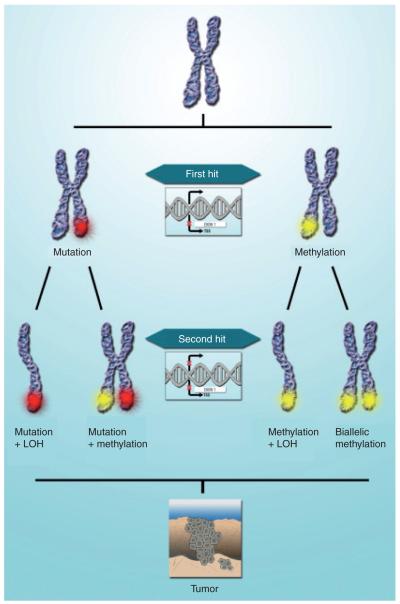
According to Knudson’s two-hit model, complete inactivation of a TSG requires both gene copies to lose their function [17]. The original theory suggested that one allele can be inactivated by a mutation and the other allele by loss of heterozygosity (LOH). LOH is commonly assumed to be caused by deletion of the appropriate genomic region in one chromosome within a neoplastic cell, but may be caused by other mechanisms, such as mitotic nondisjunction or somatic recombination, leading to uniparental heterodisomy [18].
Recently, owing to the emerging interest in epigenetics and accumulated data in the field of cancer epigenetic research, Knudson’s two-hit hypothesis has been modified (shown schematically in Figure 1). This article provides a review of the current knowledge of epigenetic alterations in ovarian carcinoma, an overview of the different technologies available for the study of epigenetics used in ovarian cancer and analysis of the translation of this knowledge into the different clinical aspects (diagnosis, treatment and prognosis) of ovarian carcinoma.
Figure 1. Two-hit hypothesis.
According to the revised Knudson’s two-hit hypothesis, in the first hit one allele can be inactivated, either by localized mutation or promoter hypermethylation, leading to silencing of one allele of the affected gene. However, for full inactivation (complete silencing) of a target gene, a second hit is essential and can occur by LOH, mutation or promoter hypermethylation. Accumulation of multiple-gene alterations by genetic or epigenetic alterations and changes in the microenvironment leads to tumor development.
LOH: Loss of heterozygosity.
Adapted with permission from [2].
Epigenetic modifications in cancer
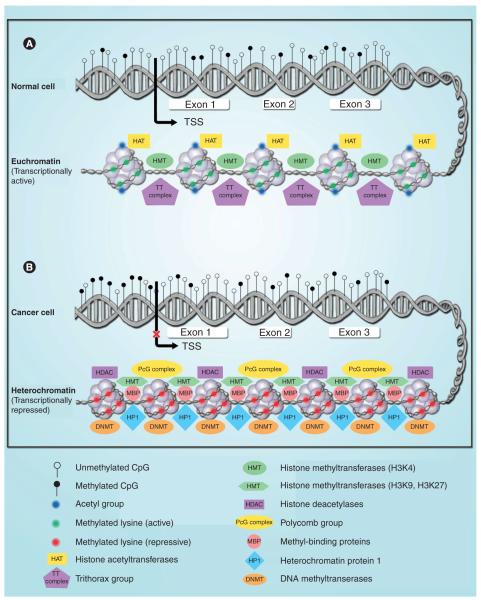
DNA methylation adds a methyl group to the 5-carbon position of a cytosine located 5′of a guanine (a CpG dinucleotide). It is a heritable epigenetic change that can modify gene expression without modifying the DNA sequence. These CpG dinucleotides (that represent approximately 100% of the human genome, and from which 70–80% are methylated) have been selectively depleted by converting the methylated cytosines to thymines via a deamination process [19]. There are areas in the genome where these CpG dinucleotides cluster (called CpG islands) and are mostly located upstream of the transcriptional start site and first exon of more than half of human genes [20]. The CpG islands found in gene promoter regions are usually unmethylated in normal cells to maintain euchromatic structure – which is the transcriptionally active form of chromatin – thus allowing gene expression. However, during cancer development, many of these genes become hypermethylated, changing the euchromatin structure to a more compact heterochromatin and repressing gene expression (Figure 2) [5–7,9,15,16]. Some of the methylated genes identified in human cancers are classic TSGs in which one mutationally inactivated allele is inherited [17]. This hypermethylation also plays an integral role in genomic imprinting, where one of the two parental alleles of a gene is silenced in order to establish mono-allelic expression, similar to X-chromosome inactivation in females [21,22].
Figure 2. Epigenetic changes (DNA methylation, histone modifications and nucleosome remodeling) in normal and cancerous cells.
Gene promoter regions are usually unmethylated in normal cells to maintain an euchromatic structure, which is the transcriptionally active form of chromatin, thus allowing gene expression (A). However, during cancer development, many of these genes become hypermethylated, changing the euchromatin structure to a more compact heterochromatin and repressing gene expression (B).
TSS: Transcription start site.
The addition of the methyl groups into the cytosines from S-adenosyl l-methionine (a co-substrate for the methylation reaction) is mediated by DNA methyltransferases (DNMTs) [23]. So far, three families of DNMTs have been described: DNMT1, -2 and -3. DNMT1 has been found to maintain the established patterns of methylation in hemimethylated genes by copying these patterns from the parent strand to the daughter [24]. The DNMT3 family (DNMT3a, -3b and -3L) lead to de novo methylation [25,26] in different cellular processes. DNMT3L lacks the ability to bind to S-adenosyl l-methionine and is responsible for increasing the binding of DNMT3a to S-adenosyl l-methionine [26,27]. DNMT2 is reported to perform a weak DNMT activity [28] and has been demonstrated to methylate both DNA and tRNA at the cytosine-5 position, where both DNA methylation and RNA methylation play important roles in human health and disease [29].
Recent studies have demonstrated that Dicer-mediated miRNA biogenesis modulates DNA methylation by regulating the expression of DNMT3 genes (Dnmt3a, -3b and -3L) [30,31]. Dicer belongs to the RNase III family of enzymes implicated in the biosynthesis of siRNAs and miRNAs [32]. In Dicer−/− cells, the miRNAs of the miR-290 cluster are depleted, and expression levels of their target retinoblastoma-like protein 2 are increased, leading to down-regulation of DNMT3 gene expression through retinoblastoma-like protein-2-mediated transcriptional repression and, in turn, causing global hypomethylation [30,31].
In normal cells, repetitive genomic sequences (e.g., centromeric satellite α-DNA and juxtacentromeric satellite DNA) are heavily methylated [6,7]. The maintenance of methylation in this repetitive DNA could be important for the protection of chromosomal integrity by preventing chromosomal rearrangements, translocations and gene disruption through the reactivation of transposable elements [7,11,33]. Besides hypermethylation of gene-associated CpG islands, hypomethylation of repetitive genomic DNA has also been identified as a specific feature in human cancers [7,34]. As mentioned earlier, several studies indicate that the global DNA hypomethylation identified in cancer cells might contribute to structural changes in chromosomes, loss of imprinting, microsatellite and chromosome instability through aberrant DNA recombination, aberrant activation of proto-oncogene expression and increased mutagenesis [7,11,35,36].
The nucleosome is composed of four core histone proteins (H2A, -2B, -3 and -4) and a stretch of DNA that wraps around, forming the basic building unit of chromatin [37]. Methylation, acetylation, phosphorylation, ubiquitylation and sumoylation are the modifications that can occur to core histone proteins, and are thus implicated in how the chromatin will be structured and whether or not gene expression will occur [38–40]. For example, acetylation at lysine 9 (K9) of H3 and K5, -8, -12 and -16 of H4, as well as methylation at K4 of H3, are involved with a euchromatic or transcriptionally active state of chromatin [6,38]. Di- and trimethylation of H3K4 is catalyzed by the trithorax group of histone methyltransferases, which are known to be involved in the transcriptional activation of developmental regulatory genes [14,41]. By contrast, when mono-, di- and tri-methylation at H3K9, H3K27 and H4K20 occurs, a closed chromatin structure (heterochromatin) is initiated and maintained, repressing gene expression [5,6,14,16]. The polycomb group of factors are known to modulate the methylation of H3K27, having an opposite effect than the trithorax group, leading to gene silencing [14,41,42]. Methylation of H3K9 allows for the binding of heterochromatin protein-1, which has been demonstrated to recruit DNMT to bind to the silenced genes [43,44]. Families of methyl-binding proteins have been associated directly or indirectly with DNMTs, histone deacetylases (HDACs) and histone methyltransferases to alter chromatin structure and suppress gene transcription (Figure 2) [45–47].
miRNAs are small (~19–25 nucleotides in length) noncoding RNAs that regulate gene expression. It has been estimated that approximately 30% of human genes are regulated by miRNAs [48]. miRNAs have been found to have regulatory roles in different biological processes, such as cell cycle, differentiation, development and metabolism [49–52], thereby playing an important role in cancer [53,54]. There are several mechanisms by which miRNAs can be deregulated in cancer. One mechanism is through genetic modifications, such as deletions, mutations and/or amplifications [55–58]. Another mechanism involved is through epigenetic modifications that regulate miRNA expression in cancer [59–61].
DNA methylation & histone modifications in ovarian carcinoma
Many different genes have been identified to be hypermethylated and silenced in ovarian carcinoma. Some of these genes are located in regions with known LOH in ovarian carcinoma and/or are epigenetically regulated in other types of malignancies, for example OPCML [62–64], DLEC1 [65], RASSF1A [63,66–69], ARLTS1 [70], ARHI [71,72] and TCEAL7 [73]. Since aberrant DNA methylation silences transcription, novel TSGs can be identified by analyzing CpG island hypermethylation. The breast cancer susceptibility gene 1 (BRCA1) has been one of the most comprehensively analyzed owing to its tumor suppressor function and its known role in inherited forms of ovarian cancer, where hypermethylation only occurs in ovarian and breast cancers [74,75]. This hypermethylation of its promoter is associated with the loss of its expression [76–78] and is predominantly detected in cancers that exhibit LOH at the BRCA1 locus [75,77]. Furthermore, the methylation-mediated silencing of BRCA1 and other repair genes, such as MGMT, could result in further inactivation of TSGs or activation of oncogenes, which further promote and drive ovarian tumorigenesis [79]. By contrast, BRCA2 promoter hypermethylation is rarely found in ovarian cancers [80,81]. Other genes found to be hypermethylated and downregulated in ovarian carcinoma demonstrate different properties: p16 [82], SPARC [83], ANGPTL2 [84] and CTGF [85] have tumor suppressor activity, LOT1 [86] and PAR-4 [87] have pro-apoptotic function, ICAM-1 [88] and CDH1 [89] participate in cell adhesion, and PEG31 [90] plays a role in imprinting. HOXA10 [91], HOXA11 [91], PALB2 [92] and TUBB3 [93] are other examples of hypermethylated genes, the latter having a contribution to taxane resistance.
As mentioned previously, global DNA hypomethylation is another feature of cancer and in ovarian epithelial neoplasms this increases with malignancy [94]. To date, hypomethylation has been demonstrated to lead to the abnormal expression of a few genes, including maspin (SERPINB5) [95], SNCG [96,97] and CLDN4 [98,99] in ovarian carcinomas. In addition, hypomethylation associated with the L1 and human endogenous retrovirus-W retrotransposons, which are repetitive sequences that are widely distributed throughout the genome, is consistent with higher expression levels that occur in malignant compared with non-malignant ovarian tissue [100]. It is hypothesized that an increase in hypomethylation promotes recombination among homologous elements, leading to chromosomal aberrations, which are associated with cancer [101,102]. Other overexpressed genes associated with promoter hypomethylation are BORIS, a cancer testis antigen family candidate oncogene [103], and IGF2, an imprinted gene implicated in other malignancies [104].
In addition, DNMT1 and −3b transcript levels have been reported to be increased in some ovarian cancer cell lines [105], as well as in primary and recurrent epithelial ovarian carcinoma [106], which could contribute to methylation-induced silencing of key TSGs, and have some correlation with clinical pathology and prognosis of epithelial ovarian carcinoma. However, in one study, DNMT3A1 and DNMT2 RNAs were significantly lower in carcinomas compared with low malignant potential (LMP) tumors, but DNMT3B1/-B2 RNA had significantly higher levels in carcinomas than in the LMP tumors [107].
Interestingly, gene methylation patterns are often associated with molecular, clinical and pathological features of ovarian carcinomas. For example, aberrant methylation of the promoters of SFN (an inhibitor of cell cycle progression), TMS1 and WT1 are more frequent events in clear-cell ovarian tumors than in other histological types [63,108–110]. Furthermore, Makarla et al. reported that RASSF1A, APC, GSTP1 and MGMT show aberrant methylation exclusively in invasive ovarian carcinomas [111] when compared with LMP tumors.
In addition, histone modifications contribute to ovarian cancer progression via the downregulation of different genes. GATA4 and −6 gene silencing was found to correlate with hypoacetylation of histones H3 and H4, and loss of histone H3 K4 tri-methylation at their promoters [112]. The cell cycle regulatory proteins cyclinB1 [113] and p21cip1/waf1 [114], and ADAM19 [115] were also found to be regulated by histone modifications.
These advances in the knowledge of the ovarian methylome strongly indicate that DNA hypermethylation plays a crucial role in initiation, promotion and maintenance of ovarian carcinogenesis, which may contribute and synergistically interact with other genetic alterations to induce the development and progression of ovarian cancer.
Current technological approaches to uncover the epigenome of ovarian cancer
A wide array of techniques has been used to detect and understand gene-specific and genome-wide epigenetic modifications in cancer. This article presents an overview of recent epigenomic technologies that have been used to discern the ovarian cancer methylome. Table 1 summarizes the technologies used to explore the ovarian cancer methylome.
Table 1.
Most commonly used technologies for DNA methylation and histone modifications assessment in ovarian cancer
| Technology | Description | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|
| Gene-specific profiling | ||||
| Bisulfite sequencing | Involves PCR amplification of bisulfite-modified DNA and subsequently sequenced directly or after cloning |
Reliable Evaluates several CpG sites at a time |
Laborious (cloning required) | [122] |
| MSP | Involves PCR amplification with two different pairs of primers complementary to the methylated or unmethylated version of the CpG site under evaluation PCR products are analyzed on an agarose gel |
Sensitive Easy to perform |
Not quantitative Only evaluates methylatable sites within the primer regions |
[116] |
| QMSP/MethyLight | Methylation assay that uses fluorescence-based real-time PCR (TaqMan®) Bisulfite-modified DNA is required in order to quantitatively detect methylated alleles |
Ability to detect minimal amounts of DNA methylation Specific and sensitive Quantitative |
The whole methylation pattern of a sample is not reflected, meaning that sequencing is usually required |
[124] |
| QM-MSP | Modified version of fluorogenic probe-based quantitative MSP assay Includes the multiplex PCR step that allows amplification of multiple target alleles PCR products are subjected to quantitative MSP assay for multiple-gene detection |
High level of sensitivity and specificity Low amounts of DNA required |
Does not take into account the existence of variable fractions of partially methylated DNA in the tissue samples |
[125] |
| Combined bisulfite restriction analysis |
Bisulfite-modified DNA is digested with restriction enzymes that distinguish methylated from unmethylated sequences |
Sensitive | Limited to specific restriction targets | [123] |
| Pyrosequencing | DNA-sequencing method in which the methylation levels of CpG sites are detected quantitatively by monitoring the incorporation of nucleotides in real-time and enzymatic conversion of released pyrophosphate into a bioluminometric signal |
Low amount of bisulfite-converted DNA required |
High cost Limited sequence range per run |
[127] |
| Genome-wide profiling: DNA methylation | ||||
| Restriction landmark genomic scanning |
DNA is radioactively labeled at unmethylated sites within methylation-sensitive restriction enzyme targets The digestion products are digested with a second restriction endonuclease that is specific for high-frequency targets and the fragments are separated in the second dimension, yielding a number of scattered hot spots of DNA methylation |
Simultaneous quantification of gene copy number and methylation status |
Reliance on specific digestion sites that are not present in all CpG islands and not all of the resulting fragments can be resolved in the electrophoresis steps |
[129] |
| Differential methylation hybridization |
DNA from the tissues of interest is digested with methylation-sensitive enzymes and the digestion products are used as templates for PCR after ligation to linkers The resulting oligonucleotides are used as probes to screen for hypermethylated sequences within a CpG-island library to identify sequences that are hypermethylated in cancer cells but not in normal control cells |
Allows the simultaneous determination of the methylation levels of a large number of CpG-island loci |
Requires a significant amount of DNA Only two samples can be compared |
[130] |
| Microarray-based gene expression profiling |
Involves comparing mRNA levels from cancer cell lines before and after treatment with a demethylating drug |
Cloning step not required | False positives | [131] |
| lllumina Infinium HumanMethylation27 BeadChip |
Detects methylation at CpG islands based on highly multiplexed genotyping of bisulfite-converted genomic DNA It uses two different bead types to detect CpG methylation: an unmethylated bead and a methylated bead Single-base extension of the probes incorporates a labeled ddNTP, which is subsequently stained with a fluorescent reagent The level of methylation can be determined by calculating the ratio of the fluorescent signals from the methylated versus unmethylated sites |
Requires as little as 500 ng of genomic DNA Analysis of 12 samples per chip Integration of data with other platforms (gene expression and miRNA) Provides genome-wide coverage Quantitative methylation measurement |
Not full coverage of the genes listed in the NCBI database Designed to analyze high-quality DNA (e.g., fresh-frozen tissue) |
[230] |
| MeDIP | Antibodies are directed against methyl-CpG-binding proteins for immunoprecipitation of methylated DNA fragments |
Ability to utilize single-stranded and highly fragmented DNA, making it potentially compatible with archival specimens (paraffin- embedded samples) Can be analyzed in a locus-specific or genome-wide manner |
Inability to pinpoint methylation changes to nucleotide resolution Relies on quality of the antibodies |
[138,231,232] |
| MIRA | Fragmented genomic DNA is incubated with the methyl-binding domain protein complex Bound methylated DNA is eluted from a glutathione affinity matrix and can be hybridized on genomic microarrays |
High specificity for methylated DNA, reducing false positives |
Methyl-binding domains preferentially bind densely methylated CpGs, requiring at least two mCpG sites for efficient pull down, thus methylated DNA obtained using MIRA is sequence dependent owing to enrichment dependence on mCpG density |
[232,233] |
| Genome-wide profiling: histone modifications | ||||
| Chromatin immunoprecipitation |
Isolation, using specific antibodies, of chromatin fragments that are bound by a particular nuclear factor or associated with a particular histone-modification signature The immunoprecipated DNA can subsequently be analyzed with specific PCR primers |
Provides information on histone modifications with particular DNA sequences |
Lack of representation of the whole human genome Relies on the quality of the antibodies |
[234] |
ddNTP: Dideoxyribonucleotide triphosphate; mCpG: Methylated CpG; MeDIP: Methylated DNA Immunoprecipitation; MIRA: Methylated CpG-island recovery assay; MSP: Methylation-specific PCR; QM-MSP: Quantitative multiplex methylation-specific PCR; QMSP: Quantitative fluorogenlc methylation-specific PCR.
Technologies for DNA methylation
Epigenetic alterations have several advantages as a means to detect and classify cancer. First, methylation analysis utilizes DNA, a more chemically stable molecule than RNA and protein. Second, aberrant DNA methylation is a binary signal, where the presence of methylation indicates the presence of malignant cells [4]. This can be detected at a low concentration in a background of excess normal DNA molecules by sensitive assays such as methylation-specific PCR (MSP) [116] and quantitative fluorogenic methylation-specific PCR [117], which allow the detection of a single methylated allele in 10,000 unmethylated alleles [118]. The third advantage of using DNA methylation to detect cancer is that assay design can focus on a single amplifiable region (e.g., CpG island) rather than scanning an entire gene for mutations. In addition, methylation biomarkers are detectable in patient serum/plasma and other bodily fluids draining or surrounding a tumor site [119]. On the other hand, studies of DNA methylation for ovarian cancer could present certain bias, which is related to sampling, technique, design and data analyses. To overcome various biases, a systematic approach is needed to identify a panel of true-positive biomarkers from the large number of biomarkers reported every year. Methylation of dozens of genes in various types of samples has been correlated with ovarian cancer, but, so far, most studies have been conducted in a single center with a limited numbers of samples. To determine the usefulness of DNA methylation in ovarian cancer detection, it is essential to conduct a specific project using a well-defined end point, the same set of blinded specimens, appropriate experimental design and data analyses, and standard technology to evaluate methylation biomarkers to generate a reliable conclusion about the usefulness of these markers for a specific clinical use in ovarian cancer.
Before the development of bisulfite treatment of DNA, the methods used for detection of methylation were performed using high-performance liquid chromatography and high-performance capillary electrophoresis [120]. Nevertheless, the study of DNA methylation was initially almost entirely based on digestion with restriction-enzyme assays that can differentiate between methylated and unmethylated recognition sites in genes of interest [121]. This approach demonstrated several disadvantages that limited its use: from incomplete restriction-enzyme cutting to limitation of the regions that can be studied. The bisulfite-conversion technique, which reproducibly changes unmethylated cytosines to uracil but leaves methylated cytosines unchanged [122] was the mainstay for the creation of several sensitive DNA methylation detection techniques (Table 1), including bisulfite sequencing, MSP and combined bisulfite restriction analysis [116,122,123]. MSP, although not a quantitative method, is very sensitive and has been the most widely used method for DNA methylation analysis on clinical samples, also partly owing to its simplicity [116]. Owing to its subjectivity, several real-time MSP methods, such as MethyLight™ [124], quantitative multiplex MSP [125,126] or pyrosequencing [127], have been developed, improving the detection of small amounts of DNA methylation in a quantitative fashion. MethyLight is a highly sensitive assay, capable of detecting methylated alleles in the presence of a 10,000-fold excess of unmethylated alleles. The assay is also highly quantitative and can very accurately determine the relative prevalence of a particular pattern of DNA methylation [124]. Pyrosequencing is a sequencing-by-synthesis method that quantitatively monitors the real-time incorporation of nucleotides through the enzymatic conversion of released pyrophosphate into a proportional light signal [127]. MSP and quantitative fluorogenic MSP technology can only detect CpG islands that are within the primer sequences and cannot detect CpG sites outside the methylation-specific primers. Pyrosequencing, in an unbiased manner, is a newly emerging method, although its intrinsic short-read sequencing (normally only up to 30 bp at a time) presents a disadvantage in comparison with DNA sequencing [127,128]. Several other methylation assays have been developed for different purposes and each has certain advantages, limitations and suitability.
Besides the gene-specific profiling methods described previously, several genome-wide techniques have been useful for the study of global DNA methylation patterns in normal and cancer cells (Table 1). Restriction-landmark genomic scanning is one of first genome-wide methylation analyses described that can evaluate the methylation status of thousands of CG-rich sequences and simultaneously obtain information on the gene copy number [129]. However, this is a laborious technique and a comparatively large amount of DNA is needed for the assay. The application of DNA microarray technology made it possible to discover new techniques that have had an important impact on cancer epigenetics. Two examples of microarray assays are differential methylation hybridization and gene expression profiling. Differential methylation hybridization was developed by Huang et al. and allows for the detection of differential methylated CpG islands between two different samples. Differential methylation hybridization has been widely used in the identification of aberrantly methylated gene promoters that are differentially expressed in various cancers [130]. Finally, gene expression profiling assesses genome-wide DNA methylation patterns, by comparing expression levels from cancer cells before and after treatment with a demethylating drug, HDAC inhibitor (HDACI) or both [131–133]. The identified candidate genes are further verified by quantitative real-time PCR and promoter methylation analyses.
Another important technique for DNA methylation analysis, useful because arbitrary primed PCR is carried out using DNA templates that have been enriched for methyl sequences, resulting in preferential amplification of CpG islands and gene-rich regions, is amplification of intermethylated sites [134,135]. However, validation by bisulfite genomic sequencing is required. In addition, other important advances made in profiling the cancer epigenome have been achieved by techniques such as HpaII tiny fragment enrichment by ligation-mediated PCR assay, which uses a modified approach to globally analyze DNA methylation patterns [136]and other methods based on chromatin immunoprecipitation (ChIP) such as methylated DNA immunoprecipitation [137–139]. Recently, Illumina released the new generation of BeadArrays for assessment of DNA methylation where 27,578 CpG loci, covering more than 14,500 genes, can be analyzed at single-nucleotide resolution. Very recently, data derived from this new generation of BeadArrays have been published [140]. The most commonly used technologies for DNA methylation are summarized in Table 1.
Technologies for histone modification analysis
As bisulfite sequencing is the gold standard for DNA methylation studies, the gold standard for accurately assessing global levels of histone modifications is mass spectrometry; however, it requires a high degree of technical expertise and is difficult to apply to the entire genome [6]. Nowadays, one of the most powerful techniques to identify and characterize the interactions of specific genomic DNA sequences associated with a target protein, such as transcription factors, is ChIP [141]. An antibody specific to the target protein is used to immunopreprecipitate the protein–DNA complexes. After the cross-link between the two is reversed, the DNA sequences are then uncovered by amplification and sequencing. Furthermore, recent techniques that combine ChIP with serial analysis of gene expression technology and high-throughput sequencing techniques, have been developed for profiling histone modifications [142–144].
Methylation profiles as ovarian cancer biomarkers
The best approach when dealing with ovarian cancer, as with other cancers, is early detection. Methylation profiling could be an important tool to evaluate the applicability of genes as potential biomarkers for cancer diagnosis, prognosis and response to therapy. For a methylation-based diagnostic assay to be reliable (i.e., sensitive and specific), it is imperative to use those potential biomarkers that are found to be hypermethylated in cancer cells/tissues but unmethylated in normal cells/tissues (Table 2). The best studied serum biomarker for ovarian cancer is CA-125, which is elevated in women with advanced disease in 80% of cases, but only in 50–60% of patients with early-stage disease [145]. The problem with CA-125 is its lack of specificity, especially in premenopausal women, where other conditions can elevate this marker (e.g., endometriosis and adenomyosis) [146]. As a result, cancer-specific hypermethylated genes are being considered as potential and promising biomarkers for early detection of ovarian cancer. In one study, tumor-specific hypermethylation of at least one of a panel of six TSG promoters, including RASSF1A, BRCA1, APC, CDKN2A and DAPK, could be detected in the serum or plasma of ovarian cancer patients with 100% specificity and 82% sensitivity, including 13 out of 17 cases of stage I disease [67]. Methylation was observed in only one peritoneal fluid sample from 15 stage IA or -B patients, but 11 out of 15 paired sera were positive for methylation [67]. Consistent with previous studies, these data indicate that circulating ovarian tumor DNA is more readily accessible in the bloodstream than in the peritoneum [147]. In another study, DAPK methylation could be detected in the peripheral blood of 14 out of 16 patients with DAPK-methylation-positive primary tumors, with the peripheral blood of ten out of ten being negative when the primary tumor was negative for DAPK methylation [148]. These studies demonstrate that it is feasible to detect specific methylation markers in the circulation of patients, thus representing a promising new screening method for the detection of early-stage ovarian cancer. All the methylation biomarker studies performed in bodily fluids for screening and prognosis purposes are small and retrospective, and clinical utility cannot be determined until they are tested in larger prospective studies. No single gene in ovarian cancer has been identified as being methylated in more than a relatively small proportion of cancers. Although new genome-wide approaches may aid in discovering such genes, it is likely that a panel of methylated genes will be necessary to detect ovarian cancer with sufficient specificity and sensitivity. A combination of genes that are commonly methylated in cancer and genes that are methylated specifically in ovarian cancer is the most likely methylation signature capable of distinguishing ovarian cancers from other type of cancers and from benign disease.
Table 2.
Hypermethylated genes in ovarian cancer†
| Gene | Tumor samples (carcinomas) |
Histology | Control (normal tissue) |
Method | Ref. |
|---|---|---|---|---|---|
| APC | 42/89 (47.2%) | E, S, M, CC | 4/16 (25%) | MSP | [235] |
| 5/23 (22%) | S, M, CC, E, UN | 0/16 (0%) | MSP | [111] | |
| 9/49 (18%) | S, E, M, CC | 1/39 (3%) | MSP | [68] | |
| BRCA1 | 12/50 (24%) | S, M, E, CC | 0/10 (0%) | MSP | [67] |
| 12/93 (13%) | S, E, M, CC, MIX | 1/18(5.5%) ADJ NLS | MSP | [236] | |
| 5/49 (10%) | S, E, M, CC | 0/39 (0%) | MSP | [68] | |
| CASP8 | 3/93 (3%) | S, E, M, CC, MIX | 0/18 (0%) ADJ NLS | MSP | [236] |
| CDH1 (E-cadherin) | 6/23 (26%) | S, M, CC, E, UN | 1/16 (6%) | MSP | [111] |
| 14/49 (29%) | S, E, M, CC | 2/39 (5%) | MSP | [68] | |
| CDH13 (H-cadherin) | 5/23 (22%) | S, M, CC, E, UN | 2/16 (13%) | MSP | [111] |
| 9/49 (18%) | S, E, M, CC | 3/39 (8%) | MSP | [68] | |
| CDKN2A | 17/89 (19.1%) | E, S, M, CC | 4/16 (25%) | MSP | [235] |
| 6/23 (26%) | S | 0/7 (0%) | Multiplex PCR | [237] | |
| 13/52 (25%) | S | 15/40 (38%) | MSP | [238] | |
| 7/23 (30%) | S, M, CC, E, UN | 0/16 (0%) | MSP | [111] | |
| 5/49 (10%) | S, E, M, CC | 3/39 (8%) | MSP | [68] | |
| CDKN2B | 17/89 (19.1%) | E, S, M, CC | 1/16 (6.3%) | MSP | [235] |
| 1/93 (1%) | S, E, M, CC, MIX | 0/18 (0%) ADJ NLS | MSP | [236] | |
| 16/52 (31%) | S | 2/40 (5%) | MSP | [238] | |
| DAPK | 20/30 (67%) | S, CC, E, M, CS, PDA | 0/1 (0%) | MSP | [148] |
| DCR1 | 10/23 (43%) | NS | 0/9 (0%) | MSP | [239] |
| GPR150 | 4/15 (26.6%) | S, E, M, CC, SCC | 0/7 (0%) END cyst | MSP | [240] |
| GSTP1 | 2/23 (9%) | S, M, CC, E, UN | 0/16 (0%) | MSP | [111] |
| 1/49 (2%) | S, E, M, CC | 0/39 (0%) | MSP | [68] | |
| HIC1 | 46/89 (51.7%) | E, S, M, CC | 2/16 (12.5%) | MSP | [235] |
| 15/93 (16%) | S, E, M, CC, MIX | 2/18 (11.1%) | MSP | [236] | |
| 17/49 (35%) | S, E, M, CC | 3/39 (8%) | MSP | [68] | |
| HOXD11 | 1/15 (6.6%) | S, E, M, CC, SCC | 0/7 (0%) END cyst | MSP | [240] |
| Htr (TERC) | 23/93 (24%) | S, E, M, CC, MIX | 0/18 (0%) ADJ NLS | MSP | [236] |
| ING1 | 21/88 (24%) | S, M, E, CC, PDA | 0/6 (0%) | MSP | [241] |
| ITGA8 | 2/15 (13.3%) | S, E, M, CC, SCC | 0/7 (0%) END cyst | MSP | [240] |
| MGMT | 2/23 (9%) | S, M, CC, E, UN | 0/16 (16%) | MSP | [111] |
| MINT25 | 15/93 (16%) | S, E, M, CC, MIX | 0/18 (0%) ADJ NLS | MSP | [236] |
| MINT31 | 45/89 (50.6%) | E, S, M, CC | 0/16 (0%) | MSP | [235] |
| 50/93 (54%) | S, E, M, CC, MIX | 5/18 (27.7%) ADJ NLS | MSP | [236] | |
| MLH1 | 9/93 (10%) | S, E, M, CC, MIX | 0/18 (0%) ADJ NLS | MSP | [236] |
| OPCML | 32/72 (44.4%) | NS | 0/20 (0%) | Restriction enzyme cut analysis | [64] |
| 20/43 (46.5%) | NS | 0/4 (0%) | MSP | [242] | |
| p73 | 9/93 (10%) | S, E, M, CC, MIX | 0/18 (0%) ADJ NLS | MSP | [236] |
| PRTFDC1 | 1/15 (6.6%) | S, E, M, CC, SCC | 0/7 (0%) END cyst | MSP | [240] |
| PTEN | 15/89 (16.9%) | E, S, M, CC | 0/16 (0%) | MSP | [235] |
| RARB | 15/89 (16.9%) | E, S, M, CC | 1/16 (6.3%) | MSP | [235] |
| 3/23 (13%) | S, M, CC, E, UN | 0/16 (0%) | MSP | [111] | |
| 1/49 (2%) | S, E, M, CC | 0/39 (0%) | MSP | [68] | |
| RASSF1 | 8/16 (50%) | NS | 0/10 (0%) | Restriction digestion by Taql | [69] |
| 25/50 (50%) | S, M, E, CC | 0/10 (0%) | MSP | [67] | |
| 39/75 (52%) | S, M, CC, TC, MIX | 0/5 (0%) | MSP | [243] | |
| 7/23 (30%) | S, M, CC, E, UN | 2/16 (13%) | MSP | [111] | |
| 20/49 (41 %) | S, E, M, CC | 6/39 (15%) | MSP | [68] | |
| 42/80 (52.5%) | S, M, E | 0/80 (0%) | MSP | [244] | |
| RIZ1 | 20/89 (22.5%) | E, S, M, CC | 3/16 (18.8%) | MSP | [235] |
| TERT | 37/124 (29.8%) | S, M, E, CC | 6/20 (30%) | QMSP | [245] |
| TMS1 (ASC) | 14/89 (15.7%) | E, S, M, CC | 2/16 (12.5%) | MSP | [235] |
| 15/80 (19%) | S, E, M, CC, UN | 0/4 (0%) | COBRA | [110] | |
| UCHL1 | 1/17 (6%) | NS | 0/5 (0%) END cyst | MSP | [246] |
Data gathered only from studies that compared methylation status in tumors versus normal tissue.
ADJ NLS: Adjacent normals; CC: Clear cell; COBRA: Combined bisulfite restriction analysis; CS: Carcinosarcoma; E: Endometroid; END: Endometrial; M: Mucinous; MIX: Mixed; MSP: Methylation-specific PCR; NS: Not specified; PDA: Poorly differentiated adenocarcinoma; QMSP: Quantitative methylation-specific PCR; S: Serous; SCC: Squamous cell carcinoma; TC: Transitional cell carcinoma; UN: Undifferentiated. Data from [247].
Besides cancer detection, DNA methylation assays might be used for risk evaluation and prognosis of ovarian cancer. Several epigenetically regulated genes have been assessed for their prognostic prediction potential in ovarian cancer. For example, IGFBP-3 hypermethylation was associated with disease progression and death in ovarian cancer, particularly in patients with early-stage disease; methylation was associated with a threefold higher risk of disease progression and a fourfold higher risk of death [149]. When IGFBP-3 methylation was combined with methylation in the promoter regions of CDKN2A, BRCA1 or MLH1, the risk of disease progression in patients with at least three methylated genes was increased sevenfold [150]. Conversely, hypermethylation of 18S and 28S ribosomal DNA is associated with prolonged progression-free survival of ovarian cancer patients [151]. Hypomethylation of certain chromosomal regions also appears to have prognostic power; patients who demonstrated little or no hypomethylation of Chr1 Sat2 or Chr1 Satα had a significantly longer relapse-free survival compared with patients with strong hypomethylation of these regions [152]. Recently, DNA methylation of SFRP1, -2, -4 and 5, SOX1, PAX1, and LMX1A was analyzed by MSP in primary tumor samples from 126 patients with ovarian cancer, 75 with a benign tumor, 14 with borderline malignancy and in the serum from 26 patients with ovarian cancer and 20 with a benign tumor [153]. Six of the seven genes analyzed had higher methylation levels in the ovarian cancer cases than in borderline malignancy or benign tumors. The methylation of SFRP1, SFRP2, SOX1 and LMX1A genes correlated with recurrence and overall survival of ovarian cancer patients [153]. Combining the data for SFRP1, SFRP2 and SOX1 genes gave a relative risk for recurrence of 3.19 (p = 0.013) in patients with at least one gene methylation, and combining the data for SFRP1, SOX1 and LMX1A gave a relative risk for cancer-related death of 6.09 (p = 0.01) [153]. Fiegl et al. identified that hypermethylation in HOXA11 (a polycomb group target) is strongly associated with the residual tumor after cytoreductive surgery and is a marker that indicates poor prognosis [91]. HOXA11 DNA methylation was independently associated with poor outcome (relative risk for death 3.4; 95% CI: 1.2–9.9; p = 0.03) [91]. Finally, another epigenetically upregulated gene strongly linked to tumor metastasis, is SNCG, also known as BCSG [97]. None of the studies that correlate hypermethylation with poor prognosis have been confirmed by subsequent studies and they need to be validated in large independent studies. In one study of cervical samples, SOX1 and HOXA11 [154] were reported to discriminate between high-grade squamous intraepithelial lesions and normal cervical controls, suggesting that these genes are also predictors of disease progression and, hence, poor prognosis. Nonetheless, as mentioned previously, it is imperative to further validate and confirm these studies, as well to analyze previously reported prognosis-related genes from other tumors into ovarian carcinoma samples. As an example, we have recently reported that TIMP3 methylation is related to poor prognosis in bladder cancer [155]. Although all these studies demonstrated promising methylated genes as prognostic markers, multicentered, blinded and standardized identical methods need to be developed before its clinical use. Even if the biological basis of a given biomarker is not elucidated, it still can be used in a clinical setting, as long as it is well validated in appropriate cases and controls (e.g., for a prognostic marker, good prognosis and poor prognosis). By contrast, it would be helpful for targeted therapy to understand the biological basis to target that molecule and/or pathways by which the molecule (marker) would be exerting its effect. A summary of ovarian cancer-specific methylated genes with their potential clinical correlation are shown in Table 3.
Table 3.
Epigenetic alterations of genes and their clinical correlation in ovarian cancer
| Gene | Type of epigenetic event | Clinical correlation | Ref. |
|---|---|---|---|
| SFN | Promoter hypermethylation | More frequent in clear-cell histology | [108] |
| TMS1 | Promoter hypermethylation | More frequent in clear-cell histology | [110] |
| WT1 | Promoter hypermethylation | More frequent in clear-cell histology | [109] |
| RASSF1A | Promoter hypermethylation | Paclitaxel-like activity Diagnosis Methylation in bodily fluids (serum/plasma) in early-stage disease |
[67,111,167] |
| APC | Promoter hypermethylation | Diagnosis Methylation in bodily fluids (serum/plasma) in early-stage disease |
[67,111] |
| GSTP1 | Promoter hypermethylation | Diagnosis Improved response to chemotherapy in late-stage disease |
[63,111] |
| MGMT | Promoter hypermethylation | Diagnosis Improved response to chemotherapy in late-stage disease |
[63,111] |
| BRCA1 | Promoter hypermethylation | Methylation in bodily fluids (serum/plasma) in early-stage disease Improved response to chemotherapy in late-stage disease |
[63,67] |
| DAPK | Promoter hypermethylation | Methylation in bodily fluids (serum/plasma) in early-stage disease |
[67,148] |
| CDKN2A | Promoter hypermethylation | Methylation in bodily fluids (serum/plasma) in early-stage disease |
[67] |
| IGFBP-3 | Promoter hypermethylation | Associated with disease progression in early-stage disease |
[149] |
| SFRP1, -2, -4, -5 | Promoter hypermethylation | Diagnosis and recurrence | [153] |
| SOX1 | Promoter hypermethylation | Diagnosis and recurrence | [153] |
| PAX1 | Promoter hypermethylation | Diagnosis | [153] |
| LMX1A | Promoter hypermethylation | Diagnosis and recurrence | [153] |
| HOXA11 | Promoter hypermethylation | Residual disease after surgery and poor prognosis |
[91] |
| SNCG | Hypomethylation | Tumor metastasis | [97] |
| hMLH1 | Hypomethylation | Reversal of drug resistance | [191] |
| HSulf-1 | Hypomethylation | Sensitization to chemotherapy | [168,169] |
| FANCF | Promoter hypermethylation | Associated with increased sensitivity to cisplatin |
[172] |
| MCJ | Hypomethylation | Upregulation induces sensitivity to cisplatin topotecan and paclitaxel |
[174] |
| TUBB3 | Promoter hypermethylation | Contributes to taxane resistance | [93] |
Chemotherapy response in ovarian cancer
Overall, 80% of patients with ovarian cancer respond to first-line chemotherapy following surgical debulking. Despite the apparent efficacy of this treatment, up to 75% of these patients will relapse within a few years. These recurrent patients may still be chemosensitive but, ultimately, the vast majority of cases succumb to chemoresistance [156]. Ovarian cancer drug resistance can be intrinsic (tumors that do not respond to first-line chemotherapy and are conferred by the genotype of pretreatment clones) or acquired (similar to intrinsic resistance, except that it is primarily caused by mutations in progeny tumor cells after the initiation of therapy) [157]. Both intrinsic and acquired mutations can manifest themselves under various temporal conditions, such as pharmacokinetic alterations (resulting in inadequate drug exposures), variations in tumor cell microenvironments (e.g., hypoxia and altered cell–cell interactions) and differential chemosensitivity during various stages of the cell cycle [158]. One phenomenon common to both intrinsic and acquired resistance is altered gene expression in the drug-resistant tumor, compared with the drug-sensitive tumor [157]. In general, ovarian tumors have been demonstrated to upregulate a number of genes, including those involved in cell proliferation, DNA repair, angiogenesis and cell migration, which may play roles in drug resistance and also the downregulation of genes associated with cell adhesion, pro-apoptotic, anti-proliferative and DNA mismatch repair proteins [157]. Epigenetic alterations represent one of the mechanisms for differential expression of genes that correlate with clinical outcome and thus, have an impact on clinical outcome. A summary of epigenetic alterations and correlation with various clinical parameters is given in Table 3.
The net effect of the platinum drugs and taxanes on sensitive cells is cell death, predominantly through the activation of apoptotic pathways. For example, taxanes stabilize tubulin, resulting in defective spindle formation, G2/M arrest and apoptosis, probably by p53-dependent cascades [159]. Similarly, platinum compounds are incorporated into DNA, inducing inter- and intra-strand platinum adducts [160]. The mismatch repair system recognizes such adducts and activates the apoptotic program [156]. Several genes in ovarian cancer, including TSGs and genes involved in apoptotic pathways, are downregulated by epigenetic mechanisms, as mentioned previously. One well-documented example is the gene encoding the DNA mismatch repair enzyme, hMLH1. Methylation-induced silencing of hMLH1 has been demonstrated in a number of tumors, including ovarian tumors [161,162]. Loss of hMLH1 expression is strongly associated with microsatellite instability [163,164], a tumor marker that has been linked to genetic hypermutability [165]. In addition, silencing of hMLH1 has been linked with resistance to platinum drugs [166], as this results in a decrease in the apoptotic response through p53 phosphorylation and subsequent activation of the MAPK pathway [160]. Another TSG found to be methylated and silenced in ovarian cancer is the gene encoding the Ras homolog RASSF1A [66,69]. RASSF1A has been reported to bind to tubulin and stabilize microtubules [167], and this protein might assist chemotherapeutics such as paclitaxel in mediating the prevention of spindle assembly.
Teodoridis et al. demonstrated that methylation of at least one of three genes involved in DNA repair/drug detoxification, BRCA1, GSTP1 and MGMT, is associated with improved response to chemotherapy of patients with late-stage epithelial ovarian tumors [63]. More recently, HSulf-1 expression has been demonstrated to influence response to chemotherapy. Patients with advanced-stage primary epithelial ovarian tumors that express high levels of HSulf-1 demonstrated an increased response rate to chemotherapy compared with patients whose tumors express low or moderate levels of HSulf-1 [168]. HSulf-1 is often downregulated in ovarian cancer by methylation-associated silencing, and this downregulation leads to the attenuation of cisplatin-induced cytoxicity [168,169].
Recent studies suggest that epigenetic inactivation of genes plays an important role in acquiring chemoresistance at disease relapse. For example, matched cell line models of acquired resistance have shown that chemotherapy can select for common patterns of CpG island methylation in vitro [170]. There is an increasing volume of evidence from clinical studies that supports this hypothesis. In the study by Wei et al., patients stratified as having a short progression-free survival with a high degree of CpG island methylation had a worse response to second-line cytotoxic therapies compared with patients with a longer progression-free survival and low CpG island methylation, suggesting that patients with high CpG island methylation acquire resistance to chemotherapy more readily [171].
Several genes involved in cell proliferation and survival have been found to be upregulated through epigenetic alterations in ovarian cancer. An example of this is FANCF, which is crucial for the activation of the DNA repair complex containing BRCA1 and −2. Inactivation of FANCF is associated with increased sensitivity to cisplatin in ovarian cancer cells with a defective BRCA2 pathway [172]. By contrast, demethylation and re-expression of FANCF, which is thought to occur early in tumor progression, is associated with acquisition of cisplatin resistance in ovarian cancer cell lines [172]. Methylation-controlled J protein (MCJ), which is required to repress the expression of the drug transporter ABCB1 (P-glycoprotein) [173], was identified as a gene that, when active, sensitized epithelial cells to cisplatin and paclitaxel, the mainstay of chemotherapy for ovarian cancer patients [174]. MCJ has been found to be methylated and silenced in normal cells, including normal ovarian surface epithelium, which is unusual for a CpG island-associated gene [175]. Nevertheless, the majority of late-stage ovarian cancers also exhibit MCJ methylation; however, many of these have undergone a partial demethylation of the MCJ gene promoter, with only 17% of cancers maintaining very high (>90%) methylation, which correlates with a poor response to chemotherapy and decreased survival [175,176]. Hence, MCJ methylation may be a useful marker of response to chemotherapy in ovarian cancer.
In terms of ovarian cancer chemoresistance and miRNAs, miR-199a, -200a and -214 were found to be upregulated in ovarian tumors, and miR-214 was demonstrated to target the tumor suppressor PTEN and to have an association with resistance to platinum therapy [177,178]. The miRNA let-7i, found to have tumor suppressor activity, was significantly downregulated in platinum-resistant ovarian tumors and when its function was restored, it led to chemosensitivity of ovarian cancer cells, thereby becoming a potential biomarker and therapeutic target candidate [179]. Other miRNAs have also been correlated with chemotherapy response [180] demonstrating that miRNAs could represent potential prognostic and diagnostic biomarkers for ovarian cancer. The study of promoter region methylation of miRNAs is an emerging field and DNA-based assays can be develop considering the dysregulation of these molecules in ovarian cancer.
In summary, in order to translate the diverse epigenetic biomarkers identified into clinics, several key points need to be considered:
■ Biomarkers should be related to early detection of disease, prognosis and/or therapeutic response;
■ The biomarker discoveries should be reproducible, consistent and supported by various laboratories;
■ Candidates should be validated through resources that could provide suitable specimens and infrastructure, such as the Early Detection Research Network;
■ There must be longitudinal follow-ups in a screening cohort;
■ The process from biomarker discovery to clinical application should follow all required processes, regulations and standards for the biomarkers to become commercially viable.
Epigenetic therapies in ovarian cancer
With the acknowledgement that epigenetic mechanisms contribute to the formation and progression of tumors, efforts have been made in order to develop novel epigenetic therapies to target cancer cells. One important characteristic of epigenetic alterations is reversibility, unlike genetic mechanisms, which are are irreversible processes. This feature has promoted the development of pharmacologic inhibitors of DNA methylation and histone deacetylation [181–184], which have been proven to demethylate DNA and inhibit histone deacetylation, in order to reverse epigenetic silencing of key genes, leading to re-expression of these genes in cancer cells and reactivation of important cellular tumor suppression pathways.
DNA methyltransferase inhibitors
The majority of these therapeutic agents are cytosine analogs, 5-azacytidine (Vidaza®) and 5-aza-2′-deoxycytidine (decitabine) being the most extensively studied DNMT inhibitors (DNMTIs). These compounds incorporate into DNA in the place of cytosines during DNA replication, covalently attaching to DNMTs [185] and cause the depletion of active DNMT enzymes. They act primarily on DNMT1 and their demethylation activity is replication dependent, requiring several cell divisions to achieve genomic demethylation [186]. The demethylating effect of decitabine is stronger than Vidaza since the former only binds DNA, unlike 5-azacytidine, which incorporates into both DNA and RNA [182]. Both compounds have been approved by the US FDA for the treatment of myelodysplastic syndrome. Zebularine, a recently developed cytosine analog-based DNMTI, has a very stable chemical property suitable for oral administration, is less toxic and has high selectivity for tumor cells than the first DNMTIs [187]. Zebularine forms a covalent complex with DNMTs (e.g., Hha I) [188] in order to deplete them (Dnmt1) or cause partial depletion (Dnmt3a and Dnmt3b) and, importantly, it has been demonstrated to reactivate hypermethylated genes in yeast models and p16INK4a in bladder cancer cells through this mechanism [189,190].
Decitabine has been demonstrated to exert its effect in several cancer cell lines, including in the ovaries and was also shown to restore the expression of several tumor suppressors, such as hMLH1 [191]. Plumb et al. demonstrated that treatment of the ovarian drug-resistant cell line A2780/CP with decitabine could restore hMLH activity and cisplatin sensitivity, in both cultured cells and mouse xenografts [191]. It has also been demonstrated to reverse lysine methylation at K9 of histone H3 [192], which represents another epigenetic silencing mechanism [193,194]. With respect to miRNA gene regulation, a group of six miRNAs clustered on chromosome 19 and seven clustered on chromosome 14, were upregulated by the DNMTI decitabine, demonstrating that miRNAs can be regulated by DNA methylation [195].
Zebularine was able to demethylate and reactivate a silenced p16 gene in vitro and in vivo [189]. However, high levels of the drug were required to achieve efficacy of zebularine and, thus, affect its potential application in a clinical setting. Other cytidine analogs, such as arabinosyl-5-aza-cytosine (fazarabine) and dihydro-5-aza-cytidine have proved disappointing in clinical trials [186].
In addition, non-nucleotide DNMTIs have recently been developed to avoid the inherent toxicity of nucleotide analogs, even though they have the same mechanism of action of binding cytosines and interfering with DNMTs. These agents include procainamide (antiarrhythmic), procaine (anesthetic), hydralazine (antihypertensive) and epigallocathechin-3-gallate derived from green tea, and the novel compound RG108 [196–199]. The therapeutic agents that specifically target one type of DNMT have also been developed, for example MG98, an antisense oligonucleotide that specifically inhibits DNMT1 function [200]. From these non-nucleotide DNMTIs, hydralazine is currently being evaluated in a randomized, double-blind Phase III clinical trial in cisplatin-resistant recurrent ovarian cancer. Table 4 shows the ongoing clinical trials of epigenetic therapies in ovarian cancer.
Table 4.
Ongoing epigenetic therapy clinical trials in ovarian cancer
| Clinical trial | Location | Drug | Phase |
|---|---|---|---|
| Neoadjuvant azacitidine with carboplatin and paclitaxel for suboptimal newly diagnosed ovarian cancer |
Loyola Univeristy Medical Center, Cardinal Bernardin Cancer Center, Maywood, IL, USA |
Azacitidine | I |
| Trial of sequential azacitidine and valproic acid plus carboplatin in the treatment of patients with platinum-resistant epithelial ovarian cancer |
MD Anderson Cancer Center, Houston, TX, USA | Azacitidine Valproic acid Carboplatin |
I II |
| Trial of decitabine as a sensitizer to carboplatin in platinum-resistant recurrent ovarian cancer |
Indiana University Cancer Center, Indianapolis, IN, USA |
Decitabine | I II |
| Trial of NY-ESO-1 protein immunization in combination with 5-aza-2′-deoxycytidine (decitabine) in patients receiving liposomal doxorubicin for recurrent epithelial ovarian or primary peritoneal carcinoma |
Roswell Park Cancer Institute, Buffalo, NY, USA | Decitabine Doxorubicin NY-ESO-1 peptide vaccine |
I |
| Randomized, double-blind trial of chemotherapy plus the transcriptional therapy hydralazine and magnesium valproate versus chemotherapy plus placebo in cisplatin-resistant recurrent ovarian cancer |
Instituto Nacional de Cancerologia, Mexico City, Mexico |
Hydralazine Magnesium Valproate |
III |
| Noncomparative study of paclitaxel plus carboplatin in combination with vorinostat in patients with advanced, recurrent epithelial ovarian cancer |
Department of Oncology, Odense University Hospital, Odense, Denmark |
Vorinostat Paclitaxel Carboplatin |
II |
| Study of combination vorinostat, carboplatin and gemcitabine plus vorinostat maintenance in women with recurrent, platinum-sensitive epithelial ovarian, fallopian tube or peritoneal cancer |
Dana-Farber Cancer Institute and Massachusetts General Hospital, Boston, MA, USA |
Vorinostat Carboplatin Gemcitabine |
IB II |
| Open-label, nonrandomized pilot study of weekly paclitaxel, and monthly carboplatin and oral vorinostat for patients newly diagnosed with stage III/IV epithelial ovarian, fallopian tube or peritoneal cancer |
Gynecologic Oncology Associates, Newport Beach, CA, USA |
Vorinostat | I II |
| Safety, pharmacodynamic and pharmacokinetic study of intravenously administered PXD101 plus carboplatin, paclitaxel or both in patients with advanced solid tumors |
Contact: Nis Nissen, TopoTarget A/S, Copenhagen, Denmark |
Belinostat Paclitaxel Carboplatin |
I II |
| Study of PXD101 in platinum-resistant epithelial ovarian tumors and micropapillary/borderline ovarian tumors |
Study Chair: Amit M Oza, Princess Margaret Hospital, Toronto, ON, Canada |
Belinostat Carboplatin |
II |
| Evaluation of belinostat and carboplatin in the treatment of recurrent or persistent platinum-resistant ovarian, fallopian tube or primary peritoneal cancer |
Study Chair: Don S Dizon, Women and Infants Hospital of Rhode Island, RI, USA |
Belinostat Carboplatin |
II |
| Open-label, dose-escalation trial of oral PXD101 in patients with advanced solid tumors |
Contact: Nis Nissen, TopoTarget A/S, Copenhagen, Denmark |
Belinostat | I |
Data taken from [301].
HDAC inhibitors
As mentioned previously, DNA-associated histone proteins are subject to modifications that modulate chromatin organization into a permissive or repressive form. Histone deacetylation is one of the modifications that is correlated with a repressive chromatin and transcriptional silencing. For this reason, HDACIs were developed to relieve gene repression and exert anticancer activity [184]. These inhibitors can upregulate specific genes, such as p21cip1/waf1, a p53-induced cyclin-dependent kinase inhibitor that causes G1 cell cycle arrest [201], and apoptotic genes, such as CD95, TRAIL, DR4, DR5, Bax, Bak, Bim, Bmf and Apaf1, involved in the extrinsic death-receptor and intrinsic mitochondrial death pathways [181]. In addition, treatment with HDACIs has been found to downregulate genes required for cell cycle progression (cyclin D1 and cyclin A), antiapoptosis (Bcl-2) and angiogenesis (VEGF and HIF-1α) [183] in cancer cells and xenograft models [181–184,202], with evidence that these antitumor effects involve both transcriptional and nontranscriptional mechanisms [181,183]. The induced hyperacetylation by HDACIs of histones and nonhistone transcription factors, such as p53, p73, E2F1, STAT1, STAT3 and NF-κB, activates or represses their target genes [181,203–206]. More importantly, cancer cells are more sensitive to growth inhibition by HDACIs compared with nontransformed cells, suggesting that HDACIs have tumor-specific properties [207]. As mentioned previously, nontranscriptional mechanisms serve as mediators of the antitumor effects of HDACIs [181,183,184]. For example, HDACIs could induce defective mitosis in tumor cells and, in turn, trigger cell death through changes in chromatin conformation caused by hyperacetylation of centromeric histones [208]. Furthermore, HDACIs deplete protein levels of many oncoproteins, whose stability is regulated by heat-shock proteins [181,183], and enhance acetylation of tubulin, increasing the effects of microtubule-stabilizing drugs such as paclitaxel [209]. This range of antitumor effects, make HDACIs a very attractive and potentially effective antineoplastic alternative.
Different HDACIs have been developed: trichostatin A and butyric acid have been used in numerous studies but showed limited clinical activity, and the high cytotoxicity of trichostatin A has limited its use in the clinic [210,211]. A member of the cyclic peptides, depsipeptide, demonstrated clinical efficacy in APL and human lymphoma xenograft models; however, it was not tested in ovarian cancer [212–215]. Vorinostat (suberoylanilide hydroxamic acid), a hydroxamate-based HDACI, is an oral drug that has demonstrated excellent bioavailability in Phase I trials but has major toxicities including anorexia, dehydration, diarrhea and fatigue [216,217]. In a Phase II trial of vorinostat as a single agent in patients with recurrent ovarian cancer, only one of 27 patients experienced a partial response [218], making it ineffective as a single-agent therapy. Vorinostat has been the only HDACI approved by the FDA for treatment of cutaneous T-cell lymphoma [219]. A recent hydroxamic acid HDACI, belinostat (PDX101), has revealed potent anti-proliferative and HDAC inhibitory activities in vitro and in xenograft ovarian and colorectal cancer models [220]. Authors of a preclinical ovarian cancer study, in which belinostat resensitized platinum-resistant xenografts in mice [221], and a Phase I trial, in which belinostat was administered intravenously in patients with advanced solid tumors, reported dose-limiting toxicities including grade 3 fatigue, diarrhea and cardiac arrhythmia, and concluded that the maximum tolerated dose was 1000 mg/m2 daily for 5 days and disease stabilization was observed in patients with different types of cancer (sarcomas, renal cancer, melanoma and thymoma) [222]. In another preclinical study, the combination of decitabine with belinostat elicited greater resensitization of platinum-resistant ovarian cancer xenografts than decitabine alone [223], making this combination a potentially effective approach to use in the clinic.
DNA methyltransferase inhibitors and HDACIs have shown promising efficacy against multiple types of cancers, both in the laboratory and in clinical trials [181–183]. Since ovarian tumorigenesis is driven by DNA methylation and chromosomal remodeling, it is reasonable to think that a combination of both DNTMIs and HDACIs could produce a greater effect in reactivating silenced TSGs and, thus, antitumor activity, than a single-agent therapy [224]. These epigenetic modifications also contribute to the silencing of genes related to chemosensitivity. Therefore, the reversal of these alterations in order to reactivate TSGs could be a potential target for ovarian cancer treatment. An example of this hypothesis is the study by Azar et al. where 5-aza-2-deoxycytidine pretreatment increased cytotoxicity of the topoisomerase inhibitor topotecan in vitro and in vivo [225]. As mentioned previously, treatment of cisplatin-resistant A2780/CP ovarian carcinoma cells with 5-aza-2-deoxycytidine induced expression of the mismatch repair enzyme hMLH1 and resensitized these cells to cisplatin in a mouse xenograft model [191,226]. Demethylating agents (e.g., decitabine), HDACIs or combinations may allow for the re-expression of silenced tumor suppressors such as hMLH1 and RASSF1A. hMLH1 plays a role in platinum resistance [191,226–228] and RASSF1A silencing may contribute to taxol resistance [167] and, thus, epigenetic re-expression of these genes might allow for resensitization of ovarian tumors to those conventional first-line therapies. Similarly, as several HDACIs enhance tubulin acetylation [209], these could conceivably augment sensitivity to taxanes. A list of the main DNMTIs and HDACIs is shown in Table 5. Radiation therapy represents a therapeutic alternative for ovarian cancer treatment. It has been demonstrated that trichostatin A could activate the ataxia telangiectasia-mutated p53 DNA damage signaling pathway, thereby enhancing ionizing-radiation-induced ataxia telangiectasia mutation activation [229], suggesting that HDACIs may override the DNA damage defense response and facilitate radiation-induced mitotic cell death.
Table 5.
List of the main DNA methyltransferases and histone deacetylase inhibitors.
| Class | Drug | Advantages | Disadvantages | Ref. |
|---|---|---|---|---|
| DNA methyltransferase inhibitors | ||||
| Nucleoside analog DNMT inhibitor |
Azacitidine (5-azacytidine) |
US FDA approved | Incorporates into both DNA and RNA (less inhibitory effect) Chemically unstable in water Toxic Induces global hypomethylation Short half-life, thus repeated daily doses are needed |
[248,249] |
| Decitabine (5-aza-2′- deoxycytidine) |
FDA approved Stronger DNMT inhibitory effect than azacitidine (only incorporates into DNA) More selective for cancer cells than normal cells Enhances the apoptotic effects of HDACIs |
Chemically unstable in water Toxic Induces global hypomethylation Demonstrated to be mutagenic in vivo Found to enhance the apoptotic effects of HDACIs Short half-life, thus repeated daily doses are needed |
[35,248–251] | |
| Zebularine | More stable in aqueous solutions Suitable for oral administration Less toxic More selective for cancer cells than normal cells |
A high level of drug is required to achieve efficacy Short half-life, thus repeated daily doses are needed |
[187,249] | |
| 5-fluoro-2′- deoxycytidine |
More stable in aqueous solutions Less toxic |
Short half-life, thus repeated daily doses are needed | [249] | |
| Non-nucleoside DNMT inhibitor |
(−)-epigallocatechin- 3-gallate |
Induces reactivation of methylation-silenced genes Represents a commonly consumed dietary constituent |
Low bioavailability Low therapeutic efficacy |
[197,252,253] |
| Hydralazine | Capable of re-expressing in vitro and in vivo the functional products of TSGs in cancer cell lines No cytotoxicity |
Induces a Lupus-like syndrome when used in high doses | [198,254] | |
| RG108 | Less toxic Specific for hypermethylated TSGs |
Its hydrophobicity makes it less valuable as an anticancer drug | [196,255] | |
| Histone deacetylase inhibitors | ||||
| Hydroxamate | Suberoylanilide hydroxamic acid (vorinostat) |
FDA approved Well tolerated and acceptable safety as monotherapy or in combination |
Side effects Limited ability to be a selective HDAC isoform inhibitor |
[256] |
| PXD101 (belinostat) | Demonstrated to suppress bladder and prostate cancer cell growth in vitro and in vivo Exhibits in vitro cytotoxicity at low micromolar concentrations Well tolerated in clinical trials |
Side effects Limited ability to be a selective HDAC isoform inhibitor |
[140, 220,222, 256,257] |
|
| Trichostatin A | Has differentiation and antiproliferative effects at nanomolar concentrations |
Side effects Limited ability to be a selective HDAC isoform inhibitor |
[256,258] | |
| Panobinostat (LBH589) |
Has shown antitumor activity in vitro and in vivo Potent inhibitory activity at low nanomolar concentrations, greater than vorinostat |
Side effects Intravenous infusions associated with QT interval prolongations Limited ability to be a selective HDAC isoform inhibitor |
[256,259] | |
| Short-chain fatty acid |
Phenylbutirate | Antitumor activity in vitro and in vivo (cell lines and xenografts) | Side effects | [260–263] |
| Valproic acid | Has been demonstrated to reduce tumor growth and metastasis Causes transformed cells to differentiate |
Side effects | [264] | |
| Cyclic peptide | Depsipeptide (romidepsin, FK228) |
Well tolerated Capable of inducing apoptosis in vitro and in vivo |
Toxicity Appears to have limited clinical activity in unselected leukemia patients |
[213] |
| Benzamide | MS-275 | Well tolerated Capable of inducing histone acetylation, p21 expression and caspase 3 activation |
Long half-life, rendering daily doses too toxic | [259] |
| Anilide | MGCD0103 | Well tolerated Has antileukemia activity Potential to regulate aberrant gene expression and restore normal growth control in malignancies |
Side effects More efficacious when combined |
[265,266] |
DMNT: DNA methyltransferase; HDACI: Histone deacetylase inhibitor; TSG: Tumor suppressor gene.
Conclusion
Ovarian cancer-specific genes discovered through the study of their DNA methylation and histone modification profiles, using the diverse technologies that have been developed, have sped up the discovery of new potential biomarkers for the diagnosis, prognosis and prediction of therapy. Through the knowledge and understanding of the ovarian cancer epigenome, it has been possible to develop epigenetic therapies that have had an enormous benefit in the prevention of chemoresistance by sensitizing tumors to conventional chemotherapeutics and abolishing cancer progression by reactivating the expression of TSGs.
Future perspective
Advances that aid in the understanding of ovarian cancer on a molecular level have provided important tools for molecular testing for high-risk populations, predictive markers for selecting patients for certain classes of drug therapies and molecular diagnostics for the noninvasive detection of early ovarian cancer. The epigenetic revolution that has come about in the field of cancer biology during the last few decades has provided valuable knowledge on how gene expression plays a key role in cancer formation and progression, and has created new insights helping us to understand the mechanisms involved in this process. Studies carried out in the field of ovarian cancer epigenetics have led to the realization that understanding the alterations of genes and pathways during the earliest steps of ovarian cancer development can aid clinical management of the patients in the near term. An understanding of the epigenetic signals that dictate the metastatic and/or drug-resistant phenotype will provide the information necessary to develop drugs to control or prevent advanced disease. Epigenetic therapy combined with chemotherapeutic agents holds significant promise for successful treatment of ovarian cancer in the future. Further epigenetic studies on ovarian cancer stem cells, along with development of more specific epigenetic drugs, may hold the key to our ability to successfully reprogram the abnormal ovarian cancer methylome. The considerable recent advances encourage us to believe that improvements in our knowledge of the epigenetic basis of ovarian cancer will continue to reduce the burden of this disease.
Executive summary.
Epigenetics & cancer
■ Epigenetic alterations do not change the primary DNA sequence but can influence the transcriptional process, leading to changes in the expression patterns of several genes.
■ The epigenetic modifications described so far involve: DNA methylation, histone modifications, and dysregulations of nucleosomes and miRNA, which mutually interact with each other to regulate gene expression.
■ The pattern seen in cancer cells is a global hypomethylation with focal hypermethylation, where global hypomethylation is linked to increased karyotypic instability and activation of oncogenes and gene-locus-specific hypermethylation can lead to the transcriptional silencing of tumor suppressor genes.
■ Histone modifications play an important role in remodeling chromatin structure, either in an active or repressive form, depending on the enzymatic machinery that catalyzes these modifications.
■ miRNAs have been found to be part of different biological processes as regulators, such as cell cycle, differentiation, development and metabolism, and, therefore, play an important role in cancer. Recently, miRNAs were found not only to be regulated by epigenetic alterations, such as DNA methylation, but also to be part of the modulation of DNA methylation in cancer.
Technologies to uncover the ovarian cancer epigenome
■ Different technologies for the study of DNA methylation and histone modifications have been developed.
■ The bisulfite-conversion technique, which reproducibly changes unmethylated cytosines to uracil but leaves methylated cytosines unchanged, was the mainstay for the creation of several sensitive DNA methylation detection techniques, including bisulfite sequencing, methylation-specific PCR, combined bisulfite restriction analysis and several real-time methylation-specific PCR methods, such as MethyLight™, quantitative multiplex methylation-specific PCR and pyrosequencing.
■ Besides the gene-specific profiling methods, several genome-wide techniques have been useful for the study of global DNA methylation patterns in normal and cancerous cells, such as restriction landmark genomic scanning, differential methylation hybridization and microarray gene expression profiling.
Epigenomic profiles as ovarian cancer biomarkers
■ The best studied serum biomarker for ovarian cancer is CA-125; however, it lacks specificity.
■ Methylation profiling has been an important tool to evaluate the applicability of genes as potential biomarkers for cancer diagnosis, prognosis and response to therapy.
■ For a methylation-based diagnostic assay to be reliable, that is, sensitive and specific, it is imperative to use those potential biomarkers that are found to be hypermethylated in cancer cells/tissues, but unmethylated in normal cells/tissues.
■ Many genes have been described in ovarian cancer to have a potential utility as biomarkers for diagnosis, prognosis and chemoresponse.
Pharmacoepigenomics in ovarian cancer
■ Owing to the fact that epigenetic alterations are reversible and that these have a profound effect in tumor initiation and progression, many efforts have been made in order to develop new epigenetic therapeutic agents to try to restore gene expression.
■ DNA methyltransferase inhibitors and histone deacetylase inhibitors have proven to be effective agents, especially when combined with each other or with other conventional chemotherapeutic agents.
Acknowledgments
MO Hoque has received support from the Flight Attendant Medical Research Institute Young Clinical Scientist Award, International Association for the Study of Lung Cancer and Career development award from SPORE in Cervical Cancer Grants (NIH grant number p50 CA098252). MO Hoque is a paid consultant to Oncomethylome Sciences, SA.
Footnotes
Financial & competing interests disclosure The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
■ of interest
■■ of considerable interest
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J. Clin. 2010 doi: 10.3322/caac.20073. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat. Genet. 1999;21(2):163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 3.Barakat RR, Markman M, Randall M. Principles and Practice Of Gynecologic Oncology. 5th Edition Wolters Kluwer/Lippincott Williams & Wilkins; PA, USA: 2009. [Google Scholar]
- 4.Barton CA, Hacker NF, Clark SJ, O’Brien PM. DNA methylation changes in ovarian cancer: implications for early diagnosis, prognosis and treatment. Gynecol. Oncol. 2008;109(1):129–139. doi: 10.1016/j.ygyno.2007.12.017. ■■ Very complete review of epigenetic changes in ovarian cancer and their importance in the detection of biomarkers for diagnosis, prognosis and therapeutic response.
- 5.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer – a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6(2):107–116. doi: 10.1038/nrc1799. ■■ Interesting article on how epigenetic alterations drive cells to certain pathways that lead to tumorigenesis.
- 6.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8(4):286–298. doi: 10.1038/nrg2005. ■■ Comprehensive review of the techniques used for the study of the DNA methylome and histone modifications, emphasizing genome-wide arrays.
- 7.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3(6):415–428. doi: 10.1038/nrg816. ■■ Excellent article on epigenetic mechanisms that alter the expression of genes.
- 8.Ting AH, McGarvey KM, Baylin SB. The cancer epigenome – components and functional correlates. Genes Dev. 2006;20(23):3215–3231. doi: 10.1101/gad.1464906. [DOI] [PubMed] [Google Scholar]
- 9.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J. Nutr. 2002;132(Suppl. 8):2401S–2405S. doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]
- 11.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300(5618):455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 12.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat. Rev. Cancer. 2004;4(2):143–153. doi: 10.1038/nrc1279. ■ Traces epigenetic alterations since their conception and reviews the advances made on this field, addressing the genetic basis of epigenetic changes.
- 13.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21(35):5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 14.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 2005;6(11):838–849. doi: 10.1038/nrm1761. ■■ Summarizes the recent advances in the understanding of how lysine methylation plays an important role in heterochromatin formation and transcriptional regulation.
- 15.Esteller M, Almouzni G. How epigenetics integrates nuclear functions. Workshop on epigenetics and chromatin: transcriptional regulation and beyond. EMBO Rep. 2005;6(7):624–628. doi: 10.1038/sj.embor.7400456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen CT, Gonzales FA, Jones PA. Altered chromatin structure associated with methylation-induced gene silencing in cancer cells: correlation of accessibility, methylation, MeCP2 binding and acetylation. Nucleic Acids Res. 2001;29(22):4598–4606. doi: 10.1093/nar/29.22.4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knudson AG. Chasing the cancer demon. Annu. Rev. Genet. 2000;34:1–19. doi: 10.1146/annurev.genet.34.1.1. [DOI] [PubMed] [Google Scholar]
- 18.Nussbaum RL, Mcinnes RR, Willard HF, Thompson MW. Thompson & Thompson Genetics in Medicine. 7th Edition Saunders/Elsevier; PA, USA: 2007. [Google Scholar]
- 19.Antequera F, Bird A, Jost IJP, Saluz . DNA Methylation: Molecular Biology and Biological Significance. Birkhauser Verlag; Basel, Switzerland: 1993. pp. 169–185. [Google Scholar]
- 20.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl Acad. Sci. USA. 2002;99(6):3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferguson-Smith AC, Surani MA. Imprinting and the epigenetic asymmetry between parental genomes. Science. 2001;293(5532):1086–1089. doi: 10.1126/science.1064020. [DOI] [PubMed] [Google Scholar]
- 22.Reik W, Lewis A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat. Rev. Genet. 2005;6(5):403–410. doi: 10.1038/nrg1602. [DOI] [PubMed] [Google Scholar]
- 23.Brenner C, Fuks F. DNA methyltransferases: facts, clues, mysteries. Curr. Top. Microbiol. Immunol. 2006;301:45–66. doi: 10.1007/3-540-31390-7_3. [DOI] [PubMed] [Google Scholar]
- 24.Tajima S, Suetake I. Regulation and function of DNA methylation in vertebrates. J. Biochem. 1998;123(6):993–999. doi: 10.1093/oxfordjournals.jbchem.a022066. [DOI] [PubMed] [Google Scholar]
- 25.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases DNMT3A and DNMT3B are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 26.Chedin F, Lieber Mr, Hsieh Cl. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by DNMT3A. Proc. Natl Acad. Sci. USA. 2002;99(26):16916–16921. doi: 10.1073/pnas.262443999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aapola U, Kawasaki K, Scott HS, et al. Isolation and initial characterization of a novel zinc finger gene, DNMT3l, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics. 2000;65(3):293–298. doi: 10.1006/geno.2000.6168. [DOI] [PubMed] [Google Scholar]
- 28.Dong A, Yoder JA, Zhang X, Zhou L, Bestor TH, Cheng X. Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA. Nucleic Acids Res. 2001;29(2):439–448. doi: 10.1093/nar/29.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schaefer M, Lyko F. DNMT2 enigma. Chromosoma. 2010;119(1):35–40. doi: 10.1007/s00412-009-0240-6. [DOI] [PubMed] [Google Scholar]
- 30.Sinkkonen L, Hugenschmidt T, Berninger P, et al. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2008;15(3):259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 31.Benetti R, Gonzalo S, Jaco I, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via RBL2-dependent regulation of DNA methyltransferases. Nat. Struct. Mol. Biol. 2008;15(9):998. doi: 10.1038/nsmb0908-998b. [DOI] [PubMed] [Google Scholar]
- 32.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005;6(5):376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 33.Ehrlich M. DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J. Nutr. 2002;132(Suppl. 8):2424S–2429S. doi: 10.1093/jn/132.8.2424S. [DOI] [PubMed] [Google Scholar]
- 34.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301(5895):89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 35.Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395(6697):89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 36.Kaneda A, Feinberg AP. Loss of imprinting of IGF2: a common epigenetic modifier of intestinal tumor risk. Cancer Res. 2005;65(24):11236–11240. doi: 10.1158/0008-5472.CAN-05-2959. [DOI] [PubMed] [Google Scholar]
- 37.Kornberg RD, Lorch Y. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell. 1999;98(3):285–294. doi: 10.1016/s0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 38.Schubeler D, Macalpine DM, Scalzo D, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18(11):1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc. Natl Acad. Sci. USA. 2003;100(23):13225–13230. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Ann. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 41.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 42.Cao R, Wang L, Wang H, et al. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 43.Fuks F, Hurd Pj, Deplus R, Kouzarides T. The DNA methyltransferases associate with hp1 and the SUV39H1 histone methyltransferase. Nucleic Acids Res. 2003;31(9):2305–2312. doi: 10.1093/nar/gkg332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410(6824):116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 45.Clouaire T, Stancheva I. Methyl-CpG binding proteins: specialized transcriptional repressors or structural components of chromatin? Cell Mol. Life Sci. 2008;65(10):1509–1522. doi: 10.1007/s00018-008-7324-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J. Biol. Chem. 2003;278(6):4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 47.Jones PL, Veenstra GJ, Wade PA, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat. Genet. 1998;19(2):187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 48.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 49.Boehm M, Slack FJ. MicroRNA control of lifespan and metabolism. Cell Cycle. 2006;5(8):837–840. doi: 10.4161/cc.5.8.2688. [DOI] [PubMed] [Google Scholar]
- 50.Carleton M, Cleary MA, Linsley PS. MicroRNAs and cell cycle regulation. Cell Cycle. 2007;6(17):2127–2132. doi: 10.4161/cc.6.17.4641. [DOI] [PubMed] [Google Scholar]
- 51.Harfe BD. MicroRNAs in vertebrate development. Curr. Opin. Genet. Dev. 2005;15(4):410–415. doi: 10.1016/j.gde.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 52.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294(5543):858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 53.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 54.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99(24):15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 57.Nakamura T, Canaani E, Croce CM. Oncogenic ALL1 fusion proteins target drosha-mediated microRNA processing. Proc. Natl Acad. Sci. USA. 2007;104(26):10980–10985. doi: 10.1073/pnas.0704559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saito Y, Liang G, Egger G, et al. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9(6):435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 59.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26(5):745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of mir-34a contributes to p53-mediated apoptosis. Mol. Cell. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 62.Sellar GC, Watt KP, Rabiasz GJ, et al. OPCMl at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat. Genet. 2003;34(3):337–343. doi: 10.1038/ng1183. [DOI] [PubMed] [Google Scholar]
- 63.Teodoridis JM, Hall J, Marsh S, et al. CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res. 2005;65(19):8961–8967. doi: 10.1158/0008-5472.CAN-05-1187. [DOI] [PubMed] [Google Scholar]
- 64.Zhang J, Ye F, Chen HZ, Ye DF, Lu WG, Xie X. Deletion of OPCMl gene and promoter methylation in ovarian epithelial carcinoma. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2006;28(2):173–177. [PubMed] [Google Scholar]
- 65.Kwong J, Lee JY, Wong KK, et al. Candidate tumor suppressor gene DLEC1 is frequently downregulated by promoter hypermethylation and histone hypoacetylation in human epithelial ovarian cancer. Neoplasia. 2006;8(4):268–278. doi: 10.1593/neo.05502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Agathanggelou A, Honorio S, Macartney DP, et al. Methylation associated inactivation of RASSF1A from region 3p21.3 in lung, breast and ovarian tumours. Oncogene. 2001;20(12):1509–1518. doi: 10.1038/sj.onc.1204175. [DOI] [PubMed] [Google Scholar]
- 67.Ibanez De Caceres I, Battagli C, Esteller M, et al. Tumor cell-specific brca1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res. 2004;64(18):6476–6481. doi: 10.1158/0008-5472.CAN-04-1529. ■ Important article showing that promoter hypermethylation can be detected in bodily fluids of patients with ovarian cancer, enhancing its early detection.
- 68.Rathi A, Virmani AK, Schorge JO, et al. Methylation profiles of sporadic ovarian tumors and nonmalignant ovaries from high-risk women. Clin. Cancer Res. 2002;8(11):3324–3331. [PubMed] [Google Scholar]
- 69.Yoon JH, Dammann R, Pfeifer GP. Hypermethylation of the CpG island of the RASSF1A gene in ovarian and renal cell carcinomas. Int. J. Cancer. 2001;94(2):212–217. doi: 10.1002/ijc.1466. [DOI] [PubMed] [Google Scholar]
- 70.Petrocca F, Iliopoulos D, Qin HR, et al. Alterations of the tumor suppressor gene ARLTS1 in ovarian cancer. Cancer Res. 2006;66(21):10287–10291. doi: 10.1158/0008-5472.CAN-06-2289. [DOI] [PubMed] [Google Scholar]
- 71.Yu Y, Fujii S, Yuan J, et al. Epigenetic regulation of ARHI in breast and ovarian cancer cells. Ann. NY Acad. Sci. 2003;983:268–277. doi: 10.1111/j.1749-6632.2003.tb05981.x. [DOI] [PubMed] [Google Scholar]
- 72.Yu Y, Xu F, Peng H, et al. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc. Natl Acad. Sci. USA. 1999;96(1):214–219. doi: 10.1073/pnas.96.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chien J, Staub J, Avula R, et al. Epigenetic silencing of TCEAL7 (Bex4) in ovarian cancer. Oncogene. 2005;24(32):5089–5100. doi: 10.1038/sj.onc.1208700. [DOI] [PubMed] [Google Scholar]
- 74.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61(8):3225–3229. ■ Interesting article showing cancer-specific methylation profiles, leading to important knowledge of the cancer epigenome.
- 75.Esteller M, Silva JM, Dominguez G, et al. Promoter hypermethylation and brca1 inactivation in sporadic breast and ovarian tumors. J. Natl Cancer Inst. 2000;92(7):564–569. doi: 10.1093/jnci/92.7.564. [DOI] [PubMed] [Google Scholar]
- 76.Baldwin RL, Nemeth E, Tran H, et al. BRCA1 promoter region hypermethylation in ovarian carcinoma: a population-based study. Cancer Res. 2000;60(19):5329–5333. [PubMed] [Google Scholar]
- 77.Chan KY, Ozcelik H, Cheung AN, Ngan HY, Khoo US. Epigenetic factors controlling the brca1 and BRCA2 genes in sporadic ovarian cancer. Cancer Res. 2002;62(14):4151–4156. [PubMed] [Google Scholar]
- 78.Wilcox CB, Baysal BE, Gallion HH, Strange MA, Deloia JA. High-resolution methylation analysis of the BRCA1 promoter in ovarian tumors. Cancer Genet. Cytogenet. 2005;159(2):114–122. doi: 10.1016/j.cancergencyto.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 79.Esteller M. Epigenetic lesions causing genetic lesions in human cancer: promoter hypermethylation of DNA repair genes. Eur. J. Cancer. 2000;36(18):2294–2300. doi: 10.1016/s0959-8049(00)00303-8. [DOI] [PubMed] [Google Scholar]
- 80.Gras E, Cortes J, Diez O, et al. Loss of heterozygosity on chromosome 13q12-q14, brca-2 mutations and lack of BRCA-2 promoter hypermethylation in sporadic epithelial ovarian tumors. Cancer. 2001;92(4):787–795. doi: 10.1002/1097-0142(20010815)92:4<787::aid-cncr1384>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 81.Hilton JL, Geisler JP, Rathe JA, Hattermann-Zogg MA, Deyoung B, Buller RE. Inactivation of BRCA1 and BRCA2 in ovarian cancer. J. Natl Cancer Inst. 2002;94(18):1396–1406. doi: 10.1093/jnci/94.18.1396. [DOI] [PubMed] [Google Scholar]
- 82.Milde-Langosch K, Ocon E, Becker G, Loning T. P16/MTS1 inactivation in ovarian carcinomas: high frequency of reduced protein expression associated with hypermethylation or mutation in endometrioid and mucinous tumors. Int. J. Cancer. 1998;79(1):61–65. doi: 10.1002/(sici)1097-0215(19980220)79:1<61::aid-ijc12>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 83.Socha MJ, Said N, Dai Y, et al. Aberrant promoter methylation of SPARC in ovarian cancer. Neoplasia. 2009;11(2):126–135. doi: 10.1593/neo.81146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kikuchi R, Tsuda H, Kozaki K, et al. Frequent inactivation of a putative tumor suppressor, angiopoietin-like protein 2, in ovarian cancer. Cancer Res. 2008;68(13):5067–5075. doi: 10.1158/0008-5472.CAN-08-0062. [DOI] [PubMed] [Google Scholar]
- 85.Kikuchi R, Tsuda H, Kanai Y, et al. Promoter hypermethylation contributes to frequent inactivation of a putative conditional tumor suppressor gene connective tissue growth factor in ovarian cancer. Cancer Res. 2007;67(15):7095–7105. doi: 10.1158/0008-5472.CAN-06-4567. [DOI] [PubMed] [Google Scholar]
- 86.Cvetkovic D, Pisarcik D, Lee C, Hamilton TC, Abdollahi A. Altered expression and loss of heterozygosity of the LOT1 gene in ovarian cancer. Gynecol. Oncol. 2004;95(3):449–455. doi: 10.1016/j.ygyno.2004.08.051. [DOI] [PubMed] [Google Scholar]
- 87.Pruitt K, Ulku AS, Frantz K, et al. Ras-mediated loss of the pro-apoptotic response protein PAR-4 is mediated by DNA hypermethylation through Raf-independent and Raf-dependent signaling cascades in epithelial cells. J. Biol. Chem. 2005;280(24):23363–23370. doi: 10.1074/jbc.M503083200. [DOI] [PubMed] [Google Scholar]
- 88.Arnold JM, Cummings M, Purdie D, Chenevix-Trench G. Reduced expression of intercellular adhesion molecule-1 in ovarian adenocarcinomas. Br. J. Cancer. 2001;85(9):1351–1358. doi: 10.1054/bjoc.2001.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuecheng Y, Hongmei L, Xiaoyan X. Clinical evaluation of E-cadherin expression and its regulation mechanism in epithelial ovarian cancer. Clin. Exp. Metastasis. 2006;23(1):65–74. doi: 10.1007/s10585-006-9020-3. [DOI] [PubMed] [Google Scholar]
- 90.Feng W, Marquez RT, Lu Z, et al. Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer. 2008;112(7):1489–1502. doi: 10.1002/cncr.23323. [DOI] [PubMed] [Google Scholar]
- 91.Fiegl H, Windbichler G, Mueller-Holzner E, et al. HOXA11 DNA methylation – a novel prognostic biomarker in ovarian cancer. Int. J. Cancer. 2008;123(3):725–729. doi: 10.1002/ijc.23563. [DOI] [PubMed] [Google Scholar]
- 92.Potapova A, Hoffman AM, Godwin AK, Al-Saleem T, Cairns P. Promoter hypermethylation of the PALB2 susceptibility gene in inherited and sporadic breast and ovarian cancer. Cancer Res. 2008;68(4):998–1002. doi: 10.1158/0008-5472.CAN-07-2418. [DOI] [PubMed] [Google Scholar]
- 93.Izutsu N, Maesawa C, Shibazaki M, et al. Epigenetic modification is involved in aberrant expression of class III β-tubulin, TUBB3, in ovarian cancer cells. Int. J. Oncol. 2008;32(6):1227–1235. doi: 10.3892/ijo_32_6_1227. [DOI] [PubMed] [Google Scholar]
- 94.Cheng P, Schmutte C, Cofer KF, Felix JC, Yu MC, Dubeau L. Alterations in DNA methylation are early, but not initial, events in ovarian tumorigenesis. Br. J. Cancer. 1997;75(3):396–402. doi: 10.1038/bjc.1997.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rose SL, Fitzgerald MP, White NO, et al. Epigenetic regulation of maspin expression in human ovarian carcinoma cells. Gynecol. Oncol. 2006;102(2):319–324. doi: 10.1016/j.ygyno.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 96.Czekierdowski A, Czekierdowska S, Wielgos M, Smolen A, Kaminski P, Kotarski J. The role of CpG islands hypomethylation and abnormal expression of neuronal protein synuclein-γ (sncg) in ovarian cancer. Neuro. Endocrinol. Lett. 2006;27(3):381–386. [PubMed] [Google Scholar]
- 97.Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J. Hypomethylation of the synuclein γ gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res. 2003;63(3):664–673. [PubMed] [Google Scholar]
- 98.Honda H, Pazin MJ, Ji H, Wernyj RP, Morin PJ. Crucial roles of Sp1 and epigenetic modifications in the regulation of the CLDN4 promoter in ovarian cancer cells. J. Biol. Chem. 2006;281(30):21433–21444. doi: 10.1074/jbc.M603767200. [DOI] [PubMed] [Google Scholar]
- 99.Litkouhi B, Kwong J, Lo CM, et al. Claudin-4 overexpression in epithelial ovarian cancer is associated with hypomethylation and is a potential target for modulation of tight junction barrier function using a C-terminal fragment of Clostridium perfringens enterotoxin. Neoplasia. 2007;9(4):304–314. doi: 10.1593/neo.07118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Menendez L, Benigno BB, Mcdonald JF. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol. Cancer. 2004;3:12. doi: 10.1186/1476-4598-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kolomietz E, Meyn MS, Pandita A, Squire JA. The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer. 2002;35(2):97–112. doi: 10.1002/gcc.10111. [DOI] [PubMed] [Google Scholar]
- 102.Symer DE, Connelly C, Szak St, et al. Human L1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110(3):327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 103.Woloszynska-Read A, James SR, Link PA, Yu J, Odunsi K, Karpf AR. DNA methylation-dependent regulation of BORIS/CTCFL expression in ovarian cancer. Cancer Immun. 2007;7:21. [PMC free article] [PubMed] [Google Scholar]
- 104.Murphy SK, Huang Z, Wen Y, et al. Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol. Cancer Res. 2006;4(4):283–292. doi: 10.1158/1541-7786.MCR-05-0138. [DOI] [PubMed] [Google Scholar]
- 105.Ahluwalia A, Hurteau JA, Bigsby RM, Nephew KP. DNA methylation in ovarian cancer. II. Expression of DNA methyltransferases in ovarian cancer cell lines and normal ovarian epithelial cells. Gynecol. Oncol. 2001;82(2):299–304. doi: 10.1006/gyno.2001.6284. [DOI] [PubMed] [Google Scholar]
- 106.Ehrlich M, Woods CB, Yu MC, et al. Quantitative analysis of associations between DNA hypermethylation, hypomethylation, and DNMT RNA levels in ovarian tumors. Oncogene. 2006;25(18):2636–2645. doi: 10.1038/sj.onc.1209145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen CL, Yan X, Gao YN, Liao QP. Expression of DNA methyltransferase 1, 3A and 3B mRNA in the epithelial ovarian carcinoma. Zhonghua Fu Chan Ke Za Zhi. 2005;40(11):770–774. [PubMed] [Google Scholar]
- 108.Kaneuchi M, Sasaki M, Tanaka Y, et al. Expression and methylation status of 14–3-3 σ gene can characterize the different histological features of ovarian cancer. Biochem. Biophys. Res. Commun. 2004;316(4):1156–1162. doi: 10.1016/j.bbrc.2004.02.171. [DOI] [PubMed] [Google Scholar]
- 109.Kaneuchi M, Sasaki M, Tanaka Y, et al. WT1 and WT1-AS genes are inactivated by promoter methylation in ovarian clear cell adenocarcinoma. Cancer. 2005;104(9):1924–1930. doi: 10.1002/cncr.21397. [DOI] [PubMed] [Google Scholar]
- 110.Terasawa K, Sagae S, Toyota M, et al. Epigenetic inactivation of TMS1/ASC in ovarian cancer. Clin. Cancer Res. 2004;10(6):2000–2006. doi: 10.1158/1078-0432.ccr-0932-03. [DOI] [PubMed] [Google Scholar]
- 111.Makarla PB, Saboorian MH, Ashfaq R, et al. Promoter hypermethylation profile of ovarian epithelial neoplasms. Clin. Cancer Res. 2005;11(15):5365–5369. doi: 10.1158/1078-0432.CCR-04-2455. [DOI] [PubMed] [Google Scholar]
- 112.Caslini C, Capo-Chichi CD, Roland IH, Nicolas E, Yeung AT, Xu XX. Histone modifications silence the GATA transcription factor genes in ovarian cancer. Oncogene. 2006;25(39):5446–5461. doi: 10.1038/sj.onc.1209533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Valls E, Sanchez-Molina S, Martinez-Balbas MA. Role of histone modifications in marking and activating genes through mitosis. J. Biol. Chem. 2005;280(52):42592–42600. doi: 10.1074/jbc.M507407200. [DOI] [PubMed] [Google Scholar]
- 114.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21WAF1 expression and gene-associated histone acetylation. Proc. Natl Acad. Sci. USA. 2000;97(18):10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chan MW, Huang YW, Hartman-Frey C, et al. Aberrant transforming growth factor-β1 signaling and SMAD4 nuclear translocation confer epigenetic repression of ADAM19 in ovarian cancer. Neoplasia. 2008;10(9):908–919. doi: 10.1593/neo.08540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93(18):9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Laird PW. The power and the promise of DNA methylation markers. Nat. Rev. Cancer. 2003;3(4):253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- 118.Rand KN, Ho T, Qu W, et al. Headloop suppression PCR and its application to selective amplification of methylated DNA sequences. Nucleic Acids Res. 2005;33(14):e127. doi: 10.1093/nar/gni120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cottrell SE, Laird PW. Sensitive detection of DNA methylation. Ann. NY Acad. Sci. 2003;983:120–130. doi: 10.1111/j.1749-6632.2003.tb05967.x. [DOI] [PubMed] [Google Scholar]
- 120.Fraga MF, Esteller M. DNA methylation: a profile of methods and applications. Biotechniques. 2002;33(3):632, 634, 636–649. doi: 10.2144/02333rv01. [DOI] [PubMed] [Google Scholar]
- 121.Bird AP, Southern EM. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J. Mol. Biol. 1978;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- 122.Frommer M, Mcdonald LE, Millar Ds, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89(5):1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25(12):2532–2534. doi: 10.1093/nar/25.12.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28(8):E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fackler MJ, Mcveigh M, Mehrotra J, et al. Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res. 2004;64(13):4442–4452. doi: 10.1158/0008-5472.CAN-03-3341. [DOI] [PubMed] [Google Scholar]
- 126.Swift-Scanlan T, Blackford A, Argani P, Sukumar S, Fackler MJ. Two-color quantitative multiplex methylation-specific PCR. Biotechniques. 2006;40(2):210–219. doi: 10.2144/000112097. [DOI] [PubMed] [Google Scholar]
- 127.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat. Protoc. 2007;2(9):2265–2275. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 128.Tost J, Gut IG. Analysis of gene-specific DNA methylation patterns by pyrosequencing technology. Methods Mol. Biol. 2007;373:89–102. doi: 10.1385/1-59745-377-3:89. [DOI] [PubMed] [Google Scholar]
- 129.Costello JF, Fruhwald MC, Smiraglia DJ, et al. Aberrant CpG-island methylation has non-random and tumour-type-specific patterns. Nat. Genet. 2000;24(2):132–138. doi: 10.1038/72785. [DOI] [PubMed] [Google Scholar]
- 130.Huang TH, Perry MR, Laux DE. Methylation profiling of CpG islands in human breast cancer cells. Hum. Mol. Genet. 1999;8(3):459–470. doi: 10.1093/hmg/8.3.459. [DOI] [PubMed] [Google Scholar]
- 131.Hoque MO, Kim MS, Ostrow KL, et al. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68(8):2661–2670. doi: 10.1158/0008-5472.CAN-07-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat. Genet. 2002;31(2):141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 133.Yamashita K, Upadhyay S, Osada M, et al. Pharmacologic unmasking of epigenetically silenced tumor suppressor genes in esophageal squamous cell carcinoma. Cancer Cell. 2002;2(6):485–495. doi: 10.1016/s1535-6108(02)00215-5. [DOI] [PubMed] [Google Scholar]
- 134.Frigola J, Ribas M, Risques RA, Peinado MA. Methylome profiling of cancer cells by amplification of inter-methylated sites (AIMS) Nucleic Acids Res. 2002;30(7):e28. doi: 10.1093/nar/30.7.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Toyota M, Ho C, Ahuja N, et al. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59(10):2307–2312. [PubMed] [Google Scholar]
- 136.Khulan B, Thompson RF, Ye K, et al. Comparative isoschizomer profiling of cytosine methylation: the help assay. Genome Res. 2006;16(8):1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Keshet I, Schlesinger Y, Farkash S, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat. Genet. 2006;38(2):149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 138.Weber M, Davies JJ, Wittig D, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37(8):853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 139.Weber M, Hellmann I, Stadler MB, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat. Genet. 2007;39(4):457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 140.Noushmehr H, Weisenberger DJ, Diefes K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kuras L. Characterization of protein-DNA association in vivo by chromatin immunoprecipitation. Methods Mol. Biol. 2004;284:147–162. doi: 10.1385/1-59259-816-1:147. [DOI] [PubMed] [Google Scholar]
- 142.Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. High-resolution genome-wide mapping of histone modifications. Nat. Biotechnol. 2004;22(8):1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- 143.Barski A, Cuddapah S, Cui K, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129(4):823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 144.Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448(7153):553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Nossov V, Amneus M, Su F, et al. The early detection of ovarian cancer: from traditional methods to proteomics. Can we really do better than serum CA-125? Am. J. Obstet. Gynecol. 2008;199(3):215–223. doi: 10.1016/j.ajog.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 146.Bast RC., Jr Status of tumor markers in ovarian cancer screening. J. Clin. Oncol. 2003;21(Suppl. 10):200S–205S. doi: 10.1200/JCO.2003.01.068. [DOI] [PubMed] [Google Scholar]
- 147.Hickey KP, Boyle KP, Jepps HM, Andrew AC, Buxton EJ, Burns PA. Molecular detection of tumour DNA in serum and peritoneal fluid from ovarian cancer patients. Br. J. Cancer. 1999;80(11):1803–1808. doi: 10.1038/sj.bjc.6690601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Collins Y, Dicioccio R, Keitz B, Lele S, Odunsi K. Methylation of death-associated protein kinase in ovarian carcinomas. Int. J. Gynecol. Cancer. 2006;16(Suppl. 1):195–199. doi: 10.1111/j.1525-1438.2006.00506.x. [DOI] [PubMed] [Google Scholar]
- 149.Wiley A, Katsaros D, Fracchioli S, Yu H. Methylation of the insulin-like growth factor binding protein-3 gene and prognosis of epithelial ovarian cancer. Int. J. Gynecol. Cancer. 2006;16(1):210–218. doi: 10.1111/j.1525-1438.2006.00299.x. [DOI] [PubMed] [Google Scholar]
- 150.Wiley A, Katsaros D, Chen H, et al. Aberrant promoter methylation of multiple genes in malignant ovarian tumors and in ovarian tumors with low malignant potential. Cancer. 2006;107(2):299–308. doi: 10.1002/cncr.21992. [DOI] [PubMed] [Google Scholar]
- 151.Chan MW, Wei SH, Wen P, et al. Hypermethylation of 18S and 28S ribosomal DNAs predicts progression-free survival in patients with ovarian cancer. Clin. Cancer Res. 2005;11(20):7376–7383. doi: 10.1158/1078-0432.CCR-05-1100. [DOI] [PubMed] [Google Scholar]
- 152.Widschwendter M, Jiang G, Woods C, et al. DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004;64(13):4472–4480. doi: 10.1158/0008-5472.CAN-04-0238. [DOI] [PubMed] [Google Scholar]
- 153.Su HY, Lai HC, Lin YW, Chou YC, Liu CY, Yu MH. An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. Int. J. Cancer. 2009;124(2):387–393. doi: 10.1002/ijc.23957. [DOI] [PubMed] [Google Scholar]
- 154.Apostolidou S, Hadwin R, Burnell M, et al. DNA methylation analysis in liquid-based cytology for cervical cancer screening. Int. J. Cancer. 2009;125(12):2995–3002. doi: 10.1002/ijc.24745. [DOI] [PubMed] [Google Scholar]
- 155.Hoque MO, Begum S, Brait M, et al. Tissue inhibitor of metalloproteinases-3 promoter methylation is an independent prognostic factor for bladder cancer. J. Urol. 2008;179(2):743–747. doi: 10.1016/j.juro.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Agarwal R, Kaye SB. Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat. Rev. Cancer. 2003;3(7):502–516. doi: 10.1038/nrc1123. [DOI] [PubMed] [Google Scholar]
- 157.Balch C, Huang TH, Brown R, Nephew KP. The epigenetics of ovarian cancer drug resistance and resensitization. Am. J. Obstet. Gynecol. 2004;191(5):1552–1572. doi: 10.1016/j.ajog.2004.05.025. [DOI] [PubMed] [Google Scholar]
- 158.Shah MA, Schwartz GK. Cell cycle-mediated drug resistance: an emerging concept in cancer therapy. Clin. Cancer Res. 2001;7(8):2168–2181. [PubMed] [Google Scholar]
- 159.Dumontet C, Sikic BI. Mechanisms of action of and resistance to antitubulin agents: microtubule dynamics, drug transport, and cell death. J. Clin. Oncol. 1999;17(3):1061–1070. doi: 10.1200/JCO.1999.17.3.1061. [DOI] [PubMed] [Google Scholar]
- 160.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22(47):7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 161.Geisler JP, Goodheart MJ, Sood AK, Holmes RJ, Hatterman-Zogg MA, Buller RE. Mismatch repair gene expression defects contribute to microsatellite instability in ovarian carcinoma. Cancer. 2003;98(10):2199–2206. doi: 10.1002/cncr.11770. [DOI] [PubMed] [Google Scholar]
- 162.Martini M, Ciccarone M, Garganese G, et al. Possible involvement of hMLH1, p16(INK4a) and PTEN in the malignant transformation of endometriosis. Int. J. Cancer. 2002;102(4):398–406. doi: 10.1002/ijc.10715. [DOI] [PubMed] [Google Scholar]
- 163.Jass JR, Walsh MD, Barker M, Simms LA, Young J, Leggett BA. Distinction between familial and sporadic forms of colorectal cancer showing DNA microsatellite instability. Eur. J. Cancer. 2002;38(7):858–866. doi: 10.1016/s0959-8049(02)00041-2. [DOI] [PubMed] [Google Scholar]
- 164.Wang L, Bani-Hani A, Montoya DP, et al. hMLH1 and hMSH2 expression in human hepatocellular carcinoma. Int. J. Oncol. 2001;19(3):567–570. [PubMed] [Google Scholar]
- 165.Atkin NB. Microsatellite instability. Cytogenet. Cell Genet. 2001;92(3–4):177–181. doi: 10.1159/000056898. [DOI] [PubMed] [Google Scholar]
- 166.Brown R, Hirst GL, Gallagher WM, et al. hMLH1 expression and cellular responses of ovarian tumour cells to treatment with cytotoxic anticancer agents. Oncogene. 1997;15(1):45–52. doi: 10.1038/sj.onc.1201167. [DOI] [PubMed] [Google Scholar]
- 167.Liu L, Tommasi S, Lee DH, Dammann R, Pfeifer GP. Control of microtubule stability by the RASSF1A tumor suppressor. Oncogene. 2003;22(50):8125–8136. doi: 10.1038/sj.onc.1206984. [DOI] [PubMed] [Google Scholar]
- 168.Staub J, Chien J, Pan Y, et al. Epigenetic silencing of HSulf-1 in ovarian cancer: implications in chemoresistance. Oncogene. 2007;26(34):4969–4978. doi: 10.1038/sj.onc.1210300. [DOI] [PubMed] [Google Scholar]
- 169.Lai J, Chien J, Staub J, et al. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J. Biol. Chem. 2003;278(25):23107–23117. doi: 10.1074/jbc.M302203200. [DOI] [PubMed] [Google Scholar]
- 170.Wei SH, Brown R, Huang TH. Aberrant DNA methylation in ovarian cancer: is there an epigenetic predisposition to drug response? Ann. NY Acad. Sci. 2003;983:243–250. doi: 10.1111/j.1749-6632.2003.tb05979.x. [DOI] [PubMed] [Google Scholar]
- 171.Wei SH, Chen CM, Strathdee G, et al. Methylation microarray analysis of late-stage ovarian carcinomas distinguishes progression-free survival in patients and identifies candidate epigenetic markers. Clin. Cancer Res. 2002;8(7):2246–2252. [PubMed] [Google Scholar]
- 172.Taniguchi T, Tischkowitz M, Ameziane N, et al. Disruption of the fanconi anemia–BRCA pathway in cisplatin-sensitive ovarian tumors. Nat. Med. 2003;9(5):568–574. doi: 10.1038/nm852. [DOI] [PubMed] [Google Scholar]
- 173.Hatle KM, Neveu W, Dienz O, et al. Methylation-controlled J protein promotes c-Jun degradation to prevent ABCD1 transporter expression. Mol. Cell. Biol. 2007;27(8):2952–2966. doi: 10.1128/MCB.01804-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Shridhar V, Bible KC, Staub J, et al. Loss of expression of a new member of the DNAJ protein family confers resistance to chemotherapeutic agents used in the treatment of ovarian cancer. Cancer Res. 2001;61(10):4258–4265. [PubMed] [Google Scholar]
- 175.Strathdee G, Davies BR, Vass JK, Siddiqui N, Brown R. Cell type-specific methylation of an intronic CpG island controls expression of the MCJ gene. Carcinogenesis. 2004;25(5):693–701. doi: 10.1093/carcin/bgh066. [DOI] [PubMed] [Google Scholar]
- 176.Strathdee G, Vass JK, Oien KA, Siddiqui N, Curto-Garcia J, Brown R. Demethylation of the MCJ gene in stage III/IV epithelial ovarian cancer and response to chemotherapy. Gynecol. Oncol. 2005;97(3):898–903. doi: 10.1016/j.ygyno.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 177.Iorio MV, Visone R, Di Leva G, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 178.Yang H, Kong W, He L, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68(2):425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 179.Yang N, Kaur S, Volinia S, et al. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68(24):10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Boren T, Xiong Y, Hakam A, et al. MicroRNAs and their target messenger RNAs associated with ovarian cancer response to chemotherapy. Gynecol. Oncol. 2009;113(2):249–255. doi: 10.1016/j.ygyno.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 181.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat. Rev. Drug Discov. 2006;5(9):769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 182.Hellebrekers DM, Griffioen AW, Van Engeland M. Dual targeting of epigenetic therapy in cancer. Biochim. Biophys. Acta. 2007;1775(1):76–91. doi: 10.1016/j.bbcan.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 183.Kim TY, Bang YJ, Robertson KD. Histone deacetylase inhibitors for cancer therapy. Epigenetics. 2006;1(1):14–23. doi: 10.4161/epi.1.1.2644. [DOI] [PubMed] [Google Scholar]
- 184.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat. Rev. Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 185.Jones PA, Taylor SM. Cellular differentiation, cytidine analogs and DNA methylation. Cell. 1980;20(1):85–93. doi: 10.1016/0092-8674(80)90237-8. [DOI] [PubMed] [Google Scholar]
- 186.Goffin J, Eisenhauer E. DNA methyltransferase inhibitors-state of the art. Ann. Oncol. 2002;13(11):1699–1716. doi: 10.1093/annonc/mdf314. [DOI] [PubMed] [Google Scholar]
- 187.Cheng JC, Yoo CB, Weisenberger DJ, et al. Preferential response of cancer cells to zebularine. Cancer Cell. 2004;6(2):151–158. doi: 10.1016/j.ccr.2004.06.023. [DOI] [PubMed] [Google Scholar]
- 188.Zhou L, Cheng X, Connolly BA, Dickman MJ, Hurd PJ, Hornby DP. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J. Mol. Biol. 2002;321(4):591–599. doi: 10.1016/S0022-2836(02)00676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cheng JC, Matsen CB, Gonzales FA, et al. Inhibition of DNA methylation and reactivation of silenced genes by zebularine. J. Natl Cancer Inst. 2003;95(5):399–409. doi: 10.1093/jnci/95.5.399. [DOI] [PubMed] [Google Scholar]
- 190.Cheng JC, Weisenberger DJ, Gonzales FA, et al. Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol. Cell. Biol. 2004;24(3):1270–1278. doi: 10.1128/MCB.24.3.1270-1278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Plumb JA, Strathdee G, Sludden J, Kaye SB, Brown R. Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res. 2000;60(21):6039–6044. [PubMed] [Google Scholar]
- 192.Nguyen CT, Weisenberger DJ, Velicescu M, et al. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002;62(22):6456–6461. [PubMed] [Google Scholar]
- 193.Fahrner JA, Eguchi S, Herman JG, Baylin SB. Dependence of histone modifications and gene expression on DNA hypermethylation in cancer. Cancer Res. 2002;62(24):7213–7218. [PubMed] [Google Scholar]
- 194.Kondo Y, Shen L, Issa JP. Critical role of histone methylation in tumor suppressor gene silencing in colorectal cancer. Mol. Cell. Biol. 2003;23(1):206–215. doi: 10.1128/MCB.23.1.206-215.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zhang L, Volinia S, Bonome T, et al. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc. Natl Acad. Sci. USA. 2008;105(19):7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Brueckner B, Boy RG, Siedlecki P, et al. Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res. 2005;65(14):6305–6311. doi: 10.1158/0008-5472.CAN-04-2957. [DOI] [PubMed] [Google Scholar]
- 197.Fang MZ, Wang Y, Ai N, et al. Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63(22):7563–7570. [PubMed] [Google Scholar]
- 198.Segura-Pacheco B, Trejo-Becerril C, Perez-Cardenas E, et al. Reactivation of tumor suppressor genes by the cardiovascular drugs hydralazine and procainamide and their potential use in cancer therapy. Clin. Cancer Res. 2003;9(5):1596–1603. [PubMed] [Google Scholar]
- 199.Villar-Garea A, Fraga MF, Espada J, Esteller M. Procaine is a DNA-demethylating agent with growth-inhibitory effects in human cancer cells. Cancer Res. 2003;63(16):4984–4989. [PubMed] [Google Scholar]
- 200.Yan L, Nass SJ, Smith D, Nelson WG, Herman JG, Davidson NE. Specific inhibition of DNMT1 by antisense oligonucleotides induces re-expression of estrogen receptor-α (ER) in ER-negative human breast cancer cell lines. Cancer Biol. Ther. 2003;2(5):552–556. doi: 10.4161/cbt.2.5.469. [DOI] [PubMed] [Google Scholar]
- 201.Gartel AL, Serfas MS, Tyner AL. p21 – negative regulator of the cell cycle. Proc. Soc. Exp. Biol. Med. 1996;213(2):138–149. doi: 10.3181/00379727-213-44046. [DOI] [PubMed] [Google Scholar]
- 202.Kelly WK, Marks PA. Drug insight: histone deacetylase inhibitors – development of the new targeted anticancer agent suberoylanilide hydroxamic acid. Nat. Clin. Pract. Oncol. 2005;2(3):150–157. doi: 10.1038/ncponc0106. [DOI] [PubMed] [Google Scholar]
- 203.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-κB action regulated by reversible acetylation. Science. 2001;293(5535):1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 204.Costanzo A, Merlo P, Pediconi N, et al. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell. 2002;9(1):175–186. doi: 10.1016/s1097-2765(02)00431-8. [DOI] [PubMed] [Google Scholar]
- 205.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408(6810):377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 206.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. STAT3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307(5707):269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 207.Dokmanovic M, Marks PA. Prospects: histone deacetylase inhibitors. J. Cell. Biochem. 2005;96(2):293–304. doi: 10.1002/jcb.20532. [DOI] [PubMed] [Google Scholar]
- 208.Cimini D, Mattiuzzo M, Torosantucci L, Degrassi F. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol. Biol. Cell. 2003;14(9):3821–3833. doi: 10.1091/mbc.E03-01-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Blagosklonny MV, Robey R, Sackett DL, et al. Histone deacetylase inhibitors all induce p21 but differentially cause tubulin acetylation, mitotic arrest, and cytotoxicity. Mol. Cancer Ther. 2002;1(11):937–941. [PubMed] [Google Scholar]
- 210.Carducci MA, Gilbert J, Bowling MK, et al. A Phase I clinical and pharmacological evaluation of sodium phenylbutyrate on an 120-h infusion schedule. Clin. Cancer Res. 2001;7(10):3047–3055. [PubMed] [Google Scholar]
- 211.Vigushin DM, Ali S, Pace PE, et al. Trichostatin A is a histone deacetylase inhibitor with potent antitumor activity against breast cancer in vivo. Clin. Cancer Res. 2001;7(4):971–976. [PubMed] [Google Scholar]
- 212.Fouladi M, Furman WL, Chin T, et al. Phase I study of depsipeptide in pediatric patients with refractory solid tumors: a children’s oncology group report. J. Clin. Oncol. 2006;24(22):3678–3685. doi: 10.1200/JCO.2006.06.4964. [DOI] [PubMed] [Google Scholar]
- 213.Klimek VM, Fircanis S, Maslak P, et al. Tolerability, pharmacodynamics, and pharmacokinetics studies of depsipeptide (romidepsin) in patients with acute myelogenous leukemia or advanced myelodysplastic syndromes. Clin. Cancer Res. 2008;14(3):826–832. doi: 10.1158/1078-0432.CCR-07-0318. [DOI] [PubMed] [Google Scholar]
- 214.Kosugi H, Ito M, Yamamoto Y, et al. In vivo effects of a histone deacetylase inhibitor, FK228, on human acute promyelocytic leukemia in NOD/Shi-scid/Scid mice. Jpn J. Cancer Res. 2001;92(5):529–536. doi: 10.1111/j.1349-7006.2001.tb01126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Sasakawa Y, Naoe Y, Inoue T, et al. Effects of FK228, a novel histone deacetylase inhibitor, on human lymphoma U-937 cells in vitro and in vivo. Biochem. Pharmacol. 2002;64(7):1079–1090. doi: 10.1016/s0006-2952(02)01261-3. [DOI] [PubMed] [Google Scholar]
- 216.Kelly WK, O’Connor OA, Krug LM, et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J. Clin. Oncol. 2005;23(17):3923–3931. doi: 10.1200/JCO.2005.14.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.O’connor OA, Heaney ML, Schwartz L, et al. Clinical experience with intravenous and oral formulations of the novel histone deacetylase inhibitor suberoylanilide hydroxamic acid in patients with advanced hematologic malignancies. J. Clin. Oncol. 2006;24(1):166–173. doi: 10.1200/JCO.2005.01.9679. [DOI] [PubMed] [Google Scholar]
- 218.Modesitt SC, Sill M, Hoffman JS, Bender DP. A Phase II study of vorinostat in the treatment of persistent or recurrent epithelial ovarian or primary peritoneal carcinoma: a gynecologic oncology group study. Gynecol. Oncol. 2008;109(2):182–186. doi: 10.1016/j.ygyno.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 219.Garber K. HDAC inhibitors overcome first hurdle. Nat. Biotechnol. 2007;25(1):17–19. doi: 10.1038/nbt0107-17. [DOI] [PubMed] [Google Scholar]
- 220.Plumb JA, Finn PW, Williams RJ, et al. Pharmacodynamic response and inhibition of growth of human tumor xenografts by the novel histone deacetylase inhibitor PXD101. Mol. Cancer Ther. 2003;2(8):721–728. [PubMed] [Google Scholar]
- 221.Qian X, Larochelle WJ, Ara G, et al. Activity of PXD101, a histone deacetylase inhibitor, in preclinical ovarian cancer studies. Mol. Cancer Ther. 2006;5(8):2086–2095. doi: 10.1158/1535-7163.MCT-06-0111. [DOI] [PubMed] [Google Scholar]
- 222.Steele NL, Plumb JA, Vidal L, et al. A Phase I pharmacokinetic and pharmacodynamic study of the histone deacetylase inhibitor belinostat in patients with advanced solid tumors. Clin. Cancer Res. 2008;14(3):804–810. doi: 10.1158/1078-0432.CCR-07-1786. [DOI] [PubMed] [Google Scholar]
- 223.Steele N, Finn P, Brown R, Plumb JA. Combined inhibition of DNA methylation and histone acetylation enhances gene re-expression and drug sensitivity in vivo. Br. J. Cancer. 2009;100(5):758–763. doi: 10.1038/sj.bjc.6604932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat. Genet. 1999;21(1):103–107. doi: 10.1038/5047. [DOI] [PubMed] [Google Scholar]
- 225.Anzai H, Frost P, Abbruzzese L. Synergistic cytotoxicity with 2′-deoxy-5-azacytidine and topotecan in vitro and in vivo. Cancer Res. 1992;52(8):2180–2185. [PubMed] [Google Scholar]
- 226.Strathdee G, Mackean MJ, Illand M, Brown R. A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene. 1999;18(14):2335–2341. doi: 10.1038/sj.onc.1202540. [DOI] [PubMed] [Google Scholar]
- 227.Branch P, Masson M, Aquilina G, Bignami M, Karran P. Spontaneous development of drug resistance: mismatch repair and p53 defects in resistance to cisplatin in human tumor cells. Oncogene. 2000;19(28):3138–3145. doi: 10.1038/sj.onc.1203668. [DOI] [PubMed] [Google Scholar]
- 228.Esteller M. Relevance of DNA methylation in the management of cancer. Lancet Oncol. 2003;4(6):351–358. doi: 10.1016/s1470-2045(03)01115-x. [DOI] [PubMed] [Google Scholar]
- 229.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 230.Thirlwell C, Eymard M, Feber A, et al. Genome-wide DNA methylation analysis of archival formalin-fixed paraffin-embedded tissue using the Illumina Infinium HumanMethylation27 BeadChip. Methods. 2010 doi: 10.1016/j.ymeth.2010.04.012. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 231.Beck S, Rakyan VK. The methylome: approaches for global DNA methylation profiling. Trends Genet. 2008;24(5):231–237. doi: 10.1016/j.tig.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 232.Thu KL, Pikor LA, Kennett JY, Alvarez CE, Lam WL. Methylation analysis by DNA immunoprecipitation. J. Cell. Physiol. 2010;222(3):522–531. doi: 10.1002/jcp.22009. [DOI] [PubMed] [Google Scholar]
- 233.Rauch T, Pfeifer GP. Methylated-CpG island recovery assay: a new technique for the rapid detection of methylated-CpG islands in cancer. Lab. Invest. 2005;85(9):1172–1180. doi: 10.1038/labinvest.3700311. [DOI] [PubMed] [Google Scholar]
- 234.Bernstein BE, Kamal M, Lindblad-Toh K, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120(2):169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 235.Ongenaert M, Van Neste L, De Meyer T, Menschaert G, Bekaert S, Van Criekinge W. PubMeth: a cancer methylation database combining text-mining and expert annotation. Nucleic Acids Res. 2008;36(Database issue):D842–D846. doi: 10.1093/nar/gkm788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Strathdee G, Appleton K, Illand M, et al. Primary ovarian carcinomas display multiple methylator phenotypes involving known tumor suppressor genes. Am. J. Pathol. 2001;158(3):1121–1127. doi: 10.1016/S0002-9440(10)64059-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Niederacher D, Yan HY, An HX, Bender HG, Beckmann MW. CDKN2a gene inactivation in epithelial sporadic ovarian cancer. Br. J. Cancer. 1999;80(12):1920–1926. doi: 10.1038/sj.bjc.6690621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Liu Z, Wang LE, Wang L, et al. Methylation and messenger RNA expression of p15INK4b but not p16INK4a are independent risk factors for ovarian cancer. Clin. Cancer Res. 2005;11(13):4968–4976. doi: 10.1158/1078-0432.CCR-04-2293. [DOI] [PubMed] [Google Scholar]
- 239.Shivapurkar N, Toyooka S, Toyooka Ko, et al. Aberrant methylation of trail decoy receptor genes is frequent in multiple tumor types. Int. J. Cancer. 2004;109(5):786–792. doi: 10.1002/ijc.20041. [DOI] [PubMed] [Google Scholar]
- 240.Cai LY, Abe M, Izumi S, Imura M, Yasugi T, Ushijima T. Identification of prtfdc1 silencing and aberrant promoter methylation of GPR150, ITGA8 and HOXD11 in ovarian cancers. Life sciences. 2007;80(16):1458–1465. doi: 10.1016/j.lfs.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 241.Shen DH, Chan KY, Khoo US, et al. Epigenetic and genetic alterations of p33ING1b in ovarian cancer. Carcinogenesis. 2005;26(4):855–863. doi: 10.1093/carcin/bgi011. [DOI] [PubMed] [Google Scholar]
- 242.Czekierdowski A, Czekierdowska S, Szymanski M, Wielgos M, Kaminski P, Kotarski J. Opioid-binding protein/cell adhesion molecule-like (OPCML) gene and promoter methylation status in women with ovarian cancer. Neuro. Endocrinol. Lett. 2006;27(5):609–613. [PubMed] [Google Scholar]
- 243.Choi YL, Kang SY, Shin YK, et al. Aberrant hypermethylation of rassf1a promoter in ovarian borderline tumors and carcinomas. Virchows Arch. 2006;448(3):331–336. doi: 10.1007/s00428-005-0091-3. [DOI] [PubMed] [Google Scholar]
- 244.Ma L, Zhang JH, Liu FR, Zhang X. Hypermethylation of promoter region of RASSF1A gene in ovarian malignant epithelial tumors. Zhonghua Zhong Liu Za Zhi. 2005;27(11):657–659. [PubMed] [Google Scholar]
- 245.Widschwendter A, Muller HM, Hubalek MM, et al. Methylation status and expression of human telomerase reverse transcriptase in ovarian and cervical cancer. Gynecol. Oncol. 2004;93(2):407–416. doi: 10.1016/j.ygyno.2004.01.036. [DOI] [PubMed] [Google Scholar]
- 246.Okochi-Takada E, Nakazawa K, Wakabayashi M, et al. Silencing of the uchl1 gene in human colorectal and ovarian cancers. Int. J. Cancer. 2006;119(6):1338–1344. doi: 10.1002/ijc.22025. [DOI] [PubMed] [Google Scholar]
- 247.Kantarjian HM, O’Brien S, Cortes J, et al. Results of decitabine (5-aza-2′deoxycytidine) therapy in 130 patients with chronic myelogenous leukemia. Cancer. 2003;98(3):522–528. doi: 10.1002/cncr.11543. [DOI] [PubMed] [Google Scholar]
- 248.Tam KF, Liu VW, Liu SS, et al. Methylation profile in benign, borderline and malignant ovarian tumors. J. Cancer Res. Clin. Oncol. 2007;133(5):331–341. doi: 10.1007/s00432-006-0178-5. [DOI] [PubMed] [Google Scholar]
- 249.Graham JS, Kaye SB, Brown R. The promises and pitfalls of epigenetic therapies in solid tumours. Eur. J. Cancer. 2009;45(7):1129–1136. doi: 10.1016/j.ejca.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 250.Chuang JC, Yoo CB, Kwan JM, et al. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2′-deoxycytidine. Mol. Cancer Ther. 2005;4(10):1515–1520. doi: 10.1158/1535-7163.MCT-05-0172. [DOI] [PubMed] [Google Scholar]
- 251.Kattan MW, Shariat SF, Andrews B, et al. The addition of interleukin-6 soluble receptor and transforming growth factor-β1 improves a preoperative nomogram for predicting biochemical progression in patients with clinically localized prostate cancer. J. Clin. Oncol. 2003;21(19):3573–3579. doi: 10.1200/JCO.2003.12.037. [DOI] [PubMed] [Google Scholar]
- 252.Choan E, Segal R, Jonker D, et al. A prospective clinical trial of green tea for hormone refractory prostate cancer: an evaluation of the complementary/alternative therapy approach. Urologic Oncol. 2005;23(2):108–113. doi: 10.1016/j.urolonc.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 253.Jatoi A, Ellison N, Burch PA, et al. A Phase II trial of green tea in the treatment of patients with androgen independent metastatic prostate carcinoma. Cancer. 2003;97(6):1442–1446. doi: 10.1002/cncr.11200. [DOI] [PubMed] [Google Scholar]
- 254.Yung R, Chang S, Hemati N, Johnson K, Richardson B. Mechanisms of drug-induced lupus. IV. Comparison of procainamide and hydralazine with analogs in vitro and in vivo. Arthritis Rheum. 1997;40(8):1436–1443. doi: 10.1002/art.1780400811. [DOI] [PubMed] [Google Scholar]
- 255.Mai A, Altucci L. Epi-drugs to fight cancer: from chemistry to cancer treatment, the road ahead. Int. J. Biochem. Cell Biol. 2009;41(1):199–213. doi: 10.1016/j.biocel.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 256.Butler KV, Kozikowski AP. Chemical origins of isoform selectivity in histone deacetylase inhibitors. Curr. Pharm. Des. 2008;14(6):505–528. doi: 10.2174/138161208783885353. [DOI] [PubMed] [Google Scholar]
- 257.Qian X, Ara G, Mills E, Larochelle WJ, Lichenstein HS, Jeffers M. Activity of the histone deacetylase inhibitor belinostat (PXD101) in preclinical models of prostate cancer. Int. J. Cancer. 2008;122(6):1400–1410. doi: 10.1002/ijc.23243. [DOI] [PubMed] [Google Scholar]
- 258.Hubbert C, Guardiola A, Shao R, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417(6887):455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 259.Marchion D, Munster P. Development of histone deacetylase inhibitors for cancer treatment. Expert Rev. Anticancer Ther. 2007;7(4):583–598. doi: 10.1586/14737140.7.4.583. [DOI] [PubMed] [Google Scholar]
- 260.Dyer ES, Paulsen MT, Markwart SM, Goh M, Livant DL, Ljungman M. Phenylbutyrate inhibits the invasive properties of prostate and breast cancer cell lines in the sea urchin embryo basement membrane invasion assay. Int. J. Cancer. 2002;101(5):496–499. doi: 10.1002/ijc.10609. [DOI] [PubMed] [Google Scholar]
- 261.Gore SD, Samid D, Weng LJ. Impact of the putative differentiating agents sodium phenylbutyrate and sodium phenylacetate on proliferation, differentiation, and apoptosis of primary neoplastic myeloid cells. Clin. Cancer Res. 1997;3(10):1755–1762. [PubMed] [Google Scholar]
- 262.Melchior SW, Brown LG, Figg WD, et al. Effects of phenylbutyrate on proliferation and apoptosis in human prostate cancer cells in vitro and in vivo. Int. J. Oncol. 1999;14(3):501–508. doi: 10.3892/ijo.14.3.501. [DOI] [PubMed] [Google Scholar]
- 263.Svechnikova I, Gray SG, Kundrotiene J, Ponthan F, Kogner P, Ekstrom TJ. Apoptosis and tumor remission in liver tumor xenografts by 4-phenylbutyrate. Int. J. Oncol. 2003;22(3):579–588. [PubMed] [Google Scholar]
- 264.Gottlicher M, Minucci S, Zhu P, et al. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 2001;20(24):6969–6978. doi: 10.1093/emboj/20.24.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Garcia-Manero G, Assouline S, Cortes J, et al. Phase I study of the oral isotype specific histone deacetylase inhibitor MGCD0103 in leukemia. Blood. 2008;112(4):981–989. doi: 10.1182/blood-2007-10-115873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Tan J, Cang S, Ma Y, Petrillo RL, Liu D. Novel histone deacetylase inhibitors in clinical trials as anti-cancer agents. J. Hematol. Oncol. 2010;3:5. doi: 10.1186/1756-8722-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
■Website
- 301.Clinical Trials (NIH) www.clinicaltrials.gov.