Abstract
The protein corona that forms around nanoparticles in vivo is a critical factor that affects their physiological response. The potential to manipulate nanoparticle characteristics such that either proteins advantageous for delivery are recruited and/or detrimental proteins are avoided offers exciting possibilities for improving drug delivery. In this work, we used nanoliquid chromatography tandem mass spectrometry to characterize the corona of five lipid formulations after incubation in mouse and human plasma with the hope of providing data that may contribute to a better understanding of the role played by both the nanoparticle properties and the physiological environment in recruiting specific proteins to the corona. Notably, we showed that minor changes in the lipid composition might critically affect the protein corona composition demonstrating that the surface chemistry and arrangement of lipid functional groups are key players that regulate the liposome-protein interactions. Notably, we provided evidence that the protein corona that forms around liposomes is strongly affected by the physiological environment, i.e., the serum type. These results are likely to suggest that the translation of novel pharmaceutical formulations from animal models to the clinic must be evaluated on a case-by-case basis.
Keywords: nanoparticles, protein corona, nanoliquid chromatography tandem mass spectrometry, liposomes, human plasma, mouse plasma
1. Introduction
The use of nanoparticles for intravenous drug delivery continues to gain increasing interest. Although much emphasis is placed on methods for producing nanoparticles possessing specific sizes and surface characteristics that facilitate prolonged circulation and delivery, it is well recognized that exposure to blood proteins typically results in significant changes in particle size and zeta-potential [1–4]. While previous studies have utilized western blots to monitor the adsorption of specific blood proteins to nanoparticles, modern mass spectrometry/proteomic methods allow the individual proteins comprising the entire corona to be identified [5–8]. Multiple studies utilizing proteomics to characterize the corona of different nanoparticles have been reported, and it is clear that alterations in the particle properties can have profound effects on the proteins that accumulate in the corona [1,9–14]. Furthermore, it is well documented that protein adsorption can play a crucial role in biodistribution and delivery efficiency. The current study utilizes modern proteomic methods to characterize the protein corona of five different liposome formulations after exposure to mouse and human plasma.
The innate immune system and complement activation are known to play a significant role in the infusion reaction that is commonly observed upon intravenous administration of liposome [15,16]. Other studies have demonstrated that the injection of nanoparticles can elicit the production of IgM and IgG antibodies that result in more rapid clearance of nanoparticles upon subsequent administration [17,18]. In each of these cases, the process is initiated by the adsorption of blood proteins onto the surface of nanoparticles upon intravenous administration. It follows that identification of proteins comprising the corona of different nanoparticles may ultimately allow researchers to predict particle biodistribution and clearance, and potentially design nanoparticles with improved surface properties to enhance drug delivery in vivo. While certain proteins (e.g., C3, C5) are known to play a key role in complement activation and clearance via the reticuloendothelial system (RES), there may be other proteins that are involved in biodistribution that have yet to be identified. In addition, the potential to alter nanoparticle characteristics such that either proteins beneficial for delivery are recruited and/or detrimental proteins are avoided offers exciting possibilities for improving drug delivery [19].
In this study we characterized the corona of five lipid formulations that have been used to deliver DNA to mammalian cells in culture. Although previous studies have shown that the coronas of these formulations differ under cell culture conditions employing fetal bovine serum, such studies are not relevant to in vivo studies conducted in mice or humans. Accordingly, we characterized protein coronas after incubation in mouse and human plasma with the hope of providing data that may contribute to a better understanding of the role of nanoparticle properties in recruiting specific proteins to the corona.
2. Materials and methods
2.1. Chemicals and standards
DOTAP (1,2-dioleoyl-3-trimethylammonium-propane), DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine), DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine), 20:0 PC (1,2-diarachidoylsn- glycero-3-phosphocholine) lipids were acquired from Avanti Polar Lipids (Alabaster, AL). D-Sphingosine and Cholesterol (Chol) were purchased by Sigma-Aldrich (Milan, Italy). All lipids were used without further refinement. The sequencing grade modified trypsin was purchased from Promega (Madison, WI, USA). Organic solvents were from Sigma Aldrich (Milan, Italy). Ultrapure water (resistivity 18.2 MΩ cm) was achieved by an Arium water purification system (Sartorius, Florence, Italy).
2.2. Mouse plasma (MP), human plasma (HP)
Lyophilized plasma from mouse was purchased by Sigma-Aldrich (Milan, Italy) and dissolved in 1 mL of ultrapure water. Human whole blood was obtained by venipuncture of ten healthy volunteers aged 20–40 years at the Department of Experimental Medicine (‘Sapienza’ University of Rome) according with the institutional bioethics code. After blood collection, K2 EDTA anticoagulant and protease inhibitors cocktail were immediately added to HP. Aliquots were stored at −80 °C in labeled Protein LoBind tubes (Eppendorf, Milan, Italy) to ensure plasma stability until use. When used, both MP and HP aliquots were thawed at 4°C left to warm at room temperature and centrifuged for 10 minutes at 18000 g to eliminate insoluble protein aggregates.
2.3. Liposomes preparation
Shingosine-Chol-DSPC (3:2:5 molar ratio), DOTAP-Chol-DPPC (3:2:5 molar ratio), DOTAP-Chol-DSPC (3:2:5 molar ratio), DOTAP-Chol-PC, (3:2:5 molar ratio) and Chol-PC (2:8) liposomes were prepared in accordance with standard procedures by dissolving appropriate amounts of lipids in chloroform. Lipid films were hydrated with ultrapure water for size and zeta-potential measurements (final lipid concentration 1 mg/mL). For proteomics experiments lipid films were hydrated with a dissolving buffer (Tris–HCl, pH 7.4, 10 mmol/L; NaCl, 150 mmol/L; EDTA, 1 mmol/L) and stored at 4°C. Liposomes were sonicated for 10 minutes with an ultratip sonicator
2.4. Size and Zeta-potential experiments
Liposomes were measured at 0, 24 and 72 hours from their preparation. All size and zeta-potential experiments were made at 37 °C on a Zetasizer Nano ZS90 (Malvern, U.K.) spectrometer equipped with a 5 mW HeNe laser (wavelength λ = 632.8 nm) and a digital logarithmic correlator. By using the CONTIN approach, the normalized intensity autocorrelation functions were analyzed in order to obtain the distribution of the particles diffusion coefficient (D). D is translated into hydrodynamic radius (RH) through the Stokes-Einstein equation RH =KBT/(6πηD), where KBT is the thermal energy and η is the solvent viscosity. The same apparatus used for size measurements was employed for zeta-potential experiments. Thhe mobility, u, was measured by means of the laser Doppler electrophoresis technique. The mobility was converted into the zeta-potential through the Smoluchowski relation (zeta-potential = uη/ε), where η and ε are the viscosity and the permittivity of the solvent phase respectively. Results are given as mean ± standard deviation of not less than five replicates.
2.5. Proteomics experiments
One hundred seventy microliters of liposomes (1 mg/mL) were incubated with 170 µL of either mouse or human plasma at 37 °C for 1 h. After incubation, liposome-protein complexes were centrifuged for 15 min at 18000 × g. Unbound proteins were removed washing pellets three times with 200 µL of the dissolving buffer. To assure statistical significance of data, three experimental replicates were performed.
2.5.1. In solution digestion and desalting
The protein pellets were resuspended in 50 µL of a denaturant buffer composed of 8 mol/L urea in 50 mmol/L NH4HCO3, 2.5 µL DTT 200 mmol/L in 50 mmol/L NH4HCO3 were added to break disulfide bonds and samples were incubated for 1 h at 37 °C. Then 10 µL of iodoacetamide 200 mmol/L in 50 mmol/L NH4HCO3 were added to alkylate thiol groups and samples were left in the dark at room temperature for 1 h. Finally any leftover alkylating reagent activity was removed by the addition of 10 µL DTT solution and incubation at 37 °C for 1 h. Then samples were diluted with 50 mmol/L NH4HCO3 to reach the final 1mol/L urea concentration and trypsin was added in order to ensure a minimum enzyme-to-substrate ratio of 1:20. Enzymatic digestion was carried out overnight at 37 °C and quenched with trifluoroacetic acid (TFA). Digested samples were desalted using an solid phase extraction C18 column (Bond Elut 1CC LRCC18, Varian, Palo Alto, CA, USA) and eluted with 0.5 mL H2O:ACN (50:50, v/v) solution containing 0.1 % TFA. After lyophilization in a Speed-Vac apparatus (mod. SC 250 Express; Thermo Savant, Holbrook, NY, USA), samples were reconstituted with 0.1% HCOOH solution to obtain a final concentration of 0.32 mg/mL, and stored at -80°C until nanoLC-MS/MS analysis.
2.5.2. Nanoliquid chromatography tandem mass spectrometry (nanoLC-MS/MS)
Tryptic peptides were analyzed by a Dionex Ultimate 3000 (Sunnyvale, CA, USA) nanoLC system connected to the hybrid mass spectrometer LTQ Orbitrap XL (Thermo Fisher Scientific Bremen, Germany), equipped with a nanoelectrospray ion source. Ten microliters of peptide mixture were on-line enriched onto a 300 µm i.d. × 5 mm Acclaim PepMap 100 C18 (5 µm particle size, 100 Å pore size) µ-precolumn (Dionex), using a premixed mobile phase H2O:ACN 98:2 (v/v) containing 0.1% HCOOH, at 10 µL min−1 flow-rate. Peptide mixtures were separated by reverse-phase chromatography on in-house manufactured 25 cm long silica micro-column, with a 75 µm i.d and packed with ReproSil-PurC18-AQ 2.2 µm resin (Dr. Maisch GmbH, Ammerbuch, Germany). Mobile phase was H2O:ACN (98:2, v/v) (A) and ACN: H2O (90:10 v/v) (B), both with 0.1% (v/v) HCOOH. After a 5 min isocratic step at 5%, B was linearly increased from 5% to 30% within 130 min and then to 45% in 10 min. After that, B was increased to 80% within 10 min and kept constant for 10 min. Then, B was decreased to 5% within 1 min and kept constant for the following 30 min to rinse the column. Separation was made at a flow rate of 300 nL min−1. MS spectra were collected over the 400–1800 m/z range at60000 resolution (FWHM), operating in the data dependent mode, thus acquiring the MS/MS spectra of the five most intense ions with a charge state greater than 1, using a dynamic exclusion of 60 s. Collision induced dissociation was performed with a normalized collision energy of 35 V. Each of the three experimental replicates was analyzed in triplicate (nine measurements in total) to assess the variation due to the experimental procedure and to increase the number of identified proteins.
2.5.3. Data analysis and protein validation
Xcalibur (v.2.07, Thermo Fisher Scientific) raw data files were submitted to Proteome Discover (1.2 version, Thermo Scientific) for database search using Mascot (version 2.3.2 Matrix Science). Data were searched against Swiss Prot database (57.15 version, 20266 sequences) using the decoy search option of Mascot. Enzymatic digestion with trypsin was selected, along with maximum 2 missed cleavages, peptide charges +2 and +3, 10 ppm precursor mass tolerance and 0.8 Da fragment mass tolerance; acetylation (N-term), oxidation (M) and deamidation (N, Q) were used as dynamic modifications; carbamidomethylation (C) was used as static modification. The Scaffold software (version 3.1.2, Proteome Software Inc.) was used to validate MS/MS-based peptide and protein identifications and for label-free relative quantitation based on spectral counting. The peptide and protein probabilities were set to minimum 95% and 99%, respectively, with at least two unique peptides for each identification. For protein quantitative analysis, Scaffold software allows the normalization of the spectral countings (normalized spectral countings, NSCs) and offers various statistical tests to identify significant abundance differences in two or more categories. The mean value of NSCs from three experimental replicates was calculated for each protein and then normalized to the protein molecular weight, MW. Finally, the relative protein abundance (RPA) of protein k in the corona, RPAk, was calculated by applying the following equation
| (1) |
This correction takes into account the protein size and evaluates the real contribution of each protein in the corona.
3. Results and Discussion
3.1. Size and zeta-potential of cationic liposomes
Size and zeta-potential of liposomes are reported in Table 1. Liposomes were measured at 0, 24, 48, 72 hours from their preparation. At 0 hr all formulations showed hydrodynamic diameter lower than 260 nm. The only exception is represented by DOTAP-Chol-DSPC liposomes, whose mean hydrodynamic diameter was about twice larger (470 ± 44 nm). Measurements at different times from the preparation showed that liposomes dispersions are quite stable in size with the exception of DOTAP-Chol-PC liposomes (100 nm size increase at 72 hours after preparation). All formulations exhibited a positive zeta-potential at t=0 (30–40 mV). This result is expected considering that four liposomal formulations are composed of ternary mixtures of cholesterol, saturated phosphocholine of varying chain length, and a cationic lipid (Sphingosine or DOTAP) that confers positive charge to the particles. In contrast, Chol-PC liposomes that do not contain cationic lipids, exhibit a slightly negative zeta-potential (−7.64 ± 1.77 mV). Zeta-potential values slightly increased over the time except for DOTAP-Chol-DSPC liposomes. This formulation showed an oscillation in zeta-potential values that, together with size results, suggest that the dispersion is not stable most likely due to particle aggregation.
Table 1.
Size and zeta-potential of all liposome formulations as a function of time up to 72 hours from their preparation.
| D (nm) | Zeta-potential (mV) | |||||||
|---|---|---|---|---|---|---|---|---|
| 0 h | 24 h | 48 h | 72 h | 0 h | 24 h | 48 h | 72 h | |
| Sphingosine-CHOL-PC | 193.4±15.2 | 203±9 | 192.7±11.8 | 203±9 | 39.9±1.8 | 40.8±2.5 | 41±3 | 33.7±2.9 |
| DOTAP-CHOL-DPPC | 209.9±25.3 | 246.3±12.2 | 204.2±16.9 | 204.3±15.3 | 32.5±1.1 | 34±2 | 37.9±2.5 | 35.5±3.2 |
| DOTAP-CHOL-DSPC | 470.3±44.4 | 421.8±30.5 | 404.9±32.4 | 427.8±30.5 | 31.1±1.1 | 37.1±2.9 | 37±3 | 41.2±1.9 |
| DOTAP-CHOL-PC | 254.6±22.9 | 244.3±20.3 | 258.4±30.5 | 351.2±20.8 | 29.7±0.8 | 45.7±5.3 | 30±4 | 35.8±1.2 |
| CHOL-PC | 191±23 | 248.8±34.5 | 265.8±36.5 | 230.5±15.5 | −7.6±1.8 | −10.9±1.1 | −8.2±0.8 | −7.7±0.9 |
3.2. Protein identification
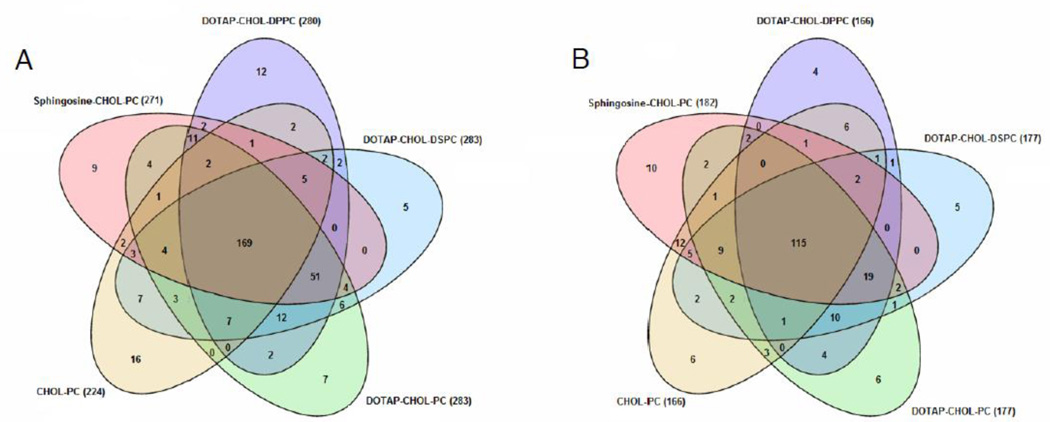
A complete comprehension of the interactions between liposomes and biological fluids is essential for the step from in vitro to in vivo experiments. To characterize and quantify the proteins adsorbed onto the liposome surface, we employed high-resolution nanoLC-MS/MS. In Tables S1 and S2 we reported all the identified proteins adsorbed on the five liposomal formulations analyzed after 1 h incubation with both MP and HP. Commons proteins among formulations are reported in Tables S3 and S4. A total of 224 proteins were reproducibly detected from Chol-PC/MP complexes, while approximtaely 280 proteins were detected from DOTAP-Chol-DPPC/MP DOTAP-Chol-DSPC/MP and DOTAP-Chol-PC/MP complexes. A total of 271 of proteins were detected for Shingosine-Chol-DSPC/MP complexes. These findings suggest that the protein corona absorption is affected by both surface charge and chemistry. Nonetheless, the number of identified proteins is in good agreement with previous studies, showing that the nanoparticle-protein corona typically consists of hundreds of proteins [14,20,21]. With the proteins being identified, we compared the composition of different formulations (Fig. 1). Venn diagrams depicting the detected proteins of Fig. 1A demonstrated that 169 proteins were in common to all formulations. The corona of liposome-HP complexes was found to be much less enriched in proteins than that of liposome-MP complexes, with a lower number of proteins (115) being in common between liposomal formulations (Fig. 1B). This observation indicates that the physiological environment is a “key” factor shaping the protein corona. The quantitative determination of the “protein corona” is a fundamental step towards a better understanding of the protein absorption process and its effect on NP behavior in vivo. Thus, in the following section identified proteins were ranked by their molecular mass, charge and physiological function
Fig. 1.
Venn diagram reporting the number of proteins identified in the Sphingosine-CHOL-PC, DOTAP-CHOL-DPPC, DOTAP-CHOL-DSPC, DOTAP-CHOL-PC and CHOL-PC containing liposomes after interaction with MP (A) and HP (B).
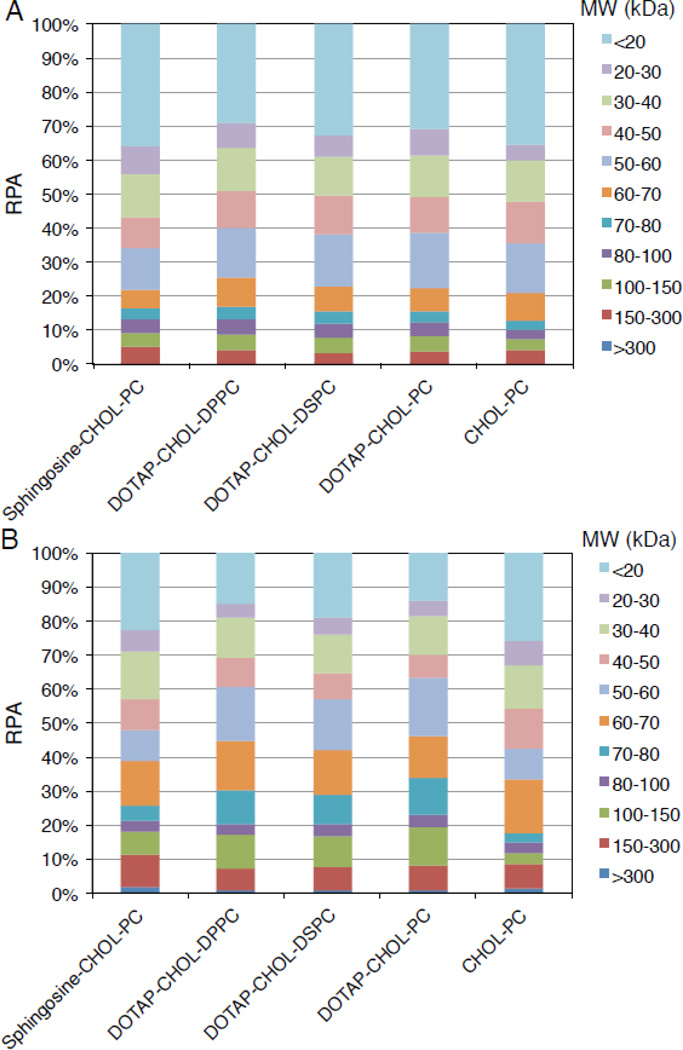
3.3. Protein classification by molecular mass
First, we classified proteins according to their molecular mass (Fig. 2). As Fig. 2 shows, both liposome-MP and liposome-HP complexes were enriched in low molecular mass proteins (< 20 kDa). Proteomics data show that proteins < 100 kDa constituted the majority (about 92%) of the corona components for each liposome-MP complex. A slight difference was found for liposome-HP complexes. Proteins < 100 kDa constituted about the 82% of the corona components except for Chol-PC liposomes (88%). These results are in contrast with previous work on inorganic nanomaterials (e.g. silica nanoparticles) where high molecular mass proteins constituted the majority of the protein corona [21]. Results reported in Fig. 2 suggest that the corona composition seems to be influenced by the biological medium (i.e. MP vs HP) to a greater extent than by lipid composition.
Fig. 2.
Relative protein abundance (RPA) of corona proteins classified according to their calculated molecular mass for Sphingosine-CHOL-PC, DOTAP-CHOL-DPPC, DOTAP-CHOL-DSPC, DOTAP-CHOL-PC and CHOL-PC liposomes after interaction with MP (A) and HP (B).
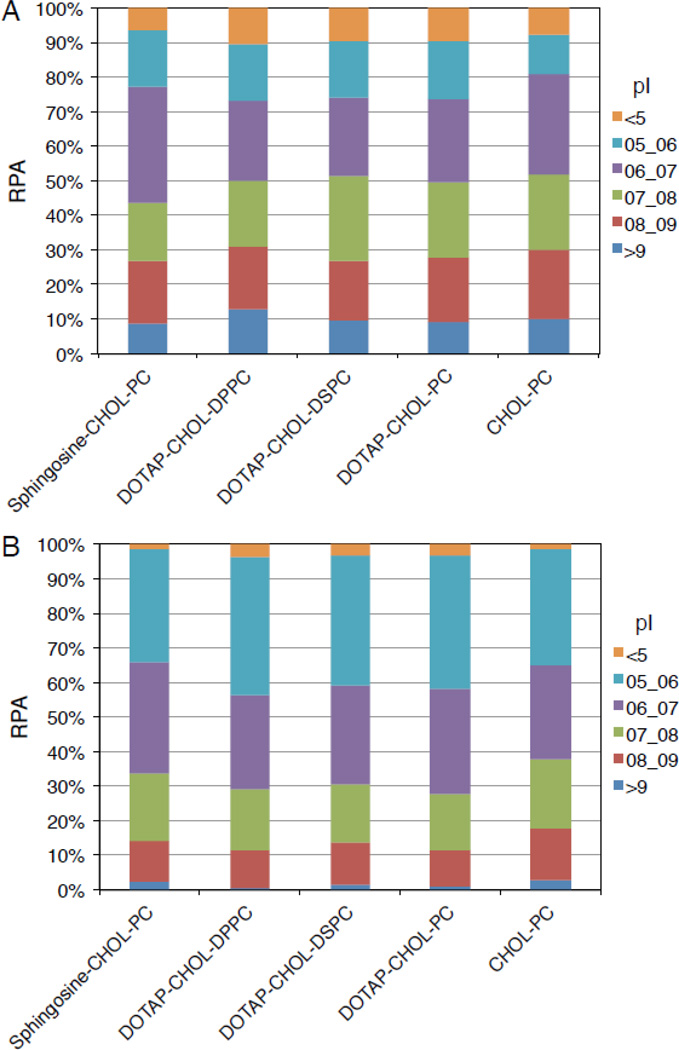
3.4. Protein classification by charge
Further bioinformatic analyses were executed on the 25 most abundant proteins, which constitute 50–60% of the whole “protein corona”. Proteins were classified based on their isoelectric point (Fig. 3). Fig. 3 shows that about 50% and 70% of the 25 hits display a negative charge (isoelectric point (pI) < 7) in liposome-MP and liposome-HP complexes, respectively. Protein absorption on cationic liposomes is generally thought to be driven by electrostatic interactions between negatively charged proteins and positively charged lipid head groups [6,22]. On the contrary, our results show that a significant portion of the cationic liposome-protein corona is made by positively charged proteins. It is noteworthy that the protein coronas of cationic and anionic liposomes are pretty similar to each other. According to what has already been described for inorganic nanoparticles [23], our data confirm that classifying liposomes based on charge is not sufficient to fully describe the liposome-protein corona composition. Protein adsorption to a liposome results from either higher protein affinity, or the presence of more protein binding sites. Evidently, the chemistry, and arrangement of surface lipid functional groups are essential determinants of liposome–protein interactions. For each formulation, significant changes between the protein coronas following incubation with MP and HP were found. Since the protein corona ultimately controls the nanoparticle physiological response (circulation times, immune response, selective targeting, etc.), a relevant implication of the present investigation is that the use of animal models does not ensure the direct extension of findings to humans [24].
Fig. 3.
Relative protein abundance (RPA) of corona proteins classified according their calculated isoelectric point for Sphingosine-CHOL-PC, DOTAP-CHOL-DPPC, DOTAP-CHOL-DSPC, DOTAP-CHOL-PC and CHOL-PC liposomes after interaction with MP (A) and HP (B).
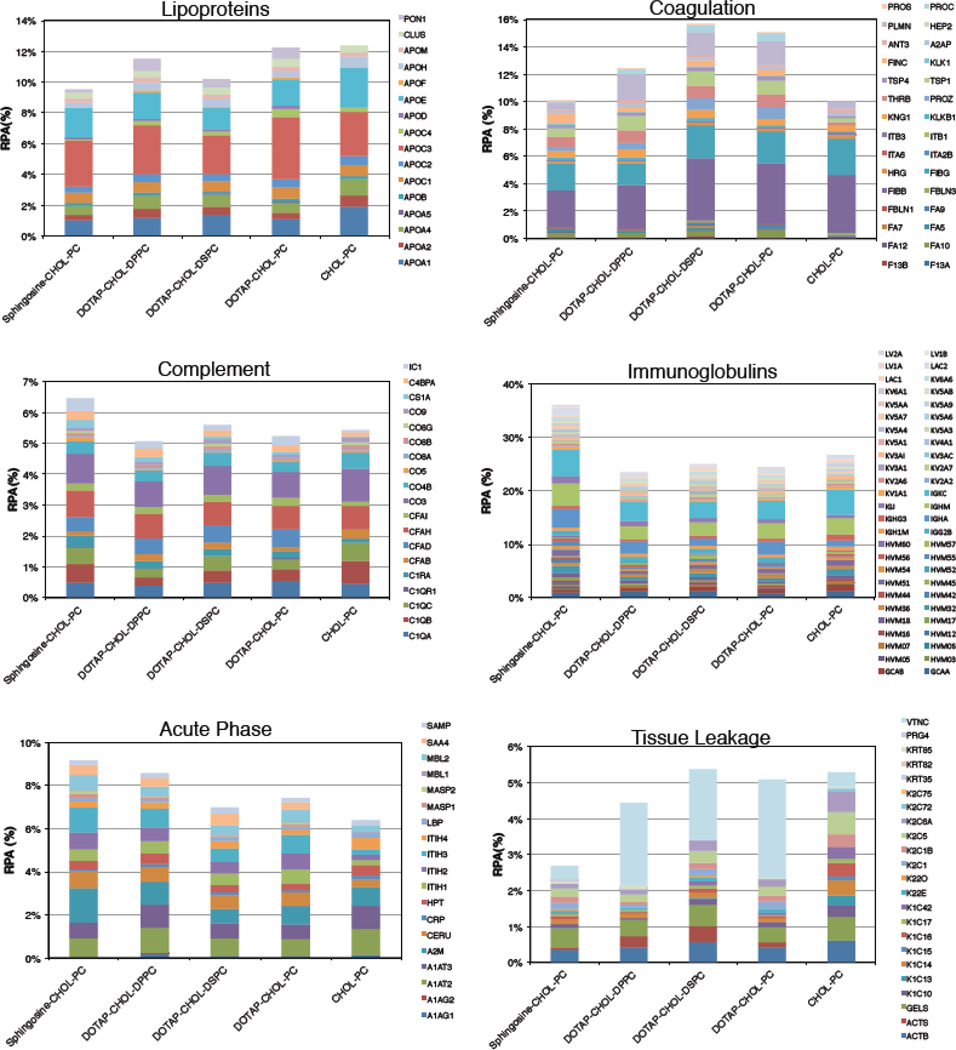
3.5. Protein classification by physiological function
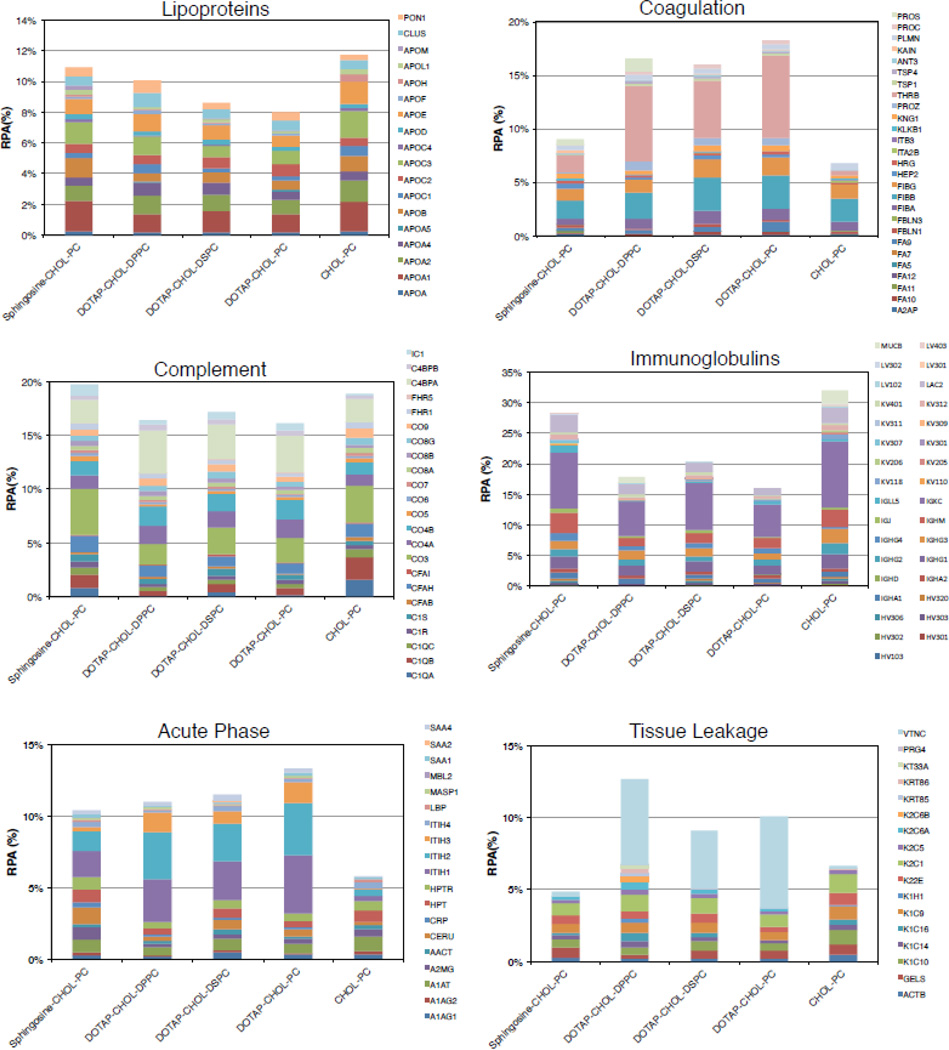
Lastly, a further bioinformatic analysis allowed us to classify proteins with respect to their physiological function (Figs. 4 and 5). RPAs of lipoproteins were found to be quite similar for liposome-MP (Fig. 4; 9.2% < RPA < 12.5%) and liposome-HP complexes (Fig. 5; 8% < RPA < 11.8%). Apo A1, Apo C3 and Apo E are the most abundant proteins and together constitute about 50% and 30% of all the lipoproteins identified in the corona of liposome-MP and liposome-HP complexes respectively. In particular, ApoC3 promotes interaction with scavenger receptor class B type 1 (SRBI) that, in turn, mediates the lipid transfer between low- and high-density lipoproteins and cells, while Apo E can affect the hepatic uptake of liposomes.
Fig. 4.
Bioinformatic classification of proteins identified in the corona of Sphingosine-CHOL-PC, DOTAP-CHOL-DPPC, DOTAP-CHOL-DSPC, DOTAP-CHOL-PC and CHOL-PC liposomes after 1-hour exposure to MP. The relative protein abundances (RPAs) of total proteins are shown. Detailed values for all individual proteins are available in Supporting Information
Fig. 5.
Bioinformatic classification of proteins identified in the corona of Sphingosine-CHOL-PC, DOTAP-CHOL-DPPC, DOTAP-CHOL-DSPC, DOTAP-CHOL-PC and CHOL-PC liposomes after 1-hour exposure to HP. The relative protein abundances (RPAs) of total proteins are shown. Detailed values for all individual proteins are available in Supporting Information.
Also, we specifically considered the coagulation system proteins, and concluded that: i) following incubation with either MP and HP, coagulation proteins were more abundant in the coronas of DOTAP-containing liposomes than in those of Sphingosine-Chol-PC and Chol-PC liposomes; (ii) the coronas of DOTAP-containing liposomes were affected by both PC length and biological medium; (iii) The enrichment was highly dependent on the biological medium. The most abundant proteins were found to be FIBG, FIBB, PLMN, TSP1 for liposome-MP complexes, while THRB PROS, FIBA, and were the most enriched for liposome-HP complexes. These findings are possibly relevant since proteins involved in coagulation not only contain factors that stimulate, but also those that counteract, coagulation cascades.
Fig. 4 shows that complement proteins were found to be poorly adsorbed on liposomes incubated with MP (Fig. 4; 5% < RPA < 6.4%) with minor differences between formulations. In contrast, complement proteins were abundant in the coronas of liposome-HP complexes (Fig. 5; 16% < RPA < 19.8%) (the most enriched proteins were CO3, CO4B, CO4A and C4BPA). This finding is interesting because complement proteins promote elimination from systemic circulation, accumulation in the liver and spleen and clearance. Therefore, the higher levels of complement proteins after incubation in HP, as compared to MP, indicate that circulation times in human patients may be shorter than that observed in mouse models.
For each cationic formulation, Igs were more enriched in the corona of liposome-MP complexes (Fig. 4; 23% < RPA < 36%) than in their human counterpart (Fig. 5; 16% < RPA < 28%). Based on these results, one may suppose a different immune response to the administration of the investigated cationic liposomes in mice and humans. However, it is noteworthy that Chol-PC liposomes exhibited the opposite trend in Ig enrichment (RPA=27.8% and 32% for liposome-MP and liposome-HP complexes, respectively). It is also notable that the coronas of Sphingosine-containing liposomes were the most enriched ones. This finding means that Sphingosine has a higher affinity for Igs than that of DOTAP. Following incubation with MP, RPAs of identified acute phase proteins were very similar to each other, with Sphingosine-CHOL-PC and CHOL-PC liposomes showing the highest and lowest affinity for this class of proteins, respectively. Comparing cationic liposome-HP complexes (Fig. 5), we note that DOTAP-containing liposomes bind more acute phase proteins than Sphingosine. Even in the case of interaction with HP, CHOL-PC liposomes exhibited the lowest affinity for acute phase proteins. Further studies would be needed to determine if these clear differences between the cationic formulations and the one anionic formulation we investigated (CHOL-PC) are due predominantly to their opposite charge.
Lastly, we considered tissue leakage proteins. DOTAP based formulations follow the same trend in contrast with CHOL-PC and Sphingosine-CHOL-PC liposomes, for both MP and HP. For all formulations, tissue leakage proteins (specially VTNC) show higher abundance in liposome-HP complexes than liposome-MP complexes. In summary, our results show that following incubation with MP: (i) Sphingosine makes the liposome-protein corona richer in Igs (IGKC and IGHM) than DOTAP; ii) DOTAP binds higher levels of coagulation (FIBB and FIBG) and tissue leakage proteins (especially VTNC) than Sphingosine. When incubated with HP, the following observations could be made: i) Lipoproteins, complement proteins and Igs bind to both cationic and slightly anionic liposomes; (ii) cationic lipid, i.e., Sphingosine or DOTAP, promoted high adsorption of acute phase proteins (especially ITH1 and ITH2); iii) Sphingosine specifically recruited complement proteins (C4BPA and CO3 are the most enriched) and Igs (mainly IGK), whereas DOTAP promotes adsorption of coagulation (THRB) and tissue leakage proteins (VTNC).
4. Conclusions
In this work, we characterized the protein corona of five lipid formulations that have been used to deliver DNA to mammalian cells in culture with the hope of enhancing our understanding of the role of nanoparticle properties in recruiting specific proteins from biological fluids. Collectively, our data confirm that classifying liposomes based on charge is not sufficient to fully describe the liposome-protein corona composition. The data suggest that the chemistry and arrangement of surface lipid functional groups are key players that regulate the liposome–protein interactions. Notably, we provided evidence that the nanoparticle–protein corona that forms around liposomes is strongly affected by the physiological environment, i.e., the type of plasma. Given the relationship between the biological identity acquired by nanoparticles in vivo and their physiological response (circulation times, immune response, selective targeting, etc.) our findings support the opinion that results obtained in mice models are not directly applicable to humans. Notably, we also showed that minor changes in the lipid composition might critically affect the protein corona composition. These results are likely to suggest that the translation of novel pharmaceutical formulations from animal models to the clinic must be evaluated on a case-by-case basis.
Supplementary Material
Significance.
In the present work nanoliquid chromatography tandem mass spectrometry was used to characterize the protein corona of five different liposome formulations after exposure to mouse and human plasma. The modern proteomic methods employed have clarified that the arrangement of lipid functional groups is a key player that regulates the liposome-protein interactions. We also clarified that the protein corona enrichment and complexity depend on the serum type. Our results suggest that the translational of novel pharmaceutical formulations from animal models to the clinic must be evaluated on a case-by-case basis.
Acknowledgments
This work was supported by grants “Futuro in Ricerca 2008” (RBFR08TLPO; “Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale”) to GC and NIH # 1RO1GM093287 to TJA. GC, DP and AL acknowledge support by the Istituto Italiano di Teconologia, Center for Life Nano Science@Sapienza.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare no conflict of interest regarding the material discussed in the manuscript.
References
- 1.Lück M, Paulke BR, Schröder W, Blunk T, Müller R. Analysis of plasma protein adsorption on polymeric nanoparticles with different surface characteristics. J Biomed Mater Res. 1998;39:478–485. doi: 10.1002/(sici)1097-4636(19980305)39:3<478::aid-jbm19>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 2.Zelphati O, Uyechi LS, Barron LG, Szoka FC., Jr Effect of serum components on the physicochemical properties of cationic lipid/oligonucleotide complexes and on their interactions with cells. BBA-Lipid Lipid Met. 1998;1390:119–133. doi: 10.1016/s0005-2760(97)00169-0. [DOI] [PubMed] [Google Scholar]
- 3.Moghimi S, Szebeni J. Stealth liposomes and long circulating nanoparticles: critical issues in pharmacokinetics, opsonization and protein-binding properties. Prog lipid res. 2003;42:463–478. doi: 10.1016/s0163-7827(03)00033-x. [DOI] [PubMed] [Google Scholar]
- 4.Caracciolo G, Callipo L, De Sanctis SC, Cavaliere C, Pozzi D, Laganà A. Surface adsorption of protein corona controls the cell internalization mechanism of DC-Chol-DOPE/DNA lipoplexes in serum. BBA-Biomembranes. 2010;1798:536–543. doi: 10.1016/j.bbamem.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Capriotti AL, Caracciolo G, Caruso G, Foglia P, Pozzi D, Samperi R, Laganà A. Differential analysis of "protein corona" profile adsorbed onto different nonviral gene delivery systems. AB. 2011;419:180–189. doi: 10.1016/j.ab.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Capriotti AL, Caracciolo G, Cavaliere C, Foglia P, Pozzi D, Samperi R, Laganà A. Do plasma proteins distinguish between liposomes of varying charge density? J Proteomics. 2012;75:1924–1932. doi: 10.1016/j.jprot.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Lai ZW, Yan Y, Caruso F, Nice EC. Emerging techniques in proteomics for probing nano–bio interactions. ACS nano. 2012;6:10438–10448. doi: 10.1021/nn3052499. [DOI] [PubMed] [Google Scholar]
- 8.Capriotti AL, Caracciolo G, Caruso G, Cavaliere C, Pozzi D, Samperi R, Laganà A. Label-free quantitative analysis for studying the interactions between nanoparticles and plasma proteins. ABC. 2013;405:635–645. doi: 10.1007/s00216-011-5691-y. [DOI] [PubMed] [Google Scholar]
- 9.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Nanoparticle size and surface properties determine the protein corona with possible implications for biological impacts. Proc Natl Acad Sci USA. 2008;105:14265–14270. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Baldelli Bombelli F, Dawson KA. Physical-chemical aspects of protein corona: relevance to in vitro and in vivo biological impacts of nanoparticles. JACS. 2011;133:2525–2534. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 11.Tenzer S, Docter D, Rosfa S, Wlodarski A, Kuharev, Rekik A, Knauer SK, Bantz C, Nawroth T, Bier C. Nanoparticle size is a critical physicochemical determinant of the human blood plasma corona: a comprehensive quantitative proteomic analysis. ACS nano. 2011;5:7155–7167. doi: 10.1021/nn201950e. [DOI] [PubMed] [Google Scholar]
- 12.Mahon E, Salvati A, Baldelli Bombelli F, Lynch I, Dawson KA. Designing the nanoparticle– bio molecule interface for "targeting and therapeutic delivery. J Control Release. 2012;161:164–174. doi: 10.1016/j.jconrel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Monopoli MP, Åberg C, Salvati A, Dawson KA. Biomolecular coronas provide the biological identity of nanosized materials. Nature Nanotech. 2012;7:779–786. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 14.Caracciolo G, Pozzi D, Capriotti AL, Cavaliere C, Laganà A. Effect of DOPE and cholesterol on the protein adsorption onto lipid nanoparticles. J Nanopart Res. 2013;15:1498. [Google Scholar]
- 15.Szebeni J, Muggia F, Gabizon A, Barenholz Y. Activation of complement by therapeutic liposomes and other lipid excipient-based therapeutic products. Prediction and prevention Adv drug deliver rev. 2011;63:1020–1030. doi: 10.1016/j.addr.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 16.Szebeni J, Bedőcs P, Rozsnyay Z, Weiszhár Z, Urbanics R, Rosivall L, Cohen R, Garbuzenko O, Báthori G, Tóth M, Bunger R, Barenholz Y. Liposome-induced complement activation and related cardiopulmonary distress in pigs: factors promoting reactogenicity of Doxil and AmBisome. Nanomed: Nanotechnol Biol Med. 2012;8:176–184. doi: 10.1016/j.nano.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Ishida T, Harada M, Wang XY, Ichihara M, Irimura K, Kiwada H. J Control Release. 2005;105:305–317. doi: 10.1016/j.jconrel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Ichihara M, Shimizu T, Imoto A, Hashiguchi Y, Uehara Y, Ishida T, Kiwada H. Anti-PEG igm response against pegylated liposomes in mice and rats. Pharmaceutics. 2010;3:1–11. doi: 10.3390/pharmaceutics3010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caracciolo G. Liposome-protein corona in a physiological environment: Challenges and opportunities for targeted delivery of nanomedicines. Nanomed Nanotech Biol Med. doi: 10.1016/j.nano.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Pozzi D, Colapicchioni V, Caracciolo G, Piovesana S, Capriotti AL, Palchetti S, De Grossi S, Riccioli A, Amenitsch H, Lagana A. Effect of polyethyleneglycol (PEG) chain length on the bio-nano-interactions between PEGylated lipid nanoparticles and biological fluids: from nanostructure to uptake in cancer cells. Nanoscale. 2014;6:2782–2792. doi: 10.1039/c3nr05559k. [DOI] [PubMed] [Google Scholar]
- 21.Tenzer S, Docter D, Kuharev J, Musyanovych A, Fetz V, Hecht R, Schlenk F, Fischer D, Kiouptsi K, Reinhardt C, Landfester K, Schild H, Maskos M, Knauer SK, Stauber RH. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. Nature nanotechnol. 2013;8:772–781. doi: 10.1038/nnano.2013.181. [DOI] [PubMed] [Google Scholar]
- 22.Caracciolo G, Pozzi D, Candeloro De Sanctis S, Capriotti AL, Caruso G, Samperi R, Laganà A. Effect of membrane charge density on the protein corona of cationic liposomes. Appl Phys Lett. 2011:99. [Google Scholar]
- 23.Walkey CD, Chan WCW. Chemical Society Reviews. 2012;41:2780–2799. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 24.Caracciolo G, Pozzi D, Capriotti AL, Cavaliere C, Piovesana S, La Barbera G, Amici A, Laganà A. The liposome-protein corona in mice and humans and its implication for in vivo delivery. J Mater Chem B. 2014;2:7419–7428. doi: 10.1039/c4tb01316f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.