Abstract
The rice blast fungus Magnaporthe oryzae is a serious pathogen that jeopardises the world’s most important food-security crop. Ten common Malaysian rice varieties were examined for their morphological, physiological and genomic responses to this rice blast pathogen. qPCR quantification was used to assess the growth of the pathogen population in resistant and susceptible rice varieties. The chlorophyll content and photosynthesis were also measured to further understand the disruptive effects that M. oryzae has on infected plants of these varieties. Real-time PCR was used to explore the differential expression of eight blast resistance genes among the ten local varieties. Blast disease has destructive effects on the growth of rice, and the findings of our study provide evidence that the Pikh, Pi9, Pi21, and Osw45 genes are involved in defence responses in the leaves of Malaysian rice at 31 h after inoculation with M. oryzae pathotype P7.2. Both the chlorophyll content and photosynthesis were reduced, but the levels of Pikh gene expression remained constant in susceptible varieties, with a developed pathogen population and mild or severe symptoms. The Pi9, Pi21, and Osw45 genes, however, were simultaneously upregulated in infected rice plants. Therefore, the presence of the Pikh, Pi9, Pi21, and Osw45 genes in the germplasm is useful for improving the resistance of rice varieties.
Introduction
Rice (Oryzasativa L.) is the second most widely cultivated crop in the world, and due to the growing global population, the production of rice is expected to increase by 40% by 2030. Hence, the production of rice varieties with potential and more stable yield is indispensable for overcoming grain yield reductions and arable land limitations [1]. However, the stability of the rice yield is affected by biotic and abiotic stresses. Rice blast,caused by Magnaporthe oryzae, is one of the most calamitous diseases for thiscrop, reducing the annual grain yield by approximately 10–30% [2]. It has injured different parts of plants such as leaf, collar, node, neck and panicle by production of spores and penetration of infection [3]. Blast disease also has destructive effects on the physiological aspects of rice growth; leaf blast disease decreases photosynthesis by reducing the green leaf area and affecting photosynthesis in the regions surrounding lesions [4]. Various control strategies have been applied to overcome rice blast disease, including the use ofresistant varieties [5]. To date, over 85 blast resistance genes and 300 QTLs have been identified, of which 20 have been cloned. Rice blast can be categorised in a gene-for-gene system, in whichone or more similar motifs are encoded byR genes[6]. These similar motifs consist of kinase domains, nucleotide binding sites (NBSs), and leucine-rich repeat segments [7]. Among the cloned blast resistance genes, only Pid2 and Pi21 encode a receptor that is a kinase [8] and proline-rich protein [9], respectively; the other genes are classified as containing nucleotide binding site-leucinerich repeats (NBS-LRRs) [10]. Pib,an NBS-LRR, is a member of a dominant small family gene and confers high resistance to blast fungi in rice plants [11]. The expression of Pib is induced at 12 and 24 h after M. Oryzae inoculation but returns to basal levels at 96 h post-inoculation [12]. In addition, the genes Pikh and Pita induce broad-spectrum resistance to blast disease [13]. Pita is a single-copy gene with low constitutive expression that is the same in both resistant and susceptible rice plants prior to inoculation with M. oryzae [14]. The Pi9 and Os11gRGA8 genes also belong to the NBS-LRR class of proteins and are found in indica rice lines, e.g., 75-1-127, and Peh-kuh-tsao-tu lines, respectively [15], and Os11gRGA8 is tightly linked to the Pia gene on chromosome 11 [16]. The majority of the cloned and mapped R genes confer resistance to only specific isolates due to race specificity, though it has been reported that cloning these genes into any rice strain does confer resistance. Nevertheless, identifying R genes that have broad-spectrum resistance or their loci will provide essential genetic resources for rice breeding programmes [17–19]. Members of the WRKY gene family are transcription factors that bind to the promoter of W-box defence genes to regulate their transcription [20,21], and many WRKY genes are expressed in response to pathogen infections, such as blast fungi and bacterial blight diseases [22–24]. Indeed, an over-expression analysis of WRKY45 and WRKY22 genes revealed the induction of a resistant phenotype following blast inoculation [25]. Although many resistant varieties are available from IRRI’s vast collection of rice germplasm, some rice varieties that were released as resistant became susceptible after only a few years of cultivation due to blast fungal evolution and adaptation [26]. Therefore, screening rice varieties based on their resistance to blast disease and identifying the most important genes involved in the defence response are essential for improving rice crops. To better investigate plant-fungal interactions, a rapid and precise measurement of fungal growth in plants is needed to properly assess both fungal pathogenicity and host resistance levels. Several methods have been used to identify and quantify fungal pathogens in the environment (plants and soil). For instance, the overall fungal biomass in the environment has been approximated by calculating the amount of fungal compounds such as ergosterol or even chitin [27–29]. However, these methods often lack specificity and usually do not correlate with the actual fungal biomass due to fungal sporulation and disturbances of enzymatic reactions in plants [30]. In addition, numerous studies have used quantitative PCR to calculate the amount of plant pathogenic fungus during plant-fungal interactions [31,32], yet this type of quantitative information is not produced directly from target DNA but instead is derived from the similarity rate between target and non-target DNA.
In this study, the expression profiles of 8 selected genes involved in defence responses to blast disease among 10 common Malaysian rice varieties were for the first time examined using real-time PCR (RT-qPCR). Two gene groups, Pikh[33], Pib[12], Pi21 [9], Pia (Os11gRGA8) [16], Pita [34,35] and Pi9 [36] (major genes confer high resistance to blast fungal), and also OsWRKY22 [37] and OsWRKY45 [24] (involving in mitogen-associated protein kinase (MAPK) [38] and salicylic acid (SA) signalling [24,39], cascades and transcriptional re-programming in plant/pathogen interactions) were selected to be investigated. MAPK and SA signalling cascades are the earliest signalling events after plant sensing of the invading pathogen as they link the perception of external stress to cellular responses [38,40]. Furthermore, we investigated correlations among gene expression levels, leaf blast severity, and photosynthesis and its components (chlorophyll a and b) in the visible lesion area of inoculated plants. The fungal population was also quantified and compared between susceptible and resistant plants using a qPCR quantification technique.
Experimental Procedure
Plant material and fungal isolate
Ten important Malaysian rice varieties and an M. oryzae strain (P7.2) were provided by the Malaysian Agricultural Research and Development Institute (MARDI) (Table 1). Rice seeds were soaked in water for three days and germinated on moist Whatman filter paper in Petri dishes. Subsequently, the seeds were planted in pots using autoclaved potting soil, transferred to a glasshouse and kept at 25–30°C for three weeks.
Table 1. Selected characteristics of ten Malaysian rice varieties.
| Variety | Yield (t/ha) | Leaf blight disease(Xanthomonasoryzea) | Brown planthopper | Sheath blight (Thanatephoruscucumeris) |
|---|---|---|---|---|
| MR159 | 3–5.4 | Resistant | Semi-resistant | Susceptible |
| PH9 | 3.8–4.7 | Susceptible | Semi-susceptible | Resistant |
| MR84 | 4–6.2 | Susceptible | Semi-susceptible | - |
| MR185 | 6–9.2 | Resistant | Semi- resistant | - |
| MR232 | 6.5–8.7 | Semi-resistant | Semi-susceptible | Susceptible |
| MR253 | 6–7.5 | Semi-resistant | Semi-resistant | Semi-resistant |
| MR219 | 6.5–10.7 | Resistant | - | - |
| MR263 | 6–7.5 | Semi-resistant | Semi-resistant | Semi-resistant |
| MR269 | 6–7.5 | Semi-resistant | Semi-resistant | Semi-resistant |
| MRQ74 | 4.5–5.5 | Semi-resistant | Semi-resistant | - |
Plant inoculation and disease severity assay
Three-week-old rice seedlings at the three-leaf stage were inoculated with an M. oryzae spore suspension containing 3×105 spores/mL.The inoculation experiments were repeated three times for all rice varieties. We measured three components of partial resistance: the blast lesion degree (BLD), blast lesion type (BLT), and percent disease leaf area (%DLA). Two traits, %DLA and BLT, were estimated and scored based on the Correa-Victoria and Zeigler methods [41]. Disease severity (Fig 1) was recorded at 7 days after inoculation (DAI) using a 0 to 9 disease assessment scale: 0, no signs of infection; 1, brown specks ˂ 0.5 mm in diameter; 3, brown spots 0.5–1 mm wide; 5, round to ovate-shaped lesions and 1–3 mm in diameter with a grey centre, 7: spindle-shaped lesions with a grey centre; and 9, combinations of small, white, grey, or bluish lesions without distinct borders. The %DLA was scored as follows: 0, highly resistant and no symptoms; 1–2, lesions 1–2 mm and no sporulation; 3, lesions 2–3 mm size and little sporulation; and 4, spindle-shaped lesions more than 3 mm in size with heavy sporulation. The BLD scoring was implemented based on the Standard Evaluation System (SES) of the International Rice Research Institute [42].
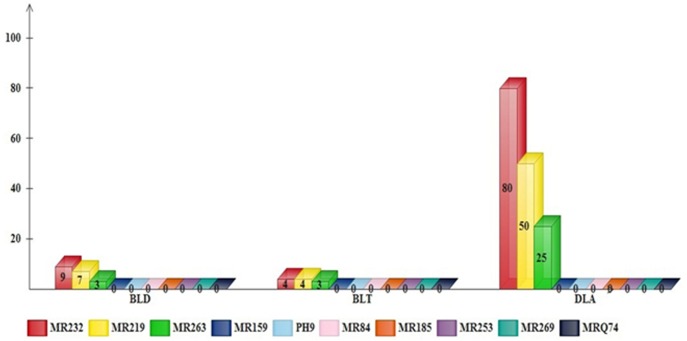
Fig 1. Blast disease symptoms caused by M. oryzae on the leaves of susceptible rice varieties.
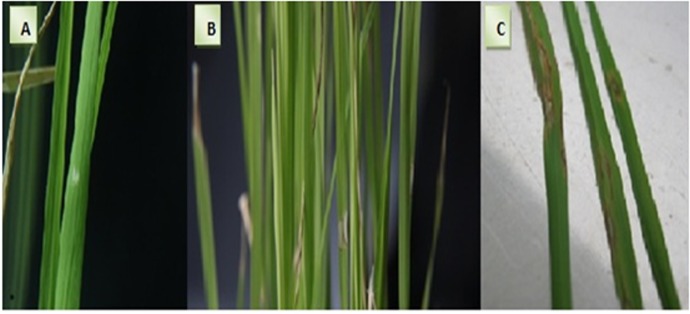
A. Symptoms at 3 days after inoculation. B and C. Disease severity in rice seedling at 7 days after inoculation.
Differentiation of pathogen population between inoculated susceptible and resistant plants using a bacterial quantification technique
Real-time PCR assay
For the real-time PCR assay, total DNA was extracted from three-week-old susceptible (MR232, MR219 and MR263) and resistant seedlings, which hadbeen inoculated with M. oryzae pathotype P7.2 at a concentration of 3×105 spores/mL, at 31, 48 and 72 h post-infection using the i-genomic BYF DNA extraction mini kit (EBN, Route d'Esneux, Dolembreux, Belgium). The absolute quantification of the M. oryzae population in inoculated rice plants was carried out based on the real-time PCR standard curve method. The standard curves were constructed using the copy number of the 28S rDNA gene plotted against the quantification cycle (Cq) obtained from 10-fold serial dilutions of the PCR products from a pure fungal culture. All preparing proceduresof standard curve, ranging from DNA extraction to the purification of specific bands (28S rDNA gene), are described below.
DNA extraction from fungal cultures
About 0.2 g of fungal mycelia previously cultured in potato dextrose broth was ground with liquid nitrogen and fungal DNA was extracted using modified CTAB method [43].
Primer design and 28SrDNA isolation from the fungal genome
A specific pair of DNA primers was designed based on the 3' end of the M. oryzae 28SrDNA gene (Table 2). More than 100 copies of the 28SrDNA gene are present in the M. oryzae genome and are constitutively and highly expressed [44,45]. The PCR programme was performed using a Taq DNA polymerase kit (Fermentas, Waltham, Massachusetts, USA) with the following parameters: initial denaturation at 95°C for 4 min, 40 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 1 min, and a final extension at 72°C for 5 min. The PCR product was isolated on a 1% agarose gel and stained with ethidium bromide. Finally, the expected 28SrDNA gene band (approximately 330 bp) was purified using a QIAprepspin miniprep kit (Qiagen, Hilden, North Rhine-Westphalia, Germany).
Table 2. Primers for targetingM. oryzae used for real-time PCR.
| Target group | Sequence 5/–3/ | |
|---|---|---|
| 28S rDNA | Forward | TACGAGAGGAACCGCTCATTCAGATAATTA |
| Reverse | TCAGCAGATCGTAACGATAAAGCTACTC | |
The purity and concentration of the 28S rDNA gene in each sample was measured using a Nanodrop ND-1000 spectrophotometer (ImplenNanoPhotometer, Munich, Bavaria, Germany). The number of copies of 28S rDNA per mL of elution buffer was calculated using the following formula (available online in the Genomics and Sequencing Centre web-based calculator of the University of Rhode Island) [46].
Because the efficiency of amplification among primers and templates can vary, the amplification efficiency (E) of each primer-template combination was determined based on the slope value of the linear regression of each standard curve calculated by the following equation:
Real-time PCR was performed using aBioRad CFX96 Real-time PCR system(BioRad, Hercules, California, USA)with optical-grade plates. The primers used in the quantification of different fungal populations are shown in Table 2. The real-time PCR reaction was performed in a total volume of 25 μL using the QiagenQuantifast SYBR Green PCR, USA. Each sample was amplified in triplicate. A no-template control (NTC) was included to rule out any cross-contamination. The real-time PCR cycling conditions consisted of 5 min at 95°C, followed by 40 cycles of 10 s at 95°C and 30 s at 60°C. Upon the completion of amplification, the specificity of the product was confirmed by melting curve analysis. The real-time PCR products were incubated by raising the temperature from 70 to 95°C in 0.5°C increments, with a hold of 5 s at each increment.
Chlorophyll content
The leaves of treated and untreated plants were collected at seven days after inoculation. The chlorophyll content was estimated using the method of Witham et al. [47]. The absorbance of the solution was recorded at 645 and 663 nm using a scanning spectrophotometer (AL800, Aqualytic, Germany). The chlorophyll content was measured as mg/g-1 of the sample using the following formulae:
where A645 and A663 are the absorbance of the solution at 645 and 663 nm, respectively, V is the volume of the solution in mL, W is the weight of the fresh leaf sample in mg, and 12.7, 2.69, 22.9, 4.86, 20.2 and 8.02 are absorption coefficients.
Photosynthesis analysis
Seven days after inoculation, the amount of photosynthesis was measured among treated and control varieties using the LI-6400XT Portable photosynthesis system (LI-COR, Lincoln, Nebraska, USA).
Real time-PCR analysis
Specific primers were designed for the rice resistance genes available in the GenBank database using the primer premier6 software (Table 3). The 18S rRNA and tubulin reference genes of O. sativa were used as expression control genes for normalisation.
Table 3. List of primers for 10 genes used for Real time-PCR.
| List of genes | Forward primer (5´- 3´) | Reverse primer (5´- 3´) |
|---|---|---|
| Pikh | AAGATTTTCGAGGCTCTTCTCTA | ATGAATCTGTTTCCTCGTCTTG |
| Pib | GGGAAAAATGGAAATGTGC | AGTAACCTTCTGCTGCCCAA |
| Pita | ATTATTGAGCTTCTTTCTTTCTCTG | GAAACAAAATTACCTCTACTCTGAAG |
| Pi21 | GGTCATCTTGGTGGACCTGCAATG | CGATGCAGTACTCCTCTTCAAGGC |
| Pi9 | TGCCCAACCTTTACCCACTGTA | AACATGAGTAGAAACAAATTAGTTTG |
| OSWRKY22 | CGGCGGAAAGACTGAGAAAG | CAGCCTCTGATGACGGTGAG |
| OSWRKY45 | ACGACGAGGTTGTCTTCGATCTG | GCCCGTGTCCATCCATGATTCTTC |
| Os11gRGA8 | CAAGCCAAGTTCCAC | TGCACTCTGCATGTTGTTCA |
| 18SrRNA | ATGATAACTCGACGGATCGC | CTTGGATGTGGTAGCCGTTT |
| α-Tubulin | GGAAATACATGGCTTGCTGCTT | TCTCTTCGTCTTGATGGTTGCA |
At 31 h after inoculation, a time when the blast fungus emerged through the cell wall and entered into the epidermal cell [48], RNA was extracted from the leaves of infected and control plants using TRIzol Reagent (Invitrogen, USA)[49]. The expression levels of eight blastresistance genes (Pikh, Pib, Pita, Pi21,Pi9,Os11gRGA8, OsWRKY22 and OsWRKY45)were evaluated by real-time PCR.
A 1-μL aliquot of RNA from each sample was used to prepare 20-μL reactions (based on the KAPA SYBER FAST One-Step RT-qPCR, Wilmington, Delaware, USA). The cycling conditions were a primary incubation of 5 min at 42°C and 5 min at 95°C, followed by 40 cycles of 95°C for 3 sec, 60°C for 30 sec, and 72°C for 3 sec. In this part of the study, all data were derived from three independent biological replicates. Amplicon specificity was checked using a melting curve analysis after 40 cycles by increasing the temperature from 60°C to 95°C.
Data analysis
The collected data were standardised and statistically analysed using the R software version 3.0.2 (R Core Team,Vienna, Austria). The comparison ofmeans was performed using the Duncan test. Cluster analysis based on similarity matrices was employed for the morphological and physiological data. The Relative Expression Software Tool(REST) (Qiagen, Hilden, North Rhine-Westphalia, Germany) was used to analyse the real-time amplification data.
Results
Severity assessment
Seven of the 10 rice varieties showed a high degree of resistance to pathotype P7.2 of rice leaf blast in glasshouse tests (Table 4).
Table 4. The ANOVA of seven physiological and morphological traits in ten rice varieties.
| Source Of Variation | DF | BLT | BLD | DLA | Dif Chlo A | Dif Chlo B | Dif T Chlo | Photo |
|---|---|---|---|---|---|---|---|---|
| Block | 2 | 0.000ns | 0.000ns | 0.000ns | 0.169** | 0.091ns | 0.043ns | 0.004ns |
| Treat | 9 | 9.633** | 34.300** | 23.741** | 5.368** | 5.770** | 5.561** | 1.706** |
| Error | 18 | 0.000 | 0.000 | 0.000 | 0.0148 | 0.062 | 0.012 | 0.008 |
| Total | 29 | - | - | - | - | - | - | - |
| CV | - | 1.81E-6 | 1.48E-06 | 0 | 14.111 | 24.615 | 12.124 | 14.947 |
ns and ** indicate not significant and a significance level of 1%, respectively. Dif Chlo A: difference in chlorophyll A between control and treated plants. Dif Chlo B: difference in chlorophyll B between control and treated plants. Dif T Chlo: difference in total chlorophyll content between control and treated plants. Photo: difference in photosynthesis between control and treated plants. BLT: Blast lesion type. BLD: Blast lesion degree. DLA: Disease leaf area (%).
However, three rice varieties exhibited different levels of susceptibility. The results of a comparison of meansusing the Duncan test showed that the highest blast lesion degree (9), blast lesion type (4), and disease leaf area (80%) were found in MR232. Furthermore, MR219 and MR263 scored 7 and 3 for the blast lesion degree and 4 and 3 for the blast lesion type, respectively (Table 5). The percentage of disease leaf area was 50% and 25% in MR219 and MR263, respectively (Fig 2).
Table 5. Mean comparison of different morphological and physiological traits in rice varieties.
| Varieties | BLT | BLD | DLA | DIFF A | DIFF B | DIFF T | PHOTO |
|---|---|---|---|---|---|---|---|
| MR159 | 0 c | 0 d | 0 d | 0.000 e | 0.146 c | 0.465 d | 0.239 d |
| PH9 | 0 c | 0 d | 0 d | 0.089 ed | 0.298 c | 0.187 e | 0.158 d |
| MR84 | 0 c | 0 d | 0 d | 0.329 d | 0.324 c | 0.144 e | 0.239 d |
| MR185 | 0 c | 0 d | 0 d | 0.137 ed | 0.241 c | 0.000 e | 0.260 d |
| MR232 | 4 a | 9 a | 80 a | 1.696 c | 3.4603 a | 4.288 a | 2.534 a |
| MR253 | 0 c | 0 d | 0 d | 0.097 ed | 0.148 c | 0.126 e | 0.346 d |
| MR219 | 4 a | 7 b | 50 b | 2.324b | 3.439 a | 1.564 c | 1.177 b |
| MR263 | 3 b | 3 c | 25 c | 3.888a | 1.854 b | 1.985 b | 0.833 c |
| MR269 | 0 c | 0 d | 0 d | 0.000 e | 0.000 c | 0.521 d | 0.184 d |
| MRQ74 | 0 c | 0 d | 0 d | 0.063 ed | 0.226 c | 0.020 e | 0.170 d |
| Mean | 1.1 | 1.9 | 15.5 | 0.8623 | 1.01363 | 0.93 | 0.614 |
| CV(%) | 1.81E-6 | 1.48E-06 | 0 | 14.111 | 24.615 | 12.124 | 14.947 |
Same letter(s) in a column denote no significant difference at P ≤ 0.05. DifChlo A: difference in chlorophyll A between control and treated plants. DifChlo B: difference in chlorophyll B between control and treated plants. Dif T Chlo: difference in total chlorophyll content between control and treated plants. Photo: difference in photosynthesis between control and treated plants. BLT: Blast lesion type. BLD: Blast lesion degree. DLA: Disease leaf area (%).
Fig 2. Differences in morphological traits (blast lesions) of rice varieties.
BLD: Blast lesion degree. BLT: Blast lesion type. DLA (%): Disease leaf area.
Real time-PCR quantification between inoculated susceptible and resistant plants
A significant variation was detected in the abundance ofM. oryzae among the inoculated susceptible and resistant plants, including all sample collected at 31, 48 and 72 hours after inoculation. The expected size of PCR amplicon was 330 bp forthe blast fungus28S rDNA (S1 Fig and Table 6). The standard curve of the target gene is shown in S2 Fig and was constructed using the plot of copy numbers of 28S rDNA gene of M. oryzae against its Cq value. The standard curves had a high correlation coefficient of R2 = 0.996, indicating that the Cq value was proportional to the copy numbers of 28S rDNA gene, for blast fungal. From the slope of the liner regression of -3.353 amplification efficiency obtained was 98.7% for M. oryzae. A good reaction should have an amplification efficiency of between 90–110% and R2 value should be ≥ 0.985,and in this study, the amplification efficiencies of 98.7%, and R2 values of 0.996 for the standard curves of M. oryzae was within the acceptable range.
Table 6. Concentration and purity of extractedM. oryzae DNA.
| Sample | Concentration (ng/μL) | Purity | |
|---|---|---|---|
| 260/280 | 260/230 | ||
| Extracted fungal DNA | 3700.2 | 2.18 | 1.12 |
| DNA extracted from the gel | 94 | - | - |
The amplification curve for M. oryzae (S3 Fig) was constructed by plotting the cycle numbers against the fluorescence signal (RFUs, relative fluorescence units). No fluorescence signals were detected from the no-template control.
The melting curve for the blast fungal is shown in S4 Fig. From the result, the melting temperature of 86.5 was detected at which the set of primer was specific for estimation of M. oryzae.
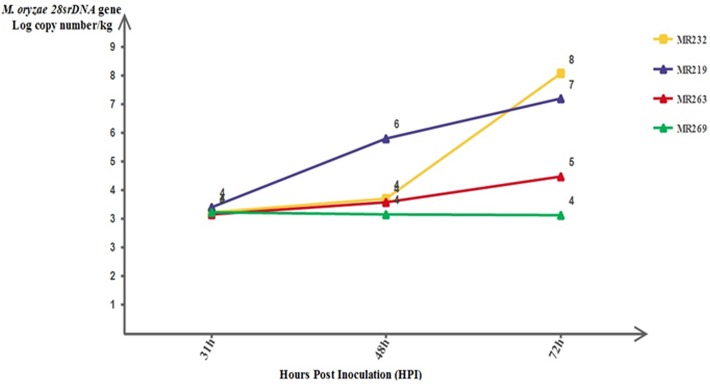
The real-time PCR quantification results for the M. oryzae populationsfrom inoculated susceptible and resistant plants at 31, 48 and 72 hours after inoculation are shown in Fig 3. The blast fungal population were significantly (P < 0.05) higher in susceptible plants compared with those observed in resistant plants at 48 and 72 hours after inoculation.
Fig 3. A comparison of M. oryzae populations between resistant and susceptible plants at 31, 48 and 72 hours after infection, as quantified using real-time PCR.
Bars represent the means of three biological samples at each time point.
The M. oryzae populations quantified by real-time PCR are shown in Fig 3. At 48 and 72 hours after inoculation, the fungal populations in the susceptible plants were significantly (P< 0.05) higher (4.116 and 7.914 in MR232; 5.944 and 7.151 in MR219; 4 and 4.781 in MR263, respectively, for the log 28S rDNA gene copy number/kg) than their populations in resistant plants (3.628 and 3.609,respectively, for thelog28S rDNA gene copy number/kg). However, at 31h of inoculation, the blast fungal population in the resistant plants (3.510 log 28S rDNA gene copy number/kg) was not significantly different from that of the susceptible plants (3.706, 3.848 and 3.628 log 28S rDNA gene copy number/kg for MR232, MR219 and MR263, respectively).
Chlorophyll content
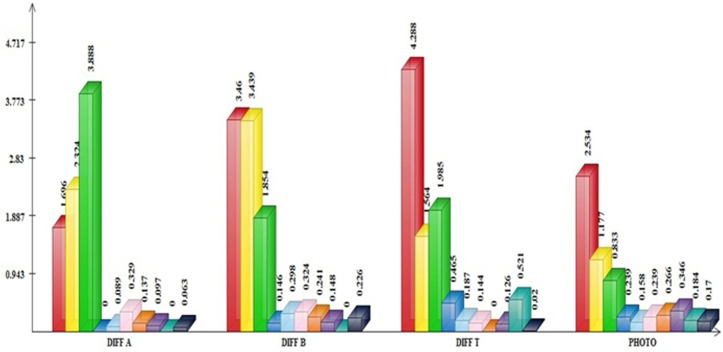
The chlorophyll contents (total, a and b) showed significant differences in 5% level of probability among ten varieties (Table 4). The highest decrease rate of chlorophyll a content was obtained in infected MR263 compared with the control. However, the maximum reduction rate of chlorophyll b and total chlorophyll contents in infected plants wasfound for MR232 (Table 5 and Fig 4).
Fig 4. Variations in physiological traits among ten rice varieties.
DIFF A: difference in chlorophyll a. DIFF B: difference in chlorophyll b. DIFF T: difference in total chlorophyll. PHOTO: difference in photosynthesis between control and treated plants.
Photosynthesis analysis
The effects of the blast pathogen in reducing leaves photosynthesis in inoculated plants were significant among the rice varieties (Table 4). The most decreased level of photosynthesis was revealed in MR232. Besides, in comparison to control plants, the diminished photosynthesis ratio was observed in inoculated MR219 and MR263, respectively (Table 5 and Fig 4). On the other hand, the variationsin photosynthesis between the treated and control plants were not significant in the remainder varieties.
Correlation analysis
Determination of the correlations between different traits, especially photosynthesis and chlorophyll components, allow for the identification of interactions amongaffected traits in rice plants infected with M. oryzae. Blast lesion type, disease leaf area and the blast lesion degree showed a negative correlation with photosynthesis and chlorophyll components. The highest correlation coefficient was observed between the blast lesion type and disease leaf area (r = 0.962). Chlorophyll b, in comparison to chlorophyll a,represented a larger proportion of the total chlorophyll content and photosynthesis (r = 0.929 and 0.801, respectively) ininfected rice leaves. The changes in blast lesion type and disease leaf area were able negatively to manage the amount of photosynthesis (r = -0.916 and -0.891) (S1 Table).
Cluster analysis
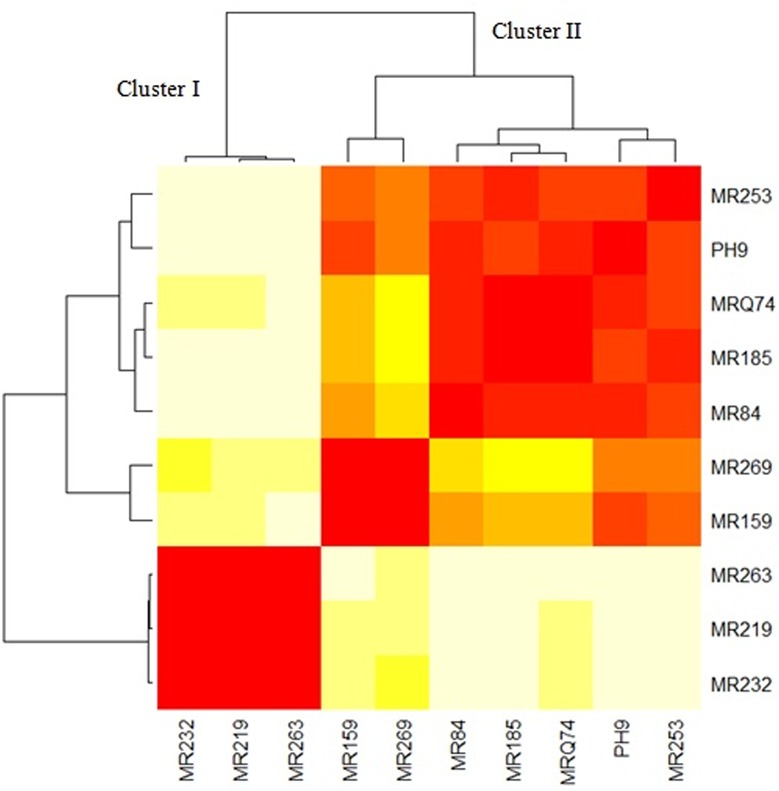
The ten Malaysian rice varieties were categorized to two clusters based on seven morphological and physiological characters (Fig 5). Cluster I is composed of the susceptible rice varieties MR232, MR263 and MR219 to blast disease. While, cluster II is collected of the resistant varieties.
Fig 5. Cluster of ten rice varieties based on morphological and physiological traits.
Gene expression study
Gene expression studyof eight blast resistance genes was performed simultaneously in an attempt to identify statistically up-regulated genes in the most common and popular rice varieties infected by M. oryzae in Malaysia.
RT-qPCR was used to explore the variabilityin gene expression between control and M. oryzae-infected rice plants. Various expression patterns of Pikh, Pib, GA8, OsW22 and OsW45 genes have been found at 31 h after inoculation of rice varieties (Table 7). Pikh gene was up-regulated significantly in MR159, PH9, MR185, MR269 and MRQ74, and down-regulated in MR84 and MR253, while relative expression of this gene was constant in MR232, MR219 and MR263 varieties. On the other hand, Pita gene showed significantly lower expression in ten infected varieties compared with control. Whereas, the transcript level of Pi9 and Pi21 genes significantly increased in the leaves of all varieties after 31 hours of inoculation (Fig 6). Furthermore, the expression level of OsW45 was higher in the MR185, MR232, MR219, MR263, and MRQ74 varieties at 31 h post-inoculation, whereas the expression of GA8, Pib and OsW22 was down-regulatedin most of the rice varieties.
Table 7. Relative expression patterns and P-values of 8 genes in 10 rice varieties.
| Genes | MR159 | PH9 | MR84 | MR185 | MR232 | MR253 | MR219 | MR263 | MR269 | MRQ74 |
|---|---|---|---|---|---|---|---|---|---|---|
| PikhP-value | UP | UP | DOWN | UP | - | DOWN | - | - | UP | UP |
| 0.000 | 0.000 | 0.000 | 0.000 | 0.650 | 0.000 | 0.161 | 0.822 | 0.000 | 0.000 | |
| PibP-value | DOWN | - | DOWN | - | DOWN | DOWN | - | DOWN | UP | UP |
| 0.000 | 0.170 | 0.000 | 0.499 | 0.000 | 0.000 | 0.501 | 0.000 | 0.000 | 0.000 | |
| GA8P-value | - | - | - | UP | - | DOWN | UP | - | - | - |
| 0.661 | 0.169 | 0.834 | 0.000 | 0.650 | 0.000 | 0.000 | 0.162 | 0.154 | 0.837 | |
| Pi9P-value | UP | UP | UP | UP | UP | UP | UP | UP | UP | UP |
| 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Osw22P-value | - | - | UP | - | DOWN | DOWN | - | DOWN | DOWN | - |
| 0.169 | 0.169 | 0.000 | 0.827 | 0.000 | 0.000 | 0.502 | 0.000 | 0.000 | 0.667 | |
| PitaP-value | DOWN | - | DOWN | DOWN | DOWN | DOWN | DOWN | - | DOWN | DOWN |
| 0.000 | 0.169 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.162 | 0.000 | 0.000 | |
| Pi21P-value | UP | UP | UP | UP | UP | UP | UP | UP | UP | UP |
| 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | |
| Osw45P-value | - | - | - | UP | UP | - | UP | UP | DOWN | UP |
| 0.339 | 0.169 | 0.164 | 0.000 | 0.000 | 0.177 | 0.000 | 0.000 | 0.000 | 0.000 |
UP: Up-regulated gene; DOWN: Down-regulated gene; Dash sings: gene expression was unchanged.
Fig 6. Relative expression levels of Pikh, Pib,Os11gRGA8(GA8), OsWRKY22 (Osw22), Pita, Pi21 and OsWRKY45 (Osw45) genes calibrated using 18SrRNA and tubulin as reference genes in infected and control rice plants using relative quantitative real-time PCR.
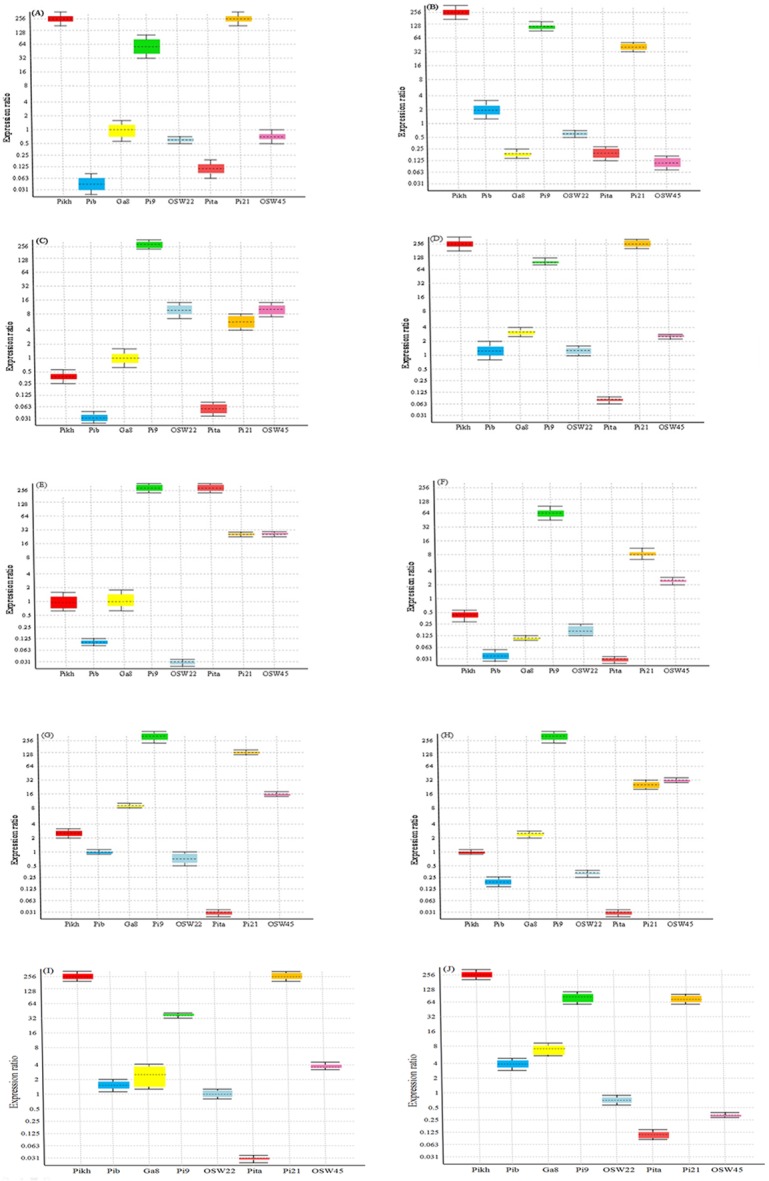
Expression levels of 8 genes in 10 infected varieties (A) MR159, (B) PH9, (C) MR84, (D) MR185, (E) MR232, (F) MR253, (G) MR219, (H) MR263, (I) MRQ74, and (J) MR269.
Discussion
The results of our severity assessment revealed different levels of resistance among the rice varieties. This evaluation confirmed the susceptibility of the MR232, MR219 and MR263 varieties to the M. oryzae pathotype P7.2. However, it should be noted that the morphological screening of rice plants provides only preliminary results that must be confirmed by further testing. Compared with the common disease scoring that is based on the number and size of blast lesions, the DNA-based real-time PCR technique enables a more precise computation of the relative growth and absolute biomass of M. oryzae. Additionally, the quantity of fungal growth in the plants may not be continuously proportional to the distribution of disease symptoms elicited by various blast fungal isolates. Thus, both the quantification of fungal growth and the morphological evaluation of disease symptoms are required to more precisely assess the aggressiveness of various blast fungal isolates. In our study, the quantification of fungal populations in inoculated susceptible and resistant plants was confirmed by a morphological analysis. The growth of the fungus and the susceptibility of MR232, MR219 and MR263 were verified, but the fungal growth was constant in the resistant varieties at 31, 48 and 72 h after inoculation. LargerM. oryzae populations (46 to 80 times) were found in seedling leaves of susceptible cultivars compared with resistant cultivars, as has been previously reported at 4 and 6 days post-inoculation using real-time PCR quantification [41]. In fact, real-time PCR is an excellent tool for M. oryzae quantification in plants and can be applied for the reliable evaluation of fungal pathogenicity [32].
We found a negative correlation between photosynthesis and its components and the degree of blast lesions (BLD, BLT and DLA) in infected rice plants. Photosynthesis produces sustained energy for a plant and must therefore be incorporated into photogenedefence mechanisms. Indeed, a decrease in chlorophyll contents and photosynthesis in response to infection is a plant defence strategy that is usedin pathogen evasion[50]. The activation of plant defences leads to a rapid decrease in non-photochemical quenching (NPQ) and thus limits the carbon availability tothe pathogen [51]. There are numerous reasons why plant contacts with pathogens can bedetrimental to photosynthesis [52]. The initiation of defence responses requires energy and photo-assimilate as carbon sources to produce defence compounds;consequently, the photosynthesis demands in the plant increase [53]. Furthermore, sugars, as carbon compounds, are consumed by pathogens. Despite the predicted increase, the infection of plants by pathogens often leads to a reduction in photosynthesis [54–58].
Quantitative real-time PCR (RT-qPCR) is a multipurpose technique for the precise, sensitive, reliable and high-throughput detection and quantification of a target sequence in various samples [59].
In this study, we performed RT-qPCR for eight defence response genes to identify resistance genes with increased expression levels in 10 commonly grown local rice varieties. Gene expression study through simultaneous measurement of the activity of several genes can elucidate how cells react to a particular treatment. The simultaneously increasing transcript levels of Pikh, Pi9, Pi21, and Osw45 among most of the rice varieties highlight their importance in immunity against M. oryzae at31 h after inoculation. However, the over-expression of Pi9, Pi21 and Osw45 is valuable when it is accompanied by the upregulation of Pikh expression. We hypothesise that the absence of resistance to blast fungus pathotype P7.2 in susceptible rice varieties is due to a lack of Pikh transcript upregulation. When the blast fungus enters the plant, the plant’s immune system is activated, and energy sources (carbon) are compromised. This process promotes catabolic pathways and reduces cell growth and proliferation by switching off protein, carbohydrate and lipid biosyntheses [60]. The activation of these genes at the same time could prevent this deficiency in resistant varieties by producing proteins that are involved in the innate immune system. The recovered immune system does not need to consume more carbon as an energy source. Therefore, and as our experiments show, the decrease in photosynthesis and chlorophyll content in resistant rice plants is not significant. However, compared with control plants, the amount of chlorophyll and photosynthesis are reduced in susceptible rice plants. The over-expression of Pikh, Pi9 and OsWRKY45 genes in transgenic rice plants after inoculation confers extremely strong resistance against blast disease [25,33,36]. Moreover, the over-expression of the Pi21 transcript successfully induces resistance against blast disease in rice lines [9]. Finally, it has to mention that the results of this study will be differed using of other isolates because of diverse effectors encoded by different isolates and their specific interactions with rice blast immunity system. Rice blast immunity is categorized in a gene-for-gene system. It explains how Avr genes within the pathogen connect to particular R genes in rice and how the absence of R genes will make rice susceptible [61]. Certain molecules, known as effectors, are produced by phytopathogens and encoded by virulence genes (Avr). These effectors are delivered directly into plant cells during the primary stage of infection and change host plant physiology. Effectors are used to promote pathogen colonization or interrupt the activation of host cells defenses. On the other hand, rice plants have consequently responded with a form of immunity that is based on the sensitivity of effectors to host resistance proteins. This immunity is called the gene-for-gene system [62].
Conclusion
The innate immune system allows plants to react to potential pathogens in an efficient manner while reducing damage and energy costs. Photosynthesis and chlorophyll contents decrease significantly in treated susceptible varieties compared with controls. These data indicate that the pathogen is accessing the plant’s carbon sources. Thus, resistance genes should play a role in the rice immune system against blast. As no significant differences were observed in photosynthesis and its components between inoculated and non-inoculated resistant varieties, it appears that energy sources are provided to both resistant and susceptible plants but that the expression of resistance genes restricts pathogen growth and accessibility to carbon. Our findings provide evidence that the expression of the Pikh, Pi9, Pi21 and Osw45 genes is involved in defence responses in the leaves of rice at 31 h after the inoculation with M. oryzae pathotype P7.2. The expression level of the Pikh gene was constant in susceptible varieties, with symptoms that had reduced chlorophyll content and photosynthesis. Nevertheless, the Pi9, Pi21 and Osw45 genes were simultaneously up-regulated in these plants. Therefore, Pi9, Pi21, and Osw45 are useful in the germplasm for the improvement of rice varieties against M. oryzae pathotype P7.2, whenever they are accompanied byup-regulation of the Pikh gene.
Supporting Information
Lane M is the 1 kb marker, and the other lanes show the isolated 28SrDNA gene from M. oryzae.
(TIF)
E: amplification efficiency. R2: correlation coefficient.
(TIF)
(TIF)
A melting temperature of 86.5°Cwas obtained.
(TIF)
(PDF)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The Long-Term Research Grant Scheme (LRGS), Food Security project, grant number 5525001, Ministry of Higher Education, Malaysia. The funders had no role in study design, data collection and analysis, but they are the authors' financial supporter.
References
- 1. Khush GS. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol Biol. 2005; 59: 1–6. [DOI] [PubMed] [Google Scholar]
- 2. Strange RN, Scott PR. Plant disease: a threat to global food security. Phytopathology. 2005; 43: 83–116. [DOI] [PubMed] [Google Scholar]
- 3. Kato H. Rice blast disease. Pesticide outlook. 2001; 12: 23–25. [Google Scholar]
- 4. Bastiaans L. Effects of leaf blast on photosynthesis of rice. 1. Leaf photosynthesis. Neth J Plant Pathol. 1993; 99: 197–203. [Google Scholar]
- 5. Huang J, Rozelle S, Pray C, Wang Q. Plant biotechnology in China. Science. 2002; 295: 674–676. [DOI] [PubMed] [Google Scholar]
- 6. Staskawicz BJ. Genetics of plant-pathogen interactions specifying plant disease resistance. Plant Physiol. 2001; 125: 73–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994; 19: 415–421. [DOI] [PubMed] [Google Scholar]
- 8. Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W,et al. AB-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006; 46: 794–804. [DOI] [PubMed] [Google Scholar]
- 9. Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K, et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009; 325: 998–1001. 10.1126/science.1175550 [DOI] [PubMed] [Google Scholar]
- 10. Ballini E, Morel JB, Droc G, Price A, Courtois B, Notteghem JL, et al. A genome-wide meta-analysis of rice blast resistance genes and quantitative trait loci provides new insights into partial and complete resistance. Mol Plant MicrobeIn. 2008;21: 859–868. 10.1094/MPMI-21-7-0859 [DOI] [PubMed] [Google Scholar]
- 11. Shinoda H, Toriyama K, Yunoki T, Ezuka A, Sakurai Y. Studies in the varietal resistance of rice to blast. 6. linkage relationship of blast resistance genes (in Japanese with English summary) Bull. Chugoku Agric Exp Stn Ser A. 1971; 20: 1–25. [Google Scholar]
- 12. Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L,Hayasaka H,et al. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 1999; 19: 55–64. [DOI] [PubMed] [Google Scholar]
- 13. Ramkumar G, Biswal A, Mohan KM, Sakthivel K, Sivaranjani A,Neeraja CN, et al. Identifying novel alleles of rice blast resistance genes Pikh and Pita through allele mining. Int Rice Res Notes. 2010; 117: 185. [Google Scholar]
- 14. Bryan GT, Wu KS, Farrall L, Jia Y, Hershey HP, Mcadams SA,et al. A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta . Plant Cell. 2000; 12: 2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu G, Lu G, Zeng L, Wang GL. Two broad-spectrum blast resistance genes, Pi9 (t) and Pi2 (t), are physically linked on rice chromosome 6. Mol Genet Genomics. 2002; 267: 472–480. [DOI] [PubMed] [Google Scholar]
- 16. Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M, Saitoh H,et al. A multifaceted genomics approach allows the isolation of the rice Pia blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011; 66: 467–479. 10.1111/j.1365-313X.2011.04502.x [DOI] [PubMed] [Google Scholar]
- 17. Liu X, Lin F, Wang L, Pan Q. The in silico map-based cloning of Pi36, a rice coiled-coil-nucleotide-binding site-leucine-rich repeat gene that confers race-specific resistance to the blast fungus. Genetics. 2007; 176: 2541–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J, Matsumoto T, et al. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance. Genetics, 2008; 180: 2267–2276. 10.1534/genetics.108.095034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Young ND. QTL mapping and quantitative disease resistance in plants. Annu Rev Phytopathol. 1996; 34: 479–501. [DOI] [PubMed] [Google Scholar]
- 20. Rushton PJ, Torres JT, Parniske M, Wernert P, Hahlbrock K,Somssich IE. Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J. 1996; 15: 5690 [PMC free article] [PubMed] [Google Scholar]
- 21. Maleck K, Levine A, Eulgem T, Morgan A, Schmid J, Lawton KA,et al. The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nature Genet. 2000; 26: 403–410. [DOI] [PubMed] [Google Scholar]
- 22. Kalde M, Barth M, Somssich IE, Lippok B. Members of the Arabidopsis WRKY group III transcription factors are part of different plant defense signaling pathways. Plant Microbe In. 2003; 16: 295–305. [DOI] [PubMed] [Google Scholar]
- 23. Wen N, Chu Z, Wang S. Three types of defense-responsive genes are involved in resistance to bacterial blight and fungal blast diseases in rice. Mol Genet Genomics. 2003; 269: 331–339. [DOI] [PubMed] [Google Scholar]
- 24. Ryu HS, Han M, Lee SK, Cho JI, Ryoo N,Heu S,et al. A comprehensive expression analysis of the WRKY gene superfamily in rice plants during defense response. Plant Cell Rep. 2006; 25: 836–847. [DOI] [PubMed] [Google Scholar]
- 25. Shimono M, Koga H, Akagi AYA, Hayashi N, Goto S,Sawada M, et al. Rice WRKY45 plays important roles in fungal and bacterial disease resistance. Molecular Plant Pathol. 2012; 13: 83–94. 10.1111/j.1364-3703.2011.00732.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuyek D. Blast, biotech and big business Jointly published by Biothai (Thailand), GRAIN, KMP (Philippines), MASIPAG (Philippines), PAN Indonesia, Philippine Greens and UBINIG (Bangladesh) Research Inc;2000. [Google Scholar]
- 27. Gardner RM, William Tindall G, Cline SM, Brown KL. Ergosterol determination in activated sludge and its application as a biochemical marker for monitoring fungal biomass. J Microbiol Methods. 1993; 17: 49–60. [Google Scholar]
- 28. Gretenkort MA, Ingram DS. The use of ergosterol as a quantitative measure of the resistance of cultured tissues of Brassica napus ssp. oleifera to Leptosphaeria maculans. J Phytopathol. 1993; 138: 217–224. [Google Scholar]
- 29. Plassard CS, Mousain DG, Salsac LE. Estimation of mycelial growth of basidiomycetes by means of chitin determination. Phytochemistry. 1982; 21: 345–348. [Google Scholar]
- 30. Bermingham S, Maltby L, Cooke RC. A critical assessment of the validity of ergosterol as an indicator of fungal biomass. Mycol Res. 1995; 99: 479–484. [Google Scholar]
- 31. Hu X, Nazar RN, Robb J. Quantification of Verticillium biomass in wilt disease development. Physiol Mol Plant. 1993; 42: 23–36. [Google Scholar]
- 32. Qi M, Yang Y. Quantification of Magnaporthe grisea during infection of rice plants using real-time polymerase chain reaction and northern blot/phospho imaging analyses. Phytopathology. 2002; 92: 870–876. 10.1094/PHYTO.2002.92.8.870 [DOI] [PubMed] [Google Scholar]
- 33. Sharma TR, Rai AK, Gupta SK, Singh NK. Broad-spectrum blast resistance gene Pi-kh cloned from rice line tetep designated as Pi54 . J Plant Biochem Biot. 2010; 19: 87–89. [Google Scholar]
- 34. Hittalmani S, Parco A, Mew TV, Zeigler RS, Huang N. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor Appl Genet. 2000; 100: 1121–1128. [Google Scholar]
- 35. Jia Y, Wang Z, Fjellstrom RG, Moldenhauer KAK, Azam MA,Correll J, et al. Rice Pi-ta gene confers resistance to the major pathotypes of the rice blast fungus in the United States. Phytopathology. 2004; 94: 296–301. 10.1094/PHYTO.2004.94.3.296 [DOI] [PubMed] [Google Scholar]
- 36. Qu S, Liu G, Zhou B, Bellizzi M, Zeng L,Dai L, et al. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site-leucine-rich repeat protein and is a member of a multigene family in rice. Genetics. 2006; 172: 1901–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abbruscato P, Nepusz T, Mizzi L, Del Corvo M, Morandini P,Fumasoni I, et al. OsWRKY22, a monocot WRKY gene, plays a role in the resistance response to blast. Mol Plant Pathol. 2012; 13: 828–841. 10.1111/j.1364-3703.2012.00795.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Taj G, Giri P, Tasleem M, Kumar A. MAPK Signaling Cascades and Transcriptional Reprogramming in Plant-Pathogen Interactions Approaches to Plant Stress and their Management: Springer;2014. pp. 297–316. [Google Scholar]
- 39. Pandey SP, Somssich IE. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009; 150: 1648–1655. 10.1104/pp.109.138990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pieterse CMJ, Van Loon LC. Salicylic acid-independent plant defence pathways. Trends Plant Sci. 1999; 4: 52–58. [DOI] [PubMed] [Google Scholar]
- 41. Correa Victoria FJ, Zeigler RS. Pathogenic variability in Pyricularia grisea at a rice blast hot spot breeding site in eastern Colombia. Plant Disease. 1993; 77: 1029–1035. [Google Scholar]
- 42. Rice INfGEo. Standard Evaluation System for Rice: IRRI, International Rice Research Institute; 1996. [Google Scholar]
- 43. Diedhiou AG, Borges WL, Sadio O, de Faria SM. Assessment of DNA extraction methods for detection of arbuscular mycorrhizal fungi in plant roots by nested-PCR. Acta Sci Biol Sci. 2014; 36: 433–441. [Google Scholar]
- 44. Sandhu GS, Kline BC, Stockman L, Roberts GD. Molecular probes for diagnosis of fungal infections. J Clin Microbiol. 1995; 33: 2913–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maleszka R, Clark Walker GD. Yeasts have a four-fold variation in ribosomal DNA copy number. Yeast. 1993; 9: 53–58. [DOI] [PubMed] [Google Scholar]
- 46.URI. Genomics and Sequencing Center of University of Rhode Island.
- 47. Witham FH, Blaydes DF, Devlin RM. Experiments in plant physiology New York: D Van Nostrand; 1971. pp. 55–58. [Google Scholar]
- 48. Parker D, Beckmann M, Enot DP, Overy DP, Rios ZC,Gilbert M, et al. Rice blast infection of Brachypodium distachyon as a model system to study dynamic host/pathogen interactions. Nat Protoc. 2008; 3: 435–445. 10.1038/nprot.2007.499 [DOI] [PubMed] [Google Scholar]
- 49. Simms D, Cizdziel PE, Chomczynski P. TRIzol: A new reagent for optimal single-step isolation of RNA. Focus. 1993; 15: 532–535. [Google Scholar]
- 50. Bolton MD. Primary metabolism and plant defense-fuel for the fire. Mol Plant Microbe In. 2009; 22: 487–497. 10.1094/MPMI-22-5-0487 [DOI] [PubMed] [Google Scholar]
- 51. Gohre V, Jones AME, Sklenar J, Robatzek S, Weber APM. Molecular crosstalk between PAMP-triggered immunity and photosynthesis. Mol Plant Microbe In. 2012; 25: 1083–1092. 10.1094/MPMI-11-11-0301 [DOI] [PubMed] [Google Scholar]
- 52. Swarbrick PJ, Schulze Lefert P, Scholes JD. Metabolic consequences of susceptibility and resistance (race-specific and broad-spectrum) in barley leaves challenged with powdery mildew. Plant Cell Environ. 2006; 29: 1061–1076. [DOI] [PubMed] [Google Scholar]
- 53. Berger S, Sinha AK, Roitsch T. Plant physiology meets phytopathology: plant primary metabolism and plant-pathogen interactions. J Exp Bot. 2007; 58: 4019–4026. 10.1093/jxb/erm298 [DOI] [PubMed] [Google Scholar]
- 54. Barriuso J, Solano BR, Gutierrez Manero FJ. Protection against pathogen and salt stress by four plant growth-promoting rhizobacteria isolated from Pinus sp. on Arabidopsis thaliana . Phytopathology. 2008; 98: 666–672. 10.1094/PHYTO-98-6-0666 [DOI] [PubMed] [Google Scholar]
- 55. Ishiga Y, Uppalapati SR, Ishiga T, Elavarthi S, Martin B, Bender CL. The phytotoxin coronatine induces light-dependent reactive oxygen species in tomato seedlings. New Phytol. 2009; 181: 147–160. 10.1111/j.1469-8137.2008.02639.x [DOI] [PubMed] [Google Scholar]
- 56. Marsh E, Alvarez S, Hicks LM, Barbazuk WB, Qiu W, Kovacs L,et al. Changes in protein abundance during powdery mildew infection of leaf tissues of Cabernet Sauvignon grapevine (Vitis vinifera L.). Proteomics. 2010; 10: 2057–2064. 10.1002/pmic.200900712 [DOI] [PubMed] [Google Scholar]
- 57. Pineda M, Sajnani C, Baron M. Changes induced by the Pepper mild mottle tobamovirus on the chloroplast proteome of Nicotiana benthamiana . Photosynth Res. 2010; 103: 31–45. 10.1007/s11120-009-9499-y [DOI] [PubMed] [Google Scholar]
- 58. Yang H, Huang Y, Zhi H, Yu D. Proteomics-based analysis of novel genes involved in response toward soybean mosaic virus infection. Mol Biol Rep. 2011; 38: 511–521. 10.1007/s11033-010-0135-x [DOI] [PubMed] [Google Scholar]
- 59. Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J,Lind K, et al. The real-time polymerase chain reaction. Mol Asp Med. 2006; 27: 95–125. [DOI] [PubMed] [Google Scholar]
- 60. Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007; 8: 774–785. [DOI] [PubMed] [Google Scholar]
- 61. Zeigler RS, Leong SA, Teng PS. Rice blast disease: Int. Rice Res. Inst; 1994. [Google Scholar]
- 62. Hammond Kosack KE, Kanyuka K. Resistance genes (R genes) in plants. Encycl Life Sci. 2007; 40: 1–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lane M is the 1 kb marker, and the other lanes show the isolated 28SrDNA gene from M. oryzae.
(TIF)
E: amplification efficiency. R2: correlation coefficient.
(TIF)
(TIF)
A melting temperature of 86.5°Cwas obtained.
(TIF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.