Abstract
Intrinsic immunity describes the set of recently discovered but poorly understood cellular mechanisms that specifically target viral pathogens. Their discovery derives in large part from intensive studies of HIV and SIV that revealed restriction factors acting at various stages of the retroviral life cycle. Recent studies indicate that some factors restrict both retroviruses and retrotransposons but surprisingly in ways that may differ. We screened known interferon-stimulated antiviral proteins previously untested for their effects on cell culture retrotransposition. Several factors, including BST2, ISG20, MAVS, MX2, and ZAP, showed strong L1 inhibition. We focused on ZAP (PARP13/ZC3HAV1), a zinc-finger protein that targets viruses of several families, including Retroviridae, Tiloviridae, and Togaviridae, and show that ZAP expression also strongly restricts retrotransposition in cell culture through loss of L1 RNA and ribonucleoprotein particle integrity. Association of ZAP with the L1 ribonucleoprotein particle is supported by co-immunoprecipitation and co-localization with ORF1p in cytoplasmic stress granules. We also used mass spectrometry to determine the protein components of the ZAP interactome, and identified many proteins that directly interact and colocalize with ZAP, including MOV10, an RNA helicase previously shown to suppress retrotransposons. The detection of a chaperonin complex, RNA degradation proteins, helicases, post-translational modifiers, and components of chromatin modifying complexes suggest mechanisms of ZAP anti-retroelement activity that function in the cytoplasm and perhaps also in the nucleus. The association of the ZAP ribonucleoprotein particle with many interferon-stimulated gene products indicates it may be a key player in the interferon response.
Author Summary
Retrotransposons are mobile DNA elements that duplicate themselves by a "copy and paste" mechanism using an RNA intermediate. They are insertional mutagens that have had profound effects on genome evolution, fostering DNA deletions, insertions and rearrangements, and altering gene expression. LINE-1 retrotransposons occupy 17% of human DNA, although it is believed that only about 100 remain competent for retrotransposition in any individual. The cell has evolved defenses restricting retrotransposition, involving in some cases interferon-stimulated genes (ISGs) that are part of the innate immune system that protects the cell from viral infections. We screened a panel of ISGs and found several to strongly limit retrotransposition in a cell culture assay. Our investigations increase understanding of how ZAP, an important restriction factor against positive- and negative-strand RNA and some DNA viruses, also interacts with human retrotransposons to prevent genome mutation. Microscopy and immunoprecipitation show a close association of ZAP protein with the L1 ribonucleoprotein particle, as well as MOV10, an RNA helicase that also inhibits retrotransposons. A detailed examination of the ZAP protein interactome reveals many other ISGs that directly bind ZAP, and suggests new directions for exploring the mechanisms of ZAP-mediated anti-retroelement activity.
Introduction
Host restriction factor proteins are part of the intrinsic immune system of the cell, forming an early line of defense against viral infection. Intrinsic immunity is triggered when viral RNAs are recognized by pattern-recognition receptors, such as Toll-like and retinoic acid-inducible gene (RIG-I)-like receptor family members, causing activation of an effector protein (for example, IRF3) and the expression of interferon (IFN) and hundreds of IFN-stimulated genes (ISGs).
Many viral restriction factors are ISGs that function by diverse mechanisms against a wide range of viral pathogens. For example, Myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) (MX1, also known as MXA)) and MX2 (MXB) are closely related members of the IFN-induced dynamin family of large GTPases. MX1 is a broad-spectrum inhibitor of many RNA and DNA viruses (reviewed in [1]). IFN-induced transmembrane protein family members (IFITM1/2/3) are also potent inhibitors of a range of viruses including HIV-1, although their mechanisms of action are unclear ([2]; reviewed in [3]). BST2 (Tetherin) is a type II transmembrane glycoprotein capable of trapping enveloped virions at the cell surface (reviewed in [4]). RSAD2 (Viperin) is an endoplasmic reticulum-associated protein that inhibits many RNA and DNA viruses at multiple stages of the viral life cycle, and which may be involved in innate immune signaling (reviewed in [5]). RNA helicases and IFIH1 interact with Mitochondrial antiviral signaling protein (MAVS), a mitochondrial outer membrane protein, activating formation of the MAVS signalosome and upregulation of NF-κB and IRF3 signaling pathways [6]. ISG20 is a 3'-5' exoribonuclease that inhibits single-strand RNA viruses including HIV-1 [7]. The transcriptional regulator TRIM28 (KAP1) also limits HIV integration by binding acetylated integrase and inducing its deacetylation by recruiting HDAC1 [8].
While the cell can be infected by a wide variety of viruses, unrestricted activity of endogenous retroelements also poses a threat to genome integrity and cell function. Long-terminal repeat (LTR) retrotransposons include the human endogenous retroviruses (HERVs) that comprise 8% of the human genome, although no HERVs capable of replication have been identified. However, increased HERV expression has been implicated in multiple sclerosis, lupus, amyotrophic lateral sclerosis, and autoimmune rheumatic disease, although whether the increase is the cause or effect of disease remains to be determined ([9]; reviewed in [10–12]).
Non-LTR retrotransposons comprise at least one-third of the human genome and remain an ongoing cause of disease [13,14]. LINE-1s (L1s) are the only active autonomous mobile DNA remaining in humans, and among ∼500,000 copies at least 100 remain potentially active for retrotransposition in any human individual [15,16]. L1s have also been responsible for the genomic insertion in trans of thousands of processed pseudogenes and a million SINEs (Alus and SVAs). The current residual activity of human retrotransposons is the background that escapes a variety of mechanisms that have evolved to limit replication of mobile DNA.
Phylogenetic analyses suggest that eukaryote non-LTR retrotransposons predate LTR retrotransposons, which in turn gave rise to the retroviruses through the acquisition of an envelope gene [17–19]. The ancient origin and interrelatedness of the major classes of retroelements predicts they will be subject to some of the same host restriction factors. On the other hand, significant differences between their modes of replication suggest that non-LTR retrotransposons and retroviruses could be affected by the same restriction factors in divergent ways. For example, Apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like 3 (APOBEC3) family members, first shown to inhibit HIV by hypermutation of minus-strand DNA, were then found to potently inhibit retrotransposition but without obvious hypermutation, suggesting a unique pathway (reviewed in [20,21]). Recently, it was proposed that APOBEC3A inhibits L1 retrotransposition by deaminating transiently exposed single-strand genomic DNA that flanks the site of L1 integration [22]. The Aicardi-Goutières syndrome (AGS)-related anti-retroviral protein SAM domain and HD domain 1 protein (SAMHD1) inhibits retroviruses in non-dividing myeloid cells and resting CD4+ T cells by depleting dNTP levels. However, SAMHD1 also limits non-LTR retrotransposition in dividing cells, suggesting an inhibitory mechanism different from nucleotide depletion [23].
Other innate restriction factors also suppress retrotransposons. LINE-1 activity is upregulated in cells deficient for TREX1, a gene associated with systemic lupus erythematosus (SLE) and, like SAMHD1, with AGS [24]. RNaseL, a member of the IFN antiviral response to dsRNA, likely restricts retrotransposons in cell culture by cleavage of their mRNAs [25]. Finally, the RNA helicase MOV10, previously reported to affect replication of several RNA viruses, also limits activity of all human retrotransposons [26–28]. MOV10 may target retroelement complexes for degradation, possibly by RNA-induced silencing complexes (RISC). Clearly, studying factors that restrict both retrotransposons and retroviruses can inform both fields.
Therefore, we overexpressed a panel of antiviral ISGs and assayed for their previously untested effects on LINE-1 retrotransposition in a cell culture assay. Several of these factors strongly inhibited retrotransposition of an L1 reporter construct. We report that the zinc finger antiviral protein ZAP (also called Zinc finger CCCH-type, antiviral 1, ZC3HAV1, and Poly (ADP-ribose) polymerase 13, PARP13) is a potent inhibitor of not only viruses but also retrotransposons. ZAP targets positive- and negative-strand RNA viruses of several families, including Retroviridae (HIV-1, MoLV, and XMRV), Filoviridae (Ebola and Marburg), Togaviridae (alpha-, sindbis, Semliki Forest, and Ross river viruses), and Hepadnaviridae (hepatitis B) [29], and has been shown to inhibit the double-stranded DNA murine gammaherpesvirus [30]. However, restriction is not universal: ZAP fails to inhibit vesicular stomatitis, poliovirus, yellow fever, and herpes simplex I viruses [31]. The presence of orthologs in fish, birds and reptiles suggests that ZAP (like MOV10) is ancient in origin [32,33].
L1 expresses a 6-kb bicistronic RNA that encodes a 40 kD RNA-binding protein (ORF1p) of essential but uncertain function for retrotransposition, and a 150 kD ORF2 protein with endonuclease and reverse transcriptase (RT) activities. In the cytoplasm, ORF1p and ORF2p preferentially bind their own encoding RNA in cis to form a functional ribonucleoprotein particle (RNP) [34,35]. We show that ZAP strongly restricts retrotransposition in cell culture with loss of L1 RNA and RNP integrity. Association of ZAP with the L1 RNP is supported by co-IP and co-localization with ORF1p in cytoplasmic granules. Mass spectrometry (MS) analyses of the ZAP proteome revealed proteins that bind ZAP, including MOV10, and suggest possible mechanisms of ZAP-mediated retroelement restriction.
Results
ISGs limit LINE1 retrotransposition in a cell culture assay
We asked if increased interferon limits L1 activity in a cell culture assay for retrotransposition. In this assay, an enhanced green fluorescent protein (EGFP) reporter gene cassette, interrupted by a backwards intron and inserted in opposite transcriptional orientation into the 3' UTR of L1-RP (a highly active L1 [36]), is expressed only when the L1 transcript is spliced, reverse-transcribed, its cDNA inserted in the genome, and the EGFP reporter gene expressed from its own promoter [37,38]. The full-length L1 and reporter cassette are cloned in a modified version of pCEP4 vector (Invitrogen) lacking a cytomegalovirus (CMV) promoter. Therefore, expression of the L1 is driven by its own 5' UTR.
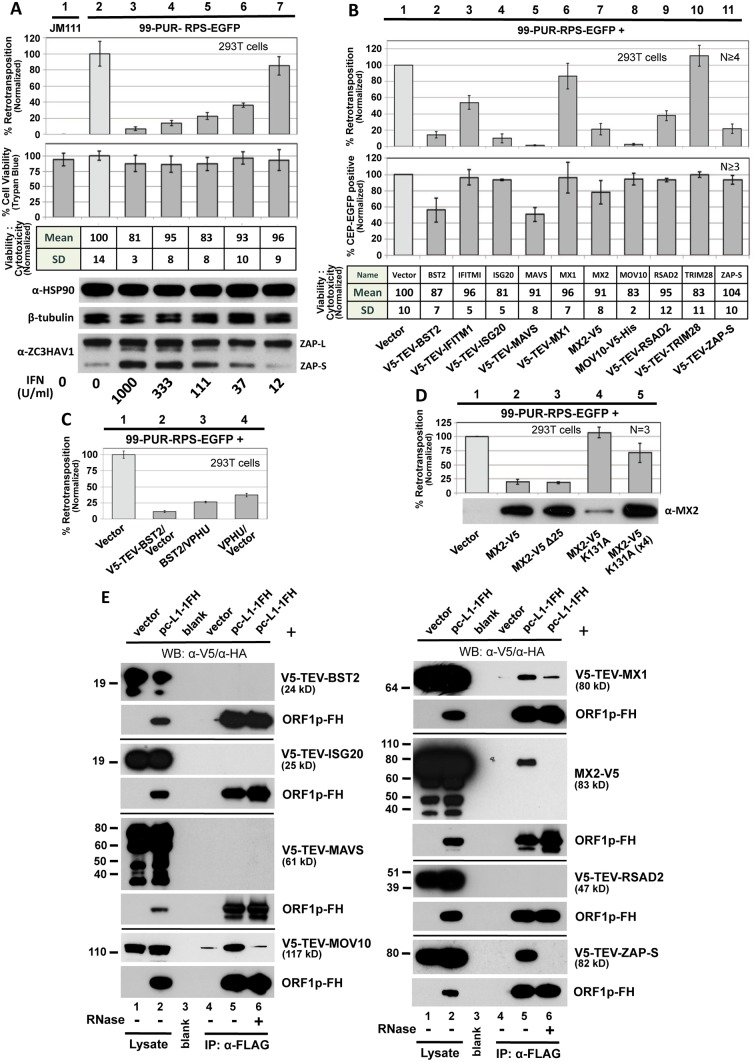
We transfected HEK 293T cells with the L1-EGFP reporter (99-PUR-RPS-EGFP) in the presence of increasing amounts of Universal Type I Interferon (IFN) alpha, and at 5 days post-transfection assayed for fluorescent (i.e. retrotransposition-positive) cells by flow cytometry. Addition of IFN reduced cell culture retrotransposition in a dose-dependent manner. At 1000 U IFN/ml, retrotransposition was over 90 percent less than non-treated controls, with a loss of cell viability of less than 20 percent as determined by trypan blue staining and the MultiTox-Fluor Multiplex Cytotoxicity Assay (Promega) (Fig 1A, compare lanes 2 and 3).
Fig 1. Selected ISG proteins strongly inhibit L1 retrotransposition in 293T cells.
(A) Type I interferon inhibits cell culture retrotransposition in a dose-dependent manner. Universal Type I Interferon (PBL Assay Science) at increasingly dilute concentrations was applied at the time of transfection with 99-PUR-RPS-EGFP and replenished once during the course of the experiment. Flow cytometry was performed on Day 5. Four replicate wells were analyzed for each concentration tested. Normalized percentages of retrotransposition-positive cells are shown (top bar chart). To assess possible cytotoxicity caused by interferon, cells were 1) stained on day 5 with trypan blue and counted using a Cellometer Auto T4 Cell Viability Counter (Nexcelom) (lower bar chart), and 2) assayed with the MultiTox-Fluor Multiplex Cytotoxicity assay (table). Western blotting (below) shows that treatment with interferon did not significantly alter expression of endogenous heat shock protein 90 (HSP90) or β-tubulin-2, but strongly induced expression of the short but not the long isoform of ZAP. For protein preparations, an equal volume of cells was removed from each of the four test wells prior to FACs analysis and pooled. JM111: construct 99-PUR-JM111-EGFP, having an ORF1p double-point mutant, was used as a negative control for retrotransposition. (B) Top panel: 99-PUR-RPS-EGFP was cotransfected with empty vector (pcDNA3 or pcDNA6 myc/his B) or test constructs expressing V5-tagged proteins. Five days later, the numbers of EGFP-positive cells were determined by flow cytometry. Each construct pair was tested in quadruplicate wells for each of 4 to 6 independent experiments. Results are normalized to empty vector control (pale shaded bar). Middle panel: To control for off-target effects, test constructs were cotransfected with CEP-EGFP, a plasmid that constitutively expresses EGFP. Four days later, cells were assayed for gain or loss of fluorescent cells. Percentages of fluorescent cells are normalized to the empty vector control. Lower table: Results of MultiTox-Fluor Multiplex Cytotoxicity assay (Promega) for potential cell toxicity caused by overexpression of test proteins. Test constructs were transfected in 293T cells in 96-well plates and assayed at 3 days. The bar chart shows ratios of live to dead cell readings normalized to empty vector control. Results of a single experiment are shown. (C) Inhibition of 99-PUR-RPS-EGFP retrotransposition by BST2 (tetherin) is partially relieved by cotransfection of a codon-optimized version of HIV-1-encoded Vpu. (D) MX2 with a K131A mutation has diminished ability to inhibit retrotransposition, but loss of its N-terminal nuclear localization signal (MX2-V5 Δ25, lane 3) maintains inhibition. Amounts of the K131A mutant plasmid cotransfected were 0.5 μg (lane 4) and 2 μg (4X; lane 5). Proteins were sampled at 5 days post-transfection and detected by α-MX2 antibody. (E) Ectopically expressed V5-tagged MOV10, MX1, MX2, and ZAP proteins associate in an RNA-dependent manner with L1 complexes expressed from pc-L1-1FH in 293T cells. Input lysate, lanes 1,2; IP on α-FLAG affinity gel, lanes 4–6: Western blotting (WB) with α-V5 antibody to detect test proteins (upper panel for each test protein); WB with α-HA antibody to detect ORF1p-FH (lower panels). IP reactions were in the absence (lanes 1–5) or presence (lane 6) of 50 μg/ml RNase. Molecular weight indicated for each protein includes the epitope tag. Exposure times for the different panels vary.
Several interferon-inducible proteins are known to restrict not only viruses but also endogenous retroelements. To extend the list of these proteins, we next screened a panel of ISGs and restriction factors with known antiviral activities for effects of their expression on L1 retrotransposition (Fig 1B, upper bar chart). To reveal differences of transfection efficiency, cytotoxicity or reporter gene expression caused by the test proteins, we followed the approach of Wei et al. [39] and in conjunction with the retrotransposition assays transfected CEP-EGFP, a vector that constitutively expresses EGFP, together with empty vector or test constructs. After 4 days we performed flow cytometry to assay for loss of green cells (Fig 1B, lower bar chart). It should be noted that the CEP-EGFP assay is a snapshot of GFP fluorescence at a single time-point, while the retrotransposition assay measures accumulated EGFP insertions that occur over 5 days. Therefore, the MultiTox-Fluor Multiplex Cytotoxicity Assay was also used as a direct assay for cell toxicity (assessed at Day 3 post-transfection). This dual-detection assay generates a ratio of live to dead cell readings, thereby normalizing for cell number (Fig 1B, table).
293T cells were cotransfected with 99-PUR-RPS-EGFP and empty vector or tagged ISG cDNA constructs. With the exception of MOV10, none of the proteins shown in Fig 1B had previously been tested for effects on cell culture retrotransposition. Six factors, MOV10 (as previously reported [26–28]), BST2, ISG20, MAVS, MX2, and ZAP, reduced plasmid-directed LINE1 retrotransposition in 293T cells by over 75 percent (Fig 1B). While minimal toxicity was detected by the MultiTox-Fluor assay for all proteins, BST2 and MAVS reduced CEP-EGFP vector expression 44 and 50 percent, respectively. However, loss of CEP-EGFP signal only partly explains the strong inhibition of cell culture retrotransposition observed for these proteins (86 and over 98 percent, respectively).
To assess specificity of BST2-induced reduction of retrotransposition, we coexpressed Vphu, a codon-optimized version of the HIV-1-encoded BST2 antagonist Vpu [40]. Vphu restored L1 retrotransposition over 2-fold (11 to 26 percent), an amount limited by the fact that overexpression of Vphu itself reduced retrotransposition to 37 percent (Fig 1C).
We also tested MX2 mutant proteins for their activity in the cell culture assay (Fig 1D). The N-terminal 25 amino acids of MX2 encode a nuclear envelope targeting domain essential for HIV-1 inhibition. Conversely, while MX2 mutated at K131A fails to bind GTP, it still inhibits HIV-1 as if wild-type [41–44]. Contrary to the results for HIV-1, deleting the first 25 residues of our MX2 construct failed to block its inhibition of retrotransposition, but the K131A mutant lost ability to restrict L1s. Because the K131A mutant is expressed at a lower level than the wild-type protein (also noted by [45]), we increased the amount of mutant MX2 plasmid transfected fourfold but still failed to restore inhibition of retrotransposition to wild-type levels. Thus, the mechanisms by which MX2 restricts retroviruses and retrotransposons appear to differ in some aspects. Unfortunately, endogenous MX2 is expressed at detectable levels in none of the cell lines we use for our retrotransposition assay (293T, HeLa, or human embryonal carcinoma 2102Ep cells) and is induced by IFN in HeLa cells only (S1 Fig). Consequently, we could not assay endogenous MX2 for effect on retrotransposition, and we pursued MX2-related experiments no further.
Finally, we tested for interaction of IGS factors with the L1 RNP (Fig 1E). Expression of pc-L1-1FH, a construct containing L1-RP with a tandem FLAG-HA tag fused to the C-terminus of its ORF1, permits immunoprecipitation (IP) of basal L1 RNP complexes [46]. pc-L1-1FH and V5 epitope-tagged ISG proteins were coexpressed in 293T cells, followed by their co-IP on α-FLAG agarose. TRIM28 and IFITM1 bound non-specifically to the affinity agarose, and their associations with L1 were inconclusive. MOV10 (as previously reported [27]), MX1, MX2, and the short isoform of ZAP (ZAP-S/PARP13.2) all co-immunoprecipitated with the L1 RNP. These interactions were lost with RNase treatment. We have not confirmed that these proteins directly bind L1 RNA. Rather, proteins might bind non-L1 RNAs or other multi-protein complexes captured within the L1 RNP. Moreover, an unknown amount of tagged ORF1p along with its bound partners, free in solution and not part of retrotransposition-competent RNPs, will have co-purified within immunoprecipitates. Only functional analyses of restriction factors associated with the L1 RNP will ascertain their relevance to L1 biology.
Thus, we have expanded the growing list of IFN-stimulated cellular restriction factors that inhibit human retrotransposition. We decided to focus on the zinc finger protein ZAP for further investigation.
The ZAP zinc finger domain is sufficient for restriction of retrotransposition
ZAP (PARP13) is a member of the poly(ADP-ribose) polymerase (PARP) family of 17 proteins, some of which are capable of poly(ADP) ribosylation (pADPr) of acceptor proteins using NAD+ as a substrate. Human ZAP is a predominantly cytoplasmic protein that exists in two alternatively spliced isoforms. Long isoform 1 (ZAP-L) possesses a defective C-terminal PARP-like domain that lacks a catalytic glutamate residue essential for pADPr (Fig 2A) [47,48]. This domain is absent in ZAP-S. It was previously demonstrated that the first 254 amino acids of rat ZAP (NZAP), comprising only the four CCCH-type zinc fingers, were sufficient to induce loss of viral mRNA and severely inhibit infection [49]. The zinc finger domain is believed to mediate binding of ZAP to long ZAP-responsive element (ZRE) sequences within the RNAs of target viruses [29,50–52]. Hayakawa et al. [53] reported that ZAP-S, but not ZAP-L, is an ISG whose endogenous expression is increased by Type I interferon, a fact we confirm for 293T cells (Fig 1A, bottom panel).
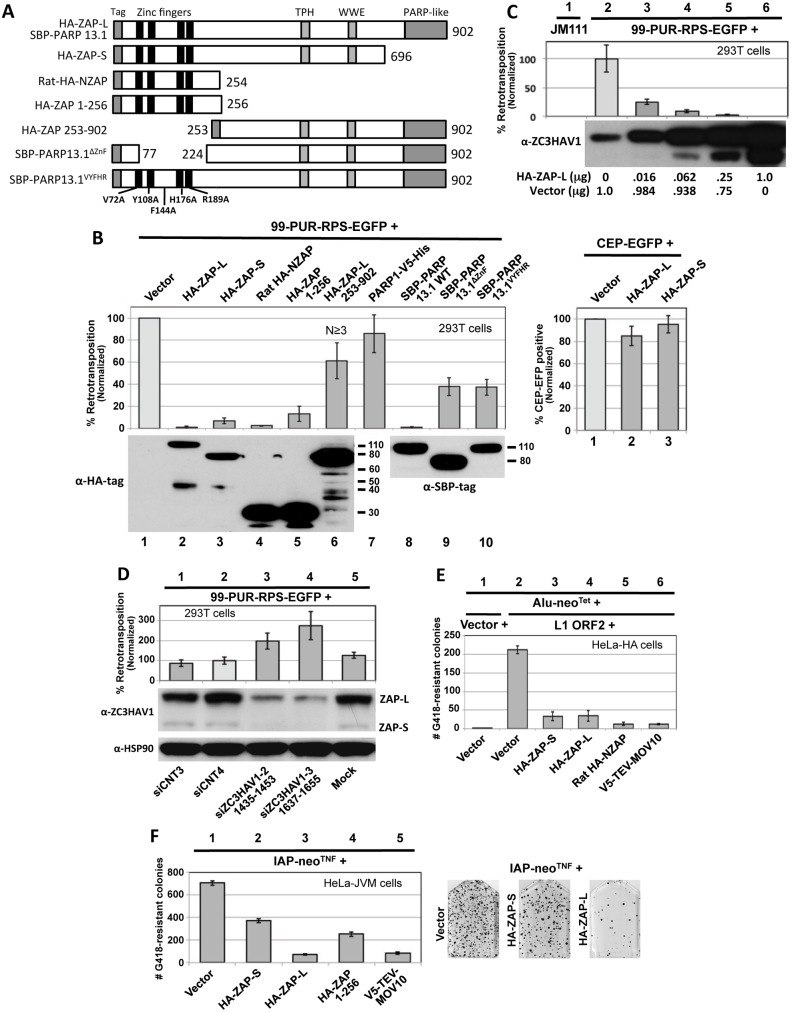
Fig 2. Evidence from cell culture assays that ZAP inhibits retrotransposition.
(A) The structure of ZAP constructs used in the retrotransposition assays. Known protein domains are indicated. (B) Left upper panel, right: 99-PUR-RPS-EGFP was cotransfected in 293T cells with empty vector or constructs expressing tagged ZAP constructs, and 5 days later the percentages of EGFP-positive cells were determined by flow cytometry. Each construct was tested in quadruplicate wells for each of at least 3 independent experiments. Results are normalized to empty vector control (lane 1). Right upper panel: to assess transfection efficiency and cell toxicity, ZAP constructs were cotransfected with CEP-EGFP and fluorescent cells were assayed 4 days later by flow cytometry. Lower panels: Western blot showing expression of tagged ZAP constructs transfected alone. Lower molecular weight bands are considered to be degradation products and are most evident for HA-ZAP-L 253–902 (lane 6). Proteins were sampled at 4 days post-transfection. (C) Expression of HA-ZAP-L inhibits retrotransposition in a dose-dependent manner. Increasing amounts of HA-ZAP-L expressing plasmid, mixed with empty vector to maintain equal DNA concentrations, were cotransfected with 99-PUR-RPS-EGFP in 293T cells and assayed for retrotransposition at 5 days. Results are normalized to empty vector control. Western blot analysis of cytoplasmic lysates (below) shows that even small amounts of exogenous ZAP-L protein can inhibit retrotransposition. Lane 2 shows the background level of endogenous ZAP expression in untransfected 293T cells. (D) Loss of endogenous ZAP expression increases retrotransposition. 293T cells were mock transfected (lane 5) or transfected with scrambled siRNAs (lanes 1 and 2), or sequences directed against ZAP (lanes 3 and 4), and tested for retrotransposition competency of 99-PUR-RPS-EGFP. Bottom panels: Western blots showing that siZC3HAV1-1(1435–1453) and siZC3HAV1-2(1637–1655) decreased endogenous ZAP-L and ZAP-S protein levels by about 90 percent in 293T cells, but had no effect on levels of HSP90. (E) Overexpression of HA-ZAP-L, HA-ZAP-S, rat HA-NZAP, and V5-TEV-MOV10 constructs strongly decreased Alu retrotransposition in HeLa-HA cells. As described in [61], a Ya5 Alu is cloned in a plasmid containing the 7SL pol III enhancer and neo Tet reporter cassette consisting of an antisense copy of a neomycin phosphotransferase gene (neo) disrupted by a self-splicing Group I intron. Upon transcription, the intron is spliced out. When this construct is co-expressed with L1 ORF2 (lanes 2 to 7) but not an empty vector (lane 1), Alu RNAs are reverse-transcribed along with the neo gene and integrated into the genome to confer neomycin resistance. Following 15 days of treatment with neomycin, resistant colonies were stained and counted. Colony counts are not normalized. (F) Expression of HA-ZAP-L and V5-MOV10 strongly inhibited (lanes 3 and 5), but HA-ZAP-S and HA-ZAP 1–256 (lanes 2 and 4) modestly inhibited mouse IAP element retrotransposition in HeLa-JVM cell culture [62]. Cells were treated with neomycin to select for retrotransposition events. Colony counts are not normalized. On the right are representative T75 flasks with Giemsa-stained IAP retrotransposition-positive colonies in the absence (left) or presence (middle and right) of HA-ZAP.
We cotransfected 99-PUR-RPS-EGFP in 293T cells, together with epitope-tagged ZAP or empty vector, and assayed for fluorescent cells at 5 days post-transfection (Fig 2A and 2B). Coexpressed human HA-tagged HA-ZAP-S and HA-ZAP-L, and SBP-PARP13.1 (ZAP-L with an N-terminal streptavidin binding protein tag [54]) caused a precipitous decrease in the number of retrotransposition-positive 293T cells relative to the empty vector control. L1 inhibition caused by HA-ZAP-S was greater than by V5-TEV-ZAP-S (Figs 1B and 2B), and it is possible that the long tag of the latter construct partially reduced ZAP-S activity. In turn, decrease in retrotransposition was greater for HA-ZAP-L and SBP-PARP13.1 than HA-ZAP-S. ZAP-L inhibited retrotransposition in a dose-dependent manner, and even the lowest levels of ectopically expressed protein caused significant loss of L1 activity (Fig 2C, lanes 3–6). Our data are in line with previous observations that ZAP-L exerts stronger activity against Semliki Forest virus, Sindbis virus, and Moloney leukemia virus (MoLV) than ZAP-S [32,55]. This increased antiviral activity of ZAP-L has been attributed to S-farnesylation within the C-terminal PARP-like domain [56]. Overexpression of PARP1, a nuclear protein, failed to inhibit retrotransposition (Fig 2B, lane 7).
We next tested for the effect of ZAP's N-terminal zinc finger and predicted RNA-binding domain. Expressing only the first 256 residues of human ZAP (HA-ZAP 1–256) reduced L1 retrotransposition to 13 percent of vector control (Fig 2B, lane 5). The analogous N-terminal domain of rat ZAP (HA-NZAP) reduced cell culture retrotransposition even more effectively (to 2.5 percent, although, it should be noted that these truncated ZAP proteins were expressed at higher levels than ZAP-S or ZAP-L; Fig 2B, lower panel).
ZAP constructions with deletions of the zinc finger domain (constructs HA-ZAP-L 253–902 and SBP-PARP13.1ΔZnF, which lacks residues 77 to 223) or mutated for five residues considered important for RNA binding (SBP-PARP13.1VYFHR) inhibited retrotransposition to a lesser but still significant degree (40–65 percent of vector control; Fig 2B, lanes 6, 9 and 10). Similar results were seen when the retrotransposition assay was performed in HeLa cells (S2A Fig). Together, our data show that the ZAP zinc finger domain alone is sufficient but not exclusive for L1 inhibition.
Tagged ZAP constructs had minimal effect on the expression of cotransfected CEP-EGFP plasmid (Fig 2B, right bar chart). As an additional test for cell toxicity, ZAP constructs or empty vector were cotransfected with a plasmid that constitutively expresses the blasticidin S-resistance gene (bsr). Coexpression of HA-ZAP-S, HA-ZAP-L, or rat HA-NZAP did not reduce the number of bsr-expressing foci remaining after 10 days of selection with blasticidin (S2B Fig).
We next cotransfected ZAP with ORFeus-HS, a synthetic human LINE-1 construct tagged with the EGFP reporter cassette and containing codon-modified ORF1 and ORF2 sequences and a CMV promoter in place of the L1 5' UTR [57]. Even though ORFeus-HS contains little authentic LINE1 DNA sequence, its retrotransposition in 293T cells was decreased by coexpression of ZAP full-length or N-terminal truncated proteins in the same manner as human L1-RP (S2C Fig). We also assayed ORFeus-Mm [58], a modified version of L1spa, a mouse L1 element with low retrotransposition activity [59], possessing both CMV promoter and mouse L1 5' UTR, and codon-optimized to boost its activity to a level similar to that of human L1-RP. ZAP also inhibited cell culture retrotransposition of ORFeus-Mm (S2D Fig). Thus, ZAP suppression of L1s is neither DNA sequence-, promoter-, nor species-specific. We also determined by HIRT preparation plasmid recovery and subsequent PCR that expression of ZAP had no effect on the stability of 99-PUR-RPS-EGFP reporter plasmid DNA (S2E Fig).
We next asked whether endogenous ZAP inhibits cell culture L1 retrotransposition. We confirmed by Western blotting that two previously described siRNA sequences [60] efficiently depleted endogenous ZAP protein when transiently transfected in 293T cells (Fig 2D). Depletion of ZAP enhanced L1 retrotransposition 2- to 3-fold compared with control siRNAs or mock-transfected cells.
We also determined the effect of ZAP expression on retrotransposition of the non-autonomous Alu SINE retrotransposon. Using the assay of Dewannieux et al. [61], we cotransfected in HeLA-HA cells an Alu reporter construct tagged with the neomycin phosphotransferase gene (Alu-neo Tet), an ORF2 construct (pCEP-5′UTR-ORF2-No-Neo) to drive retrotransposition, and ZAP constructs or empty vector. Expression of HA-ZAP-L, HA-ZAP-S, rat HA-NZAP, and MOV10 reduced the number of neomycin-resistant retrotransposition-positive colonies more than 80 percent compared with empty vector control. pCEP-5′UTR-ORF2-No-Neo consists of CMV promoter, the L1 5' UTR, and ORF2 sequences only, suggesting that L1 ORF1 or 3' UTR sequence is not essential for ZAP inhibition of retrotransposition.
All human endogenous retroviruses are thought to be incapable of retrotransposition due to inactivating mutations. However, mouse intracisternal A particle (IAP) LTR retrotransposons actively retrotranspose and cause new mutations. Using an established cell culture assay [62], we found that overexpression of HA-ZAP-L and V5-TEV-MOV10 strongly restricted insertion of neo-tagged IAP elements in HeLa-JVM cells (Fig 2F). Inhibition by HA-ZAP-S and HA-ZAP 1–256 was less severe, consistent with our results for L1 retrotransposition.
Thus, expression of ZAP inhibits not only retroviruses but also both LTR and non-LTR retrotransposons, including mobile DNA currently active in the human genome.
ZAP and the L1 RNP associate and colocalize in cytoplasmic granules
We examined the subcellular distribution of ZAP and its association with the L1 RNP. When expressed from a full-length L1 construct, ORF1p is present as an RNP in cytoplasmic stress granules (SGs) together with L1 RNA, ORF2p, and many other RNA-binding proteins. Granules are also detected in cells that express endogenous ORF1p at high levels, and their formation is not dependent upon external stress applied to the cell [63–65]. Stress granules (SGs) are cytoplasmic aggregates that contain stalled 48S pre-initiation complexes and are induced by a range of stresses. P-bodies (PBs) are constitutively expressed cytoplasmic granules rich in factors of RNA decay, including those of RISC [66]. ORF1p granules generally do not overlap, but may juxtapose PBs [63]. Interestingly, we reported [64], and others have confirmed [67], that ORF2p is detected in only a minor subset of ORF1p-positive cells when the two proteins are coexpressed from an L1 construct. The reason for this is unknown.
Both ZAP-L and ZAP-S have been reported to colocalize in the cytoplasm with markers of PBs and SGs [52,54,68,69]. Epitope-tagged ZAP-S, coexpressed with ORF1-GFP-L1-RP (a construct with CMV promoter, ORF1 C-terminally tagged with EGFP and intact downstream L1 sequence [64]) and detected by immunofluoresence (IF) of fixed 293T cells, strongly colocalizes with ORF1-GFP and SG marker proteins eIF3 (Fig 3A) and TIA-1. Endogenous ZAP and ORF1p similarly colocalize in the cytoplasm of 293T and 2102Ep cells (Fig 3B and 3C).
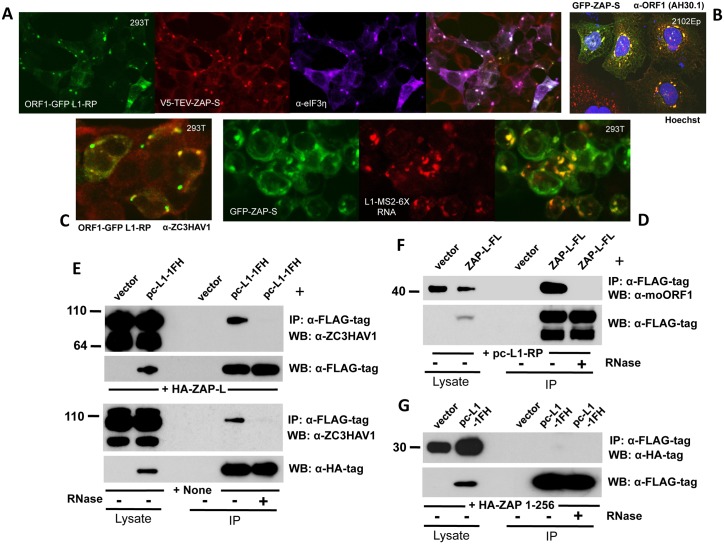
Fig 3. ZAP colocalizes with L1 ORF1 protein in cytoplasmic granules and interacts with its RNP in an RNA-dependent manner.
(A) EGFP-tagged ORF1p expressed from the construct ORF1-GFP L1-RP colocalizes with V5- tagged ZAP-S in the cytoplasm and SGs of 293T cells. DyLight 650-conjugated secondary antibody was used to detect endogenous eIF3η, a marker of SGs. (B) GFP-tagged ZAP-S (construct GFP-PARP13.2) colocalizes with endogenous ORF1p in cytoplasmic granules of 2102Ep cells. Cell nuclei were stained with Hoechst 33342. (C) EGFP-tagged ORF1p colocalizes with endogenous ZAP in the cytoplasm of 293T cells. (D) L1 RNA tagged with MS2-repeats and expressed from construct 99-PUR-L1-RP MS2-6X colocalizes with GFP-tagged ZAP-S when detected by antisense RNA-FISH probe Cy3-MS2. (E) Exogenously expressed HA-ZAP-L (top panels) and endogenous ZAP (bottom panels) associate with pc-L1-1FH immunoprecipitates from 293T cells. The associations are lost with RNase treatment. (F) FLAG-tagged ZAP-L (ZAP-L-FL) co-IPs untagged ORF1p expressed from construct pc-L1-RP in 293T cells. (G) Human HA-ZAP 1–256, comprising the N-terminal zinc-finger domain only, fails to co-IP with pc-L1-1FH-encoded ORF1p.
To track L1 RNA in fixed cells we used construct 99-PUR-L1-RP-MS2-6X [64], consisting of L1-RP tagged at its C-terminus with a tandem array of six 19-bp stem loop sequences that bind bacteriophage MS2 coat protein. This construct was cotransfected with GFP-tagged ZAP-S and its RNA was detected in fixed cells with a fluorescent in situ hybridization (FISH) probe to the MS2 repeats. ZAP-S colocalized with L1 RNA in cytoplasmic granules (Fig 3D).
As noted above, V5-TEV-tagged ZAP-S interacts with L1 RNPs in an RNA-dependent manner (Fig 1D). HA-ZAP-L and endogenous full-length ZAP also co-IP with L1 RNPs expressed from pc-L1-1FH (Fig 3E, upper and lower panels, respectively). Shorter ZAP products, presumed to be degradants, were not recovered in the immunoprecipates. Conversely, FLAG-tagged ZAP-L (ZAP-L-FL) coimmunoprecipitates untagged ORF1p expressed from a full-length L1 construct (Fig 3F). Unexpectedly, despite the ability of the zinc-finger domain alone to strongly inhibit retrotransposons (Fig 2B), we failed to co-IP human HA-ZAP 1–256 (Fig 3G) or rat HA-NZAP with pc-L1-1FH RNPs. Chen et al. [52] proposed that some residues in the zinc finger domain are involved in ZAP's interaction with protein factors rather than RNA. Perhaps an unknown non-L1 protein or RNA can recruit truncated ZAP to the L1 RNP, although this requires further investigation.
In summary, L1 RNPs and ZAP associate and are directed to the same cytoplasmic compartments.
The ZAP protein interactome
We previously identified 96 proteins associated with the L1 ORF1p RNP and confirmed a subset of these by direct co-IP and subcellular colocalization experiments [46]. We wished to determine the ZAP protein interactome and identify its members that are shared with that of the L1. We transfected ZAP-L-FL and empty vector in parallel in 293T cells and performed IP from whole cell lysates in the presence or absence of RNase. Purification was highly efficient with relatively few proteins identified in vector-only lanes (Fig 4A). Following IP, complex samples were analyzed by tandem mass spectrometry (MS). Excluding 36 ribosomal proteins and likely contaminants (such as keratins), 78 proteins satisfied three criteria: 1) predicted by 3 or more peptides, 2) present in two independent replicate IPs, and 3) unique to ZAP-L-FL isolates. Eleven proteins were shared with the L1 ORF1p RNP interactome defined in our previous paper (Table 1, S1 Table).
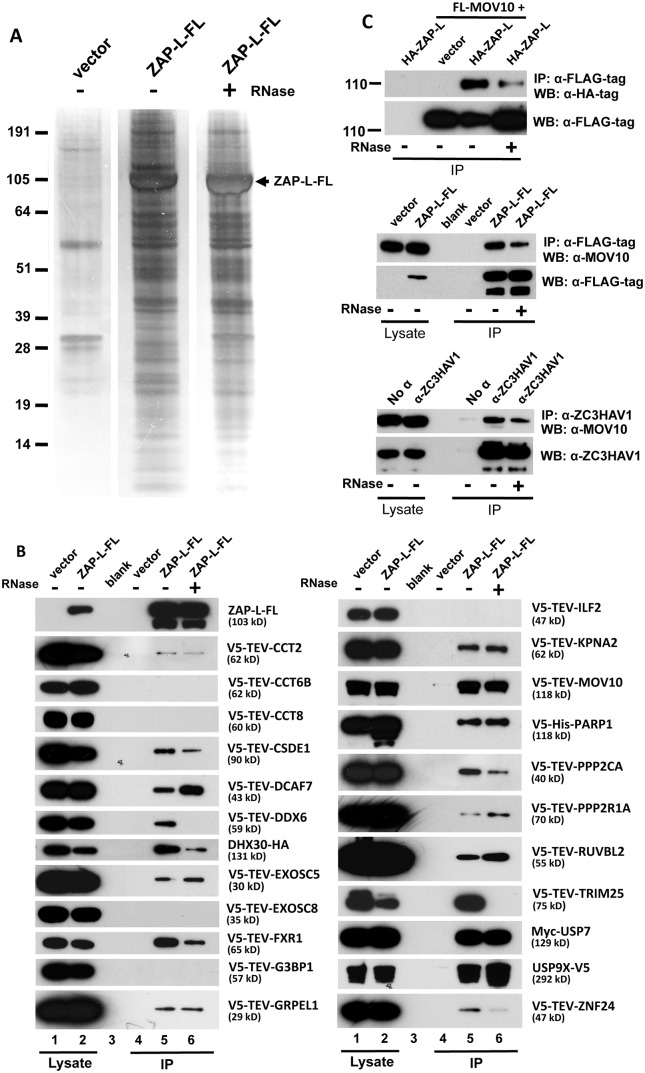
Fig 4. Detection of proteins in the ZAP RNP interactome.
(A) ZAP-L-FL or empty vector was immunoprecipitated with α-FLAG agarose from 293T whole cell lysates and resolved on silver-stained polyacrylamide gels. IP reactions were in the presence or absence of RNase. (B) Selected proteins detected in the ZAP interactome were tagged and many were found to specifically co-IP from 293T cells with ZAP-L-FL but not empty vector. Approximately 1% of the input lysate (lanes 1, 2) and 30% of the immunoprecipitate (lanes 4–6) were loaded in gels. All proteins tested for interaction with ZAP-L-FL are shown. IP reactions were in the presence or absence of 50 μg/ml RNase. All test proteins were detected by α-V5 antibody, except USP7 and DHX30, which were detected by α-Myc and α-HA antibodies, respectively. The top-most left panel is representative of tagged ZAP-L-FL present in the input and IP fractions (detected by α-FLAG antibody) and confirms that RNase treatment did not affect ZAP immunoprecipitation on α-FLAG agarose. (Similar results for all of the IP reactions are shown in S4 Fig). Indicated protein molecular weights include the epitope tag. (C) Both endogenous and exogenously expressed ZAP and MOV10 co-IP even in the presence of RNase. Top panels: HA-ZAP-L co-IPs with FLAG-tagged MOV10 on α-FLAG agarose. Middle panel: co-IP of endogenous MOV10 from 293T cells by ZAP-L-FL on α-FLAG agarose. Bottom: Protein G affinity matrix pull-down from 2012Ep cells of endogenous MOV10 and ZAP proteins by α-ZC3HAV1 antibody. The α-ZC3HAV1 antibody (ProteinTech) recognizes the first 351 amino acids of ZAP.
Table 1. Summary of mass spectrometry analysis of proteins detected with ZAP-L-FL RNP complexes.
| Category | Protein Name | Gene Symbol | Accession Number | #Unique Peptides | ||
|---|---|---|---|---|---|---|
| Rnase | ||||||
| - | + | |||||
| 1 | Zinc finger CCCH-type, antiviral 1 | ZC3HAV1 | NM_020119 | 72 | 84 | |
| 2 | Post-translational | Ubiquitin specific peptidase 9, X-linked | USP9X | NM_001039590 | 24 | 27 |
| 3 | modifiers | Protein phosphatase 2, regulatory subunit A, alpha | PPP2R1A | NM_014225 | 13 | 16 |
| 4 | Protein phosphatase 2, regulatory subunit A, beta | PPP2R1B | NM_001177562 | 9 | 10 | |
| 5 | Protein phosphatase 2, catalytic subunit, alpha isozyme | PPP2CA | NM_002715 | 6 | 7 | |
| 6 | Tripartite motif containing 25/E3 ubiquitin/ISG15 ligase TRIM25 | TRIM25 | NM_005082 | 6 | 2 | |
| 7 | Ubiquitin specific peptidase 7 (herpes virus-associated) | USP7 | NM_003470 | 2 | 3 | |
| 8 | Poly [ADP-ribose] polymerase 1 | PARP1 | NM_001618 | 0 | 3 | |
| 9 | Chaperonin | Chaperonin containing TCP1, subunit 2 (beta) | CCT2 | NM_001198842 | 17 | 16 |
| 10 | Chaperonin containing TCP1, subunit 6A (zeta 1) | CCT6A | NM_001762 | 10 | 6 | |
| 11 | T-complex protein 1, alpha subunit | CCT1 | NM_030752 | 9 | 6 | |
| 12 | Chaperonin containing TCP1, subunit 3 (gamma) | CCT3 | NM_005998 | 9 | 6 | |
| 13 | Chaperonin containing TCP1, subunit 4 (delta) | CCT4 | NM_006430 | 7 | 7 | |
| 14 | Chaperonin containing TCP1, subunit 7 (eta) | CCT7 | NM_006429 | 6 | 8 | |
| 15 | DnaJ (Hsp40) homolog, subfamily A, member 1 | DNAJA1 | NM_001539 | 5 | 7 | |
| 16 | Heat shock 105kDa/110kDa protein 1 | HSPH1 | NM_006644 | 2 | 4 | |
| 17 | Heat shock 70kDa protein 4 | HSPA4 | NM_002154 | 1 | 7 | |
| 18 | DnaJ (Hsp40) homolog, subfamily A, member 2 | DNAJA2 | NM_005880 | 1 | 4 | |
| 19 | RNA Helicase | DEAH (Asp-Glu-Ala-His) box polypeptide 30 | DHX30 | NM_014966 | 23 | 6 |
| 20 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 6 | DDX6 | NM_004397 | 5 | 0 | |
| 21 | DEAH (Asp-Glu-Ala-Asp/His) box polypeptide 57 | DHX57 | NM_198963 | 4 | 0 | |
| 22 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 | DDX18 | NM_006773 | 3 | 0 | |
| 23 | Moloney leukemia virus 10, homolog (mouse) | MOV10 | NM_020963 | 3 | 1 | |
| 24 | YTH domain containing 2 | YTHDC2 | NM_022828 | 3 | 0 | |
| 25 | SG/PB proteins | Fragile X mental retardation, autosomal homolog 1 | FXR1 | NM_005087 | 7 | 7 |
| 26 | ELAV (embryonic lethal, abnormal vision, Drosophila)-like 1 (Hu antigen R) | ELAVL1 | NM_001419 | 3 | 1 | |
| 27 | GTPase activating protein (SH3 domain) binding protein 1 | G3BP1 | NM_198395 | 3 | 0 | |
| 28 | GTPase activating protein (SH3 domain) binding protein 2 | G3BP2 | NM_203504 | 3 | 0 | |
| 29 | Fragile X mental retardation, autosomal homolog 2 | FXR2 | NM_004860 | 1 | 3 | |
| 30 | mRNA decay | 5'-3' exoribonuclease 2 | XRN2 | NM_012255 | 12 | 6 |
| 31 | Exosome component 8 | EXOSC8 | NM_181503 | 3 | 2 | |
| 32 | YTH domain family, member 2 | YTHDF2 | NM_016258 | 3 | 2 | |
| 33 | Chromatin | Chromodomain helicase DNA binding protein 4 | CHD4 | NM_001273 | 6 | 7 |
| 34 | binding/ | Histone deacetylase 1 | HDAC1 | NM_004964 | 3 | 2 |
| 35 | remodelling | Heterochromatin protein 1-binding protein 3 | HP1BP3 | NM_016287 | 3 | 0 |
| 36 | RuvB-like 1 (E. coli) | RUVBL1 | NM_003707 | 2 | 3 | |
| 37 | RuvB-like 2 (E. coli) | RUVBL2 | NM_006666 | 2 | 3 | |
| 38 | Zinc finger | Zinc finger protein 326, transcript variant 1 | ZNF326 | NM_182976 | 4 | 9 |
| 39 | transcription | Zinc finger RNA binding protein | ZFR | NM_016107 | 4 | 0 |
| 40 | factors | Zinc finger protein 24 | ZNF24 | NM_006965 | 2 | 3 |
| 41 | Others | Sjogren syndrome antigen B (autoantigen La) | SSB | NM_003142 | 20 | 0 |
| 42 | Ribosomal L1 domain containing 1 | RSL1D1 | NM_015659 | 19 | 0 | |
| 43 | Endoplasmic reticulum protein 44 | ERP44 | NM_015051 | 15 | 18 | |
| 44 | Carbamoyl-phosphate synthetase 2, aspartate transcarbamylase | CAD | NM_004341 | 11 | 31 | |
| 45 | Interleukin enhancer-binding factor 2 | ILF2 | NM_004515 | 11 | 7 | |
| 46 | Pentatricopeptide repeat domain 3 | PTCD3 | NM_017952 | 11 | 11 | |
| 47 | GrpE-like 1, mitochondrial (E. coli) | GRPEL1 | NM_025196 | 10 | 11 | |
| 48 | Guanine nucleotide binding protein-like 3 (nucleolar) /nucleostemin | GNL3 | NM_014366 | 9 | 2 | |
| 49 | GTP binding protein 4 | GTPBP4 | NM_012341 | 9 | 0 | |
| 50 | General transcription factor IIi | GTF2I | NM_032999 | 8 | 15 | |
| 51 | Insulin-like growth factor 2 mRNA binding protein 3 | IGF2BP3 | NM_006547 | 8 | 0 | |
| 52 | Glutamyl-prolyl-tRNA synthetase | EPRS | NM_004446 | 7 | 9 | |
| 53 | KRI1 homolog (S. cerevisiae) | KRI1 | NM_023008 | 7 | 0 | |
| 54 | Heterogeneous nuclear ribonucleoprotein U-like 1 | HNRPUL1 | NM_007040 | 6 | 0 | |
| 55 | Ly1 antibody reactive | LYAR | NM_017816 | 6 | 0 | |
| 56 | NOP2 nucleolar protein | NOP2 | NM_001258308 | 6 | 0 | |
| 57 | Ribonuclease/angiogenin inhibitor 1 | RNH1 | NM_002939 | 6 | 0 | |
| 58 | Apoptosis-inducing factor, mitochondrion-associated, 1 | AIFM1 | NM_001130846 | 5 | 4 | |
| 59 | La ribonucleoprotein domain family, member 1 | LARP1 | NM_015315 | 5 | 0 | |
| 60 | Reticulocalbin 2, EF-hand calcium binding domain | RCN2 | NM_002902 | 5 | 4 | |
| 61 | Cell cycle associated protein 1 | CAPRIN1 | NM_005898 | 4 | 0 | |
| 62 | FtsJ homolog 3 (E. coli) | FTSJ3 | NM_017647 | 4 | 0 | |
| 63 | Heterogeneous nuclear ribonucleoprotein A0 | HNRNPA0 | NM_006805 | 4 | 0 | |
| 64 | La ribonucleoprotein domain family, member 7 | LARP7 | NM_015454 | 4 | 0 | |
| 65 | MYB binding protein (P160) 1a | MYBBP1A | NM_014520 | 4 | 3 | |
| 66 | Ribosomal RNA processing 1B | RRP1B | NM_015056 | 4 | 0 | |
| 67 | Coiled-coil domain containing 86 | CCDC86 | NM_024098 | 3 | 0 | |
| 68 | Cold shock domain containing E1, RNA-binding | CSDE1 | NM_001130523 | 3 | 0 | |
| 69 | Karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | KPNA2 | NM_002266 | 3 | 3 | |
| 70 | Staufen double-stranded RNA binding protein 1 | STAU1 | NM_004602 | 3 | 0 | |
| 71 | Topoisomerase (DNA) I | TOP1 | NM_003286 | 3 | 0 | |
| 72 | A kinase (PRKA) anchor protein 8 | AKAP8 | NM_014371 | 2 | 3 | |
| 73 | Tu translation elongation factor, mitochondrial | TUFM | NM_003321 | 0 | 6 | |
| 74 | DDB1- and CUL4-associated factor 7 | DCAF7 | NM_005828 | 0 | 3 | |
| 75 | General transcription factor IIIC, polypeptide 1, alpha 220kDa | GTF3C1 | NM_001520 | 0 | 3 | |
| 76 | General transcription factor IIIC, polypeptide 3, 102kDa | GTF3C3 | NM_012086 | 0 | 3 | |
| 77 | Pyrroline-5-carboxylate reductase-like | PYCRL | NM_023078 | 0 | 3 | |
| 78 | RNA binding motif protein 14 | RBM14 | NM_006328 | 0 | 3 | |
* Only non-ribosomal proteins predicted by three or more unique peptides, present in two independent co-IP reactions, and not detected in empty vector control isolates are shown. Proteins are organized within selected functional categories and ordered within each category by number of unique peptides identified in the IP reaction not treated with RNase. Detailed MS data are presented in S1 Table.
Gene Symbol: Proteins shared with the L1 ORF1p interactome [46].
To confirm protein interactions, a subset of cDNAs of proteins identified by MS were subcloned with an N-terminal V5-TEV-epitope tag or were obtained as gifts. Following cotransfection in 293T cells, 18 of 23 proteins tested co-IPed with ZAP-L-FL on α-FLAG agarose (Fig 4B). A majority of these interactions were resistant to RNase digestion, suggesting direct protein-protein binding. In contrast, our previous work characterizing the ORF1p interactome found almost all of 41 confirmed protein associations to be lost upon RNase treatment [46].
Our analyses identified most previously described ZAP-interacting proteins. We confirmed RNA-independent binding of ZAP and PPR2R1A (PR65A) [70], a component of the protein phosphatase 2 (PP2A) complex, and show for the first time RNase-resistant binding of its catalytic subunit, PPP2CA, as well (Fig 4B). Wang et al. [70] reported that knockdown of PPR2R1A by shRNAs reduced ZAP suppression of an MoLV reporter. It has been reported that several serine residues immediately downstream of the rat ZAP zinc-finger domain are phosphorylated by glycogen synthase kinase 3β (GSK3β), and that overexpression of GSK3β reduces, and its inhibition increases ZAP activity against MoLV [71]. These data are all consistent with inhibition of ZAP antiviral function by phosphorylation. Our MS analyses of human ZAP protein sequence (65% coverage) confirmed two phosphorylated residues (S257 and S284) homologous to those detected by Sun et al. [71] in rats, and identified four additional phosphorylated sites (S335, T375, S378, S796). However, a role for phosphorylation or PP2A-mediated dephosphorylation in modulating ZAP inhibition of retrotransposition remains to be determined.
It has been proposed that ZAP recruits the 3'-5' exosome to degrade target viral RNAs in cytoplasmic granules [51,72]. We identified exosome component EXOSC8 (RRP43) in ZAP RNP immunoprecipitates, although we could not confirm its direct binding with ZAP-L-FL (consistent with [72]). According to previous reports, EXOSC5 (RRP46) binds rat but not human ZAP [50,72]. However, when tested, we found EXOSC5 to bind weakly with human ZAP-L-FL in the presence or absence of RNase. We also identified as a strong ZAP interactor DHX30, an RNA helicase believed to be recruited by ZAP to unwind viral RNAs and facilitate their exosome-mediated degradation in stress granules [69,73]. We confirmed colocalization of DHX30 with both ZAP-S and ORF1-GFP in cytoplasmic granules, and RNA-independent binding of DHX30 with ZAP-L-FL (Figs 4B and 5D and S3A).
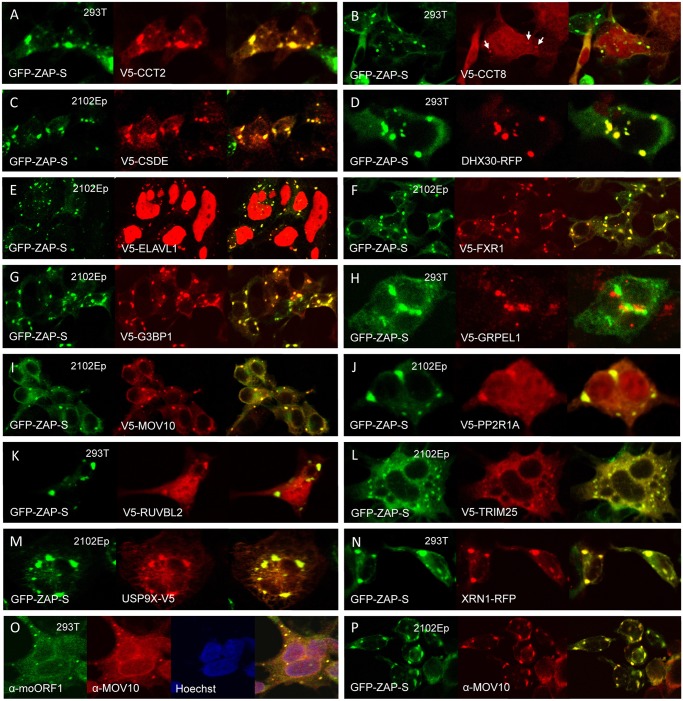
Fig 5. Many members of the ZAP interactome colocalize with GFP-tagged ZAP-S in cytoplasmic granules of unstressed 293T cells.
(A–N) Construct GFP-PARP13.2 was co-transfected with V5-tagged proteins in all cases except (D) DHX30 and (N) XRN1, which were tagged with red fluorescent protein (RFP). The cell type is indicated in each panel. (O) Endogenous ORF1p and MOV10 co-localize in cytoplasmic granules. (P) Endogenous MOV10 protein co-localizes with GFP-ZAP-S.
In addition to the 3'-5' degradation exosome, 5′-3′ degradation enzymes, including 5'-3' Exoribonuclease 1 (XRN1) and Poly(A)-Specific Ribonuclease (PARN), have been reported to bind ZAP and augment antiviral function [51]. We detected the XRN1 paralog XRN2 in the ZAP interactome. XRN2 not being available, we tested XRN1 tagged with RFP and found it to colocalize with GFP-tagged ZAP-S in cytoplasmic granules (Fig 5N). XRN1 is a known SG and PB component and also colocalizes with ORF1p-GFP in SGs [63,74].
We also discovered novel ZAP-associated proteins. In addition to PP2A complex members, ZAP RNPs contain several other proteins involved in post-translational modification (PTM). TRIM25, which functions as an E3 ubiquitin and ISG15 ligase and defends against viruses by mediating ubiquitination of RIG-1/DDX58 [75], associates with ZAP in the presence of RNA (Fig 4B). Even after RNase digestion, ZAP-L strongly binds two ubiquitin carboxyl-terminal peptidases, USP7 and USP9X. Our MS analyses predicted two ubiquitinated lysines (K226, K783) and the UbPred and CKSAA_UBSITE algorithms [76,77] predicted 6 additional sites of ubiquitination within ZAP-L. Perhaps ubiquitin peptidases associate with the ZAP RNP to enhance its anti-retroelement activity by limiting ubiquitin-mediated degradation, although that remains to be determined.
While its own PARP domain is likely catalytically inactive, ZAP-L itself is ADP-ribosylated and known to recruit other PARPs that are capable of pADPr, such as PARP5 and PARP12 [68]. PARP1 is the founding member of the PARP family. We report for the first time the direct binding of ZAP and PARP1 independent of RNase digestion. It is not known if PARP1 ribosylates ZAP.
Also identified within the ZAP RNP were almost all components of the cytosolic chaperonin-containing TCP1 (CCT) complex. We tested several CCT complex members and confirmed weak binding of CCT2, but not CCT6B or CCT8, with ZAP-L-FL (Fig 4B). The CCT complex was first discovered for its critical role in the folding of actin and tubulin, and subsequently many other CCT binding proteins were reported [78,79]. This is the first report of the association of CCT and ZAP. CCT4 and CCT6 were also detected in the ORF1p interactome [46].
The ZAP interactome includes several canonical components of SGs (FXR1, FXR2, G3BP1, and ELAVL1) and PBs (DDX6). We confirmed the association of these proteins with ZAP by colocalization in cytoplasmic granules and/or co-IP (Figs 4B and 5). Indeed, 65 percent (12/18) of the proteins tested that directly immunoprecipitated with ZAP-L-FL (Fig 4B) also colocalized with GFP-tagged ZAP-S in cytoplasmic granules of 293T cells (Fig 5). To the best of our knowledge, CCT2, CCT8, PPP2R1A, TRIM25, and USP9X have not previously been reported in granules (Figs 5A, 5B, 5J, 5L and 5M).
We screened test proteins for colocalization in cytoplasmic granules with ZAP-S rather than ZAP-L (Fig 5), and it is possible that additional ZAP-associated proteins, bound only by ZAP-L's PARP-like domain, escaped detection. While ZAP-L and ZAP-S colocalize in cells, it is of interest that overexpression of ZAP-L induces fewer and larger cytoplasmic aggregates, and apparently binds ZAP-S to cause its redistribution to these large foci when the two isoforms are coexpressed; ZAP is known to dimerize (S3B Fig; [52]). Lee et al. [69] reported that murine ZAP-S colocalizes in the cytoplasm with markers of PBs and SGs, but not with markers of mitochondria, endosomes, peroxisomes, or lysosomes. On the other hand, Charron et al. [56], found that C-terminal S-farnesylation of mouse ZAP-L caused its partial redistribution to lysosomes and late endosomes. Reasons for the differing patterns of ZAP-S and ZAP-L bear further investigation.
We previously reported that endogenous L1 ORF1p and antiviral protein MOV10 associate in an RNA-dependent manner and colocalize in SGs of cells (Fig 5O; [27]). We now show that MOV10 is also a component of the ZAP interactome. Recently, Gregersen et al. [80] also detected ZAP by SILAC (stable isotope labeling by amino acids in cell culture) analyses of MOV10-interacting proteins. Both endogenous and exogenously expressed ZAP and MOV10 co-IP in a manner partially resistant to RNase digestion (Fig 4C). We could not detect binding of MOV10 with HA-ZAP-L 1–256 or rat HA-NZAP. ZAP and MOV10 proteins closely colocalize in cytoplasmic granules (Fig 5I and 5P).
In summary, we present for the first time a comprehensive analysis of the ZAP protein interactome. Most of its components we tested directly bound and/or colocalized in RNA granules with ZAP. ZAP may recruit many cellular proteins to these cytoplasmic structures (or vice versa).
ZAP restricts L1 RNPs
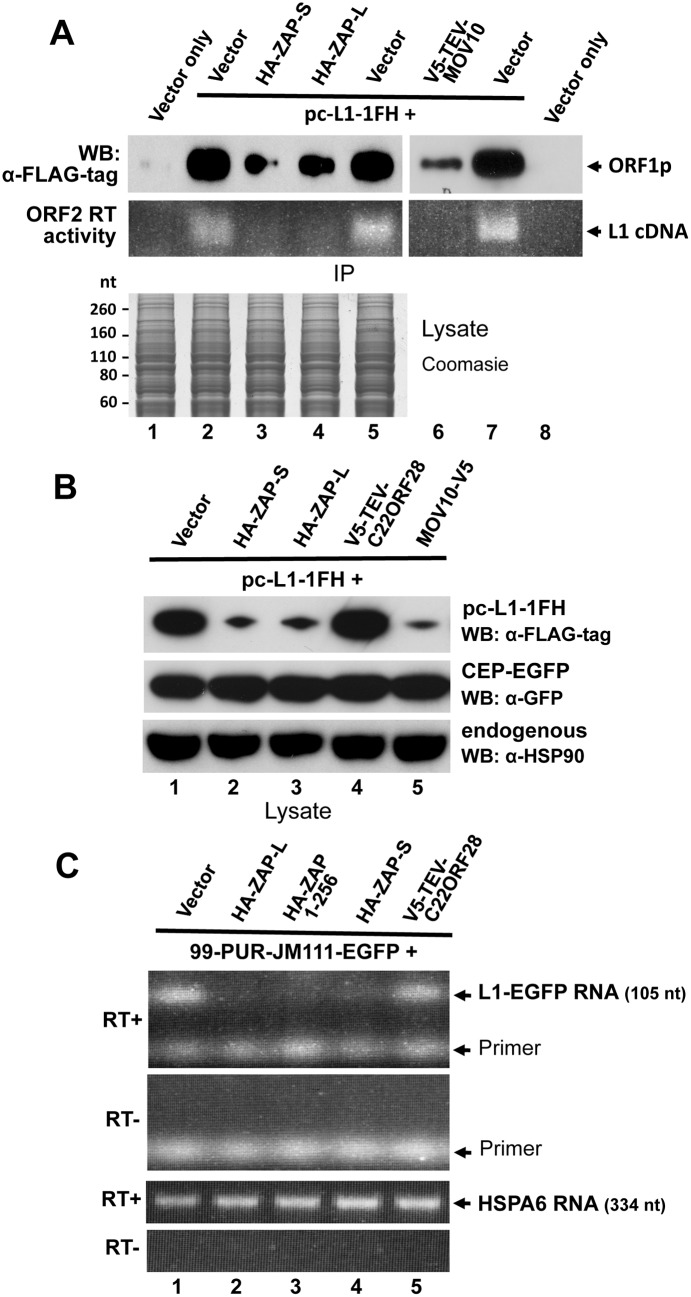
Previously, we demonstrated that overexpression of MOV10 strongly reduces the steady state number of L1-encoded RNA and proteins in transfected cells, although the mechanism of this loss remained uncertain [64]. We similarly assayed the effects of ZAP on L1 expression. As with MOV10, levels of ORF1 protein expressed from pc-L1-1FH were significantly reduced in L1 RNPs immunoprecipitated from cytoplasmic extracts in the presence of cotransfected ZAP-L and ZAP-S. ORF2 activity was almost undetectable in the LEAP assay for L1 reverse transcription (Fig 6A, lanes 3 and 4; [81]. In whole cell lysates, ORF1p expressed from pc-L1-1FH was significantly lower in the presence of ZAP-S, ZAP-L, or MOV10 but not empty vector or an unrelated protein (Fig 6B). This was not a general protein effect, as overexpression of ZAP did not affect levels of coexpressed EGFP or endogenous heat shock protein 90 (Fig 6B), and was without obvious effect on global protein expression detected by Coomassie blue staining of cell lysates (Fig 6A, lower panel).
Fig 6. ZAP inhibits exogenous L1 RNP levels in cells.
(A) Expression of HA-ZAP-L, HA-ZAP-S, and V5-TEV-MOV10 reduces the amount of ORF1 protein (top IP panel) and ORF2p RT activity (bottom IP panel) in α-FLAG antibody purified pc-L1-1FH immunoprecipitates. RT activity was determined by the LEAP assay [81]. Below: Coomasie-stained gel of cell lysates prior to IP showed no obvious loss of global protein levels in the presence of ZAP. (B) Analysis of 293T cell lysates showing that ORF1 protein from pc-L1-1FH is reduced in the presence of ZAP and MOV10, but not empty vector or an unrelated protein, C22ORF28 (top panel). Panels below show by Western blotting that neither ZAP nor MOV10 alter expression of endogenous HSP90 or GFP protein expressed from cotransfected CEP-EGFP. Lysates were prepared from two pooled wells of a six-well plate. (C) Expression of ZAP causes loss of L1 RNA. Top panels: RT-PCR of L1 RNA expressed from 99-PUR-JM111-EGFP, which expresses L1-RP with an ORF1 mutation that prevents genomic insertion. PCR primers spanned the intron of the EGFP reporter cassette to distinguish spliced RNA products from contaminating plasmid DNA (generating a 105 nt band vs a 1 kb band). PCR reactions are shown in the presence or absence of RT. Bottom panels: Levels of endogenous HSPA6 RNA were the same in the presence or absence of ZAP.
We next examined L1 RNA transcribed from construct 99-PUR-JM111-EGFP in the presence or absence of ZAP. This construct contains a mutation in L1 ORF1 that prevents genomic insertions. PCR primers flanked the intron of the EGFP reporter cassette allowing products amplified from spliced cDNA to be distinguished from those of contaminating plasmid DNA. Paralleling the loss of ORF1 protein, diminished levels of L1 RNA were detected by RT-PCR in whole cell lysates cotransfected with HA-ZAP-L HA-ZAP-S, or HA-ZAP 1–256, but not with empty vector or an unrelated protein. Analysis of endogenous HSPA6 RNA showed no such effect (Fig 6C). We failed to determine whether or not ZAP affects expression of endogenous L1s in the genome. The observed reduction of exogenous L1 RNA in the presence of ZAP protein is consistent with previous reports that ZAP inhibits infecting viruses by causing loss of their RNAs at a post-transcriptional step [29,49,51,72,82].
Discussion
Restriction factor proteins are part of the innate immune defense system of the cell, which often detects infection by receptors that recognize viral nucleic acids. Many of these factors target retroviruses, but some, such as ZAP, act against a wide range of viral families. In certain cell types expression of antiviral genes is induced by type I or type II interferons. We have expanded the list of IFN-stimulated genes that limit LINE-1 retrotransposition when overexpressed, including BST2, ISG20, MAVS, MX2, and ZAP (Fig 1B).
Most dramatic was an almost complete loss of retrotransposition in the presence of MAVS protein, which is only partly explained by cytotoxicity. MAVS acts downstream of the RIG-I and IFIH1 cytoplasmic receptors for viral dsRNAs. Receptor activation causes multimerization of MAVS to trigger a signaling cascade and production of type I IFNs [83]. Recent evidence indicates that upon viral detection peroxisomal-localized MAVS rapidly induces interferon-independent expression of defense factors to provide short-term protection. Mitochondrial MAVS signaling occurs later in infection, triggering IFN expression and induction of ISGs, and sustaining the immune response [84]. Thus, the profound inhibition of retrotransposition caused by overexpressed MAVS is likely due to the combined action of a number of interferon-induced genes.
ISG20 strongly inhibited L1 retrotransposition without obvious cytotoxicity caused by its overexpression. ISG20 is a 3'-5' exonuclease that inhibits replication of several human and animal RNA viruses, including HIV-1 [7,85,86]. Its mechanism of action is unknown, although one might assume that it degrades viral or retrotransposon RNA
Inhibition of L1 retrotransposition by the dynamin-like GTPase MX2, but not by its closely related paralog MX1, parallels recent reports that MX1, a broad-spectrum inhibitor of many RNA and DNA viruses including influenza virus, fails to inhibit HIV retrovirus. On the other hand, MX2 does not inhibit infuenza virus but inhibits multiple strains of HIV1 at a late post-entry step by targeting the viral capsid and preventing accumulation of viral cDNA in the nucleus. [41–43,45]. Our mutation analyses suggest that the mechanism of MX2 inhibition of L1 retrotransposition may differ in some aspects from retroviral inhibition, requiring GTP binding but not nuclear localization.
Like MX1, overexpression of TRIM28/KAP1 had little effect on retrotransposition (Fig 1B). However, in mouse ES cells TRIM28 strongly silences expression of multiple classes of endogenous LTR retroelements and modestly suppresses L1s by recruiting chromatin-remodeling factors [87,88]. Thus, TRIM28 may inhibit human retrotransposition by epigenetic modification of endogenous L1 elements, but remain ineffective in our cell culture assay against L1s expressed from a plasmid.
Importantly, we showed that transient expression of either the long or short isoform of a general protein inhibitor of viral infection, ZAP, potently restricts genomic insertion of both non-LTR and LTR retrotransposons. Furthermore, siRNA-mediated knockdown of endogenous ZAP increased L1 retrotransposition 2- to 3-fold in 293T cells. Association with the L1 complex is confirmed by close colocalization of ZAP protein with ORF1p and L1 RNA in cytoplasmic stress granules, and by the detection of ZAP in RNP particles captured by immunoprecipitation of a tagged L1 construct.
The CCCH-type zinc finger domain of ZAP recognizes MoLV and other viral transcripts and induces their degradation by recruiting RNA decay proteins [50,51,69,72,73]. Selected cellular RNAs are also targeted, including the TRAIL receptor, TRAILR4 [54]. Our evidence suggests that RNA degradation is also a characteristic of ZAP-associated loss of retrotransposition. Levels of exogenously expressed L1 RNA and protein are reduced in cell lysates in the presence of ZAP (Fig 6C). Loss of ZAP binding in the L1 RNP upon RNase treatment, and the fact that deletion of the zinc finger domain or mutation of residues considered important for its RNA binding significantly reduced inhibition of retrotransposition, suggests that ZAP binds the L1 RNA to promote loss of RNP integrity. However, we cannot exclude the possibility that ZAP binds some other RNA that itself is recruited to the L1 RNP. Many non-L1 RNA species have been found in association with the L1 ORF1p complex, including mRNAs, Alu, SVA, and small cytoplasmic and nuclear RNAs [46,89]. ZAP binds ZREs with a minimum known length of 500 bp, and no common motif or secondary structure has been found [29,50,51]. A detailed investigation of how ZAP binds L1 RNA is required. The fact that deletion or mutation of the zinc finger RNA-binding domain reduced but did not abolish ZAP inhibition of retrotransposition (Fig 2B), suggests that a second RNA-binding domain may exist, or that protein-protein interactions are also important for retrotransposon inhibition.
Our data cannot exclude the possibility that ZAP may also inhibit L1s at the protein level (perhaps by binding L1 RNA to interfere with ORF translation). Effects of ZAP on viral translation have been described [31,51]. Inhibition of GSK3β phosphorylation increases ZAP's ability to repress target mRNA translation without increased mRNA degradation [71]. And recently, Zhu et al. [82] demonstrated that ZAP represses translation of HIV-1 and Sindbis virus reporter constructs independently of mRNA decay by directly binding translation initiation factor eIF4A and interfering with its interaction with eIF4G.
MOV10 is an RNA helicase that also strongly inhibits retrotransposition in cell culture assays [26,27,90]. Li et al. [28] showed that overexpression of MOV10 strongly reduced levels of exogenously expressed IAP and L1 RNA at a post-transcriptional step, while inhibition of endogenous MOV10 increased L1 RNA. On the other hand, Lu et al. [90] found that MOV10 decreases IAP RT products but not levels of IAP RNA and protein. The facts that MOV10 and ZAP bind each other independently of RNA, colocalize in cytoplasmic granules, associate with the L1 RNP, and promote similar loss of L1 RNP integrity and retrotransposition in cells, suggest that the two proteins may function in a common pathway, a notion worthy of further investigation.
In this study, we also present for the first time a detailed analysis of the ZAP protein interactome, confirming most of its previously known interacting proteins and identifying new member proteins whose association with the ZAP RNP is consistent with its known cellular functions. For example, close colocalization in cytoplasmic granules of many of these proteins with ZAP, including L1 ORF1p, is not surprising in light of ZAP's dual roles in the assembly of RNA granules and the control of microRNA silencing. While itself catalytically inactive, ZAP recruits other PARPs active for poly(ADP)-ribosylation which is critical for SG formation [68,91]. Furthermore, overexpression of ZAP causes loss of microRNA silencing by targeting Ago2 (a binding partner of ZAP, although not one detected by our study) for pADPr [91,92].
We detected several PTM-related proteins within the ZAP RNP, including protein phosphatase 2A complex members (PPP2R1A/B and PPP2CA), two related deubiquitinating enzymes (USP7 and USP9X), and TRIM25 (an E3 ubiquitin/ISG15 ligase). In addition to pADPr [68], Sun et al. [71] reported phosphorylation of ZAP by GSK3β, and Charron et al. [56] found that S-farnesylation at the C-terminus of ZAP-L enhanced its restriction of Sindbis virus. Thus, PTMs likely modulate ZAP antiviral activity and may explain why ZAP efficiently restricts some classes of virus but not others.
We also identified epigenetic modifying enzymes in ZAP-L-FL immunoprecipitates (Table 1). RUVBL1 and RUVBL2 are conserved AAA+ protein ATPases present in histone acetyltransferase complexes NuA4 and Tip60, chromatin remodelling complexes Ino80 and SWR-C, and the telomerase complex [93]. HDAC1 and CHD4 are components of the NuRD (nucleosome remodeling and histone deacetylase) complex, along with HP1, SETDB1 and TRIM28 (KAP1). While we detected TRIM28 in ZAP immunprecipitates, it was also present in vector-only samples (11 vs 4 peptides, respectively) and is excluded from Table 1. TRIM28 is targeted to specific DNA sequences via its interactions with various zinc finger proteins, and mediates gene silencing by recruiting NuRD to target promoters [94,95]. Epigenetic modifications involving TRIM28 and SETDB1 have been implicated in silencing both endogenous retroviruses and retrotransposons, including multiple classes of LTR retroelements and L1s [88,89,96–100]. Reichman et al. [101] also showed in silico that HDAC1 silences LTR retrotransposons in mouse mES cells. A possible link between ZAP and epigenetic silencers deserves investigation. Although ZAP is predominantly a cytoplasmic protein it can also function in the nucleus [29,102].
Macdonald et al. [103] showed that an unknown IFN-induced factor(s) synergizes with ZAP to control viral infection. Karki et al [104] identified 16 IFN-stimulated genes that act synergistically with ZAP to reduce alphavirus infectivity, including IFIH1 and SAMHD1. Furthermore, ZAP-S stimulates RIG-1-mediated production of Type I interferon [53]. It is remarkable that 34 of the 78 proteins of the ZAP interactome shown in Table 1 are identified in the Interferome v2.01 Database of about 2000 ISGs as being induced at least 2-fold by interferon in either mice or humans (S1 Table; [105]). This is a four-fold enrichment of ISG proteins in the ZAP interactome over what would be expected by chance. Likely we would have detected an even greater number of ISG products within the ZAP-L-FL interactome if prior to IP we had first stimulated their expression with interferon. MX1 and, MX2, for example, while not detected by our MS analyses of endogenous ZAP-interacting proteins, when overexpressed strongly bind ZAP-L-FL in a manner resistant to RNase treatment (S5 Fig). The association of ZAP with many ISG products suggests it may be a key player in the interferon response. This idea is supported by a recent study showing that ZAP-L-depleted HeLa cells are strongly enriched for expression of genes in the interferon immune response pathway [54].
In addition to MOV10, other IFN-stimulated proteins associated with the ZAP RNP complex also play roles in viral control. Overexpression of DHX30, for example, strongly enhances expression of HIV-1, while restricting its RNA packaging [106]. G3BP1 and G3BP2 regulate expression of ISGs known to have broad antiviral activity, and are required for an IFN response against dengue and yellow fever viruses [107]. Chaperone HSPA4 (HSP70) inhibits viral gene expression and replication [108]. Its co-chaperone DNAJ (HSP40) has been associated with both activation and inhibition of viruses, including HIV [109]. Ubiquitination by TRIM25 increases the ability of RIG-1 to initiate antiviral signaling by facilitating its interaction with MAVS. Influenza A virus nonstructural protein 1 specifically inhibits TRIM25-mediated RIG-I ubiquitination, thereby suppressing its antiviral activity [75,110]. USP7 was originally identified by its interaction with the HSV-1-encoded E3 ubiquitin ligase ICP0 [111]. PARP1 is required for efficient replication and integration of HIV-1 [112,113], and has been implicated in repressing Epstein Bar virus, Kaposi's sarcoma-associated herpesvirus, and Drosophila retrotransposons [114–116]. RUVBL2 inhibits influenza virus replication, apparently by interfering with oligomerization of the viral nucleoprotein [117]. Future investigations of the associations of these proteins with the ZAP complex could yield new insights into antiviral restriction.
As we showed in Fig 1A, application of Type I interferon represses retrotransposition in cell culture. While this paper was under review, another study was published showing that increased endogenous L1 expression in the testes of MOV10L-deficient mice, or transfection of L1 constructs in mouse embryonic fibroblasts, is marked by increased levels of INFβ [118]. MOV10L is a restriction factor that represses retroelements in the male germline [119]. Conversely, treatment of human cells with INFβ suppressed L1 replication. Functional loss of an anti-retroelement restriction factor can alter the normal metabolism of retrotransposon RNA or its reverse-transcribed cDNA with possible consequences for the organism. Such may be the case with Type I interferonopathies, which include AGS, SLE, spondyloenchondrodysplasia, and STING-associated vasculopathy [120 121]. Indeed, it has been proposed that misregulated nucleic acids, deriving from an as yet unknown endogenous source, but possibly retrotransposons, accumulate and are recognized by sensors, triggering an interferon response and causing Aicardi—Goutières Syndrome [122,123].
AGS is a severe Mendelian inflammatory disorder that affects particularly the brain and frequently causes death in childhood. The disease is characterized by progressive encephalopathy, psychomotor regression, and lesions of the skin, together with increased levels of Type I IFN in the cerebrospinal fluid and serum, and induction of ISGs detectable in peripheral blood. AGS is associated with mutations in seven genes, all involved with nucleic acid metabolism or signaling: TREX1, RNASEH2 A/B/C, SAMHD1, ADAR1 and IFIH1 [124–126]. Most of these genes also have been linked with the innate immune system that restricts retroviral infection and suppresses endogenous retrotransposons [23,123,127,128]. We predict mutations in additional anti-retroelement genes, perhaps even including ZAP or MOV10, will be linked to inflammatory diseases involving interferon overexpression.
Materials and Methods
Plasmid and RNAi constructs
Constructs CEP-EGFP, pc-L1-1FH, pc-L1-RP, ORF1-GFP L1-RP, 99-PUR-L1-RP-MS2-6X, pCEP-5′UTR-ORF2-No-Neo, and XRN1-RFP have been described [63,64]. ZAP-L-FL with a C-terminal FLAG-tag, and HA-ZAP 1–256 and HA-ZAP 253–902 with N-terminal HA-tags were generated by PCR and cloned in pcDNA6 myc/hisB vector. Ultimate ORF cDNA clones (Invitrogen) were cloned with V5-epitope tags and tobacco etch virus (TEV) protease cleavage sites on their N-termini by shuttling them from pENTR221 vector into pcDNA3.1/nV5-DEST vector using Gateway Technology (Invitrogen). Ultimate ORF Clone ID numbers are shown in S1 Table.
Clones obtained as gifts included Alu-neoTet and IAP-neoTNF (M. Dewannieux, Institut Gustave Roussy, Villejuif [61,62]), DHX30(v2)-HA and DHX30(v2)-RFP (C. Liang, Lady Davis Institute-Jewish General Hospital, Montreal [106]), pcDNA3.1-V5-His-MOV10 (Y.-H. Zheng, Michigan State University, East Lansing [129]), ORFeus-Mm (WA-125) (W. An, Washington State Univ. [130]), ORFeus-HS (WA117) (L. Dai, Johns Hopkins School of Medicine, Baltimore [57]), pCDNA3.1-V5-His full-length PARP1 (J. Pascal, Thomas Jefferson Univ., Philadelphia [131]), GFP-PARP13.2 (GFP-ZAP-S; A Leung, Johns Hopkins School of Medicine [89]), SBP-PARP13.1, SBP-PARP13.1ΔZnF and SBP-PARP13.1VYFHR (P Chang, MIT, Cambridge [54]), pDest51-USP9X-V5 (R. Hughes, Buck Institute for Research on Aging, Novato [132]), Myc-USP7 (Y. Sheng, York University, Toronto [133]), pcDNA-Vphu (a codon-optimized version of the native Vpu gene; NIH AIDS Reagent Program [40]), pCEP-5′UTR-ORF2-No-Neo (J.L. García-Pérez, GENYO, Spain) [134]), and HA-ZAP-L, HA-ZAP-S, and Rat HA-NZAP (H. Malik, Fred Hutchinson Cancer Research Center, Seattle [32]). An altered amino acid (M201K) was restored to consensus in HA-ZAP-L, and the change was found to have no effect on L1 retrotransposition. pEasiLV-MCS MX2-Flag WT was obtained from M. Malim (King's College, London [45]) and the MX2 gene was amplified with C-terminal V5-tag by PCR and recloned in the vector pcDNA3. The K131A mutant was generated by the Quikchange Site-Directed Mutagenesis method (Agilent Technologies).
siRNAs were generated by Sigma-Aldrich based on the following sense sequences:
siCNT3 AUGUAUUGGCCUGUAUUAG[dT][dT],
siZC3HAV1-2 1435–1453 UUGGGUCAGCAUCAUCUGC[dT][dT],
siZC3HAV1-3 1637–1655 AUGUGCUCAAAGUCCGUCC[dT][dT] [60], and
siCNT4 UAAGGCUAUGAAGAGAUAC[dT][dT].
Immunoprecipitation and MS sequencing
For MS sequence determination, HEK 293T cells were transfected in T75 flasks with 15 μg of ZAP-L-FL or pcDNA6 myc/hisB (Invitrogen) empty vector and expanded for approximately 45 hr, followed by whole cell lysate preparation by sonication. IP and sample recovery were as previously described [46]. Treatment of samples with 30 μg/ml DNase-free RNase (Roche) was in the absence of RNase inhibitors. MS sequencing and database analyses was performed by the Johns Hopkins Mass Spectrometry and Proteomics Facility as previously described [46].
For each co-IP, extracts from approximately 6×106 293T cells in T75 flasks transfected with ZAP-L-FL and test protein constructs were prepared in 750 μl of lysis buffer supplemented with protease, phosphatase, and RNase inhibitors, and immunoprecipitated as previously described [46]. Lysates containing test proteins of predominantly nuclear localization were sonicated. RNase-treated reactions contained 25 μg/ml RNase, DNase-free HC (Roche) and 25 μg/ml RNaseA (Invitrogen) and no RNase inhibitors.
Cell culture and retrotransposition assay
Human 2102Ep embryonal carcinoma cells (a gift from P. K. Andrews, University of Sheffield), HeLa-HA and HeLa-JVM cells ([135]; gifts from J.L. García-Pérez, GENYO, Spain), and HEK 293T cells (ATCC) were grown in Dulbecco’s modified Eagle’s medium with 10% FBS (Hyclone), GlutaMax and Pen-Strep (Invitrogen). Plasmid and siRNA transfections used FuGENE HD (Promega) and Lipofectamine RNAiMAX (Life Technologies) reagents, respectively.
The EGFP L1 cell culture retrotransposition assay was conducted as previously described [27,38]. 2.5×105 HeLa or 293T cells/well were seeded in 6-well dishes. The following day, 1.0 μg of 99-PUR-RPS-EGFP, a plasmid containing L1-RP and the EGFP retrotransposition reporter cassette, was cotransfected with 0.5 μg of empty vector (pcDNA3 or pcDNA6 myc/hisB, Invitrogen) or test plasmid. All transfections were in quadruplicate wells. Five days post-transfection, cells having a retrotransposition event, and hence expressing EGFP, were assayed by flow cytometry. Gating exclusions were based on background fluorescence of plasmid 99-PUR-JM111-EGFP, an L1 construct containing two point mutations in ORF1 that abolish retrotransposition [37]. Within each experiment, results were normalized to fluorescence of 99-PUR-RPS-EGFP cotransfected with empty vector.
The Alu retrotransposition assay was carried out essentially as described in Dewannieux et al. [61]. Retrotransposition construct Alu-neoTet was cotransfected in HeLa-HA cells with pcDNA6 myc/hisB empty vector or retrotransposition driver plasmid pCEP-5′UTR-ORF2-No-Neo, together with test plasmids. Eighteen hours post-transfection, HeLa-HA cells were expanded from six-well plates to T75 flasks, and three days later selection for retrotransposition events with 550 μg/ml of G418 was begun. After 15 days of selection, cells were fixed, stained with Giemsa, and colonies were counted.
Similarly, 1.0 μg of the IAP element reporter plasmid, IAP-neoTNF [62], was cotransfected with 0.5 μg empty vector or test plasmid in HeLa-JVM cells, selected with G418, and colony numbers were counted.
Assessment of toxicity
To reveal any differences in transfection efficiencies of test proteins or off-target effects on EGFP reporter expression, we followed the strategy of Wei et al. (39). Each test plasmid (0.5 μg) was co-transfected in quadruplicate wells of 12-well plates with CEP-EGFP (0.5 μg), a construct that constitutively expresses EGFP from a CMV promoter. Four days post-transfection, EGFP fluorescence was determined by flow cytometry, as previously described [46].
To determine potential cell toxicity caused by test proteins, 18,000 293T cells were seeded in 75 μl of Dulbecco’s modified Eagle’s complete medium in 96-well plates. The next day, transfection reactions prepared with 70 ng of test plasmid, 0.2 μl of Fugene HD and 25 μl of Opti-MEM Reduced Serum Medium (Invitrogen) were added to each well. After 3 or 4 days, a MultiTox-Fluor Multiplex Cytotoxicity Assay kit (Promega) was used to assay cell toxicity, as previously described [46].
To further test potential toxicity from expression of ZAP, we co-transfected in HeLa cells pcDNA6 myc/his B, a bsr expression vector, together with either empty vector (pcDNA3) or ZAP expression constructs. On day 2, cells were expanded to T75 flasks and selection with 2 μg/ml blasticidin was begun. After 12 days, cells were fixed, stained and colonies were counted. Cytotoxicity will reduce total colony counts compared with empty vector control (S2B Fig).
Immunofluorescence and western blotting
Commercial antibodies included mouse (ms) α-V5-tag (Invitrogen), ms α-FLAG-tag (Sigma), rabbit (rb) α-HA-tag (C29F4), rb α-HSP90 and rb α-Myc-tag (71D10)(Cell Signaling Technology), goat (gt) α- anti-eIF3η (N-20), gt α-MX2 (C-20), ms α-SBP (SB19-C4), and gt α-TIA1 (C-20)(Santa Cruz Biotechnology), rb α-β-tubulin-2 (Pierce), and rb α-MOV10 and rb α-ZC3HAV1 (ProteinTech). Donkey Cy3-, Cy5-, DyLight 488-, or DyLight 549-conjugated, and HRP-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories. Purified polyclonal α-ORF1p (AH40.1) and monoclonal α-ORF1p (α-moORF1) antibodies were gifts from M. Singer (Carnegie Institution of Washington [136]) and K. Burns (Johns Hopkins School of Medicine [137]), respectively.
Western blotting, IF, and FISH were performed as described [63,64].
Reverse transcriptase and RNA analyses
L1 ORF2p reverse transcriptase analysis followed the LEAP protocol [81]. Primers used were:
3′RACE adapter NV: GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN
3′RACE outer: GCGAGCACAGAATTAATACGACT
bORF2-end2, GATGAGTTCATATCCTTTGTAGGG
The sequence of the antisense RNA-FISH probe Cy2-MS2 was,
Cy3-GTCGACCTGCAGACATGGGTGATCCTCATGTTTTCTAGGCAATTA.
Cells were lysed and their RNA initially extracted with Trizol (Life Technologies), followed by further purification using an RNeasy Mini Kit (Qiagen). Residual DNA was removed by Turbo DNA-free Kit DNase treatment (Ambion), and cDNA was generated from the RNA using the SuperScript III First Strand Synthesis System (Invitrogen) and a polyT primer. Subsequent PCR used GoTaq DNA polymerase (Promega). RT-PCR primers were:
1EGFPcass5P TGTTCTGCTGGTAGTGGTCG
2EGFPcass3P TATATCATGGCCGACAAGCAG,
which span the intron of the 99-PUR-JM111-EGFP reporter cassette, and
13HSPA6for CAAAATGCAAGACAAGTGTCG
14HSPA6rev TTCTAGCTTTGGAGGGAAAG,
which amplify HSPA6 (Accession No. NM_002155).
Supporting Information
Cells were treated with 1000 U/ml Universal Type I Interferon Alpha (PML Assay Science) or left untreated for 48 hours. Plasmid-expressed MX2-V5 is shown in lane 7.
(TIF)
(A) Similar to 293T cells, ectopic expression of ZAP restricts retrotransposition of 99-PUR-RPS-EGFP in HeLa-JVM cells. (B) Overexpressed ZAP constructs have minimal effect on cell viability. HeLa-JVM cells were cotransfected with 0.05 μg of pcDNA6 myc/hisB and 0.5 μg of plasmid containing ZAP or unrelated proteins (C22ORF28 or KIF11) in six-well plates, and after two days were expanded to T75 flasks. Cells were then selected for two weeks with 2 μg/ml blasticidin, fixed, and stained with giemsa. Colony counts are normalized to empty vector transfection. Standard deviation is for three separate flasks in a single experiment. (C) Expression of ZAP constructs restrict cell culture retrotransposition of an EGFP reporter-tagged and codon-optimized human L1 (ORFeus-HS, WA117 [57]). Results for a single experiment are show (quadruplicate wells). (D) Similarly, ZAP constructs inhibit retrotransposition of ORFeus-Mm (WA125 [129]), a codon-optimized version of mouse L1spa [59] cloned downstream of a CMV promoter. Results are for a single experiment. (E) Expression of ZAP does not affect the integrity of a contransfected L1 reporter construct. Vector alone, HA-ZAP-S or HA-ZAP-L was cotransfected with 99-PUR-JM111-EGFP, containing a mutant L1 defective for retrotransposition. At 3 days post-transfection, 293T cells were harvested, HIRT DNA extraction was performed, and plasmid DNA was analyzed using PCR and primers that flank the intron of the L1 EGFP reporter cassette.
(TIF)
(A) ZAP-interacting helicase DHX30 tagged with RFP colocalizes with GFP-tagged ORF1p in cytoplasmic granules. (B) ZAP-L and ZAP-S isoforms colocalize in large cytoplasmic foci.
(TIF)
ZAP-L-FL protein immunoprecipates efficiently in the presence of test proteins and RNase.
(TIF)
The interactions are resistant to digestion by RNase. V5-TEV-ISG20 also weakly interacts with ZAP-L-FL in the absence of RNase. IP conditions were as described for Fig 4B.
(TIF)
(PDF)
Acknowledgments
The authors appreciate reagents provided by researchers listed in the Methods section. Special thanks to the Mass Spectrometry and Proteomics Facility, ChemCore, Synthesis & Sequencing Facility, Ross Flow Cytometry Core Facility, and the Sidney Kimmel Comprehensive Cancer Center Flow Cytometry Core of Johns Hopkins University School of Medicine. We thank Drs. A. Leung and Y. Ando (JHU) for helpful advice and reagents and G. Abril for technical assistance.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by: 1R03NS087290-01 (JLG) : National Institute of Neurological Disorders and Stroke, National Institutes of Health; ALS Therapy Alliance (JLG); 1R01GM099875-01 (HHK): National Institute of General Medical Studies, National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Haller O, Kochs GJ (2011) Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity. Interferon Cytokine Res 31: 79–87. Review. 10.1089/jir.2010.0076 [DOI] [PubMed] [Google Scholar]
- 2. Lu J, Pan Q, Rong L, Liu SL, Liang C (2011) The IFITM proteins inhibit HIV-1 infection. J Virol 85: 12881–12889. 10.1128/JVI.05633-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Diamond MS, Farzan M (2013) The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13: 46–57. 10.1038/nri3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Neil SJ (2013) The antiviral activities of tetherin. Curr Top Microbiol Immunol 371: 67–104 10.1007/978-3-642-37765-5_3 [DOI] [PubMed] [Google Scholar]
- 5. Helbig KJ, Beard MR (2014) The role of viperin in the innate antiviral response. J Mol Biol 426: 1210–1219. 10.1016/j.jmb.2013.10.019 [DOI] [PubMed] [Google Scholar]
- 6. Koshiba T (2013) Mitochondrial-mediated antiviral immunity. Biochim Biophys Acta 1833: 225–232. Review. 10.1016/j.bbamcr.2012.03.005 [DOI] [PubMed] [Google Scholar]
- 7. Degols G, Eldin P, Mechti N (2007) ISG20, an actor of the innate immune response. Biochimie 89: 831–835. [DOI] [PubMed] [Google Scholar]
- 8. Allouch A, Di Primio C, Alpi E, Lusic M, Arosio D, Giacca M et al. (2011) The TRIM family protein KAP1 inhibits HIV-1 integration. Cell Host Microbe 9: 484–495. 10.1016/j.chom.2011.05.004 [DOI] [PubMed] [Google Scholar]
- 9. Katoh I, Kurata SI (2013) Association of endogenous retroviruses and long terminal repeats with human disorders. Front Oncol 3: 234 10.3389/fonc.2013.00234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alfahad T, Nath A (2013) Retroviruses and amyotrophic lateral sclerosis. Antiviral Res 99: 180–187. 10.1016/j.antiviral.2013.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cusick MF, Libbey JE, Fujinami RS (2013) Multiple sclerosis: autoimmunity and viruses. Curr Opin Rheumatol 25: 496–501. 10.1097/BOR.0b013e328362004d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tugnet N, Rylance P, Roden D, Trela M, Nelson P (2013) Human endogenous retroviruses (HERVs) and autoimmune rheumatic disease: is there a link? Open Rheumatol J 7: 13–21. 10.2174/1874312901307010013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cordaux R, Batzer MA (2009) The impact of retrotransposons on human genome evolution. Nat Rev Genet 10: 691–703. 10.1038/nrg2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hancks DC, Kazazian HH Jr (2012) Active human retrotransposons: variation and disease. Curr Opin Genet Dev 22: 191–203. 10.1016/j.gde.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, et al. (2003) Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA 100: 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, et al. (2010) LINE-1 retrotransposition activity in human genomes. Cell 141: 1159–1170. 10.1016/j.cell.2010.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Doolittle RF, Feng DF (1992) Tracing the origin of retroviruses. Curr Top Microbiol Immunol 176: 195–211. [DOI] [PubMed] [Google Scholar]
- 18. Malik HS, Eickbush TH (2001) Phylogenetic analysis of ribonuclease H domains suggests a late, chimeric origin of LTR retrotransposable elements and retroviruses. Genome Res 11: 1187–1197. [DOI] [PubMed] [Google Scholar]
- 19. Malik HS, Henikoff S, Eickbush TH (2000) Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res 10: 1307–1318. [DOI] [PubMed] [Google Scholar]
- 20. Schumann GG (2007) APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans 35: 637–642. [DOI] [PubMed] [Google Scholar]
- 21. Arias JF, Koyama T, Kinomoto M, Tokunaga K (2012) Retroelements versus APOBEC3 family members: no great escape from the magnificent seven. Front Microbiol 3: 275 10.3389/fmicb.2012.00275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richardson SR, Narvaiza I, Planegger RA, Weitzman MD, Moran JV (2014) APOBEC3A deaminates transiently exposed single-strand DNA during LINE-1 retrotransposition. Elife 3: e02008 10.7554/eLife.02008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhao K, Du J, Han X, Goodier JL, Li P, Zhou X, et al. (2013) Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutières syndrome-related SAMHD1. Cell Reports 4: 1108–1115 10.1016/j.celrep.2013.08.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stetson DB, Ko JS, Heidmann T, Medzhitov R (2008) Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134: 587–598. 10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang A, Dong B, Doucet AJ, Moldovan JB, Moran JV, Silverman RH (2013) RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res 42: 3803–2820. 10.1093/nar/gkt1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Arjan-Odedra S, Swanson CM, Sherer NM, Wolinsky SM, Malim MH (2012) Endogenous MOV10 inhibits the retrotransposition of endogenous retroelements but not the replication of exogenous retroviruses. Retrovirology 9: 53 10.1186/1742-4690-9-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goodier JL, Cheung LE, Kazazian HH Jr (2012) MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet 8: e1002941 10.1371/journal.pgen.1002941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li X, Zhang J, Jia R, Cheng V, Xu X, Qiao W, et al. (2013) The MOV10 helicase inhibits LINE-1 mobility. J Biol Chem 288: 21148–21160. 10.1074/jbc.M113.465856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mao R, Nie H, Cai D, Zhang J, Liu H, Yan R, et al. (2013) Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog 9: e1003494 10.1371/journal.ppat.1003494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xuan Y, Gong D, Qi J, Han C, Deng H, Gao G (2013) ZAP inhibits murine gammaherpesvirus 68 ORF64 expression and is antagonized by RTA. J Virol 87: 2735–2743. 10.1128/JVI.03015-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR (2003) Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol 77: 11555–11562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kerns JA, Emerman M, Malik HS (2008) Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet 4: e21 10.1371/journal.pgen.0040021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Goossens KE, Karpala AJ, Ward A, Bean AG (2014) Characterisation of chicken ZAP. Dev Comp Immunol 46: 373–381. 10.1016/j.dci.2014.05.011 [DOI] [PubMed] [Google Scholar]
- 34. Esnault C, Maestre J, Heidmann T (2000) Human LINE retrotransposons generate processed pseudogenes. Nat Genet 24: 363–367. [DOI] [PubMed] [Google Scholar]
- 35. Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, et al. (2001) Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol 21: 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH Jr (1999) Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum Mol Genet 8: 1557–1560. [DOI] [PubMed] [Google Scholar]
- 37. Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr (1996) High frequency retrotransposition in cultured mammalian cells. Cell 87: 917–927. [DOI] [PubMed] [Google Scholar]
- 38. Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH Jr (2000) Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res 28: 1418–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wei W, Morrish TA, Alisch RS, Moran JV (2000) A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal Biochem 284: 435–438. [DOI] [PubMed] [Google Scholar]
- 40. Nguyen K-L, Llano M, Akari H, Miyagi E, Poeschla EM, Strebel K, et al. (2004) Codon optimization of the HIV-1 vpu and vif genes stabilizes their messenger RNA and allows for highly efficient Rev-independent expression. Virology 319: 163 [DOI] [PubMed] [Google Scholar]
- 41. King MC, Raposo G, Lemmon MA (2004) Inhibition of nuclear import and cell-cycle progression by mutated forms of the dynamin-like GTPase MxB. Proc Natl Acad Sci USA 101: 8957–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kane M, Yadav SS, Bitzegeio J, Kutluay SB, Zang T, Wilson SJ, et al. (2013) MX2 is an interferon-induced inhibitor of HIV-1 infection. Nature 502: 563–6. 10.1038/nature12653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu Z, Pan Q, Ding S, Qian J, Xu F, Zhou J, et al. (2013) The interferon-inducible MxB protein inhibits HIV-1 infection. Cell Host Microbe 14: 398–410. 10.1016/j.chom.2013.08.015 [DOI] [PubMed] [Google Scholar]
- 44. Goujon C, Moncorgé O, Bauby H, Doyle T, Barclay WS, Malim MH (2014) Transfer of the amino-terminal nuclear envelope targeting domain of human MX2 converts MX1 into an HIV-1 resistance factor. J Virol 88: 9017–9026. 10.1128/JVI.01269-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Goujon C, Moncorgé O, Bauby H, Doyle T, Ward CC, Schaller T, et al. (2013) Human MX2 is an interferon-induced post-entry inhibitor of HIV-1 infection. Nature 502: 559–562. 10.1038/nature12542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Goodier JL, Cheung LE, Kazazian HH Jr (2013) Mapping the LINE1 ORF1 protein interactome reveals associated inhibitors of human retrotransposition. Nucleic Acids Res 41: 7401–7419. 10.1093/nar/gkt512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Marsischky GT, Wilson BA, Collier RJ (1995) Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J Biol Chem 270: 3247–3254. [DOI] [PubMed] [Google Scholar]
- 48. Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, et al. (2008) Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell 32: 57–69. 10.1016/j.molcel.2008.08.009 [DOI] [PubMed] [Google Scholar]
- 49. Gao G, Guo X, Goff SP (2002) Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297: 1703–1706. [DOI] [PubMed] [Google Scholar]
- 50. Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G (2004) The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol 78: 12781–12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu Y, Chen G, Lv F, Wang X, Ji X, Xu Y, et al. (2011) Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci USA 108: 15834–15839. 10.1073/pnas.1101676108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen S, Xu Y, Zhang K, Wang X, Sun J, Gao G, Liu Y (2012) Structure of N-terminal domain of ZAP indicates how a zinc-finger protein recognizes complex RNA. Nat Struct Mol Biol 19: 430–435. 10.1038/nsmb.2243 [DOI] [PubMed] [Google Scholar]
- 53. Hayakawa S, Shiratori S, Yamato H, Kameyama T, Kitatsuji C, Kashigi F, et al. (2011) ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol 12: 37–44. 10.1038/ni.1963 [DOI] [PubMed] [Google Scholar]
- 54. Todorova T, Bock FJ, Chang P. (2014) PARP13 regulates cellular mRNA post-transcriptionally and functions as a pro-apoptotic factor by destabilizing TRAILR4 transcript. Nat Commun 5: 5362 10.1038/ncomms6362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gläsker S, Töller M, Kümmerer BM (2014) The alternate triad motif of the poly(ADP-ribose) polymerase-like domain of the human zinc finger antiviral protein is essential for its antiviral activity. J Gen Virol 95(Pt 4): 816–822 10.1099/vir.0.060988-0 [DOI] [PubMed] [Google Scholar]
- 56. Charron G, Li MM, MacDonald MR, Hang HC (2013) Prenylome profiling reveals S-farnesylation is crucial for membrane targeting and antiviral activity of ZAP long-isoform. Proc Natl Acad Sci USA 110: 11085–11090. 10.1073/pnas.1302564110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. An W, Dai L, Niewiadomska AM, Yetil A, O’Donnell KA, Han JS, Boeke JD (2011) Characterization of a synthetic human LINE-1 retrotransposon ORFeus-Hs. Mobile DNA 2: 2 10.1186/1759-8753-2-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Han JS, Boeke JD (2004) A highly active synthetic mammalian retrotransposon. Nature 429: 314–318. [DOI] [PubMed] [Google Scholar]
- 59. Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, et al. (1998) An actively retrotransposing, novel subfamily of mouse L1 elements. EMBO J 17: 590–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bebek F (2012) Towards the identification of novel interferon-alpha induced anti-hepatitis C virus effectors. Ph.D. thesis, School of Biotechnology and Biomolecular Sciences, University of New South Wales, Australia.
- 61. Dewannieux M, Esnault C, Heidmann T (2003) LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35: 41–48 [DOI] [PubMed] [Google Scholar]
- 62. Dewannieux M, Dupressoir A, Harper F, Pierron G, Heidmann T (2004) Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat Genet 36: 534–539. [DOI] [PubMed] [Google Scholar]
- 63. Goodier JL, Zhang L, Vetter MR, Kazazian HH Jr (2007) LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol 27: 6469–6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goodier JL, Mandal PK, Zhang L, Kazazian HH Jr (2010) Discrete subcellular partitioning of human retrotransposon RNAs despite a common mechanism of genome insertion. Hum Mol Genet 19: 1712–1725. 10.1093/hmg/ddq048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, et al. (2010) Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet 6 pii: e1001150 10.1371/journal.pgen.1001150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Adjibade P, Mazroui R (2014) Control of mRNA turnover: Implication of cytoplasmic RNA granules. Semin Cell Dev Biol 34:15–23. 10.1016/j.semcdb.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 67. Taylor MS, Lacava J, Mita P, Molloy KR, Huang CR, Li D, et al. (2013) Affinity proteomics reveals human host factors implicated in discrete stages of LINE-1 retrotransposition. Cell 155: 1034–1048. 10.1016/j.cell.2013.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Leung A, Todorova T, Ando Y, Chang P (2011) Poly(ADP-ribose) regulates post-transcriptional gene regulation in the cytoplasm. RNA Biol 9: 542–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lee H, Komano J, Saitoh Y, Yamaoka S, Kozaki T, Misawa T, et al. (2013) Zinc-finger antiviral protein mediates retinoic acid inducible gene I-like receptor-independent antiviral response to murine leukemia virus. Proc Natl Acad Sci USA 110: 12379–12384. 10.1073/pnas.1310604110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang ZF, Wang XL, Gao GX (2012) PR65A regulates the activity of the zinc-finger antiviral protein. Prog Biochem Biophysics 39: 431–437. [Google Scholar]
- 71. Sun L, Lv F, Guo X, Gao G (2012) Glycogen synthase kinase 3β (GSK3β) modulates antiviral activity of zinc-finger antiviral protein (ZAP). J Biol Chem 287: 22882–22888. 10.1074/jbc.M111.306373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Guo X, Ma J, Sun J, Gao G (2007) The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci USA 104: 151–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ye P, Liu S, Zhu Y, Chen G, Gao G (2010) DEXH-Box protein DHX30 is required for optimal function of the zinc-finger antiviral protein. Protein Cell 1: 956–964. 10.1007/s13238-010-0117-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sheth U, Parker R (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, et al. (2007) TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446: 916–920. [DOI] [PubMed] [Google Scholar]
- 76. Chen Z, Zhou Y, Song J and Zhang Z (2013) hCKSAAP UbSite: Improved prediction of human ubiquitination site by exploiting amino acid pattern and properties. Biochim Biophys Acta 1834: 1461–1467. 10.1016/j.bbapap.2013.04.006 [DOI] [PubMed] [Google Scholar]
- 77. Radivojac P, Vacic V, Haynes C, Cocklin RR, Mohan A, Heyen JW, et al. (2010) Identification, analysis and prediction of protein ubiquitination sites. Proteins 78: 365–380. 10.1002/prot.22555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Willison KR, Grantham J (2001) The roles of the cytosolic chaperonin, CCT, in normal eukaryotic cell growth In Molecular Chaperones in the Cell, Lund P (ed), pp 90–118. Oxford: Oxford University Press. [Google Scholar]
- 79. Dekker C, Stirling PC, McCormack EA, Filmore H, Paul A, et Brost RL, al. (2008) The interaction network of the chaperonin CCT. EMBO J 27: 1827–1839. 10.1038/emboj.2008.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gregersen LH, Schueler M, Munschauer M, Mastrobuoni G, Chen W, Kempa S, et al. (2014) MOV10 is a 5' to 3' RNA helicase contributing to UPF1 mRNA target degradation by translocation along 3' UTRs. Mol Cell 54: 573–585. 10.1016/j.molcel.2014.03.017 [DOI] [PubMed] [Google Scholar]
- 81. Kulpa DA, Moran JV (2006) Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol 13: 655–660. [DOI] [PubMed] [Google Scholar]
- 82. Zhu Y, Wang X, Goff SP, Gao G. (2012) Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J 31: 4236–4246. 10.1038/emboj.2012.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Jacobs JL and Coyne CB (2013) Mechanisms of MAVS regulation at the mitochondrial membrane. J Mol Biol 425: 5009–5019. 10.1016/j.jmb.2013.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Chiang JJ, Davis ME, Gack MU (2014) Regulation of RIG-I-like receptor signaling by host and viral proteins. Cytokine Growth Factor Rev 25: 491–505. 10.1016/j.cytogfr.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Espert L, Degols G, Lin Y-L, Vincent T, Benkirane M, Mechti N (2005) Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J Gen Virol 86: 2221–2229 [DOI] [PubMed] [Google Scholar]
- 86. Zhou Z, Wang N, Woodson SE, Dong Q, Wang J, Liang Y, et al. (2011) Antiviral activities of ISG20 in positive-strand RNA virus infections. Virology 409: 175–188. 10.1016/j.virol.2010.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, et al. (2010) KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240. 10.1038/nature08674 [DOI] [PubMed] [Google Scholar]
- 88. Wolf D, Goff SP (2007) TRIM28 mediates primer binding site- targeted silencing of murine leukemia virus in embryonic cells. Cell 131: 46–57. [DOI] [PubMed] [Google Scholar]
- 89. Mandal PK, Ewing AD, Hancks DC, Kazazian HH Jr (2013) Enrichment of processed pseudogene transcripts in L1-ribonucleoprotein particles. Hum MolGenet 22: 3730–3748. 10.1093/hmg/ddt225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lu C, Luo Z, Jäger S, Krogan NJ, Peterlin BM (2012) Moloney leukemia virus type 10 inhibits reverse transcription and retrotransposition of intracisternal a particles. J Virol 86: 10517–10523 10.1128/JVI.00868-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Leung AK, Calabrese JM, Sharp PA (2006) Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci USA 103: 18125–18130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Leung AK, Vyas S, Rood JE, Bhutkar A, Sharp PA, Chang P (2011) Poly (ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm. Mol Cell 42: 489–499. 10.1016/j.molcel.2011.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nano N, Houry WA (2013) Chaperone-like activity of the AAA+ proteins Rvb1 and Rvb2 in the assembly of various complexes. Philos Trans R Soc Lond B Biol Sci 368: 20110399 10.1098/rstb.2011.0399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Wolf D, Goff SP (2009) Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204. 10.1038/nature07844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Iyengar S, Ivanov AV, Jin VX, Rauscher FJ 3rd, Farnham PJ, Friedli M, et al. (2011) Functional analysis of KAP1 genomic recruitment. Mol Cell Biol 31: 1833–1847. 10.1128/MCB.01331-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, et al. 2014. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev 28: 1397–1409. 10.1101/gad.241661.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, et al. (2014) SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun 5: 5011 10.1038/ncomms6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, et al. (2010) Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931. 10.1038/nature08858 [DOI] [PubMed] [Google Scholar]
- 99. Karimi MM, Goyal P, Maksakova IA, Bilenky M, Leung D, Tang JX, et al. (2011) DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 8: 676–687. 10.1016/j.stem.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Maksakova IA, Thompson PJ, Goyal P, Jones SJ, Singh PB, Karimi MM, et al. (2013) Distinct roles of KAP1, HP1 and G9a/GLP in silencing of the two-cell-specific retrotransposon MERVL in mouse ES cells. Epigenetics Chromatin 6: 15 10.1186/1756-8935-6-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Reichmann J, Crichton JH, Madej MJ, Taggart M, Gautier P, Garcia-Perez JL, et al. (2012) Microarray analysis of LTR retrotransposon silencing identifies Hdac1 as a regulator of retrotransposon expression in mouse embryonic stem cells. PLoS Comput Biol 8: e1002486 10.1371/journal.pcbi.1002486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Liu L, Chen G, Ji X, Gao G (2004) ZAP is a CRM1-dependent nucleocytoplasmic shuttling protein. Biochem Biophys Res Commun 321: 517–523 [DOI] [PubMed] [Google Scholar]
- 103. MacDonald MR, Machlin ES, Albin OR, Levy DE (2007) The zinc finger antiviral protein acts synergistically with an interferon-induced factor for maximal activity against alphaviruses. J Virol 81: 13509–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Karki S, Li MM, Schoggins JW, Tian S, Rice CM, MacDonald MR (2012) Multiple interferon stimulated genes synergize with the zinc finger antiviral protein to mediate anti-alphavirus activity. PLoS One 7:e37398 10.1371/journal.pone.0037398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Rusinova I, Forster S, Yu S, Kannan A, Masse M, Chapman R, et al. (2013) INTERFEROME v2. 0: an updated database of annotated interferon-regulated genes. Nucl Acids Res 41 (database issue): D1040–D1046. 10.1093/nar/gks1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Zhou Y, Ma J, Bushan Roy B, Wu JY, Pan Q, Rong L, Liang C (2008) The packaging of human immunodeficiency virus type 1 RNA is restricted by overexpression of an RNA helicase DHX30. Virology 372: 97–106. [DOI] [PubMed] [Google Scholar]
- 107. Bidet K, Dadlani D, Garcia-Blanco MA (2014) G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog 10: e1004242 10.1371/journal.ppat.1004242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kumar M, Rawat P, Khan SZ, Dhamija N, Chaudhary P, Ravi DS, et al. (2011) Reciprocal regulation of human immunodeficiency virus-1 gene expression and replication by heat shock proteins 40 and 70. J Mol Biol 410: 944–958. 10.1016/j.jmb.2011.04.005 [DOI] [PubMed] [Google Scholar]
- 109. Urano E, Morikawa Y, Komano J (2013) Novel role of HSP40/DNAJ in the regulation of HIV-1 replication. J Acquir Immune Defic Syndr 64: 154–162 10.1097/QAI.0b013e31829a2ef8 [DOI] [PubMed] [Google Scholar]
- 110. Gack MU, Albrecht RA, Urano T, Inn KS, Huang IC, Carnero E, et al. (2009) Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5: 439–449. 10.1016/j.chom.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Meredith M, Orr A, Everett RD (1994) Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology 100: 457–469. [DOI] [PubMed] [Google Scholar]
- 112. Ha HC, Juluri K, Zhou Y, Leung S, Hermankova M, Snyder SH (2001) Poly(ADP-ribose) polymerase-1 is required for efficient HIV-1 integration. Proc Natl Acad Sci USA 98: 3364–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kameoka M, Nukuzuma S, Itaya A, Tanaka Y, Ota K, Ikuta K, et al. (2004) RNA interference directed against Poly(ADP-Ribose) polymerase 1 efficiently suppresses human immunodeficiency virus type 1 replication in human cells. J Virol 78: 8931–8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Tulin A, Stewart D, Spradling AC (2002) The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev 16: 2108–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ohsaki E, Ueda K, Sakakibara S, Do E, Yada K, Yamanishi K (2004) Poly(ADP-ribose) polymerase 1 binds to Kaposi's sarcoma-associated herpesvirus (KSHV) terminal repeat sequence and modulates KSHV replication in latency. J Virol 78: 9936–9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Tempera I, Deng Z, Atanasiu C, Chen CJ, D'Erme M, Lieberman PM (2010) Regulation of Epstein-Barr virus OriP replication by poly(ADP-ribose) polymerase 1. J Virol 84: 4988–4997. 10.1128/JVI.02333-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Kakugawa S, Shimojima M, Neumann G, Goto H, Kawaoka Y (2009) RuvB-like protein 2 is a suppressor of influenza A virus polymerases. J Virol. 83: 6429–6434. 10.1128/JVI.00293-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yu Q, Carbone CJ, Katlinskaya YV, Zheng H, Zheng K, Luo M, Wang PJ, Greenberg RA, Fuchs SY (2015) Type I interferon controls propagation of Long Interspersed Element-1. J Biol Chem: 290: 10191–10199. 10.1074/jbc.M114.612374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN (2010) MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci USA 107: 11847–52. 10.1073/pnas.1007158107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Crow YJ (2011) Type I interferonopathies: a novel set of inborn errors of immunity. Ann NY Acad Sci 1238: 91–98. 10.1111/j.1749-6632.2011.06220.x [DOI] [PubMed] [Google Scholar]
- 121. Liu Y, Jesus AA, Marrero B, Yang D, Ramsey SE, Montealegre Sanchez GA, et al. (2014) Activated STING in a vascular and pulmonary syndrome. N Engl J Med 371: 507–518. 10.1056/NEJMoa1312625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Crow YJ, Rehwinkel J (2009) Aicardi-Goutieres syndrome and related phenotypes: linking nucleic acid metabolism with autoimmunity. Hum Mol Genet 18(R2): R130–6. 10.1093/hmg/ddp293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Volkman HE, Stetson DB (2014) The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol 15: 415–422. 10.1038/ni.2872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Rice GI, Forte GM, Szynkiewicz M, Chase DS, Aeby A, Abdel-Hamid MS, et al. (2013) Assessment of interferon-related biomarkers in Aicardi-Goutières syndrome associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, and ADAR: a case-control study. Lancet Neurol 12: 1159–1169. 10.1016/S1474-4422(13)70258-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Rice GI, del Toro Duany Y, Jenkinson EM, Forte GM, Anderson BH, Ariaudo G, et al. (2014) Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet 46: 503–509. 10.1038/ng.2933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Oda H, Nakagawa K, Abe J, Awaya T, Funabiki M, Hijikata A, et al. (2014) Aicardi-Goutières syndrome is caused by IFIH1 mutations. Am J Hum Genet 95: 121–125. 10.1016/j.ajhg.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Nishikura K (2007) Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat Genet 7: 919–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Stetson DB, Ko JS, Heidmann T, Medzhitov R (2008) Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134: 587–598. 10.1016/j.cell.2008.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Wang X, Han Y, Dang Y, Fu W, Zhou T, Ptak RG, et al. (2010) Moloney leukemia virus 10 (MOV10) protein inhibits retrovirus replication. J Biol Chem 285: 14346–55. 10.1074/jbc.M110.109314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. An W, Han JS, Wheelan SJ, Davis ES, Coombes CE, Ye P, et al. (2006) Active retrotransposition by a synthetic L1 element in mice. Proc Natl Acad Sci USA 103: 18662–18667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Langelier MF, Planck JL, Roy S, Pascal JM (2011) Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: structural and functional insights into DNA-dependent PARP-1 activity. J Biol Chem 286: 10690–10701. 10.1074/jbc.M110.202507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Agrawal P, Chen YT, Schilling B, Gibson BW, Hughes RE (2012) Ubiquitin-specific peptidase 9, X-linked (USP9X) modulates activity of mammalian target of rapamycin (mTOR). J Biol Chem 287: 21164–21175. 10.1074/jbc.M111.328021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Sarkari F, Wheaton K, La Delfa A, Mohamed M, Shaikh F, Khatun R, et al. (2013) Ubiquitin-specific protease 7 is a regulator of ubiquitin-conjugating enzyme UbE2E1. J Biol Chem 288: 16975–16985. 10.1074/jbc.M113.469262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV (2006) Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev 20: 210–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Hulme AE, Bogerd HP, Cullen BR, Moran JV (2007) Selective inhibition of Alu retrotransposition by APOBEC3G. Gene 390: 199–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Leibold DM, Swergold GD, Singer MF, Thayer RE, Dombroski BA, Fanning TG (1990) Translation of LINE-1 DNA elements in vitro and in human cells. Proc Natl Acad Sci USA 87: 6990–6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Rodić N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, et al. (2014) Long interspersed element-1 protein expression is a hallmark of many human cancers. Am J Pathol 184: 1280–1286. 10.1016/j.ajpath.2014.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cells were treated with 1000 U/ml Universal Type I Interferon Alpha (PML Assay Science) or left untreated for 48 hours. Plasmid-expressed MX2-V5 is shown in lane 7.
(TIF)
(A) Similar to 293T cells, ectopic expression of ZAP restricts retrotransposition of 99-PUR-RPS-EGFP in HeLa-JVM cells. (B) Overexpressed ZAP constructs have minimal effect on cell viability. HeLa-JVM cells were cotransfected with 0.05 μg of pcDNA6 myc/hisB and 0.5 μg of plasmid containing ZAP or unrelated proteins (C22ORF28 or KIF11) in six-well plates, and after two days were expanded to T75 flasks. Cells were then selected for two weeks with 2 μg/ml blasticidin, fixed, and stained with giemsa. Colony counts are normalized to empty vector transfection. Standard deviation is for three separate flasks in a single experiment. (C) Expression of ZAP constructs restrict cell culture retrotransposition of an EGFP reporter-tagged and codon-optimized human L1 (ORFeus-HS, WA117 [57]). Results for a single experiment are show (quadruplicate wells). (D) Similarly, ZAP constructs inhibit retrotransposition of ORFeus-Mm (WA125 [129]), a codon-optimized version of mouse L1spa [59] cloned downstream of a CMV promoter. Results are for a single experiment. (E) Expression of ZAP does not affect the integrity of a contransfected L1 reporter construct. Vector alone, HA-ZAP-S or HA-ZAP-L was cotransfected with 99-PUR-JM111-EGFP, containing a mutant L1 defective for retrotransposition. At 3 days post-transfection, 293T cells were harvested, HIRT DNA extraction was performed, and plasmid DNA was analyzed using PCR and primers that flank the intron of the L1 EGFP reporter cassette.
(TIF)
(A) ZAP-interacting helicase DHX30 tagged with RFP colocalizes with GFP-tagged ORF1p in cytoplasmic granules. (B) ZAP-L and ZAP-S isoforms colocalize in large cytoplasmic foci.
(TIF)
ZAP-L-FL protein immunoprecipates efficiently in the presence of test proteins and RNase.
(TIF)
The interactions are resistant to digestion by RNase. V5-TEV-ISG20 also weakly interacts with ZAP-L-FL in the absence of RNase. IP conditions were as described for Fig 4B.
(TIF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.