Abstract
We show here that the class I human cytoplasmic isoleucyl-tRNA synthetase is an exceptionally large polypeptide (1266 aa) which, unlike its homologues in lower eukaryotes and prokaryotes, has a third domain of two repeats of an approximately 90-aa sequence appended to its C-terminal end. While extracts of Escherichia coli do not aminoacrylate mammalian tRNA with isoleucine, expression of the cloned human gene in E. coli results in charging of the mammalian tRNA substrate. The appended third domain is dispensable for detection of this aminoacylation activity and may be needed for assembly of a multisynthetase complex in mammalian cells. Alignment of the sequences of the remaining two domains shared by isoleucyl-tRNA synthetases from E. coli to human reveals a much greater selective pressure on the domain needed for tRNA acceptor helix interactions and catalysis than on the domain needed for interactions with the anticodon. This result may have implications for the historical development of an operational RNA code for amino acids.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Biou V., Yaremchuk A., Tukalo M., Cusack S. The 2.9 A crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNA(Ser). Science. 1994 Mar 11;263(5152):1404–1410. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Buechter D. D., Schimmel P. Dissection of a class II tRNA synthetase: determinants for minihelix recognition are tightly associated with domain for amino acid activation. Biochemistry. 1993 May 18;32(19):5267–5272. doi: 10.1021/bi00070a039. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Rees B., Ruff M., Thierry J. C., Moras D. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993 Mar 11;362(6416):181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- Cerini C., Kerjan P., Astier M., Gratecos D., Mirande M., Sémériva M. A component of the multisynthetase complex is a multifunctional aminoacyl-tRNA synthetase. EMBO J. 1991 Dec;10(13):4267–4277. doi: 10.1002/j.1460-2075.1991.tb05005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csank C., Martindale D. W. Isoleucyl-tRNA synthetase from the ciliated protozoan Tetrahymena thermophila. DNA sequence, gene regulation, and leucine zipper motifs. J Biol Chem. 1992 Mar 5;267(7):4592–4599. [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Edwards J. B., Delort J., Mallet J. Oligodeoxyribonucleotide ligation to single-stranded cDNAs: a new tool for cloning 5' ends of mRNAs and for constructing cDNA libraries by in vitro amplification. Nucleic Acids Res. 1991 Oct 11;19(19):5227–5232. doi: 10.1093/nar/19.19.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englisch U., Englisch S., Markmeyer P., Schischkoff J., Sternbach H., Kratzin H., Cramer F. Structure of the yeast isoleucyl-tRNA synthetase gene (ILS1). DNA-sequence, amino-acid sequence of proteolytic peptides of the enzyme and comparison of the structure to those of other known aminoacyl-tRNA synthetases. Biol Chem Hoppe Seyler. 1987 Aug;368(8):971–979. doi: 10.1515/bchm3.1987.368.2.971. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Dirheimer G., Gangloff J. Cysteinyl-tRNA synthetase: determination of the last E. coli aminoacyl-tRNA synthetase primary structure. Nucleic Acids Res. 1991 Jan 25;19(2):265–269. doi: 10.1093/nar/19.2.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R., Puglisi J. D., Florentz C. tRNA structure and aminoacylation efficiency. Prog Nucleic Acid Res Mol Biol. 1993;45:129–206. doi: 10.1016/s0079-6603(08)60869-7. [DOI] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., Shiba K., Mottes C., Schimmel P. Sequence determination and modeling of structural motifs for the smallest monomeric aminoacyl-tRNA synthetase. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):976–980. doi: 10.1073/pnas.88.3.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hountondji C., Dessen P., Blanquet S. Sequence similarities among the family of aminoacyl-tRNA synthetases. Biochimie. 1986 Sep;68(9):1071–1078. doi: 10.1016/s0300-9084(86)80181-x. [DOI] [PubMed] [Google Scholar]
- Jenal U., Rechsteiner T., Tan P. Y., Bühlmann E., Meile L., Leisinger T. Isoleucyl-tRNA synthetase of Methanobacterium thermoautotrophicum Marburg. Cloning of the gene, nucleotide sequence, and localization of a base change conferring resistance to pseudomonic acid. J Biol Chem. 1991 Jun 5;266(16):10570–10577. [PubMed] [Google Scholar]
- Lazard M., Mirande M. Cloning and analysis of a cDNA encoding mammalian arginyl-tRNA synthetase, a component of the multisynthetase complex with a hydrophobic N-terminal extension. Gene. 1993 Oct 15;132(2):237–245. doi: 10.1016/0378-1119(93)90201-d. [DOI] [PubMed] [Google Scholar]
- Martindale D. W., Gu Z. M., Csank C. Isolation and complete sequence of the yeast isoleucyl-tRNA synthetase gene (ILS1). Curr Genet. 1989 Feb;15(2):99–106. doi: 10.1007/BF00435455. [DOI] [PubMed] [Google Scholar]
- Mead D. A., Szczesna-Skorupa E., Kemper B. Single-stranded DNA 'blue' T7 promoter plasmids: a versatile tandem promoter system for cloning and protein engineering. Protein Eng. 1986 Oct-Nov;1(1):67–74. doi: 10.1093/protein/1.1.67. [DOI] [PubMed] [Google Scholar]
- Mirande M. Aminoacyl-tRNA synthetase family from prokaryotes and eukaryotes: structural domains and their implications. Prog Nucleic Acid Res Mol Biol. 1991;40:95–142. doi: 10.1016/s0079-6603(08)60840-5. [DOI] [PubMed] [Google Scholar]
- Mirande M., Lazard M., Martinez R., Latreille M. T. Engineering mammalian aspartyl-tRNA synthetase to probe structural features mediating its association with the multisynthetase complex. Eur J Biochem. 1992 Feb 1;203(3):459–466. doi: 10.1111/j.1432-1033.1992.tb16570.x. [DOI] [PubMed] [Google Scholar]
- Moras D. Structural and functional relationships between aminoacyl-tRNA synthetases. Trends Biochem Sci. 1992 Apr;17(4):159–164. doi: 10.1016/0968-0004(92)90326-5. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Nureki O., Niimi T., Muramatsu T., Kanno H., Kohno T., Florentz C., Giegé R., Yokoyama S. Molecular recognition of the identity-determinant set of isoleucine transfer RNA from Escherichia coli. J Mol Biol. 1994 Feb 25;236(3):710–724. doi: 10.1006/jmbi.1994.1184. [DOI] [PubMed] [Google Scholar]
- Ohashi M., Aizawa S., Ooka H., Ohsawa T., Kaji K., Kondo H., Kobayashi T., Noumura T., Matsuo M., Mitsui Y. A new human diploid cell strain, TIG-1, for the research on cellular aging. Exp Gerontol. 1980;15(2):121–133. doi: 10.1016/0531-5565(80)90083-2. [DOI] [PubMed] [Google Scholar]
- Park S. J., Schimmel P. Evidence for interaction of an aminoacyl transfer RNA synthetase with a region important for the identity of its cognate transfer RNA. J Biol Chem. 1988 Nov 15;263(32):16527–16530. [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Saks M. E., Sampson J. R., Abelson J. N. The transfer RNA identity problem: a search for rules. Science. 1994 Jan 14;263(5144):191–197. doi: 10.1126/science.7506844. [DOI] [PubMed] [Google Scholar]
- Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- Schimmel P., Giegé R., Moras D., Yokoyama S. An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci U S A. 1993 Oct 1;90(19):8763–8768. doi: 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P., Shepard A., Shiba K. Intron locations and functional deletions in relation to the design and evolution of a subgroup of class I tRNA synthetases. Protein Sci. 1992 Oct;1(10):1387–1391. doi: 10.1002/pro.5560011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard A., Shiba K., Schimmel P. RNA binding determinant in some class I tRNA synthetases identified by alignment-guided mutagenesis. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9964–9968. doi: 10.1073/pnas.89.20.9964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K., Schimmel P. Functional assembly of a randomly cleaved protein. Proc Natl Acad Sci U S A. 1992 Mar 1;89(5):1880–1884. doi: 10.1073/pnas.89.5.1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba K., Schimmel P. Tripartite functional assembly of a large class I aminoacyl tRNA synthetase. J Biol Chem. 1992 Nov 15;267(32):22703–22706. [PubMed] [Google Scholar]
- Shigesada K., Itamura S., Kato M., Hatanaka M., Imai M., Tanaka M., Masuda N., Nagai J., Nakashima K. Construction of a new plasmid vector that can express cloned cDNA in all translational reading frames. Gene. 1987;53(2-3):163–172. doi: 10.1016/0378-1119(87)90004-7. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
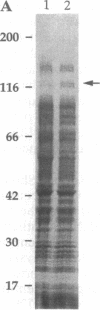
- Stanbridge E. J., Flandermeyer R. R., Daniels D. W., Nelson-Rees W. A. Specific chromosome loss associated with the expression of tumorigenicity in human cell hybrids. Somatic Cell Genet. 1981 Nov;7(6):699–712. doi: 10.1007/BF01538758. [DOI] [PubMed] [Google Scholar]
- Starzyk R. M., Webster T. A., Schimmel P. Evidence for dispensable sequences inserted into a nucleotide fold. Science. 1987 Sep 25;237(4822):1614–1618. doi: 10.1126/science.3306924. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Webster T., Tsai H., Kula M., Mackie G. A., Schimmel P. Specific sequence homology and three-dimensional structure of an aminoacyl transfer RNA synthetase. Science. 1984 Dec 14;226(4680):1315–1317. doi: 10.1126/science.6390679. [DOI] [PubMed] [Google Scholar]
- Zu Y. L., Shigesada K., Nishida E., Kubota I., Kohno M., Hanaoka M., Namba Y. 65-kilodalton protein phosphorylated by interleukin 2 stimulation bears two putative actin-binding sites and two calcium-binding sites. Biochemistry. 1990 Sep 11;29(36):8319–8324. doi: 10.1021/bi00488a017. [DOI] [PubMed] [Google Scholar]