Summary
How the highly stereotyped morphologies of individual neurons are genetically specified is not well understood. We identify six transcription factors (TFs) expressed in a combinatorial manner in seven post-mitotic adult leg motor neurons (MNs) that are derived from a single neuroblast in Drosophila. Unlike TFs expressed in mitotically active neuroblasts, these TFs do not regulate each other's expression. Removing the activity of a single TF resulted in specific morphological defects, including muscle targeting and dendritic arborization, and in a highly specific walking defect in adult flies. In contrast, when the expression of multiple TFs was modified nearly complete transformations in MN morphologies were generated. These results show that the morphological characteristics of a single neuron are dictated by a combinatorial code of morphology TFs (mTFs). mTFs function at a previously unidentified regulatory tier downstream of factors acting in the NB, but independently of factors that act in terminally differentiated neurons.
Introduction
Neurons are the most morphologically diverse cell types in the animal kingdom, providing animals with the means to sense their environment and move in response. In Drosophila, neurons are generated by neuroblasts (NBs), specialized stem cells dedicated to the generation of neurons and glia (Doe and Skeath, 1996; Prokop and Technau, 1991; Truman and Bate, 1988). As they divide, NBs express a temporal sequence of transcription factors (TFs) that contribute to the generation of neuronal diversity. For example, in the embryonic ventral nerve cord (VNC) most NBs express a sequence of five TFs (Hunchback, Krüppel, Pdm1/Pdm2, Castor and Grainyhead) (Isshiki et al., 2001; Li et al., 2013a), while in medulla NBs and intermediate neural progenitors of the Drosophila larval brain a different series of TFs have been described (Bayraktar and Doe, 2013; Li et al., 2013b). In vertebrates, analogous strategies are likely used by neural stem cells, e.g. in the cerebral cortex and retina, suggesting that this regulatory logic is evolutionarily conserved (Jacob et al., 2008; Okano and Temple, 2009). Nevertheless, although temporally expressed NB TFs play an important role in generating diversity, this strategy cannot be sufficient to explain the vast array of morphologically distinct neurons present in nervous systems (Fishell and Heintz, 2013). For example, in the Drosophila optic lobe there is estimated to be ∼40,000 neurons, classified into ∼70 morphologically distinct types, each making unique connections within the fly's visual circuitry (Fischbach and Dittrich, 1989).
A second class of TFs has been proposed to specify subtypes of neurons (Briscoe and Novitch, 2008; Dasen and Jessell, 2009; Landgraf and Thor, 2006). For example, in the vertebrate spinal cord, all motor neurons (MNs) express a common set of TFs at the progenitor stage (Olig2, Nkx6.1/6.2 and Pax6) and a different set of TFs after they become post-mitotic (Hb9, Islet1/2 and Lhx3) (Dasen and Jessell, 2009). Hox6 at brachial and Hox10 at lumbar levels further distinguish MNs that target muscles in the limbs instead of body wall muscles. Subsequently, limb-targeting MNs are further refined into pools, where all MNs in a single pool target the same muscle. Each pool is molecularly defined by the expression of pool-specific TFs, including a unique combination of Hox TFs (Dasen and Jessell, 2009; Philippidou and Dasen, 2013). In Drosophila embryos, subclassess of MNs are also specified by unique combinations of TFs: evenskipped (eve) and grain are expressed in six MNs that target dorsal body wall muscles (Fujioka et al., 2003; Garces and Thor, 2006; Landgraf et al., 1999), and Hb9, Nkx6, Islet, Lim3 and Olig2 are required for ventral-targeting MNs (Broihier et al., 2004; Broihier and Skeath, 2002; Certel and Thor, 2004; Oyallon et al., 2012; Thor et al., 1999; Thor and Thomas, 1997). However, each neuronal subtype defined by these TFs includes multiple morphologically distinct neurons, leaving open the question of how individual neuronal morphologies are specified.
A third class of TFs suggested to be important for neuronal identity is encoded by terminal selector genes (Allan et al., 2005; Eade et al., 2012; Hobert, 2011). Initially defined in C. elegans, these factors maintain a neuron's terminally differentiated characteristics by, for example, regulating genes required for the production of a particular neurotransmitter or neuropeptide. Consequently, these TFs must be expressed throughout the lifetime of a terminally differentiated neuron. Notably, as with neurons that are from the same subtype, neurons that share terminal characteristics, and are therefore likely to share the same terminal selector TFs, can have distinct morphological identities. For example, in C. elegans two terminal selector TFs, Mec-3 and Unc-86, function together to maintain the expression of genes required for a mechanosensory fate in six morphologically distinct touch sensitive neurons (Duggan et al., 1998).
In contrast to the logic revealed by these three classes of TFs, very little is known about how individual neurons, each with their own stereotyped dendritic arbors and synaptic targets, obtain their specific morphological characteristics. Here we address this question by focusing on how individual MNs that target the adult legs of Drosophila obtain their morphological identities. The adult leg MNs of Drosophila offer several advantages for understanding the genetic specification of neuronal morphology. For one, all eleven NB lineages that generate the ∼50 legtargeting MNs in each hemisegment have been defined (Baek and Mann, 2009; Brierley et al., 2012). More than two-thirds of these MNs are derived from only two lineages, Lin A (also called Lin 15) and Lin B (also called Lin 24), which produce 28 and 7 MNs, respectively, during the second and third larval stages (Baek and Mann, 2009; Truman et al., 2004). Second, each legtargeting MN has been morphologically characterized – both dendrites and axons – at the single cell level (Baek and Mann, 2009). In the adult VNC, the leg MN cell bodies in each thoracic hemisegment (T1, T2, and T3) are clustered together (Figure 1A, B, movie S1). Each MN extends a highly stereotyped array of dendrites into a dense neuropil within the VNC and a single axon into the ipsilateral leg where it forms synapses onto one of fourteen muscles in one of four leg segments: coxa (Co), trochanter (Tr), femur (Fe), and tibia (Ti) (Baek and Mann, 2009; Soler et al., 2004) (Figure 1C, D). Not only does each MN target a specific region of a muscle, the pattern of dendritic arbors of each MN is also stereotyped and correlates with axon targeting. The tight correlation between axon targeting and dendritic morphology has been referred to as a myotopic map (Baek and Mann, 2009; Brierley et al., 2009; Mauss et al., 2009). The stereotyped morphology exhibited by each MN suggests that it is under precise genetic control that is essential to its function.
Figure 1. Organization of Lin B MNs.
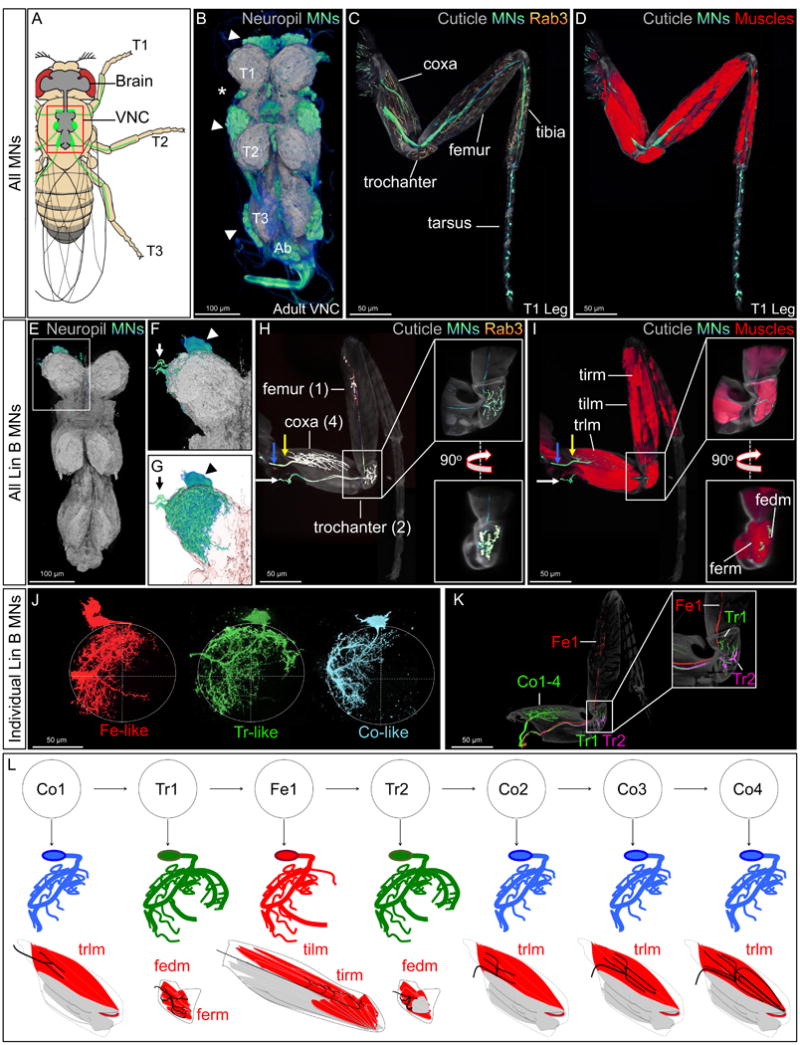
(A): Drawing of an adult fly showing the position of the CNS (grey brain and VNC) and leg MNs (green cell bodies in the VNC and axons in the legs). Red box indicates the VNC imaged in B.
(B): Adult VNC expressing mCD8::GFP (blue/green, depending on intensity) under the control of VGlut-Gal4 and the neuropil marker Bruchpilot (BRP; grey) (Wagh et al., 2006). Arrowheads and asterisk indicate leg and wing MN cell bodies, respectively. (T1-T3, thoracic segments 1-3, ab: abdominal segments).
(C,D): T1 leg expressing mCD8::GFP and the synaptic marker rab3::YFP (yellow) under the control of VGlut-Gal4 and Mhc (Myosin heavy chain)-RFP (D; red) (Gajewski and Schulz, 2010).
(E-G): Lin B MARCM clone in the T1 segment of an adult VNC labeled with mCD8::GFP (green) and rab3::YFP (yellow) under the control of VGlut-Gal4), and BRP (grey). (F) Enlargement of boxed region in (E). (G) Same as (F), but with a transparent neuropil; arrowheads and arrows point to Lin B MN cell bodies and axons exiting the VNC, respectively.
(H,I): Lateral view of a right T1 leg containing a Lin B MARCM clone stained for axons (green), rab3::YFP (yellow), and Mhc (Myosin heavy chain)-RFP (red). Axons of Co1-4 (blue arrow) and Fe1, Tr1, and Tr2 (white arrow) are indicated; white boxes magnify the trochanter. The five muscles (tirm, tilm, trlm, fedm, and ferm) innervated by Lin B MNs are labeled. See also Figure S1 and movies 1-3 for 3D images. Muscle nomenclature is from Soler et al., 2004.
(J,K): Single Lin B MNs visualized using the MARCMbow technique (see Experimental Procedures and Figure S5). In the VNC (J), the seven Lin B MNs exhibit three characteristic morphologies: Fe-like (red), Tr-like (green), and Co-like (blue). In the adult leg (K), all seven Lin B MNs have distinct axon targeting properties. In this example, Tr2 and Fe1 were independently labeled (magenta and red, respectively) and all other Lin B MNs (Co1-4 and Tr1) were labeled in green.
(L) Top: birth order of the seven Lin B MNs (Baek and Mann, 2009); middle: schematic of the three patterns of dendritic aborization (Co-like, blue; Tr-like, green; Fe-like, red); bottom: schematic summarizing muscle (red) targeting of the seven Lin B MNs.
Here we demonstrate that individual post-mitotic MNs express a unique combination of TFs that endows them with their specific morphological properties. We focused on Lin B, which generates seven MNs, and identified six TFs that can account for most of the morphological diversity within this lineage. Interestingly, these TFs do not cross-regulate each other, and are not required for other attributes of MN identity, such as their choice of neurotransmitter (glutamine) or whether their axons target muscles in the periphery, i.e. they remain terminally differentiated leg motor neurons. Consistent with the existence of a combinatorial code, when two or three, but not individual, TFs were simultaneously manipulated nearly complete transformations in morphology were observed. However, removing the function of a single TF, which is expressed in only three Lin B MNs, resulted in a highly specific walking defect that suggests a dedicated role for these neurons in fast walking. Together, these findings reveal the existence of a regulatory step downstream of temporal NB factors in which combinations of morphology TFs (mTFs) control individual neuron morphologies, while leaving other terminal characteristics of neuronal identity unaffected.
Results
Dendritic organization and axon targeting of wild type Lin B MNs
In wild type (WT) flies, Lin B produces seven MNs (Figure 1). Although Lin B generates the same set of MNs in all three thoracic segments, we focused on the prothoracic (T1) segment from which intact legs that include the most proximal coxa segment can be readily dissected.
Each Lin B MN has a specific morphology defined by its unique pattern of dendritic arbors and axonal targeting (Figure. 1E-L) (Baek and Mann, 2009; Brierley et al., 2012). In the leg, the seven Lin B MNs target muscles in the coxa (4 neurons; Co1-4), trochanter (2 neurons; Tr1-2) and femur (1 neuron; Fe1) (Figure. 1H, I) (Baek and Mann, 2009). As a baseline for any mutant analyses, we characterized the highly stereotyped targeting of each of the seven MNs onto specific muscles in the leg. All seven Lin B axons exit the VNC at the same position but enter the T1 leg at three different points. Co1 enters the T1 leg at an anterior region of the coxa, Co2 and Co3 enter the T1 leg at the antero-lateral region of the coxa, while Tr1, Tr2 and Fe1 enter the leg at a postero-medial region of the coxa. Co1-4 innervate the trochanter levator muscle (trlm), Tr1, Tr2 innervate the femur reductor (ferm) and femur depressor (fedm) muscles, and Fe1 innervates the distal part of the tibia levator (tilm) and tibia reductor (tirm) muscles (Figure. 1I, Fig. S1, movies S2-5). A 3D analysis of a T1 segment in the VNC with a GFP-labeled Lin B clone shows that most Lin B dendrites are localized in the lateral region of the neuropil (Figure 1E-G,). An analysis of Lin B MNs at the single cell level revealed that these seven neurons exhibit three types of dendritic morphologies, Fe-like (Fe1), Tr-like (T1 and Tr2), and Co-like (Co1-4), respectively, and seven distinct axonal targeting morphologies (Figure 1J-L) (Baek and Mann, 2009). As with most neuronal morphologies, how the exquisitely stereotyped dendritic arbors and axon trajectories of these seven MNs are genetically encoded remains an unsolved question.
A combinatorial code of transcription factors expressed in Lin B MNs
In Drosophila, most adult neurons, including MNs, obtain their final morphologies during the pupal stage when axons find their targets and dendrites elaborate their arbors. We reasoned that just prior to pupation, at the end of the third instar larval stage (L3), each MN is likely to express a unique set of TFs that will control each of these morphological features. To identify such TFs in Lin B MNs, we screened antibodies against ∼230 different TFs, for immunoreactivity in GFP-labeled Lin B clones in L3 using the MARCM technique (see Experimental Procedures). For comparison, we also screened GFP-labeled Lin A MARCM clones. We found six TFs expressed in subsets of Lin B MNs: Empty spiracle (Ems; four MNs), the Zinc finger homeodomain factors 1 and 2 (Zfh1 and Zfh2; six and two MNs, respectively), the Hox TF Proboscipedia (Pb; three MNs), the Pax6 ortholog Twin of Eyeless (Toy; three MNs), and Prospero (Pros; two MNs) (Figure 2, Figure S2). Three of these TFs – Pb, Toy and Ems – are not expressed in Lin A.
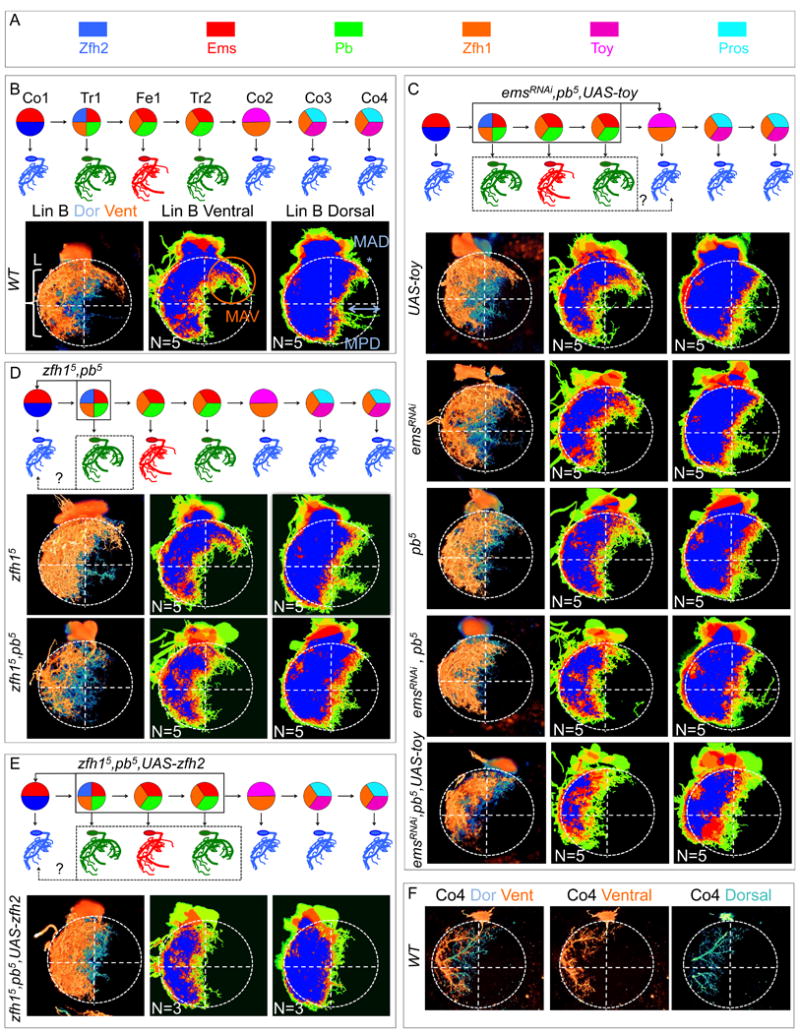
Figure 2. Combinatorial expression of TFs in Lin B.
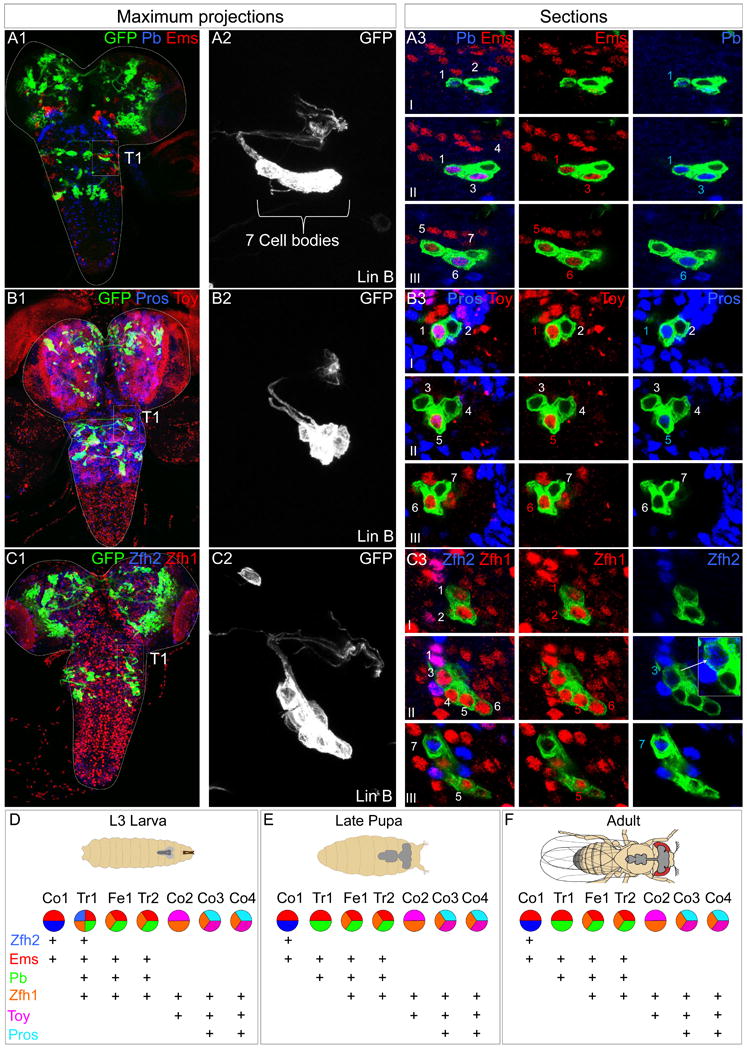
(A-C): L3 CNSs with Lin B MARCM clones in T1 (boxed) expressing mCD8::GFP under the control of VGlut-Gal4. (A1, B1, C1) Ventral views of maximal confocal projections immunostained with anti-Pb (blue) and anti-Ems (red) (A1), anti-Toy (blue) and anti-Pb (red) (B1) and anti-Pros (blue) and anti-Toy (red) (C1). (A2, B2, C2) show magnifications of the GFP+ clones in A1, B1, C1, respectively. (A3, B3, C3) show confocal sections of each clone from ventral (I) to dorsal (III). Note: One of the two Zfh2+ cells consistently expresses lower levels of this TF, as illustrated by the inset in C3 that shows a higher magnification and intensity of this cell and a non-expressing cell for comparison. Pros levels are also consistently higher in one of the two Pros-expressing cells (B3).
(D-F): Summary of the post-mitotic TF combinatorial code in Lin B in the L3 CNS (D), late pupa (E), and adult (F); (see also Figure S2, S3 and S4). Note that although the current code does not discriminate between Fe1 and Tr2 or Co3 and Co4, differing TF levels (e.g. Pros in Co3 and Co4) may contribute to their distinct identities.
Next, we stained for all pairwise combinations of these six TFs in Lin B MARCM clones (Figure 2A-C, Figure S2). Altogether, Lin B MNs exhibit five distinct TF signatures that are highly stereotyped between segments and animals, suggesting the existence of a combinatorial TF code (Figure 2D). For such a code to be valuable, it must uniquely define individual or small groups of closely related cell types. This is indeed the case for the Lin B code: while each of these TFs are expressed elsewhere in the VNC, the three Lin B MNs that coexpress Ems and Pb are the only cells with this signature in the entire L3 central nervous system (CNS) (Figure 2A, D). Moreover, the single Lin B MN that coexpresses Ems, Pb, and Zfh2 is the only cell in the entire L3 CNS with this TF signature (Figure 2A, C, D, Figure S2).
To establish the relationship between individual post-mitotic Lin B MNs in L3 larvae, defined by different combinations of TFs, and the seven morphologically distinct Lin B MNs in the adult, we searched for Gal4 driver lines that report the expression of these TFs in Lin B (see Experimental Procedures). Most informatively, pb492-Gal4 labeled the three Pb+ Lin B MNs in L3 and lineage tracing these cells into the adult showed that they correspond to Tr1, Tr2, and Fe1 (Figure S3). Second, an enhancer trap into zfh2 is expressed in Co1 and Tr1 (Figure S3). Together, these data suggest the correspondence between L3 and adult stages summarized in Figure 2D.
We also analyzed the expression of these six TFs at the pupal and adult stages. For six of the seven Lin B MNs, the expression of these TFs remains the same at all three stages, while one, Tr1, loses the expression of Zfh1 and Zfh2 at the later two stages (Figure 2D-F, Figure S4).
These observations support the idea that a post-mitotic combinatorial code of TFs controls the morphologies of individual adult leg MNs. Below, we test this hypothesis, first by examining the consequences of removing the function of a single TF and, subsequently, by manipulating the expression of multiple TFs at the same time.
pb is required for Tr1, Tr2 and Fe1 dendrite patterning
pb is a Hox gene that controls the identity of the maxillary palps and proboscis (Aplin and Kaufman, 1997; Cribbs et al., 1995; Kaufman, 1978; Kaufman et al., 1990; Percival-Smith et al., 1997; Pultz et al., 1988). Although previous work described Pb expression in thoracic neurons (Baek et al., 2013), it has not been shown to play a role in neurogenesis. According to the combinatorial code summarized above, removing the function of pb should not result in the transformation of one MN identity into another, because the resulting codes (Ems+Zfh1 in Fe1 and Tr2; and Ems+Zfh1+Zfh2 in Tr1) are not observed in wild type Lin B MNs in L3 larva (Figure 2D). Instead, these experiments test the role of a single TF in directing MN morphologies.
We used the MARCM technique and a novel combination of MARCM (Lee and Luo, 2001) and Flybow (Hadjieconomou et al., 2011) (MARCMbow, see Methods) that we developed to analyze the function of pb in Lin B. Importantly, in pb mutant or Pb-overexpressing Lin B clones the expression of Ems, Zfh1, Zfh2, and Toy was unaffected, suggesting that Pb does not regulate, either by repression or activation, the expression of the other four TFs (Figure S5). These results contrast sharply with the cross-regulation between temporal TFs that has been observed in NBs (Li et al., 2013a).
Although WT Lin B dendrites are generally localized on the lateral side of the neuropil, the medial neuropil is colonized by three groups of branches: the most prominent is localized medio-antero-ventrally (MAV) while the other two are localized medio-antero-dorsally (MAD) and medio-postero-dorsally (MPD) (Figure 3A, Figure S6). Using the MARCMbow method to obtain single cell resolution (Figure S7), we found that the Lin B dendrites colonizing the medial region of the neuropil are solely derived from the three MNs expressing Pb: Fe1, Tr1 and Tr2 (Figure 3B-D, movies S6 and S7). The MAV Lin B dendrites are from Tr1 and Tr2, MAD dendrites are from Tr1, Tr2 and Fe1 while MPD dendrites are primarily from Fe1 (Figure 3B-D, movies S6 and S7. In contrast, coxa-targeting Lin B MNs only colonized the lateral region of the neuropil (Figure 1J, L and data not shown).
Figure 3. Pb shapes the dendritic arbors of Tr1, Tr2, and Fe1.
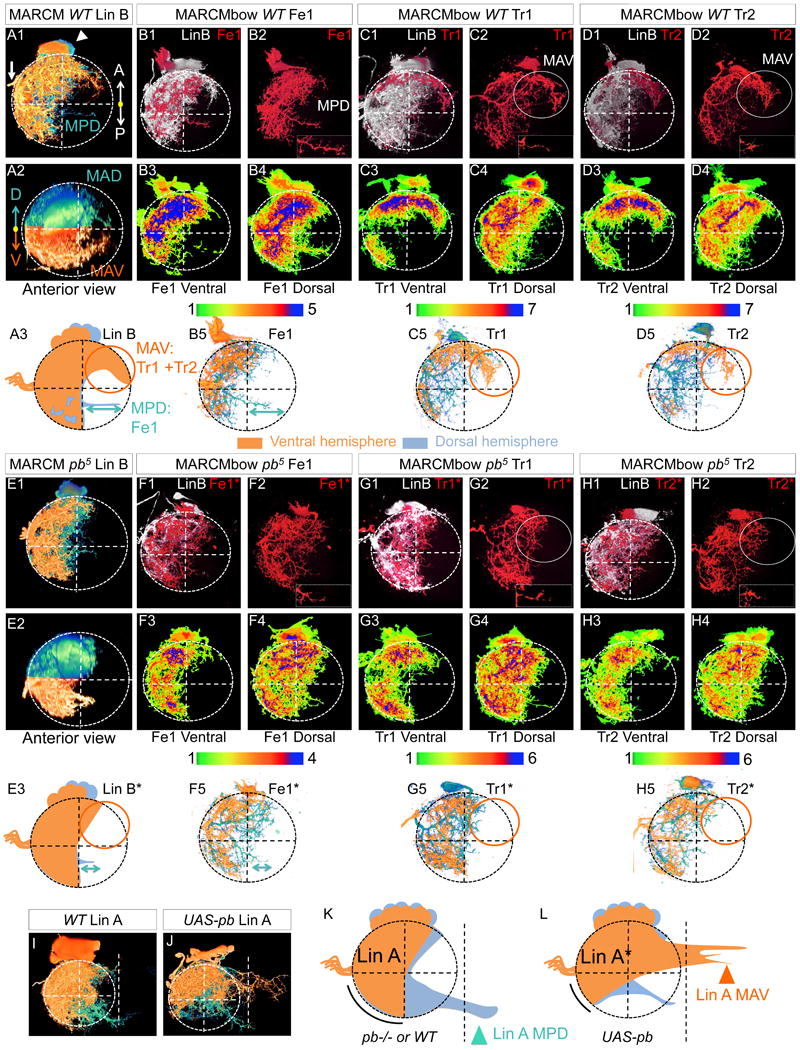
(A, E): WT (A) and pb mutant (E) Lin B MARCM clones labeled with mCD8::GFP under the control of VGlut-Gal4 in the right T1 hemisegments of adult VNCs. Shown are single Lin B clones in which the ventral and dorsal hemispheres were pseudo-colored in orange and blue, respectively. A1 and E1 are ventral views; A2 and E2 are anterior views; A3 and E3 summarize the phenotypes. The circles and double arrows indicate the most affected region. MAV, medial-anterior-ventral; MAD, medial-anterior-dorsal; MPB, medial-posterior-dorsal. See movie 4 for 3D representation and Figure S4 for examples in T2 with the same pb allele and in T1 with a different pb allele.
(B-D and F-H): WT (B-D) and pb mutant (F-H) Lin B MARCMbow clones in the T1 segment of adult VNCs in which Fe1 (B, F), Tr1 (C, G), or Tr2 (D, H) were individually labeled with mCherry (red). In each example, the top panels shows the entire Lin B (GFP+) MARCM clone in white and the individually labeled MN in red; the bottom panels show heat maps of ventral and dorsal hemispheres, illustrating the degree of overlap for independent samples (N>4) with the same individually labeled MN. See Figure S5 and movie 5 for 3D images of MARCMbow clones. B5, C5 and D5 (WT) and F5, G5 and H5 (pb mutant) show representative single MNs in which the ventral and dorsal hemispheres were pseudo-colored in orange and blue, respectively. Note that the dendritic arborizations of these three MNs are readily distinguished in the WT (compare B5, C5, D5) but appear nearly identical when mutant for pb (compare F5, G5, H5).
(I-L): WT (I, K) and Pb overexpressing (J, L) Lin A MARCM clones labeled with mCD8::GFP under the control of VGlut-Gal4 in the right T1 hemisegments of adult VNCs. I and J show single Lin A clones in which the ventral and dorsal hemispheres were pseudo-colored in orange and blue, respectively. K and L summarize these phenotypes; arrowheads and curved lines indicate regions most affected when pb function is altered. The white dashed lines indicate the position of the VNC midline.
In pb mutant MARCM clones the area covered by MAV and MAD was reduced and MPD dendrites were shorter (Figure 3E, Figure S5). These phenotypes are not the result of a change in cell number since all WT and pb mutant Lin B clones had seven cells. To characterize the dendrites of pb mutant Tr1, Tr2 and Fe1 at the single cell level, we analyzed MARCMbow clones in which these three MNs were individually labeled and their axon targeting remained intact. The highly stereotyped lateral branches typical of Fe1, Tr1 and Tr2 were reorganized in the absence of Pb, and the MAV, MAD and MPD dendrites of these MNs were all strongly reduced (Figure 3F-H, movies S6 and S7).
If Pb instructs Tr1, Tr2, and Fe1 dendrites to colonize the more medial regions of the neuropil, it may also have the ability to redirect the dendrites of Lin A, which normally does not express this TF. In the adult VNC, WT Lin A gives rise to 28 MNs, some of which, unlike Lin B dendrites, cross the midline from the postero-dorsal region of the neuropil (Figure 3I, K). Although the number of Lin A MNs was reduced upon ectopic Pb expression in postmitotic MNs (from 28 to ∼20 MNs), those that survive showed a striking phenotype. The MAV region of the neuropil, which is poorly innervated by WT Lin A dendrites, was fully colonized (Figure 3J, L). In contrast, the regions normally innervated by WT Lin A dendrites were poorly targeted (Figure 3J, L). Thus, expression of Pb in Lin A caused the relocalization of dendrites to the MAV region of the neuropil, the same region that is highly innervated by WT Lin B neurons Fe1, Tr1 and Tr2. Interestingly, ectopic expression of Pb did not prevent Lin A dendrites from crossing the midline. Instead, Pb changed the position in the neuropil where Lin A dendrites crossed, from postero-dorsal to antero-ventral. Thus, on its own, Pb controls a subset of MN characteristics (e.g. where dendrites colonize) independently of other characteristics (e.g. competence to cross the midline).
pb is required for Fe1, Tr1, Tr2 axonal targeting
MARCMbow analysis of axon targeting by pb-/- Lin B MNs displayed several phenotypes that could not be revealed by standard MARCM analysis. Most samples, which we group into three phenotypic classes (see below for details), showed defects in targeting by Fe1, Tr1, or Tr2. The three classes differed in whether there was a complete switch in targeting from the trochanter to the femur (class I; 14/37 samples), whether a single MN targeted both the trochanter and femur (class II; 7/37 samples), or whether overall targeting was normal, but the terminal branches of Tr2 and Tr1 were aberrant (class III; 5/37 samples) (Figure 4 and Figure S7). In the remaining 11/37 samples the labeled axons appeared WT.
Figure 4. Single cell analysis of pb axon targeting phenotypes.
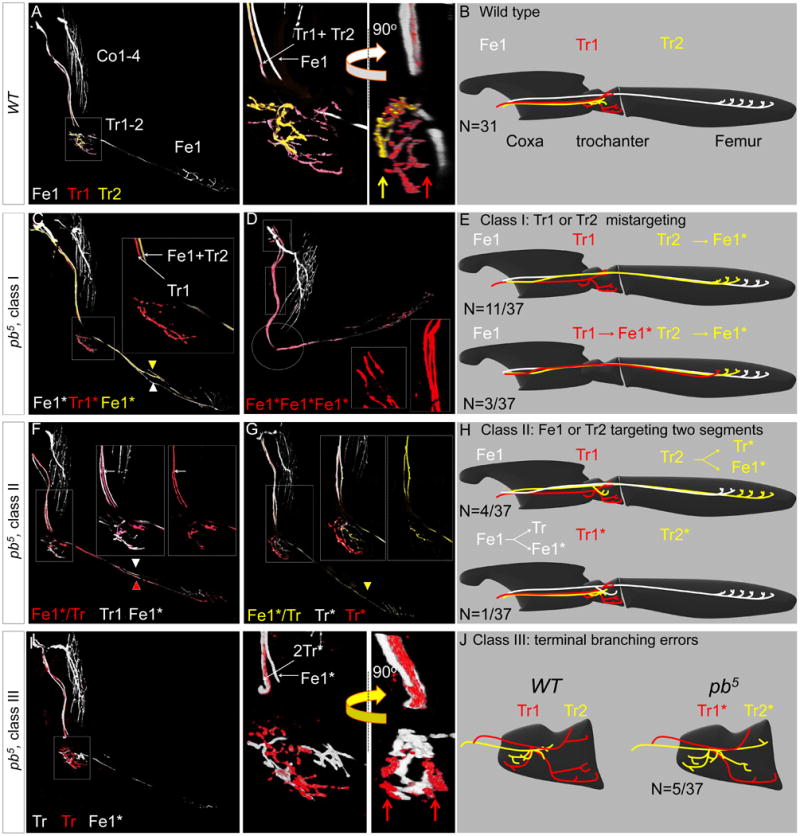
(A, B): Axons of a WT MARCMbow Lin B clone (A) and schematic (B) in which Tr1 and Tr2 were individually labeled. Note the stereotyped separation of the Tr1+Tr2 and Fe1 axons in the magnified image of the trochanter; the 90° rotation shows the medial and lateral positions of the Tr1 (red arrow) and Tr2 (yellow arrow) terminal branches, respectively.
(C-E): Two examples (C, D) and schematics (E) of class I pb mutant phenotypes. In the first example (C), Tr2 targeted the femur instead of the trochanter. In the second example, both Tr1 and Tr2 targeted the femur instead of the trochanter; note the presence of three femur-targeting axons in the magnified (boxed) regions, and the absence of any branching in the trochanter (circles).
(F-H): Two examples (F, G) and schematics (H) of class II pb mutant phenotypes. In both examples, a single labeled MN targeted both the trochanter and the femur. In the first example (F) two MNs targeted the femur, and one of these also targeted the trochanter. In the second example (G), only one MN targeted the femur, and it also targeted the trochanter. Because Tr1 and Tr2 still targeted the trochanter, we can unambiguously determine that Fe1 is the bifurcating MN.
(I, J): In class III pb mutant clones, two MNs target the trochanter, but their terminal branches do not resemble either Tr1 or Tr2. Compare with (A, B). Red arrows point to lateral and medial terminal branches of a single MN, a pattern never observed for wild type Tr1 or Tr2 (compare with panel A).
In class I pb MARCMbow clones, Tr2 (11/37) or both Tr1 and Tr2 (3/37) no longer targeted the trochanter, and instead targeted the femur, resulting in two or three femur-targeting MNs (Figure 4C-E). Despite having more than one femur-targeting MN, the terminal branches synapsing onto tirm and tilm appeared indistinguishable from WT samples with only a single MN targeting femur muscles (Figure 4C-E).
In class II pb MARCMbow clones (N=7/37) a single MN axon split in the coxa and targeted two leg segments. In two cases (N=2/7), one of the bifurcations terminated in the coxa without defasciculating and in five cases (N=5/7) one branch targeted the trochanter while the other targeted the femur (Figure 4F, H). In one of these five samples Tr2 targeting was not affected, allowing us to unambiguously conclude that Fe1 bifurcated to target both the femur and the trochanter (Figure 4G, H).
In class III pb MARCMbow clones (N=5/37), although pb mutant Tr1 and Tr2 still targeted the trochanter, their highly stereotyped pattern of terminal branches was disrupted. Thus, it was not possible to distinguish between Tr1 and Tr2. However, the sum of the innervations by these axon terminals was indistinguishable from the sum of WT Tr1 and Tr2 innervations (Figure 4I-J, movie S8). These results suggest that there is a competition between Tr1 and Tr2 that remains intact even in the absence of pb function.
In summary, the analysis at the single cell level reveals that in the absence of Pb, Tr1, Tr2, and Fe1 remain glutamatergic MNs that target the leg, but exhibit specific targeting defects.
Abnormal walking of fast flies with pb mutant Lin B clones
The leg motor system in Drosophila is used for several adult behaviors, including walking, flight take-off, and grooming (Dickinson et al., 2000). Although each of these behaviors has been characterized in detail, in only a few cases (Bidaye et al., 2014) has a specific behavioral phenotype been described that results from interfering with the function of a small number of neurons within a motor circuit. In particular, very little is known about the role of the trochanter muscles and Tr-innervating MNs in walking. To address these questions, we tested if there was a behavioral consequence of removing the function of pb in Lin B using a quantitative assay that measures many aspects of fly walking (Mendes et al., 2013). For this experiment, we analyzed flies that each contained a single Lin B pb-/- clone in the left T2 hemisegment and compared them to sibling flies generated in parallel that did not have pb-/- Lin B clones (see Methods). Strikingly, most walking parameters, including average walking speed and the percentage of time flies use the tripod gait, were indistinguishable between control and experimental animals (Figure 5 and Figure S8). However, the stance linearity parameter, which measures how linear the path of a fly's body is relative to its leg stance, was affected: flies containing T2 pb-/- Lin B clones walked with significantly more wobble than control flies (Figure 5A-C). Interestingly, this difference was only observed at fast walking speeds (>34 mm/sec); no difference in this parameter was observed between control and experimental flies at slow or medium speeds.
Figure 5. Specific walking defects of flies with pb mutant Lin B MARCM clones.
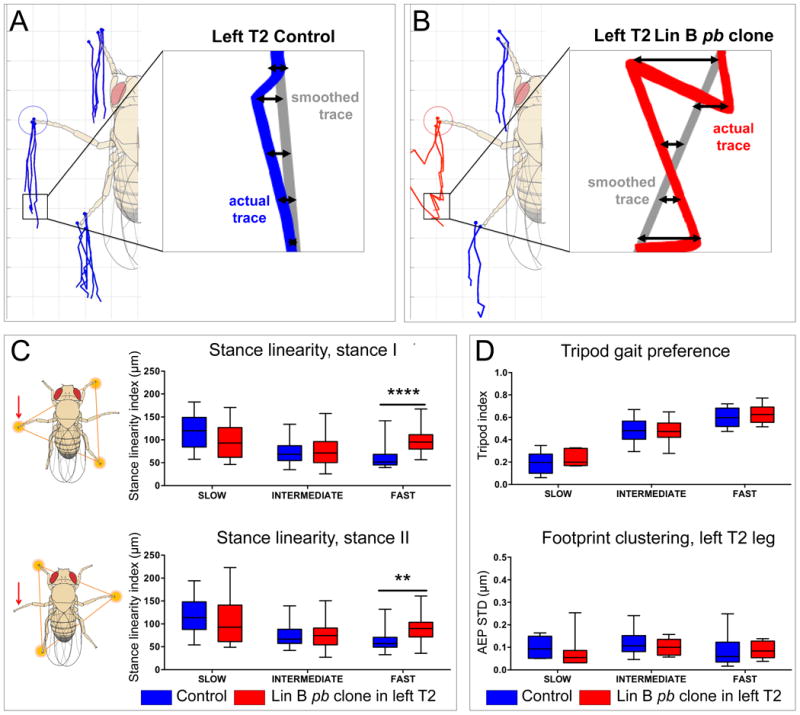
(A, B) Examples of stance traces, which mark the footprint position relative to the center of the fly's body as the fly moves forward, for representative control (A, blue lines) and experimental (B, red lines) flies. Stance linearity is the average distance (double arrows) between the actual stance trace and a ‘smoothed’ trace (gray lines) generated using every five frames, and is thus a measure of how straight the fly's path is; flies that walk perfectly straight would have a stance linearity index of 0 (Mendes et al., 2013). Insets show portions of actual traces from left T2 legs. (C) Stance linearity values averaged for the three legs in tripod stances I (top) and II (bottom) and calculated for both control (blue) and experimental (red) flies that were binned into three speed cohorts. Stance linearity values only differed in the fast speed group, and the difference was larger for tripod stance I, which includes the leg innervated by mutant pb Lin B MNs (red arrow, see Methods) (****p = 0.0001 and **p = 0.004, 2-way ANOVA Sidak's multiple comparisons test).
(D) No differences in tripod index or footprint clustering were observed between control and experimental animals. Tripod index is the fraction of video frames that an animal spends in a tripod configuration (see schematics in (C)). Footprint clustering is the standard deviation from the average of the anterior extreme positions (AEPs; see blue and red circles in A, B) for all steps in a single video (AEP STD). See Mendes et al., 2013 for details.
These results suggest that there is a specialized requirement for the three pb-expressing Lin B MNs, Tr1, Tr2, and Fe1, for stable walking at high speeds. Further, they suggest that the precise morphologies of these MNs, which are disrupted in the absence of pb activity, are critical for their function.
Testing the combinatorial TF code
While the above results establish a critical role for pb in directing specific morphological features of Fe1, Tr1 and Tr2, they fall short of testing the idea that the TF code (Figure 2D) instructs individual MN morphologies. The prediction is that changing the code of one MN to another MN should cause a nearly complete transformation of morphological identity. We tested this prediction in three experiments: (1) by changing the Tr1, Tr2 and Fe1 code (Pb+Ems+Zfh1) into the Co2 code (Toy+Zfh1), (2) by converting the Tr1 code (Pb+Ems+Zfh1+Zfh2) into Co1 code (Ems+Zfh2), and (3) by converting the Tr1, Tr2 and Fe1 code (Pb+Ems+Zfh1) into cox1 code (Ems+Zfh2).
To convert Pb+Ems+Zfh1 MNs (Fe1, Tr1, Tr2) to Toy+Zfh1 MNs (Co2-like) we generated pb-/- MARCM clones that also expressed emsRNAi and UAS-toy. Consistent with a cell fate transformation, the dendrites of these clones appeared Co-like (Figure 6B, C). This transformation was not observed in UAS-toy or emsRNAi MARCM clones but was partially observed in pb-/- emsRNAi MARCM clones (Figure 6B, C). In addition to this highly penetrant dendritic phenotype, pb-/- emsRNAi MARCM clones resulted in axons that target the coxa instead of the femur and trochanter (Figure 7C; N=4/17). This phenotype was never observed in pb-/- or emsRNAi MARCM clones (Figure 7B and data not shown). Expressing Toy in pb-/- emsRNAi MARCM clones did not increase coxa targeting. Together, these results suggest that while Toy plays a role in dendritic arborization, it has little or no function in axon targeting. Consistent with these observations, coxa-targeting MNs could be converted into femur-targeting MNs by expressing Pb and Ems in all Lin B progeny (N=4/7) (Figure 7D). This phenotype was never observed when Pb or Ems were expressed individually in Lin B MARCM clones.
Figure 6. Testing the Lin B TF code: dendritic arborization.
(A): TF color key.
(B): Top, birth order, TF code, and schematic of dendritic arbors for the seven wild type Lin B MNs. Bottom left, WT Lin B MARCM clone in the T1 hemisegment of an adult VNC in which the ventral (orange) and dorsal (blue) hemispheres are pseudo-colored; bottom right, heat maps of ventral and dorsal hemispheres illustrating the degree of overlap for five independent samples.
(C): From top to bottom: schematic of expected transformation in emsRNAi, pb-/-, UAS-toy MARCM clones; UAS-toy; emsRNAi; pb-/-; emsRNAi, pb-/- and emsRNAi, pb-/-, UAS-toy Lin B MARCM clones.
(D): From top to bottom: schematic of expected transformation in zfh1-/-, pb-/- MARCM clones; zfh1-/- and zfh1-/-, pb-/- Lin B MARCM clones.
(E): Top: schematic of expected transformation in zfh1-/-, pb-/-, UAS-zfh2 Lin B MARCM clones; bottom: zfh1-/-, pb-/-, UAS-zfh2 Lin B MARCM clone.
(F): WT Co4 MN labeled with MARCMbow in which the ventral (orange) and dorsal (blue) hemispheres are pseudo-colored. Note the similarity to the arborization pattern at bottom of panels C, D and E.
Figure 7. Testing the Lin B TF code: axonal targeting.
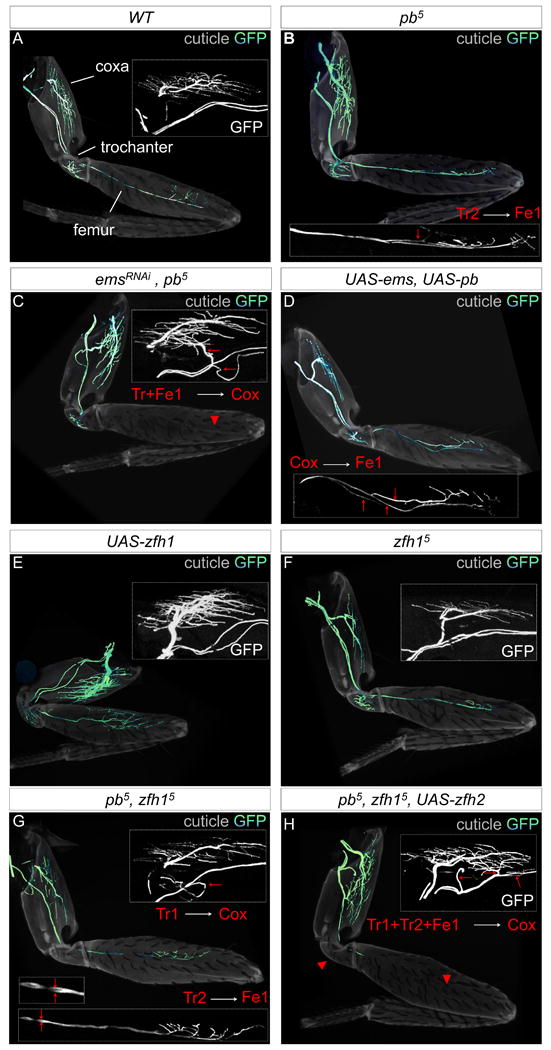
(A-H): Axon targeting phenotypes of WT (A); pb-/- (B); emsRNAi, pb-/- (C); UAS-ems, UAS-pb (D); UAS-zfh1 (E); zfh1-/- (F); zfh1-/-, pb-/- (G); and zfh1-/-, pb-/-, UAS-zfh2 (H) Lin B MARCM clones. The cuticle is light grey and the axons green; insets show leg regions most affected, red arrowheads and arrows point to either the absence of or aberrant targeting, respectively.
To convert a Pb+Ems+Zfh1+Zfh2 MN (Tr1) into an Ems+Zfh2 MN (Co1), we generated pb-/- zfh1-/- Lin B MARCM clones. Consistent with a transformation of cell fate, the dendrites of these MNs closely resemble a Co-like pattern (Figure 6D). In contrast, the dendrites of zfh1-/- clones appeared wild type (8/10) or exhibited a few misplaced medial dendrites (2/10) (Figure 6D). In the leg, Tr1, but not Fe1 or Tr2, was observed to target the coxa in several pb-/- zfh1-/- clones (Figure 7G) (N=3/15; the remaining samples were similar to pb-/- or zfh1-/- clones). Although MN axons targeted the correct muscles in zfh1-/- clones, the amount of terminal axon branching in the coxa and femur was dramatically reduced; this branching phenotype was also observed in pb-/- zfh1-/- clones. Conversely, branching increased when Zfh1 was expressed in all Lin B MNs (Figure 7E-G). Notably, these branching phenotypes were reminiscent of a previously documented zfh1-/- phenotype in the larval neuromuscular junction (Vogler and Urban, 2008).
Third, to reprogram Pb+Ems+Zfh1 MNs (Tr1, Tr2 and Fe1) into Ems+Zfh2 MNs (Co1), we generated pb-/- zfh1-/- MARCM clones that also express Zfh2. As predicted by the code, the dendrites of all Lin B MNs appeared Co-like (Figure 6E). Moreover, in the leg, Fe1 and either one or both of the Tr MNs targeted the coxa in these clones (Figure 7H) (N=3/3). This axon targeting phenotype was never observed in Lin B clones overexpressing Zfh2 or in pb-/- zfh1-/-Lin B clones (Figure 7G and data not shown). Taken together, these results strongly support the idea that different combinations of these TFs determine the morphological identities of Lin B MNs because when the TF code was altered in specific ways, predictable transformations in MN morphology were observed.
Discussion
In this study we tested the hypothesis that the unique morphological identity of each postmitotic neuron is, at least in part, defined cell intrinsically by distinct sets of TFs. We identified six TFs expressed in different combinations that can account for most of the morphological distinctions of seven MNs that all target the adult leg of Drosophila. Because of the role they play in specifying both dendritic and axonal morphology, without affecting other aspects of MN identity, we refer to these TFs as morphology TFs (mTFs). Below we discuss the implications of these findings and the relationship of mTFs to other classes of TFs known to be important in specifying neuronal identity.
A combinatorial code of mTFs
Inherent in the concept of a combinatorial TF code is the idea that removing or ectopically expressing a single TF will only generate a transformation of fate when a different wild type code is generated. Consistent with this notion, only when we simultaneously manipulated the expression of two or three mTFs were we able to partially mimic a distinct mTF code and, as a result, transform the identity of one Lin B MN into another. In contrast, manipulating single TFs typically resulted in aberrant or neo-codes that are not observed in wild type flies. For example, removing pb function from Lin B resulted in two MNs with a code (Ems+Zfh1) and MN morphology that are not observed in wild type Lin A and Lin B lineages. Analogously, ectopic Pb expression in Lin A, which normally does not express this TF, generated aberrant codes and MN morphologies. This latter experiment was particularly informative because although Pb redirected a subset of Lin A dendrites to grow in an anterior region of the neuropil, it did not alter the ability of these dendrites to cross the midline. Thus, the dendrites of these MNs had characteristics of both Pb-expressing Lin B MNs (occupying an antero-ventral region) and Pb-non-expressing Lin A MNs (competence to cross the midline). Axon targeting of these MNs was also aberrant: although they still targeted leg muscles, Pb-expressing Lin A MNs frequently terminated in the coxa, which is not a normal characteristic of Pb-expressing Lin B MNs or of any Lin A MN (data not shown). These observations suggest that the final morphological identity of a neuron is a consequence of multiple TFs executing functions that comprise a complete morphological signature. Some functions, such as the ability to occupy the antero-ventral region of the neuropil, can be directed by a single TF (e.g. Pb), while other functions, such as the ability to accurately target the distal femur, require multiple TFs (e.g. Pb+Ems). Further, because it was possible to generate MNs that have both Lin B and Lin A morphological characteristics, our results argue against the idea that there are lineage-specific mTFs shared by all progeny derived from the same lineage. Instead, our data are more consistent with the idea that the final morphological identity of a MN depends on its mTF code.
mTFs, temporal TFs, and terminal selector TFs
As summarized in the Introduction, Drosophila NBs, and perhaps vertebrate neural stem cells, express a series of TFs that change over time and have therefore been referred to as temporal TFs (Bayraktar and Doe, 2013; Brody and Odenwald, 2000; Isshiki et al., 2001; Li et al., 2013b). For Lin B, we do not know the sequence of these factors, in part because the Lin B NB is not easily identified in the 2nd larval instar VNC, the time at which it is generating MNs (Baek and Mann, 2009; Brierley et al., 2012). Nevertheless, each MN derived from Lin B and Lin A has a stereotyped birth order (Baek and Mann, 2009; Brierley et al., 2012), consistent with the idea that temporal TFs play an important role in directing the identities of MNs derived from these lineages and, therefore, the mTFs they express. For Lin B, this birth order is Co1→Tr1 →Fe1→Tr2→Co2→Co3→Co4 (Baek and Mann, 2009). Interestingly, according to the mTF code proposed here, each of these MNs differs by at most two mTFs in any successive step (Figure 7E). For example, Tr1 has the code [Zfh1, Ems, Pb, Zfh2] while Fe1, the next MN to be born, has the code [Zfh1, Ems, Pb] (Figure 7E). Thus, we posit that the sequence of temporal TFs acting in the NB is responsible for directing each successive change in mTF expression in postmitotic MNs (e.g., in the Tr1→Fe1 step, repression of zfh2) (Figure 8). Although a link between temporal TFs and TFs expressed in postmitotic neurons has been proposed in Drosophila, the role of these TFs in conferring neuron morphologies is not known (Baumgardt et al., 2009; Li et al., 2013b). Further, there may be additional diversity-generating mechanisms in lineages that produce many more neurons than the seven MNs generated by Lin B. One additional source of diversity may come from NB identity TFs, which distinguish lineages based on their position (Broadus et al., 1995). Such spatial information could in principle allow the same temporal TFs to regulate different sets of mTFs in different NB lineages. It is also likely that differences in the levels of some mTFs may contribute to neuronal identities. Consistent with this idea, the levels of Zfh2 and Pros differ in the Lin B MNs expressing these TFs (Figure 2B,C), differences that are consistent in all three thoracic segments and between animals. Further, Zfh1 levels vary between Lin B MNs and its levels control the amount of terminal axon branching (Figure 7E,F). Previous studies also demonstrated that TF levels are important for neuron morphology, including Antp in adult leg MNs derived from Lin A (Baek et al., 2013) and Cut in the control of dendritic arborization complexity in multidendritic neurons (Grueber et al., 2003). If the levels of mTFs are important, it may provide a partial explanation for why the transformations of morphological identity generated here with the MARCM technique, which cannot control levels, are typically only partially penetrant.
Figure 8. TF control of individual neural identities.
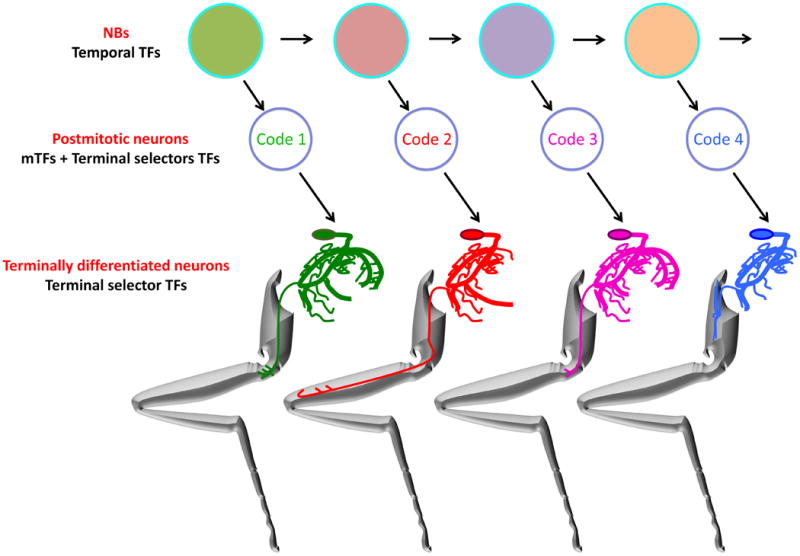
NBs express a series temporal TFs, represented here by different colored circles. The blue outlines indicate TFs that may be shared throughout the lineage. NBs divide asymmetrically to eventually give rise to post-mitotic neurons that have both unique mTF codes, which specify individual dendritic and axonal morphologies, and terminal selector TF codes, which specify other terminal characteristics such as choice of neurotransmitter. Once differentiated, terminal selector TFs maintain the characteristics of terminally differentiated neurons.
Another distinction between temporal TFs and mTFs is that we have not observed any evidence of cross-regulation between mTFs. In situations when we either removed (e.g. pb-/-; emsRNAi) or ectopically expressed (e.g. UAS-pb + UAS-ems) mTFs in postmitotic Lin B MARCM clones, the expression of the remaining mTFs was unchanged (Figure S3). In contrast, when a NB lineage is mutant for a temporal TF, the prior TF in the series typically continues to be expressed (Li et al., 2013a). These observations suggest that the choice of mTF expression is made in the NB, and that once the postmitotic code is established it is not further influenced by coexpressed mTFs.
Our data further suggest that mTFs are distinct from terminal selector TFs (Figure 8). In mutants for the mTFs studied here the resulting neurons remain glutamatergic leg motor neurons: they continue to express VGlut, which encodes a vesicular glutamate transporter, expressed by all Drosophila MNs, and they still exit the VNC to target and synapse onto muscles in the adult legs. Thus, whereas terminal selector TFs maintain the terminal characteristics of fully differentiated neurons, mTFs are required transiently to execute functions required for each neuron's specific morphological characteristics. Together, we suggest that the combined activities of terminal selector TFs and mTFs specify and maintain the complete identity of each post-mitotic neuron (Figure 8).
Although the mTFs defined here, e.g. Ems, Pb, and Toy, do not fit the criteria for a terminal selector TF, it is plausible that some TFs function both as mTFs and terminal selector TFs. One example may be Apterous, a TF that is expressed in six interneurons in the thoracic embryonic segments, and that functions with other TFs to control the terminal differentiation state of these neuropeptide expressing neurons (Allan et al., 2005). In addition to the loss of neuropeptide expression, these neurons display axon pathfinding defects in the absence of apterous (Lundgren et al., 1995). Despite the potential for overlapping functions, it is conceptually valuable to consider the specification of neuronal morphologies as distinct from other terminal characteristics, as some mTFs regulate morphology without impacting these other attributes. It is also plausible that some of the TFs that have been previously designated as determinants of subtype identity may also be part of mTF codes. For example, eve is required for the identity of dorsally-directed MNs in Drosophila embryogenesis (Landgraf et al., 1999), but the TFs required for distinguishing the individual morphologies of these neurons are not known. It may be that Eve is one component of the mTF code, and that it functions together with other mTFs to dictate the specific morphologies of these neurons.
Precise dendritic morphologies are essential for specific motor functions
Flies containing a single pb mutant Lin B clone exhibited a highly specific walking defect: when walking at high speed, these flies were significantly more unsteady compared to control flies. The restriction of this defect to high speeds suggests that the Pb-dependent characteristics of these MNs may be specifically required when the walking cycle is maximally engaged, raising the possibility that Tr1, Tr2, and Fe1 are analogous to so-called fast MNs described in other systems (Ritzmann and Buschges, 2007). Further, these data support the idea that the highly stereotyped morphology of these MNs is critical to the wild type function of the motor circuit used for walking. In particular, the precise dendritic arborization pattern exhibited by these MNs, which is disrupted in the pb mutant, is likely to be essential for their function. Although we cannot exclude that other pb-dependent functions contribute to this walking defect, these observations provide strong evidence that the myotopic map, in which MNs that target similar muscle types have similar dendritic arborization patterns, is important for the fly to execute specific adult behaviors.
Experimental procedure
Fly stocks
Unless otherwise noted, fly stocks were obtained from the Bloomington Stock Center: P{TRiP.JF03153}attP2 (RNAiems, Drosophila RNAi Screening Center at Harvard Medical School); UAS-toy (Kyoto stock center), y w hs-Flp1.22 and FRT82B tub-Gal80 (Gary Struhl), UAS-mCD8::GFP, UAS-YFP::rab3, Mhc-RFP, VGlut-Gal4 (also called OK371-Gal4) (Mahr and Aberle, 2006), FRT82B Ki, pb5, pp, ry506/Tm6B and UAS-pb (David Cribbs), FRT82B pb27 (Anthony Percival-Smith), UAS-FB1.1, UAS-mFlp5 (this work), VGlut>stop,y+>lexAvp16 (this work), lexO-mCD8:GFP (this work), UAS-Flp, Pb-Gal4492(VT37492 from Vienna stock center).
MARCM
y w hs-Flp1.22; VGlut-Gal4, UAS-mCD8::GFP, Mhc-RFP//; FRT82B tub-Gal80// flies were crossed with
y w hs-Flp1.22; VGlut-Gal4, UAS-YFP::rab3, Mhc-RFP//; FRT82B// flies to generate WT clones and to visualize axons, synaptic boutons and muscles.
y w hs-Flp1.22; VGlut-Gal4, UAS-mCD8::GFP//; FRT82B tub-Gal80// flies were crossed with:
y w hs-Flp1.22; UAS-mCD8::GFP//; FRT82B // flies to generate WT clones,
y w hs-Flp1.22; UAS-mCD8::GFP//; FRT82B Ki, pb5 p,p ry,506/Tm6B flies or
y w hs-Flp1.22; UAS-mCD8::GFP//; FRT82B pb27 /Tm6B flies to generate pb mutant clones,
y w hs-Flp1.22; UAS-mCD8::GFP//; P{TRiP.JF03153}attP2 FRT82B// flies to generate RNAiems expressing clones,
y w hs-Flp1.22; UAS-pb, UAS-ems//; FRT82B // flies to generate Pb+Ems-overexpressing clones
y w hs-Flp1.22; UAS-mCD8::GFP//; P{TRiP.JF03153}attP2 FRT82B Ki pb5 /TM6B flies to generate pb mutant clones and RNAiems expressing clones,
y w hs-Flp1.22; UAS-pb//; FRT82B // flies to generate Pb-overexpressing clones,
y w hs-Flp1.22; UAS-mCD8::GFP//; FRT82B Ki, pb5 zfh15/Tm6B flies to generate zfh1 mutant clones,
y w hs-Flp1.22; UAS-mCD8::GFP//; FRT82B Ki, pb5 zfh15/Tm6B flies to generate pb, zfh1 double mutant clones,
y w hs-Flp1.22; VGlut-Gal4, UAS-mCD8::GFP/cyo ; FRT82B tub-Gal80/+, UAS-zfh2/ In(4)ciD, ciD panciD flies were crossed with:
y w hs-Flp1.22; VGlut-Gal4, UAS-mCD8::GFP/Cyo ; FRT82B /Tm6b flies to generate zfh2-overexpressing clones
y w hs-Flp1.22; VGlut-Gal4, UAS-mCD8::GFP/Cyo ; FRT82B Ki, pb5 zfh15/Tm6B flies to generate pb, zfh1 double mutant clones overexpressing zfh2
y w hs-Flp1.22; VGlut-Gal4, UAS-mCD8::GFP// ; FRT82B tub-Gal80/MKRS, UAS-toy flies were crossed with:
y w hs-Flp1.22; UAS-mCD8::GFP//; FRT82B // flies to generate Toy-overexpressing clones,
y w hs-Flp1.22; UAS-mCD8::GFP//; P{TRiP.JF03153}attP2, FRT82B Ki, pb5 /TM6B flies to generate pb mutant, RNAiems expressing and Toy-overexpressing clones.
To generate MARCM clones, embryos were collected for 12 hours in vials and incubated for 24 hours at 25°C. First instar larvae were heat shocke d at 37°C for 30 minutes for clonal analysis in L3 larvae and at 35°C for 15 minutes for clonal analysis in adults. Under these conditions one out of ten flies (37°C) or one out of twenty flies (35°C) feature on average a single Lin B or Lin A MARCM clone.
MARCMbow technique
A previous combination of MARCM and flybow used heat shock promoters and a single heat shock to induce expression of both Flp and mFlp5 (Hadjieconomou et al., 2011). As a result, entire MARCM clones were typically labeled with a single flybow color. To express multiple flybow colors in individual cells within single MARCM clones we generated transgenes where mFlp5 was under the control of Gal4 (UAS-mFlp5) so that it would be expressed only in the MARCM clone, independently of heat shock. UAS-mFlp5, VGlut-Gal4, UAS-mCD8::GFP//; FRT82B tub-gal80// flies were crossed with:
y w hs-Flp1.22; UAS-FB1.1//; FRT82B// flies to generate WT clones
y w hs-Flp1.22; UAS-FB1.1//; FRT82B Ki pb5 pp ry506/Tm6B flies to generate pb mutant clones.
To generate MARCMbow clones, embryos were collected for 12 hours and incubated for 24 hours at 25°C. First instar larvae were heat shocke d at 37°C for 13 minutes.
Lineage tracing
pb-Gal4492 flies were crossed with:
VGlut>stop,y+>lexAvp16, lexO-mCD8:GFP/FM7; UAS-Flp, VGlut>stop,y+>lexAvp16, lexO-CD8:GFP/CyO; VGlut >stop,y+>lexAvp16, lexO-mCD8:GFP// flies and incubated at 29°C . About 300 flies were analyzed.
Immunohistochemistry of L3 Larval CNS
The initial set of antibodies were mostly generated by the modENCODE project and provided to us by C. Desplan (Li et al., 2013b). About 230 antibodies were used and ∼10 CNSs were analyzed for each antibody. Positives were confirmed by independent primary antibodies: rabbit anti-Pb (e9, 1:100) (Cribbs et al., 1992), rat anti-Ems (1:400) (Walldorf and Gehring, 1992), guinea pig anti-Toy (1:400) (Uwe Walldorf), guinea pig anti-Zfh1 (1:500) (Jim Skeath), rat anti-Zfh2 (1:500) and mouse anti-Pospero (Chris Doe). Secondary antibodies used were: AlexaFluor555 and AlexaFluor647 conjugates (Molecular Probes). See Supplemental Information for L3 VNC, adult VNC and leg preparation details.
Microscopy and 2D imaging
Multiple 1-μm-thick sections in the z-axis (dorsoventral for L3 larval CNS or adult VNC and mediolateral for adult legs) were imaged with a Leica TCS SP5 II confocal microscope. Binary images for Z-stack images were generated using NIH Image J and Photoshop (Adobe Systems).
3D dendrite analysis and 3D Leg analysis
Amira 3D software. has been used to visualize dendrites in the neuropil. See Supplemental Information for details.
Quantification of dendrite arborization phenotypes
To quantify the dendritic phenotypes, we created heat maps that measured the degree of overlap between multiple, aligned samples. Ventral and dorsal Z-stacks were generated by NIH Image J. Z-stacks were aligned based on the structure of the T1 neuromere using Photoshop (Adobe Systems). The intensity of each Z-stack was normalized (maximum intensity). All the Z-stacks were subsequently flattened and used to generate a heat map using the gradient map option in Photoshop.
Plasmid constructions and transgenic lines
UAS-mFlp5: A KpnI-mFlp5-XbaI fragment from hs-mFlp5 (Hadjieconomou et al., 2011) was inserted into a pUAS-attB vector. See Supplemental Information for details.
VGlut->y+stop>-LexAVP16: The 5.9 kb sequence upstream of VGlut was cloned using a NotI-LexAVP16-XbaI fragment from pBS-LexAVP16 (Lai and Lee, 2006) and a >y+stop> fragment (Gary Struhl) into pCasper-nucLacZ from which nucLacZ was removed. See Supplemental Information for additional details.
Quantification of walking parameters
pb mutant MARCM animals were generated by crossing
y, w, hs-Flp1.22; VGlut-Gal4, UAS-mCD8::GFP// ; FRT82B tub-Gal80// to
y, w, hs-Flp1.22; UAS-mCD8::GFP// ; FRT82B Ki, pb5, pp, ry506/Tm6b.
First instar F1 larvae from this cross were heat shocked at 35°C for 30 minutes. 24 to 48 hours after eclosion, single animals were selected for correct genotype under CO2 anesthesia, assigned an individual identification number, and allowed to recover for ∼1 hour. Following recovery, 5 to 10 videos of individual flies' walking behavior were recorded and analyzed as described (Mendes et al., 2013). After the videos were acquired, individual flies were screened for the presence of GFP+ Lin B MARCM clones in the legs; flies without Lin B clones were included in the ‘control’ group (N=29 videos). Flies in the ‘experimental’ group (N=29 videos) all had a single MARCM clone in Lin B in the left T2 hemisegment. This group was chosen due to the higher frequency of flies containing this type of clone.
Supplementary Material
Highlights.
7 leg MNs from the same neuroblast express different combinations of 6 TFs
Changing the TF code results in predictable changes in MN morphology
One TF, Pb, is required for the precise morphological identities of three MNs
When these MNs are mutant for pb flies exhibit a highly specific walking defect
eTOC.
Enriquez et al describe a set of six transcription factors that are expressed in a combinatorial manner to direct the individual morphologies of seven adult leg motor neurons that all come from the same neuroblast stem cell in Drosophila. Changing the transcription factor code results in predictable changes in motor neuron morphology and axon targeting. Small morphological defects in a few motor neurons result in highly specific walking defects.
Acknowledgments
We are very grateful to D. Cribbs, C. Desplan, B. Dickson, C. Doe, L. Luo, A. Percival-Smith, I. Salecker, J. Skeath, G. Struhl, and U. Walldorf for antibodies and fly stocks; T. Jessell, W. Grueber, J. Dasen and O. Hobert for comments on the manuscript and our two fly media facility managers, C. Allain and A. Rainey. This work was supported by RO1NS070644 to R.S.M., fellowships from the Association Française contre le Myopathies and the Philippe Foundation to J.E., and a Gatsby fellowship to M.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature cited
- Allan DW, Park D, St Pierre SE, Taghert PH, Thor S. Regulators acting in combinatorial codes also act independently in single differentiating neurons. Neuron. 2005;45:689–700. doi: 10.1016/j.neuron.2005.01.026. [DOI] [PubMed] [Google Scholar]
- Aplin AC, Kaufman TC. Homeotic transformation of legs to mouthparts by proboscipedia expression in Drosophila imaginal discs. Mechanisms of development. 1997;62:51–60. doi: 10.1016/s0925-4773(96)00649-1. [DOI] [PubMed] [Google Scholar]
- Baek M, Enriquez J, Mann RS. Dual role for Hox genes and Hox co-factors in conferring leg motoneuron survival and identity in Drosophila. Development. 2013;140:2027–2038. doi: 10.1242/dev.090902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M, Mann RS. Lineage and birth date specify motor neuron targeting and dendritic architecture in adult Drosophila. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2009;29:6904–6916. doi: 10.1523/JNEUROSCI.1585-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgardt M, Karlsson D, Terriente J, Diaz-Benjumea FJ, Thor S. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell. 2009;139:969–982. doi: 10.1016/j.cell.2009.10.032. [DOI] [PubMed] [Google Scholar]
- Bayraktar OA, Doe CQ. Combinatorial temporal patterning in progenitors expands neural diversity. Nature. 2013;498:449–455. doi: 10.1038/nature12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidaye SS, Machacek C, Wu Y, Dickson BJ. Neuronal control of Drosophila walking direction. Science. 2014;344:97–101. doi: 10.1126/science.1249964. [DOI] [PubMed] [Google Scholar]
- Brierley DJ, Blanc E, Reddy OV, Vijayraghavan K, Williams DW. Dendritic targeting in the leg neuropil of Drosophila: the role of midline signalling molecules in generating a myotopic map. PLoS biology. 2009;7:e1000199. doi: 10.1371/journal.pbio.1000199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley DJ, Rathore K, VijayRaghavan K, Williams DW. Developmental origins and architecture of Drosophila leg motoneurons. The Journal of comparative neurology. 2012;520:1629–1649. doi: 10.1002/cne.23003. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Novitch BG. Regulatory pathways linking progenitor patterning, cell fates and neurogenesis in the ventral neural tube. Philosophical transactions of the Royal Society of London Series B, Biological sciences. 2008;363:57–70. doi: 10.1098/rstb.2006.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadus J, Skeath JB, Spana EP, Bossing T, Technau G, Doe CQ. New neuroblast markers and the origin of the aCC/pCC neurons in the Drosophila central nervous system. Mechanisms of development. 1995;53:393–402. doi: 10.1016/0925-4773(95)00454-8. [DOI] [PubMed] [Google Scholar]
- Brody T, Odenwald WF. Programmed transformations in neuroblast gene expression during Drosophila CNS lineage development. Developmental biology. 2000;226:34–44. doi: 10.1006/dbio.2000.9829. [DOI] [PubMed] [Google Scholar]
- Broihier HT, Kuzin A, Zhu Y, Odenwald W, Skeath JB. Drosophila homeodomain protein Nkx6 coordinates motoneuron subtype identity and axonogenesis. Development. 2004;131:5233–5242. doi: 10.1242/dev.01394. [DOI] [PubMed] [Google Scholar]
- Broihier HT, Skeath JB. Drosophila homeodomain protein dHb9 directs neuronal fate via crossrepressive and cell-nonautonomous mechanisms. Neuron. 2002;35:39–50. doi: 10.1016/s0896-6273(02)00743-2. [DOI] [PubMed] [Google Scholar]
- Certel SJ, Thor S. Specification of Drosophila motoneuron identity by the combinatorial action of POU and LIM-HD factors. Development. 2004;131:5429–5439. doi: 10.1242/dev.01418. [DOI] [PubMed] [Google Scholar]
- Cribbs DL, Benassayag C, Randazzo FM, Kaufman TC. Levels of homeotic protein function can determine developmental identity: evidence from low-level expression of the Drosophila homeotic gene proboscipedia under Hsp70 control. The EMBO journal. 1995;14:767–778. doi: 10.1002/j.1460-2075.1995.tb07055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribbs DL, Pultz MA, Johnson D, Mazzulla M, Kaufman TC. Structural complexity and evolutionary conservation of the Drosophila homeotic gene proboscipedia. The EMBO journal. 1992;11:1437–1449. doi: 10.1002/j.1460-2075.1992.tb05188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasen JS, Jessell TM. Hox networks and the origins of motor neuron diversity. Current topics in developmental biology. 2009;88:169–200. doi: 10.1016/S0070-2153(09)88006-X. [DOI] [PubMed] [Google Scholar]
- Dickinson MH, Farley CT, Full RJ, Koehl MA, Kram R, Lehman S. How animals move: an integrative view. Science. 2000;288:100–106. doi: 10.1126/science.288.5463.100. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Skeath JB. Neurogenesis in the insect central nervous system. Current opinion in neurobiology. 1996;6:18–24. doi: 10.1016/s0959-4388(96)80004-3. [DOI] [PubMed] [Google Scholar]
- Duggan A, Ma C, Chalfie M. Regulation of touch receptor differentiation by the Caenorhabditis elegans mec-3 and unc-86 genes. Development. 1998;125:4107–4119. doi: 10.1242/dev.125.20.4107. [DOI] [PubMed] [Google Scholar]
- Eade KT, Fancher HA, Ridyard MS, Allan DW. Developmental transcriptional networks are required to maintain neuronal subtype identity in the mature nervous system. PLoS genetics. 2012;8:e1002501. doi: 10.1371/journal.pgen.1002501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach KFD, Dittrich APM. The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 1989;258:441–475. [Google Scholar]
- Fishell G, Heintz N. The neuron identity problem: form meets function. Neuron. 2013;80:602–612. doi: 10.1016/j.neuron.2013.10.035. [DOI] [PubMed] [Google Scholar]
- Fujioka M, Lear BC, Landgraf M, Yusibova GL, Zhou J, Riley KM, Patel NH, Jaynes JB. Even-skipped, acting as a repressor, regulates axonal projections in Drosophila. Development. 2003;130:5385–5400. doi: 10.1242/dev.00770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski KM, Schulz RA. CF2 represses Actin 88F gene expression and maintains filament balance during indirect flight muscle development in Drosophila. PloS one. 2010;5:e10713. doi: 10.1371/journal.pone.0010713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces A, Thor S. Specification of Drosophila aCC motoneuron identity by a genetic cascade involving even-skipped, grain and zfh1. Development. 2006;133:1445–1455. doi: 10.1242/dev.02321. [DOI] [PubMed] [Google Scholar]
- Grueber WB, Jan LY, Jan YN. Different levels of the homeodomain protein cut regulate distinct dendrite branching patterns of Drosophila multidendritic neurons. Cell. 2003;112:805–818. doi: 10.1016/s0092-8674(03)00160-0. [DOI] [PubMed] [Google Scholar]
- Hadjieconomou D, Rotkopf S, Alexandre C, Bell DM, Dickson BJ, Salecker I. Flybow: genetic multicolor cell labeling for neural circuit analysis in Drosophila melanogaster. Nature methods. 2011;8:260–266. doi: 10.1038/nmeth.1567. [DOI] [PubMed] [Google Scholar]
- Hobert O. Regulation of terminal differentiation programs in the nervous system. Annual review of cell and developmental biology. 2011;27:681–696. doi: 10.1146/annurev-cellbio-092910-154226. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Pearson B, Holbrook S, Doe CQ. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell. 2001;106:511–521. doi: 10.1016/s0092-8674(01)00465-2. [DOI] [PubMed] [Google Scholar]
- Jacob J, Maurange C, Gould AP. Temporal control of neuronal diversity: common regulatory principles in insects and vertebrates? Development. 2008;135:3481–3489. doi: 10.1242/dev.016931. [DOI] [PubMed] [Google Scholar]
- Kaufman TC. Cytogenetic Analysis of Chromosome 3 in DROSOPHILA MELANOGASTER: Isolation and Characterization of Four New Alleles of the Proboscipedia (pb) Locus. Genetics. 1978;90:579–596. doi: 10.1093/genetics/90.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman TC, Seeger MA, Olsen G. Molecular and genetic organization of the antennapedia gene complex of Drosophila melanogaster. Advances in genetics. 1990;27:309–362. doi: 10.1016/s0065-2660(08)60029-2. [DOI] [PubMed] [Google Scholar]
- Lai SL, Lee T. Genetic mosaic with dual binary transcriptional systems in Drosophila. Nature neuroscience. 2006;9:703–709. doi: 10.1038/nn1681. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Roy S, Prokop A, VijayRaghavan K, Bate M. even-skipped determines the dorsal growth of motor axons in Drosophila. Neuron. 1999;22:43–52. doi: 10.1016/s0896-6273(00)80677-7. [DOI] [PubMed] [Google Scholar]
- Landgraf M, Thor S. Development and structure of motoneurons. International review of neurobiology. 2006;75:33–53. doi: 10.1016/S0074-7742(06)75002-4. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends in neurosciences. 2001;24:251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Li X, Chen Z, Desplan C. Temporal patterning of neural progenitors in Drosophila. Current topics in developmental biology. 2013a;105:69–96. doi: 10.1016/B978-0-12-396968-2.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Erclik T, Bertet C, Chen Z, Voutev R, Venkatesh S, Morante J, Celik A, Desplan C. Temporal patterning of Drosophila medulla neuroblasts controls neural fates. Nature. 2013b;498:456–462. doi: 10.1038/nature12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren SE, Callahan CA, Thor S, Thomas JB. Control of neuronal pathway selection by the Drosophila LIM homeodomain gene apterous. Development. 1995;121:1769–1773. doi: 10.1242/dev.121.6.1769. [DOI] [PubMed] [Google Scholar]
- Mahr A, Aberle H. The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene expression patterns : GEP. 2006;6:299–309. doi: 10.1016/j.modgep.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Mauss A, Tripodi M, Evers JF, Landgraf M. Midline signalling systems direct the formation of a neural map by dendritic targeting in the Drosophila motor system. PLoS biology. 2009;7:e1000200. doi: 10.1371/journal.pbio.1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes CS, Bartos I, Akay T, Marka S, Mann RS. Quantification of gait parameters in freely walking wild type and sensory deprived Drosophila melanogaster. eLife. 2013;2:e00231. doi: 10.7554/eLife.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano H, Temple S. Cell types to order: temporal specification of CNS stem cells. Current opinion in neurobiology. 2009;19:112–119. doi: 10.1016/j.conb.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Oyallon J, Apitz H, Miguel-Aliaga I, Timofeev K, Ferreira L, Salecker I. Regulation of locomotion and motoneuron trajectory selection and targeting by the Drosophila homolog of Olig family transcription factors. Developmental biology. 2012;369:261–276. doi: 10.1016/j.ydbio.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percival-Smith A, Weber J, Gilfoyle E, Wilson P. Genetic characterization of the role of the two HOX proteins, Proboscipedia and Sex Combs Reduced, in determination of adult antennal, tarsal, maxillary palp and proboscis identities in Drosophila melanogaster. Development. 1997;124:5049–5062. doi: 10.1242/dev.124.24.5049. [DOI] [PubMed] [Google Scholar]
- Philippidou P, Dasen JS. Hox genes: choreographers in neural development, architects of circuit organization. Neuron. 2013;80:12–34. doi: 10.1016/j.neuron.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokop A, Technau GM. The origin of postembryonic neuroblasts in the ventral nerve cord of Drosophila melanogaster. Development. 1991;111:79–88. doi: 10.1242/dev.111.1.79. [DOI] [PubMed] [Google Scholar]
- Pultz MA, Diederich RJ, Cribbs DL, Kaufman TC. The proboscipedia locus of the Antennapedia complex: a molecular and genetic analysis. Genes & development. 1988;2:901–920. doi: 10.1101/gad.2.7.901. [DOI] [PubMed] [Google Scholar]
- Ritzmann RE, Buschges A. Adaptive motor behavior in insects. Current opinion in neurobiology. 2007;17:629–636. doi: 10.1016/j.conb.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Soler C, Daczewska M, Da Ponte JP, Dastugue B, Jagla K. Coordinated development of muscles and tendons of the Drosophila leg. Development. 2004;131:6041–6051. doi: 10.1242/dev.01527. [DOI] [PubMed] [Google Scholar]
- Thor S, Andersson SG, Tomlinson A, Thomas JB. A LIM-homeodomain combinatorial code for motor-neuron pathway selection. Nature. 1999;397:76–80. doi: 10.1038/16275. [DOI] [PubMed] [Google Scholar]
- Thor S, Thomas JB. The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron. 1997;18:397–409. doi: 10.1016/s0896-6273(00)81241-6. [DOI] [PubMed] [Google Scholar]
- Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Developmental biology. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131:5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
- Vogler G, Urban J. The transcription factor Zfh1 is involved in the regulation of neuropeptide expression and growth of larval neuromuscular junctions in Drosophila melanogaster. Developmental biology. 2008;319:78–85. doi: 10.1016/j.ydbio.2008.04.008. [DOI] [PubMed] [Google Scholar]
- Wagh DA, Rasse TM, Asan E, Hofbauer A, Schwenkert I, Durrbeck H, Buchner S, Dabauvalle MC, Schmidt M, Qin G, et al. Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron. 2006;49:833–844. doi: 10.1016/j.neuron.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Walldorf U, Gehring WJ. Empty spiracles, a gap gene containing a homeobox involved in Drosophila head development. The EMBO journal. 1992;11:2247–2259. doi: 10.1002/j.1460-2075.1992.tb05284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


