Abstract
Aptamers are composed of short RNA or single-stranded DNA sequences that, when folded into their unique three-dimensional conformation, can specifically bind to their cognate targets with high specificity and affinity. Although functionally similar to protein antibodies, oligonucleotide aptamers offer several advantages over protein antibodies in biomedical and clinical applications. Additionally, through the enhanced permeability and retention (EPR) effect, nanomedicines can improve the therapeutic index of a treatment and reduce side effects by enhancing accumulation at the disease site. However, this EPR effect is “passive targeting” to tumors and thus, may not be an ideal approach for targeted cancer therapy. To construct ligand-directed “active targeting” nano-based delivery systems, aptamer technology has been widely studied. The aptamer-equipped nanomedicines have been tested for in vitro diagnosis, in vivo imaging, targeted cancer therapy, theranostic approaches, sub-cellular molecule detection, food safety, and environment monitoring. This review will focus on the development of aptamer-conjugated nanomedicines and their application for in vivo imaging, targeted therapy, and theranostics. In some applications, aptamers can also be used as drug carriers or ON/OFF switches. Herein, some outstanding therapeutic approaches are also discussed on a case-by-case basis, such as an “on-command” release system and a combinational therapy strategy.
Keywords: aptamers, nanomedicine, active targeting, on-command release system, combinational therapy
1. Introduction
Aptamers were introduced by two independent groups in 1990[1, 2] and have since been developed as a class of novel, multifunctional oligonucleotide ligands with various biomedical applications. In essence, aptamers are a class of small, single-stranded RNA or DNA nucleic acids with unique three-dimensional structures that can recognize and bind to their cognate targets with high specificity and affinity, dissociation constants are typically within the pico to nanomolar range.[3] Due to their functional similarity, aptamers are often referred to as “chemical antibodies” and can be utilized as activating ligands, antagonists, or for specific therapeutic applications.
Although protein antibodies are currently more widely used, most suffer from high immunogenicity and are expensive and time consuming to produce, which can limit their clinical applicability. However, based on the unique properties of nucleic acids, aptamers offer several advantages over protein antibodies in terms of application and production, such as faster tissue penetration, virtual non-immunogenicity, thermal stability, ease of conjugation or modification with different functional moieties, simple and low-cost chemical synthesis, and wide range of potential biological targets (See Table 1 for a detailed list).[4-10] Thus, aptamers represent an excellent alternative to protein antibodies.
Table 1.
Comparison of aptamers and antibodies
| characteristics | aptamers | antibodies |
|---|---|---|
| specificity and affinity | high, pico- to nano-molar | high, pico- to nano-molar |
| molecular weight | 8-25 kDa | ~ 150 kDa |
| tissue barrier penetration | +++, due to smaller size | + |
| immunogenicity | - | +++, even with humanized antibodies |
| thermal stability | ++ | - |
| modification potential | easy, unlimited | difficult, limited |
| manufacture | chemical synthesis | mammalian cell systems |
| batch-to-batch variation | low | high |
| production time | hours | days to months |
| production cost | $ | $$$ |
| ranges of targets | proteins and non-protein molecules | limited to immunogenic proteins |
Despite these advantages, aptamers also have some inherent limitations to their in vivo applicability. As nucleic acids, they are vulnerable to degradation by the ubiquitous nucleases present in biological environments, this is especially true for RNA-based aptamers.[11] Despite this, after the initial introduction of aptamer technology, RNA-based aptamers were more widely developed. This was due to the fact that RNA oligonucleotides could fold into a more diverse repertoire of 3D structures owing to the presence of the 2’-OH group and non-Watson-Crick base pairing. Additionally, the increased flexibility of RNA sequences made it easier to successfully achieve high-affinity and -specificity aptamers.[12] To overcome the obstacle of nuclease sensitivity, several effective chemical modification strategies have been developed in recent years. These include: (i) substitution of the 2’-OH RNA functional group by 2’-fluoro, 2’-amino, or 2’-O-methoxy motifs;[13-15] (ii) substitution of the phosphodiester backbone with boranophosphate or phosphorothioate;[16] (iii) simultaneous substitutions with phosphorodithioate and 2’-O-methyl in one nucleotide;[17] (iv) locked nucleic acid (LNA) technology;[18, 19] (v) and the generation of “mirror” RNA sequences (termed Spiegelmers).[20]
Concomitant with the effort to increase the in vivo stability of RNA aptamers, the use of DNA-based aptamers was increasingly explored, as DNA molecules are naturally resistant to 2’-endonucleases due to lack of the 2’-OH group. As a proof-of-concept study, our group recently developed a DNA-based aptamer specific for CD30, a biomarker that is over-expressed in Hodgkin and anaplastic large cell lymphomas, which is stable under physiological conditions. Functional analysis demonstrated that the CD30-specific aptamer was stable in human serum for up to 8 hours and had high binding affinity for CD30 biomarker (as low as 2 nM). Conversely, the RNA-based CD30-specific aptamer was digested within 10 minutes under the same conditions.[21] AS1411 is another well-characterized DNA-based aptamer that specifically targets nucleolin, which is a biomarker over-expressed on a variety of tumor cell types. AS1411 is currently undergoing clinical evaluation as a novel anti-cancer therapy in patients with solid tumors and acute myeloid leukemia.[22]
The methodology used to generate aptamers is known as SELEX (Systematic Evolution of Ligands by EXponential enrichment) and entails several repetitive rounds of amplification and enrichment after exposure to the target ligand or cell type of interest. In brief, the basic SELEX process contains the following steps: (i) mixing the target of interest with a random oligonucleotide library; (ii) separating target-bound sequences from unbound species; (iii) amplifying the target-bound sequences to generate an enriched library; (iv) mixing the target of interest with the newly amplified library for the next round of enrichment; and finally (v) sequencing the enriched aptamer sequences after a number of rounds of selection. For a more detailed description of the SELEX process see [23].
In recent years, some strategies have been employed to improve the outcome of the SELEX process, and have aimed to: (i) maximize affinity and specificity, and improve the speed of selection and success rate, such as bead-based selection,[24, 25] Slow Off-rate Modified Aptamer (SOMAmer),[26, 27] Magnetic-Assisted Rapid Aptamer Selection (MARAS),[28] and Monoclonal Surface Display SELEX (MSD-SELEX);[29] (ii) impart additional properties to the selected aptamers, such as X-Aptamers[30] and internalized Cell-SELEX;[31-37] (iii) select aptamers that can recognize the target in its native conformational state under physiological conditions, such as Cell-SELEX,[9, 38] hybrid-SELEX,[21] FACS-SELEX,[39, 40] in vivo-SELEX,[41] and tissue-based SELEX,[42] and so on. To date, more than a thousand specific aptamers have been developed[26, 27] and extensively characterized in various biomedical assays, such as bioassays, drug development, cell detection, tissue staining, in vitro and in vivo imaging, targeted therapy, and nanomedicine.
In the past decades, the field of nanomedicine has grown, especially for treatment of cancer and infectious diseases. Conceptually, the term “nanomedicine” refers to nano-scale molecular “devices” (10-200 nm) that can be used in disease diagnosis, prevention, and therapy.[43-45] In particular applications, nanomedicines include nanopharmaceutics, nanoimaging agents, and more popular multifunctional entities, termed theranostics, which combine real-time diagnostic and therapeutic functions.
Compared to traditional small-molecule drugs that can suffer from low solubility, rapid metabolic clearance, severe off-target side effects, and narrow therapeutic index,[46, 47] drug delivery via nanomedicine exhibits several improvements. For example, they can (i) improve solubility and reduce metabolic degradation of active drugs, which can prolong circulating half-life and significantly improve bioavailability; (ii) more easily transport active drugs across the tissue or cell barriers through different endocytosis pathways; (iii) carry two different active drugs simultaneously for synergistic therapy; and most importantly, (iv) improve therapeutic index and reduce side effects through the enhanced permeability and retention (EPR) effect.[48-50] There is data to support that the improved therapeutic index of nanomedicines is, in fact, primarily dependent on the EPR effect. It has been demonstrated that use of nano-based delivery systems improves accumulation of the drug at the desired site, owing to the high permeability of neo-vasculature and poor lymphatic drainage of diseased tissues, especially in instances of cancer or inflammation.[51, 52] To date, dozens of nanomedicines have been approved by the Food and Drug Administration (FDA) and have shown to be well tolerated in patients, many more are in the pipeline of clinical development.[53, 54]
However, most nanomedicines on the market are considered as first-generation nanomedicines, which utilize a “passive targeting” principle through the EPR effect to improve their therapeutic index. Although they can enhance accumulation in disease sites verses healthy tissue, passive targeting cannot specifically distinguish between diseased and healthy tissues. Thus, the therapeutic outcomes of a particular nanomedicine are strongly influenced by several other factors such as the structure of the neo-vasculature, blood pressure, and disease type and location, which inherently differ between patients and/or stages of disease,[45] hence the therapeutic outcomes are varied and often times disappointing. To further improve the therapeutic index of first-generation nanomedicines, a series of high-affinity and high-specificity ligands, such as antibodies, peptides, small molecules, and aptamers, are now being conjugated to nano-based delivery systems to create “active targeting” nanomedicines.
Aptamer-equipped nanomedicines provide several outstanding advantages over first-generation passive delivery systems, and have been used in various biomedical applications. This review will focus on applications of aptamer technology in the context of aptamer-conjugated nanomedicines and their potential for in vivo disease imaging, targeted therapy, and theranostics. In certain applications, aptamers can also be used as drugs carriers, or as an ON/OFF switches for “on-command” release systems, or in combination therapy strategies, these will be discussed on a case-by-case basis.
2. Aptamer-Based Therapeutic Nanomedicines
Similar to therapeutic protein antibodies, aptamers can be functionally used as therapeutic antagonists when their targets have specific biological functions in disease development. Up to now, a series of therapeutic aptamers are being evaluated with their potentials for specific disease therapy in clinical trials, an detailed summary of therapeutic aptamers undergoing clinical trials please see [23]. However, as discussed above, aptamers are a class of small-sized nucleic acids. If therapeutic, unmodified aptamers are introduced directly into biological conditions, they have a high renal clearance rate that leads to the rapid removal from systemic circulation. Alternatively, they can also be rapidly digested by various nucleases, which then results in a short circulating half-life.[11] Although chemical modifications or development of biostable DNA-based aptamers can improve nuclease-resistance to some extent, they are not sufficient for therapeutic in vivo applications. To overcome these limitations, an effective strategy could be to develop therapeutic aptamer-based nanomedicines by conjugating aptamers with high molecular weight bioavailable nanomaterials, which will (i) improve nuclease-resistance and reduced renal clearance rate to prolong circulating half-life, (ii) enhance aptamer affinity for their targets based on the multivalent effect which create by multiple aptamers packing on the surface of nanomaterials, and (iii) acquire higher internalization efficiency due to variations in endocytosis pathway.[55]
As the first aptamer-based drug approved by the FDA, Macugen (Pegaptanib sodium) is the most successful example to explain how develop a therapeutic aptamer from bench to bedside translation. Based on the solid evidences that vascular endothelial growth factor (VEGF) is a key regulator in age-related macular degeneration (AMD) development, and inhibiting of VEGF is a promising approach for AMD therapy,[56] NeXstar Pharmaceuticals developed a 28-mer RNA-based aptamer with high-affinity and -specificity for VEGF by their patented SELEX technology.[15] For the specific purpose of clinical application, Macugen was modified by a two-step strategy in the basic and post SELEX process, including chemical modification to enhance nuclease-resistance and addition of high molecular weight bioavailabe nanomaterial to prolong circulating half-life. To get nuclease-resistant aptamer, Macugen was initially selected in a 2′-fluoro pyrimidine-modified library, and other modifications included: (i) substitution of 12 of the 14 ribopurines with 2′-O-methyl purines; (ii) an inverted nucleotide was capped at the 3′-terminus. To further improve circulating half-life, a 40-kDa polyethylene glycol (PEG) molecule, well-characterized polymer approved by FDA for clinical application, was conjugated at the 5′-terminus of the aptamer sequence.[57] This strategy is also termed PEGylation, which has been extensively used in many small-sized drugs modification. It has been shown that PEGylated aptamer can not only prolong its circulating half-life but also enhance its stability and decrease its toxic accumulation in nontarget tissues.[58, 59] After these modifications, Macugen still possesses high affinity and specificity for VEGF, but the mean terminal half-life is prolonged significantly up to 10 days in humans.[60, 61] Based on the choice of VEGF as therapeutic target and effective modification of the selected aptamer for improving its clinical applicability, the nano-sized Macugen was approved by FDA for AMD therapy in 2004.[62]
In recent years, to prolong therapeutic aptamers efficacy, other aptamer-based nanomedicines have been developed based on various bioavailable nanomaterials. For example, a DNA aptamer specific against transform growth factor receptor was conjugated with the biodegradable polymer chitosan, a widely used drug carrier due to its biocompatibility, low immunogenicity, especially good macro-adhesion and effective protection of encapsulated nucleic acid from nuclease degradation. The resulting aptamer-chitosan conjugates showed significant biostability for up to 48 h with a 20:30 molar ratio of chitosan:aptamer, and bound the transform growth factor receptor II resulting in inhibition of transform growth factor induced cell proliferation.[63] In another study, a specific DNA aptamer against human immunodeficiency virus-1 (HIV-1) reverse transcriptase was conjugated to 13 nm gold nanoparticles (NPs), another well-known metal NPs which can effectively stabilize DNA due to its strong steric effect and highly ionic charges, although it is considered as non-degradable nanomaterial in vivo application. These formed aptamer-gold NPs showed significant inhibition of HIV-1 reverse transcriptase, compared with the DNA aptamer alone, even in DNase I-containing buffer. The authors argued that the increased biostability was due to the highly negatively-charged surfaces of aptamer-gold NPs combined with a high local salt concentrations that could minimize access of DNase I to the surface-exposed aptamers of gold NPs.[64] All of these studies demonstrate that conjugating aptamers to NPs can significantly improve the biostability of RNA- and DNA-based therapeutic aptamers and thus their applicability and utility for clinical applications.
In addition to increasing biostability, aptamer-NP conjugation can enhance affinity for the desired target. Wu et al. developed an aptamer-micelle, in which an aptamer (named TD05) specific for immunoglobulin heavy mu chain, which is over-expressed on B-cell lymphoma Ramos cells, was conjugated to a lipid tail through a PEG linker and then shown to self assemble into aptamer-micelles. Cell binding studies showed that at 4°C, TD05 aptamer had high affinity and specificity for Ramos cells, which was lost at 37°C. Surprisingly, TD05 aptamer-micelles showed high affinity and specificity for Ramos cells at both 4°C and 37°C, presumably due to multivalent effect of multiple TD05 aptamers on one micelle. As described by the authors, the binding affinity was improved about 750 folds.[65] This study provides strong evidence that multivalent aptamers conjugated with nanomaterials can enhance the affinity for their targets.
3. Aptamer-Targeted Nanomedicines
Currently, a rapid development of nanomedicines depends on the development and characterization of bioavailable inorganic and organic nanomaterials, such as gold nanomaterials, magnetic nanomaterials, singe-walled carbon nanotubes (SWCNTs), mesoporous silica nanoparticles (MSN), quantum dots (QDs), liposomes, copolymers, nucleic acid-based and protein-based nanomaterials. In general, these nanomaterials possess several common characteristics, such as a large surface area for loading aptamers or drugs, a uniform size and shape for excellent bio-distribution profile. In addition, nanomaterials also possess unique composite-dependent features for specific diagnostic and therapeutic functions, such as magnetic imaging of magnetic nanomaterials, optical imaging of QDs, SWCNTs and gold nanomaterials, and photothermal effects of SWCNTs and gold nanomaterials. These unique physicochemical properties, combined with oligonucleotide aptamer targeting specificity, have been wildly used to develop specific aptamer-targeted nanomedicines. Consequently, data show that coating of nanomaterial surfaces with aptamers furnishes the resultant nanomedicines with novel characteristics, such as: (i) reduced non-specific aggregation and improved stability; (ii) enhanced biocompatibility in vivo; and (iii) acquisition of the “active targeting” function.[66, 67] In the section below, we will select several representative models and discuss their applications for in vivo imaging, targeted therapy, and theranostics.
3.1 Aptamer-Targeted Nanoimaging Agents
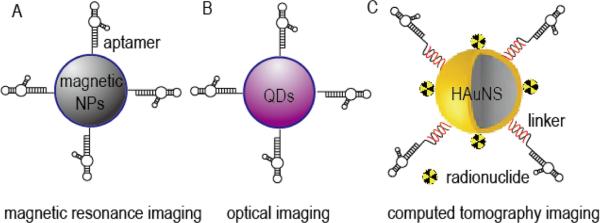
For specific in vivo imaging, aptamer-targeted nanoimaging agents can be constructed by combining aptamers with imaging nanomaterials or radionuclide probes (Figure 1), such as the case with magnetic nanomaterials used in magnetic resonance imaging. Magnetic nanocrystal is considered as excellent imaging probe with strong enhancement of proton relaxation resultant in lower detection limit, although always suffers from low water solubilization. Recently, Kim et al. constructed VEGF receptor 2-specific aptamer-magnetic nanocrystal conjugates for glioblastoma diagnosis (Figure 1A), in which magnetic nanocrystal was enveloped by carboxyl polysorbate 80 for improving water solubilization and conjugation of targeting aptamer. In their study, the formed conjugates showed no cytotoxicity even at high concentrations, while effectively binding with VEGF receptor 2-positive cells in vitro. Moreover, the formed conjugates were further validated for sensitive magnetic resonance imaging of glioblastoma-bearing mice.[68] In a subsequent study, the same group also developed epithelial cell adhesion molecule-specific aptamer-magnetic nanocrystal conjugates for diagnosis of gastric carcinomas. In vitro, the formed conjugates showed excellent biocompatibility and targeting ability, and was then evaluated in an orthotopic model of gastric cancer. In these in vivo studies, the formed conjugates effectively targeted epithelial cell adhesion molecule-positive tumor tissues and served as a highly sensitive magnetic resonance imaging probe, improving the performance of the unmodified magnetic nanocrystal.[69]
Figure 1.
Representative schematic diagrams of aptamer-targeted nanoimaging agents: (A) Aptamer-targeted magnetic NPs for magnetic resonance imaging, modified from reference [68, 69]; (B) Aptamer-targeted QDs for optical imaging, modified from reference [70]; (C) Aptamer-targeted HAuNS loaded with radionuclide for computed tomography imaging, modified from reference [71].
In spite of undesirable biocompatibility and cellular toxicity, QDs offer superior fluorescence properties as compared to traditional fluorophores, and are thus studied extensively for developing imaging agents. In a recently published study, in order to improve biocompatibility and reducing toxicity, Zhang et al. described a simple one-pot synthesis strategy for constructing aptamer-QDs, where Zn2+ doped CdTe QDs were coated with DNA aptamer and optimized for cancer-related in vitro and in vivo imaging (Figure 1B). The authors used MUC1 to develop a targeting aptamer, which was then conjugated with QDs through phosphorothioates linkers. The resultant aptamer-QD compounds exhibited excellent photo-stability and reduced toxicity, compared to QDs alone. This reduced toxicity was achieved through Zn doping, while the DNA aptamers improved QD biocompatibility. In in vitro studies, aptamer-QDs achieved superb cell binding and internalization efficiency specific to MUC1-positive cells, and showed strong fluorescence imaging capabilities in xenograft mouse models.[70]
Radionuclides, or radioisotopes, are atoms with unstable nuclei that can release their excess energy and thus, are excellent candidates for use as imaging probe. In a recent report, aptamers targeting the human epidermal growth factor receptor were tethered with hollow gold nanospheres (HAuNS) through complementary DNA linkers and labeled with 111In to develop computed tomography imaging probes (Figure 1C). These newly formed aptamer-HAuNS particles combined high aptamers loading capacity (about 250 aptamer molecules per particle), excellent bio-stability in plasma (up to 48 hours), and high binding and internalization into epidermal growth factor receptor over-expressing oral tumor cells. In tumor-bearing mice, in addition to the expected bio-distribution profile and pharmacokinetics, 111In-labeled aptamer-HAuNS particles exhibited a greater uptake into the tumor cells than did the 111In-labeled antibodies conjugated to HAuNS.[71] In addition to the example described above, other aptamer-targeted nanoimaging agents have also been developed and revealed excellent diagnostic sensitivity. These include an aptamer specific to tenascin-C, a biomarker over-expressed on tumor cells, that was conjugated with carbon nanodots for optical imaging of cervical cancer;[72] and sgc8 aptamer specific to protein tyrosine kinase 7, a biomarker of leukemia T cells, that was conjugated with dendrimer for targeted cells labeling or temperature-responsive targeted cells catching and dissociation;[73, 74] and AS1411-silica NPs labeled with 64Cu radioisotope for identification of lymph nodes in patients with metastatic tumors by positron emission tomography imaging.[75] A detailed summary of aptamer-targeted nanoimaging agents described in this section is listed in Table 2.
Table 2.
Summary of aptamer-targeted nanoimaging agents
| target | aptamer | nanomaterial | active molecule | applications | reference |
|---|---|---|---|---|---|
| VEGF receptor 2 | DNA | magnetic nanocrystal | magnetic nanocrystal | magnetic resonance imaging | [68] |
| epithelial cell adhesion molecule | DNA | magnetic nanocrystal | magnetic nanocrystal | magnetic resonance imaging | [69] |
| MUC 1 | DNA | QDs | QDs | optical imaging | [70] |
| epidermal growth factor receptor | RNA | HAuNS | radionuclide 111In | computed tomography imaging | [71] |
| tenascin-C | RNA | carbon nanodots | carbon nanodots | optical imaging | [72] |
| protein tyrosine kinase 7 | DNA | dendrimer | fluorescein cadaverine | targeted cells labeling | [73, 74] |
| nucleolin | DNA | silica NPs | radioisotope 64Cu | positron emission tomography imaging | [75] |
3.2 Aptamer-Targeted Therapeutic Nanomedicines
In order to develop successful “active targeting” nanomedicines, they should possess three important properties: be (i) biologically stable under normal physiological conditions, (ii) able to rapidly release their therapeutic cargo at their target site, and (iii) highly selective for their intended targets in order to minimize/eliminate unwanted toxicity. With this focus, a number of “active targeting” nanomedicines have been developed in recent year. Currently, there are over a hundred papers describing aptamer-targeted therapeutics that deliver drugs or gene silencing tools for specific disease therapy. As an example, aptamers that contain paired GC/CG sites in their three-dimensional structure can be used as drug carrier to incorporate doxorubicin. In another example, aptamers can change their conformation upon binding to their targets under certain conditions, thereby becoming “switchable”, which means that they can be used as ON/OFF switches for strictly controlled drug release. Below, we describe other strategies that utilized aptamer-targeted (Figure 2) or stimuli-triggered drug delivery systems (Figure 3).
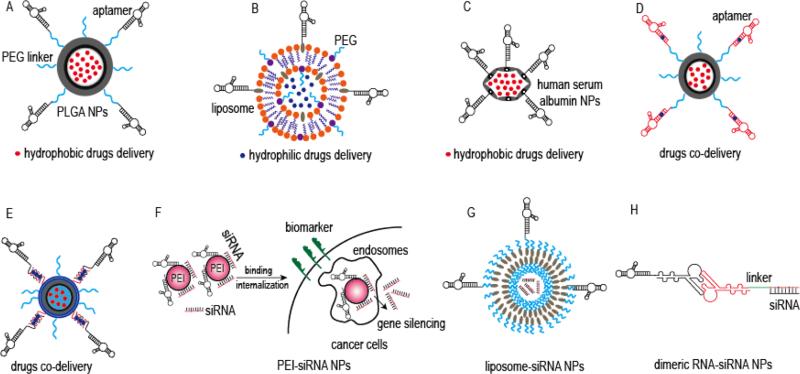
Figure 2.
Representative schematic diagrams of aptamer-targeted drug delivery systems: (A) Aptamer-targeted PLGA NPs for chemical drugs delivery, modified from reference [76]; (B) Aptamer-targeted liposome NPs for chemical drugs delivery, modified from reference [77]; (C) Aptamer-targeted human serum albumin NPs for chemical drugs delivery, modified from reference [78]; (D) Aptamer-targeted PLGA NPs for hydrophilic and hydrophobic drug co-delivery, in which aptamer also can be used as hydrophilic drugs carrier, and hydrophobic drugs were encapsulated in copolymer PLGA NPs, modified from reference [80]; (E) Aptamer-targeted PLGA NPs for chemical drugs co-delivery, in which hydrophilic drugs are intercalated into complementary DNA linkers containing multiple paired GC/CG sites, modified from reference [81]; (F) Aptamer-targeted PEI nanocomplexes for siRNA delivery, modified from reference [82]; (G) Aptamer-targeted liposome NPs for siRNA delivery, modified from reference [83]; (H) Aptamer-targeted packing RNA NPs for siRNA delivery, modified from reference [87].
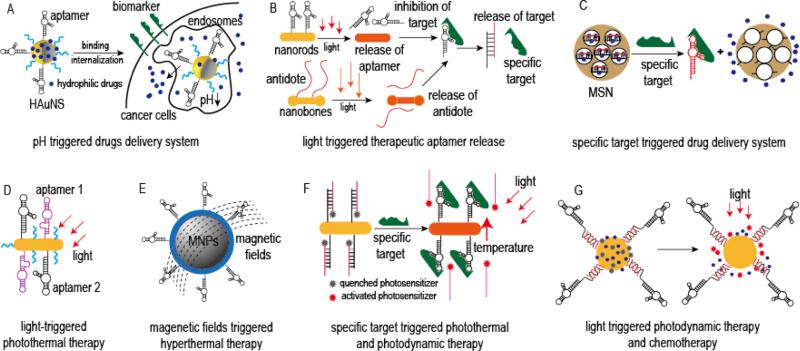
Figure 3.
Representative schematic diagrams of stimuli-triggered drug delivery systems: (A) pH-triggered aptamer-targeted HAuNS for chemical drugs delivery, modified from reference [88]; (B) Light-triggered therapeutic aptamer controlled release system for blood clotting therapy, modified form reference [90]; (C) Specific targets triggered, switchable aptamer-targeted MSN drug delivery system, modified from reference [93]; (D) Light triggered, aptamer-targeted photothermal therapy model, modified from reference [96]; (E) Magnetic fields triggered, aptamer-targeted magnetic-hyperthermia model, modified from reference [97]; (F) Specific targets triggered, aptamer-targeted gold nanorods loaded with photosensitizer for combinational photothermal and photodynamic therapy, modified from reference [98]; (G) Light triggered, aptamer-targeted gold NPs loaded with photosensitizer and chemotherapeutic drugs for combinational photodynamic therapy and chemotherapy, modified from reference [99].
3.2.1 Aptamer-Targeted Drug Delivery System
With excellent biocompatibility and biodegradability, copolymer and liposome are considered as most outstanding nanomaterials which can produce rapid distribution and enhance circulating time of encapsulated active drugs, and thus, successfully approved in first-generation nanomedicines for clinical applications. In one of the first reports, Farokhzad et al. described their pioneering work on the development of prostate cancer-specific, aptamer-targeted copolymer NPs for docetaxel delivery. In their study, an aptamer specific for prostate specific membrane antigen (PSMA) was conjugated with polylactic-co-gycolic-acid (PLGA)-PEG functional groups (Figure 2A). After optimization and docetaxel loading, the resultant aptamer-NP conjugates bound to and showed improved in vitro cellular toxicity in prostate cancer cells, indicating targeted drug delivery. More importantly, a single intratumoral injection of the aptamer-NP conjugates resulted in a significantly reduced tumor burden and very low systemic toxicity in vivo.[76] Similarly, Xing et al. developed AS1411 aptamer-targeted liposome NPs for breast cancer specific doxorubicin therapy (Figure 2B). In vitro, these aptamer-liposome NPs exhibited excellent tumor cells targeting abilities and high internalization efficiency. Moreover, treatment of mice bearing xenografted breast cancer tumors with the aptamer-liposome NPs also showed enhanced tumor tissue penetration and superb antitumor efficacy.[77]
Using the same AS1411 aptamer, Wu et al. developed a therapeutic aptamer-human serum albumin conjugate to deliver paclitaxel for breast cancer therapy. In this approach, paclitaxel is encapsulated in the human serum albumin, which is similar to nanomedicine Abraxane approved by FDA, and then, AS1411 aptamers were covalently conjugated with the formed NPs (Figure 2C). The published report describes these new conjugates as highly biostable particles that are effectively internalized by breast cancer cells, resulting in a significant killing effect.[78] In addition to drug delivery, with the specific anti-tumor activity of AS1411 aptamer, an encouraging multivalent AS1411 aptamer-pyramid DNA NPs model was also reported recently, which could inhibit nucleolin-positive HeLa cells growth significantly, even without carrying any chemical drugs.[79] Foreseeably, this model can be further extended its therapeutic index when eqquipped with other active drugs concurrently.
Recent advances in pharmacology and pharmacokinetics suggest that an effective cancer therapy, especially in chemotherapy-resistant tumors, is best accomplished by combining two different drugs in order to achieve a synergistic effect and improve therapeutic index. However, on a practical note, co-delivery of two different drugs using the same drug delivery system is extremely difficult, especially when hydrophilic and hydrophobic drugs must be used concurrently. To overcome this obstacle, Zhang et al. developed a PSMA-specific aptamer conjugated with PLGA-PEG NPs for treatment of prostate tumors. These nanoparticles delivered hydrophilic doxorubicin and hydrophobic docetaxel chemotherapeutics simultaneously, and the PSMA-specific aptamer was used not only as a targeting ligand, but also as doxorubicin carrier because it contained paired GC/CG sites. Simultaneously, PLGA was used to encapsulate docetaxel and achieve hydrophilic state (Figure 2D). The aptamer preloaded with doxorubicin was then covalently conjugated with the PLGA-encapsulated docetaxel NPs through a PEG linker. In the subsequent in vitro studies, the formed aptamer-NP conjugates selectively bound to and were internalized by PSMA-positive LNCaP cells, but not by the off-target PC3 prostate cancer cells, resulting in doxorubicin and docetaxel release and cell death. Importantly, utilization of the doxorubicin and docetaxel co-delivery system resulted in a significantly higher level of cell death than treatment with comparable concentrations of either drug alone.[80] In a more recent study, Huang et al. constructed sgc8 aptamer-PLGA-lecithin-PEG self-assemble NPs to deliver doxorubicin and paclitaxel simultaneously (Figure 2E). The resultant self-assembling NPs achieved an improved killing effect.[81] Although the approaches used in the two studies were similar, Zhang's model had a low doxorubicin loading capacity because the aptamer possessed a single doxorubicin intercalation site. Conversely, Huang's model modified the sgc8 aptamer by adding repetitive 5’-GCA-3’ sequences in order to generate multiple paired GC/CG sites and improve doxorubicin loading capacity.
Although different from traditional chemotherapeutic approaches, gene silencing tools, such as small interfering RNA (siRNA), may present more effective therapeutic strategies. However, due to their nuclease sensitivity and lack of cell/tissue specificity for in vivo delivery, their clinical applicability has been extremely limited. To address these problems, our group developed a functionalized siRNA nanocomplex comprised of a CD30-specific aptamer conjugated with an anaplastic lymphoma kinase gene specific siRNA and nano-sized polyethylenimine (PEI) polymer carriers (Figure 2F). The reasons for taking PEI as the carrier are that, PEI exhibits high cell transfection efficiency, protection siRNA from nuclease degradation, strong buffering capacity, and an ability to release functional nucleic acids from endosomes into the cytoplasm by inducing osmotic endosomal rupture. Our study showed that the functional siRNA/aptamer nanocomplexes preferentially bound to and were internalized by the CD30-expressing lymphoma cells, while displaying no binding to off-target cells, and resulted in down-regulation of cellular anaplastic lymphoma kinase gene expression, growth inhibition, and apoptosis. Importantly, this nanocomplexes is reduced cytotoxicity due to low PEI dosage.[82] In addition to PEI, cationic liposomes may also serve as good gene carriers. For example, Li et al. developed a functionalized AS1411 aptamer-PEG-liposome NP that delivers BRAF gene specific siRNA for melanoma therapy (Figure 2G). Their results indicated that these formed NPs selectively bound with and were internalized into necleolin-positive cancer cells, causing silencing of BRAF gene and down-regulating its protein expression to inhibit cells proliferation. In the subsequent xenograft mouse models, the formed NPs also demonstrated rapid accumulation within tumors and resulted in significant BRAF gene silencing.[83]
Due to their good biocompatibility, biodegradability and programmability, RNA nanotechnology could be used for delivering siRNAs to target other diseases. For example, Guo group recently reported a series of outstanding methods to construct multifunctional RNA NPs based on bacteriophage phi29 packing RNA, including dimeric-, trimeric- and tetra-structure RNA NPs.[84-86] In particular, Zhou et al. developed a gp120-specific aptamer containing dimeric RNA NPs to deliver anti-tet/rev siRNA for HIV therapy (Figure 2H). Their study results showed that the dimeric RNA-NP conjugates were resistant to RNase degradation and selectively bound to gp120-expressing cells, resulting in tet/rev gene silencing and inhibiting viral replication. More importantly, these NPs inhibited infection of human peripheral blood mononuclear cells when challenged by HIV.[87]
3.2.2 Stimuli-Triggered Drug Delivery System
In order to improve therapeutic outcomes and reduce side effect, stimuli-triggered drug release systems that release their therapeutic loads “on-command” at specific disease sites have also been developed. Some of the stimuli include pH, temperature, ionic strength, redox reagents, enzymes, specific disease targets, light, magnetic field, and so on. Recently, our group developed a pH-triggered, CD30-specific aptamer-HAuNS drug delivery system for doxorubicin lymphoma therapy (Figure 3A). In this system, aptamers and PEG molecules are sequentially chemically conjugated with HAuNS by thiol-gold linkers. Then, doxorubicin is loaded through the charge force. The resultant aptamer-HAuNS conjugates are uniform in size and have a doxorubicin payload efficiency of >90%, which equals to approximately 30% w/w. Careful analysis of the consequent conjugates revealed two important features: (i) the drug release profile is ultrasensitive to pH changes, such that conjugates are stable at pH 7.4, but release 80% of their drug payloads within 2 hours when pH is reduced to 5.0, similar to lysosomal conditions; (ii) aptamer-HAuNS particles are highly selective for their target cells.[88] In a similar fashion, Taghdisi et al. constructed a pH-triggered drug delivery system based on the sgc8 aptamer and SWCNTs for leukemia therapy. In their study, the sgc8 DNA aptamer and a chemotherapeutic agent daunorubicin were non-covalently and sequentially conjugated with SWCNTs through pi-pi interactions. The study results showed that the formed nanomedicine complexes were taken up and induced apoptosis only in their target protein tyrosine kinase 7-positive cells. Similar to our report, daunorubicin release profile was pH-sensitive, and 60% of daunorubicin was released from the formed nanomedicine complexes within 72 hours of pH 5.5. However, incubation of the nanomedicine complexes with their target cells at pH 7.0 caused little to no drug release.[89]
As opposed to using pH-triggered aptamers, de Puig et al. developed a light-triggered therapeutic aptamer drug delivery system targeting thrombin and designed for blood clotting therapy (Figure 3B). Thrombin-specific aptamers were grafted onto the gold nanorods, while the complementary DNA sequences designed to serve as antidotes were grafted onto the gold nanobones. Utilizing irradiation with light of different wavelengths, the thrombin-specific aptamer and its antidote could be released separately and act as an ON/OFF switch for blood clotting. Consequently, inhibition and restoration of the blood clotting time could be precisely controlled.[90] In yet another light-triggered approach, Cohen & Bergkvist took advantage of virus-like particles and used them as nanocarriers. Consequently, they developed functional AS1411 aptamer-virus-like particles that delivered porphyrin photosensitizer for breast cancer photodynamic therapy. Their results show that these newly formed NPs achieved an excellent cell killing effect following tumor irradiation, with almost 100% of tumor cells undergoing apoptosis. In comparison, they used an immortalized mammary gland cell line treated under similar conditions, with virtually no cell death observed.[91]
Different from external stimuli that could initiate aptamer's cargo release, various intracellular stimuli can regulate cargo release in a more precise manner. For example, Zhu et al. developed a novel target-triggered photodynamic therapy model based on a switchable thrombin-specific aptamer and SWCNTs, an efficient quencher of fluorescence probe and drug carrier, and aimed at controlling singlet oxygen generation. In this model, a Ce6 photosensitizer was covalently labeled with a thrombin-specific, DNA-based aptamer, which was then wrapped on the surface of SWCNTs via pi-stacking interaction. In the absence of thrombin, Ce6 and SWCNTs are located in a close proximity of one another, and Ce6 is quenched by SWCNTs, resulting in inhibition of singlet oxygen generation and reducing side effects. However, when thrombin is present, the aptamer confirmation changes and it is released from the SWCNTs surface, resulting in restoration of singlet oxygen generation.[92]
In another approach, He et al. developed an adenosine triphosphate (ATP)-triggered drug release system, in which MSN, with unique properties of high drugs loading capacity, easy modifications and tunable pore size, were used as drugs carriers and switchable ATP-specific aptamers were used as locks that controlled drug release (Figure 3C). After drugs were loaded into the MSN, the two-armed complementary DNA was hybridized with an ATP-specific aptamer to form a sandwich-type structure. Then, these DNA structures were grafted onto the MSN pore outlets through the click chemistry approach, the MSN pores locked, and leakage of the drugs inhibited. In the presence of ATP, the ATP-specific aptamer has a higher affinity and tighter binding to ATP, as compared to its binding to the complementary DNA arm. Consequently, when ATP is present, the aptamer falls off, exposing MSN surface pores and allowing drug release. In proof-of-principal studies using Ru(bipy)32+ as a guest molecule, this drug delivery system showed high loading capacity and acted in an “on-command” release pattern. Since ATP is a multifunctional nucleotide for all living organisms, this type of drug delivery system will have very extensive applications.[93] A similar drug delivery system was also constructed by Zhu et al., with the difference being that the switchable ATP-specific aptamer was covalently conjugated with Au NPs, which then served as the cap.[94] In another model, AS1411 aptamer was modified, such that it not only blocked drug leakage from the MSN, but also served as a targeting ligand. Subsequent studies showed that this simple, effective drug delivery system can be used for therapeutic targeting of nucleolin-expressing tumors.[95]
Another simple model for the development of therapeutic aptamer-nanomaterials conjugates is based on the unique therapeutic functions of nanomaterials, such as photothermal effects afforded by the gold nanomaterials and magnetic hyperthermia associated with the use of magnetic nanomaterials. In one example, Wang et al. constructed a photothermal model by conjugating two aptamers, CSC1 aptamer that targeted prostate cancer cells and CSC13 aptamer that was specific against a subpopulation of cancer stem cells, on the surface of gold nanorods (Figure 3D). In their in vitro studies, the authors showed that a 10-minute irradiation with a near-infrared laser results in 36% and 47% reduced viability of non-stem and stem cancer cells respectively.[96] In another study, Pala et al. constructed a HER2-specific aptamer conjugated with superparamagnetic NPs to induce selective hyperthermia in human breast adenocarcinoma tumors (Figure 3E). In their study, the HER2-specific aptamers were conjugated with dextran-coated ferric oxide NPs. Following incubation with the cancer cells and magnetic field treatment, these aptamer-magnetic NP conjugates induced significant cell death in HER2-positive target cells at doses 90-fold lower than that of the control magnetic NPs, and had little cytotoxicity in the off-target cells. Although the results were promising, the authors noted that hyperthermia alone is not sufficient for effective tumor elimination, and should therefore only be used in an adjuvant setting.[97]
Because of their tremendous promise, photodynamic therapy approaches are continuously modified. For example, Tan group recently combined photothermal therapy with photodynamic therapy (Figure 3F). In their study, a sgc8 DNA aptamer was first conjugated with gold nanorods through thiol-gold linkers, and then a complementary DNA reporter pre-labeled with the Ce6 photosensitizer was hybridized with the sgc8 aptamer. As shown in the report, upon sgc8 aptamer bounding with its target cells, the Ce6-labeled reporter sequence was released. Under irradiation with near-infrared laser, Ce6 produced singlet oxygen and gold nanorods induced a photothermal effect. This combination of photothermal and photodynamic therapies resulted in more effective therapeutic outcomes than either modality alone.[98] In another set of studies, a photodynamic-triggered release system for co-delivery of TMPyP4 photosensitizer and doxorubicin was developed (Figure 3G). In this nanoplatform, the AS1411 aptamer was conjugated with gold nanoparticles, which used as nanocarriers for TMPyP4 and doxorubicin. Upon binding with their target cancer cells followed by irradiation, TMPyP4 and doxorubicin were released to achieve simultaneous photodynamic therapy and chemotherapies. Utilization of this therapeutic platform induced significant levels of cell death in nucleolin-positive and doxorubicin-resistant cells, indicated that this innovate approach could be used for targeting chemotherapy-resistant tumors.[99]
A detailed summary of aptamer-targeted therapeutic nanomdicines described in this section is listed in Table 3.
Table 3.
Summary of aptamer-targeted therapeutic nanomedicines
| target | aptamer | nanomaterial | active molecule | applications reference | |
|---|---|---|---|---|---|
| PSMA | RNA | PLGA NPs | docetaxel | targeted drugs delivery | [76] |
| doxorubicin and docetaxel | targeted drugs co-delivery | [80] | |||
| nucleolin | DNA | liposome NPs | doxorubicin | targeted drugs delivery | [77] |
| human serum albumin | paclitaxel | targeted drugs delivery | [78] | ||
| virus-like particles | porphyrin | light-triggered, photodynamic therapy | [91] | ||
| DNA NPs | AS1411 aptamer | targeted therapy | [79] | ||
| liposome NPs | siRNA | targeted siRNA delivery | [83] | ||
| gold NPs | TMPyP4 and doxorubicin | light-triggered photodynamic therapy and chemotherapy | [99] | ||
| MSN | / | targets-triggered delivery system | [95] | ||
| protein tyrosine kinase 7> | DNA | PLGA NPs | doxorubicin and paclitaxel | targeted drugs co-delivery | [81] |
| SWCNTs | daunorubicin | pH-triggered, targeted drugs delivery | [89] | ||
| gold nanorods | gold nanorods and Photosensitizer Ce6 | light-triggered photothermal and photodynamic therapy | [98] | ||
| CD30 | RNA | PEI | siRNA | targeted siRNA delivery | [82] |
| HAuNS | doxorubicin | pH-triggered, targeted drugs delivery | [88] | ||
| gp120 | RNA | RNA NPs | siRNA | targeted siRNA delivery | [87] |
| thrombin | DNA | gold nanorods | thrombin aptamer | light-triggered, therapeutic aptamer delivery | [90] |
| SWCNTs | Ce6 | target-triggered, photodynamic therapy | [92] | ||
| ATP | DNA | MSN | / | target-triggered delivery system | [93, 94] |
| prostate cancer and stem cells | DNA | gold nanorods | gold nanorods | light-triggered photothermal therapy | [96] |
| HER2 | DNA | superparamagnetic NPs | superparamagnetic NPs | magnetic field induced hyperthermia | [97] |
3.3 Aptamer-Targeted Theranostic Nanomedicines
As the demand for personalized medicine increases, a completely novel, multifunctional therapeutic modality has been developed, termed “theranostics”. As illustrated in Figure 4, theranostics combines the real-time diagnostic and therapeutic functions. For instance, Wang et al. constructed a multifunctional theranostic agent that is based on the PSMA-specific aptamer conjugated to superparamagnetic iron oxide nanoparticles for prostate cancer imaging and therapy (Figure 4A). In this multimodality, the PSMA-specific aptamers were used not only as targeting ligands, but also as doxorubicin carriers. Aptamers were conjugated with thermally cross-linked superparamagnetic iron oxide nanoparticles, doxorubicin was intercalated into the aptamer sequences, which were then loaded onto the thermally cross-linked superparamagnetic iron oxide nanoparticles through charge interaction. These formed NPs showed excellent imaging capacity, and exhibited very potent killing of PSMA-positive cells.[100] A similar theranostic agent was also developed by Yu et al. recently.[101]
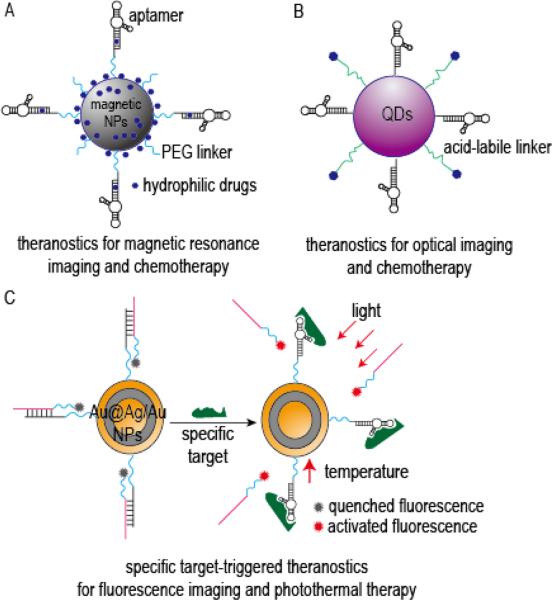
Figure 4.
Representative schematic diagrams of aptamer-targeted theranostic nanomedicines: (A) Aptamer-targeted superparamagnetic NPs loaded with chemical drugs for cancer magnetic resonance imaging diagnosis and chemotherapy, modified from reference [100]; (B) Aptamer-targeted QDs loaded with chemical drugs for cancer optical-imaging and chemotherapy, modified from reference [104]; (C) Switchable aptamer-targeted Au@Ag/Au NPs loaded with fluorescence for cancer fluorescence imaging and photothermal therapy, modified from reference [105].
As already discussed above, QDs possess excellent imaging properties and are widely used in the development of novel theranostics. For example, PSMA-specific aptamer-QD-doxorubicin conjugates for diagnosis and therapy of prostate cancer,[102] and MUC1-specific aptamer-QD-daunorubicin conjugates for delivering and monitoring daunorubicin release in prostate cancer cells[103] have been recently described. Moreover, Savla et al. developed MUC1-specific aptamer-QD-doxorubicin conjugates for imaging and therapeutic targeting of ovarian carcinoma, in which DNA-based MUC1-specific aptamers were chemically conjugated with QDs, and doxorubicin was conjugated with QDs through the acid-labile hydrazone bond (Figure 4B). The study results showed that the formed nanoparticles could carry up to 46 doxorubicin molecules per QD particle, had high binding and internalization efficiency in target cells, and displayed a pH-sensitive doxorubicin release profile even in doxorubicin-resistant ovarian cancer cells. In vivo, treatment of tumor-bearing mice with the formed conjugates resulted in its accumulation in tumor tissues and allowed for sensitive diagnostic imaging.[104]
In a slightly different approach, Shi et al. developed a theranostic model based on a switchable aptamer and a fluorescence-labeled complementary DNA sequence as imaging elements (Figure 4C). In this model, a switchable S6 aptamer specific for A549 lung cancer cells was first hybridized with a fluorescence-labeled complementary DNA sequence, and then grafted onto the surface of Au@Ag/Au NPs, which were used as photothermal agents and fluorescence quenchers. In the absence of A549 target cells, fluorescence was quenched by Au@Ag/Au NPs due to their close proximity to the DNA. However, upon the aptamer binding to its target cells, its conformation changed and fluorescence was activated. Evaluation of these conjugates in in vivo models of lung cancer revealed that treatment mice with theranostics resulted in a prominent tumor fluorescence 5 minutes after the injection, and subsequent photothermal therapy resulted in a significant level of tumor necrosis.[105] A detailed summary of aptamer-targeted theranostic nanomdicines described in this section is listed in Table 4.
Table 4.
Summary of aptamer-targeted theranostic nanomedicines
| target | aptamer | nanomaterial | active molecule | application | reference |
|---|---|---|---|---|---|
| PSMA | RNA | superparamagnetic iron oxide NPs | superparamagnetic iron oxide NPs and doxorubicin | targeted magnetic resonance imaging and chemotherapy | [100, 101] |
| QDs | QDs and doxorubicin | targeted optical imaging and chemotherapy | [102] | ||
| MUC1 | DNA | QDs | QDs and daunorubicin | targeted optical imaging and chemotherapy | [103] |
| QDs and doxorubicin | targeted optical imaging and chemotherapy | [104] | |||
| A549 cells | DNA | Au@Ag/Au NPs | Au@Ag/Au NPs and fluorescence | Target-triggered and light-induced optical imaging and photothermal therapy | [105] |
4. Conclusions
In summary, “passive targeting” nanomedicines can achieve somewhat improved therapeutic outcomes and reduce side effect, but they are affected by various external factors that can lead to disappointing therapeutic outcomes. Therefore, it is necessary to develop ligand-directed “active targeting” nanomedicines. Although antibodies exhibit high target specificity and affinity, their potential in immunogenicity and high production costs are of great concern, as these attributes limit their clinical applicability. Aptamers are novel multifunctional oligonucleotide ligands that also possess high specificity and affinity for their targets. Aptamers also offer several advantages over protein antibodies and are considered as excellent alternative to replace or supplement protein antibodies. To date, several approaches had been developed with aptamers used as targeting ligands, therapeutics, drugs carriers, and even ON/OFF switches. This functional adaptability had allowed the aptamer technology to be applied in vitro diagnostics, in vivo imaging, targeted therapy, theranostics, subcellular molecule detection, food safety, and environment monitoring, just to name a few.
However, despite their great potential, only one PEGylated aptamer (Macugen) had been approved by the FDA for clinical applications. As the patent of SELEX technology is expired now, more and more aptamers and related applications will be developed for specific clinical use, and as expected, many other aptamer-based nanomedicines are in the pipeline for clinical development to date. As discussed in this review, development of successful aptamer-based nanomedicines is contingent upon several multidisciplinary considerations, such as the virtue of the disease, aptamer structure and biochemical properties, and selection of appropriate nanomaterials and drugs. Specifically, some important parameters that must be considered are: (i) how can aptamer be modified in order to maintain its correct conformational structure on the surface of NPs? (ii) How many aptamers should be conjugated with each NP to achieve effective binding with target cells? (iii) How will the size and shape of the selected nanomaterials affect the nanomedicine bio-distribution? (iv) Can the preparation method be easily and cost-effectively scaled up for industrial production?
Although there are great progress of aptamer technology and nanomedicine technology in recent years, other challenges to these two outstanding technologies should be considered continuously. For instance, the main challenges of aptamer technology are that: (v) how can rapidly select an aptamer with high-specificity and -affinity to the disease-relevant targets? (vi) How can improve the biostability of aptamer in vivo applications? (vii) What are the best strategies for aptamer to achieve effective drug delivery? The main discussions of nanomedicine technology will be focused on: (viii) how can improve physicochemical stability of different nanomaterials when the specific nanomedicines are prepared? (ix) How can reduce the toxicity of different nanomaterials in vivo applications, especially for non-degradable nanomaterials? (x) What are the exact mechanisms of nanomedicines to work in vivo? Some of these issues have been reviewed in [23, 43, 48, 53, 106, 107].
Taken together, aptamers are attractive multifunctional tools that can be adapted to development of nanomedicines. However, some challenges will have to be overcome in order to gain useful preclinical and clinical data, which will then lead to translation of sophisticated, highly effective aptamer-based nanomedicines from bench to bedside.
References
- 1.Ellington AD, Szostak JW. Nature. 1990;346:818. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 2.Tuerk C, Gold L. Science. 1990;249:505. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 3.Nimjee SM, Rusconi CP, Sullenger BA. Annu Rev Med. 2005;56:555. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- 4.Parekh P, Tang Z, Turner PC, Moyer RW, Tan W. Anal Chem. 2010;82:8642. doi: 10.1021/ac101801j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sefah K, Tang ZW, Shangguan DH, Chen H, Lopez-Colon D, Li Y, Parekh P, Martin J, Meng L, Phillips JA, Kim YM, Tan WH. Leukemia. 2009;23:235. doi: 10.1038/leu.2008.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayrac AT, Sefah K, Parekh P, Bayrac C, Gulbakan B, Oktem HA, Tan W. ACS Chem Neurosci. 2011;2:175. doi: 10.1021/cn100114k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruno JG, Kiel JL. Biosens Bioelectron. 1999;14:457. doi: 10.1016/s0956-5663(99)00028-7. [DOI] [PubMed] [Google Scholar]
- 8.Kirby R, Cho EJ, Gehrke B, Bayer T, Park YS, Neikirk DP, Mcdevitt JT, Ellington AD. Anal Chem. 2004;76:4066. doi: 10.1021/ac049858n. [DOI] [PubMed] [Google Scholar]
- 9.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc Natl Acad Sci U S A. 2006;103:11838. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Z, Parekh P, Turner P, Moyer RW, Tan W. Clin Chem. 2009;55:813. doi: 10.1373/clinchem.2008.113514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Osborne SE, Matsumura I, Ellington AD. Curr Opin Chem Biol. 1997;1:5. doi: 10.1016/s1367-5931(97)80102-0. [DOI] [PubMed] [Google Scholar]
- 12.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, John M, Kesavan V, Lavine G, Pandey RK, Racie T, Rajeev KG, Rohl I, Toudjarska I, Wang G, Wuschko S, Bumcrot D, Koteliansky V, Limmer S, Manoharan M, Vornlocher HP. Nature. 2004;432:173. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 13.Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, Mccauley TG, Kurz JC, Epstein DM, Wilson C, Keefe AD. Chem Biol. 2005;12:25. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 14.Proske D, Gilch S, Wopfner F, Schatzl HM, Winnacker EL, Famulok M. Chembiochem. 2002;3:717. doi: 10.1002/1439-7633(20020802)3:8<717::AID-CBIC717>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 15.Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD, Claesson-Welsh L, Janjic N. J Biol Chem. 1998;273:20556. doi: 10.1074/jbc.273.32.20556. [DOI] [PubMed] [Google Scholar]
- 16.Keefe AD, Cload ST. Curr Opin Chem Biol. 2008;12:448. doi: 10.1016/j.cbpa.2008.06.028. [DOI] [PubMed] [Google Scholar]
- 17.Wu SY, Yang X, Gharpure KM, Hatakeyama H, Egli M, Mcguire MH, Nagaraja AS, Miyake TM, Rupaimoole R, Pecot CV, Taylor M, Pradeep S, Sierant M, Rodriguez-Aguayo C, Choi HJ, Previs RA, Armaiz-Pena GN, Huang L, Martinez C, Hassell T, Ivan C, Sehgal V, Singhania R, Han HD, Su C, Kim JH, Dalton HJ, Kovvali C, Keyomarsi K, Mcmillan NA, Overwijk WW, Liu J, Lee JS, Baggerly KA, Lopez-Berestein G, Ram PT, Nawrot B, Sood AK. Nat Commun. 2014;5:3459. doi: 10.1038/ncomms4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forster C, Zydek M, Rothkegel M, Wu Z, Gallin C, Gessner R, Lisdat F, Furste JP. Biochem Biophys Res Commun. 2012;419:60. doi: 10.1016/j.bbrc.2012.01.127. [DOI] [PubMed] [Google Scholar]
- 19.Schmidt KS, Borkowski S, Kurreck J, Stephens AW, Bald R, Hecht M, Friebe M, Dinkelborg L, Erdmann VA. Nucleic Acids Res. 2004;32:5757. doi: 10.1093/nar/gkh862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eulberg D, Klussmann S. Chembiochem. 2003;4:979. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]
- 21.Parekh P, Kamble S, Zhao N, Zeng Z, Portier BP, Zu Y. Biomaterials. 2013;34:8909. doi: 10.1016/j.biomaterials.2013.07.099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ireson CR, Kelland LR. Mol Cancer Ther. 2006;5:2957. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 23.Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y. Mol Ther Nucleic Acids. 2014;3:e182. doi: 10.1038/mtna.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Bassett SE, Li X, Luxon BA, Herzog NK, Shope RE, Aronson J, Prow TW, Leary JF, Kirby R, Ellington AD, Gorenstein DG. Nucleic Acids Res. 2002;30:e132. doi: 10.1093/nar/gnf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang X, Li X, Prow TW, Reece LM, Bassett SE, Luxon BA, Herzog NK, Aronson J, Shope RE, Leary JF, Gorenstein DG. Nucleic Acids Res. 2003;31:e54. doi: 10.1093/nar/gng054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, Carter J, Dalby AB, Eaton BE, Fitzwater T, Flather D, Forbes A, Foreman T, Fowler C, Gawande B, Goss M, Gunn M, Gupta S, Halladay D, Heil J, Heilig J, Hicke B, Husar G, Janjic N, Jarvis T, Jennings S, Katilius E, Keeney TR, Kim N, Koch TH, Kraemer S, Kroiss L, Le N, Levine D, Lindsey W, Lollo B, Mayfield W, Mehan M, Mehler R, Nelson SK, Nelson M, Nieuwlandt D, Nikrad M, Ochsner U, Ostroff RM, Otis M, Parker T, Pietrasiewicz S, Resnicow DI, Rohloff J, Sanders G, Sattin S, Schneider D, Singer B, Stanton M, Sterkel A, Stewart A, Stratford S, Vaught JD, Vrkljan M, Walker JJ, Watrobka M, Waugh S, Weiss A, Wilcox SK, Wolfson A, Wolk SK, Zhang C, Zichi D. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraemer S, Vaught JD, Bock C, Gold L, Katilius E, Keeney TR, Kim N, Saccomano NA, Wilcox SK, Zichi D, Sanders GM. PLoS One. 2011;6:e26332. doi: 10.1371/journal.pone.0026332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai JC, Hong CY. ACS Comb Sci. 2014;16:321. doi: 10.1021/co5000272. [DOI] [PubMed] [Google Scholar]
- 29.Zhu Z, Song Y, Li C, Zou Y, Zhu L, An Y, Yang CJ. Anal Chem. 2014;86:5881. doi: 10.1021/ac501423g. [DOI] [PubMed] [Google Scholar]
- 30.He W, Elizondo-Riojas MA, Li X, Lokesh GL, Somasunderam A, Thiviyanathan V, Volk DE, Durland RH, Englehardt J, Cavasotto CN, Gorenstein DG. Biochemistry. 2012;51:8321. doi: 10.1021/bi300471d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilner SE, Wengerter B, Maier K, De Lourdes Borba Magalhaes M, Del Amo DS, Pai S, Opazo F, Rizzoli SO, Yan A, Levy M. Mol Ther Nucleic Acids. 2012;1:e21. doi: 10.1038/mtna.2012.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan A, Levy M. Methods Mol Biol. 2014;1103:241. doi: 10.1007/978-1-62703-730-3_18. [DOI] [PubMed] [Google Scholar]
- 33.Gourronc FA, Rockey WM, Thiel WH, Giangrande PH, Klingelhutz AJ. Virology. 2013;446:325. doi: 10.1016/j.virol.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou J, Bobbin ML, Burnett JC, Rossi JJ. Front Genet. 2012;3:234. doi: 10.3389/fgene.2012.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiel WH, Bair T, Peek AS, Liu X, Dassie J, Stockdale KR, Behlke MA, Miller FJ, Jr., Giangrande PH. PLoS One. 2012;7:e43836. doi: 10.1371/journal.pone.0043836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camorani S, Esposito CL, Rienzo A, Catuogno S, Iaboni M, Condorelli G, De Franciscis V, Cerchia L. Mol Ther. 2014;22:828. doi: 10.1038/mt.2013.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiel KW, Hernandez LI, Dassie JP, Thiel WH, Liu X, Stockdale KR, Rothman AM, Hernandez FJ, Mcnamara JO, 2nd, Giangrande PH. Nucleic Acids Res. 2012;40:6319. doi: 10.1093/nar/gks294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. Proc Natl Acad Sci U S A. 2003;100:15416. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer G, Ahmed MS, Dolf A, Endl E, Knolle PA, Famulok M. Nat Protoc. 2010;5:1993. doi: 10.1038/nprot.2010.163. [DOI] [PubMed] [Google Scholar]
- 40.Avci-Adali M, Metzger M, Perle N, Ziemer G, Wendel HP. Oligonucleotides. 2010;20:317. doi: 10.1089/oli.2010.0253. [DOI] [PubMed] [Google Scholar]
- 41.Mi J, Liu Y, Rabbani ZN, Yang Z, Urban JH, Sullenger BA, Clary BM. Nat Chem Biol. 2010;6:22. doi: 10.1038/nchembio.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li S, Xu H, Ding H, Huang Y, Cao X, Yang G, Li J, Xie Z, Meng Y, Li X, Zhao Q, Shen B, Shao N. J Pathol. 2009;218:327. doi: 10.1002/path.2543. [DOI] [PubMed] [Google Scholar]
- 43.Duncan R. Curr Opin Biotechnol. 2011;22:492. doi: 10.1016/j.copbio.2011.05.507. [DOI] [PubMed] [Google Scholar]
- 44.Zhong Y, Meng F, Deng C, Zhong Z. Biomacromolecules. 2014;15:1955. doi: 10.1021/bm5003009. [DOI] [PubMed] [Google Scholar]
- 45.Rink JS, Plebanek MP, Tripathy S, Thaxton CS. Curr Opin Oncol. 2013;25:646. doi: 10.1097/CCO.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Langer R. Nature. 1998;392:5. [PubMed] [Google Scholar]
- 47.Tong R, Gabrielson NP, Fan TM, Cheng J. Curr Opin Solid State Mater Sci. 2012;16:323. doi: 10.1016/j.cossms.2013.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Z, Farokhzad OC. ACS Nano. 2012;6:3670. doi: 10.1021/nn301869z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Davis ME, Chen ZG, Shin DM. Nat Rev Drug Discov. 2008;7:771. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 50.Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Chem Soc Rev. 2012;41:2971. doi: 10.1039/c2cs15344k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nehoff H, Parayath NN, Domanovitch L, Taurin S, Greish K. Int J Nanomedicine. 2014;9:2539. doi: 10.2147/IJN.S47129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka T, Shiramoto S, Miyashita M, Fujishima Y, Kaneo Y. Int J Pharm. 2004;277:39. doi: 10.1016/j.ijpharm.2003.09.050. [DOI] [PubMed] [Google Scholar]
- 53.Duncan R, Gaspar R. Mol Pharm. 2011;8:2101. doi: 10.1021/mp200394t. [DOI] [PubMed] [Google Scholar]
- 54.Weissig V, Pettinger TK, Murdock N. Int J Nanomedicine. 2014;9:4357. doi: 10.2147/IJN.S46900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liang H, Zhang XB, Lv Y, Gong L, Wang R, Zhu X, Yang R, Tan W. Acc Chem Res. 2014 doi: 10.1021/ar500078f. 10.1021/ar500078f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng EW, Adamis AP. Can J Ophthalmol. 2005;40:352. doi: 10.1016/S0008-4182(05)80078-X. [DOI] [PubMed] [Google Scholar]
- 57.Que-Gewirth NS, Sullenger BA. Gene Ther. 2007;14:283. doi: 10.1038/sj.gt.3302900. [DOI] [PubMed] [Google Scholar]
- 58.Tan L, Neoh KG, Kang ET, Choe WS, Su X. Macromol Biosci. 2011;11:1331. doi: 10.1002/mabi.201100173. [DOI] [PubMed] [Google Scholar]
- 59.Taghdisi SM, Danesh NM, Sarreshtehdar Emrani A, Tabrizian K, Zandkarimi M, Ramezani M, Abnous K. J Drug Target. 2013;21:739. doi: 10.3109/1061186X.2013.812095. [DOI] [PubMed] [Google Scholar]
- 60.Macugen AMDSG, Apte RS, Modi M, Masonson H, Patel M, Whitfield L, Adamis AP. Ophthalmology. 2007;114:1702. doi: 10.1016/j.ophtha.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 61.Basile AS, Hutmacher M, Nickens D, Nielsen J, Kowalski K, Whitfield L, Masayo O, Nakane M. J Clin Pharmacol. 2012;52:1186. doi: 10.1177/0091270011412961. [DOI] [PubMed] [Google Scholar]
- 62.Ng EW, Adamis AP. Ann N Y Acad Sci. 2006;1082:151. doi: 10.1196/annals.1348.062. [DOI] [PubMed] [Google Scholar]
- 63.Chen X, Zhu X, Li L, Xian G, Wang W, Ma D, Xie L. Int J Pharm. 2013;456:499. doi: 10.1016/j.ijpharm.2013.08.028. [DOI] [PubMed] [Google Scholar]
- 64.Shiang YC, Ou CM, Chen SJ, Ou TY, Lin HJ, Huang CC, Chang HT. Nanoscale. 2013;5:2756. doi: 10.1039/c3nr33403a. [DOI] [PubMed] [Google Scholar]
- 65.Wu Y, Sefah K, Liu H, Wang R, Tan W. Proc Natl Acad Sci U S A. 2010;107:5. doi: 10.1073/pnas.0909611107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Javier DJ, Nitin N, Levy M, Ellington A, Richards-Kortum R. Bioconjug Chem. 2008;19:1309. doi: 10.1021/bc8001248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Latorre A, Posch C, Garcimartin Y, Celli A, Sanlorenzo M, Vujic I, Ma J, Zekhtser M, Rappersberger K, Ortiz-Urda S, Somoza A. Nanoscale. 2014;6:7436. doi: 10.1039/c4nr00019f. [DOI] [PubMed] [Google Scholar]
- 68.Kim B, Yang J, Hwang M, Choi J, Kim HO, Jang E, Lee JH, Ryu SH, Suh JS, Huh YM, Haam S. Nanoscale Res Lett. 2013;8:399. doi: 10.1186/1556-276X-8-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Heo D, Lee E, Ku M, Hwang S, Kim B, Park Y, Han Lee Y, Huh YM, Haam S, Cheong JH, Yang J, Suh JS. Nanotechnology. 2014;25:275102. doi: 10.1088/0957-4484/25/27/275102. [DOI] [PubMed] [Google Scholar]
- 70.Zhang C, Ji X, Zhang Y, Zhou G, Ke X, Wang H, Tinnefeld P, He Z. Anal Chem. 2013;85:5843. doi: 10.1021/ac400606e. [DOI] [PubMed] [Google Scholar]
- 71.Melancon MP, Zhou M, Zhang R, Xiong C, Allen P, Wen X, Huang Q, Wallace M, Myers JN, Stafford RJ, Liang D, Ellington AD, Li C. ACS Nano. 2014;8:4530. doi: 10.1021/nn406632u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee CH, Rajendran R, Jeong MS, Ko HY, Joo JY, Cho S, Chang YW, Kim S. Chem Commun (Camb) 2013;49:6543. doi: 10.1039/c3cc42752h. [DOI] [PubMed] [Google Scholar]
- 73.Zhou J, Soontornworajit B, Martin J, Sullenger BA, Gilboa E, Wang Y. Macromol Biosci. 2009;9:831. doi: 10.1002/mabi.200900046. [DOI] [PubMed] [Google Scholar]
- 74.Zhou J, Soontornworajit B, Wang Y. Biomacromolecules. 2010;11:2087. doi: 10.1021/bm100450k. [DOI] [PubMed] [Google Scholar]
- 75.Tang L, Yang X, Dobrucki LW, Chaudhury I, Yin Q, Yao C, Lezmi S, Helferich WG, Fan TM, Cheng J. Angew Chem Int Ed Engl. 2012;51:12721. doi: 10.1002/anie.201205271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Farokhzad OC, Cheng J, Teply BA, Sherifi I, Jon S, Kantoff PW, Richie JP, Langer R. Proc Natl Acad Sci U S A. 2006;103:6315. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xing H, Tang L, Yang X, Hwang K, Wang W, Yin Q, Wong NY, Dobrucki LW, Yasui N, Katzenellenbogen JA, Helferich WG, Cheng J, Lu Y. J Mater Chem B Mater Biol Med. 2013;1:5288. doi: 10.1039/C3TB20412J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wu J, Song C, Jiang C, Shen X, Qiao Q, Hu Y. Mol Pharm. 2013;10:3555. doi: 10.1021/mp300686g. [DOI] [PubMed] [Google Scholar]
- 79.Charoenphol P, Bermudez H. Mol Pharm. 2014;11:1721. doi: 10.1021/mp500047b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang L, Radovic-Moreno AF, Alexis F, Gu FX, Basto PA, Bagalkot V, Jon S, Langer RS, Farokhzad OC. ChemMedChem. 2007;2:1268. doi: 10.1002/cmdc.200700121. [DOI] [PubMed] [Google Scholar]
- 81.Huang F, You M, Chen T, Zhu G, Liang H, Tan W. Chem Commun (Camb) 2014;50:3103. doi: 10.1039/c3cc49003c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhao N, Bagaria HG, Wong MS, Zu Y. J Nanobiotechnology. 2011;9:2. doi: 10.1186/1477-3155-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li L, Hou J, Liu X, Guo Y, Wu Y, Zhang L, Yang Z. Biomaterials. 2014;35:3840. doi: 10.1016/j.biomaterials.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 84.Guo S, Tschammer N, Mohammed S, Guo P. Hum Gene Ther. 2005;16:1097. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shu D, Shu Y, Haque F, Abdelmawla S, Guo P. Nat Nanotechnol. 2011;6:658. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Haque F, Shu D, Shu Y, Shlyakhtenko LS, Rychahou PG, Evers BM, Guo P. Nano Today. 2012;7:245. doi: 10.1016/j.nantod.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhou J, Shu Y, Guo P, Smith DD, Rossi JJ. Methods. 2011;54:284. doi: 10.1016/j.ymeth.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhao N, You J, Zeng Z, Li C, Zu Y. Small. 2013;9:3477. doi: 10.1002/smll.201202694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taghdisi SM, Lavaee P, Ramezani M, Abnous K. Eur J Pharm Biopharm. 2011;77:200. doi: 10.1016/j.ejpb.2010.12.005. [DOI] [PubMed] [Google Scholar]
- 90.De Puig H, Cifuentes Rius A, Flemister D, Baxamusa SH, Hamad-Schifferli K. PLoS One. 2013;8:e68511. doi: 10.1371/journal.pone.0068511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cohen BA, Bergkvist M. J Photochem Photobiol B. 2013;121:67. doi: 10.1016/j.jphotobiol.2013.02.013. [DOI] [PubMed] [Google Scholar]
- 92.Zhu Z, Tang Z, Phillips JA, Yang R, Wang H, Tan W. J Am Chem Soc. 2008;130:10856. doi: 10.1021/ja802913f. [DOI] [PubMed] [Google Scholar]
- 93.He X, Zhao Y, He D, Wang K, Xu F, Tang J. Langmuir. 2012;28:12909. doi: 10.1021/la302767b. [DOI] [PubMed] [Google Scholar]
- 94.Zhu CL, Lu CH, Song XY, Yang HH, Wang XR. J Am Chem Soc. 2011;133:1278. doi: 10.1021/ja110094g. [DOI] [PubMed] [Google Scholar]
- 95.Hernandez FJ, Hernandez LI, Pinto A, Schafer T, Ozalp VC. Chem Commun (Camb) 2013;49:1285. doi: 10.1039/c2cc37370j. [DOI] [PubMed] [Google Scholar]
- 96.Wang J, Sefah K, Altman MB, Chen T, You M, Zhao Z, Huang CZ, Tan W. Chem Asian J. 2013;8:2417. doi: 10.1002/asia.201300375. [DOI] [PubMed] [Google Scholar]
- 97.Pala K, Serwotka A, Jelen F, Jakimowicz P, Otlewski J. Int J Nanomedicine. 2014;9:67. doi: 10.2147/IJN.S52539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang J, You M, Zhu G, Shukoor MI, Chen Z, Zhao Z, Altman MB, Yuan Q, Zhu Z, Chen Y, Huang CZ, Tan W. Small. 2013;9:3678. doi: 10.1002/smll.201202155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shiao YS, Chiu HH, Wu PH, Huang YF. ACS Appl Mater Interfaces. 2014 doi: 10.1021/am5026243. 10.1021/am5026243. [DOI] [PubMed] [Google Scholar]
- 100.Wang AZ, Bagalkot V, Vasilliou CC, Gu F, Alexis F, Zhang L, Shaikh M, Yuet K, Cima MJ, Langer R, Kantoff PW, Bander NH, Jon S, Farokhzad OC. ChemMedChem. 2008;3:1311. doi: 10.1002/cmdc.200800091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu MK, Kim D, Lee IH, So JS, Jeong YY, Jon S. Small. 2011;7:2241. doi: 10.1002/smll.201100472. [DOI] [PubMed] [Google Scholar]
- 102.Bagalkot V, Zhang L, Levy-Nissenbaum E, Jon S, Kantoff PW, Langer R, Farokhzad OC. Nano Lett. 2007;7:3065. doi: 10.1021/nl071546n. [DOI] [PubMed] [Google Scholar]
- 103.Lin Z, Ma Q, Fei X, Zhang H, Su X. Anal Chim Acta. 2014;818:54. doi: 10.1016/j.aca.2014.01.057. [DOI] [PubMed] [Google Scholar]
- 104.Savla R, Taratula O, Garbuzenko O, Minko T. J Control Release. 2011;153:16. doi: 10.1016/j.jconrel.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 105.Shi H, Ye X, He X, Wang K, Cui W, He D, Li D, Jia X. Nanoscale. 2014;6:8754. doi: 10.1039/c4nr01927j. [DOI] [PubMed] [Google Scholar]
- 106.Jiao Q, Li L, Mu Q, Zhang Q. Biomed Res Int. 2014;2014:426028. doi: 10.1155/2014/426028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Oh N, Park JH. Int J Nanomedicine. 2014;9(Suppl 1):51. doi: 10.2147/IJN.S26592. [DOI] [PMC free article] [PubMed] [Google Scholar]