Abstract
Background
Genes involved in angiopoietin and pericyte pathways may become escape mechanisms under anti-VEGF therapy. We investigated whether variations within genes in these pathways are associated with clinical outcome in patients with colorectal liver metastases (CLM) undergoing liver resection and perioperative bevacizumab-based chemotherapy.
Methods
Single nucleotide polymorphisms (SNP) in nine genes (ANGPT1, ANGPT2, TEK, PDGFB, PDGFRB, IGF1, TGFB1, RALBP1, RGS5) were analyzed in genomic DNA from 149 patients, and evaluated for associations with clinical outcome.
Results
RALBP1 rs329007 showed a significant difference in recurrence-free survival (RFS) (A/A 14.0 months, A/G or G/G 9.2 months; hazard ratio (HR) 1.60; P=0.024). PDGFB rs1800818 was associated with 3-year overall survival (OS) rates (A/A 78%, A/G 69%, G/G 53%; HR 1.37 and 2.12; P=0.048). In multivariate analysis, RALBP1 rs329007 remained significant (HR 1.99; P=0.002). PDGFB rs1800818 and RALBP1 rs329007 correlated with radiological response (A/A or A/G 86%, G/G 71%; P=0.042 and A/A 78%, A/G or G/G 94%; P=0.018, respectively). RALBP1 rs329007 showed significantly different rates for histological response (A/A 35% MjHR, 34% PHR, 30% NHR; A/G or G/G 46%, 13%, 41%; P=0.029). Recursive partitioning revealed that ANGPT2 rs2442599 and RALBP1 rs329007 were the main SNPs to predict histological response and RFS, whereas PDGFB rs1800818 was the leading SNP to predict OS. ANGPT2 rs2916702 and rs2442631 were significantly associated with probability of cure.
Conclusions
Our data suggest that variations in genes involved in the angiopoietin and pericyte pathways may be predictive and/or prognostic biomarkers in patients with resected CLM treated with bevacizumab-based chemotherapy.
Keywords: Angiopoietin, bevacizumab, colorectal liver metastases, neoadjuvant chemotherapy, pericytes, single nucleotide polymorphisms
Introduction
Colorectal cancer (CRC) is a common malignancy in both males and females1. Approximately 20% of the patients are diagnosed with metastatic disease and a significant number of patients with stage II and III disease will develop metastases at a later time2, 3. Patients with resectable liver-limited metastases who undergo liver resection have prolonged survival and can even be cured4. In a pivotal study investigating a multidisciplinary treatment approach in patients with resectable CLM, perioperative chemotherapy plus surgery was associated with prolonged progression-free survival compared with surgery alone5. Furthermore, the monoclonal antibody bevacizumab that inhibits angiogenesis by targeting the Vascular Endothelial Growth Factor-A (VEGF-A) is commonly used in combination with chemotherapy in the metastatic setting and can been safely administered in the perioperative setting6–8.
While approximately 25% of the patients with CLM can be cured with liver resection, most patients experience disease recurrence demonstrating the need for biomarkers that help to guide treatment decisions and predict clinical outcome5. In patients with resectable CLM receiving perioperative bevacizumab-based chemotherapy, angiogenesis-related factors may serve as potential biomarkers. So far, pro-angiogenic factors, such as IL-8, Fibroblast Growth Factor, Hepatocyte Growth Factor and others have been associated with compensatory upregulation under anti-VEGF targeted therapy, which may facilitate alternative angiogenic signaling9. However, none has been established as a validated biomarker in clinical practice.
The Angiopoietin/TIE2 axis is involved in blood vessel maturation via Angiopoietin 1 and in early angiogenesis via Angiopoietin 210. In addition, the Platelet-Derived Growth Factor Beta (PDGFB)/Platelet-Derived Growth Factor Receptor Beta (PDGFRB) pericyte pathway is involved in pericyte attachment to immature blood vessels to facilitate vessel maturation11. As the Angiopoietin/TIE2 pathway and the pericyte pathways are playing a significant role in early angiogenesis and vessel maturation independent from the VEGF pathway, they have been proposed to be potential molecular escape mechanisms in patients receiving anti-VEGF targeted therapy9. The clinical significance of these pathways in patients with resectable CLM undergoing perioperative bevacizumab-based chemotherapy and liver resection remains unknown.
The goal of treatment is to cure patients with resectable CLM and predictive and prognostic biomarkers are urgently needed to guide treatment strategies. Therefore, we investigated the predictive and prognostic value of SNPs within genes involved in the Angiopoietin/TIE2 and pericyte pathways that may act as potential escape mechanism under anti-VEGF targeted therapy. We analyzed a comprehensive panel of genes involved in the Angiopoietin/TIE2 axis, such as Angiopoietin 1 (ANGPT1), Angiopoietin 2 (ANGPT2) and Tyrosine Kinase, Endothelial (TEK, coding for TIE2), involved in the pericyte attachment on immature vessels, such as Platelet-Derived Growth Factor Beta (PDGFB), Platelet-Derived Growth Factor Receptor Beta (PDGFRB), Insulin-like Growth Factor 1 (IGF1) and Transforming Growth Factor, Beta 1 (TGFB1), as well as genes involved in the pericyte maturation, such as RalA binding Protein 1 (RALBP1) and Regulator of G-protein signaling 5 (RGS5).10–16
Methods
One hundred forty-nine patients who underwent 3 months of neoadjuvant and 3 months of adjuvant fluoropyrimidine-based combination chemotherapy (oxaliplatin (n=124), irinotecan (n=18), both (n=7)) and bevacizumab and had their liver metastases resected at the Medical University Vienna were included in this study (2005–2011). The last preoperative dose of bevacizumab was administered 5 weeks before the liver resection. Clinical and treatment data were obtained from a prospectively maintained database. Eighty-seven (58.4%) patients were male. The median age was 62 years (range 30–80). The median follow-up in this study was 3.9 years (range 0.02–7.7). The median RFS was 11.7 months (95% confidence interval 10.3, 15.1) and the 3-year OS rate was 71% (standard error (SE) ±4%). Patient’s baseline demographic and tumor-related data are presented in Table 1. The study was approved by the local institutional review board.
Table 1.
Patient demographic and tumor related data (n=149)
| n | % | |
|---|---|---|
| Sex | ||
| Male | 87 | 58.4 |
| Female | 62 | 41.6 |
| Age | ||
| Median (range) | 62 (30–80) | |
| < 65 years | 90 | 60.4 |
| ≥ 65 years | 59 | 39.6 |
| Primary tumor site* | ||
| Right colon | 39 | 26.4 |
| Left colon | 60 | 40.5 |
| Rectum | 49 | 33.1 |
| Number of metastases* | ||
| 1–2 | 88 | 59.5 |
| >2 | 60 | 40.5 |
| Size of metastases* | ||
| 1–50mm | 127 | 85.8 |
| >50mm | 21 | 14.2 |
| Timing of metastases | ||
| Metachronous | 55 | 36.9 |
| Synchronous | 94 | 63.1 |
| Distribution of metastases | ||
| Unilobar | 76 | 51.0 |
| Bilobar | 73 | 49.0 |
1 patient had missing information
Response Evaluation Criteria In Solid Tumors (RECIST 1.1) was used to assess radiological response to neoadjuvant chemotherapy by two radiologists using computed tomography (CT)17. Patients with disease progression did not undergo liver resection and were not included in this study.
To assess histological response, the classification proposed by Rubbia-Brandt and colleagues was used. This classification defines five tumor regression grades (TRG) and comprises major histological response (MjHR; TRG 1 and 2), partial histological response (PHR; TRG 3) and no histological response (NHR; TRG 4 and 5)18.
Follow-up investigations to detect disease recurrence were performed every three months for the first three years and then every six months. Examinations comprised of: clinical examination, 3-phase CT scan of thorax, abdomen and liver and blood tests including carcinoembryonic antigen (CEA).
SNP selection and genotyping
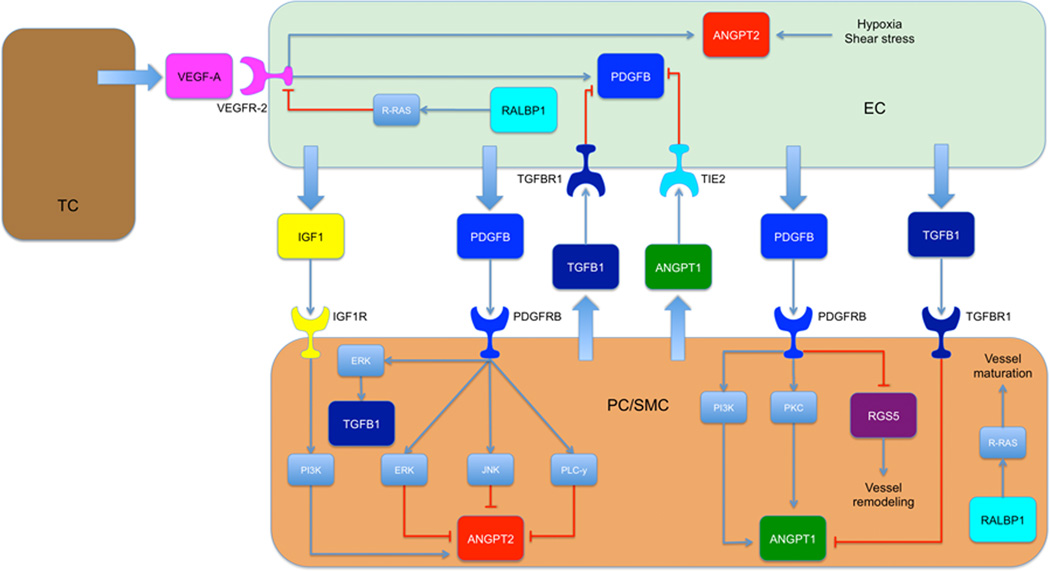
Variations in genes involved in the Angiopoietin/TIE2 and the pericyte pathways were selected according to their functional or potential functional relevance, as described the literature, or if they were Tag SNPs representing a larger set of other SNPs (Supplementary Table S1)19–22. In addition, the SNP’s functional relevance was assessed by Queen’s University F-SNP and National Institute of Environmental Health Sciences SNP Function Prediction tools (Supplementary Table S1)23, 24. Figure 1 gives the network of interaction of the investigated genes. Candidate SNPs with a minor allele frequency of ≥10% in Caucasians according to the Ensembl database were selected for analyses in this study25.
Figure 1.
Network of interaction of the investigated genes (tumor cell, TC; endothelial cell, EC; pericyte/smooth muscle cell, PC/SMC; Transforming Growth Factor Beta Receptor 1, TGFBR1; Insulin-like Growth Factor 1 Receptor, IGF1R; Extracellular-signal Regulated Kinase, ERK; Phosphatidylinositide 3-kinase, PI3K; Jun N-terminal kinase, JNK; Phospholipase C-γ, PLC-γ; Protein kinase C, PKC)
Genomic DNA was extracted from formalin fixed paraffin embedded (FFPE) specimen using the QIAamp DNAeasy Kit (Qiagen, Hilden Germany) and analyzed by direct Sanger sequencing. Primers used for polymerase chain reaction (PCR) are given in Supplementary Table S1. DNA sequences were analyzed using the ABI Sequencing Scanner v1.0 (Applied Biosystems). Investigators involved in SNP analyses were blinded to patients’ clinical data.
Statistical analyses
The predictive and prognostic value of variations in genes involved in the Angiopoietin/TIE2 and the pericyte pathways for patients with resected CLM treated with perioperative 5-FU-based chemotherapy including bevacizumab was investigated in this study. To test if the allelic distribution of the polymorphisms was within Hardy-Weinberg equilibrium, a 1-degree-freedom χ2 test was used. The primary endpoint was RFS, and the secondary endpoints were radiological and histological response and OS. RFS was defined as the interval from the day of liver resection to the day of disease recurrence or death. RFS was censored on the date of the last CT scan, if none of the two events were observed. OS was calculated from the date of liver resection to death or censored on the date last known to be alive. The associations of the genetic variations with radiological and histological response, RFS, and OS were investigated in univariate analyses. Kaplan-Meier estimations and log-rank tests were used in univariate analysis to test if genetic variations were associated with RFS and OS. Cox proportional hazards regression model were used in multivariate analysis and adjusted for age (<65 vs. 65+), number of metastases (1–2, vs. >2), distribution (unilobar vs. bilobar), and timing of liver metastases (metachronous vs. synchronous) due to association with RFS or OS at a 0.10 significance level. The associations of genetic variations with radiological and histological response were analyzed with χ2 tests in the univariate analysis and with logistic regression models in the multivariable analysis to control for potential predictive variables. Recursive partitioning was used to analyze polymorphism patterns associated with radiological and histological response, RFS, and OS.
Cure rate by individual SNPs was estimated by mixture cure model26. The patterns of recurrence were estimated using cumulative incidence of intrahepatic only or extrahepatic (±intrahepatic) recurrence using competing risks model.
SAS/STAT 12.3 (SAS Institute, Cary, NC, USA), a SAS macro (%pspmcm), along with rpart and cmprsk functions in R package were used for all analyses. 2-sided tests were used for all analyses. P values <0.05 were significant and not adjusted for multiple hypothesis testing.
Results
Gene variants and radiological response
With respect to radiological response, the homozygous variant of PDGFB rs1800818 A>G was associated with a lower response rate in this study, as 71% of the patients harboring a G/G genotype responded compared to 86% of the patients with a A/G or A/A genotypes (P=0.042). RALBP1 rs329007 A>G was also associated with a significant difference in radiological response: for patients with genotypes containing at least one variant allele G, the response rate was 94% compared to 78% for the patients with an A/A genotype (P=0.018). Data on radiological response are presented in Supplementary Table S2.
Gene variants and histological response
With respect to histological response, variant genotypes of RALBP1 rs329007 A>G were associated with a higher MjHR rate in this study. In patients with an A/A genotype, MjHR, PHR and NHR rates were 36%, 34% and 30%, respectively, compared to 46%, 13% and 41% for patients carrying A/G or G/G genotypes (P=0.029). Variant genotypes (T/C or C/C) of ANGPT2 rs2442599 T>C were associated with a higher MjHR rate. MjHR, PHR and NHR rates were 43%, 33% and 24% in patients with T/C or C/C genotypes compared to 34%, 23% and 43% for T/T (P=0.046). Data on histological response are presented in Supplementary Table S2.
Gene variants and recurrence-free survival
The variant genotypes (A/G or G/G) of RALBP1 rs329007 A>G were associated with a lower median RFS (Supplementary Figure S1). Median RFS was 14.0 months for A/A compared to 9.2 months for A/G or G/G. The HR was 1.60 (1.06, 2.40; P=0.024) in univariate analysis and 1.99 (1.28, 3.09; P=0.002) in multivariate analysis. Data on RFS are presented in Supplementary Table S3.
Gene variants and overall survival
The variant genotypes of PDGFB rs1800818 A>G were associated with shorter OS in this study (Supplementary Figure S2). The 3-year OS rate was 53% (±11%) for the G/G genotype, 69% (±8%) for the A/G genotype and 78% (±7%) for A/A. The HR was 2.12 (0.99, 4.53) for G/G and 1.37 (0.68, 2.78) for A/G in univariate analysis (P=0.048). In multivariate analysis, PDGFB rs1800818 A>G did not remain significantly associated with OS (HR 1.86 (0.85, 4.06) and 1.16 (0.56, 2.40); P=0.25). Data on OS are presented in Supplementary Table S3.
Gene variants and probability of cure
ANGPT2 rs2916702 C>T and ANGPT2 rs2442631 G>A predicted the probability of cure after liver resection. For ANGPT2 rs2916702 C>T, patients with a T/T genotype had a 2.88 times higher chance of cure than those with a C/T genotype. Patients with a C/T genotype had a 2.88 times higher probability of cure than those with C/C (odds ratio (OR) 0.347, 95% CI 0.180, 0.668; P=0.002). For ANGPT2 rs2442631 G>A, patients with a A/A genotype had 2.87 times higher probability of cure than those with G/A. Patients with a G/A genotype had a 2.87 times higher probability of cure than those with G/G (OR 0.349, 95% CI 0.175, 0.695; P=0.003). The results for these polymorphisms were similar, as they were in linkage disequilibrium (D’=0.946, r2=0.66).
Recursive partitioning for radiological and histological response, recurrence-free survival and overall survival
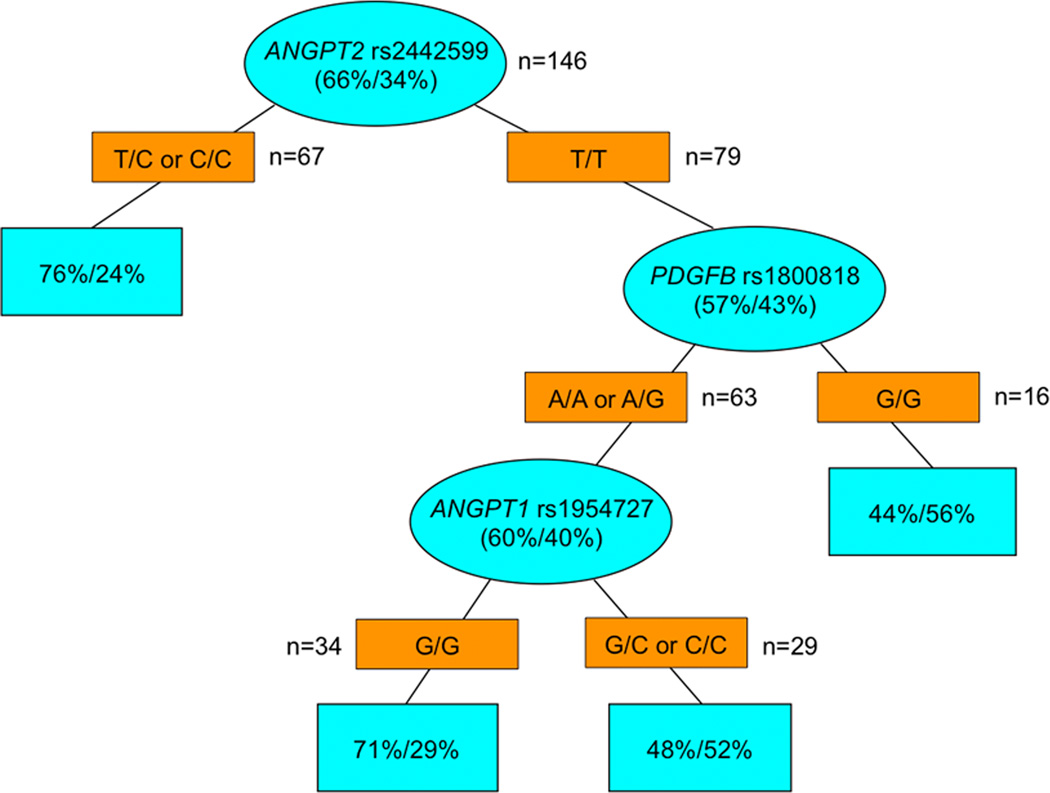
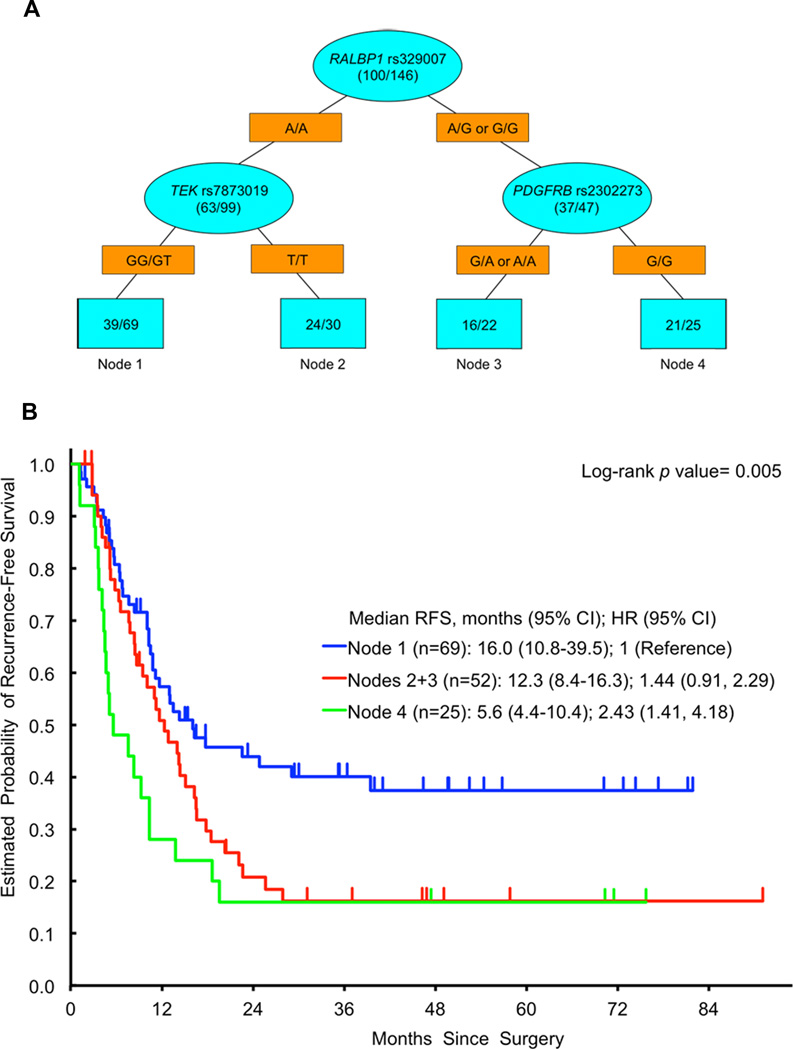
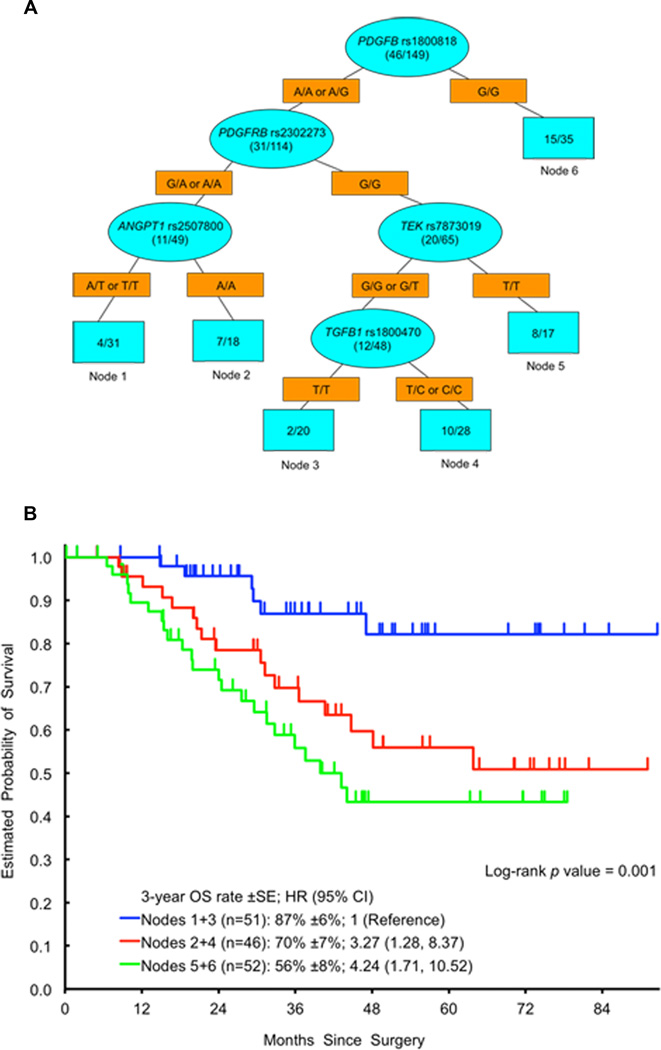
ANGPT2 rs2442599 T>C was the most important SNP to predict any histological (major or partial) response upon recursive partitioning (Figure 2). Other SNPs predicting any histological response in subgroups were PDGFB rs1800818 A>G and ANGPT1 rs1954727 G>C. RALBP1 rs329007 A>G was the most important SNP to predict RFS upon recursive partitioning (Figure 3). Other SNPs predicting significantly different RFS in subgroups were TEK rs7873019 G>T and PDGFRB rs2302273 G>A. PDGFB rs1800818 A>G was the dominant SNP to predict OS upon recursive partitioning (Figure 4). Other SNPs predicting significantly different OS in subgroups were PDGFRB rs2302273 G>A, ANGPT1 rs2507800 A>T, TEK rs7873019 G>T and TGFB1 rs1800470 T>C. None of the SNPs predicted radiological response upon recursive partitioning.
Figure 2.
Recursive Partitioning for histological response. Blue ovals represent intermediate subgroups; blue squares represent terminal nodes. Yellow rectangles indicate predictive polymorphism. Fractions within nodes indicate patients who had major or partial response/no response with that node.
Figure 3.
A Recursive Partitioning for RFS. Blue ovals represent intermediate subgroups; blue squares represent terminal nodes. Yellow rectangles indicate predictive polymorphism. Fractions within nodes indicate patients who relapsed/total patients with that node. Node 1 represents low risk patients; Node 2, 3 represent intermediate risk patients. Node 4 represents high risk patients. B Kaplan Meier curves for RFS nodes
Figure 4.
A Recursive Partitioning for OS. Blue ovals represent intermediate subgroups; blue squares represent terminal nodes. Yellow rectangles indicate predictive polymorphism. Fractions within nodes indicate patients who died/total patients with that node. Node 1, 3 represent low risk patients; Node 2, 4 represent intermediate risk patients. Node 5, 6 represent high risk patients. B Kaplan Meier curves for OS nodes
Gene variants and recurrence patterns
Disease recurrence after liver resection was only intrahepatic in 15 and extrahepatic (±intrahepatic) in 85 patients. The homozygous wild-type genotype of TEK rs1360773 G>T (G/G) was associated with a higher 2-year incidence of extrahepatic recurrence than the variant genotypes (G/T or T/T). The incidences were 72% (SE ±9%) and 54% (±5%), respectively (P=0.033). Other SNPs were not associated with extrahepatic or intrahepatic recurrence patterns (data not shown).
Discussion
Our goal was to identify biomarker for cure in a unique cohort of patients with liver-limited metastatic colorectal cancer who received perioperative bevacizumab-based chemotherapy and liver resection in curative intent. Although a substantial number of patients can be cured by this multidisciplinary treatment approach, most patients experience recurrence of disease and finally die. At present, there are no biomarkers available that predict cure and favorable clinical outcome for these patients. All patients in this study were treated with perioperative anti-VEGF targeted therapy, however we have no validated biomarkers of anti-VEGF efficacy. Recent data suggest that VEGF-independent pathways involved in angiogenesis and vessel maturation may act as potential escape mechanism to anti-VEGF targeted therapy. The ANGPT/TIE2 axis is one of these pathways, which has been suggested as an escape mechanism to anti-VEGF therapy27. Our comprehensive approach on investigating variants in genes involved in early angiogenesis, and pericyte and vessel maturation demonstrated significant clinical associations with response, RFS, OS and cure rate in patients with CLM undergoing bevacizumab-based chemotherapy.
One of the key genes of angiogenesis and vessel maturation is RALBP1, which is affecting Ras family GTPase Related Ras (R-Ras)14. R-Ras leads to vessel maturation and inhibition of angiogenesis by endothelial cell quiescence and reduced expression of the Vascular Endothelial Growth Factor Receptor 2 (VEGFR-2)15. In non-small cell lung cancer, RalA, an upstream activator of RALBP1, has been shown to be upregulated in cancer stemness-related CD44+ cells, suggesting a role of this pathway in disease initiation and maintenance28. These different functions of RALBP1 might be one explanation for the different associations found for RALBP1 rs329007 A>G for response and RFS in this study. We hypothesize that response may be related to the effect of RALBP1 in endothelial cells and pericytes upon anti-VEGF targeted treatment, while recurrence may be influenced by the cancer stem cell associated function of RALBP1.
PDGFB is a key regulator of vessel maturation by facilitating pericyte coverage of vessels via the PDGFB/PDGFRB pathway. In pericytes, PDGFB increases the expression of ANGPT1 via the PI3K and PKC pathways and inhibits the expression of ANGPT2 via the ERK, JNK and PLC-γ pathways, leading to reduced angiogenesis and increased vessel maturation12, 13. The PDGFB rs1800818 A>G SNP may be functionally relevant as in a previous study it was associated with cardiac allograft vasculopathy29. Thereby, we hypothesize that in patients with variant genotypes of PDGFB rs1800818 A>G, PDGFB could have impaired activity in the normalization of the blood vessels phenotype in tumors30. This reduced normalization could affect the adequate delivery of chemotherapy, explaining the inferior response and OS in this study.
ANGPT2 is a pro-angiogenic growth factor, mainly released by endothelial cells experiencing hypoxia. ANGPT2 binds to the TIE2 receptor and inhibits ANGPT1 signaling related to vessel maturation10. A previous study correlated low serum ANGPT2 levels with better response, progression-free survival and OS to anti-VEGF targeted treatment in metastatic CRC suggesting ANGPT2 to be a potential biomarker31. As the main goal of treatment in patients with resectable CLM is to achieve cure, we used a novel statistical method to investigate if any of the SNPs was associated with the probability of cure. In this analysis, ANGPT2 rs2916702 C>T and rs2442631 G>A (in high linkage disequilibrium, r2=0.66) were significantly associated with the probability of cure, but not with RFS and OS. The median RFS was similar regardless of the genotype due to the steep decline of the RFS curves within the first two years. However, a different plateau was found for each of the genotypes reflecting different cure rates: the presence of a variant allele increased the probability of cure. However, the higher cure rates did not translate into longer median OS in this study. One explanation is that patients with a more favorable genotype regarding cure had fewer recurrences, but they have a poor prognosis when they recur (data not shown). Although ANGPT2 is considered to be pro-angiogenic, its biological mechanisms of action under anti-VEGF targeted therapy are still unclear, due to the fact that ANGPT2 also acts anti-angiogenic in the absence of VEGF32.
ANGPT2 rs2442599 T>C and RALBP1 rs329007 A>G were associated with histological response. However, in the T/T group for ANGTP2 rs2442599 T>C, and in the A/G or G/G group for RALBP1 rs329007 A>G, the partial responders were less frequent than the major or non responders. This may be explained by fact that partial responders comprise only one TRG group (TRG 3), while major and non responders comprise two groups (TRG 1+2 and TRG 4+5, respectively). Nevertheless, it cannot be ruled out that it is related to the patient number in this study.
While some SNPs are relevant for the overall population, others may be only important for a subgroup of patients based on their biological function. To investigate this issue in an unbiased, hierarchical manner, we performed recursive partitioning analyses for response and clinical outcome. These analyses suggest biomarkers associated with response, RFS and OS are not the same and the subgroup patterns differ significantly. The different patterns of these three decision trees suggest that genes influencing response may be different than those relevant for RFS or OS, which is easily explained by the fact that markers predicting tumor shrinkage are not the same than the one predicting tumor growth and that survival is a mixture of both. These provocative data raise the question how we more successfully identify biomarker in the future and not neglecting biomarkers only associated with response or with PFS. Interestingly, trees were only created for histological response, RFS and OS, but not for radiological response. Although RECIST is the gold standard to assess radiological response, it may not be the best method, especially when targeted agents are used. This may explain why no tree was created for this endpoint. Other methods, such as morphological response, are currently investigated to overcome this potential limitation.33 Although recursive partitioning is a valuable tool to put biomarkers in a hierarchical context, they may be currently not ready to be used in clinical practice to guide treatment decisions. However, it may be contribute to establish comprehensive genetic profiling panels that require prospective investigation.
Finally, we investigated if any of the SNPs predicted the site of recurrence to identify patients with only intrahepatic recurrence who may be candidates for repeat liver resection. None of the SNPs was associated with intrahepatic recurrence, but TEK rs1360773 G>T was associated with extrahepatic recurrence. However, the value for clinical practice to predict extrahepatic recurrence still needs to be elucidated.
A limitation of this study is that the results were not validated in an independent patient cohort, due to the unique nature of the cohort. Besides, the results were not corrected for multiple testing.
In conclusion, the present study identified SNPs that may act as potential escape mechanisms to anti-VEGF targeted therapy and therefore could have a potential role as clinical biomarkers in patients undergoing perioperative chemotherapy including bevacizumab and liver resection. The goal of liver resection in patients with CLM is cure. The results of this study may contribute to the efforts to identify patients who do not benefit from this invasive and complication-associated treatment approach and should not undergo surgery. However, their role in guidance of treatment in this context needs to be further investigated. In addition, this study identified genes and pathways, which may be potential targets for novel treatment strategies to inhibit escape from VEGF blockade.
Supplementary Material
Acknowledgments
Financial support: Stefan Stremitzer is a recipient of an Erwin Schrödinger fellowship of the Austrian Science Fund. Ana Sebio is a recipient of a Rio Hortega Research Grant from the Insituto de Salud Carlos III (CM11/00102). Sebastian Stintzing is supported by a postdoctoral fellowship from the German Cancer Aid (Mildred-Scheel Foundation). Thomas Gruenberger receives financial support by Roche, Merck-Serono, Sanofi-Aventis, Bayer and Amgen (all Vienna, Austria). Heinz-Josef Lenz receives financial support by the 5P30CA014089-27S1 NIH grant, and the Daniel Butler Research Fund.
Conflict of interest: Thomas Gruenberger is member of the advisory boards of Roche, Merck-Serono and Sanofi-Aventis (all Vienna, Austria). Heinz-Josef Lenz is member of the advisory board of Genentech (San Francisco, CA).
References
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, et al. SEER Cancer Statistics Review, 1975–2010. Bethesda, MD: National Cancer Institute; 2013. Apr, based on November 2012 SEER data submission, posted to the SEER web site. [Google Scholar]
- 3.Andre T, Boni C, Mounedji-Boudiaf L, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med. 2004;350:2343–2351. doi: 10.1056/NEJMoa032709. [DOI] [PubMed] [Google Scholar]
- 4.Kopetz S, Chang GJ, Overman MJ, et al. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol. 2009;27:3677–3683. doi: 10.1200/JCO.2008.20.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nordlinger B, Sorbye H, Glimelius B, et al. Perioperative chemotherapy with FOLFOX4 and surgery versus surgery alone for resectable liver metastases from colorectal cancer (EORTC Intergroup trial 40983): a randomised controlled trial. Lancet. 2008;371:1007–1016. doi: 10.1016/S0140-6736(08)60455-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 2004;350:2335–2342. doi: 10.1056/NEJMoa032691. [DOI] [PubMed] [Google Scholar]
- 7.Saltz LB, Clarke S, Diaz-Rubio E, et al. Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. J Clin Oncol. 2008;26:2013–2019. doi: 10.1200/JCO.2007.14.9930. [DOI] [PubMed] [Google Scholar]
- 8.Gruenberger B, Tamandl D, Schueller J, et al. Bevacizumab, capecitabine, and oxaliplatin as neoadjuvant therapy for patients with potentially curable metastatic colorectal cancer. J Clin Oncol. 2008;26:1830–1835. doi: 10.1200/JCO.2007.13.7679. [DOI] [PubMed] [Google Scholar]
- 9.Tejpar S, Prenen H, Mazzone M. Overcoming resistance to antiangiogenic therapies. Oncologist. 2012;17:1039–1050. doi: 10.1634/theoncologist.2012-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Augustin HG, Koh GY, Thurston G, Alitalo K. Control of vascular morphogenesis and homeostasis through the angiopoietin-Tie system. Nat Rev Mol Cell Biol. 2009;10:165–177. doi: 10.1038/nrm2639. [DOI] [PubMed] [Google Scholar]
- 11.Betsholtz C. Insight into the physiological functions of PDGF through genetic studies in mice. Cytokine Growth Factor Rev. 2004;15:215–228. doi: 10.1016/j.cytogfr.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Phelps ED, Updike DL, Bullen EC, Grammas P, Howard EW. Transcriptional and posttranscriptional regulation of angiopoietin-2 expression mediated by IGF and PDGF in vascular smooth muscle cells. Am J Physiol Cell Physiol. 2006;290:C352–C361. doi: 10.1152/ajpcell.00050.2005. [DOI] [PubMed] [Google Scholar]
- 13.Nishishita T, Lin PC. Angiopoietin 1, PDGF-B, and TGF-beta gene regulation in endothelial cell and smooth muscle cell interaction. J Cell Biochem. 2004;91:584–593. doi: 10.1002/jcb.10718. [DOI] [PubMed] [Google Scholar]
- 14.Goldfinger LE, Ptak C, Jeffery ED, Shabanowitz J, Hunt DF, Ginsberg MH. RLIP76 (RalBP1) is an R-Ras effector that mediates adhesion-dependent Rac activation and cell migration. J Cell Biol. 2006;174:877–888. doi: 10.1083/jcb.200603111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawada J, Urakami T, Li F, et al. Small GTPase R-Ras regulates integrity and functionality of tumor blood vessels. Cancer Cell. 2012;22:235–249. doi: 10.1016/j.ccr.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berger M, Bergers G, Arnold B, Hammerling GJ, Ganss R. Regulator of G-protein signaling-5 induction in pericytes coincides with active vessel remodeling during neovascularization. Blood. 2005;105:1094–1101. doi: 10.1182/blood-2004-06-2315. [DOI] [PubMed] [Google Scholar]
- 17.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Rubbia-Brandt L, Giostra E, Brezault C, et al. Importance of histological tumor response assessment in predicting the outcome in patients with colorectal liver metastases treated with neo-adjuvant chemotherapy followed by liver surgery. Ann Oncol. 2007;18:299–304. doi: 10.1093/annonc/mdl386. [DOI] [PubMed] [Google Scholar]
- 19.Dai J, Wan S, Zhou F, et al. Genetic polymorphism in a VEGF-independent angiogenesis gene ANGPT1 and overall survival of colorectal cancer patients after surgical resection. PLoS One. 2012;7:e34758. doi: 10.1371/journal.pone.0034758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andraweera PH, Dekker GA, Thompson SD, et al. The interaction between the maternal BMI and angiogenic gene polymorphisms associates with the risk of spontaneous preterm birth. Mol Hum Reprod. 2012;18:459–465. doi: 10.1093/molehr/gas016. [DOI] [PubMed] [Google Scholar]
- 21.Pooja S, Francis A, Rajender S, et al. Strong impact of TGF-beta1 gene polymorphisms on breast cancer risk in Indian women: a case-control and population-based study. PLoS One. 2013;8:e75979. doi: 10.1371/journal.pone.0075979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winder T, Zhang W, Yang D, et al. Germline polymorphisms in genes involved in the IGF1 pathway predict efficacy of cetuximab in wild-type KRAS mCRC patients. Clin Cancer Res. 2010;16:5591–5602. doi: 10.1158/1078-0432.CCR-10-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee PH, Shatkay H. F-SNP: computationally predicted functional SNPs for disease association studies. Nucleic Acids Res. 2008;36:D820–D824. doi: 10.1093/nar/gkm904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Z, Taylor JA. SNPinfo: integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009;37:W600–W605. doi: 10.1093/nar/gkp290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flicek P, Amode MR, Barrell D, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corbiere F, Joly P. A SAS macro for parametric and semiparametric mixture cure models. Comput Methods Programs Biomed. 2007;85:173–180. doi: 10.1016/j.cmpb.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 27.Erber R, Thurnher A, Katsen AD, et al. Combined inhibition of VEGF and PDGF signaling enforces tumor vessel regression by interfering with pericyte-mediated endothelial cell survival mechanisms. FASEB J. 2004;18:338–340. doi: 10.1096/fj.03-0271fje. [DOI] [PubMed] [Google Scholar]
- 28.Male H, Patel V, Jacob MA, et al. Inhibition of RalA signaling pathway in treatment of non-small cell lung cancer. Lung Cancer. 2012;77:252–259. doi: 10.1016/j.lungcan.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Tambur AR, Pamboukian S, Costanzo MR, Heroux A. Genetic polymorphism in platelet-derived growth factor and vascular endothelial growth factor are significantly associated with cardiac allograft vasculopathy. J Heart Lung Transplant. 2006;25:690–698. doi: 10.1016/j.healun.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 30.Jain RK. Normalization of tumor vasculature: an emerging concept in antiangiogenic therapy. Science. 2005;307:58–62. doi: 10.1126/science.1104819. [DOI] [PubMed] [Google Scholar]
- 31.Goede V, Coutelle O, Neuneier J, et al. Identification of serum angiopoietin-2 as a biomarker for clinical outcome of colorectal cancer patients treated with bevacizumab-containing therapy. Br J Cancer. 2010;103:1407–1414. doi: 10.1038/sj.bjc.6605925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lobov IB, Brooks PC, Lang RA. Angiopoietin-2 displays VEGF-dependent modulation of capillary structure and endothelial cell survival in vivo. Proc Natl Acad Sci U S A. 2002;99:11205–11210. doi: 10.1073/pnas.172161899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shindoh J, Loyer EM, Kopetz S, et al. Optimal morphologic response to preoperative chemotherapy: an alternate outcome end point before resection of hepatic colorectal metastases. J Clin Oncol. 2012;30:4566–4572. doi: 10.1200/JCO.2012.45.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.