Summary
We recently published genetic lineage tracing experiments using the Fezf2 and Cux2 loci. These experiments demonstrated that at both the clonal and population levels Fezf2+ RGCs are multipotent, and that at the population level Cux2+ RGCs are multipotent. Here, we extend our work on the lineages of Fezf2+ and Cux2+ RGCs. Clonal analysis of E10.5 neocortical progenitors suggests that most, if not all, Cux2+ and Fezf2+ RGCs generate diverse projection neuron subtypes located throughout layers 2–6. These results support our previous conclusion that both Fezf2+ and Cux2+ RGCs are multipotent neocortical progenitors. This Matters Arising Response paper addresses the Gil-Sanz et al. (2015) Matters Arising paper, published concurrently in Neuron.
Introduction
Recent work proposes that the neocortex contains distinct classes of radial glial cells (RGCs) which are intrinsically predisposed to generate either deep- or upper-layer projection neurons. This model is based upon genetic lineage tracing experiments, which concluded that early (E10.5) RGCs labeled by Cux2-Cre or Cux2-CreERT2 mediated recombination (Cux2+) are fated to generate corticocortial projection neurons that reside primarily within upper layers (Franco et al., 2012).
We recently published a study in which we tested this model using genetic lineage tracing with the Fezf2 and Cux2 loci. This work demonstrated that: (1) Fezf2-expressing (Fezf2+) RGCs are multipotent and sequentially generate deep- and upper-layer cortical projection neurons followed by macroglia and (2) RGCs marked by Cux2-CreERT2 mediated recombination are not fate-restricted to generate only corticocortical projection neurons. Collectively, these results do not support the existence of early laminar-restricted RGCs within either the Fezf2+ or Cux2+ RGC lineages. Thus, although our results did not exclude their existence, they demonstrated that neither Fezf2 nor Cux2 expression alone is sufficient to identify fate-restricted RGCs.
Here, in response to challenges raised by Gil-Sanz et al. (2015), we extend our work on the lineages of Fezf2+ and Cux2+ RGCs. We show that the Cux2-Cre allele does not accurately report the lineage of neocortical RGCs. Moreover, clonal analysis of E10.5 RGCs using Cux2-CreERT2 and Fezf2-CreERT2 mice indicates that most, if not all, early RGCs labeled by these alleles generate multiple subtypes of cortical projection neurons located in layers 2–6. These results reinforce our previous conclusion that both Fezf2+ and Cux2+ RGCs are multipotent neocortical progenitors (Guo et al., 2013).
Results
The Cux2-Cre allele is not suitable for lineage tracing of neocortical RGCs
A significant difference between our previous study (Guo et al., 2013) and the work of Franco et al. (2012) and Gil-Sanz et al. (2015) is that the latter two studies relied extensively on the Cux2-Cre allele to analyze the lineages of RGCs. This allele expresses a constitutively active form of CRE that, when utilized in combination with Cre-dependent reporters, permanently labels any cell in which the Cux2 locus has been, or is currently, active. In addition, any cell derived from a Cux2+ progenitor will be permanently labeled.
We reported that at E15.5, Cux2-Cre labeled RGCs, basal progenitors and postmitotic neurons, thus masking the true lineage of Cux2+ RGCs (Guo et al., 2013). To demonstrate this further, we bred heterozygous Cux2-Cre mice to homozygous RCE-GFP and Ai9 reporter mice and examined the brains of double heterozygous mice at postnatal day 28 (P28) (Figures S1A–S1B and data not shown). We observed strong fluorescence throughout all cortical layers. Although in some cases labeling of cell bodies was evident (Figure S1B), high levels of reporter expression in neurites complicated the analysis of labeled cell bodies.
To circumvent this, we examined publicly available data from the Allen Brain Atlas (ABA), which includes extensive analysis of the same Cux2-Cre and Cux2-CreERT2 alleles that were used in previous lineage studies (Franco et al., 2012; Guo et al., 2013). The ABA datasets include in situ hybridization (ISH) experiments for Cre, Cux2 and tdTomato, which facilitate high-resolution analysis of cell bodies labeled by the Cre-dependent tdTomato reporter Ai14 (Madisen et al., 2010). Analysis of P58 Cux2-Cre; Ai14 mice indicated that Cre expression closely mimicked Cux2 expression (Figures S1C–S1F). Both Cux2 and Cre transcripts were highly enriched within upper cortical layers (L2–L4) with sparse expression in deep layers (L5 and L6). P56 Cux2-CreERT2; Ai14 mice, that received daily administration of tamoxifen (TM) from P45–P49, had many tdTomato+ cells in L2–L4, with sparse cell labeling in deep layers (Figures S1G–S1H). This expression of tdTomato mRNA indicates that CRE recombinase driven by the Cux2 locus is active in postmitotic neurons.
Compared to Cux2 and Cre expression however, tdTomato (Figure S1I–S1J) was more broadly expressed throughout both deep and upper cortical layers in P56 Cux2-Cre; Ai14 mice (compare Figures S1C–S1F with Figures S1I–S1J). Indeed, while double fluorescence in situ hybridization (dFISH) for Cux2 and tdTomato demonstrated significant co-expression in upper-layer cells, many tdTomato+ cells in L5 and L6 did not express Cux2 (Figures S1K–S1O). This suggests that these deep-layer tdTomato+ cells originated from Cux2+ progenitors or immature neurons but subsequently reduced their Cux2 expression in adult stages.
To further evaluate the distribution of tdTomato+ cells in P56 Cux2-Cre; Ai14 brains, we analyzed ABA experiments No. 100144051 (which is the same experiment shown in Supplementary Figure 1 of Gil-Sanz et al. (2015)) and No. 113098686. Mice analyzed in these experiments were generated by breeding Cux2-Cre heterozygous mice with Ai14 homozygous reporter mice. We examined multiple cortical areas including the somatosensory, somatomotor, primary motor, and visual cortices, as well as the anterior cingulate and posterior parietal areas. We found that tdTomato was robustly expressed throughout both deep and upper cortical layers (Figures S1I–S1J and S2). However, in some cortical areas, particularly the anterior cingulate area, there were fewer tdTomato+ cells in deep layers (Compare Figures S2D2 and S2H2 with S2B–S2D1 and S2F–S2H1). This indicates that a thorough examination of reporter expression across multiple cortical areas is required to fully understand the extent of cell labeling by the Cux2-Cre allele, as it has a complex temporal and spatial expression pattern (Figure 1A).
Figure 1.
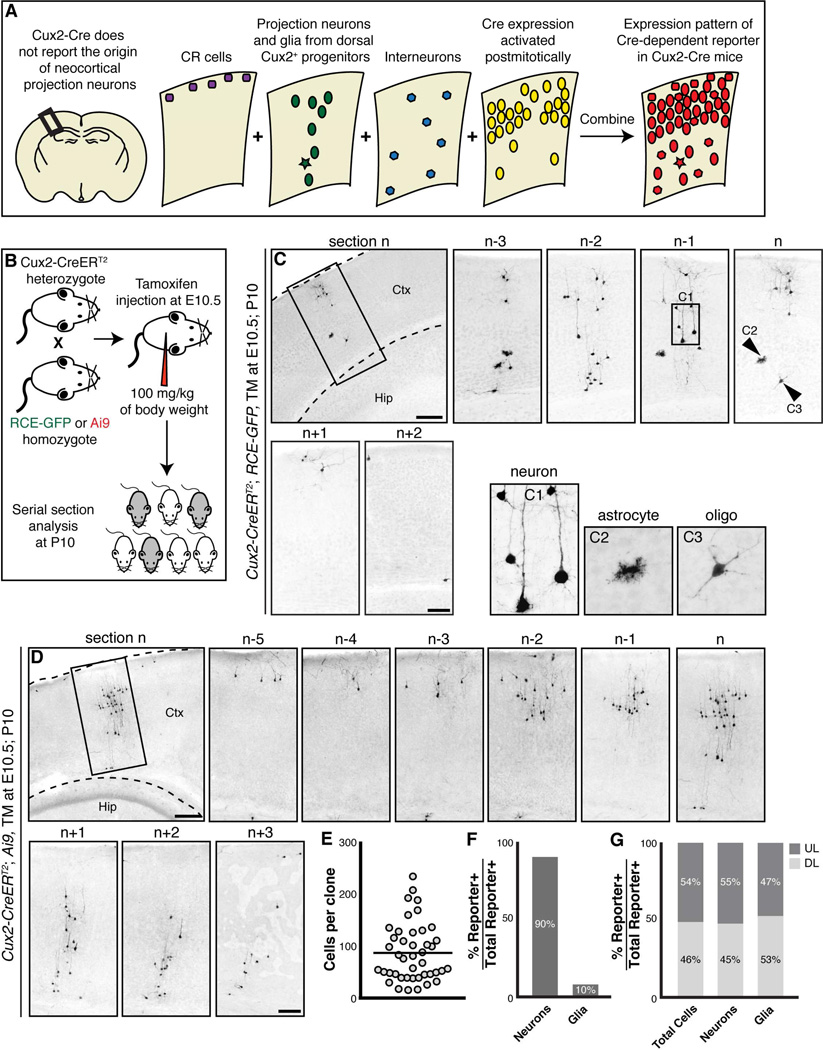
Individual E10.5 Cux2+ RGCs generate cortical projection neurons and macroglia located throughout cortical layers 2–6. (A) The constitutive activity of the CRE recombinase makes the Cux2-Cre allele unsuitlable for lineage analysis of neocortical RGCs. (B) Experimental methodology for lineage analysis of Cux2+ RGCs using the Cux2-CreERT2 allele. (C–D) Examples of TM at E10.5, P10 Cux2+ RGC clones. For each clone, a lower magnification image is shown first to illustrate the position of the clone in the cerebral cortex, followed by higher magnification images of seriel sections that span the entire clone. Panels (C1), (C2) and (C3) show neurons, an astrocyte, and an oligodendrocyte found within the clone shown in (C). (E) Scatter plot of the number of cells for each of the 43 clones analyzed. (F) Quantification of the percentages of neurons and glia generated from E10.5 Cux2+ RGCs based upon the analysis of 43 fully reconstructed clones. (G) Quantification of the percentages of all lineage traced cells, neurons, and glia that were located in deep or upper layers. Abbreviations: CR, Cajal–Retzius cells; Ctx, cerebral cortex; DL, deep cortical layers (layers 5 and 6); Hip, hippocampus; UL, upper cortical layers (layers 2–4). Scale bars: (C, D) 500 µm, (Inserts) 250 µm.
The preceding analysis demonstrates that, in the adult brain, the Cux2-Cre allele labels cells that are present in both deep and upper cortical layers, including cells that no longer express Cux2 at adult stages. Importantly, the recombinase activity of Cux2-Cre in both progenitor cells and postmitotic neurons makes it unsuitable for lineage analysis of neocortical RGCs, because the possibility cannot be excluded that Cux2-Cre-mediated recombination was initiated in postmitotic neurons (Figure 1A). A projection neuron labeled by Cux2-Cre may have originated from a Cux2-Cre+ RGC, a Cux2-Cre+ basal progenitor, or it may have turned on Cre expression postmitotically. Accordingly, the expression of reporter alleles in Cux2-Cre mice is not reflective of the developmental origin of the cells that express the reporter (Figure 1A).
E10.5 RGCs marked by Cux2-CreERT2 generate neocortical projection neurons in both deep and upper layers and macroglia
Our previous work demonstrated that at the population level, Cux2+ RGCs are not fate-restricted to generate only corticocortical projection neurons (Guo et al., 2013). To extend our analysis we performed lineage-tracing experiments using the Cux2-CreERT2 allele, which expresses an inducible form of CRE that only induces recombination of a reporter allele for approximately 24–48 hours following tamoxifen administration. This enables labeling of Cux2+ RGCs during a short temporal window in order to track their progeny. In these experiments we crossed heterozygous Cux2-CreERT2 mice to homozygous RCE-GFP (Sousa et al., 2009) or Ai9 (Madisen et al., 2010) reporter mice. A single dose of tamoxifen was injected at E10.5, a time at which postmitotic neurons and intermediate progenitors are largely absent in the developing cortex. Brains from Cux2-CreERT2; Ai9 and Cux2-CreERT2; RCE-GFP mice were collected at P10 (TM at E10.5; P10), after cortical projection neurons have migrated to their final laminar position (Chen et al., 2008; Miyoshi and Fishell, 2012) (Figure 1B). We injected tamoxifen at 100 mg per kg of body weight, a concentration that reliably generated sparse labeling in the cortices of these mice. Whole mount examination of the brains revealed sparse columns of reporter+ cells (Figure S3A), suggesting these cells may be clonally related. Although we refer to the spatially isolated columns of labeled cells as clones, we cannot exclude the possibility that some of the clones may consist of progeny from more than one RGC. This caveat however does not change the interpretation of our analysis with regard to the lack of lineage restriction within the Cux2+ RGC lineage.
To examine the progeny of early Cux2+ RGCs we analyzed serial brain sections and reconstructed 43 clones from 6 mice (Figures 1C–1D and S3C and Table S1). We focused our analysis on spatially isolated columns of cells that predominantly consisted of projection neurons and macroglia. We excluded individual cells that were sparsely distributed throughout the cortex, as they were likely labeled interneurons. The number of cells in each clone varied from 13 to 227 with an average of 83 cells per clone (Figure 1E and Table S1). Cells in the clones spanned an average of 350 µm along the rostral-caudal axis (Table S1). The number of cells per clone and physical dispersion of the cells in the clones is similar to that reported for E10.5 RGC clones in a recent study that utilized Mosaic Analysis with Double Markers (MADM) to label RGC progeny at clonal density (Gao et al., 2014). Of the 43 reconstructed clones, 30 (70%) contained both neurons and glia, while 13 (30%) consisted of only neurons (Table S1). Strikingly, 100% of the E10.5 Cux2-CreERT2-labeled clones that we examined contained projection neurons located in cortical layers 2–6 (Figures 1C–D and S3C). The 43 reconstituted clones contained a total of 3,562 lineage-traced cells of which 90% exhibited a characteristic neuronal morphology while 10% exhibited glial morphologies (Figure 1F and Table S1). Analysis of the 3,188 lineage-traced neurons demonstrated that their distribution in deep- and upper-layers was roughly equivalent (55% upper-layer neurons versus 45% deep-layer neurons) (Figure 1G and Table S1). This distribution of lineage-traced neurons is similar to that observed in lineage tracing experiments of E10.5 RGCs using the Nestin-CreERT2 (Franco et al., 2012), Emx1-CreERT2 (Gao et. al. 2014), and Fezf2-CreERT2 alleles (Figure 3 and Table S1). These results provide strong evidence that most, if not all, individual Cux2+ RGCs are multipotent, and generate projection neurons in cortical layers 2–6 and macroglia.
Figure 3.
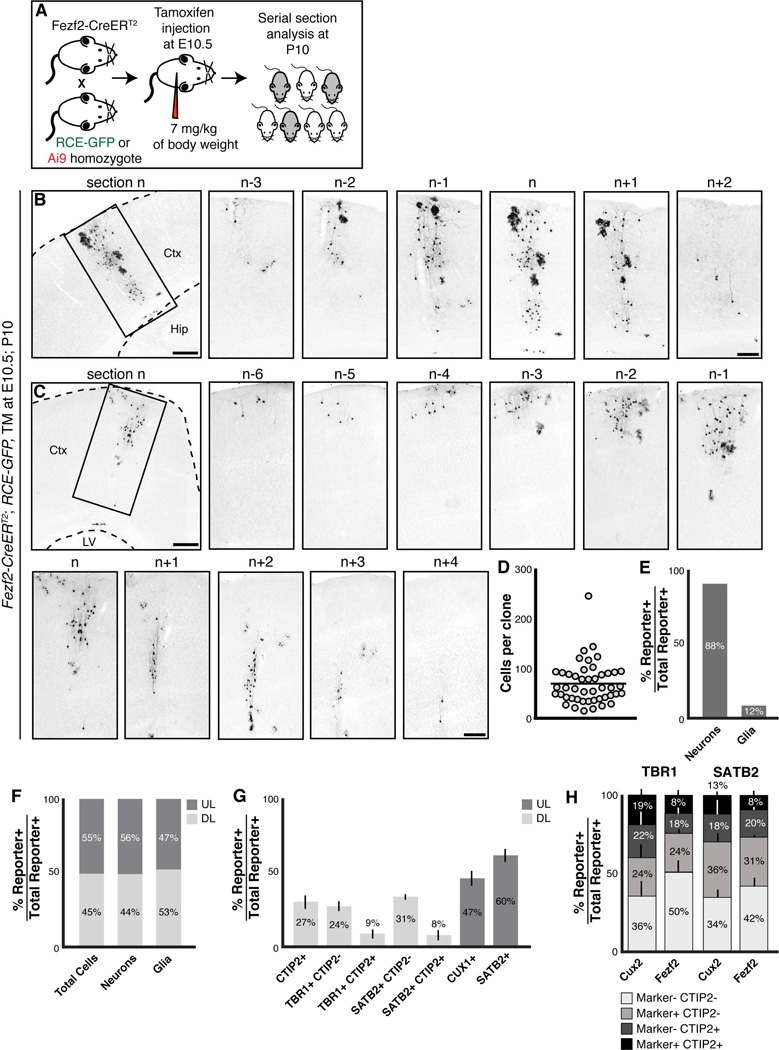
Similar to early Cux2+ RGCs, E10.5 Fezf2+ RGCs generate diverse subtypes of cortical projection neurons and glia. (A) Experimental methodology. (B–C) Two examples of clones generated from E10.5 Fezf2+ RGCs. For each clone, a lower magnification image is shown first to illustrate the position of the clone in the cerebral cortex, followed by higher maginification images of seriel sections that span the full clone. (D) Scatter plot of the number of cells for each of the 46 clones analyzed. (E) Quantification of the percentages of lineage-traced cells that were neurons or glia. (F) Quantification of the percentages of lineage-traced cells, neurons, and glia that were located in deep and upper cortical layers. (G) Percentages of lineage-traced deep-layer and upper-layer neurons that expressed the indicated markers. (H) Percentages of deep-layer neurons generated from E10.5 Cux2+ or E10.5 Fezf2+ RGCs that expressed combinations of either CTIP2/TBR1 or CTIP2/SATB2. Error bars represent standard deviation. Abbreviation: Ctx, cerebral cortex; Hip, hippocampus; LV, lateral ventricle. Scale bars: (A, B, C) 500 µm, (Inserts) 250 µm.
Molecular analysis of the Cux2 lineage
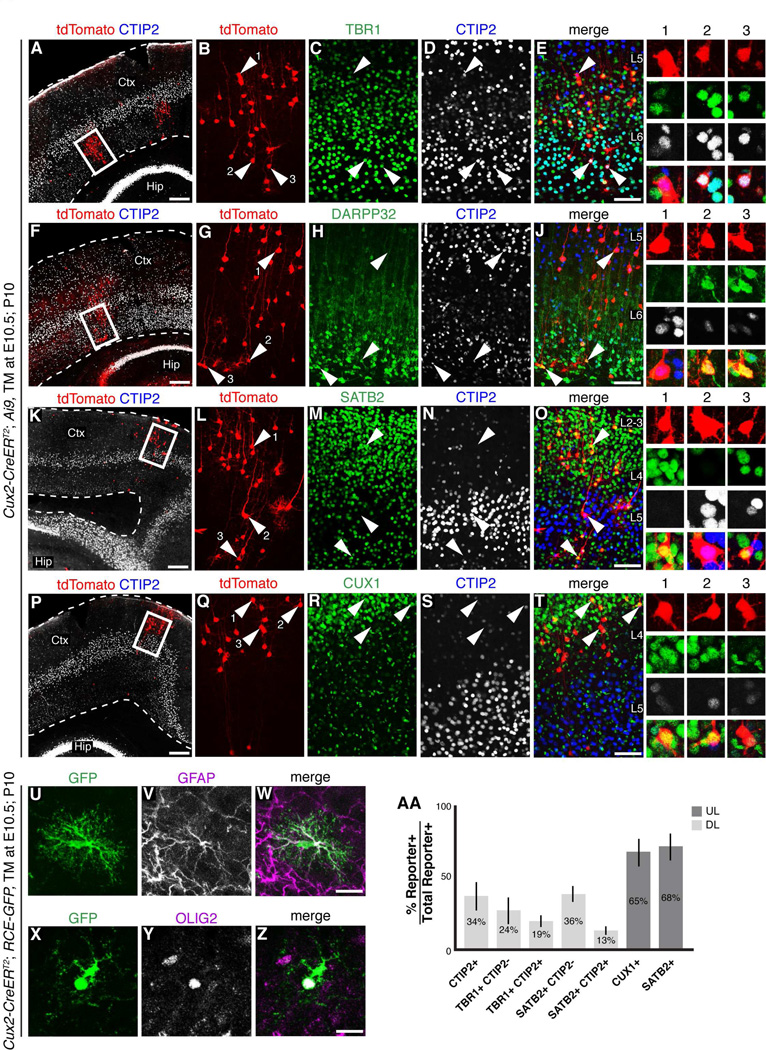
We next analyzed the molecular expression profiles of tdTomato+ and GFP+ cells in TM at E10.5; P10, Cux2-CreERT2; Ai9 and Cux2-CreERT2; RCE-GFP mice. To restrict our analysis to the progeny of neocortical Cux2+ RGCs, we focused only on projection neurons and glia that were part of putative RGC clones and excluded the sparsely distributed individual cells that were likely interneurons. We analyzed 6,258 neocortical cells from 4 animals. Among 2,583 lineage-traced deep-layer neurons, 33.9 ± 9.5% expressed the subcerebral neuron marker CTIP2 (McKenna et al., 2011; Molyneaux et al., 2005) (Figures 2A–T, 2AA). When lineage-traced, deep-layer neurons were analyzed by double labeling with antibodies for CTIP2 and TBR1, a marker for corticothalamic neurons (Bedogni et al.; Han et al.; Hevner et al., 2001; McKenna et al., 2011), 23.7 ± 8.7% expressed TBR1 alone, and 18.8 ± 3.8% expressed both TBR1 and CTIP2 (Figures 2A–E, 2AA). Similar results were observed with another corticothalamic neuron marker DARPP32 (McKenna et al., 2011), which was co-expressed in TdTomato+ or GFP+ deep-layer neurons (Figures 2F–2J). We also investigated molecular expression profiles using a combination of antibodies against CTIP2 and SATB2, another projection neuron marker (Alcamo et al., 2008; Britanova et al., 2008). Within L5 and L6 35.8 ± 5.7% of lineage-traced neurons expressed SATB2, and 13.2 ± 2.5% expressed both SATB2 and CTIP2 (Figures 2K–2O and 2AA). Thus, within deep cortical layers, projection neurons generated from E10.5 Cux2+ RGCs express four molecular markers previously associated with different projection neuron subtypes.
Figure 2.
Cux2+ RGCs generate diverse, molecularly defined subtypes of cortical projection neurons and macroglia. All images are of brain sections from P10 Cux2-CreERT2; Ai9 (A–T) or P10 Cux2-CreERT2; RCE-GFP (U–Z) mice that received a single tamoxifen injection at E10.5 (TM at E10.5; P10). Lineage traced neurons expressed TBR1 (A–E), DARPP32 (F–J), SATB2 (K–O), CUX1 (P–T), or CTIP2 (A–T). Panels (B–E), (G–J), (L–O),and (Q–T) show the higher maginification images of the boxed areas in panels (A), (F), (K) and (P) respectively. The arrowheads in (B–E), (G–J), (L–O) and (Q–T) point to cells that are shown in higher magnification to the right. (U–Z) E10.5 lineage-traced Cux2+ RGCs generated astrocytes that expressed GFAP (U–W) and oligodendrocytes that expressed OLIG2 (X–Z). (AA) Percentages of lineage-traced deep-layer and upper-layer neurons that expressed the indicated markers. Error bars represent standard deviation. Abbreviation: Ctx, cerebral cortex; Hip, hippocampus. Scale bars: (A, F, K, P) 500 µm, (E, J, O, T) 100 µm, (W, Z) 25 µm.
We examined 3,064 tdTomato+ or GFP+ neurons within the upper layers (L2–L4) of TM at E10.5; P10 Cux2-CreERT2; Ai9 or Cux2-CreERT2; RCE-GFP mice. As expected, significant numbers of these cells expressed CUX1 (64.6 ± 8.2%) (Cubelos et al., 2010) or SATB2 (68.3 ± 10.4%) (Alcamo et al., 2008; Britanova et al., 2008), two markers previously associated with upper-layer callosal projection neurons (Figures 2K–2T and 2AA). In addition, some lineage-traced cells exhibited characteristic astrocyte or oligodendrocyte morphology and expressed GFAP or OLIG2 respectively (Figures 2U–2Z). Thus, results from molecular expression analyses further strengthen the evidence that early Cux2+ RGCs are not lineage-restricted to generate only upper-layer or corticocortical projection neurons. Rather, these data support the conclusion that Cux2+ RGCs generate a diversity of projection neuron subtypes and macroglia that reside within both deep and upper cortical layers.
Cux2+ RGCs and Fezf2+ RGCs generate similar progeny
We previously demonstrated that early Fezf2+ RGCs are multipotent and sequentially generate projection neurons and macroglia (Guo et al., 2013). To extend our clonal analysis of Fezf2+ RGCs, we bred Fezf2-CreERT2 animals to RCE-GFP or Ai9 reporter mice, administered a single low dose of tamoxifen at E10.5, and analyzed brains at P10 (Figure 3A). We reconstituted 46 clones from 3 animals (Figures 3B–3C, S3D and Table S1). The number of cells in each clone varied from 13 to 242 with an average of 67 cells per clone, and the clones spanned an average of 300 µm along the rostral-caudal axis (Figure 3D and Table S1). Twenty-eight percent of the clones contained only neurons, and 72% contained both neurons and glia. Similar to E10.5 Cux2+ RGCs, 100% of Fezf2+ RGC clones contained neurons located throughout cortical layers 2–6 (Figures 3B–3C and S3D). We quantified the distribution of the 3,100 lineage-traced cells in the 46 E10.5 Fezf2 clones and found that, comparable to results from clonal analysis using the Cux2-CreERT2 allele, Fezf2+ RGCs generated projection neurons (88%) and glia (12%) (Figure 3E and Table S1). Lineage-traced neurons were located in both deep (44%) and upper layers (56%) (Figures 3B–3C and 3F, and Table S1).
We examined 2,328 deep-layer lineage-traced neurons from 3 brains which consisted of both fully and partially reconstructed clones and found that 26.8 ± 5.0% of these neurons expressed CTIP2 (Figure 3G). When lineage-traced neurons were analyzed using antibodies for CTIP2 and TBR1, 23.5 ± 3.0% of deep-layer neurons expressed TBR1 alone, and 8.8 ± 2.9% expressed both CTIP2 and TBR1 (Figure 3G). We also analyzed reporter+ neurons in L5 and L6 using a combination of CTIP2 and SATB2 antibodies and found that 30.8 ± 1.4% expressed SATB2 alone, and 7.6 ± 3.3% expressed both SATB2 and CTIP2 (Figure 3G). Within upper cortical layers, lineage-traced neurons expressed CUX1 (47.1 ± 5.9%) or SATB2 (59.7 ± 4.7%) (Figure 3G). Notably, the percentage of lineage-traced neurons that expressed each of these markers is similar to that observed with TM at E10.5; P10 Cux2-CreERT2 mice (Figures 2AA, 3G and 3H). Collectively, our clonal and molecular analyses suggest that both Fezf2+ RGCs and Cux2+ RGCs are multipotent, and generate multiple subtypes of projection neurons and macroglia.
Retrograde tracing confirms that Cux2+ RGCs generate subcerebral projection neurons
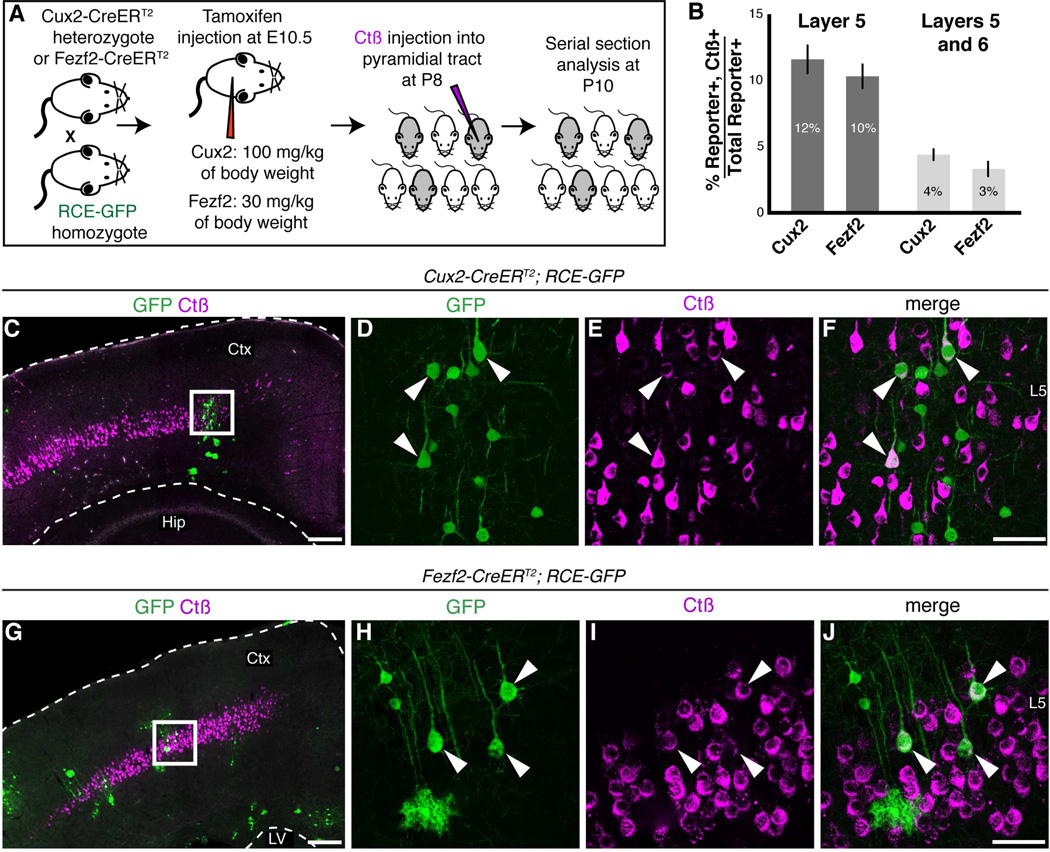
Thus far we have provided evidence that Cux2+ RGCs generate neurons located in deep cortical layers that also express molecular markers for subcerebral projection neurons. To confirm that these neurons extend axons to subcerebral targets, we combined Cre-dependent lineage analysis with retrograde tracing (Figure 4A). We bred heterozygous Cux2-CreERT2 mice to homozygous RCE-GFP reporter mice, and administered a single dose of tamoxifen at E10.5 (100 mg/kg), to mark the progeny of Cux2+ RGCs. We then injected cholera toxin beta (Ctβ) into the pyramidal tract at P8 (Figures 4A and S3B).
Figure 4.
Retrograde labeling of the Cux2+ and Fezf2+ lineages. (A) Experimental setup. (B) Precentages of GFP+ cells in layer 5 or in layers 5 and 6 that are also Ctβ positive. Error bars represent standard deviation (n=2 brains for Cux2 and 6 brains for Fezf2). (C–F) Example of brain section from Cux2-CreERT2; RCE-GFP mice that received TM at E10.5 and were injected with Ctβ at P8. Note the dense band of Ctβ labeling in layer 5. Panels (D–F) show an enlarged view of boxed region in (C). Arrowheads indicate GFP+ cells originating from a Cux2+ RGC that extend subcerebral projections. (G–J) Example of brain section from Fezf2-CreERT2; RCE-GFP mouse that received TM at E10.5 and was injected with Ctβ at P8. (H–J) Enlarged view of boxed regions shown in G. Arrowheads indicate GFP; Ctβ double positive cells. Abbreviation: Ctx, cerebral cortex; Hip, hippocampus; LV, lateral ventricle. Scale bars: (C, G) 500 µm, (F, J) 100 µm.
Ctβ injection into the pyramidal tract labeled a subset of subcerebral projection neurons located in layer 5 (Figure 4C). Notably, we observed GFP; Ctβ double positive cells in Cux2-CreERT2; RCE-GFP mice, indicating that these subcerebral neurons were generated from Cux2+ RGCs (Figure 4D–F). We quantified the percentage of GFP+ cells derived from Cux2+ RGCs that projected subcerebral axons in Cux2-CreERT2; RCE-GFP brains. We found that 11.5 ± 1.1% of GFP+ cells in layer 5 and 4.4 ± 0.4% of GFP+ cells in layers 5 and 6 were retrogradely traced (Figure 4B).
We performed a similar analysis using the Fezf2-CreERT2 allele (Figures 4A, 4G–J), which was previously shown to label RGCs that generate neurons which project axons to the corpus callosum, thalamus and spinal cord (Guo et al., 2013). Strikingly, both Fezf2-CreERT2; RCE-GFP and Cux2-CreERT2; RCE-GFP brains contained similar percentages of GFP; Ctβ double positive cells (10.3 ± 0.9% L5 and 3.3 ± 0.6% L5 and L6 for Fezf2-CreERT2 allele) (Figure 4B). These results confirm that both Cux2+ and Fezf2+ RGCs produce subcerebral projection neurons, and demonstrate that early Cux2+ RGCs are not lineage restricted to generate only corticocortical projection neurons.
Discussion
Our previous study demonstrated that at both the clonal and population levels Fezf2+ RGCs are multipotent, and that at the population level Cux2+ RGCs are multipotent (Guo et al., 2013). This result suggested that either all Cux2+ RGCs are multipotent or, alternatively, Cux2+ RGCs might represent a heterogeneous population with different lineage potentials. Herein, our lineage analysis of individual Cux2+ RGCs provides evidence that most, if not all, Cux2+ RGCs are multipotent and generate diverse projection neuron subtypes located in cortical layers 2–6. Further, we found that the laminar distribution and molecular expression profile of neurons generated from early Cux2+ RGCs was similar to that of neurons generated from early Fezf2+ RGCs (Figure 3), Nestin-CreERT2+ RGCs (Franco et al., 2012), and Emx1-CreERT2+ RGCs (Gao et al., 2014).
Using the Cux2-Cre and Cux2-CreERT2 alleles, Franco et al. (2012) reported that Cux2+ RGCs generated only neurons, and not glia. They also reported that among the progeny generated by E10.5 Cux2-CreERT2-labeled RGCs, 90% of labeled cells were located in upper cortical layers while 10% were in deep layers. These data are significantly different from our previous and current results. By using two Cre reporter lines, RCE-GFP and Ai9, analyzing 6,258 lineage-traced cells, and reconstructing 43 RGC clones, we observed that at P10 roughly 10% of cells generated from E10.5 Cux2+ RGCs were astrocytes or oligodendrocytes. Moreover we found that every E10.5 RGC clone we examined contained neurons located in both deep and upper cortical layers, and that there was not a strong bias of the labeled cells to occupy upper cortical layers.
Both the current study and Gil-Sanz et al. (2015) demonstrate that Cux2+ RGCs generate a greater percentage of SATB2+ than CTIP2+ neurons. However, Gil-Sanz et al. (2015) interpret the prevalence of SATB2+ neurons labeled by the Cux2-Cre and Cux2-CreERT2 alleles as evidence that Cux2+ RGCs are lineage-restricted. SATB2 is expressed in a greater number of neocortical cells compared to CTIP2 (Figure S4; see also Figure 3E of Gil-Sanz et al. (2015)). Thus it is not surprising that a multipotent RGC, which generates diverse subtypes of projection neurons, can produce a higher percentage of SATB2+ than CTIP2+ progeny. This highlights the need to define projection neuron subtype identity using multiple parameters, including additional molecular markers and axonal projections. Towards this, we found that a similar percentage of neurons with subcerebral projections were born from E10.5 Cux2+ RGCs and E10.5 Fezf2+ RGCs (Figure 4).
Gil-Sanz et al. (2015) attribute the difference in results between experiments preformed in the Müller laboratory (Franco et al., 2012; Gil-Sanz et al., 2015) and our work (Guo et al., 2013 and current study) to breeding strategy and genetic background. In particular, the authors propose that the low recombination efficiency we observe in Cux2-CreERT2 mice at E10.5, using a TM dosage of 100 mg/kg of body weight, is an artifact of genetic background. Furthermore, they propose that our lineage analysis results with the Cux2-CreERT2 allele are highly influenced by genetic background. Gil-Sanz et al. (2015) report that the Cux2-CreERT2 allele previously induced more widespread recombination at a TM dosage of 50 mg/kg than in our studies. We disagree; our interpretation of Supplemental Figure 2 from Franco et al. (2012) is that sparse recombination was prevalent at this dosage, similar to results from our laboratory. Moreover, in Gil-Sanz et al. (2015) the authors injected 4-hydroxytamoxifen (4-OHT) at a dosage of 50 mg/kg of body weight (or 1 mg/20 g), to achieve sparse labeling of E10.5 Cux2+ RGCs. However, 4-OHT has a much greater affinity (50–100×) than TM for binding estrogen receptors (Malet et al., 1988), and is significantly more potent at inducing recombination (Hans et al., 2009). Thus the recombination efficiency of the Cux2-CreERT2 allele in studies from the Müller laboratory does not appear to be significantly higher than that observed in our laboratory (compare Figures 1 and 2 in the current study with Figures 3 and S3D–S3I of Gil-Sanz et al., 2015)).
One explanation for the divergent conclusions reached by our work and that of Franco et al. (2012) and Gil-Sanz et al. (2015) is that the latter two studies relied heavily on Cux2-Cre mice, which are not suitable for lineage analysis of neocortical RGCs. Here we present evidence that explains why the lineage analysis of neocortical RGCs is confounded by the complex temporal and spatial expression pattern of the Cux2-Cre allele. The recombinase activity of CRE driven from the Cux2 locus is constitutively active and thus recombination can occur in RGCs, basal progenitor cells, or postmitotic neurons. Accordingly, a reporter+ neuron in the cortical plate may not have originated from a Cux2+ RGC. Within the neocortex, Cux2 is preferentially expressed in upper cortical layers. Therefore, a higher percentage of reporter-expressing cells within upper versus deep cortical layers in a small subset of cortical areas (e.g. the anterior cingulate area) in Cux2-Cre mice is not unexpected based upon the expression pattern of Cux2, and does not indicate that Cux2+ RGCs preferentially generate upper-layer neurons.
Recently, Gao et al. (2014) performed lineage analyses of individual RGCs using MADM. Through low-density labeling of RGCs with the Emx1-CreERT2 and Nestin-CreERT2 alleles, they reported that 100% of clones generated from RGCs labeled at E10.5 contained neurons that occupied cortical layers 2–6. Notably, despite the differences in recombination efficiency between the MADM strategy and our current study, the average numbers of cells per clone, and rostral-caudal dispersion of cells in the clones are similar between the two studies. Moreover, by analyzing subtype-specific markers, Gao et al. (2014) found that individual clones contained cells expressing markers for multiple subclasses of neocortical projection neurons. Combined with the current study, this work provides important evidence for the prevalence of multipotent RGCs within the early neocortical progenitor pool.
Collectively, our results demonstrate that early Cux2+ and Fezf2+ RGCs are multipotent neocortical progenitors. However, these findings do not exclude the existence of fate-restricted RGCs during early cortical development. Rather, they indicate that neither Fezf2 nor Cux2 expression alone is sufficient to identify these cells, and that appropriate molecular or genetic markers await future investigation.
Experimental Procedures
All experiments were carried out in accordance with the protocols approved by the IACUC at University of California at Santa Cruz and institutional and federal guidelines. The day of vaginal plug detection was designated as E0.5. The day of birth was designated as P0. Immunohistochemistry was performed as previously described (Eckler et al., 2011). Extended experimental details are included in Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Individual E10.5 Cux2+ RGCs generate both deep- and upper-layer neurons and glia.
Cux2+ RGCs generate diverse subtypes of molecularly defined projection neurons.
Neurons generated by E10.5 Cux2+ RGCs project subcerebral axons.
Acknowledgements
We thank Dr. Arturo Alvarez-Buylla for advice on experiments. We thank Drs. Chris Doe, Songhai Shi, Susan McConnell, Pushkar Joshi, and Jake Kirkland for critical reading and advice on the manuscript. We thank Dr. Ben Abrams and the UCSC Microscopy Center for help with image acquisition, and Dr. Tzvia Abramson (San Jose State University) for her support of this research. This work was funded by grant R01MH094589 from the National Institutes of Health (to B.C.), New Faculty Award RN1-00530-1 from the California Institute of Regenerative Medicine (CIRM; to B.C.), Bridges Grant TB1-01195 from CIRM to San Jose State University (to T.D.N.), Training Grant TG2-001157 from CIRM (to W.L.M. and C.G.) and grants R01MH049428 and R01NS34661 from the National Institutes of Health (to J.L.R.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
References
- Alcamo EA, Chirivella L, Dautzenberg M, Dobreva G, Farinas I, Grosschedl R, McConnell SK. Satb2 regulates callosal projection neuron identity in the developing cerebral cortex. Neuron. 2008;57:364–377. doi: 10.1016/j.neuron.2007.12.012. [DOI] [PubMed] [Google Scholar]
- Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, Beyer RP, Bammler TK, Rubenstein JL, Hevner RF. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci U S A. 107:13129–13134. doi: 10.1073/pnas.1002285107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britanova O, de Juan Romero C, Cheung A, Kwan KY, Schwark M, Gyorgy A, Vogel T, Akopov S, Mitkovski M, Agoston D, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57:378–392. doi: 10.1016/j.neuron.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Chen B, Wang SS, Hattox AM, Rayburn H, Nelson SB, McConnell SK. The Fezf2-Ctip2 genetic pathway regulates the fate choice of subcortical projection neurons in the developing cerebral cortex. Proc Natl Acad Sci U S A. 2008;105:11382–11387. doi: 10.1073/pnas.0804918105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Sebastian-Serrano A, Beccari L, Calcagnotto ME, Cisneros E, Kim S, Dopazo A, Alvarez-Dolado M, Redondo JM, Bovolenta P, et al. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. 2010;66:523–535. doi: 10.1016/j.neuron.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckler MJ, McKenna WL, Taghvaei S, McConnell SK, Chen B. Fezf1 and Fezf2 are required for olfactory development and sensory neuron identity. J Comp Neurol. 2011;519:1829–1846. doi: 10.1002/cne.22596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco SJ, Gil-Sanz C, Martinez-Garay I, Espinosa A, Harkins-Perry SR, Ramos C, Muller U. Fate-restricted neural progenitors in the mammalian cerebral cortex. Science. 2012;337:746–749. doi: 10.1126/science.1223616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Postiglione MP, Krieger TG, Hernandez L, Wang C, Han Z, Streicher C, Papusheva E, Insolera R, Chugh K, et al. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C, Eckler MJ, McKenna WL, McKinsey GL, Rubenstein JL, Chen B. Fezf2 expression identifies a multipotent progenitor for neocortical projection neurons, astrocytes, and oligodendrocytes. Neuron. 2013;80:1167–1174. doi: 10.1016/j.neuron.2013.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Kwan KY, Shim S, Lam MM, Shin Y, Xu X, Zhu Y, Li M, Sestan N. TBR1 directly represses Fezf2 to control the laminar origin and development of the corticospinal tract. Proc Natl Acad Sci U S A. 2011;108:3041–3046. doi: 10.1073/pnas.1016723108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans S, Kaslin J, Freudenreich D, Brand M. Temporally-controlled site-specific recombination in zebrafish. PLoS One. 2009;4:e4640. doi: 10.1371/journal.pone.0004640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, Bulfone A, Goffinet AM, Campagnoni AT, Rubenstein JL. Tbr1 regulates differentiation of the preplate and layer 6. Neuron. 2001;29:353–366. doi: 10.1016/s0896-6273(01)00211-2. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malet C, Gompel A, Spritzer P, Bricout N, Yaneva H, Mowszowicz I, Kuttenn F, Mauvais-Jarvis P. Tamoxifen and hydroxytamoxifen isomers versus estradiol effects on normal human breast cells in culture. Cancer Res. 1988;48:7193–7199. [PubMed] [Google Scholar]
- McKenna WL, Betancourt J, Larkin KA, Abrams B, Guo C, Rubenstein JL, Chen B. Tbr1 and Fezf2 regulate alternate corticofugal neuronal identities during neocortical development. J Neurosci. 2011;31:549–564. doi: 10.1523/JNEUROSCI.4131-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi G, Fishell G. Dynamic FoxG1 expression coordinates the integration of multipolar pyramidal neuron precursors into the cortical plate. Neuron. 2012;74:1045–1058. doi: 10.1016/j.neuron.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- Sousa VH, Miyoshi G, Hjerling-Leffler J, Karayannis T, Fishell G. Characterization of Nkx6-2-derived neocortical interneuron lineages. Cereb Cortex. 2009;19(Suppl 1):i1–i10. doi: 10.1093/cercor/bhp038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.