Abstract
In contrast to the wealth of structural data available for the mature p66/p51 heterodimeric human immunodeficiency virus type 1 reverse transcriptase (RT), the structure of the homodimeric p66 precursor remains unknown. In all X-ray structures of mature RT, free or complexed, the processing site in the p66 subunit, for generating the p51 subunit, is sequestered into a β-strand within the folded ribonuclease H (RNH) domain and is not readily accessible to proteolysis, rendering it difficult to propose a simple and straightforward mechanism of the maturation step. Here, we investigated, by solution NMR, the conformation of the RT p66 homodimer. Our data demonstrate that the RNH and Thumb domains in the p66 homodimer are folded and possess conformations very similar to those in mature RT. This finding suggests that maturation models which invoke a complete or predominantly unfolded RNH domain are unlikely. The present study lays the foundation for further in-depth mechanistic investigations at the atomic level.
Keywords: NMR, enzyme, protein, structure, proteolysis, maturation, dynamics
INTRODUCTION
Reverse transcriptase (RT) plays a central role in the replication of all retroviruses and related retrotransposons.1–5 In the human immunodeficiency virus type 1 (HIV-1) life cycle, RT is expressed as part of the Gag-Pol polypeptide, which is processed by retroviral protease into several proteins. The mature RT enzyme is a heterodimer, composed of two subunits, p66 and p51 (Fig. 1A).6–9 The RT p51 subunit is generated by removal of the C-terminal RNase H (RNH) domain from p66.10–15 In vivo processing of the Gag-Pol polyprotein is complex, and the detailed mechanism of RT maturation into the heterodimer is still unclear. Based on data obtained from model systems, cleavage at the p51-RNH processing site is assumed to occur in a p66 homodimer.15–20 However, in all known RT X-ray structures, as well as those of the isolated RNH domain, the p51-RNH cleavage site is located within the folded RNH domain, sequestered into the center of a β-sheet, and thus seemingly inaccessible to the protease (Fig. 1B).16, 21–25 No significant motions were observed at the p51-RNH processing site in the isolated RNH domain,26, 27 which may have suggested partial accessibility of the site. In addition, the lack of structural information on the “immature” p66 RT precursor renders any mechanistic explanation(s) tentative. We, therefore, embarked on studies aimed at providing the foundation for structurally elucidating RT processing.
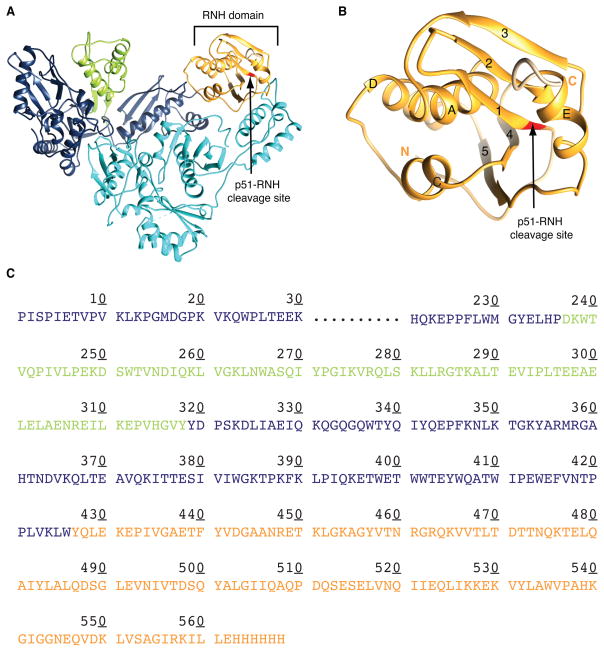
Figure 1.
Ribbon representation of the structures of (A) p66/p51 RT heterodimer and (B) the RNH domain, indicating the p51-RNH processing site (arrow), and (C) amino acid sequence of p66. In (A)-(C), the Thumb and RNH domains in the p66 subunit are shown in green and orange, respectively; the p51 subunit in cyan. Structures in (A) and (B) were drawn using PDB:1DLO.
To evaluate protein conformation in solution, Nuclear Magnetic Resonance (NMR) spectroscopy provides powerful approaches,28–30 since it permits the investigation of conformational equilibria and protein dynamics at the amino acid residue level.31–33 However, NMR studies of HIV RT are challenging, given the protein’s large molecular mass (117 kDa); to date, NMR of RT has been mostly limited to observing methyl groups of side chains, such as in methionine or isoleucine.34–36 Although methyl resonances are valuable probes for obtaining general qualitative information about a protein’s conformation in solution,37, 38 they report only on a limited number of positions and, therefore, cannot inform on the secondary and tertiary structural details that are mirrored by a protein’s backbone chemical shifts.39
Here, we present an investigation of the p66 homodimeric RT. The p66 dimer possesses enzymatic activity40–42 and is widely considered to function as the RT precursor.15–20, 43 We took advantage of the extreme sensitivity of backbone amide resonance frequencies to assess conformational similarities between different protein constructs. In the 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum of the p66 homodimer, over 240 resonances were observed. Comparison of the p66 spectrum with the spectra of the isolated domains revealed that greater than 60% of the isolated Thumb domain and more than 40% of the isolated RNH domain resonances, respectively, are in very similar positions. In contrast, only18% of the Finger-Palm domain resonances match those of the p66 homodimer. This establishes that both the Thumb and RNH domains are stably folded in the immature p66 homodimeric RT and exhibit essentially the same structures as in the isolated domains. With these findings in mind, the question arises how HIV-1 protease gains access to the p51-RNH processing site in the p66 homodimer. Our data suggest that maturation models, which invoke a complete unfolded or predominantly unfolded RNH domain17, 21, 22 are unlikely, and suggest that p51-RNH processing may involve selection of a minor conformation or a protease-binding induced structure, which is cleaved during maturation.
MATERIALS AND METHODS
Sample preparation
The coding sequence for the RT p66 subunit was amplified from the p6HRT-PROT vector, kindly provided by Dr. Sluis-Cremer, using 5′-acc gca cat atg ccc att agc cct att gag act gta-3′ and 5′-gca gat ctc gag tag tat ttt cct gat tcc agc act gac-3′ as forward and backward primers, respectively. The amplified product was inserted into the pET21a(+) vector (Invitrogen, Carlsbad, CA), which encodes a six histidine tag at the C-terminus of the protein construct. After initial expression and purification trials, a codon-optimized C280S/C38V double cysteine version was created for increased protein expression in E. coli (DNA 2.0 gene synthesis, Menlo Park, CA). The coding sequence for the p51 subunit with an N-terminal Strep-tag was created by amplification of the appropriate region (coding for residues 1-440) of the p66 RT codon optimized sequence, using 5′-cc gca tcc atg gat tgg agt cac ccg cag ttc gag aaa cca atc agc cca atc gaa acg gtc cc -3′ and 5′-ccg cat ctc gag tta gaa cgt ttc cgc gcc aac aat cgg ttc ttt ctc c-3′ as forward and reverse primers, respectively, and the amplified product was then inserted into a pET28a+ vector (Invitrogen). Constructs coding for residues 1-216 (Finger-Palm domain), 237-318 (Thumb domain) and 427-556 (RNH domain) were amplified from the p66 sequence using 5′-gca gct cat atg cca atc agc cca atc gaa acg gtc cc-3′ and 5′-gca gat ctc gag ggt cgt cag acc cca acg cag cag atg c-3′ as forward and backward primers, respectively, for the finger-palm domain, 5′-cgt acg cat atg gat aaa tgg aca gta cag cct ata gtg ctg cc-3′ and 5′-cgt acg ctc gag ata cac tcc atg tac tgg ttc ttt tag aat ctc-3′ as forward and backward primers, respectively, for the thumb domain, and 5′-gca gat cat atg tat caa ctg gag aaa gaa ccg att gtt ggc-3′ and 5′-gca gat ctc gag cag gat ttt gcg aat acc tgc gct cac c-3′ as the forward and backward primers, respectively, for the RNH domain. Each amplified product was inserted into a pET21a(+) vector, which resulted in a hexa-histidine tag at the C-terminus of each protein construct. The sequences of all constructs were verified by DNA sequencing, using primers for the T7 promoter and terminator (Genewiz, South Plainfield, NJ).
All proteins were produced in E. coli BL21 (DE3) gold cells (Agilent Technologies, Santa Clara, CA), using 0.5 mM IPTG for induction over 16 hours at 27°C. Uniform 15N- labeling of the isolated Finger-Palm, Thumb, and RNH domains was achieved using modified minimal media containing 15NH4Cl as the nitrogen source. Uniform 15N- and 13C- labeling of the Thumb and RNH domains, for assignment purposes, was carried out, using 15NH4Cl and 13C6-glucose as sole nitrogen and carbon sources, respectively. Uniform 15N- and 2H- labeling of the p66 and the p51 protein was achieved in modified minimum medium containing 2H2O and 15NH4Cl. Cells were harvested by centrifugation at 5887 g, re-suspended in lysis buffer containing 25 mM sodium phosphate (pH 7.5), 25 mM imidazole and 500 mM NaCl, and lysed using a microfluidizer. Cell debris was removed by centrifugation at 34530 g and the supernatant was applied to a 5 mL HisTrap (GE healthcare Life Sciences, Piscataway, NJ) column, equilibrated in lysis buffer. Thumb, RNH, and p66 proteins were eluted using a linear gradient of 0.025 – 0.5 M imidazole. Protein-containing fractions were further purified over a 5 mL HiTrap SP column (GE healthcare), equilibrated with 25 mM sodium phosphate (pH 6.5) and eluted using a linear gradient of 0 – 0.5 M NaCl. Final purification of the p66 protein involved separation on a HiLoad 26/60 Superdex 200 gel filtration column (GE Healthcare), in phosphate-buffered saline.
For the p51 protein, which contained and N-terminal Strep-tag, the supernatant after cell lysis and centrifugation was loaded onto a 5 mL HisTrap column. The flow through was collected and applied to a 5 mL StrepTrap column equilibrated with 25 mM sodium phosphate (pH 7.5), 6 mM KCl, 280 mM NaCl, and eluted in equilibration buffer that contained 3 mM d-Desthiobiotin (Sigma-Aldrich, St. Louis, MO). Protein containing fractions were further purified over a 5 mL HiTrap SP (GE healthcare) column, equilibrated with 25 mM sodium phosphate (pH 6.5), and eluted using a linear gradient of 0–0.5 M NaCl. Protein fractions were pooled and concentrated in an Amicon Ultra concentrator (EMD Millipore, Billerica, MA) to ~10 μM. 50% v/v glycerol was added and samples were stored at −80 °C.
NMR experiments
Samples were buffer-exchanged into NMR buffer (25 mM sodium phosphate, pH 6.8, 100 mM NaCl) in an Amicon Ultra concentrator (EMD Millipore). In the final exchange step, the sample was concentrated to 350 μL and supplemented with 10% (v/v) D2O. Final protein concentrations for NMR experiments were ~200 μM (in monomer). All NMR spectra were recorded at 303 K on Bruker 600 and 900 MHz AVANCE spectrometers, equipped with 5-mm triple-resonance, z-axis gradient cryoprobes. For p66 and p51 proteins, a TROSY version the 1H-15N HSQC NMR experiments was used.44, 45 For backbone assignments of the Thumb and RNH domains 3D CBCAONH, HNCACB, and 1H-15N NOESY-HSQC 46–49 spectra were recoded. All data were processed with NMRPipe and analyzed with CCPN.50, 51 Chemical shifts of amide resonances, as defined by Eq. (1), were deemed identical if the picked peaks in two spectra resided within ± 0.03 ppm.,
| (1) |
Here, δ indicates chemical shift in ppm units, and γ indicates gyromagnetic ratios of N and H resonances, respectively. Secondary structure elements were delineated using CSI and TALOS+.39, 52, 53 Chemical shifts for the Thumb domain spectrum were calculated using the coordinates from the crystal structure for residues 237 to 318 of p66 in RT p66/p51 (PDB ID: 1DLO), with the program Sparta.54
Multi-angle Light Scattering
Size-exclusion chromatography/multi-angle light scattering (SEC-MALS) data were obtained at room temperature using an analytical Superdex 200 (10 × 300 mm, GE Healthcare) column with in-line multi-angle light scattering, refractive index (Wyatt Technology, Inc., Santa Barbara, CA) and UV (Agilent Technologies) detectors. 100 μL of a protein solution (10.9 – 59.9 μM for p66 and 19.3 – 58.6 μM for p51) was loaded onto the column, pre-equilibrated and eluted with NMR buffer at a flow rate of 0.5 mL/min. The molecular masses of the eluted protein species were determined using the ASTRA V.5.3.4 program (Wyatt Technologies). The dimer association constant for p66/p66 was extracted from peak intensities at the monomer and dimer molecular masses.
RESULTS
Amide backbone resonances of homodimeric p66
To obtain insight into the conformation of the immature p66 homodimeric RT in solution, 1H-15N TROSY HSQC spectra of perdeuterated p66 were recorded (Fig. 2A). Excluding resonances that likely arise from amino acid side chains, at least 247 resonances were resolved. Although this number is fewer than half of all possible amide resonances (~600), it is still remarkable that such a large number is observed, given the large molecular mass of the p66 homodimer and concomitant line broadening in TROSY HSQC spectra.38, 55 Note that the p66 sample is predominantly a homodimer (>80%) at the concentration used (~200 μM), based on multi-angle light scattering (MALS) analysis (Fig. S1) and a previously reported dissociation constant, KD, of 4 μM.12, 56, 57
Figure 2.
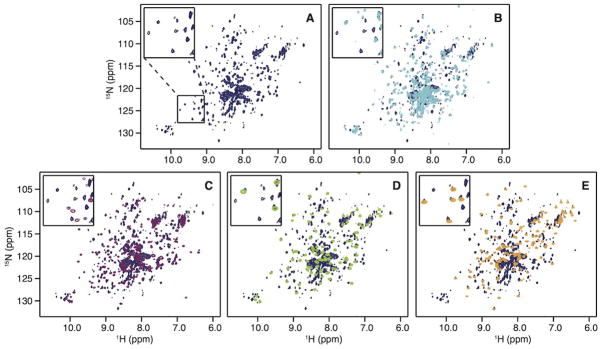
1H-15N HSQC spectra of (A) p66 (dark blue), (B) p51 (cyan), (C) Palm-Finger (purple), (D) Thumb (green), and (E) RNH (orange). In (B)-(E), individual isolated domain spectra are superimposed on the p66 spectrum (dark blue). The insets depict expanded regions of the spectra as indicated in panel (A).
For comparison, a 1H-15N TROSY HSQC spectrum was also recorded for the p51 sample (Fig. 2B), which contained less than 60% dimer, based on MALS analysis (Fig. S1) and a reported KD of 230 μM.12, 56, 57 Given that both our p66 and p51 NMR samples contain monomeric and dimeric protein, but give rise to only one set of resonances, the monomer and dimer forms are likely in fast exchange on the chemical shift scale. For p51, with a KD ≫1 μM and an estimated on-rate >106 s−1/M,58, 59 this is not surprising. For p66, with a KD ~4 μM, the observed chemical shift positions are most likely those of the major dimer species (>80%), irrespective of the exchange régime.
Despite the different proportion of dimerization in the p66 and p51 samples, a significant number of amide resonances exhibit the same resonance frequencies in the p51 and p66 NMR spectra; this is easily seen upon superposition of the spectra (Fig. 2B and Table I). Since NMR chemical shifts are extremely sensitive to local environments, those associated with residues that experience a different environment in two states, for example when located at domain interfaces in multimers, are expected not to be the same. Thus, the observed high similarity of the spectra, irrespective of the difference in dimer population, suggests that the observed signals represent residues that reside in identical local structures of p51 and p66. Interestingly, the fact that we observe a large number of resonances in the 1H-15N TROSY HSQC spectrum for the p66 homodimer, despite its large molecular mass, suggests that maybe some of the smaller domains exhibit a certain degree of independent motion, faster than the overall rotational molecular diffusion.
Table I.
Percentage of amide resonances in the spectra of individual isolated domains that reside at identical frequencies in the p66 or p51 spectra
| Isolated domains | p51 | RNH | Thumb | Finger-Palm |
|---|---|---|---|---|
| Total number of counted resonances in each spectrum | 219 | 119 | 83 | 179 |
|
| ||||
| Percentage of signals in the individual domain spectra that coincide with those in p66 | 62% | 40% | 59% | 18% |
| Percentage of signals in the individual domain spectra that coincide with those in p51 | - | 18% | 55% | 18% |
Resonances were assumed to be identical if they resonated within 0.03 ppm (see the method).
The RNH and Thumb domains are independently folded domains in the p66 homodimer
The RNH domain of HIV-RT can exist as a stably folded, individual domain with a structure similar to that in the intact protein.21, 25, 26 It is not known, however, whether other RT domains can also exist as stable folded sub-structures. We, therefore, prepared the Finger-Palm domain (residues1 to 216), the Thumb domain (residues 237 to 318) and the RNH domain (residues 427 to 556, see Fig. 1C). The 1H-15N HSQC spectra for all three isolated domains exhibit well-dispersed resonances, indicative of stably folded structures (Fig. 2C–2E). To qualitatively assess whether the domain structures within p66 and in isolation are identical, the isolated domain spectra were individually superimposed onto the spectrum of p66 (Fig. 2C–2E). Amide resonances that were considered identical in frequencies (0.03 ppm in the combined chemical shift in Eq. (1)) were counted: of the three domains, the isolated Thumb and RNH domains exhibited large degrees of identity with the intact p66 protein (>40%), while for the Finger-Palm domain it was low (18%, Table I). When comparing the spectra of the individual domains with their counterparts in p51, a high degree of resonance frequency identity was observed for the Thumb domain (55%). We also evaluated whether, by chance, any spectral similarity exists between the structurally diverse RNH and Thumb domains; gratifyingly no overlap was noted, as can be appreciated from the superposition of their spectra onto the p66 spectrum (compare the orange and green resonances in Fig. 3A). Likewise, p51 and RNH domain resonances do not coincide (18% identity, compare the cyan and orange resonances), whereas significant overlap occurs for p51 and p66 (compare the cyan and dark blue resonances in Fig. 3B). Counting these coinciding cross-peaks in the p51 and RNH spectra provides a rough estimate for the percentage of amide resonances that coincidentally reside at the same 15N and 1H frequencies, amounting to < 20%.
Figure 3.
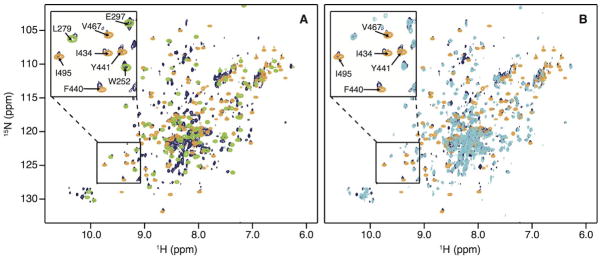
Superposition of 1H-15N HSQC correlation (A) of the Thumb domain (green) and of the RNH domain (orange) onto the p66 spectrum (dark blue) and (B) of the p51 domain (cyan) and the RNH domain (orange) onto the p66 spectrum (dark blue). Several resonances are labeled with amino acid names and numbers in the insets.
Since no structure is available for the isolated Thumb domain and it was not clear a priori whether the isolated domain has the same conformation as the Thumb domain in the p66 homodimer, we used NMR to investigate this question. Backbone assignments were obtained for a sample of the isolated domain and qualitatively assessed using CSI and TALOS+.39, 52, 53 The derived secondary structure elements (α-helices) are consistent with an overall domain structure similar to the one observed in the p66/p51 RT crystal. Likewise, Cα chemical shifts that were predicted using Sparta,54 based on the coordinates of the Thumb domain in the RT heterodimer crystal structure (PDB ID,1DLO), match the experimental ones of the isolated Thumb domain (Fig. S2). The similarity in secondary structure elements, and probably the tertiary structure, for the Thumb domain alone and that in the p66 homodimer is further inferred from the resonance overlap noted above (59%, Table I).
The combined results imply that the Thumb and RNH domains within the p66 homodimer and the Thumb domain in the p51 exhibit the same overall structures as the isolated domains. This, however, is not the case for the Finger-Palm region (Table I). For this domain, notable chemical shift differences were observed in the p66 homodimer, possibly due to local structural changes or altered domain-domain or subunit-subunit interactions.
Conservation of the p51-RNH processing site conformation in p66
The conformation of the p51-RNH processing site (F440↓Y441) in the p66 homodimer was assessed based on the comparison of the amide resonance frequencies in the 1H-15N HSQC spectrum of the p66 homodimer and those in the isolated RNH domain.25, 60 Backbone amide resonances from residues in the β-1 strand, which contains the processing site in the isolated RNH domain, were essentially identical (amino acids 438 to 447) and several of these (I434, F440, and Y441) are labeled in the Fig. 3B inset. The fact that no chemical shift differences were noted for these processing site residues suggests that they are similarly located in a β-strand within the p66 homodimer (Fig. S3). Since NMR resonance frequencies are highly sensitive to local electronic and conformational influences, the close match between amide resonances for the p51-RNH processing site in the p66 homodimer and the isolated RNH domain indicates that the site is similarly structured in both. We also did not observe a significant number of random coil resonances in the p66 spectrum, making it very unlikely that one or both of the RNH domains is unfolded or disordered in the RT p66 homodimer. Since the intensities of the observed RNH resonances are similar or slightly larger than those of the observed TrpNεH resonances of the p51 domain in the p66 spectrum (Fig. 3), it is very unlikely that another RNH conformation exists in the p66 homodimer, different from the one we identified. Even with possible exchange between an unfolded and folded RNH domain in p66, any unstructured fraction would be <5% of total, given the close match between resonance frequencies for the bona-fide folded, isolated RNH domain and the RNH domain in p66. Given all of the above, it is strongly suggested that the processing mechanism has to involve the selection of a very minor conformer in the overall conformational ensemble by the protease or a local conformational change upon protease binding.
DISCUSSION
Numerous p66/p51 RT structures are available, with and without substrate, inhibitors and other ligands, allowing for a detailed elucidation of the conformational transitions that are possible in the RT heterodimer upon ligand interaction. In all of these structures, differences in domain orientations between the two subunits are observed.16, 22, 24, 61, 62 In contrast, structural information for the p66 homodimer, which is assumed to be the precursor of mature RT,15–20, 43 is not available. Therefore, any conformational changes that accompany processing of the homodimer into the heterodimer remain ambiguous.
Here, we investigated the solution conformation of the p66 homodimer, aimed at providing the basis for elucidating RT heterodimer formation. We showed that the 1H-15N TROSY HSQC spectrum of the p66 homodimer contains a very large number of backbone amide resonances, a rather surprising finding for a protein with a molecular mass of 132 kDa. This suggests that some of the smaller domains exhibit a certain degree of independent motion, faster than the overall rotational molecular diffusion. Our study also establishes that the overall major structures of the Thumb and RNH domains within the p66 homodimer are very similar to those of the isolated domains. In addition, the high similarity in chemical shifts for the p51-RNH processing site in the isolated RNH domain and the p66 homodimeric RT indicates that the cleavage site in the p66 homodimer exhibits essentially the same conformation as the isolated RNH domain, seemingly buried in the β-structure of the domain.
The mechanistic details involved in the formation of the mature RT p66/p51 heterodimer are not well understood, but several models have been proposed. One prevailing model assumes that cleavage of the immature p66 homodimer occurs in an unstructured RNH domain (Fig. 1C),21, 22 with subsequent folding of the remaining p66-RNH domain into the structure of the mature heterodimeric RT. This hypothesis is based primarily on the assumption that p51-RNH processing would be very inefficient if the processing site were located within a structured domain.17 Another proposal assumes that the p66 homodimer exists in an asymmetric conformation.11, 18 Indeed, the p51 domain conformation in the p66 subunit differs from that in the p51 subunit of the mature RT heterodimer, and this could be true in the p66 precursor as well. A third proposal suggests cleavage of p66 monomers, followed by folding of both p66 and p51 monomers, and formation of a mature p66/p51 heterodimeric RT (a concerted model).12, 13, 15 While our NMR results do not allow us to propose an exclusive model, they do provide insight into the conformational characteristics in the RT homodimer, thereby limiting possible maturation scenarios. First, the observation that the p66 NMR spectrum is a composite of those of the folded sub-domains indicates that p66, whether monomer or homodimer, is folded in solution. Second, we found that the RNH domain is structured in the p66 homodimer, with a conformation essentially identical to that seen in the isolated RNH domain, not altered by domain-domain contacts. Third, none of our data suggest that there are different conformations of RNH in the two p66 subunits of the homodimer.
Our observation of a structured RNH domain in the p66 homodimer is consistent with previous biochemical results. Protease predominantly processes p66 at the p51-RNH processing site (F440-Y441), compared to other processing sites within the RNH domain in the p66 precursor (Y483-L484, N494-I495, and Y532-L533), and removal of the RNH domain does not involve multiple cleavages.10, 17, 63–65 Since our data show that the RNH in the p66 homodimer is folded, this explains why other protease cleavage sites are not used and only the authentic p51-RNH site is cleaved.
In the current study, NMR signals in the Finger-Palm and Connection domains were not assigned. Therefore, our data cannot rule out a model in which the p66 homodimer exists in an asymmetric conformation. However, any model in which an overall different conformation of the RNH domain is proposed for the two subunits in the p66 homodimer is very unlikely, given our data. Even if in solution a small free energy difference between the two RNH domains could exist that may give rise to two different conformations (conformational substrates) for very short times, the overall conformational equilibrium will result a single average conformation over longer time scales. Thus, both domains will adopt the same average energy state, even if two energetically distinct conformations, separated by a low energy barrier, exist. While our manuscript was in review, Zheng, et al. published a study in which they evaluated Ile methyl resonances, and suggested selective unfolding of one of the RNH domains in the p66/p66 homodimer66. Based on our data, we believe that major disruption of the RNH structure is unlikely, although local or temporal unfolding of a small region around the processing site cannot be ruled out.
CONCLUSION
Our results support maturation models that involve folded, rather than unfolded or disordered RNH domains (Fig. 4). This is somewhat surprising since the p51-RNH cleavage site has to be processed by the protease, thus needs to be available for binding. However, as pointed out above, if a minor conformation (<5%) were present, this may have eluded detection, given the limited sensitivity and signal overlap in the spectra. Indeed, at present we suggest that such a minor conformation of the p51-RNH processing site can be selected or induced by the protease.
Figure 4.
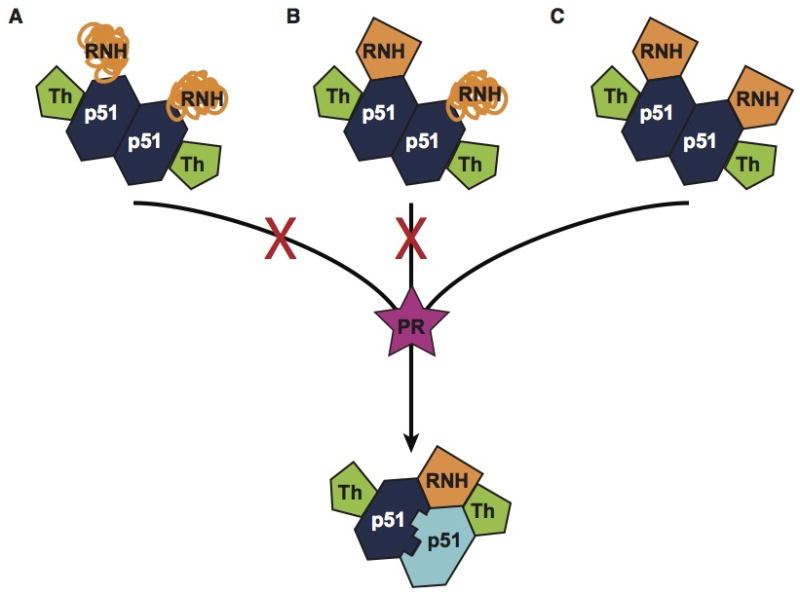
Possible RNH conformations in the p66 homodimers as maturation precursors: (A) the RNH domains are unfolded or disordered, (B) there is conformational difference between the two RNH domains, (C) both RNH domains in the two subunits are folded. Our data support (C).
Supplementary Material
Acknowledgments
This study was supported by grants from the National Institutes of Health (R01 GM105401 to R.I., P50GM082251 to R.I. and A.M.G., and P50GM103368 to M.A.P.), the National Science Foundation (Graduate Research Fellowship Grant, 1247842, to N.G.S.), and start-up funds from the University of Pittsburgh School of Medicine. We thank Teresa Brosenitsch for critical reading of the manuscript, Mike Delk for NMR support, Justine, H. Jou, Atticus Huberts, and Jinwoo Ahn for protein expression and purification support, and Nicolas Sluis-Cremer for the original RT DNA construct. We also thank Stuart F.J. Le Grice, Mary D. Barkley, and Robert E. London for helpful discussions.
Footnotes
Supplemental Information. Three figures: (Figure S1) Multi-angle Light scattering (MALS) profiles of p66 and p51 protein samples; (Figure S2) Thumb domain structure; (Figure S3) Backbone secondary structure of the region around the p51-RNH processing site in RNH with a subset of hydrogen bonds highlighted.
References
- 1.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 2.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 3.Temin HM. Retrovirus variation and reverse transcription: abnormal strand transfers result in retrovirus genetic variation. Proc Natl Acad Sci U S A. 1993;90:6900–6903. doi: 10.1073/pnas.90.15.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilhelm M, Wilhelm FX. Reverse transcription of retroviruses and LTR retrotransposons. Cell Mol Life Sci. 2001;58:1246–1262. doi: 10.1007/PL00000937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eickbush TH, Jamburuthugoda VK. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008;134:221–234. doi: 10.1016/j.virusres.2007.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katz RA, Skalka AM. The retroviral enzymes. Ann Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 7.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 8.Hizi A, Herschhorn A. Retroviral reverse transcriptases (other than those of HIV-1 and murine leukemia virus): a comparison of their molecular and biochemical properties. Virus Res. 2008;134:203–220. doi: 10.1016/j.virusres.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 9.Herschhorn A, Hizi A. Retroviral reverse transcriptases. Cell Mol Life Sci. 2010;67:2717–2747. doi: 10.1007/s00018-010-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chattopadhyay D, Evans DB, Deibel MR, Vosters AF, Eckenrode FM, Einspahr HM, Hui JO, Tomasselli AG, Zurcher-Neely HA, Heinrikson RL, Sharma SK. Purification and characterization of heterodimeric human immunodeficiency virus type 1 (HIV-1) reverse transcriptase produced by in vitro processing of p66 with recombinant HIV-1 protease. J Biol Chem. 1992;267:14227–14232. [PubMed] [Google Scholar]
- 11.Sharma SK, Fan N, Evans DB. Human immunodeficiency virus type 1 (HIV-1) recombinant reverse transcriptase. Asymmetry in p66 subunits of the p66/p66 homodimer. FEBS Lett. 1994;343:125–130. doi: 10.1016/0014-5793(94)80303-x. [DOI] [PubMed] [Google Scholar]
- 12.Divita G, Rittinger K, Geourjon C, Deleage G, Goody RS. Dimerization kinetics of HIV-1 and HIV-2 reverse transcriptase: A two step process. J Mol Biol. 1995;245:508–521. doi: 10.1006/jmbi.1994.0042. [DOI] [PubMed] [Google Scholar]
- 13.Venezia CF, Meany BJ, Braz VA, Barkley MD. Kinetics of association and dissociation of HIV-1 reverse transcriptase subunits. Biochemistry. 2009;48:9084–9093. doi: 10.1021/bi9010495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cabodevilla JF, Odriozola L, Santiago E, Martínez-Irujo JJ. Factors affecting the dimerization of the p66 form of HIV-1 reverse transcriptase. Eur J Biochem. 2001;268:1163–1172. doi: 10.1046/j.1432-1327.2001.01939.x. [DOI] [PubMed] [Google Scholar]
- 15.Sluis-Cremer N, Arion D, Abram ME, Parniak MA. Proteolytic processing of an HIV-1 pol polyprotein precursor: insights into the mechanism of reverse transcriptase p66/p51 heterodimer formation. Int J Biochem Cell Biol. 2004;36:1836–1847. doi: 10.1016/j.biocel.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 16.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3. 5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 17.Tomasselli AG, Sarcich JL, Barrett LJ, Reardon IM, Howe WJ, Evans DB, Sharma SK, Heinrikson RL. Human immunodeficiency virus type-1 reverse transcriptase and ribonuclease H as substrates of the viral protease. Protein Sci. 1993;2:2167–2176. doi: 10.1002/pro.5560021216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Smerdon SJ, Jager J, Kohlstaedt LA, Rice PA, Friedman JM, Steitz TA. Structural basis of asymmetry in the human immunodeficiency virus type 1 reverse transcriptase heterodimer. Proc Natl Acad Sci U S A. 1994;91:7242–7246. doi: 10.1073/pnas.91.15.7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tachedjian G, Moore KL, Goff SP, Sluis-Cremer N. Efavirenz enhances the proteolytic processing of an HIV-1 pol polyprotein precursor and reverse transcriptase homodimer formation. FEBS Lett. 2005;579:379–384. doi: 10.1016/j.febslet.2004.11.099. [DOI] [PubMed] [Google Scholar]
- 20.Wapling J, Moore KL, Sonza S, Mak J, Tachedjian G. Mutations that abrogate human immunodeficiency virus type 1 reverse transcriptase dimerization affect maturation of the reverse transcriptase heterodimer. J Virol. 2005;79:10247–10257. doi: 10.1128/JVI.79.16.10247-10257.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies JFn, Hostomska Z, Hostomsky Z, Jordan SR, Matthews DA. Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science. 1991;252:88–95. doi: 10.1126/science.1707186. [DOI] [PubMed] [Google Scholar]
- 22.Jacobo-Molina A, Arnold E. HIV reverse transcriptase structure-function relationships. Biochemistry. 1991;30:6351–6356. doi: 10.1021/bi00240a001. [DOI] [PubMed] [Google Scholar]
- 23.Hostomska Z, Matthews DA, Davies JFn, Nodes BR, Hostomsky Z. Proteolytic release and crystallization of the RNase H domain of human immunodeficiency virus type 1 reverse transcriptase. J Biol Chem. 1991;266:14697–14702. [PubMed] [Google Scholar]
- 24.Jacobo-Molina A, Ding J, Nanni RG, Clark ADJ, Lu X, Tantillo C, Williams RL, Kamer G, Ferris AL, Clark P, Hizi A, Hughes SH, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3. 0 A resolution shows bent DNA. Proc Natl Acad Sci U S A. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pari K, Mueller GA, DeRose EF, Kirby TW, London RE. Solution structure of the RNase H domain of the HIV-1 reverse transcriptase in the presence of magnesium. Biochemistry. 2003;42:639–650. doi: 10.1021/bi0204894. [DOI] [PubMed] [Google Scholar]
- 26.Powers R, Clore GM, Stahl SJ, Wingfield PT, Gronenborn AM. Analysis of the backbone dynamics of the ribonuclease H domain of the human immunodeficiency virus reverse transcriptase using nitrogen-15 relaxation measurements. Biochemistry. 1992;31:9150–9157. doi: 10.1021/bi00153a006. [DOI] [PubMed] [Google Scholar]
- 27.Mueller GA, Pari K, DeRose EF, Kirby TW, London RE. Backbone dynamics of the RNase H domain of HIV-1 reverse transcriptase. Biochemistry. 2004;43:9332–9342. doi: 10.1021/bi049555n. [DOI] [PubMed] [Google Scholar]
- 28.Wüthrich K. Protein structure determination in solution by NMR spectroscopy. J Biol Chem. 1990;265:22059–22062. [PubMed] [Google Scholar]
- 29.Clore GM, Gronenborn AM. NMR of Proteins. London: The Macmillan Press Ltd; 1993. [Google Scholar]
- 30.Clore GM, Gronenborn AM. NMR structure determination of proteins and protein complexes larger than 20 kDa. Curr Opin Struct Biol. 1998;2:564–570. doi: 10.1016/s1367-5931(98)80084-7. [DOI] [PubMed] [Google Scholar]
- 31.Palmer AG., 3rd Probing molecular motion by NMR. Curr Opin Struct Biol. 1997;7:732–737. doi: 10.1016/s0959-440x(97)80085-1. [DOI] [PubMed] [Google Scholar]
- 32.Kay LE. NMR studies of protein structure and dynamics. J Magn Reson. 2005;173:193–207. doi: 10.1016/j.jmr.2004.11.021. [DOI] [PubMed] [Google Scholar]
- 33.Igumenova TI, Frederick KK, Wand AJ. Characterization of the fast dynamics of protein amino acid side chains using NMR relaxation in solution. Chemical Reviews. 2006;106:1672–1699. doi: 10.1021/cr040422h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng X, Mueller GA, DeRose EF, London RE. Solution characterization of [methyl-(13)C]methionine HIV-1 reverse transcriptase by NMR spectroscopy. Antiviral Res. 2009;84:205–214. doi: 10.1016/j.antiviral.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng X, Mueller GA, Cuneo MJ, Derose EF, London RE. Homodimerization of the p51 subunit of HIV-1 reverse transcriptase. Biochemistry. 2010;49:2821–2833. doi: 10.1021/bi902116z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider A, Peter D, Schmitt J, Leo B, Richter F, Rösch P, Wöhrl BM, Hartl MJ. Structural requirements for enzymatic activities of foamy virus protease-reverse transcriptase. Proteins. 2014;82:375–385. doi: 10.1002/prot.24394. [DOI] [PubMed] [Google Scholar]
- 37.Gardner KH, Kay LE. Production and incorporation of N-15, C-13, H-2 (H-1-delta 1 methyl) isoleucine into proteins for multidimensional NMR studies. J Am Chem Soc. 1997;119:7599–7600. [Google Scholar]
- 38.Kay LE, Gardner KH. Solution NMR spectroscopy beyond 25 kDa. Curr Opin Struct Biol. 1997;7:722–731. doi: 10.1016/s0959-440x(97)80084-x. [DOI] [PubMed] [Google Scholar]
- 39.Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 40.Restle T, Müller B, Goody RS. Dimerization of human immunodeficiency virus type 1 reverse transcriptase. A target for chemotherapeutic intervention. J Biol Chem. 1990;265:8986–8988. [PubMed] [Google Scholar]
- 41.Fletcher RS, Holleschak G, Nagy E, Arion D, Borkow G, Gu Z, Wainberg MA, Parniak MA. Single-step purification of recombinant wild-type and mutant HIV-1 reverse transcriptase. Protein Expr Purif. 1996;7:27–32. doi: 10.1006/prep.1996.0004. [DOI] [PubMed] [Google Scholar]
- 42.Sluis-Cremer N, Kempner E, Parniak MA. Structure-activity relationships in HIV-1 reverse transcriptase revealed by radiation target analysis. Protein Sci. 2003;12:2081–2086. doi: 10.1110/ps.03130503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Marko RA, Liu HW, Ablenas CJ, Ehteshami M, Götte M, Cosa G. Binding kinetics and affinities of heterodimeric versus homodimeric HIV-1 reverse transcriptase on DNA-DNA substrates at the single-molecule level. J Phys Chem B. 2013;117:4560–4567. doi: 10.1021/jp308674g. [DOI] [PubMed] [Google Scholar]
- 44.Pervushin K, Riek R, Wider G, Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weigelt J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J Am Chem Soc. 1998;120:10778–10779. [Google Scholar]
- 46.Grzesiek S, Bax A. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J Magn Reson. 1992;99:201–207. [Google Scholar]
- 47.Grzesiek S, Bax A. Correlating backbone amide and side-chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 48.Muhandiram DR, Kay LE. Gradient-Enhanced Triple-Resonance Three-Dimensional NMR Experiments with Improved Sensitivity. J Magn Reson. 1994;103:203–316. [Google Scholar]
- 49.Zhang O, Kay LE, Olivier JP, Forman-Kay JD. Backbone 1H and 15N resonance assignments of the N-terminal SH3 domain of drk in folded and unfolded states using enhanced-sensitivity pulsed field gradient NMR techniques. J Biomol NMR. 1994;4:845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]
- 50.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 51.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinás M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN Data Model for NMR Spectroscopy: Development of a Software Pipeline. Proteins: Structure, Function, and Bioinformatics. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 52.Wishart DS, Sykes BD. The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J Biomol NMR. 1994;4:171–180. doi: 10.1007/BF00175245. [DOI] [PubMed] [Google Scholar]
- 53.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen Y, Bax A. Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. J Biomol NMR. 2007;38:289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- 55.Pervushin K. Impact of transverse relaxation optimized spectroscopy (TROSY) on NMR as a technique in structural biology. Q Rev Biophys. 2000;33:161–197. doi: 10.1017/s0033583500003619. [DOI] [PubMed] [Google Scholar]
- 56.Sluis-Cremer N, Dmietrinko GI, Balzarini J, Camarasa M-J, Parniak MA. Human immunodeficiency virus type-1 reverse transcriptase dimer destabilization by 1-[Spiro[4″-amino-2″,2″ -dioxo-1″,2″ -oxathiole-5″,3′-[2′, 5′-bis-O-(tert-butyldimethylsilyl)-beta-D-ribofuranosyl]]]-3-ethylthymine. Biochemistry. 2000;39:1427–1433. doi: 10.1021/bi991682+. [DOI] [PubMed] [Google Scholar]
- 57.Venezia CF, Howard KJ, Ignatov ME, Holladay LA, Barkley MD. Effects of efavirenz binding on the subunit equilibria of HIV-1 reverse transcriptase. Biochemistry. 2006;45:2779–2789. doi: 10.1021/bi051915z. [DOI] [PubMed] [Google Scholar]
- 58.Schlosshauer M, Baker D. Realistic protein-protein association rates from a simple diffusional model neglecting long-range interactions, free energy barriers, and landscape ruggedness. Protein Sci. 2004;13:1660–1669. doi: 10.1110/ps.03517304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schreiber G, Haran G, Zhou HX. Fundamental aspects of protein-protein association kinetics. Chem Rev. 2009;109:839–860. doi: 10.1021/cr800373w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Christen MT, Menon L, Myshakina NA, Ahn J, Parniak MA, Ishima R. Structural Basis of the Allosteric Inhibitor Interaction on the HIV-1 Reverse Transcriptase RNase H domain. Chem Biol Drug Des. 2012;80:706–716. doi: 10.1111/cbdd.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jäger J, Smerdon S, Wang J, Boisvert DC, Steitz TA. Comparison of three different crystal forms shows HIV-1 reverse transcriptase displays an internal swivel motion. Structure. 1994;2:869–876. doi: 10.1016/s0969-2126(94)00087-5. [DOI] [PubMed] [Google Scholar]
- 62.Unge T, Knight S, Bhikhabhai R, Lövgren S, Dauter Z, Wilson K, Strandberg B. 2. 2 A resolution structure of the amino-terminal half of HIV-1 reverse transcriptase (fingers and palm subdomains) Structure. 1994;2:953–961. doi: 10.1016/s0969-2126(94)00097-2. [DOI] [PubMed] [Google Scholar]
- 63.Schulze T, Nawrath M, Moelling K. Cleavage of the HIV-1 p66 reverse transcriptase/RNase H by the p9 protease in vitro generates active p15 RNase H. Arch Virol. 1991;118:179–188. doi: 10.1007/BF01314028. [DOI] [PubMed] [Google Scholar]
- 64.Abram ME, Parniak MA. Virion instability of human immunodeficiency virus type 1 reverse transcriptase (RT) mutated in the protease cleavage site between RT p51 and the RT RNase H domain. J Virol. 2005;79:11952–11961. doi: 10.1128/JVI.79.18.11952-11961.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pettit SC, Lindquist JN, Kaplan AH, Swanstrom R. Processing sites in the human immunodeficiency virus type 1 (HIV-1) Gag-Pro-Pol precursor are cleaved by the viral protease at different rates. Retrovirology. 2005;2:66. doi: 10.1186/1742-4690-2-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng X, Pedersen LC, Gabel SA, Mueller GA, Cuneo MJ, Derose EF, Krahn JM, London RE. Selective unfolding of one Ribonuclease H domain of HIV reverse transcriptase is linked to homodimer formation. Nucleic Acids Res. 2014 doi: 10.1093/nar/gku143. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.