Abstract
Mass spectrometry (MS)-based proteomics has become the most utilized tool to characterize histone post-translational modifications (PTMs). Since histones are highly enriched in lysine and arginine residues, lysine derivatization has been developed to prevent the generation of short peptides (3-6 residues) during trypsin digestion. One of the most adopted protocols applies propionic anhydride for derivatization. However, the propionyl group is not sufficiently hydrophobic to fully retain the shortest histone peptides in reversed-phase liquid chromatography, and such procedure also hampers the discovery of natural propionylation events. In this work we tested 12 commercially available anhydrides, selected based on their safety and hydrophobicity. Performance was evaluated in terms of yield of the reaction, MS/MS fragmentation efficiency and drift in retention time by using the following samples: (i) a synthetic unmodified histone H3 tail, (ii) synthetic modified histone peptides and (iii) a histone extract from cell lysate. Results highlighted that 7 of the selected anhydrides increased peptide retention time as compared to propionic, and several anhydrides such as benzoic and valeric led to high MS/MS spectra quality. However, propionic anhydride derivatization still resulted in our opinion as the best protocol to achieve high MS sensitivity and more even ionization efficiency among the analyzed peptides.
Keywords: anhydride, bottom-up proteomics, histones, mass spectrometry, protein derivatization
Introduction
Histone proteins are modified by a large number of post-translational modifications (PTMs) including methylation, acetylation and phosphorylation [1]. Histone PTMs affect chromatin structure, which influences enzyme recruitment, gene regulation, DNA repair and chromosome condensation [2]. Their fundamental biological function led to a large number of dedicated studies, revealing their involvement in numerous cellular events or disease [1, 3, 4]. However, due to the high diversity, dynamic range and combinatorial patterns of such PTMs still much needs to be revealed about their role in the cell. Because of this, research has been also focusing on developing methods to precisely identify, map and quantify single and combinatorial PTMs on histone proteins [5].
Mass spectrometry (MS) is currently the technique of choice to characterize protein PTMs, and different MS-based strategies have been developed for histone PTM analysis. The most widely adopted is the bottom-up strategy, where histones are digested into short peptides (<20 residues) prior to liquid chromatography – tandem mass spectrometry (LC-MS/MS) analysis. Analysis of short peptides facilitates both LC separation and high resolution MS detection. Trypsin is the most adopted enzyme in bottom-up proteomics, due to its high specificity (cleaves after lysine (K or Lys) and arginine (R or Arg) unless they are followed by a proline (P or Pro)) and high efficiency [6]. However, histones are highly enriched in Lys and Arg residues, leading trypsin digestion to the generation of peptides that are too short (2-4 aa) for LC-MS analysis. Because of this, derivatization of Lys side chains has been implemented to avoid trypsin cleavage after Lys [7-9]. This allows generation of most of the peptides that carry the most studied PTMs of adequate length (6-20 aa), which are suitable for both LC separation and MS identification.
Propionylation is one of the most commonly adopted derivatization strategies, and it has been used by several research groups (e.g. [10-20]). Propionic anhydride is mixed in solution with intact histones; the ε-nitrogen of the Lys side chain reacts with the anhydride leading to propionylation on unmodified and mono-methylated Lys residues. After protein digestion, propionylation of the peptide N-terminal is also performed to increase hydrophobicity of histone peptides, which enhances LC retention, separation and resolution. However, such derivatization method does not add sufficient hydrophobicity to short peptides (e.g. aa 3-8 of histone H3 when it is di- or tri-methylated), which signal is often lost due to poor retention (Unpublished, Garcia et al.). Moreover, different peptides are characterized by different ionization efficiency [21]. This is mostly caused by the different properties of the peptide sequence and endogenous modifications [21, 22]. Finally, propionylation is also a natural PTM, thus such derivatization precludes its investigation.
In this work we compared propionic anhydride to 11 other different anhydrides that can be used to derivatize the Lys side chain and perform ArgC-like digestion with trypsin. Results illustrated that propionic anhydride derivatization was the best compromise in terms of sensitivity of the analysis, ease of use and accuracy of the quantification. Propionylation achieved high peptide MS signal when analyzing a digestion of unmodified histone tail, suggesting highly efficient peptide digestion and ionization. Most of the analyzed anhydrides led to increased hydrophobicity of the peptides, achieving higher retention time as compared to propionic anhydride. Moreover, anhydrides such as benzoic and valeric led to high MS/MS spectra quality. We selected propionic, valeric and benzoic anhydride for a more in depth study with synthetic modified peptides and HeLa cells histone extract. Final results highlighted that propionic anhydride derivatization was the most accurate and sensitive protocol.
Materials and methods
Sample preparation of synthetic peptides and HeLa cell culture
The following anhydrides were all purchased from Sigma-Aldrich: benzoic, glutaric, phtalic, methacrylic, valeric, maleic, chloroacetic, diphenic, succinic, heptafluorobutyric, butyric and propionic. The synthetic histone H3 tail (aa 1-51) was purchased from GenScript with a purity >95%. Aliquotes of 10 μg each were prepared for the derivatization protocols. The derivatization / digestion was performed as previously described [23]. Briefly, histones were dissolved in 30 μL of 50 mM NH4HCO3, pH 8.0. Derivatization reagent was prepared by mixing the anhydride with 2-propanol in the ratio 1:3 (v/v) and such reagent was mixed with the histone sample in the ratio of 1:2 (v/v) for 15 min at 37 degrees. This reaction was performed twice. Histones were then digested with trypsin (enzyme:sample ratio 1:20, 6 hours, room temperature) in 50 mM NH4HCO3. After digestion the derivatization reaction was performed again twice to derivatize peptide N-termini. Benzoic anhydride derivatization was then optimized by testing (i) standard derivatization protocol, (ii) derivatization only of Lys side chains (only before digestion) with two cycles of reaction (like standard), (iii) derivatization only of Lys side chains with one cycle of reaction and (iv) derivatization of Lys side chains with benzoic anhydride (two cycles) and peptide N-termini with propionic anhydride (two cycles). Samples were then desalted by using C18 Stage-tips [24].
93 synthetic peptides representing various histone ArgC-like digested peptides were synthesized by Cell Signaling Technology (CST)® Protein Aqua™. In this library isobaric peptides were labeled with different isotopes to allow discrimination at the MS level. Single peptide concentration was measured by amino acid analysis. For the derivatization test one 0.27 pmol/μL stock was divided in three parts of 37 μL each (10 pmol) for the comparison of propionic, valeric and benzoic anhydride. Derivatization was performed with propionic, benzoic and valeric anhydride using the standard protocol [23], with a minor adjustment for benzoic anhydride, where only one cycle of derivatization was performed. Samples were then desalted by using C18 Stage-tips.
For the cell lysate study, HeLa S3 cells were grown and harvested as previously described [25]. Nuclei were isolated and histone proteins were extracted as described in the protocol of Lin and Garcia with minor adjustments [23]. Briefly, histones were acid extracted from nuclei with 0.2 M H2SO4 for 2 hours and precipitated with 33% trichloroacetic acid (TCA) overnight. Protein concentration was calculated using the Bradford assay. The resulting pellets were redissolved in 100 mM NH4HCO3 and derivatized / digested with either propionic, valeric or benzoic anhydride using the standard protocol [23], with a minor adjustment for benzoic anhydride, where only one cycle of derivatization of only the Lys side chains was performed. Samples were then desalted by using C18 Stage-tips.
NanoLC−MS/MS
Samples were analyzed by using a nanoLC-MS/MS setup. nanoLC was configured with a 75 μm ID x 17 cm Reprosil-Pur C18-AQ (3 μm; Dr. Maisch GmbH, Germany) nano-column using an EASY-nLC nanoHPLC (Thermo Scientific, Odense, Denmark). The HPLC gradient was 0-35% solvent B (A = 0.1% formic acid; B = 95% MeCN, 0.1% formic acid) over 10 or 40 min and from 34% to 100% solvent B in 7 minutes at a flow-rate of 250 nL/min. 40 min gradient was used for analysis of the 10 different anhydride protocols and the HeLa cells analysis, 10 min gradient was used for the optimization of the benzoic anhydride reaction and the synthetic library analysis. LC was coupled with an LTQ-Orbitrap Elite or an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Full scan MS spectrum (m/z 290−1650) was performed in the Orbitrap with a resolution of 30,000 (at 400 m/z) with an AGC target of 1x10e6. The 10 most intense ions at each cycle were selected for MS/MS performed with higher-energy collisional dissociation (HCD) with normalized collision energy of 35, an AGC target of 2x10e4 and a maximum injection time of 200 ms. MS/MS data were collected in centroid mode in the orbitrap mass analyzer. Precursor ion charge state screening was enabled and all unassigned charge states as well as singly charged species were rejected. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 30 s.
Data Analysis
The mass shift of the anhydride derivatization was determined applying a large peptide mass tolerance (± 300 Da) during database searching performed with pFind [26]. Once the mass of the anhydride was determined MS/MS database searching was performed by setting anhydride derivatization on the peptide N-terminus as fixed modification (unless otherwise specified), and the Lys derivatization as variable modification. The peptide and fragment mass tolerance were set as 10 ppm and 0.02 Da, respectively. pBuild was used to estimate the false discovery rate (FDR) of peptide-spectrum matches (PSMs), and results were filtered for FDR <1%. pLabel was used to show the confident PSMs. pProfile was used to extract the retention time and area under curve for each identified peptide. The peptide relative ratio was calculated by considering all peptides that share the same aa sequence as 100%. All tools were downloaded from the pFind Studio website (http://pfind.ict.ac.cn/software.html).
Results
We took into consideration 12 different anhydrides, including propionic, to derivatize histone proteins and perform bottom-up proteomics for PTM analysis, of which only 10 (Fig. 1A) provided comparable results in this work. Diphenic and heptafluorobutyric anhydride protocols did not achieve any detectable signal at the LC-MS analysis. The remaining 10 different anhydrides (Fig. 1A) were tested on an unmodified version of histone H3 N-terminal tail. We found valeric and benzoic anhydride had the best performance in terms of increasing peptide hydrophobicity, quality of MS/MS spectra, reduced variation in ionization efficiency between peptides and ease of use. Propionic, valeric and benzoic anhydride protocols were thus selected as the most suitable reagents for histone peptide derivatization, and they were tested on a synthetic library of modified peptides [21] and an extract of histones from HeLa cells.
Figure 1. comparison of 10 anhydride protocols on a synthetic histone H3 unmodified tail.
(A) List of the anhydrides used to perform this study. The table contains their monoisotopic mass, their adduct masses imparted to the peptide, and their formula. (B) Workflow of derivatization and digestion of the histone tail. Two rounds of derivatization are performed; before digestion, to label Lys side chains, and after digestion, to label peptide N-termini. (C) Average LC retention time drift of the four peptides as compared to propionylated ones. (D) Absolute intensity of the four peptides derivatized with the different protocols, considering 10 μg of starting material and all processed with the same workflow. The total area of the graph for each anhydride graphically represents the yield of the sample preparation.
Comparison of all anhydrides using synthetic unmodified peptides
A synthetic histone H3 N-terminal tail without any PTM was derivatized and digested using the standard protocol [23] for each of the 12 investigated anhydrides. The mass of the derivatization group was theoretically calculated and then verified by MS data analysis. Most of the anhydrides react with the peptide generating an amide and a carboxylic acid as a leaving group. On the other hand, glutaric, maleic, phtalic and succinic anhydrides open their ring structure for the reaction, generating no leaving groups. The derivatization / digestion protocol applied to the synthetic histone H3 N-terminal tail generated 4 peptides (aa 3-8, 9-17, 18-26 and 27-40) modified with 2, 3, 3 and 4 anhydride groups, respectively (Fig. 1B). The order of peptide hydrophobicity was aa 3-8 < aa 9-17 < aa 27-40 < aa 18-26 (Fig. S1).
Each anhydride derivatization experiment was analyzed by injecting 1/20th of the starting sample material (10 μg). As we started from the same total amount, the absolute signal intensity of the four peptides indicated the yield after the reaction and the ionization efficiency of the differently derivatized peptides. As expected, almost all anhydrides analyzed led to an increased retention time for the peptides as compared to the propionylation protocol (Fig. 1C). Phtalic (+10.2 min), valeric (+13.6 min) and benzoic (+17.4 min) derivatization increased LC retention of more than 10 minutes in a 40 min gradient. Succinic was the only anhydride with a consistent decrease in LC retention as compared to propionic. Methyacrylic and maleic derivatization protocols achieved the best signal intensities for the four peptides analyzed (Fig. 1D). However, the anhydrides that generate the most intense signals showed large differences between peptide signals, present in the sample at the same stoichiometry. Methacrylic-derivatized peptides were the most different between each other, as they were observed with up to ~21-fold change difference in terms of absolute abundance (Fig. 1D). This indicated that such anhydride increased the differences in ionization efficiency between the peptides as compared to propionic anhydride derivatization. Conversely, benzoic-derivatized peptides were the most homogeneous, with a maximum of only 2.4-fold change difference between the peptide aa 18-26 and aa 3-8, indicating that benzoic derivatization does not exacerbate differences in ionization efficiencies. We thus ruled out succinic and glutaric anhydrides from the 12 remaining candidates due to their low retention time drift, methacrylic anhydride due to its large bias in ionization efficiency, and chloroacetic anhydride due to the very low signal intensity produced. Moreover, we discarded diphenic and heptafluorobutyric anhydrides as they led to no LC-MS peptide signal (not shown).
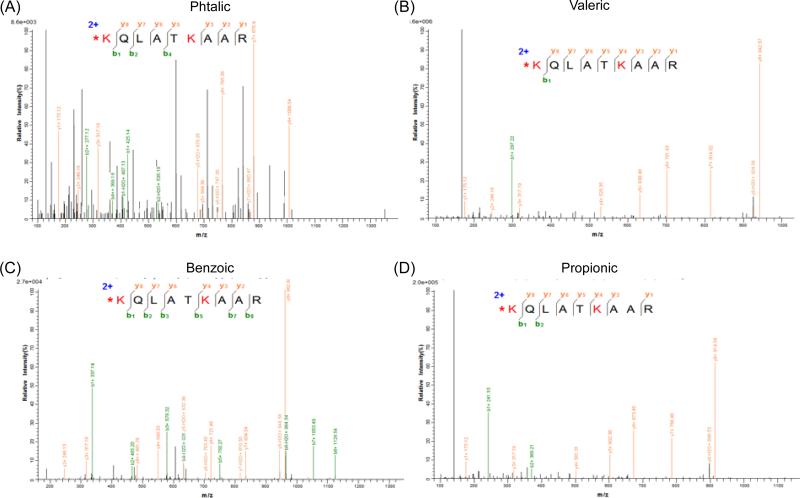
Next, we analyzed the MS/MS spectra quality of the unmodified peptides after anhydride derivatization. Phtalic, maleic and chloroacetic anhydride generated the MS/MS spectra with the lowest, the 2nd lowest and the 3rd lowest score after database searching, respectively (Fig. S2). This was due to the large number of fragment ions present in the spectrum, including several non-backbone fragments (Fig 2A and S2). Therefore, we did not consider them as appropriate candidates for histone derivatization, as they might prevent confident identification during data dependent MS experiments. Valeric and benzoic anhydride derivatization led to high quality MS/MS spectra, highly comparable with propionic derivatization (Fig. 2B, 2C and 2D).
Figure 2. MS/MS spectra of the peptide aa 18-26 after derivatization.
The best peptide-spectrum match for the peptide KQLATKAAR was analyzed for the derivatization protocols using (A) phtalic anhydride, (B) valeric anhydride, (C) benzoic anhydride and (D) propionic anhydride.
Finally, we ruled out butyric anhydride for the following reasons: (i) butyrylation is a natural PTM in histones [27], thus the use of such derivatization strategy would prevent its study; (ii) the average retention time shift (+ 6.3 min) was not as dramatic as other anhydride treatments, such as benzoic and valeric anhydride (Fig. 1C); (iii) the yield of the reaction was among the lowest in terms of recovery of the four targeted peptides (Fig. 1D). After these preliminary experiments we concluded that valeric and benzoic anhydrides were the best candidates to be compared with the traditional propionylation protocol, due to their high retention time shift, their MS/MS spectra quality and the small relative intensity change between the four analyzed peptides.
Optimization of benzoic anhydride derivatization
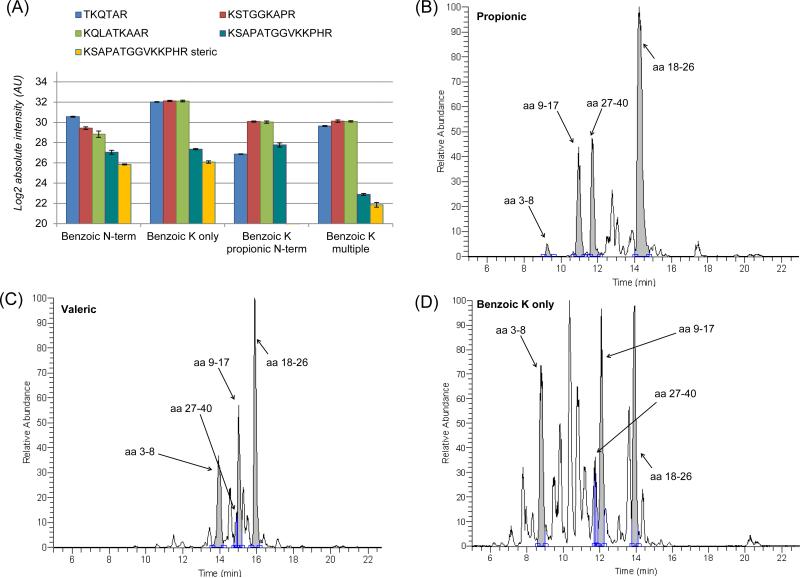
Even though benzoic anhydride proved to be one of the best candidates for histone derivatization, we observed a large number of side products and incomplete reaction in the analysis of the unmodified histone H3 tail (Sup. Fig.) By performing the standard protocol with benzoic anhydride (2 cycles of derivatization of Lys side chains and peptide N-termini) we observed that about 30% of the peptide aa 27-40 carried one less benzoic group as compared to the total 27-40 peptide detected (data not shown). Moreover, since benzoic anhydride is solid in its natural state, it leaves a thick layer of salts after the solution is dried for the following steps of the protocol. Thus, we tested a faster derivatization protocol, in which we performed as few as possible derivatization cycles. Specifically, we tested benzoic derivatization in the following protocols: (i) performing derivatization of both Lys side chains and peptide N-terminal (named benzoic N-term, reaction performed with only one cycle of derivatization before and one after digestion); (ii) derivatization of only Lys side chains (named benzoic K only, one cycle before digestion only); (iii) derivatization of Lys side chain with benzoic anhydride and propionylation at the peptide N-terminal (named benzoic K propionic N-term), to verify whether propionylation of peptide N-termini can be performed with fewer issues of steric hindrance and thus incomplete reaction; (iv) derivatization of Lys side chains by performing two cycles of the reaction (named benzoic K multiple). We did not include the standard protocol in this comparison (2 cycles of derivatization of Lys side chains and peptide N-termini), as it provided similar results to the protocol (i) (not shown) and led to an excessive amount of salts at the end of the sample preparation.
Results evidenced a significant difference between the different protocols (Fig. 3A). The protocol named “benzoic K only” produced comparable intensities for the peptides aa 3-8, aa 9-17 and aa 18-26 (average absolute signal 4.5x10e9 ± 1.8x10e8), but the peptide aa 27-40 was significantly less intense (1.7x10e8). This was also due to the incomplete derivatization of the peptide, as about 30% of the peptide signal was quantified with one less benzoic group. Most likely, the steric hindrance of the anhydride led to this effect, as we could observe it only for the peptide aa 27-40, which presents two nearby Lys (aa 36 and 37). We selected the protocol with only one cycle of derivatization only on the Lys side chains (named benzoic K only), as it produced the most intense and comparable signals for benzoic anhydride derivatization. Then, we tested if the remaining benzoic anhydride after sample drying continued to react with peptide N-termini during and after digestion. Our analysis showed that the percentage of peptide derivatized at the N-termini did not exceed 3%, which we considered as sufficiently low to not affect the final results (Fig. S4). Taken together, we considered protocol (ii) as the most suitable for the following experiments. However, the chromatogram of the benzoic derivatized sample (Fig. 3D) was significantly more complex than propionic and valeric (Fig. 3B and 3C), indicating that we could not avoid side reactions even when only Lys side chain derivatization was performed.
Figure 3. optimization of benzoic anhydride reaction and comparison with valeric and propionic.
(A) Log2 signal intensity of the four peptides from histone H3 synthetic unmodified tail after derivatization with benzoic anhydride using four different protocols: (i) Lys side chains and peptide N-terminal derivatization; (ii) derivatization of only Lys side chains; (iii) derivatization of Lys side chain with benzoic anhydride and propionylation at the peptide N-terminal: (iv) derivatization of Lys side chains with two cycles of reaction. The peptide aa 27-40 named “steric” represents the peptide with one less benzoic group attached. Error bars represent standard deviation between LCMS technical replicates. (B) LC-MS chromatogram of the processed histone H3 synthetic and unmodified N-terminal tail with propionyl anhydride, (C) valeric anhydride and (D) benzoic anhydride. This last chromatogram displays the results of the selected protocol for benzoic anhydride derivatization.
Ionization efficiency calculated with synthetic modified peptides
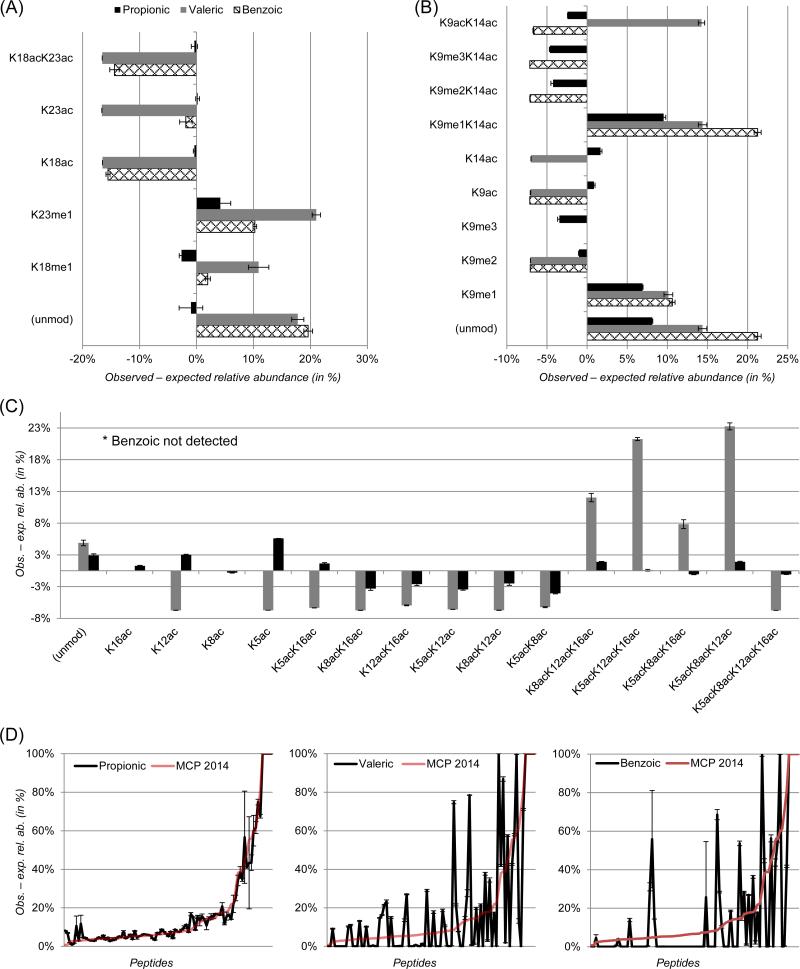
Recently, our group analyzed the ionization efficiency of most of the commonly seen peptide during histone PTM analysis by using a library of 93 AQUA synthetic peptides [21]. In the current study, we derivatized such library with propionic, valeric and benzoic groups to assess the variation between observed and expected signal intensity of the peptides. All 93 synthetic peptides were mixed with equal molarity, and 100 fmol of sample was analyzed with LC-MS/MS. Each peptide was quantified from the LC-MS/MS runs and compared with its theoretical relative abundance. Results indicated that both valeric and benzoic anhydrides led to peptide signal intensity more divergent to the expected intensity as compared to propionic (Fig. 4 and Table S1). Propionic anhydride derivatization allowed for the quantification of all forms of the peptide aa 3-8, including H3K4me2/me3 and T3ph. By using valeric anhydride derivatization we did not detect H3K4me3, while with benzoic we did not detect H3K4me2/me3. In figure 4A we focused on the peptide KQLATKAAR (histone H3, aa 18-26) as unmodified and modified in K18ac, K23ac, K18me1, K23me1 and K18acK23ac. Both benzoic and valeric derivatization generated significantly higher deviations from the expected intensity as compared to propionic. Similar observations were made for the peptide KSTGGKAPR (histone H3 aa 9-17). Moreover, the modification states K9me3K14ac, K9me2K14ac and K9me3 were not detected in the sample derivatized with valeric anhydride, while the peptides K14ac and K9me3 were not detected in the benzoic anhydride sample (Fig. 4B). None of the acetylated peptides from histone H4 were detected after benzoic derivatization (Fig. 4C), while valeric showed an overall higher deviation from theoretical intensity as compared to propionic. Specifically, singly, doubly and quadruply acetylated histone H4 peptides were all underestimated as compared to the expected amount spiked in, while triply acetylated peptides were dramatically overestimated after valeric derivatization.
Figure 4. comparison of observed and expected peptide signals using library of AQUA synthetic peptides.
(A) PTMs analyzed for the histone H3 peptide aa 18-26 KQLATKAAR. The relative abundance of the observed peptides was calculated by LC peak area integration, while the theoretical one was calculated by considering each peptide isoform as 1:6 of the total peptide. Error bars represent standard deviation between LC-MS technical replicates. (B) Analysis of PTMs of the histone H3 peptide aa 9-17 KSTGGKAPR. Missing bars represent undetected peptides. (C) Analysis of PTMs of the histone H4 peptide aa 3-17 GKGGKGLGKGGAKR. (D) Comparison of the observed relative abundances for the three anhydrides to a reference dataset [21]. The y-axis represents the observed relative abundance subtracted by the expected relative abundance. The x-axis contains the peptide list, sorted by an increasing difference between observed and expected values of the reference dataset. For the reference dataset no error bars are reported. Gaps in the line graph represent unquantified peptides.
We sorted the calculated errors between observed and expected intensity for all peptides, and we compared them with the results from the previously published study [21] (Fig. 4D). As expected, the 93 detected peptides derivatized with propionic anhydride were highly comparable with the previous study, as the same type of derivatization was used. On the other hand, valeric and benzoic derivatization presented large differences from the prior referenced study. Moreover, only 73 and 36 peptides could be detected after valeric and benzoic derivatization, respectively. Collectively, our results indicated that valeric and benzoic derivatization led to the identification of fewer peptides from the library of AQUA synthetic ones, and their deviation with the expected relative intensity was on average higher than by using propionic anhydride.
Analysis of HeLa cell histone extract
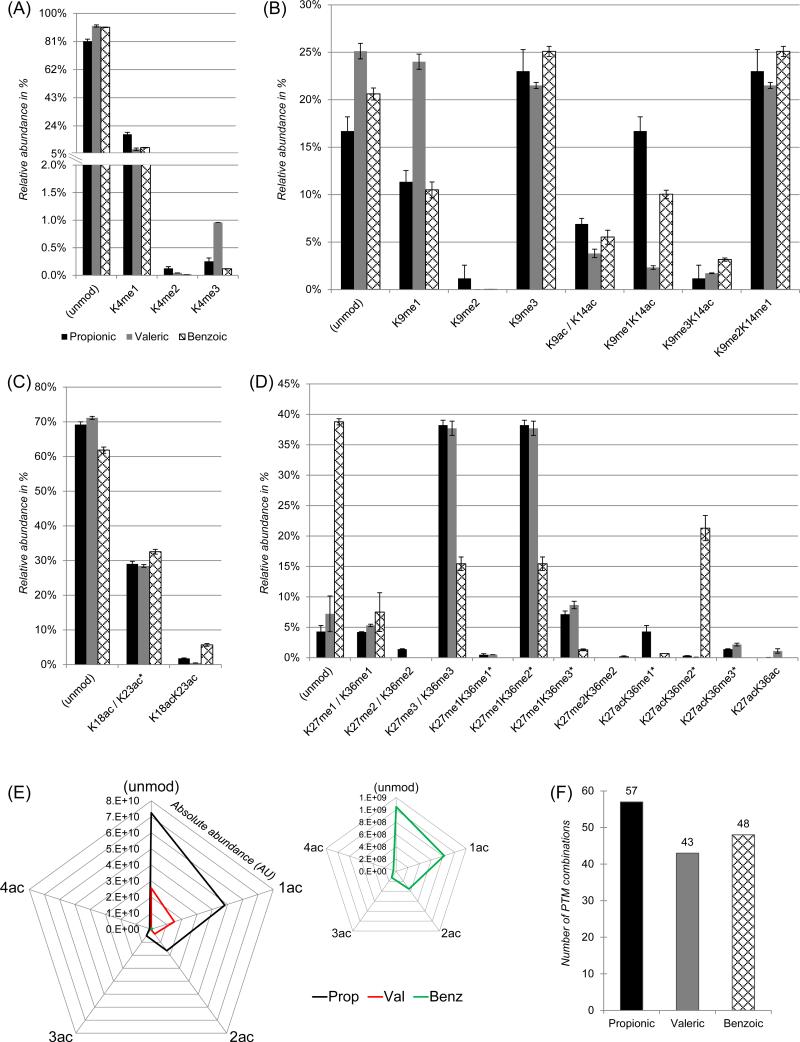
Finally, we tested the performance of the different derivatization protocols on an extract of histones from HeLa cells. This test aimed to be more exhaustive than the synthetic peptide analysis, as it included inevitable biases during the digestion step, the issue of detecting differently modified peptides with a wide dynamic range of intensities, and the presence of natural contamination from a standard histone extraction protocol. With all strategies we could detect low abundance H3K4me2/me3 (Fig. 5A), which are known to be retained with low efficiency by LC with propionic anhydride derivatization (unpublished, Garcia et al.). As well, the peptide aa 9-17 from histone H3 was detected in all of its analyzed forms with high sensitivity (Fig. 5B). However, the results from the three experiments did not provide consistent quantification among the anhydrides demonstrating, together with the synthetic peptide library analysis (Fig. 4), that the signal from histone peptides is biased not only through peptide sequence and PTM types, but also the derivatization group. Such difference was present mostly in the quantification of the peptide aa 9-17, while for the peptide aa 18-26 we observed a more consistent quantification between the three protocols (Fig. 5C). The analysis of the peptide aa 27-40 highlighted an evident bias with benzoic anhydride derivatization, as results were clearly in disagreement with the other two protocols. For instance, the quantification of the unmodified peptide was 4.3% and 38.8% when quantified with propionic and benzoic protocols, respectively.
Figure 5. peptide quantification from HeLa cells prepared with the three derivatization protocols.
(A) Relative abundance of H3K4 methylated forms. (B) Relative abundance of differently modified histone H3 peptides for the sequence aa 9-17. (C) Relative abundance of unmodified and acetylated histone H3 peptide for the sequence aa 18-26. H3K18ac was no discriminated from H3K23ac, as we did not perform MS/MS level quantification. Thus, the bars represent the sum of the two species. (D) Relative abundance of unmodified and acetylated histone H3 peptide for the sequence aa 27-40. Forms marked with an asterisk were not discriminated because they are isobaric. (E) Absolute intensity of the differently acetylated histone H4 peptide aa 4-17 GKGGKGLGKGGAKR. (F) Total number of identified histone peptides for the three derivatization protocols.
PTMs belonging to histone H4 and H2A were also identified and quantified. For instance, the histone H4 peptide aa 4-17 was detected in all known forms, from unmodified to tetra-acetylated (Fig. 5E). In this case the three different protocols led to comparable results in terms of relative intensity of the different forms, but propionylation provided the highest sensitivity. Collectively, by using the propionylation protocol we identified and quantified 57 different combinations of PTMs in ArgC-like peptides using a standard LC-MS/MS bottom-up proteomics strategy, while with valeric and benzoic derivatization we identified only 43 and 48 combinations of PTMs, respectively (Fig. 5F). This revealed that propionic anhydride derivatization is still the most sensitive strategy between the ones we investigated.
Discussion
The aim of our study was to test anhydrides alternative to propionic, as peptide propionylation has limits related to the poor LC retention of short peptides such as histone H3 aa 3-8 and investigation of the propionylation as endogenous PTM. After searching into chemical databases we selected 11 anhydrides alternative to propionic for the study, according to hydrophobicity and safety documentations. For instance, we did not include acetic and trifluoroacetic anhydrides, as safety documentation specifies that they react violently with water. Diphenic and heptafluorobutyric anhydride could not provide observable signals at the LC-MS analysis, maybe due to their large molecular structure and thus incomplete derivatization due to steric hindrance. The 9 other anhydrides were first compared in terms of hydrophobic properties and LC-MS signal intensity. As we started with the same sample amount for each of the tested protocols the signal intensity provided an estimation of the reaction yield. Benzoic anhydride derivatization was further tested using four different protocols, and we found the best signal by using only one cycle of derivatization exclusively at the Lys side chains (Fig. 3A). This could be explained as benzoic anhydride is solid in its natural state; such anhydride was easily solubilized in acetonitrile, but it produced a consistent amount of salt at each drying cycle. Thus, one single cycle of derivatization led to less salt present in the sample tube and lower sample losses. However, derivatizing the Lys side chain only led to the generation of peptides less hydrophobic as compared to the standard derivatization protocol. The advantage of the dramatic retention time drift in case of full benzoic anhydride derivatization was thus lost and led to similar elution time to propionic derivatized peptides (Fig. 3B and 3D). In future studies it would be wise to optimize also temperature of the reaction and reaction time, as the current guidelines for derivatization are optimized for propionic anhydride.
By using the selected protocol we compared benzoic, valeric and propionic anhydride derivatization by quantifying synthetic peptides present in an equimolar mixture. The analysis of the synthetic peptides clearly highlighted the issues of valeric and benzoic derivatization, as the analysis revealed peptide quantification up to 75% different from the expected value. Even though a bias regarding differently modified peptides and relative quantification is already known from previous studies [21, 22], valeric and benzoic derivatization largely exceeded this bias in terms of quantification accuracy as compared to propionic derivatization. We might speculate that once such bias has been characterized the signal of the peptides could be corrected for subsequent analyses. However, it is concerning that the signal of the synthetic peptides was dramatically different from the expected signal. Moreover, less than half of the synthetic peptides identified and quantified with propionic anhydride derivatization were also found in the experiment with benzoic anhydride, demonstrating that benzoic derivatization achieved significantly lower yield. Surprisingly, in the HeLa cell extract analysis we detected more peptides for benzoic anhydride as compared to the synthetic library analysis and compared to the valeric derivatization protocol (Fig. 5E). However, this result could be due to the fact that both Lys side chains and N-termini must be modified in the synthetic library since such peptides have free N-termini, whereas we determined that benzoic derivatization yields the best signal when only side chains are modified (Fig. 3A). Therefore, a large number of synthetic peptide signals were probably suppressed by N-termini derivatization.
In summary, our work demonstrated that peptide propionylation is still the most efficient derivatization protocol for general histone PTM analysis under the tested conditions. Other anhydrides generate consistent shifts in retention time and increase MS/MS fragmentation efficiency. Thus, they might be adopted in dedicated experiments, in presence of high amount of starting material or when natural propionylation is being investigated. However, our database searching did not detect any endogenous propionylation in the HeLa cells sample derivatized with valeric and benzoic anhydride (data not shown). Protocols adopting combinations of anhydrides, e.g. for differential derivatization of Lys side chains and peptide N-termini, could lead to more efficient analyses. We did not perform such study, as we aimed to find an alternative procedure that required a sample preparation as simple as our reference, namely the propionylation protocol. Moreover, the use of different anhydrides in combination might lead to incomplete quenching of the reactive anhydride during derivatization of Lys side chains, and thus two different anhydrides competing for peptide N-termini derivatization after digestion. Therefore, we do not recommend using different anhydrides to derivatize histones.
Finally, particular attention must be taken when considering relative amounts of PTMs within single experiments, as differently modified peptides might present dramatically different ionization efficiencies. Analysis of larger polypeptides or intact proteins, such as the middle-down or the top-down strategy, might overcome the issue of ionization variability upon presence of PTMs. However, bottom-up is currently the strategy that provides the highest throughput and sensitivity.
Supplementary Material
Acknowledgements
BAG acknowledges funding from an NIH Innovator grant (DP2OD007447) from the Office of the Director, and NIH grant R01GM110174.
Abbreviations
- ac
acetylation
- ArgC
endoproteinase Arginine-C me1/me2/me3, mono/di/tri-methylation
- ph
phosphorylation
References
- 1.Cheung P, Allis CD, Sassone-Corsi P. Signaling to chromatin through histone modifications. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 4.Cosgrove MS. Histone proteomics and the epigenetic regulation of nucleosome mobility. Expert review of proteomics. 2007;4:465–478. doi: 10.1586/14789450.4.4.465. [DOI] [PubMed] [Google Scholar]
- 5.Sidoli S, Cheng L, Jensen ON. Proteomics in chromatin biology and epigenetics: Elucidation of post-translational modifications of histone proteins by mass spectrometry. Journal of proteomics. 2012;75:3419–3433. doi: 10.1016/j.jprot.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 6.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Analytical chemistry. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 7.Garcia BA, Mollah S, Ueberheide BM, Busby SA, et al. Chemical derivatization of histones for facilitated analysis by mass spectrometry. Nature protocols. 2007;2:933–938. doi: 10.1038/nprot.2007.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mackeen MM, Kramer HB, Chang KH, Coleman ML, et al. Small-molecule-based inhibition of histone demethylation in cells assessed by quantitative mass spectrometry. Journal of proteome research. 2010;9:4082–4092. doi: 10.1021/pr100269b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao R, Wu H, Deng H, Yu Y, et al. Specific and efficient N-propionylation of histones with propionic acid N-hydroxysuccinimide ester for histone marks characterization by LC-MS. Analytical chemistry. 2013;85:2253–2259. doi: 10.1021/ac303171h. [DOI] [PubMed] [Google Scholar]
- 10.Mandava V, Fernandez JP, Deng H, Janzen CJ, et al. Histone modifications in Trypanosoma brucei. Mol Biochem Parasit. 2007;156:41–50. doi: 10.1016/j.molbiopara.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouvry-Patat SA, Schey KL. Characterization of antimicrobial histone sequences and posttranslational modifications by mass spectrometry. J Mass Spectrom. 2007;42:664–674. doi: 10.1002/jms.1200. [DOI] [PubMed] [Google Scholar]
- 12.Robin P, Fritsch L, Philipot O, Svinarchuk F, Ait-Si-Ali S. Post-translational modifications of histones H3 and H4 associated with the histone methyltransferases Suv39h1 and G9a. Genome Biol. 2007:8. doi: 10.1186/gb-2007-8-12-r270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drogaris P, Wurtele H, Masumoto H, Verreault A, Thibault P. Comprehensive profiling of histone modifications using a label-free approach and its applications in determining structure-function relationships. Analytical chemistry. 2008;80:6698–6707. doi: 10.1021/ac800739d. [DOI] [PubMed] [Google Scholar]
- 14.Anderson DC, Green GR, Smith K, Selker EU. Extensive and Varied Modifications in Histone H2B of Wild-Type and Histone Deacetylase 1 Mutant Neurospora crassa. Biochemistry-Us. 2010;49:5244–5257. doi: 10.1021/bi100391w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darwanto A, Curtis MP, Schrag M, Kirsch W, et al. A Modified “Cross-talk” between Histone H2B Lys-120 Ubiquitination and H3 Lys-79 Methylation. Journal of Biological Chemistry. 2010;285:21868–21876. doi: 10.1074/jbc.M110.126813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilczek C, Chitta R, Woo E, Shabanowitz J, et al. Protein Arginine Methyltransferase Prmt5-Mep50 Methylates Histones H2A and H4 and the Histone Chaperone Nucleoplasmin in Xenopus laevis Eggs. Journal of Biological Chemistry. 2011;286:42221–42231. doi: 10.1074/jbc.M111.303677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan W, Xu M, Huang C, Liu N, et al. H3K36 Methylation Antagonizes PRC2-mediated H3K27 Methylation. Journal of Biological Chemistry. 2011;286:7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peach SE, Rudomin EL, Udeshi ND, Carr SA, Jaffe JD. Quantitative Assessment of Chromatin Immunoprecipitation Grade Antibodies Directed against Histone Modifications Reveals Patterns of Co-occurring Marks on Histone Protein Molecules. Molecular & Cellular Proteomics. 2012;11:128–137. doi: 10.1074/mcp.M111.015941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xie ZY, Dai JBA, Dai LZ, Tan MJ, et al. Lysine Succinylation and Lysine Malonylation in Histones. Molecular & Cellular Proteomics. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng YP, Sweet SMM, Popovic R, Martinez-Garcia E, et al. Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:13549–13554. doi: 10.1073/pnas.1205707109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S, Wein S, Gonzales-Cope M, Otte GL, et al. Stable Isotope labeled histone peptide library for histone post-translational modification and variant quantification by mass spectrometry. Molecular & cellular proteomics : MCP. 2014 doi: 10.1074/mcp.O113.036459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Analytical chemistry. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 23.Lin S, Garcia BA. Examining Histone Posttranslational Modification Patterns by High-Resolution Mass Spectrometry. Nuclesomes, Histones & Chromatin, Pt A. 2012;512:3–28. doi: 10.1016/B978-0-12-391940-3.00001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Analytical chemistry. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 25.Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: A bird's eye view. Journal of proteome research. 2006;5:240–247. doi: 10.1021/pr050266a. [DOI] [PubMed] [Google Scholar]
- 26.Wang LH, Li DQ, Fu Y, Wang HP, et al. pFind 2.0: a software package for peptide and protein identification via tandem mass spectrometry. Rapid communications in mass spectrometry : RCM. 2007;21:2985–2991. doi: 10.1002/rcm.3173. [DOI] [PubMed] [Google Scholar]
- 27.Chen Y, Sprung R, Tang Y, Ball H, et al. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Molecular & cellular proteomics : MCP. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.