Abstract
Oxidative stress has been implicated as a component of various pathologies including ischemia/reperfusion injury (IRI) and neurodegenerative diseases such as Parkinson's disease (PD) and schizophrenia. Similarly, regulator of G-protein signaling 4 (RGS4) has been implicated as an important player in each of these pathologies. RGS4, like other RGS proteins, is responsible for temporally regulating G-protein coupled receptor signaling by increasing the intrinsic GTPase activity of Gα subunit of the heterotrimeric signaling complex. In this study we evaluated whether modification by 4-hydroxy-2-nonenal (4HNE), a common lipid peroxidation product, inhibits RGS4. Using immunoprecipitation, we first determined RGS4 modification was occurring in cells at concentrations of 4HNE within reported physiological conditions. Following this determination, we evaluated modification of RGS4 by 4HNE by both Western blot and mass spectrometry (MS). Once it was established that covalent modification occurred only on cysteine containing constructs, tryptic digest followed by mass spectrometry analysis revealed modification occurs at cysteine residues 71, 148, and 183. In order to determine the effect 4HNE had on RGS4 activity, a steady-state colorimetric assay was used to analyze the GAP activity of Δ51-RGS4 as well as the cysteine null mutant. From the data, we determined that RGS4 activity can be modulated by 4HNE through modification at cysteine residues similar to previously reported small molecule inhibition of RGS4.
Introduction
Oxidative stress has been suggested as an important component of several pathologies including ischemia/reperfusion injury (IRI) and neurodegenerative diseases, such as Parkinson's disease (PD) and schizophrenia.1–3 Oxidative stress is the result of an imbalance in reductive and oxidative reactions in the cell leading to the production of reactive oxygen species (ROS). Cellular oxidative stress processes produce a wide array of reactive biomolecules, such as the lipid peroxidation product 4-hydroxy-2-nonenal (4HNE), one of the most prevalent and well-studied lipid peroxidation products. Basal levels of 4HNE are typically around 0.1 μM in plasma but increase to 1 μM in pathological conditions.4 In response to oxidative stress, previous research has suggested that 4HNE can accumulate in stressed membranes at concentrations of 10 μM to 5 mM.5 4HNE is capable of modifying a variety of intracellular targets at cysteine, histidine, and lysine residues. Of these, cysteine modification is considered to be the most relevant modification for modulation of enzymes. 4HNE has also been shown to modulate G-protein coupled receptor (GPCR) signaling through direct modification of Gαq/11.6
While many enzymes involved in xenobiotic metabolism such as glutathione S-transferase and select cytochrome P450s have been intensely studied for modification by 4HNE, other important cellular systems have remained under-investigated. In this article, we focus on regulator of G-protein signaling 4 (RGS4), a modulator of G-protein signaling. GPCRs are a class of receptors containing seven trans-membrane helices. When activated, these receptors facilitate the release of GDP to allow the binding of GTP to the Gα subunit of the heterotrimeric G- protein complex This nucleotide exchange results in the dissociation of both the α- and βγ-heterodimer subunits from the receptor, and each continues on to potentiate a variety of downstream targets. Termination of G-protein signaling is dependent upon hydrolysis of GTP to GDP, via intrinsic GTPase activity of the Gα-subunit followed by re-association of the Gα and βγ subunits.7 The rate of intrinsic hydrolysis of GTP to GDP in this system occurs too slowly to account for normal cellular signal transduction. These GTPase accelerating proteins (GAPs) function on inhibitory G-proteins, including Gαo, Gαi, and Gαq but not on the stimulatory Gαs. RGS proteins maintain this GAP activity through a highly conserved, 120 amino acid alphahelical RGS domain. RGS4, in particular, is a relatively small RGS protein containing a short N-terminal amphipathic helix and a RGS domain. Palmitoylation of RGS4 is used to direct the protein to the plasma membrane.8 This membrane-associated protein has been the target of several high-throughput screens to develop inhibitors.9–11 These studies have identified several small molecule inhibitors of RGS4, some of which inhibit via covalent modification at two cysteine residues, C132 and C148, with the latter affording a greater degree of inhibition.10
RGS4 has emerged as an interesting drug target due to its role in several pathologies. Increased RGS4 activity has been linked with improved renal function after induced renal IRI.15 RGS4 modulation of cholinergic signaling by regulation of M4 muscarinic autoreceptors has been implicated in the motor symptoms of PD.16 Further interrogation of RGS4 in PD has revealed that deletion of RGS4 ablates motor deficits in 6-hydroxydopamine treated mice.17 RGS4 has been implicated as a susceptibility gene in schizophrenia, although its importance is controversial.18 With the known roles of oxidative stress in each of these disease states, we set out to investigate the role of oxidative stress in modulating RGS4 activity using 4HNE as a model. We hypothesized that the sensitivity of RGS4 inhibition by exogenous small molecules with mechanisms of action being cysteine modification may be recapitulated by modification of those cysteine residues by reactive biomolecules generated during oxidative stress. This could also reveal an important role in normal physiology (or pathology) for these cysteine residues in modulating RGS4 activity.
In this study, we investigated the sensitivity of RGS4 modification by 4HNE, a common lipid peroxidation product. This study represents the first attempt to evaluate 4HNE as a potential modulator of RGS activity. When cells transiently expressing RGS4 were exposed to 4HNE, RGS4 modification by 4HNE was readily detectable using immunoprecipitation. When compared to the cysteine null mutant (designated henceforth as Δ7) in both Western blot and mass spectrometry (MS), only the wild-type (WT) construct contained detectable adducts. The RGS4(Δ7) construct has been previously characterized and been found to be similar to RGS4(WT) in both activity and association with its native binding partner Gαo.11 Further examination of RGS4(WT), by tryptic digest/MS revealed modification occurred at C71, C148, and C183. When challenged in a steady-state malachite green based activity assay, 4HNE was found to inhibit RGS4 at concentrations of 4HNE observed during oxidative stress. We propose that lipid peroxidation products inhibit RGS4 during oxidative stress through cysteine residues.
Experimental Procedures
Cell Culture and Treatment
Human embryonic kidney cells transfected with the SV40 large T antigen (HEK293T) were obtained from ATCC (Manassas, VA). The cells were grown as an adherent monolayer in T-75 tissue culture flasks in high glucose DMEM (Life Technologies; Grand Island, NY) supplemented with 10% fetal bovine serum (Fisher Scientific; Waltham, MA) and 1% penicillin/streptomycin (Life Technologies; Grand Island, NY). Following transfection, HEK293T cells were treated with increasing concentrations of 4HNE (Cayman Chemical; Ann Arbor, MI), 1, 5, and 10 μM, in phenol red free DMEM (Life Technologies; Grand Island, NY) containing 0.02% ethanol. Following a 10 min incubation at 37 °C, stressing media was removed and cells were washed with PBS. Cells were lysed in Lysis Buffer (10 mM KH2PO4 pH 7.5 and 0.1% Triton X-100) and homogenized using a Sonic Dismembrator (Fisher Scientific; Waltham, MA). Samples were cleared by centrifugation at 13 000g for 15 min, and the supernatant was collected. Crude protein concentration was determined by DC protein assay (Bio-Rad; Hercules, CA) for use in later experiments.
Plasmid Construct and Transfection
A construct containing a N-terminal HA tagged RGS4, with cysteine 2 mutated to serine to avoid degradation, (C2S) in pCDNA3.1(+) was used for transfection.19 For live cell imaging experiments, a construct containing a N-terminal green fluorescent protein (GFP) tagged RGS4 in pAcGFP-C1 and a construct containing Gαo in pCDNA3.1(+) was used. First, cells were plated into 12-well dishes at 2 × 105 cells/well or 96-well glass bottom ViewPlate (Perkin-Elmer; Waltham, MA) at 2 × 104 cells/well. Cells were transfected the following day using Lipofectamine 2000 (Life Technologies; Grand Island NY) according to manufacturer's protocol. After 6 h, media was exchanged with fresh media and allowed to incubate for 48 h before being stressed with 4HNE.
Immunoprecipitation
Four micrograms of goat anti-4HNE antibody, ab46544 (Abcam; Cambridge, MA) was added to 500 μg of sample and was diluted to 500 μL for a final concentration of 1 mg/mL sample. The sample was incubated for 16 h at 4 °C under light agitation using a HULA Shaker (Life Technologies; Grand Island, NY). 50 microliters of Protein-G Magna Beads (Life Technologies; Grand Island, NY) were added to each sample and incubated for 2 h at 25 °C. Beads were washed three times for 5 min in PBS. Beads were then suspended in 30 μL of Denaturation Buffer (200 mM Glycine pH 2.6) and incubated for 10 min at 55 °C.
Live Cell Imaging
Single cells were selected prior to treatment for GFP-RGS4 localization to the plasma membrane. After the addition of 1, 5, or 10 μM 4HNE as described earlier, cells were imaged at 15 min intervals for 1 h post-treatment using a LSM 510 confocal microscope (Zeiss; Oberkochen, Germany).
Protein Expression and Purification
Tobacco etch virus (TEV) protease was expressed and purified as a His-tagged protein in E. coli, BL21-pRIL (Stratagene; La Jolla, CA), in the pRK793 vector as previously described.20 Rat RGS4, sharing 97% sequence identity with human RGS4, (WT) and the cysteine to alanine (Δ7) mutant were expressed as fusion proteins of maltose binding protein (MBP), a 10× His tag, and a TEV protease recognition site to the N-terminus of an RGS4 construct lacking the first 51 amino acids, in the vector pMALC2H10T.21 The fusion protein was induced using 100 μM IPTG and expressed in LB at 37 °C for 4 h. The bacteria was pelleted and suspended in RGS4 buffer (50 mM HEPES at pH 8, 100 mM NaCl, 5 mM β-mercaptoethanol). The cells were lysed using 0.5 mg of lysozyme (Fisher Scientific; Waltham, MA) for every 1 mL of pellet. The pellet was cleared of DNA by the addition of 1 mg of DNAase1 (Fisher Scientific; Waltham, MA) and incubated on ice until consistency became fluid. The samples were then centrifuged at 100 000g for 1 h to clear the sample. After centrifugation, the resulting supernatant was filtered (0.45 μm) and loaded onto an amylose column (New England Biolabs; Ipswich, MA), 1.5 mL of resin for every 1 L of culture. The protein of interest was eluted using 10 mM maltose, and analysis via SDS-PAGE showed a fusion protein of >95% purity, with the minor contaminant being free MBP. Fractions containing the protein of interest were pooled and incubated with TEV protease at a molar ratio of 10:1 (fusion protein/TEV protease) overnight at 4 °C. The cleaved Δ51-RGS4 was then isolated by purification over ANX column (GE Healthcare Life Sciences; Uppsala, Sweden) in 50 mM HEPES at pH 6.8 and 50 mM NaCl. The flow through, containing the Δ51-RGS4, was then collected and concentrated using a YM-10 centrifugal concentrator (Millipore; Billerica, MA). The rate-altered variant of Human Gαi1 (R178M, A326S), described in the literature, was expressed in Terrific Broth (TB) media as a 6× His labeled protein in the pQE80 vector.13 Protein expression was inducted at OD600 nm of 1.0 using 100 μM IPTG at 30 °C. At 16 h after induction, the bacteria were pelleted at 3600g for 15 min. Pellets were lysed, centrifuged, and filtered similar to as described above for RGS4, but in Gαi buffer (50 mM HEPES at pH 7.5, 500 mM NaCl, 1 mM β-mercaptoethanol, and 20 μM GDP). After loading onto a Ni-NTA column (Qiagen; Hilden Germany) containing 3 mL of resin for every 1 L media, the column was washed with 2 column volumes of Gαi buffer supplemented with 25 mM imidazole. The protein of interest was eluted from the column using Gαi buffer supplemented with 300 mM imidazole. Fractions containing Gαi, as determined by SDS-PAGE, were pooled and dialyzed for 12 h against Gαi dialysis buffer (50 mM HEPES at pH 7.5, 25 mM NaCl, 1 mM β-mercaptoethanol, and 20 μM GDP). The pooled fractions were then loaded onto a Q-sepharose column (GE Healthcare Life Sciences; Uppsala, Sweden) and eluted along a salt gradient from 50 mM NaCl to 1 M NaCl in Gαi buffer. Fractions containing Gαi, determined by SDS-PAGE, were pooled and concentrated using an Amicon stirred cell concentrator (Millipore; Billerica, MA) with a YM-10 filter (Millipore; Billerica, MA). Gαi activity was assayed using the [35S] GTPγS binding assay.
4HNE Treatment of Purified RGS4
Δ51-RGS4(WT) and Δ51-RGS4(Δ7) were treated with increasing concentrations of 4HNE for 30 min at 37 °C. Two micrograms of cleaved Δ51-RGS4 was treated with 0, 6, 60, or 600 μM 4HNE, which corresponds to 1:0, 1:1, 1:10, and 1:100 molar ratios (RGS4/4HNE).
SDS-PAGE and Western Blotting
Samples were then loaded into SDS-PAGE gels for analysis by Western blot. After transfer of the samples to Immobilon-P transfer membrane (Millipore; Billerica, MA) according to established protocols, the membrane was then blocked, overnight, using tris buffered saline Tween-20 (TBST; 50 mM Tris pH 7.4, 150 mM NaCl, 0.1% Tween-20) supplemented with 3% bovine serum albumin (BSA). To detect RGS4, samples were first probed for 4 h at 4 °C using U1079 (rabbit anti-RGS4) at a 1:10 000 dilution in 3% BSA TBST. The blot was then probed using a 1:20 000 dilution of goat antirabbit secondary, conjugated to horseradish peroxidase (Protein Biosystems; Pelham, Alabama), for 1.5 h at 25 °C. The blot was developed using WestPico chemiluminescent substrate (Thermo Scientific, Waltham, MA) and imaged using a UVP Biospectrum Imaging system (Upland, CA). To detect 4HNE, the blot was probed using 0.1 μg/mL ab46544 anti-4HNE antibody (Abcam, Cambridge, MA) for 4 h at 4 °C and subsequently detected utilizing an anti-Goat HRP conjugated secondary (ab753, Abcam, Cambridge, MA), at a dilution of 1:20 000, for 1.5 h at 25 °C. The blot was developed using WestPico chemiluminescent substrate (Thermo Scientific, Waltham, MA) and imaged using a UVP Biospectrum Imaging system (Upland, CA).
LC/MS Analysis of RGS4
Δ51-RGS4(WT) and Δ51-RGS4(Δ7) were treated for 30 min at 37 °C. Then, 1.5 μg of RGS4 was treated with 50 μM 4HNE in a final volume of 15 μL in reaction buffer (50 mM HEPES pH 8.0, 100 mM NaCl). Following treatment, the samples were diluted 1:100 in doubly deionized water and loaded into LC-ESI-IT-TOF (Shimadzu; Kyoto, JP) similarly to as described previously.23 Ten microliters of the diluted sample was injected onto a Jupiter C-18 column (Phenomenex; Torrance, CA) and eluted at a gradient of 20–80% acetonitrile in 0.05% formic acid over 30 min after a 5 min step at 20% acetonitrile to remove excess salt. The results were analyzed using MagTran software. To determine sites of modification, RGS4 was treated with 4HNE, as described above, and digested to peptide fragments for MS analysis. Following the 30 min incubation at 37 °C, the samples were quenched using a molar excess of cysteine. The samples were then digested 10 h at 25 °C with trypsin (Sigma Alderich; St Louis, MO) at a ratio of 20:1 (RGS4/trypsin) in reaction buffer. The sample was the diluted 1:100 as before and injected onto the LC-ESI-IT-TOF utilizing the previously described protocol.23 Ten microliters of the diluted sample was injected onto a Jupiter C-18 column (Phenomenex; Torrance, CA) and eluted at a gradient of 5– 90% acetonitrile in 0.05% formic acid over 30 min after a 5 min step at 5% acetonitrile to remove excess salt. The results were then analyzed using MagTran software.
Steady-State GTPase Accelerating (GAP) Assay
A steady-state analysis of RGS4 GAP activity was performed largely as described previously.24 In a 384-well plate, 10 μL of each component and 5 μL Gαi1 was added sequentially with a 5 min centrifugation at 100g after each addition. Stock solutions of 4HNE (4mM, 1mM final) and vehicle treatment were prepared and added to appropriate wells. To each well, 800 nM RGS4 (200 nM RGS4) was added to each well and allowed to incubate 30 min at 25 °C. The reaction was quenched by the addition of 5 μL of 80 mM cysteine and incubated for 5 min after centrifugation. Five microliters of 40 μM Gαi1 (5 μM final) was added to each well. After a 5 min incubation, 600 μM GTP (150 μM final) was added to each well. Developing solution (DS) was prepared on the day of use by creating a 50:12.5:1 ratio of the following: 0.12% malachite green (w/v) dissolved in 17% H2SO4 (v/v), 7.5% ammonium molybdate tetrahydrate, and 11% Tween-20 (v/v). After a 75 min incubation, DS was added to each well. After a 50 min incubation, the plate was read using an EnVision plate reader (PerkinElmer; Waltham, MA).
Statistical Analysis
Steady-state assay data was analyzed using GraphPad Prism software (La Jolla, CA). Treated samples were compared to control samples using an unpaired t-test for statistical significance. Values of p < 0.05 were considered significant.
Results
Detection of RGS4 Modification in HEK293T Cells
Initially, we set out to identify whether RGS4, known to be sensitive to cysteine modification, was capable of being modified in cells.23 Forty-eight hours after transient transfection with the HA-RGS4(C2S) construct, which evades proteasomal degradation, cells were treated with 0–10 μM 4HNE for 10 min and analyzed for 4HNE modification of HA-RGS4(C2S) by immuno-precipitation.19 While no modification was determined for 1 μM, a significant amount of 4HNE-modified RGS4 was detected at 5 μM as quantified by densitometry. Nearly twice as much modified RGS4 was detected when the cells were exposed to 10 μM 4HNE versus 5 μM 4HNE, as shown in Figure 1a. The assay was shown to be specific for 4HNE modified RGS4, failing to detect either 4HNE alone or RGS4 alone (Figure 1b).
Figure 1.
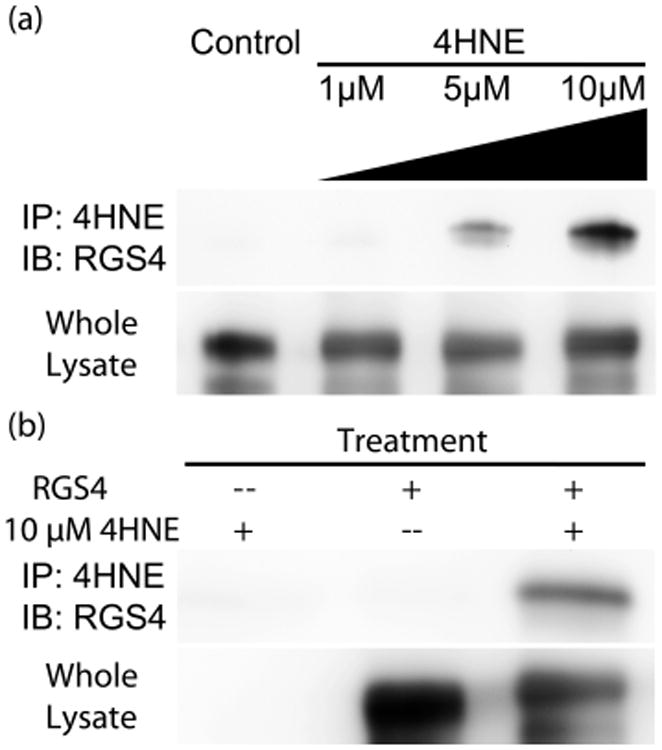
Immuno-precipitation of 4HNE-modified RGS4. Immuno-detection of RGS4 via Western blot after immune-precipitation of 4HNE labeled protein. (a) HEK293T cells, transfected with HA-(C2S)RGS4, were treated with increasing concentrations of 4HNE from 0–10 μM. No detectable RGS4 was seen at 0 μM, but a faint band appears at 1 μM, accounting for 0.5 ± 0.1% of total RGS4, and significantly increases at 5 μM, accounting for 4.6 ± 0.6%. At 10 μM, 7.8 ± 0.6% of total RGS4 was modified. (b) Immunoprecipitation specificity for 4HNE modified RGS4. In the absence of RGS4 expression, no modified protein was detected. Similarly, mock treatment with vehicle produced no detectable band. Only in cells both transfected with RGS4 and treated with 4HNE produced a detectable band in the Western blot. The images shown are representative of n = 3 experiments.
RGS4 Treatment with 4HNE and Detection of Products
2 μg of Δ51-RGS4(WT) and Δ51-RGS4(Δ7) were treated with variable amounts of 4HNE, and a Western blot was performed to determine whether modification occurs on either construct. Increasing concentrations of 4HNE resulted in extensive protein modification by 4HNE as shown by the increasing intensity of the band corresponding to 4HNE modified RGS4 in Figure 2. While the modification was detectable at even the 1:1 molar ratio in the Δ51-RGS4(WT), no modification was detected on the Δ51-RGS4(Δ7) construct, even at molar ratios of 1:100.
Figure 2.
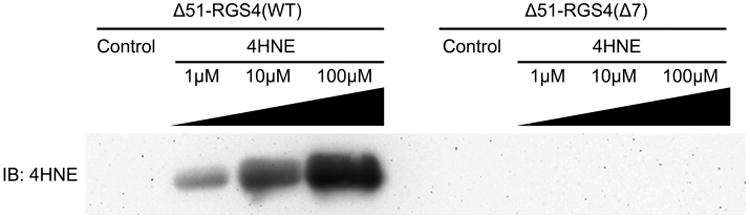
Immuno-detection of 4HNE-modified Δ51-RGS4(WT) and Δ51-RGS4(Δ7). Purified protein exposed to increasing ratios of 4HNE, from 0–100-fold. Δ51-RGS4(WT) was clearly detected by Western blot at even 1:1 ratio of 4HNE, while Δ51-RGS4(Δ7) was not detected even at ratios of 1:100 4HNE. The image shown is representative of n = 3 experiments.
MS Quantification of 4HNE Adducts on RGS4
Δ51-RGS4(WT) and Δ51-RGS4(Δ7) were treated with 4HNE and separated by RP-HPLC using a C18 column with a gradient from 20–80% ACN in 0.5% formic acid. The Δ51-RGS4 constructs eluted at 10–11 min in the protocol described in the methods. The unadducted Δ51-RGS4(WT) species was detected with a deconvoluted m/z of 18090.8, as shown in Figure 3a. In treated samples, another peak coeluting with the unmodified protein contained a protein with a deconvoluted mass of 18558.1. The difference in the mass, 467.3 (3 × 155.8), corresponds to Michael addition of three 4HNE molecules. Upon repeating experimentation with Δ51-RGS4(Δ7) protein (Figure 3b), untreated samples produced a distribution of peaks that deconvoluted to m/z of 17869. In samples treated with 4HNE, the only detectable species corresponded to the unadducted protein, with a deconvoluted mass of 17869. These results confirm that Cys residues are being preferentially modified on RGS4.
Figure 3.
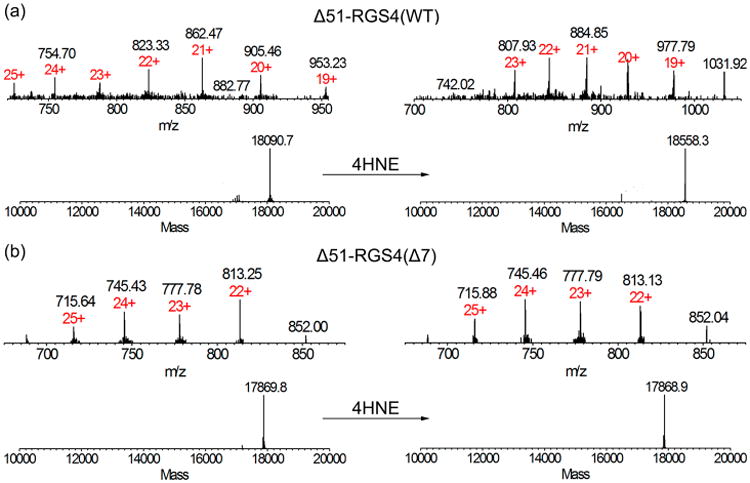
MS detection of 4HNE modification in Δ51-RGS4(WT) and Δ51-RGS4(Δ7). (a) Injection of 10 μL of RGS4, about 0.7 pmol, was analyzed by LC-ESI-IT-TOF. Expected MW for the deconvoluted singly charged molecular weight was 18090.7 and a detected deconvoluted signal was 18090.8. Treatment with 4HNE revealed a deconvoluted mass was 18558.1. The calculated difference in the mass, 467.3, accounts for three adduction sites for 4HNE. (b) Expected MW for the deconvoluted Δ51-RGS4(Δ7). Singly charged molecular weight was 17869, as detected. After 4HNE treatment, the detected deconvoluted mass was 17869, indicating no detectable modification. The data shown are representative of n = 3 experiments.
Tryptic Digest/MS Analysis of 4HNE Adduction Sites on RGS4
Tryptic digest of RGS4 yields a manageable nine detectable tryptic fragments containing no more than two Cys residues per peptide as shown in Table 1. After treatment with 1:10 molar excess 4HNE (5 μM RGS4 to 50 μM 4HNE), Δ51-RGS4(WT) was digested with trypsin and the resulting fragments were analyzed by MS. Initial analysis of the tryptic fragments revealed the expected sequence coverage of 75%, due predominantly to many tryptic fragments less than four amino acids in length as shown in Figure 4a. In the total ion chromatogram, the adducted fragments were detected as either doubly or triply charged species eluting later than their unadducted counterparts (Figure 4b). For fragment 2 (WAE-SLENLIN HECGLAAFK), the adducted fragment was detected as a triply charged species with a m/z of 767.99, shown in Figure 4c. For fragments 6 (NMLEPTITCF DAEQK) and 8 (FYLDLTNPSS CGAEK), both were detected as doubly charged species with a m/z of 948.88, in Figure 4d, and 901.37, in Figure 4e, respectively. These adducted fragments correspond to the three adduct sites on RGS4 identified in the whole protein MS analysis. Fragments 2, 6, and 8 correspond to specific cysteine residues within RGS4: C71, C148, and C183 respectively.
Table 1. Tryptic Digest of Δ51-RGS4(WT) Constructa.
| fragment position | sequence | position | predicted mass | experimental mass | adducted mass |
|---|---|---|---|---|---|
| 1 | SQEEVK | 52–57 | 719.77 | 719.82 | |
| 2 | WAESLENLINHECGLAAFK | 59–77 | 2146.44 | 2145.87 | 2303.98 |
| 3 | SEYSEENIDFWISCEEYK | 82–99 | 2272.42 | 2272.76 | |
| 4 | IYNEFISVQATK | 114–125 | 1413.62 | 1413.63 | |
| 5 | EVNLDSCTR | 126–134 | 1037.14 | 1037.40 | |
| 6 | NMLEPTITCFDEAQK | 140–154 | 1741.00 | 1740.66 | 1897.77 |
| 7 | IFNLMEK | 156–162 | 895.12 | 895.40 | |
| 8 | FYLDLTNPSS CGAEK | 173–187 | 1645.84 | 1646.64 | 1802.74 |
| 9 | SSADCTSLVPQCA | 193–205 | 1282.44 | 1281.77 |
This table shows the individual detected tryptic fragments of RGS4, the corresponding cysteine residues, and their detected MW. The segregation of individual cysteine residues between fragments allowed for simple identification of modified residues.
Figure 4.
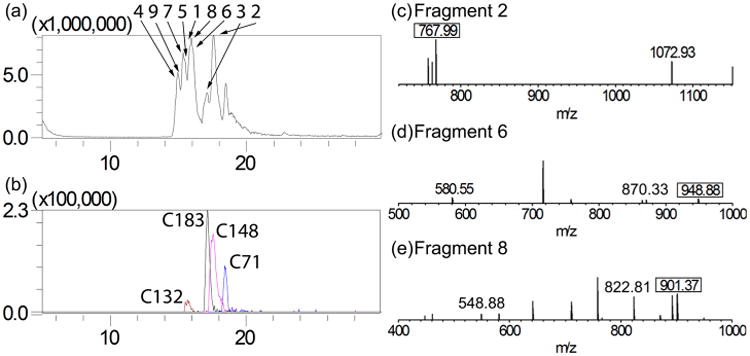
MS detection of 4HNE treated Δ51-RGS4(WT) tryptic digest. The elution gradient of 5–90% ACN allowed for separation of tryptic peptides for analysis. (a) Outlined is the total ion chromatogram as well as the base peak for each fragment. Peaks are identified according to fragment number from Table 1. (b) The elution peak for each of the adducted fragments. (c) Fragment 2, corresponding to C71, was detected containing a 4HNE adduct. The adducted fragment was detected as a triply charged species with an m/z of 767.99. (d) Fragment 6, corresponding to C148, was detected containing a 4HNE adduct. The adducted fragment was detected as a doubly charged species with an m/z of 948.88. (e) Fragment 8, corresponding to C183, was detected containing a 4HNE adduct. The adducted fragment was detected as a doubly charged species with an m/z of 901.37. The data shown are representative of n = 3 experiments.
Steady State Analysis of 4HNE Modified RGS4
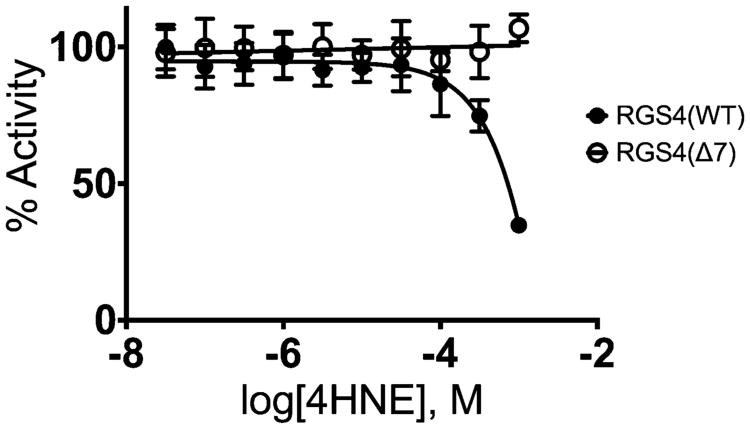
4HNE modulation of RGS4 activity was evaluated using the malachite green based steady-state GAP activity assay. In Figure 5, we show that 1 mM 4HNE inhibits Δ51-RGS4(WT). In contrast, Δ51-RGS4(Δ7) was not inhibited by 4HNE.
Figure 5.
Steady-state analysis of 4HNE modified Δ51-RGS4(WT) and Δ51-RGS4(Δ7). Steady-state analysis of RGS4 GAP activity was evaluated using Gαi1 as the GTPase. 4HNE was shown to inhibit RGS4(WT) with a predicted IC50 of 7.5 mM. RGS4(Δ7) was not inhibited at all by 4HNE under similar conditions. The data shown are the result of n = 3 experiments.
RGS4 Membrane Localization Analysis of 4HNE Treated Cells
4HNE modulation of RGS4 localization was evaluated using a membrane localization based assay. When overexpressed, Gαo, a native binding partner of RGS4, can cause localization to the plasma membrane.26 In Figure 6a,d,g, we show pretreatment RGS4 localization. Treatment with 1 μM 4HNE was not sufficient to reverse GFP-RGS4 localization (Figure 6b). This was quantified as pixel intensity across a cross-section of the cell, shown in Figure 6c. In Figure 6e, we show that after 1 h treatment with 5 μM 4HNE results in RGS4 delocalizing from the membrane. This comparison was quantified as pixel intensity, shown in Figure 6f. In Figure 6h, we show RGS4 delocalization in response to treatment with 10 μM 4HNE. This comparison was quantified as pixel intensity across a cross-section of the cell, shown in Figure 6i.
Figure 6.
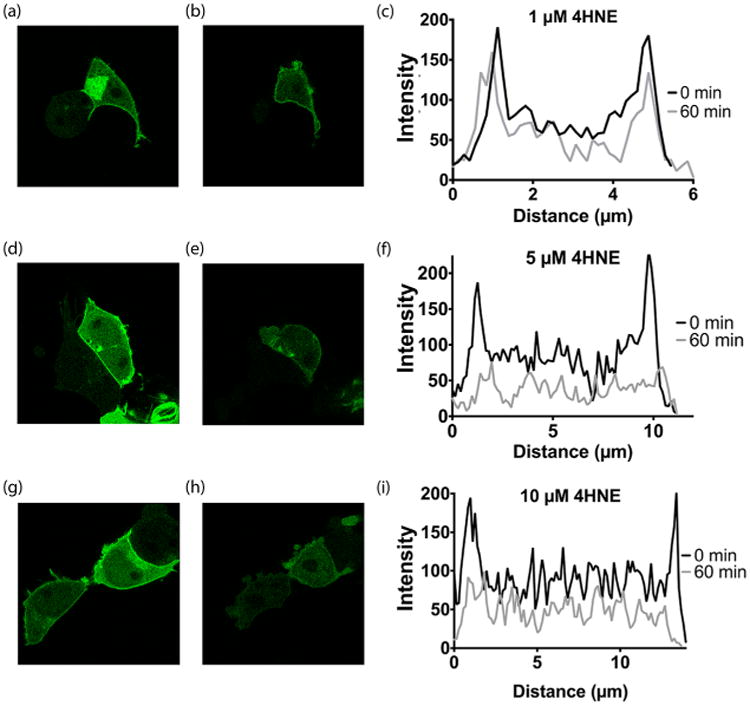
Subcellular localization of RGS4 during 4HNE treatment. RGS4 interaction with its native binding partner, Gαo, was evaluated by monitoring subcellular localization of GFP-RGS4 in HEK293T. (a) Prior to treatment with 1 μM 4HNE, GFP-RGS4 strongly associates with the plasma membrane. (b) After 1 h treatment with 1 μM 4HNE, GFP-RGS4 showed no change in localization. (c) Quantification of GFP-RGS4 fluorescence in a cross-section of both pretreatment (a) and post-treatment with 4HNE (b). Prior to treatment, the pixel intensity of GFP-RGS4 at the plasma membrane was 280% of the cell body. After 1 h of treatment with 1 μM 4HNE, the pixel intensity ratio of GFP-RGS4 at the plasma membrane as compared to the cell body remained unchanged. (d) Prior to treatment with 5 μM 4HNE, GFP-RGS4 strongly associates with the plasma membrane. (e) After 1 h treatment with 5 μM 4HNE, GFP-RGS4 localization to the plasma membrane is completely reversed. (f) Quantification of GFP-RGS4 fluorescence in a cross-section of both pretreatment (d) and post-treatment with 4HNE (e). Prior to treatment, the pixel intensity of GFP-RGS4 at the plasma membrane was 260% of the cell body. After 1 h of treatment with 5 μM 4HNE, the pixel intensity of GFP-RGS4 at the plasma membrane fell to within 50% above the mean intensity of the plasma membrane. (g) Prior to treatment with 10 μM 4HNE, GFP-RGS4 strongly associates with the plasma membrane. (h) After 1 h treatment with 10 μM 4HNE, GFP-RGS4 localization to the plasma membrane is completely reversed. (i) Quantification of GFP-RGS4 fluorescence in a cross-section of both pretreatment (g) and post-treatment with 4HNE (h). Prior to treatment, the pixel intensity of GFP-RGS4 at the plasma membrane was 250% of the cell body. After 1 h of treatment with 10 μM 4HNE, the pixel intensity of GFP-RGS4 at the plasma membrane fell to within 50% above the mean intensity of the plasma membrane. The images shown are representative of n = 3 experiments.
Discussion
4HNE is a common lipid peroxidation product and a signal of oxidative stress at the cell membrane. Oxidative stress has been implicated as an important component of the pathology of various neurological disorders including Parkinson's disease.27 In a disease with similar pathology, manganism, excess manganese is capable of activating microglia and stimulating the release of hydrogen peroxide.28 The direct production of lipid peroxidation products at the plasma membrane can result in alteration of various signaling pathways. RGS4 plays a critical role in regulating M4 autoreceptor activity in striatal neurons.16 As a membrane associated protein, RGS4 is an interesting target for 4HNE modification during oxidative stress. 4HNE modification of RGS4 could alter signaling from stressed cells. Using a variety of techniques, we show that RGS4 can be modified by 4HNE, and the resulting modification can disrupt RGS4 activity.
Initially, we set out to detect whether RGS4 was modified by 4HNE within cells. Western blot analysis of RGS4 was used to determine whether or not modification by 4HNE was possible. In whole cell lysate, RGS4 was consistently modified by 4HNE in treated cells. This modification was expected due to high accumulation of 4HNE in the plasma membrane and the membrane association of RGS4 when modulating signaling pathways.5,8 In PD, RGS4 activity is hypothesized to play a critical role in mediating the motor symptoms. Oxidative stress modulation of RGS4 signaling by covalent modification may act to attenuate this effect.
4HNE has been shown to modify cysteine, histidine, and lysine residues.29 In comparison to the cysteine-null mutant, which did not show detectable modification in the Western blot, Δ51-RGS4(WT) was modified by increasing concentrations of 4HNE. We did not expect to detect much modification at other residues due to the significantly increased reactivity of 4HNE with cysteine residues over both histidine and lysine residues.29 The construct used contains seven cysteine residues, four of which are within the RGS domain of RGS4.30 By mass spectrometry, we show that 4HNE readily modifies Δ51-RGS4(WT), but not the Δ51-RGS4(Δ7) mutant. This marks the first report of a lipid peroxidation product modifying an RGS protein. Based on previous experiments, we expected this to be the case. RGS4 has been previously shown to be sensitive to thiol modification at cysteine residues 132 and 148, suggesting those locations as modification sites for 4HNE as well.23 Further analysis of Δ51-RGS4(WT) revealed consistent modification at tryptic fragments corresponding to three residues: C71, C148, and C183. The detection of modification at C148 suggested a mode of inhibition of RGS4 by 4HNE similar to the previously reported small molecule inhibitors, such as inhibitor CCG-4986.23
In order to analyze modulation of RGS4 activity, a label free activity assay was required. A steady-state colorimetric assay was utilized to determine the effect of 4HNE on RGS4 GAP activity. At the concentrations examined, the free cysteine quenching of excess 4HNE protected Gαi, allowing for the facile analysis of 4HNE modification on RGS4 activity. 4HNE was found to inhibit Δ51-RGS4(WT). 1 mM 4HNE inhibited Δ51-RGS4(WT) at 50% of mock treated controls. The cysteine null mutant, Δ51-RGS4(Δ7), was not inhibited by 4HNE at similar concentrations. The resistance of Δ51-RGS4(Δ7) to inhibition by 4HNE indicates that cysteine modification accounts for the relevant modification of RGS4, similar to previously described mechanisms of action of known RGS4 inhibitors.23
To evaluate the ability of 4HNE to inhibit RGS4 directly within the cell, a subcellular localization assay was also employed. This assay successfully monitored the activity of novel RGS4 inhibitors in thte literature.31 In cells expressing both RGS4 and Gαo, its native binding partner, RGS4, is recruited to the plasma membrane. This is presumably due to interaction between the two, and the amount of RGS4 at the plasma membrane would represent the functional RGS4 within the cell. 1 μM 4HNE failed to delocalize RGS4 after 1 h treatment. However, 5 and 10 μM 4HNE solutions were able to reverse membrane localization of RGS4, indicating a loss of RGS4 activity. This results in a 100-fold difference in activity of 4HNE in the steady-state assay in comparison to the localization assay. This is most likely due to localization of 4HNE within plasma membranes resulting in a significantly higher local concentration of 4HNE in comparison to the media. Similar to previously identified small molecule inhibitors, 4HNE inhibits RGS4 through ablation of its interaction with its native binding partners, Gαi and Gαo.
In conclusion, we have shown RGS4 to be a target for covalent modification and inhibition by 4HNE, a common lipid peroxidation product. Specifically, in cultured cells exposed to 4HNE, RGS4 was readily identified as a target for modification. In this study, we show RGS4 is susceptible to modification at particular cysteine residues, including C148, which has previously been shown to be a target for covalent modification. With the identification of C148 as a modification target for 4HNE, this allows for the possibility for 4HNE to act as an internal control for aberrant signaling due to excess RGS4 activity in a variety of pathologies where oxidative stress is a strong component, such as PD. 4HNE inhibition of RGS4 could, in early stages of PD, act as a neuroprotector through minimizing RGS4 mediated motor deficits.17 Future studies will focus on determining the effects of 4HNE modification on RGS4, and other RGS proteins, on downstream signaling events critical to GPCR signal transduction.
Acknowledgments
We would like to give thanks to Dr. D. J. Murry in the University of Iowa College of Pharmacy for the use of his LC/ESI-IT-TOF instrument, the University of Iowa Central Microscopy Core for use of their confocal microscope, and Christopher R. Bodle, University of Iowa, for technical assistance and for manuscript preparation.
Funding: This work was supported by the grant NIHT32 GM067795 (to C.A.M.) and the University of Iowa College of Pharmacy Dissertation Fellowship (to C.A.M.).
Abbreviations
- 4HNE
4-hydroxy-2-nonenal
- BSA
bovine serum albumin
- GAP
GTPase accelerating protein
- GPCR
G-protein coupled receptor
- IRI
ischemia/reperfusion injury
- MBP
maltose binding protein
- MS
mass spectrometry
- PD
Parkinson's disease
- RGS4
regulator of G-protein signaling 4
- ROS
reactive oxygen species
Footnotes
Notes: The authors declare no competing financial interest.
References
- 1.Dauer W, Przedborski S. Parkinson's disease:mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 2.Gottlieb RAR. Cell death pathways in acute ischemia/reperfusion injury. J Cardiovasc Pharmacol Ther. 2011;16:233–238. doi: 10.1177/1074248411409581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yao JK, Keshavan MS. Antioxidants, redox signaling, and pathophysiology in schizophrenia: an integrative view. Antioxid Redox Signaling. 2011;15:2011–2035. doi: 10.1089/ars.2010.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siems W, Grune T. Intracellular metabolism of 4-hydroxynonenal. Mol Aspects Med. 2003;24:167–175. doi: 10.1016/s0098-2997(03)00011-6. [DOI] [PubMed] [Google Scholar]
- 5.Uchida K. 4-Hydroxy-2-nonenal: a product and mediator of oxidative stress. Prog Lipid Res. 2003;42:318–343. doi: 10.1016/s0163-7827(03)00014-6. [DOI] [PubMed] [Google Scholar]
- 6.Blanc EME, Kelly JFJ, Mark RJR, Waeg GG, Mattson MPM. 4-Hydroxynonenal, an aldehydic product of lipid peroxidation, impairs signal transduction associated with muscarinic acetylcholine and metabotropic glutamate receptors: possible action on G alpha(q/11) J Neurochem. 1997;69:570–580. doi: 10.1046/j.1471-4159.1997.69020570.x. [DOI] [PubMed] [Google Scholar]
- 7.Neubig RR, Connolly MP, Remmers AE. Rapid kinetics of G protein subunit association: a rate-limiting conformational change? FEBS Lett. 1994;355:251–253. doi: 10.1016/0014-5793(94)01212-1. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasa SP, Bernstein LS, Blumer KJ, Linder ME. Plasma membrane localization is required for RGS4 function in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:5584–5589. doi: 10.1073/pnas.95.10.5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young KH, Wang Y, Bender C, Ajit S, Ramirez F, Gilbert A, Nieuwenhuijsen BW. Yeast-based screening for inhibitors of RGS proteins. Meth Enzymol. 2004;389:277–301. doi: 10.1016/S0076-6879(04)89017-7. [DOI] [PubMed] [Google Scholar]
- 10.Roman DL, Talbot JN, Roof RA, Sunahara RK, Traynor JR, Neubig RR. Identification of small-molecule inhibitors of RGS4 using a high-throughput flow cytometry protein interaction assay. Mol Pharmacol. 2007;71:169–175. doi: 10.1124/mol.106.028670. [DOI] [PubMed] [Google Scholar]
- 11.Roman DL, Ota S, Neubig RR. Polyplexed flow cytometry protein interaction assay: A novel high-throughput screening paradigm for RGS protein inhibitors. J Biomol Screening. 2009;14:610–619. doi: 10.1177/1087057109336590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roof RA, Roman DL, Clements ST, Sobczyk-Kojiro K, Blazer LL, Ota S, Mosberg HI, Neubig RR. A covalent peptide inhibitor of RGS4 identified in a focused one-bead, one compound library screen. BMC Pharmacol. 2009;9:9. doi: 10.1186/1471-2210-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zielinski T, Kimple AJ, Hutsell SQ, Koeff MD, Siderovski DP, Lowery RG. Two Gαi1 rate-modifying mutations act in concert to allow receptor-independent, steady-state measurements of RGS protein activity. J Biomol Screening. 2009;14:1195–1206. doi: 10.1177/1087057109347473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blazer LL, Roman DL, Chung A, Larsen MJ, Greedy BM, Husbands SM, Neubig RR. Reversible, allosteric small-molecule inhibitors of regulator of G protein signaling proteins. Mol Pharmacol. 2010;78:524–533. doi: 10.1124/mol.110.065128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Siedlecki AM, Jin X, Thomas W, Hruska KA, Muslin AJ. RGS4, a GTPase activator, improves renal function in ischemia-reperfusion injury. Kidney Int. 2011;80:263–271. doi: 10.1038/ki.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding J, Guzman JN, Tkatch T, Chen S, Goldberg JA, Ebert PJ, Levitt P, Wilson CJ, Hamm HE, Surmeier DJ. RGS4-dependent attenuation of M4 autoreceptor function in striatal cholinergic interneurons following dopamine depletion. Nat Neurosci. 2006;9:832–842. doi: 10.1038/nn1700. [DOI] [PubMed] [Google Scholar]
- 17.Lerner TN, Kreitzer AC. RGS4 is required for dopaminergic control of striatal LTD and susceptibility to Parkinsonian motor deficits. Neuron. 2012;73:347–359. doi: 10.1016/j.neuron.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwab SG, Wildenauer DB. Update on key previously proposed candidate genes for schizophrenia. Curr Opin Psychiatry. 2009;22:147–153. doi: 10.1097/YCO.0b013e328325a598. [DOI] [PubMed] [Google Scholar]
- 19.Davydov IV, Varshavsky A. RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J Biol Chem. 2000;275:22931–22941. doi: 10.1074/jbc.M001605200. [DOI] [PubMed] [Google Scholar]
- 20.Kapust RB, Tözsér J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 21.Shankaranarayanan A, Thal DM, Tesmer VM, Roman DL, Neubig RR, Kozasa T, Tesmer JJG. Assembly of high order Gαq-effector complexes with RGS proteins. J Biol Chem. 2008;283:34923–34934. doi: 10.1074/jbc.M805860200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sternweis PC, Robishaw JD. Isolation of two proteins with high affinity for guanine nucleotides from membranes of bovine brain. J Biol Chem. 1984;259:13806–13813. [PubMed] [Google Scholar]
- 23.Roman DL, Blazer LL, Monroy CA, Neubig RR. Allosteric inhibition of the regulator of G protein signaling-Gα protein-protein interaction by CCG-4986. Mol Pharmacol. 2010;78:360–365. doi: 10.1124/mol.109.063388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monroy CA, Mackie DI, Roman DL. A High Throughput Screen for RGS Proteins Using Steady State Monitoring of Free Phosphate Formation. PLoS One. 2013;8:e62247. doi: 10.1371/journal.pone.0062247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baykov AA, Evtushenko OA, Avaeva SM. A malachite green procedure for orthophosphate determination and its use in alkaline phosphatase-based enzyme immunoassay. Anal Biochem. 1988;171:266–270. doi: 10.1016/0003-2697(88)90484-8. [DOI] [PubMed] [Google Scholar]
- 26.Roy AA, Lemberg KE, Chidiac P. Recruitment of RGS2 and RGS4 to the plasma membrane by G proteins and receptors reflects functional interactions. Mol Pharmacol. 2003;64:587–593. doi: 10.1124/mol.64.3.587. [DOI] [PubMed] [Google Scholar]
- 27.Elkon H, Melamed E, Offen D. Oxidative stress, induced by 6-hydroxydopamine, reduces proteasome activities in PC12 cells: implications for the pathogenesis of Parkinson's disease. J Mol Neurosci. 2004;24:387–400. doi: 10.1385/JMN:24:3:387. [DOI] [PubMed] [Google Scholar]
- 28.Zhang P, Hatter A, Liu B. Manganese chloride stimulates rat microglia to release hydrogen peroxide. Toxicol Lett. 2007;173:88–100. doi: 10.1016/j.toxlet.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doorn JA, Petersen DR. Covalent modification of amino acid nucleophiles by the lipid peroxidation products 4-hydroxy-2-nonenal and 4-oxo-2-nonenal. Chem Res Toxicol. 2002;15:1445–1450. doi: 10.1021/tx025590o. [DOI] [PubMed] [Google Scholar]
- 30.Tesmer JJG, Berman DM, Gilman AG, Sprang SR. Structure of RGS4 bound to AlF4−-activated Giα1: Stabilization of the transition state for GTP hydrolysis. Cell. 1997;89:251–261. doi: 10.1016/s0092-8674(00)80204-4. [DOI] [PubMed] [Google Scholar]
- 31.Blazer LL, Zhang H, Casey EM, Husbands SM, Neubig RR. A nanomolar-potency small molecule inhibitor of regulator of G-protein signaling proteins. Biochemistry. 2011;50:3181–3192. doi: 10.1021/bi1019622. [DOI] [PMC free article] [PubMed] [Google Scholar]