Abstract
Objective
In this report, we present a male patient with no family history of hearing loss, in whom we identified a novel de novo mutation in the POU3F4 gene.
Methods
One hundred ninety-four (194) Japanese subjects from unrelated and families were enrolled in this study. We used targeted genomic enrichment and massively parallel sequencing of all known non-syndromic hearing loss genes for identifying the genetic causes of hearing loss.
Results
A novel de novo frameshift mutation of POU3F4, to c.727_728insA (p.N244KfsX26) was identified. The patient was a 7-year-old male with congenital progressive hearing loss and inner ear deformity. Although the patient had received a cochlear implant, auditory skills were still limited. The patient also exhibited developmental delays similar to those previously associated with POU3F4 mutation.
Conclusion
This is the first report of a mutation in POU3F4 causing hearing loss in a Japanese patient without a family history of hearing loss. This study underscores the importance of comprehensive genetic testing of patients with hearing loss for providing accurate prognostic information and guiding the optimal management of patient rehabilitation.
Keywords: Hearing loss, genetics, POU3F4, cochlear implant, massively parallel sequencing
INTRODUCTION
The majority of genetic hearing loss is autosomal inherited (autosomal recessive: approximately 75%, autosomal dominant: approximately 20%), and X-linked hearing loss is estimated to account for 1%–5 % of genetic causes.1 To date, five loci and four genes have been implicated in X linked non-syndromic hearing loss (DFNX; Hereditary Hearing Loss Homepage, http://hereditaryhearingloss.org/). The most common cause of X linked hearing loss is mutation in the POU domain class 3 transcription factor 4 (POU3F4), which was mapped at DFNX2 loci on chromosome Xq21, and previous reports have described clinically heterogeneous disease phenotypes. 2–4 Hearing loss is severe—profound sensorineural hearing loss (SNHL)—which varies from congenital to late onset, is often progressive, and may include a conductive hearing loss component in some cases. Anatomically, computed tomography (CT) of the temporal bone in such cases reveals an enlarged internal auditory canal that coalesces with the basal turn of the cochlea along with partial hypoplasia.5 With these anatomical features, perilymphatic gusher is known to occur upon stapes surgery for correcting stapedial fixation, as well as during a cochlear implant surgery.6, 7
Based solely on frequency, if a male patient has hearing loss with an apparent X-linked form of inheritance segregating in the family, and/or if a temporal bone CT abnormality were found, POU34 gene could be the most likely cause. However, sporadic cases of SNHL with no family history can be difficult to recognize as a candidate and move on to the sequencing of entire POU3F4 gene. Recent advances in targeted genomic enrichment with massively parallel sequencing (TGE+MPS) have facilitated the simultaneous sequencing of all known causative genes.8, 9
Here, we describe a male with no family history of hearing loss in whom we identified a novel de novo mutation in the POU3F4 gene. This is the first report of a diagnosis of hearing caused by POU3F4 in a patient with no family history of hearing loss and highlights the importance of comprehensive genetic testing for optimal diagnostic rates for non-syndromic hearing loss.
SUBJECTS and METHODS
Subjects
One hundred ninety-four (194) Japanese subjects (114 females) from unrelated and non-consanguineous families were ascertained through 33 otolaryngology clinics in 28 prefectures across Japan. All subjects had presumed non-syndromic hearing loss. For each proband, informed consent was obtained to participate in this study, which was approved by the human subjects ethical committee associated with each clinic.
Clinical information and blood samples were obtained from each proband and from all consenting affected and unaffected relatives.
Targeted Genomic Enrichment and Massively Parallel Sequencing
Genomic DNA was assessed for quality by gel electrophoresis and spectrophotometry (Nanodrop 1000; Thermo Fisher Scientific, Waltham, MA; 260/280 ratio of 1.8–2.2) and quantity by fluorometry (Qubit 2.0 Fluorometer; Life Technologies, Carlsbad, CA). TGE of all exons of all genes implicated in SNHL, was completed as described, targeting 89 genes as part of the OtoSCOPE® v5 platform. Libraries were prepared using a modification of the solution-based Agilent SureSelect target enrichment system (Agilent Technologies, Santa Clara, CA).10
In brief, 3μg gDNA was randomly fragmented to an average size of 250 bp (Covaris Acoustic Solubilizer; Covaris Inc., Woburn, MA), fragment ends were repaired, A-tails were added, and sequencing adaptors were ligated before the first amplification. Solid phase reverse immobilization purifications were performed between each enzymatic reaction. Hybridization and capture with RNA baits were followed by a second amplification before pooling for sequencing. Minimal amplification was used – typically 8 cycles for the pre-hybridization PCR (range 8–10 cycles) using NEB Phusion HF Master Mix (New England BioLabs Inc, Ipswich, MA) and 14 cycles for the post-hybridization PCR (range 12–16 cycles) using Agilent Herculase II Fusion DNA Polymerase. All samples were barcoded and multiplexed before sequencing on either an Illumina MiSeq or HiSeq (Illumina Inc, San Diego, CA) in pools of 4–6 or 48, respectively, using 100-bp paired-end reads.
Bioinformatics Analysis
Data were analyzed as described using a local installation of the open-source Galaxy software (http://galaxyproject.org) and the following open-source tools: BWA11 for read mapping, Picard for duplicate removal, GATK12 for local re-alignment and variant calling and NGSRich13 for enrichment statistics9. We reported and annotated variants with custom software.
Variant Confirmation
All pathogenic variants were confirmed by Sanger sequencing and segregation analysis with exon-specific custom primers.
RESULTS
We identified one case with a causative mutation in the POU3F4 in the cohort of this study (194 hearing loss patients).
Case Details
The affected patient was a 7-year-old male with no particular perinatal events but failed newborn hearing screening. He was referred to Niigata University Hospital, Department of Otolaryngology for further examinations at the age of 2 months. An auditory brainstem response (ABR) revealed a bilateral hearing loss of approximately 70 dBnHL in both ear, and at least, clear responses were observed at 100 dBnHL. Behavioral observation audiometry demonstrated thresholds of 30 to 50dB between 500 and 2000Hz. Bilateral otittis media with effusion was observed with otoscopic findings. Bilateral high frequency sloping and mild-moderate SNHL was suspected at the age of 1 year. CT findings of the middle and inner ear showed partial cochlear hypoplasia and dilatation of the fundus of the internal auditory canal, which was also incompletely separated from the basal turn of the cochlea. At 2 years of age, his parents noticed that he did not respond to their voices and had only spoken a few words. Therefore, congenital and progressive SNHL was suspected, and the patient was promptly fitted for bilateral hearing aids. However, his hearing subsequently deteriorated, and ABR was absent at the age of 3 years 6months; the hearing aids were insufficient for adequate hearing.
Therefore, a cochlear implant (CI) surgery was performed in the left ear at the age of 4 years 3 months. Perilymphatic gusher occurred during cochleostomy, which was performed using a conventional method. The patient underwent implantation with a Nucleous 24 device and the strait array (Cochlear Ltd. Lane Cove, Australia); finally, electrode insertion was accomplished.
One year after CI, the patient’s sound field threshold was 30–40 dB at low and mid frequencies, but he still could not perceive high frequency sounds with the electrode at the basal turn of the cochlea. He continued to exhibit delayed speech and low ability to interpret Japanese at 2 years after CI (age: 6 years 6 months). His limited perceptual and communicative abilities led to a diagnosis of pervasive developmental disorder (PDD).
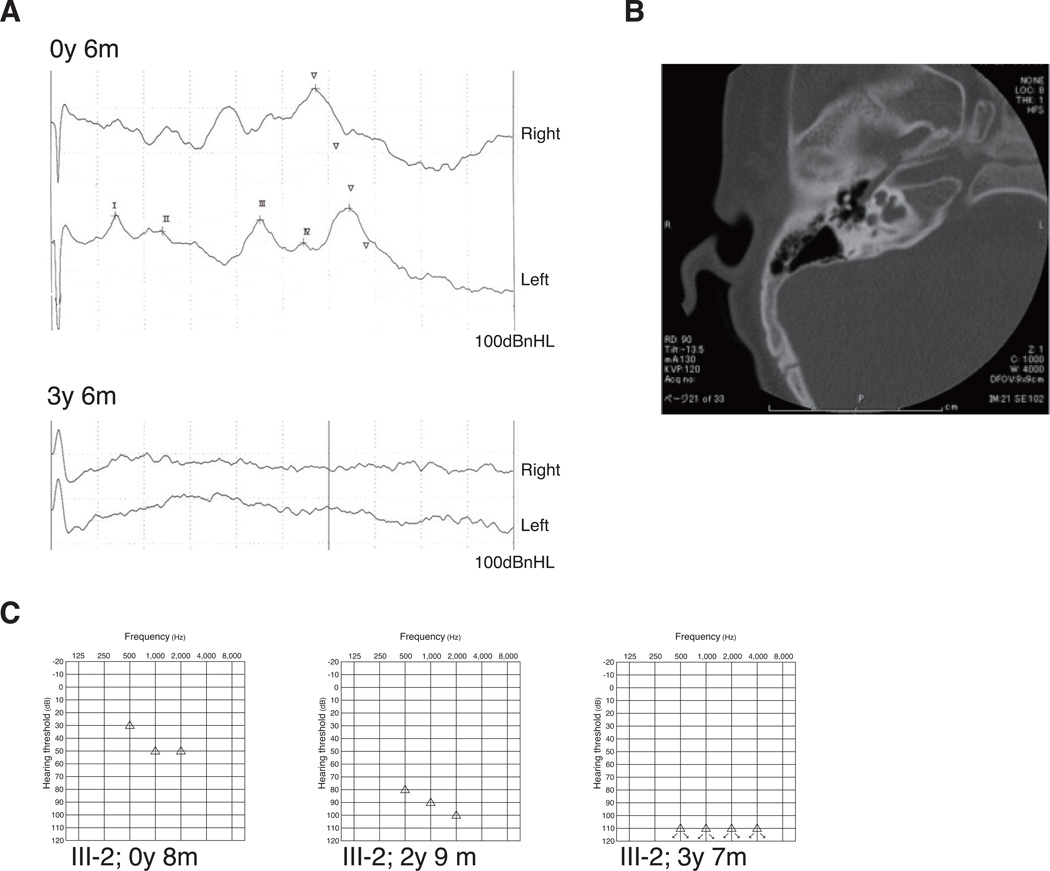
His parents and two siblings had normal hearing, and there was no family history of hearing loss or other cognitive disorders. The patient’s audiological assessment results, CT findings, and pedigree are shown in Figure 1.
Figure 1.
Clinical findings of the patient (ID 4750) (A) Auditory brain stem response (ABR) at the age of 6 months and 3 years with click stimuli at 100 dBnHL. A clear ABR was observed at the age of 6 months; however, ABR was absent in the following year. (B) Temporal bone computed tomography revealed partial cochlear hypoplasia, dilatation of the internal auditory canal fundus, and incomplete separation to the basal turn of the cochlea. (C) Behavioral observation audiometry (BOA) revealed deterioration of the threshold from 50 to 100 dB during the subsequent 3 years, suggesting progressive hearing loss.
Mutation analysis
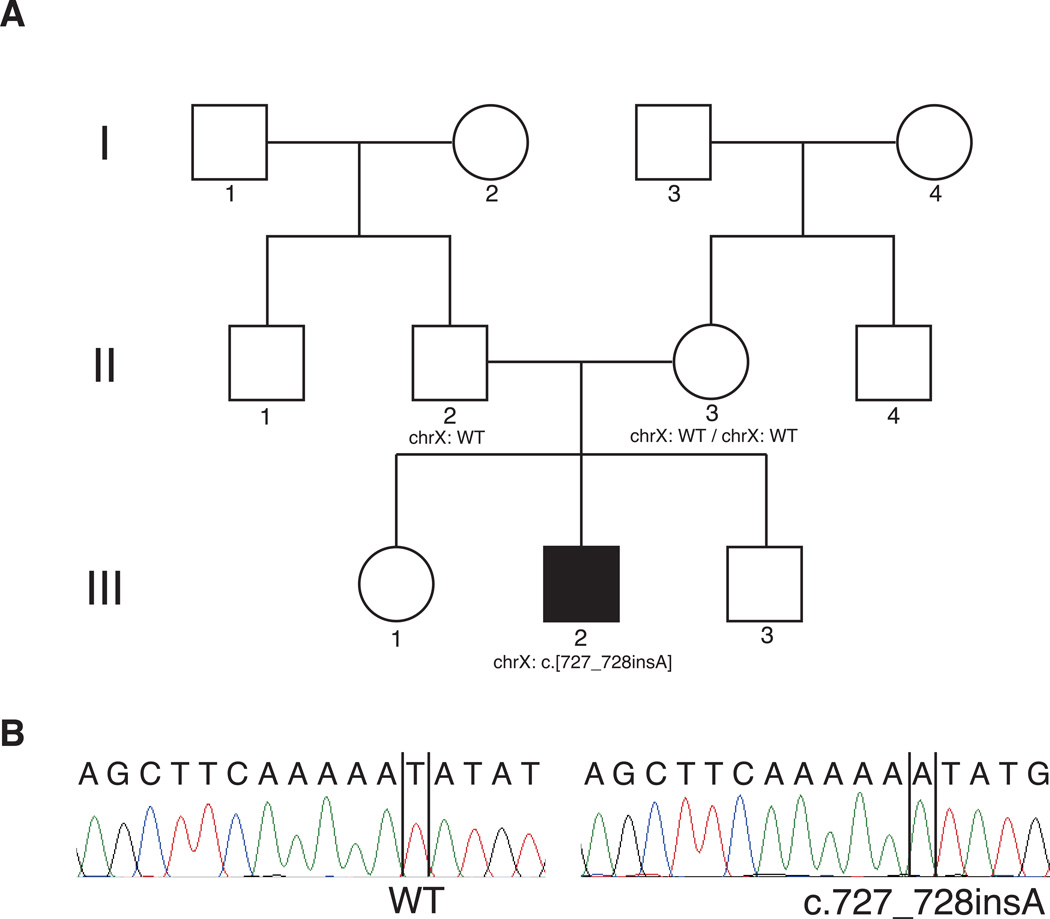
We performed comprehensive genetic testing using TGE + MPS of all known non-syndromic hearing loss genes as well as non-syndromic hearing loss mimic genes, as previously described.9 We identified a novel frameshift mutation in the POU3F4 gene, in which one adenine nucleotide was inserted. This mutation corresponded to c.727_728insA (NM_000307), and led to frameshift mutation and truncation (p.N244KfsX26). We also performed Sanger sequencing for family segregation study, and confirmed the gene variant in the proband. As shown in Figure 2, the Sanger sequencing revealed that the parents had no mutations, although his mother had been expected to the carrier of the variant. Therefore, we diagnosed the patient with a de novo mutation that had resulted in hearing loss with inner ear deformity.
Figure 2.
Pedigree of the patient (ID 4750) (A) Pedigree indicated a sporadic case in this family. (B) Electropherogram revealed a mutation only this patient (on the right) Parents (4751 and 4752) did not carry the mutation.
DISCUSSION
In this study, we identified a novel de novo POU3F4 in a case of sporadic hearing loss. TGE + MPS allowed us to identify the causative mutation, based upon all the known hearing loss genes screened. Although there has been only one previous report of a POU3F4 mutation in the Japanese population,14 suggesting a very rare event, many such cases have been reported in the Korean population, which harbors a very similar genetic background.15–17 Thus, there may be more Japanese patients harboring POU3F4 mutations even in sporadic cases that are not identified as having X-linked inheritance mode.
As shown in Table 1, more than 30 mutations, point mutations and genomic rearrangements involving in large deletions and inversions on the upstream gene regulatory element have been reported.3, 18–20 However, it is difficult to detect such large genomic alterations using conventional PCR-based DNA sequencing. Anger et al. have reported normal genetic sequencing results in a DFNX2 case with chromosomal rearrangements in the vicinity of POU3F4, suggesting a failure of Sanger sequencing to identify mutations because of genetic causes.21 In contrast, it is possible to identify such genetic alterations using MPS instead of tests such as array comparative genomic hybridization and fluorescence in situ hybridization.
Table 1.
Known mutations in POU3F4 gene and associated phenotypes.
| Nucleotide change | Amino acid change | Inheritance | CT findings | HL on set | Type of HL | Progression | Other features | Perilymphatic gusher | Year | Authors |
|---|---|---|---|---|---|---|---|---|---|---|
| Large delation | Inherited | IP3 | Congenital | Mixed | Progressive | gusher | 2005 | Vore | ||
| Large delation | Inherited | Mondini | Congenital | SNHL | Progressive | 2000 | Arellano | |||
| Large delation | Inherited | Mixed | Mental retardation | gusher | 1996 | de Kok | ||||
| Large delation | Inherited | IP3 | Congenital | Progressive | Developmental delay, MR | 2010 | Song | |||
| Large delation | De novo (Sporadic) | IP3 | 2014 | Choi | ||||||
| Deletion (Upstream and gene) | Inherited | Bony defect | Mixed | gusher | 1996 | de Kok | ||||
| Upstream delation | Inherited | Bony defect | Mixed | 1996 | de Kok | |||||
| Upstream delation | IP3 | Congenital | Mixed | 1996 | de Kok | |||||
| Upstream delation | Inherited | Congenital | SNHL | Progressive | gusher | 2010 | Naranjo | |||
| Upstream delation | De novo (Sporadic) | IP3 | Early | no | 2010 | Song | ||||
| Upstream duplication | Inherited | IP3 | Mixed | 1995 | de Kok | |||||
| Regulatory region deletion | Inherited | Bony defect | Early | Mixed | no | 2014 | Stanton | |||
| Regulatory region inversion | Inherited | IP3 | Mixed | Progressive | Developmental delay | 2013 | Anger | |||
| c.235C>T | p.G79X | De novo (Sporadic) | 2013 | Parzefall | ||||||
| c.346delG | p.A116fs | Inherited | IP3 | Congenital | Mixed | Limited verval comminication | 2009 | Lee PhyGen | ||
| c.383delG | p.G128fs | Inherited | IP3 | Early | SNHL | Progressive | 2009 | Lee | ||
| c.499C>T | p.R167X | Inherited | IP3 | Congenital | Mixed | no | Attention disorders, Learning delay | gusher | 2010 | Stankovic |
| c.540C>A | p.C180X | De novo (Sporadic) | IP3 | 2014 | Choi | |||||
| c.601-606del | p.201-202del | Inherited | Bony defect | Early | Mixed | no | gusher | 1998 | Hagiwara | |
| c.603delA | p.K202X | Inherited | Bony defect | SNHL | 1995 | de Kok | ||||
| c.623T>A | p.L208X | Inherited | IP3 | Early | SNHL | Severe | Mental retardation | 2009 | Lee | |
| c.623T>A | p.L208X | Inherited | IP3 | SNHL | 2013 | Choi | ||||
| c.632C>T | p.T211M | Inherited | IP3 | Mixed | 2013 | Choi | ||||
| c.647G>A | p.G216E | Inherited | IP3 | Congenital | SNHL | Progressive | 2010 | Li | ||
| c.648delG | p.D215X | Bony defect | Mixed | gusher | 1995 | de Kok | ||||
| c.683C>T | p.S228L | Inherited | IP3 | Congenital | Mixed | Progressive | Developmental delay | 2005 | Vore | |
| c.686A>G | p.G229R | Inherited | IP3 | SNHL | 2013 | Choi | ||||
| c.689C>T | p.T230I | Bony defect | Mixed | gusher | 1997 | Friedman | ||||
| c.727_728insA | p.N244KfsX26 | De novo (Sporadic) | IP3 | Congenital | SNHL | Progressive | Pervasive developmental disorder | gusher | 2014 | This report |
| c.853-854del | p.I285Rfs43 | 2013 | Parzefall | |||||||
| c.862-866del | p.fs | Inherited | Early | Mixed | gusher | 1995 | Binter-Glindzicz | |||
| c.895delA | p.L298X | Mixed | gusher | 1995 | de Kok | |||||
| c.910C>A | p.P303H | Inherited | IP3 | Congenital | 2014 | Choi | ||||
| c.925T>C | p.S309P | Inherited | IP3 | Congenital | Mixed | no | 2006 | Wang | ||
| c.927-929del | p.S310del | Inherited | IP3 | Early | Mixed | 2009 | Lee PhyGen | |||
| c.927-929del | p.S310del | unknown (Adopted) | IP3 | Early | Mixed | no | Learning delay | gusher | 2010 | Stankovic |
| c.935C>T | p.A312V | Inherited | Bony defect | Congenital | SNHL | Learning difficulty | 1995 | Binter-Glindzicz | ||
| c.950dupT | p.L317FfsX12 | Inherited | IP3 | Mixed | 2013 | Choi | ||||
| c.950T>G | p.L317W | Bony defect | Mixed | 1995 | de Kok | |||||
| c.967C>G | p.A323G | Mixed | gusher | 1997 | de Kok | |||||
| c.973T>A | p.W325R | Inherited | IP3 | Early | SNHL | Progressive | gusher | 2011 | Schild | |
| c.985C>G | p.R329G | IP3 | Mixed | gusher | 1997 | Friedman | ||||
| c.986G>C | p.R329G | Inherited | IP3 | Early | Mixed | Progressive | 2009 | Lee PhyGen | ||
| c.990A>T | p.A330S | Bony defect | SNHL | Progressive | Growth retardation | gusher | 1997 | de Kok | ||
| c.1000A>G | p.K334E | Mixed | Progressive | 1995 | de Kok | |||||
| c.1069delA | p.T354GfsX115 | Inherited | IP3 | SNHL | 2013 | Choi | ||||
| c.1084T>C | p.X362RexfX113 | De novo(Sporadic) | IP3 | SNHL | 2013 | Choi |
Abbreviation: IP3, incomplete partition type III; SNHL, sensorineural hearing loss. Empty rows indivate unspecified in the reports.
POU3F4 encodes a transcription factor that binds DNA using two specific domains: POU-specific domain and POU homeodomain. These domains play essential roles in inner ear development and are expressed in both the brain and neural tube. Several studies of POU3F4 knockout and mutant mouse models have been published; Parzefall et al. have clearly described an observed shortened cochlea duct as a possible Mondini malformation in a mouse harboring a POU3F4 mutation22. A case involving DFNX2 with POU3F4 mutations was characterized as exhibiting an inner ear deformity of incomplete partition type III.17 Our presented case harbored the same cochlear deformity as described in previous reports and was consistent with a de novo mutation in POU3F4 that led to congenital progressive hearing loss.
Regarding phenotypic features, previously reported manifestations and clinical histories are presented in Table 1. However, we did not observe considerable variations in the genotype-associated phenotypes. Therefore, it is difficult to determine whether these genotype–phenotype correlations exist as described in previous reports (Table 1). Cochlear deformities were observed in most cases, demonstrating mixed hearing losses that were independent of middle ear function and perilymphatic gusher. Several DFNX2 cases with large genomic deletions or chromosomal rearrangements (even single nucleotide variants) have exhibited developmental delays in addition to hearing loss (Table 1).7, 23, 24 PDD was diagnosed in our patient, who was suspected to exhibit some syndromic features. In general, hearing loss may impact early childhood development, and differences in hearing levels, social background, and parental factors may affect language acquisition and learning abilities. However, hearing loss-associated POU3F4 mutation should be additionally considered as a cause of developmental delays involving communication skills, particularly in CI recipients.
Prior to cochlear implantation, we were unable to provide either genetic testing results or counseling for this patient as TGE+ MPS technologies were unavailable for common clinical usage at that time. However, these technologies are currently applicable, and the further evolution of genetic testing will facilitate the accurate diagnosis of hearing loss. Furthermore, we must also rigorously establish phenotypes with respect to the hearing loss level, progression and other manifestations. Regarding hearing loss caused by mutations in POU3F4, all clinicians and audiologists should provide optimal CI rehabilitation management and applicable educational supports based on the phenotypic features described herein and elsewhere. This study supports the use of comprehensive genetic diagnosis for SNHL to provide the highest chance of diagnostic success, particularly in sporadic cases.
ACKNOWLEDGEMENTS
This work was supported by NIDCD RO1s DC003544, DC002842 and DC012049 to RJHS. We thank Mr. Jim George for help with manuscript preparation.
Footnotes
DECLARATION OF CONFLICTING INTERESTS
All authors have declared no competing financial interests.
REFERENCES
- 1.Petersen MB, Wang Q, Willems PJ. Sex-linked deafness. Clin. Genet. 2008;73(1):14–23. doi: 10.1111/j.1399-0004.2007.00913.x. [DOI] [PubMed] [Google Scholar]
- 2.Phelps PD, Reardon W, Pembrey M, Bellman S, Luxom L. X-linked deafness, stapes gushers and a distinctive defect of the inner ear. Neuroradiology. 1991;33(4):326–330. doi: 10.1007/BF00587816. [DOI] [PubMed] [Google Scholar]
- 3.de Kok YJ, Merkx GF, van der Maarel SM, et al. A duplication/paracentric inversion associated with familial X-linked deafness (DFN3) suggests the presence of a regulatory element more than 400 kb upstream of the POU3F4 gene. Hum. Mol. Genet. 1995;4(11):2145–2150. doi: 10.1093/hmg/4.11.2145. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Cheng J, Lu Y, et al. Identification of a novel mutation in POU3F4 for prenatal diagnosis in a Chinese family with X-linked nonsyndromic hearing loss. J Genet Genomics. 2010;37(12):787–793. doi: 10.1016/S1673-8527(09)60096-5. [DOI] [PubMed] [Google Scholar]
- 5.Friedman RA, Bykhovskaya Y, Tu G, et al. Molecular analysis of the POU3F4 gene in patients with clinical and radiographic evidence of X-linked mixed deafness with perilymphatic gusher. Ann. Otol. Rhinol. Laryngol. 1997;106(4):320–325. doi: 10.1177/000348949710600411. [DOI] [PubMed] [Google Scholar]
- 6.Lee HK, Lee SH, Lee KY, et al. Novel POU3F4 mutations and clinical features of DFN3 patients with cochlear implants. Clin. Genet. 2009;75(6):572–575. doi: 10.1111/j.1399-0004.2009.01181.x. [DOI] [PubMed] [Google Scholar]
- 7.Stankovic KM, Hennessey AM, Herrmann B, Mankarious LA. Cochlear implantation in children with congenital X-linked deafness due to novel mutations in POU3F4 gene. Ann. Otol. Rhinol. Laryngol. 2010;119(12):815–822. doi: 10.1177/000348941011901205. [DOI] [PubMed] [Google Scholar]
- 8.Shearer AE, DeLuca AP, Hildebrand MS, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc. Natl. Acad. Sci. U. S. A. 2010;107(49):21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shearer AE, Black-Ziegelbein EA, Hildebrand MS, et al. Advancing genetic testing for deafness with genomic technology. J. Med. Genet. 2013;50(9):627–634. doi: 10.1136/jmedgenet-2013-101749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shearer AE, Hildebrand MS, Smith RJ. Solution-based targeted genomic enrichment for precious DNA samples. BMC Biotechnol. 2012;12(1):20. doi: 10.1186/1472-6750-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frommolt P, Abdallah AT, Altmuller J, et al. Assessing the enrichment performance in targeted resequencing experiments. Hum. Mutat. 2012;33(4):635–641. doi: 10.1002/humu.22036. [DOI] [PubMed] [Google Scholar]
- 14.Hagiwara H, Tamagawa Y, Kitamura K, Kodera K. A new mutation in the POU3F4 gene in a Japanese family with X-linked mixed deafness (DFN3) Laryngoscope. 1998;108(10):1544–1547. doi: 10.1097/00005537-199810000-00022. [DOI] [PubMed] [Google Scholar]
- 15.Lee HK, Song MH, Kang M, et al. Clinical and molecular characterizations of novel POU3F4 mutations reveal that DFN3 is due to null function of POU3F4 protein. Physiol Genomics. 2009;39(3):195–201. doi: 10.1152/physiolgenomics.00100.2009. [DOI] [PubMed] [Google Scholar]
- 16.Song MH, Lee HK, Choi JY, Kim S, Bok J, Kim UK. Clinical evaluation of DFN3 patients with deletions in the POU3F4 locus and detection of carrier female using MLPA. Clin. Genet. 2010;78(6):524–532. doi: 10.1111/j.1399-0004.2010.01426.x. [DOI] [PubMed] [Google Scholar]
- 17.Choi JW, Min B, Kim A, et al. De Novo Large Genomic Deletions Involving POU3F4 in Incomplete Partition Type III Inner Ear Anomaly in East Asian Populations and Implications for Genetic Counseling. Otol Neurotol. 2014 doi: 10.1097/MAO.0000000000000343. [DOI] [PubMed] [Google Scholar]
- 18.de Kok YJ, Vossenaar ER, Cremers CW, et al. Identification of a hot spot for microdeletions in patients with X-linked deafness type 3 (DFN3) 900 kb proximal to the DFN3 gene POU3F4. Hum. Mol. Genet. 1996;5(9):1229–1235. doi: 10.1093/hmg/5.9.1229. [DOI] [PubMed] [Google Scholar]
- 19.Vore AP, Chang EH, Hoppe JE, et al. Deletion of and novel missense mutation in POU3F4 in 2 families segregating X-linked nonsyndromic deafness. Arch. Otolaryngol. Head Neck Surg. 2005;131(12):1057–1063. doi: 10.1001/archotol.131.12.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naranjo S, Voesenek K, de la Calle-Mustienes E, et al. Multiple enhancers located in a 1-Mb region upstream of POU3F4 promote expression during inner ear development and may be required for hearing. Hum. Genet. 2010;128(4):411–419. doi: 10.1007/s00439-010-0864-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anger GJ, Crocker S, McKenzie K, et al. X-linked deafness-2 (DFNX2) phenotype associated with a paracentric inversion upstream of POU3F4. Am J Audiol. 2014;23(1):1–6. doi: 10.1044/1059-0889(2013/13-0018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parzefall T, Shivatzki S, Lenz DR, et al. Cytoplasmic mislocalization of POU3F4 due to novel mutations leads to deafness in humans and mice. Hum. Mutat. 2013;34(8):1102–1110. doi: 10.1002/humu.22339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hildebrand MS, de Silva MG, Tan TY, et al. Molecular characterization of a novel X-linked syndrome involving developmental delay and deafness. Am J Med Genet A. 2007;143A(21):2564–2575. doi: 10.1002/ajmg.a.31995. [DOI] [PubMed] [Google Scholar]
- 24.Stanton SG, Griffin A, Stockley TL, et al. X-linked hearing loss: two gene mutation examples provide generalizable implications for clinical care. Am J Audiol. 2014;23(2):190–200. doi: 10.1044/2014_AJA-13-0040. [DOI] [PubMed] [Google Scholar]