Abstract
Macroautophagy (hereafter autophagy), literally defined as a type of self-eating, is a dynamic cellular process in which cytoplasm is sequestered within a unique compartment termed the phagophore. Upon completion, the phagophore matures into a double-membrane autophagosome that fuses with the lysosome or vacuole, allowing degradation of the cargo. Nonselective autophagy is primarily a cytoprotective response to various types of stress; however, the process can also be highly selective. Autophagy is involved in various aspects of cell physiology, and its dysregulation is associated with a range of diseases. The regulation of autophagy is complex, and the process must be properly modulated to maintain cellular homeostasis. In this review, we focus on the current state of knowledge concerning transcriptional, post-transcriptional, and post-translational regulation of autophagy in yeast and mammals.
Keywords: lysosome, phagophore, stress, vacuole, yeast
Introduction
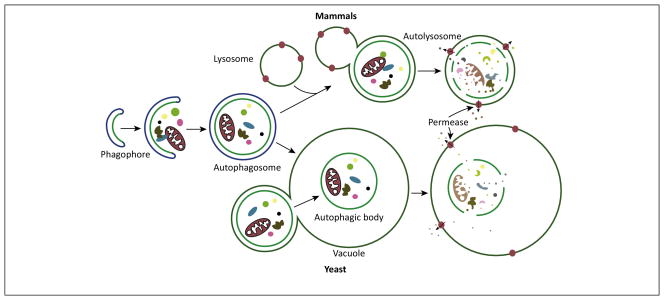
Autophagy (see Glossary) is highly conserved from yeast to mammals, both morphologically and with regard to the protein constituents that make up the core autophagy machinery. Autophagy can be selective or nonselective, depending on the type of cargo that is sequestered. Upon starvation or other stress conditions, autophagy is induced. Cytoplasm and superfluous or dysfunctional organelles are sequestered by the expanding phagophore, leading to the formation of the double-membrane autophagosome (Figure 1). The autophagosome subsequently fuses with the vacuole membrane in yeast, or the lysosome in mammalian cells; in yeast, the inner autophagosome vesicle becomes an autophagic body in the vacuole lumen, whereas in mammalian cells fusion generates an autolysosome. Eventually, the sequestered cargos are degraded or processed by hydrolases. Following degradation, the breakdown products are released back into the cytosol through permeases. This allows recycling of the macromolecular constituents as building blocks, to generate energy to maintain cell viability under unfavorable conditions and to protect the cell during various types of stress [1–4].
Figure 1.
The morphology of autophagy in yeast and mammals. The primary membrane structures involved in autophagy are depicted. The phagophore is the initial sequestering compartment that expands into the double-membrane autophagosome. The latter fuses with the vacuole (yeast) or the lysosome (mammals) to allow access to, and degradation of, the cargo (the image depicts the sequestration of random cytoplasm including an organelle). The resulting breakdown products are subsequently released back into the cytosol through integral membrane permeases for reuse.
Genetic screens for autophagy-defective mutants in yeast have led to the identification of almost 40 autophagy-related (ATG) genes, many of which have orthologs in higher eukaryotes. Among these ATG genes, one subgroup, consisting of approximately 18 genes, is shared among the various types of autophagy including nonselective macroautophagy, the cytoplasm-to-vacuole-targeting pathway (a biosynthetic autophagy-like pathway), mitophagy (the selective autophagic degradation of mitochondria), and pexophagy (the selective autophagic degradation of peroxisomes). More specifically, the corresponding gene products of this subgroup are required for autophagosome formation, and thus are termed the core autophagy machinery. The core Atg proteins can be divided into different functional subgroups: (i) the Atg1 complex (Atg1, Atg13, Atg17, Atg29, and Atg31) is the initial complex that regulates the induction of autophagosome formation, which occurs at the phagophore assembly site (PAS); (ii) Atg9 and its cycling system (Atg23, Atg27, Atg2, and Atg18) may play a role in delivery of at least some of the membrane used to expand the phagophore after the assembly of the Atg1 complex at the PAS; (iii) the phosphatidylinositol (PtdIns) 3-kinase (PtdIns3K) complex (Vps34, Vps30/Atg6, Vps15, Atg14, and Atg38) acts at the stage of vesicle nucleation, and is involved in the recruitment of PtdIns 3P-binding proteins to the PAS; (iv) two ubiquitin-like conjugation systems involving Atg12 (Atg5, Atg7, Atg10, Atg12, and Atg16) and Atg8 (Atg3, Atg4, Atg7, and Atg8) play roles in vesicle expansion [1].
Autophagy is involved in normal aspects of cell development and physiology, and defects in this process are associated with a range of diseases in humans. Accordingly, there has been tremendous interest in manipulating autophagy for therapeutic purposes. One fundamental issue, however, is that either too little or too much autophagy can be deleterious. Therefore, the cell, and clinicians interested in modulating this process, need to ensure tight regulation of the induction and magnitude of autophagy. Thus, a thorough understanding of the mechanisms involved is crucial to allow the manipulation of autophagy for the treatment of disease. In this review, we discuss the transcriptional, post-transcriptional, and post-translational regulation of autophagy.
Transcriptional regulation
Autophagy can be stimulated by several intracellular and extracellular cues. In this review, unless stated otherwise, we refer to the autophagic response during nutrient deprivation because it is the best-characterized physiological situation for inducing autophagy. TOR (target of rapamycin) kinase has long been known as the master regulator of autophagy, and as part of TOR complex 1, it controls a range of transcription factors in response to starvation; however, the relevant downstream targets with regard to autophagy regulation have not been well understood. Only recently have researchers started to identify an entire network of transcription factors that are involved in controlling autophagy (Table 1). Indeed, several transcription factors including TP53 (tumor protein p53), STAT3 (signal transducer and activator of transcription 3), and NFKB/NF-κB (nuclear factor of kappa light polypeptide gene enhancer in B-cells) play a dual role in autophagy regulation, through both transcriptional (nuclear interaction) and transcriptional-independent (cytoplasmic interaction) mechanisms, acting as both activators and repressors [5–7]. Below, we discuss the complex transcriptional network involved in autophagy regulation, highlighting those factors that are currently the most characterized with regard to their role in this process.
Table 1.
Transcriptional factors of mammalian autophagy
| Transcription factor | Target | Effecta | Refs |
|---|---|---|---|
| ATF4 | ATG5, LC3, and ULK1 | + | [96–99] |
| ATF5 | MTOR | − | [100] |
| CEBPB | BNIP3, LC3, and ULK1 | + | [101] |
| DDIT3/CHOP | ATG5 and LC3 | + | [96] |
| E2F1 | ATG5, BNIP3, LC3, and ULK1 | + | [22,24,102] |
| FOXO1 | ATG5, ATG12, ATG14, BECN1, BNIP3, LC3, and PIK3C3/VPS34 | + | [10,16,24] |
| FOXO3 | ATG4, ATG12, BECN1, BNIP3, LC3, ULK1, ULK2, and PIK3C3/VPS34 | + | [9,12,15,103,104] |
| GATA1 | LC3 | + | [105] |
| JUN | BECN1 and LC3 | + | [106–108] |
| NFKB | BCL2, BECN1, and BNIP3 | + or − | [24,29,109] |
| TP53 | ATG2, ATG4, ATG7, ATG10, BCL2, and ULK1 | Cytosol −; nucleus + | [110,111] |
| SOX2 | ATG10 | + | [34,112] |
| SREBF2/SREBP2 | ATG4 and LC3 | + | [113] |
| STAT1 | ATG12 and BECN1 | − | [114] |
| STAT3 | ATG3, BCL2, and BNIP3 | − | [115,116] |
| TFEB | ATG4, ATG9, BCL2, LC3, and WIPI | + | [117] |
| ZKSCAN3 | LC3, ULK1, and WIPI | − | [118] |
+, enhanced autophagy; −: suppressed autophagy.
FOXO family
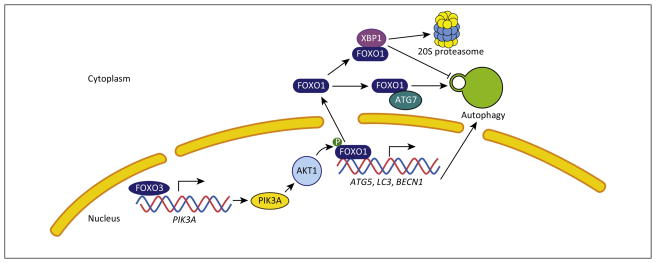
FOXO (forkhead box O) family proteins contain a characteristic DNA binding motif and function primarily by controlling the expression of genes involved in cell growth and differentiation. The regulation of autophagy by FOXO proteins has been described in various cell types, after first being demonstrated in cardiomyocytes [8–13]. FOXO proteins shuttle between the nucleus and cytoplasm, where they carry out distinct functions, and this translocation is mainly regulated by phosphorylation. Nutrient deprivation leads to FOXO3 nuclear localization (Figure 2); activated FOXO3 promotes autophagy by increasing the expression of ATG genes (Table 1), especially those of the core machinery that are involved in phagophore and autophagosome formation [8]. FOXO3 also induces transcription-dependent autophagy, in which FOXO1, another member of the FOXO protein family, is required [14]. FOXO1 regulates autophagy by two distinct mechanisms: it stimulates the transcription of various ATG genes [8,15,16], and it induces autophagy in the cytosol through direct binding of acetylated FOXO1 to ATG7 [17]. FOXO3 also dramatically increases the expression of the class I phosphoinositide 3-kinase catalytic subunit PIK3CA (phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha), leading to an increase in AKT1 (v-akt murine thymoma viral oncogene homolog 1) activity. In turn, AKT1 phosphorylates FOXO1 leading to its nuclear export and an increase in FOXO1-induced autophagy [14,17–19].
Figure 2.
The regulation of autophagy in mammalian cells through FOXO (forkhead box O) proteins. FOXO1 and FOXO3 shuttle between the cytosol and nucleus, controlling autophagy through transcriptional regulation, and by directly binding to ATG7 (autophagy related 7).
FOXO1 also affects autophagy through its connection with the unfolded protein response (UPR). In the UPR, the cell responds to ER stress (e.g., due to the accumulation of unfolded proteins within the lumen) by upregulating chaperones, and also via transcriptional changes. The stress sensor ERN1/IRE1α(endoplasmic reticulum to nucleus signaling 1), which regulates the UPR, catalyzes the splicing of XBP1 (X-box binding protein 1) mRNA to generate XBP1s, which in turn binds to and inhibits FOXO1. In addition, the unspliced variant of XBP1, XBP1u, promotes the proteasomal degradation of FOXO1 by recruiting it to the 20S proteasome; the absence of XBP1u results in a sustained level of FOXO1 and the persistent activation of autophagy (Figure 2) [20,21].
E2F family
The E2F family proteins constitute a group of transcription factors that have been best characterized for their function in cell cycle regulation. Proteins in this family act either as transcriptional activators or repressors. E2F1 (E2F transcription factor 1) is a key activator, which targets and upregulates the expression of various ATG genes (Table 1). Conversely, reducing endogenous E2F1 expression inhibits DNA damage-induced autophagy [22]. E2F transcription factors can form a complex with RB1 (retinoblastoma 1), resulting in transcriptional repression. For example, E2F1-RB1 targets the promoter of BNIP3(BCL2/adenovirus E1B 19kDa interacting protein 3), a positive regulator of autophagy, and attenuates the induction of BNIP3 by HIF1A [hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)]; BNIP3 regulates hypoxia-induced autophagy by disrupting the interaction between BCL2 (B-cell CLL/lymphoma 2) and BECN1 (beclin 1, autophagy related) [23]. Recent studies have found that the transcription factor NFKB provides a molecular switch that determines whether E2F1 promotes cell proliferation or death under physiological conditions. Under basal conditions, NFKB constitutively occupies and transcriptionally silences Bnip3 gene transcription by competing with E2F1 for Bnip3 promoter binding. Conversely, in the absence of NFKB, basal Bnip3 gene transcription is activated leading to upregulated autophagy and potentially cell death [24].
NFKB
NFKB is a transcription factor that is most closely linked to the immune response, for example, it plays a role in cytokine production. The NFKB system and autophagy are intricately interconnected via various mechanisms. Several autophagic stimuli, including nutrient deprivation, which leads to inhibition of MTOR (mechanistic TOR, the mammalian homolog of yeast TOR), require components of the canonical NFKB activation pathway such as CHUK/IKKα (conserved helix-loop-helix ubiquitous kinase) and IKBKB/IKKβ (inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta) [25–28]. Additionally, there is a reciprocal relationship between the NFKB system and autophagy. While several components of the NFKB signaling pathway contribute to optimal autophagic responses (e.g., NFKB is an upregulator of ATG genes such as BECN1 [29]), autophagy appears to be required for the activation of NFKB[30].
TP53
TP53 (tumor protein p53) is a transcription factor that regulates the cell cycle; mutations in the TP53 gene are associated with a large number of cancers. In addition to its well-characterized role as a key tumor suppressor, other aspects of TP53 function have been elucidated. Certain proteins that are under the control of TP53 [e.g., DRAM1 (DNA-damage regulated autophagy modulator 1)] stimulate autophagy flux, whereas others (e.g., AMP-activated protein kinase, AMPK, a central sensor of the cellular energy level) inhibit MTOR [31] pointing to TP53 as a positive regulator of autophagy. However, TP53 can modulate autophagy in a dual fashion, depending on its subcellular localization. In the nucleus, TP53 acts as a transcription factor that transactivates ATG genes, such as ATG2, ATG4, ATG7, and ATG10. Conversely, cytosolic TP53 can suppress autophagy [32,33]. Finally, TP63 and TP73 can substitute for TP53 during autophagy regulation, because they bind a similar set of autophagic target genes [34].
Ume6 complex
Following autophagy induction, Atg8 (the yeast homolog of mammalian LC3, microtubule-associated protein 1 light chain 3) is the most dramatically upregulated of the core machinery proteins. The amount of Atg8 correlates with the size of autophagosomes and therefore plays a critical role in modulating the magnitude of the autophagic response [35]. In yeast, Ume6 (Unscheduled Meiotic gene Expression) is part of a complex that includes the corepressor Sin3 (Switch IN dependent) and the histone deacetylase Rpd3 (Reduced Potassium Dependency), which acts as a negative regulator of ATG8 transcription. Deletion of any of the corresponding genes leads to an increase in Atg8 and autophagy activity. This regulatory mechanism is partly conserved in mammals, where homologs of Rpd3 and Sin3 are present [35].
Pho23
Pho23 (PHOsphate metabolism) negatively controls the mRNA levels, and subsequent protein levels, of many components of the autophagy core machinery, including Atg1, Atg7, Atg8, Atg9, and Atg14. Elevated autophagy activity and increased frequency of autophagosome formation is observed in the absence of Pho23. Transcription of ATG9 is most strongly affected by Pho23 [36]. Atg9 has a role in regulating the frequency of autophagosome formation; cells expressing lower levels of Atg9 have reduced numbers of autophagosomes. At this time, the regulation of autophagy through Pho23 has only been demonstrated in yeast and it remains unclear as to whether a similar mechanism exists in mammals.
Post-transcriptional regulation of autophagy: focus on miRNA
Different steps of autophagy that utilize the core machinery are regulated by various post-transcriptional mechanisms, including the action of noncoding microRNAs (miRNAs; summarized in Table 2) [37–40]. miRNAs are a class of small endogenous regulatory RNAs that control gene expression at the post-transcriptional level, typically by translational arrest or mRNA cleavage, through interaction mainly at the 3′-untranslated regions (UTRs) of the target mRNAs. miRNAs play important roles in the regulation of development, growth, and metastasis of cancer, and in determining the response of tumor cells to anticancer therapy. Here, we highlight some of the recent data concerning the involvement of miRNAs in regulating different steps of the autophagy core machinery.
Table 2.
miRNAs involved in regulating the autophagy core machinery
| miRNA | Target | Refs | |
|---|---|---|---|
| Mir20a, Mir106b | Ulk1 | Autophagy induction | [41] |
| MIR885-3p | ULK2 | [42] | |
| MIR10A | RB1CC1 | [119] | |
| Mir195 | Atg14 | Phagophore nucleation | [43] |
| MIR30A, MIR30D, MIR376B, MIR519A | BECN1 | [45,46,120,121] | |
| MIR101, MIR376B | ATG4 | Phagophore elongation | [46,47] |
| MIR30A, MIR30D, MIR181A, MIR374A | ATG5 | [45,120,122] | |
| MIR17, MIR375 | ATG7 | [109,123] | |
| MIR519A | ATG10 | [120] | |
| MIR30D, MIR630 | ATG12 | [45,120] | |
| MIR519A, MIR885-3p | ATG16L1 | [42,120] | |
| MIR204 | LC3 | [124] | |
| MIR30D, MIR130A | ATG2 | Cycling system | [45,53] |
| MIR34A | ATG9A | [52] | |
The ULK1/2 (unc-51 like autophagy activating kinase 1/2; yeast Atg1) kinase complex, which includes ATG13, RB1CC1 (RB1-inducible coiled-coil 1) and ATG101, acts at an early step in the induction of autophagy, although its role is not fully understood. Mir20a and Mir106b, two members of the Mir17 microRNA family, suppress ULK1 expression in C2C12 myoblasts [41]. In addition, MIR885-3p regulates autophagy through its putative recognition sequence present in the 5′-UTR of ULK2, which results in ULK2 downregulation [42].
Nucleation of the phagophore involves the phosphatidylinositol 3-kinase complex that includes PIK3C3/VPS34-PIK3R4/VPS15 (phosphoinositide-3-kinase, regulatory subunit 4)-BECN1-ATG14. Mir195 inhibitor treatment increases autophagy activation and suppresses neuron inflammation in vivo and in vitro, which suggests a repressive role for this miRNA in autophagy [43]. Furthermore, Atg14 was identified as the functional target of Mir195. Another miRNA, MIR205 inhibits BCL2 expression, thus modulating a key negative regulator of BECN1 function in autophagy [44]. Moreover, the 3′-UTR of BECN1 binds MIR30A, which negatively regulates BECN1 expression, resulting in decreased autophagic activity. Based on bioinformatics and miRNA target prediction, MIR30D is thought to regulate multiple genes in the autophagy pathway including BECN1, BNIP3L (BCL2/adenovirus E1B 19kDa interacting protein 3-like), ATG2, ATG5, and ATG12. MIR30D directly suppresses the expression of the corresponding core proteins in the autophagy pathway, inhibiting autophagosome formation and conversion of LC3B-I (the proteolytically processed form) to LC3B-II (the lipidated form) [45]. MIR376B also controls autophagy by regulating the intracellular levels of two autophagy proteins, ATG4C and BECN1 [46]. MIR101 is a potent inhibitor of basal, etoposide-, and rapamycin-induced autophagy, at least in part through downregulation of RAB5A (RAB5A, member RAS oncogene family), STMN1 (stathmin 1), and ATG4D [47].
ATG9 is a transmembrane protein required for expansion of the phagophore. Studies in yeast suggest that this protein traffics between donor membranes and the site of phagophore formation [48–51]. Caenorhabditis elegans at g-9 contains a binding sequence for mir-34, and this miRNA inhibits ATG-9 expression at the post-transcriptional level in vitro; the C. elegans mir-34 mutation extends life span by enhancing autophagic flux, and in mammalian cells MIR34A represses autophagy by directly inhibiting the expression of ATG9A [52]. MIR130A reduces autophagosome formation, an effect mediated by downregulation of the ATG2B and DICER1 (dicer 1, ribonuclease type III) genes [53]; DICER1 is a key component of the cellular miRNA machinery, suggesting that its downregulation is part of a feedback loop that maintains optimal levels of autophagy. MIR130A thus modulates cell survival by controlling autophagic flux.
Post-translational regulation
Post-translational regulation is essential for modulating autophagy in order to adapt to different types of environmental stress, both with regard to the amplitude of autophagy activity and its duration. With regard to autophagy, post-translational modifications (PTMs) can be classified into three categories: phosphorylation, ubiquitination, and acetylation [54]. These modifications help to regulate autophagy on multiple levels. For example, autophagy core components can be directly modified, either being activated or inhibited. In addition, PTMs can also indirectly regulate autophagy by modulating regulators upstream of the Atg/ATG proteins. Here, we focus on the direct post-translational regulation of proteins that are part of the autophagy core machinery.
Phosphorylation
Phosphorylation of ATG proteins is the most well studied PTM. Phosphorylation of ATG proteins either alters the functional activity of the protein or changes the structure of the protein to modulate its affinity to binding partners. The regulation of the ULK1 complex, which is important for autophagy initiation, is a good example of PTMs altering protein function.
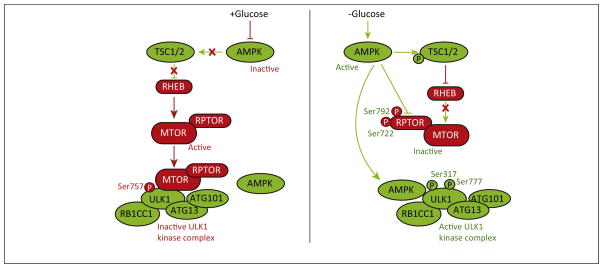
In mammalian cells, the ULK1 complex has key functions in autophagy initiation (Figure 3). The activity of the complex largely depends on the phosphorylation status of ULK1. Previous studies have established that TOR/MTOR kinase is a negative regulator of autophagy [55,56], whereas AMPK acts in a positive manner [57,58]. When glucose is present, AMPK activity is suppressed resulting in an inactive TSC(tuberous sclerosis)1–TSC2 complex. In these conditions, MTOR is activated by RHEB (Ras homolog enriched in brain; in its GTP-bound form) and phosphorylates ULK1 at the inhibitory Ser757 site. This phosphorylation disrupts the interaction of AMPK and ULK1, preventing activation of the latter. Conversely, under glucose-depleted conditions, AMPK directly phosphorylates ULK1 at Ser317 and Ser777, leading to ULK1 activation [58]. AMPK also activates autophagy by suppressing MTOR. First, AMPK phosphorylates and activates TSC2, which in turns inhibits RHEB [59]. AMPK also directly phosphorylates the MTOR binding partner RPTOR (regulatory associated protein of MTOR, complex 1) at Ser722 and Ser792, reducing MTOR kinase activity [60]. Phosphorylation and inactivation of RPTOR involves binding to a YWHA/14-3-3 (tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein) protein. Under conditions of mitochondrial stress, AMPK phosphorylates ULK1 at Ser555, and this phosphorylation is required for mitophagy [61]. Thus, the AMPK–MTOR system can regulate ULK1 kinase activity to finely tune the autophagy process.
Figure 3.
Coordinated regulation of ULK1 (unc-51 like autophagy activating kinase 1) by AMPK (AMP-activated protein kinase) and MTOR (mechanistic target of rapamycin). Phosphorylation-dependent regulation of ULK1 by AMPK and MTOR plays crucial roles in modulating autophagy activity. Arrows and bars indicate stimulation and inhibition, respectively. Green or red arrows and bars (or proteins) indicate events (or components) that promote or inhibit autophagy, respectively. Modified from Figure 8 of [58].
In yeast, the Atg1 complex consists of the Atg1 kinase, its regulatory partner Atg13, and the Atg17–Atg31–Atg29 complex [1]. TOR participates in the regulation of Atg1 through the direct phosphorylation of the regulatory Atg13 subunit, which is also inhibited by PKA (cAMP-dependent protein kinase A). Under nutrient-rich conditions, PKA and TOR phosphorylate Atg13 at distinct sites [62,63]. The phosphorylation of Atg13 affects both the nature of its interaction with Atg1 and its localization to the phagophore assembly site. Atg1 is also a direct substrate of PKA, and phosphorylation regulates Atg1 localization to the PAS. The Atg17–Atg31–Atg29 complex also modulates Atg1 kinase and autophagy activity. Atg29 is phosphorylated along its C terminus and this modification is necessary for interaction with the scaffold protein Atg11 and PAS localization and function [64].
Phosphorylation of the PtdIns3K complex, another part of the autophagy core machinery, which is responsible for producing PtdIns3P, also regulates autophagy [54,65]. In mammals, PIK3C3/VPS34 is the catalytic subunit of a class III PtdIns3K that provides the core function of the complex. PIK3C3 is functional in autophagy when it interacts with BECN1, which in turn is inhibited by anti-apoptotic proteins such as BCL2 [1]. Thus, any PTM that disrupts the interaction between BECN1 and its inhibitory partners has the potential for stimulating autophagy and vice versa. For example, DAPK (death-associated protein kinase) phosphorylates BECN1 and MAPK8/ JNK1 (mitogen-activated protein kinase 8) phosphorylates BCL2 [10,54,66]. In both cases the interaction between BECN1 and BCL2 is disrupted, allowing BECN1 to associate with PIK3C3 and induce autophagy. Conversely, STK4/MST1 (serine/threonine kinase 4) phosphorylates BECN1 to enhance its interaction with BCL2 or BCL2L1 (BCL2-like 1), resulting in autophagy inhibition [67].
The protein kinase AKT is also involved in BECN1 phosphorylation. AKT enhances the interaction of BECN1 with YWHA/14-3-3 proteins, thus negatively regulating autophagy [68]. There are multiple PtdIns3K complexes that function in various pathways including autophagy, both stimulating and inhibiting, and their regulation is complex. For example, AMPK phosphorylation inhibits the non-autophagic function of PIK3C3; whereas under glucose starvation conditions, ATG14 functions to decrease the AMPK-dependent phosphorylation of PIK3C3 and increase the phosphorylation of BECN1, which lead to autophagy induction [69]. This differential regulation acts in part to promote activation of the autophagy-specific PIK3C3 complex under stress conditions.
Many other ATG proteins are phosphorylated. For example, LC3-II can be phosphorylated by protein kinase A or protein kinase C [70], or by STK3 and STK4 [71]; the phosphorylated form has reduced function in autophagosome formation or is required for the fusion of autophagosomes with the lysosome, respectively. Yeast Atg9 is a direct target of Atg1 [72]; phosphorylation of Atg9 is required for recruitment of Atg8 to the PAS, thus ensuring normal expansion of the phagophore.
Ubiquitination
Ubiquitination is another type of PTM that regulates autophagy. In general ubiquitination can affect the stability of Atg proteins and their upstream regulators. However, ubiquitin can also serve as a signaling molecule, recruiting autophagy receptors. In either case, specific E3 ligases play an important role in ubiquitin function [73].
PARK2/parkin (parkin RBR E3 ubiquitin protein ligase), an E3 ligase that is important for mitochondrial homeostasis, is involved in mitophagy [73,74]. Once activated by PINK1 (PTEN induced putative kinase 1), PARK2 ubiquitinates multiple mitochondrial proteins on the surface of depolarized mitochondria [75]. Although the precise mechanism is not clear, these events may initiate a cascade that results in the clearance of damaged mitochondria. Conversely, PARK2 has a negative role in nonselective autophagy. PARK2 stabilizes BCL2 via monoubiquitination, which strengthens the interaction between BCL2 and BECN1, thus suppressing autophagy [76].
Ubiquitination of the autophagy core machinery can either affect protein function or stability. TRAF6 (TNF receptor-associated factor 6, E3 ubiquitin protein ligase), a RING finger-containing ligase, can ubiquitinate both BECN1 and ULK1 [77,78]. The Lys63 (K63)-linked ubiquitination of BECN1 by TRAF6 is required for the induction of autophagy by TLR4 (toll-like receptor 4), a protein that plays a role in activation of the innate immune system in response to bacterial antigen. By contrast, the deubiquitinating enzyme TNFAIP3/A20 (tumor necrosis factor, alpha-induced protein 3) antagonizes TRAF6 regulation by deubiquitinating BECN1 and repressing autophagy. TRAF6 also ubiquitinates K63 of ULK1 when it interacts with AMBRA1 (autophagy/beclin-1 regulator 1), a positive regulator of autophagy. The ubiquitination of ULK1 leads to stabilization and activation of the protein. RNF5 (ring finger protein 5, E3 ubiquitin protein ligase), another E3 ligase, controls ATG4B; ubiquitination promotes its degradation [79]. Decreased levels of ATG4B result in limited proteolytic processing of LC3 and autophagy suppression.
Although E3 ligases regulate autophagy primarily through their ligation function, these proteins may also regulate autophagy in a ligase-independent manner. A recent study shows a general role of the tripartite motif (TRIM) protein family. TRIM proteins contain an N-terminal RING domain; however, these proteins regulate autophagy mainly through the C-terminal substrate-binding domain [80]. TRIM proteins can bind ATG proteins such as BECN1, ULK1, and LC3 family proteins, as well as receptors including SQSTM1/p62 (sequestosome 1). These findings suggest that TRIM proteins serve as a platform for the association of different ATG proteins.
Acetylation
In addition to the epigenetic regulation of autophagy by histone acetylation (discussed below), ATG proteins are also regulated by direct acetylation [81]. During serum starvation, the acetyltransferase KAT5/TIP60 [K(lysine) acetyltransferase 5] is activated by GSK3 (glycogen synthase kinase 3) and it directly acetylates ULK1 [82]. This acetylation is required for ULK1 activation and autophagy initiation. Either inhibition of KAT5 or the presence of an acetylation-insensitive ULK1 mutant reduces autophagy activity. KAT5 has a yeast ortholog, Esa1, which acetylates Atg3 on K19 and K48 [83]. Blocking this acetylation attenuates Atg8–Atg3 interaction and Atg8 lipidation. The Rpd3 deacetylase counters the effect of Esa1. Rpd3 deletion results in increased acetylation of Atg3 and upregulated autophagy.
Another example of regulatory acetylation/deacetylation in autophagy is seen with EP300/p300 (E1A binding protein p300) and SIRT1 (sirtuin 1). The EP300 acetyltransferase acetylates a variety of autophagy proteins including ATG5, ATG7, LC3, and ATG12. However, acetylation appears to inhibit the function of these proteins because reduced EP300 stimulates autophagy activity [84]. Conversely, the NAD-dependent deacetylase SIRT1 deacetylates ATG5, ATG7, and LC3 [85]. Deletion of Sirt1 results in increased acetylation of ATG proteins, negatively affecting autophagosome formation. In addition to the core machinery, acetylation also regulates autophagy by modulating transcription factors upstream of the ATG genes. One example is seen with FOXO3, which activates several ATG genes as discussed above [12,86]. Acetylation of FOXO3 causes its dissociation from DNA [87]. SIRT1/2 deacetylate FOXO3 under stress conditions, and this is coordinated with the induction of autophagy [85,88].
Epigenetic regulation
PTMs can also modulate histones and epigenetically regulate autophagy [89]. Histone H3 hyperacetylation on lysine 9, 14, and 18 is accompanied by the repression of ATG gene transcription and autophagy activity [90,91]. Similarly, KAT8/hMOF [K(lysine) acetyltransferase 8]-dependent acetylation of histone H4 Lys16 (H4K16) downregulates ATG gene transcription [92,93]. Combining those data suggests that acetylation on histones generally interferes with ATG gene transcription. Histone methylation also regulates the outcome of ATG gene transcription. EHMT2/G9a (euchromatic histone-lysine N-methyltransferase 2) associates with ATG genepromoters and methylates histone H3 in nutrient-rich conditions [94]. This methylation leads to repression of ATG gene transcription. During starvation, these marks are removed allowing the induction of ATG gene expression. Inhibition of EHMT2, therefore, results in the upregulated expression of ATG genes.
Autophagy regulation through protein–protein interactions
Besides directly modifying ATG proteins, many regulators can control autophagy by binding these proteins to modulate their function [65]. One example is TP53. As discussed above, TP53 acts as both a positive and negative regulator of autophagy. Nuclear TP53 affects autophagy in a positive manner. By contrast, cytosolic TP53 is a negative regulator of autophagy and, by interacting with RB1CC1, it can inhibit the function of the ULK1 complex in normal conditions [31]. Phosphorylation of TP53 results in its translocation to the nucleus, which decreases the amount of TP53 present in the cytosolic pool, and permits autophagy induction under stress conditions.
Concluding remarks
Autophagy plays a central role in cell physiology and during the past 15 years substantial progress has been made in identifying and characterizing the components that comprise the core machinery involved in autophagosome formation. By comparison, we know much less about the complex network of regulatory factors that control this process (Box 1). For example, the number of known ATG genes has steadily increased, but this has far outpaced our understanding of their transcriptional regulation. One aspect of regulation that has received considerable attention is the role of miRNAs in the post-transcriptional control of ATG gene expression. Cells employ many other methods to prevent translation including the decapping and degradation of unwanted mRNAs, and storing mRNAs in an inaccessible/inactive state in stress granules; however, it has not been determined whether these or similar post-transcriptional mechanisms regulate autophagy. Although increasing details about the post-translational regulation of autophagy have been revealed in recent years, many questions remain to be answered. Modulation of proteins by phosphorylation and ubiquitination is a dynamic process; both conjugation and removal of the modulating groups are crucial to maintaining normal autophagy flux. However, information about dephosphorylation and deubiquitination is relatively limited. For example, the relevant phosphatases that affect the autophagy core machinery proteins are still unknown. Similarly, the role of deubiquitinating enzymes in the regulation of autophagy needs further investigation. In addition to the post-translational modification of components of the autophagy core machinery, the PTM of receptor proteins involved in autophagy has also received substantial attention. For example, SQSTM1/p62 is phosphorylated during selective autophagy [95], but the detailed mechanism and the importance of this PTM require further investigation. Finally, as we continue to expand our knowledge regarding the transcriptional, post-transcriptional, and post-translational regulation of autophagy, it is likely that we will see an expanded use of therapeutics that target autophagy in clinical applications.
Box 1. Outstanding questions.
Which ATG genes are transcriptionally regulated? Which transcription factors are involved in this regulation?
Does the transcriptional regulation of particular ATG genes, aside from ATG8 and ATG9, play an important role in controlling the autophagic response?
Is there a role for mRNA degradation in modulating autophagy induction or in downregulating autophagy?
Are the mRNAs encoding ATG genes localized to specific subcellular sites, which might keep them inactive in growing conditions?
Which phosphatases are involved in controlling the activity and/or localization of the Atg/ATG proteins? What other enzymes affect the PTMs of these proteins?
How do transcriptional, post-transcriptional, and/or post-translational mechanisms function to downregulate autophagy under inducing conditions (i.e., to prevent excessive autophagy)?
Acknowledgments
This work was supported by NIH grant GM053396 to DJK.
Glossary
- Atg
autophagy-related proteins comprise the functional machinery of macroautophagy; the core machinery refers to those Atg proteins required for autophagosome formation
- Autophagosome
the double-membrane vesicle that is the terminal cytoplasmic structure of macroautophagy; the autophagosome contains the sequestered cytoplasmic cargo that is destined for degradation
- Autophagy
there are three primary types of autophagy or self-eating including chaperone-mediated autophagy, microautophagy, and macroautophagy, which differ in the substrates and mechanism of sequestration; in each case, cytosolic proteins or parts of the cytoplasm are degraded within the lysosome or vacuole
- Phagophore
the initial sequestering compartment of macroautophagy that expands and matures into the autophagosome
- Phagophore assembly site (PAS)
the location in the cell where the phagophore nucleates; the PAS is peri-vacuolar and most of the Atg proteins are transiently localized at this site
- TOR
the target of rapamycin is a serine/threonine protein kinase that is a negative regulator of autophagy; it acts in part by phosphorylating Atg1 and Atg13
- Vacuole
the yeast analog of the lysosome; a degradative organelle that also has other functions including pH homeostasis, osmotic regulation, and amino acid storage
References
- 1.Feng Y, et al. The machinery of macroautophagy. Cell Res. 2014;24:24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr Opin Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12 (Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreuzaler PA, et al. Stat3 controls lysosomal-mediated cell death in vivo. Nat Cell Biol. 2011;13:303–309. doi: 10.1038/ncb2171. [DOI] [PubMed] [Google Scholar]
- 6.Guerriero JL, et al. Non-apoptotic routes to defeat cancer. Oncoimmunology. 2012;1:94–96. doi: 10.4161/onci.1.1.17885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang L, et al. Decision making by p53: life versus death. Mol Cell Pharmacol. 2010;2:69–77. [PMC free article] [PubMed] [Google Scholar]
- 8.Sengupta A, et al. FoxO transcription factors promote autophagy in cardiomyocytes. J Biol Chem. 2009;284:28319–28331. doi: 10.1074/jbc.M109.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mammucari C, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Xu P, et al. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25:310–322. doi: 10.1101/gad.1984311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warr MR, et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature. 2013;494:323–327. doi: 10.1038/nature11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Mammucari C, et al. Downstream of Akt: FoxO3 and mTOR in the regulation of autophagy in skeletal muscle. Autophagy. 2008;4:524–526. doi: 10.4161/auto.5905. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, et al. FOXO3 induces FOXO1-dependent autophagy by activating the AKT1 signaling pathway. Autophagy. 2012;8:1712–1723. doi: 10.4161/auto.21830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiong X, et al. The autophagy-related gene 14 (Atg14) is regulated by forkhead box O transcription factors and circadian rhythms and plays a critical role in hepatic autophagy and lipid metabolism. J Biol Chem. 2012;287:39107–39114. doi: 10.1074/jbc.M112.412569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu HY, et al. Hepatic autophagy is suppressed in the presence of insulin resistance and hyperinsulinemia: inhibition of FoxO1-dependent expression of key autophagy genes by insulin. J Biol Chem. 2009;284:31484–31492. doi: 10.1074/jbc.M109.033936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Y, et al. Cytosolic FoxO1 is essential for the induction of autophagy and tumour suppressor activity. Nat Cell Biol. 2010;12:665–675. doi: 10.1038/ncb2069. [DOI] [PubMed] [Google Scholar]
- 18.Hariharan N, et al. Deacetylation of FoxO by Sirt1 plays an essential role in mediating starvation-induced autophagy in cardiac myocytes. Circ Res. 2010;107:1470–1482. doi: 10.1161/CIRCRESAHA.110.227371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Medema RH, Jaattela M. Cytosolic FoxO1: alive and killing. Nat Cell Biol. 2010;12:642–643. doi: 10.1038/ncb0710-642. [DOI] [PubMed] [Google Scholar]
- 20.Vidal RL, Hetz C. Unspliced XBP1 controls autophagy through FoxO1. Cell Res. 2013;23:463–464. doi: 10.1038/cr.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Y, et al. XBP-1u suppresses autophagy by promoting the degradation of FoxO1 in cancer cells. Cell Res. 2013;23:491–507. doi: 10.1038/cr.2013.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Polager S, et al. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene. 2008;27:4860–4864. doi: 10.1038/onc.2008.117. [DOI] [PubMed] [Google Scholar]
- 23.Tracy K, et al. BNIP3 is an RB/E2F target gene required for hypoxia-induced autophagy. Mol Cell Biol. 2007;27:6229–6242. doi: 10.1128/MCB.02246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw J, et al. Antagonism of E2F-1 regulated Bnip3 transcription by NF-kappaB is essential for basal cell survival. Proc Natl Acad Sci USA. 2008;105:20734–20739. doi: 10.1073/pnas.0807735105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Criollo A, et al. Inhibition of autophagy by TAB2 and TAB3. EMBO J. 2011;30:4908–4920. doi: 10.1038/emboj.2011.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Criollo A, et al. The IKK complex contributes to the induction of autophagy. EMBO J. 2010;29:619–631. doi: 10.1038/emboj.2009.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Criollo A, et al. IKK connects autophagy to major stress pathways. Autophagy. 2010;6:189–191. doi: 10.4161/auto.6.1.10818. [DOI] [PubMed] [Google Scholar]
- 28.Comb WC, et al. IKK-dependent, NF-kappaB-independent control of autophagic gene expression. Oncogene. 2011;30:1727–1732. doi: 10.1038/onc.2010.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Copetti T, et al. p65/RelA modulates BECN1 transcription and autophagy. Mol Cell Biol. 2009;29:2594–2608. doi: 10.1128/MCB.01396-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Criollo A, et al. Autophagy is required for the activation of NFkappaB. Cell Cycle. 2012;11:194–199. doi: 10.4161/cc.11.1.18669. [DOI] [PubMed] [Google Scholar]
- 31.Tasdemir E, et al. Regulation of autophagy by cytoplasmic p53. Nat Cell Biol. 2008;10:676–687. doi: 10.1038/ncb1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Green DR, Kroemer G. Cytoplasmic functions of the tumour suppressor p53. Nature. 2009;458:1127–1130. doi: 10.1038/nature07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiuri MC, et al. Autophagy regulation by p53. Curr Opin Cell Biol. 2010;22:181–185. doi: 10.1016/j.ceb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 34.Kenzelmann Broz D, et al. Global genomic profiling reveals an extensive p53-regulated autophagy program contributing to key p53 responses. Genes Dev. 2013;27:1016–1031. doi: 10.1101/gad.212282.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bartholomew CR, et al. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci USA. 2012;109:11206–11210. doi: 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin M, et al. Transcriptional regulation by Pho23 modulates the frequency of autophagosome formation. Curr Biol. 2014;24:1314–1322. doi: 10.1016/j.cub.2014.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee RC, et al. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 38.Wightman B, et al. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 39.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 40.Yu Y, et al. microRNA 30A promotes autophagy in response to cancer therapy. Autophagy. 2012;8:853–855. doi: 10.4161/auto.20053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu H, et al. MiR-20a and miR-106b negatively regulate autophagy induced by leucine deprivation via suppression of ULK1 expression in C2C12 myoblasts. Cell Signal. 2012;24:2179–2186. doi: 10.1016/j.cellsig.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 42.Huang Y, et al. Phospho-DeltaNp63alpha/miR-885-3p axis in tumor cell life and cell death upon cisplatin exposure. Cell Cycle. 2011;10:3938–3947. doi: 10.4161/cc.10.22.18107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi G, et al. Increased miR-195 aggravates neuropathic pain by inhibiting autophagy following peripheral nerve injury. Glia. 2013;61:504–512. doi: 10.1002/glia.22451. [DOI] [PubMed] [Google Scholar]
- 44.Verdoodt B, et al. MicroRNA-205, a novel regulator of the anti-apoptotic protein Bcl2, is downregulated in prostate cancer. Int J Oncol. 2013;43:307–314. doi: 10.3892/ijo.2013.1915. [DOI] [PubMed] [Google Scholar]
- 45.Yang X, et al. mir-30d Regulates multiple genes in the autophagy pathway and impairs autophagy process in human cancer cells. Biochem Biophys Res Commun. 2013;431:617–622. doi: 10.1016/j.bbrc.2012.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korkmaz G, et al. miR-376b controls starvation and mTOR inhibition-related autophagy by targeting ATG4C and BECN1. Autophagy. 2012;8:165–176. doi: 10.4161/auto.8.2.18351. [DOI] [PubMed] [Google Scholar]
- 47.Frankel LB, et al. microRNA-101 is a potent inhibitor of autophagy. EMBO J. 2011;30:4628–4641. doi: 10.1038/emboj.2011.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reggiori F, et al. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 49.He C, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–935. doi: 10.1083/jcb.200606084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamamoto H, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Legakis JE, et al. A cycling protein complex required for selective autophagy. Autophagy. 2007;3:422–432. doi: 10.4161/auto.4129. [DOI] [PubMed] [Google Scholar]
- 52.Yang J, et al. MiR-34 modulates Caenorhabditis elegans lifespan via repressing the autophagy gene atg9. Age (Dordr) 2013;35:11–22. doi: 10.1007/s11357-011-9324-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kovaleva V, et al. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012;72:1763–1772. doi: 10.1158/0008-5472.CAN-11-3671. [DOI] [PubMed] [Google Scholar]
- 54.Boya P, et al. Emerging regulation and functions of autophagy. Nat Cell Biol. 2013;15:713–720. doi: 10.1038/ncb2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jung CH, et al. mTOR regulation of autophagy. FEBS Lett. 2010;584:1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chang YY, et al. Nutrient-dependent regulation of autophagy through the target of rapamycin pathway. Biochem Soc Trans. 2009;37 (Pt 1):232–236. doi: 10.1042/BST0370232. [DOI] [PubMed] [Google Scholar]
- 57.Rubinsztein DC, et al. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim J, et al. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoki K, et al. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115:577–590. doi: 10.1016/s0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 60.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Egan DF, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011;331:456–461. doi: 10.1126/science.1196371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamada Y, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–1058. doi: 10.1128/MCB.01344-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stephan JS, et al. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci USA. 2009;106:17049–17054. doi: 10.1073/pnas.0903316106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mao K, et al. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci USA. 2013;110:E2875–E2884. doi: 10.1073/pnas.1300064110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marino G, et al. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol. 2014;15:81–94. doi: 10.1038/nrm3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zalckvar E, et al. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep. 2009;10:285–292. doi: 10.1038/embor.2008.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maejima Y, et al. Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl-2. Nat Med. 2013;19:1478–1488. doi: 10.1038/nm.3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang RC, et al. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science. 2012;338:956–959. doi: 10.1126/science.1225967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim J, et al. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cherra SJ, 3rd, et al. Regulation of the autophagy protein LC3 by phosphorylation. J Cell Biol. 2010;190:533–539. doi: 10.1083/jcb.201002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wilkinson DS, et al. Phosphorylation of LC3 by the Hippo Kinases STK3/STK4 is essential for autophagy. Mol Cell. 2015;57:55–68. doi: 10.1016/j.molcel.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Papinski D, et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuang E, et al. Emerging roles of E3 ubiquitin ligases in autophagy. Trends Biochem Sci. 2013;38:453–460. doi: 10.1016/j.tibs.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Narendra D, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008;183:795–803. doi: 10.1083/jcb.200809125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Geisler S, et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat Cell Biol. 2010;12:119–131. doi: 10.1038/ncb2012. [DOI] [PubMed] [Google Scholar]
- 76.Chen D, et al. Parkin monoubiquitinates Bcl-2 and regulates autophagy. J Biol Chem. 2010;285:38214–38223. doi: 10.1074/jbc.M110.101469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nazio F, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol. 2013;15:406–416. doi: 10.1038/ncb2708. [DOI] [PubMed] [Google Scholar]
- 78.Shi CS, Kehrl JH. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1. Autophagy. 2010;6:986–987. doi: 10.4161/auto.6.7.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuang E, et al. Regulation of ATG4B stability by RNF5 limits basal levels of autophagy and influences susceptibility to bacterial infection. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1003007. http://dx.doi.org/10.1371/journal.pgen1003007. Published online October 18, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mandell MA, et al. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Dev Cell. 2014;30:394–409. doi: 10.1016/j.devcel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Banreti A, et al. The emerging role of acetylation in the regulation of autophagy. Autophagy. 2013;9:819–829. doi: 10.4161/auto.23908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin SY, et al. GSK3-TIP60-ULK1 signaling pathway links growth factor deprivation to autophagy. Science. 2012;336:477–481. doi: 10.1126/science.1217032. [DOI] [PubMed] [Google Scholar]
- 83.Yi C, et al. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–477. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- 84.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009;284:6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee IH, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Attaix D, Bechet D. FoxO3 controls dangerous proteolytic liaisons. Cell Metab. 2007;6:425–427. doi: 10.1016/j.cmet.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 87.Daitoku H, et al. Regulation of FoxO transcription factors by acetylation and protein-protein interactions. Biochim Biophys Acta. 2011;1813:1954–1960. doi: 10.1016/j.bbamcr.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 88.Wang F, et al. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell. 2007;6:505–514. doi: 10.1111/j.1474-9726.2007.00304.x. [DOI] [PubMed] [Google Scholar]
- 89.Fullgrabe J, et al. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat Rev Mol Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- 90.Eisenberg T, et al. A histone point mutation that switches on autophagy. Autophagy. 2014;10:1143–1145. doi: 10.4161/auto.28767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eisenberg T, et al. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab. 2014;19:431–444. doi: 10.1016/j.cmet.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fullgrabe J, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Fullgrabe J, et al. Histone post-translational modifications regulate autophagy flux and outcome. Autophagy. 2013;9:1621–1623. doi: 10.4161/auto.25803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Artal-Martinez de Narvajas A, et al. Epigenetic regulation of autophagy by the methyltransferase G9a. Mol Cell Biol. 2013;33:3983–3993. doi: 10.1128/MCB.00813-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ichimura Y, et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol Cell. 2013;51:618–631. doi: 10.1016/j.molcel.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 96.Rouschop KM, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Milani M, et al. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69:4415–4423. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- 98.Pike LR, et al. Transcriptional upregulation of ULK1 by ATF4 contributes to cancer cell survival. Biochem J. 2013;449:389–400. doi: 10.1042/BJ20120972. [DOI] [PubMed] [Google Scholar]
- 99.Pike LR, et al. ATF4 orchestrates a program of BH3-only protein expression in severe hypoxia. Mol Biol Rep. 2012;39:10811–10822. doi: 10.1007/s11033-012-1975-3. [DOI] [PubMed] [Google Scholar]
- 100.Sheng Z, et al. BCR-ABL suppresses autophagy through ATF5-mediated regulation of mTOR transcription. Blood. 2011;118:2840–2848. doi: 10.1182/blood-2010-12-322537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ma D, et al. Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J. 2011;30:4642–4651. doi: 10.1038/emboj.2011.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yurkova N, et al. The cell cycle factor E2F-1 activates Bnip3 and the intrinsic death pathway in ventricular myocytes. Circ Res. 2008;102:472–479. doi: 10.1161/CIRCRESAHA.107.164731. [DOI] [PubMed] [Google Scholar]
- 103.Sanchez AM, et al. AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J Cell Biochem. 2012;113:695–710. doi: 10.1002/jcb.23399. [DOI] [PubMed] [Google Scholar]
- 104.Schips TG, et al. FoxO3 induces reversible cardiac atrophy and autophagy in a transgenic mouse model. Cardiovasc Res. 2011;91:587–597. doi: 10.1093/cvr/cvr144. [DOI] [PubMed] [Google Scholar]
- 105.Kang YA, et al. Autophagy driven by a master regulator of hematopoiesis. Mol Cell Biol. 2012;32:226–239. doi: 10.1128/MCB.06166-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun T, et al. c-Jun NH2-terminal kinase activation is essential for upregulation of LC3 during ceramide-induced autophagy in human nasopharyngeal carcinoma cells. J Transl Med. 2011;9:161. doi: 10.1186/1479-5876-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jia G, et al. Insulin-like growth factor-1 and TNF-alpha regulate autophagy through c-jun N-terminal kinase and Akt pathways in human atherosclerotic vascular smooth cells. Immunol Cell Biol. 2006;84:448–454. doi: 10.1111/j.1440-1711.2006.01454.x. [DOI] [PubMed] [Google Scholar]
- 108.Li DD, et al. The pivotal role of c-Jun NH2-terminal kinase-mediated Beclin 1 expression during anticancer agents-induced autophagy in cancer cells. Oncogene. 2009;28:886–898. doi: 10.1038/onc.2008.441. [DOI] [PubMed] [Google Scholar]
- 109.Ling J, et al. KrasG12D-induced IKK2/beta/NF-kappaB activation by IL-1alpha and p62 feed forward loops is required for development of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;21:105–120. doi: 10.1016/j.ccr.2011.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Xie Z, et al. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell. 2008;19:3290–3298. doi: 10.1091/mbc.E07-12-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yee KS, et al. PUMA- and Bax-induced autophagy contributes to apoptosis. Cell Death Differ. 2009;16:1135–1145. doi: 10.1038/cdd.2009.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rosenbluth JM, Pietenpol JA. mTOR regulates autophagy-associated genes downstream of p73. Autophagy. 2009;5:114–116. doi: 10.4161/auto.5.1.7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seo YK, et al. Genome-wide localization of SREBP-2 in hepatic chromatin predicts a role in autophagy. Cell Metab. 2011;13:367–375. doi: 10.1016/j.cmet.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.McCormick J, et al. STAT1 deficiency in the heart protects against myocardial infarction by enhancing autophagy. J Cell Mol Med. 2012;16:386–393. doi: 10.1111/j.1582-4934.2011.01323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Dauer DJ, et al. Stat3 regulates genes common to both wound healing and cancer. Oncogene. 2005;24:3397–3408. doi: 10.1038/sj.onc.1208469. [DOI] [PubMed] [Google Scholar]
- 116.Lipinski MM, et al. A genome-wide siRNA screen reveals multiple mTORC1 independent signaling pathways regulating autophagy under normal nutritional conditions. Dev Cell. 2010;18:1041–1052. doi: 10.1016/j.devcel.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Settembre C, et al. TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat Cell Biol. 2013;15:647–658. doi: 10.1038/ncb2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moresi V, et al. Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. Proc Natl Acad Sci USA. 2012;109:1649–1654. doi: 10.1073/pnas.1121159109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bryant A, et al. miR-10a is aberrantly overexpressed in Nucleophosmin 1 mutated acute myeloid leukaemia and its suppression induces cell death. Mol Cancer. 2012;11 doi: 10.1186/1476-4598-11-8. http://dx.doi.org/10.1186/1476-4598-11-8. Published online February20, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Huang Y, et al. Phospho-Delta Np63 alpha-dependent regulation of autophagic signaling through transcription and micro-RNA modulation. Cell Cycle. 2012;11:1247–1259. doi: 10.4161/cc.11.6.19670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhu H, et al. Regulation of autophagy by a beclin 1-targeted microRNA, miR-30a, in cancer cells. Autophagy. 2009;5:816–823. doi: 10.4161/auto.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yu Y, et al. Targeting microRNA-30a-mediated autophagy enhances imatinib activity against human chronic myeloid leukemia cells. Leukemia. 2012;26:1752–1760. doi: 10.1038/leu.2012.65. [DOI] [PubMed] [Google Scholar]
- 123.Comincini S, et al. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol Ther. 2013;14:574–586. doi: 10.4161/cbt.24597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mikhaylova O, et al. VHL-regulated MiR-204 suppresses tumor growth through inhibition of LC3B-mediated autophagy in renal clear cell carcinoma. Cancer Cell. 2012;21:532–546. doi: 10.1016/j.ccr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]