Abstract
RecQ helicases are a family of highly conserved proteins that maintain genomic stability through their important roles in replication restart mechanisms. Cellular phenotypes of RECQ1 deficiency are indicative of aberrant repair of stalled replication forks, but the molecular functions of RECQ1, the most abundant of the five known human RecQ homologs, have remained poorly understood. We show that RECQ1 associates with FEN-1 in nuclear extracts and exhibits direct protein interaction in vitro. Recombinant RECQ1 significantly stimulated FEN-1 endonucleolytic cleavage of 5’-flap DNA substrates containing nontelomeric or telomeric repeat sequence. RECQ1 and FEN-1 were constitutively present at telomeres and their binding to the telomeric chromatin was enhanced following DNA damage. Telomere residence of FEN-1 was dependent on RECQ1 since depletion of RECQ1 reduced FEN-1 binding to telomeres in unperturbed cycling cells. Our results confirm a conserved collaboration of human RecQ helicases with FEN-1, and suggest both overlapping and specialized roles of RECQ1 in the processing of DNA structure intermediates proposed to arise during replication, repair and recombination.
Keywords: RecQ, helicase, flap endonuclease, replication, DNA repair, telomere
INTRODUCTION
The RecQ helicase family is a group of highly conserved DNA unwinding enzymes critical in guarding genome stability in all kingdoms of life [1-2]. The known human RecQ homologs include RECQ1, BLM, WRN, RECQL4, and RECQ5β. Mutations in BLM, WRN and RECQL4 are associated with distinct genetic disorders of Bloom, Werner, and Rothmund-Thomson syndromes, respectively. Distinct clinical phenotypes argue against substantial redundancy, however, a common feature of these syndromes is genomic instability. Though the five human RecQ proteins are similar in their catalytic core and share several biochemical properties in vitro [3], they are likely to participate in distinct aspects of DNA metabolism in human cells under basal and genotoxic stress conditions [4]. A systematic analysis of the molecular interactions and cellular functions of each RecQ homolog is likely to reveal aspects of RecQ functions that are important for genome maintenance. Identification of the specialized functions of individual RecQ proteins will help explain phenotypic differences whereas identifying fundamental similarities should provide a unifying theme elucidating conserved functions of RecQ proteins.
We are investigating RECQ1, also known as RECQL or RECQL1, the most abundant but yet poorly characterized human RecQ homolog. RECQ1 is essential for chromosomal stability [5-6]. Studies so far have suggested an important role of RECQ1 in the repair of DNA damage during cellular replication [7]. RECQ1 is an integral component of the replication complex in unperturbed dividing cells [8]. Association of RECQ1 with replication origins during normal replication is significantly enhanced when cells encounter replication stress [8-9]. RECQ1 deficiency is characterized by spontaneously elevated sister chromatid exchanges [10-11] reminiscent of aberrant repair of stalled replication forks. Indeed, RECQ1-deficient cells accumulate DNA damage and display increased sensitivity to DNA damaging agents that induce stalled and collapsed replication forks [9-10, 12-13]. Consistent with this, RECQ1 interacts physically and functionally with proteins involved in replication and repair. The single strand DNA binding protein RPA interacts with RECQ1 and stimulates its helicase activity [14] while inhibiting strand annealing [15]. Importantly, physical and functional interaction with RPA is a conserved feature of human RecQ proteins [1]. RECQ1 also associates with topoisomerase IIIα, an interaction that is conserved with yeast sgs1 [16] and human BLM [17]. Physical and functional interactions of RECQ1 with mismatch repair proteins and human exonuclease-1 (EXO-1) have been proposed to be relevant for suppressing promiscuous recombination [18] and may also be important in dealing with stalled replication forks [19].
Flap endonuclease-1 (FEN-1) and EXO-1 belong to the Rad2 family of structure-specific nucleases and share a core nuclease domain that is conserved from yeast to mammals [20]. Genetic studies have identified overlapping and distinct roles for EXO-1 and FEN-1 in replication, recombination, repair and maintenance of telomeres [21-22]. FEN-1 cleaves 5′ flaps of the branched DNA structures and possesses double-strand-specific 5′-3′ exonuclease activity [23-25]. The endonuclease activity of FEN-1 is required for processing the 5′ ends of Okazaki fragments in lagging strand DNA synthesis and also participates in base excision repair (BER) by removing 5′ flap structures formed during gap-filling DNA synthesis [23, 26]. FEN-1 is involved in maintenance of simple repeats and prevention of strand slippage [23, 27]. Moreover, FEN-1 is critical for telomeric lagging strand DNA synthesis [28] and contributes to telomere stability [29]. FEN-1 and EXO-1 interact both physically and functionally with WRN and BLM [30-34]. Interactions of FEN-1 with RECQL4 [35] and RECQ5β [36] have been implicated in the processing of oxidative DNA damage.
Faithful and efficient replication of DNA is critical for genome maintenance. We postulate that RecQ helicases assume the shared responsibility of cooperating with Rad2 family structure-specific nucleases for accurate processing of intermediate DNA structures and ensure efficient progression of replication. Emerging evidence implies that similar to the prominent RecQ proteins such as WRN and BLM, RECQ1 also plays a role in the processing of DNA replication and repair intermediates. Here, we identify that RECQ1 interacts with FEN-1 and stimulates its 5’-flap endonucleolytic activity in vitro. We show that RECQ1-depletion reduces FEN-1 binding to telomeres in replicating cells. Interaction with FEN-1 expands the potential repertoire of RECQ1 functions and extends the pattern of conserved collaboration between FEN-1 and RecQ family helicases that contributes to genome maintenance in human cells.
EXPERIMENTAL
Recombinant proteins
E. coli BL21DE3 cells (Agilent Technologies) were transformed with pET-RECQ1, pET-FEN-1, pGSTag-RECQ1 constructs (full length (residues 1-649), the N-terminal domain (residues 1-63), the helicase domain (residues 63-418), the RQC domain (residues 418-592), and the C-terminal domain (residues 592-649) or pGEX-4T-1-FEN-1 expression vectors and grown overnight at 37°C in 10 ml of TB medium (1% tryptone, 0.5% yeast extract,0.5 % NaCl) and 50 μg/ml kanamycin (for pET vectors) or ampicillin (for pGSTag and pGEX vectors)). 1% overnight culture was used to seed TB medium, including 50 μg/ml kanamycin or ampicillin and 20 ml of ethanol. The culture was grown at 37°C to OD600 = 2.5. The temperature was reduced to 18°C, and IPTG was added to 1 mM, followed by overnight incubation at 18°C. The bacteria were collected by centrifugation at 8,000g for 10 min, and the cell pellet was washed by suspension in ice-cold PBS, centrifuged at 8,000g for 10 min, and frozen at −80°C. Recombinant FEN-1 was purified as described [31]. Recombinant human RECQ1 protein was purified as previously described with minor modification [37]. Frozen cell pellets were extracted by sonication (8 × 10 sec) in lysis buffer (50 mM HEPES (pH 7.5), 0.5 M NaCl, 5% glycerol, 10 mM imidazole, 1 mM TCEP supplemented with protease inhibitor (Roche) and benzonase (Novagen)). 0.15% polyethyleneimine solution was added to remove nucleic acids and cell debris by centrifugation at 16,000g for 30 min. The clarified supernatant was then loaded on a 5 mL HisTrap Crude FF column (GE Healthcare) at 1 ml/min by using AKTA purifier system. The column was washed 10 times with lysis buffer (50 mM HEPES (pH 7.5), 0.5 M NaCl, 5% glycerol, 30 mM imidazole) and eluted with elution buffer (50 mM HEPES (pH 7.5), 0.5 M NaCl, 5% glycerol, 250 mM imidazole, 1mM TCEP). Fractions containing RECQ1 protein were identified by SDS-PAGE, combined, and concentrated using centrifugal ultrafiltration (Centricon). Aliquots of recombinant proteins were frozen in liquid nitrogen and stored at −80°C. The purified recombinant proteins were judged to be 98% pure from analysis on Coomassie-stained SDS polyacrylamide gels. Protein concentrations were determined by the Bio-Rad DC Protein Assay using BSA as a standard. Recombinant PCNA was purchased from ProSpec (PRO-615).
Cell culture, and knockdown (KD) of RECQ1 or FEN-1
Human HeLa (ATCC) were grown in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone Laboratories), 100 U/ml penicillin and 100 μg/ml streptomycin (Invitrogen). Cells were grown in a humidified 5% CO2 incubator at 37°C. Stable downregulation of RECQ1 was achieved by transducing HeLa cells with lentiviral shRNA as described [13]. Control HeLa cells were similarly transduced with a shRNA targeting the gene encoding Luciferase. FEN-1 was transiently knocked down in HeLa cells using a synthetic siRNA against FEN-1 (Qiagen, SI02663451), or a non-silencing control siRNA (Qiagen, SI03650325). Cells were reverse transfected with siRNA (20 nM) using Lipofectamine RNAiMAX (Invitrogen) as instructed by the manufacturer.
Immunoprecipitation (IP)
HeLa nuclear extract was prepared as previously described [13]. Extracts were incubated with Protein A-Dynal beads coupled with polyclonal antibody against human RECQ1 (A300-450A, Bethyl Lab), FEN-1 (A300-255A, Bethyl Lab) or normal rabbit IgG (Vector Labs) at 4°C for 90 min, and the immunecomplexes were eluted with 2x SDS-sample buffer following three washes with lysis buffer. Where indicated, nuclear extract was pre-incubated with benzonase (Sigma, 50 U/ml, 2 h at 4°C), a general endonuclease for DNA and RNA. Proteins were resolved by 8-16% SDS-PAGE, transferred onto PVDF membrane and subjected to Western detection of RECQ1 (1:750, sc-25547, Santa Cruz Biotech), FEN-1 (1:1000, Bethyl Lab).
Immunofluorescence assay
HeLa cells grown on glass coverslips to about 70% confluence were untreated or treated with 2 mM hydroxyurea (HU) or 15 μg/ml methyl methanesulfonate (MMS) for 16 h, fixed with 3.7% paraformaldehyde for 10 min, and permeabilized in 0.5% Triton X-100 solution for 10 min at room temperature. Cells were blocked with 3% BSA in PBS and incubated with rabbit polyclonal anti-RECQ1 antibody (1:500, Santa Cruz Biotech) and/or mouse monoclonal FEN-1 antibody (1:500, GTX70185 (4E7), GeneTex) for 1 h at 37°C. After washes in PBS with 0.1% Tween-20, cells were incubated with Alexa Fluor 488 goat anti-rabbit IgG (1:400; Invitrogen) and Alexa Fluor 568 goat anti-mouse IgG (1:400, Invitrogen) secondary antibodies for 1 h at 37°C. Cells were washed four times with PBS containing 0.1% Tween-20, mounted with Prolong Gold containing DAPI (Invitrogen), and analyzed by confocal microscopy (Olympus).
GST pull-down assays
GST-RECQ1 pull-down experiments were performed as described [38]. GST-FEN-1-Sepharose affinity pull-down experiments were performed with minor modification as described previously [39]. The frozen bacterial cell pellet was thawed on ice-cold water and sonicated in lysis buffer (PBS containing 10% glycerol and 0.4% Triton X-100) and the lysate was clarified by centrifugation at 15,000g for 1 h at 4°C. Approximately 1 ml of the resulting supernatant was incubated with 100 μl of glutathione S-transferase beads (50% v/v) for 1 h at 4°C. The beads were washed three times with 1 ml lysis buffer, and split into two aliquots, one for binding experiments and one for determination of expression by Coomassie Blue staining. For binding experiments, protein-bound beads were incubated for 2 h at 4°C with 200 ng of recombinant FEN-1 or RECQ1. The beads were subsequently washed five times with 1 ml of lysis buffer and eluted by boiling with 2x SDS-sample buffer. Eluted proteins were electrophoresed on 8-16% polyacrylamide SDS gels and either stained with Coomassie Blue to demonstrate protein loading or transferred onto PVDF membranes for Western detection. FEN-1 bound to GST-RECQ1 proteins or RECQ1 bound to GST-FEN-1 proteins was detected using anti-FEN-1 or anti-RECQ1 antibody, respectively.
ELISA for RECQ1-FEN-1 interaction
ELISA was performed as described previously [39]. Appropriate wells of a 96-well microtiter plate were coated with purified recombinant wild-type RECQ1 protein or BSA (1μg/ml) in carbonate buffer and incubated at 4°C overnight. Following blocking with 3% BSA in PBS containing 0.5% Tween 20; wells were incubated with indicated concentration of FEN-1 was diluted in binding buffer (50 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 5 mM ATP, 100 μg/ml BSA, and 50 mM NaCl), and incubated for 1 h at 30°C. In parallel reactions, benzonase (5 U/ml) was included in the incubation with FEN-1 during the binding step. Wells were washed five times before incubation with anti-FEN-1 antibody (1:1000, Bethyl) for 1 h at 30°C followed by HRP-conjugated anti-rabbit secondary antibody (1:5000) for 30 min at 30°C. After washing five times, any FEN-1 bound to the immobilized RECQ1 was detected using OPD (o-Phenylenediamine dihydrochloride) (SIGMAFAST™, Sigma-Aldrich) and absorbance readings were taken at 490 nm. The absorbance was corrected for the background signal in the presence of BSA.
DNA substrates for various assays
PAGE-purified oligonucleotides (Midland Certified Reagent Co.) used for preparation of DNA substrates were as described [15, 40-41]. Briefly, oligonucleotides were 5’ end-labeled with γ32P-dATP using T4 polynucleotide kinase (NEB) and free nucleotides were removed using G25 spin column (GE Healthcare). Fork duplex substrate consisting of flap26 and TSTEM25 was generated as published [15]. For the preparation of 5’-flap substrates, the radiolabeled downstream oligonucleotide was annealed to the appropriate template oligonucleotide (1:4 ratio) by heating in a boiling water bath for 10 min followed by slow cooling to room temperature overnight. An upstream oligonucleotide was then added to the duplex substrate (1:4:20 ratio) by incubation at 37°C for 1 h followed by slow cooling to room temperature over 3-5 h. The oligonucleotide sequences used for preparing various substrates in this study are specified in Table 1; the telomeric repeat sequences are underlined.
Table 1.
Oligonucleotide sequences for DNA substrates (5’ to 3’)
| Name | Length (nt) | Sequence |
|---|---|---|
| Template | ||
| TSTEM25 | 44 | GCACTGGCCGTCGTTTTACGGTCGTGACTGGGAAAACCCTGGCG |
| TTPL | 72 | CTACACTCAAGCTCGGTCTCGAGTCAGGATGATTGTCCCTAACCCTAAC CCTAACCCTAAGCTGCAGATTAG |
|
| ||
| Upstream Primer | ||
| TUS | 36 | CTAATCTGCAGCTTAGGGTTAGGGTTAGGGTTAGGG |
| FUS25 | 25 | CGCCAGGGTTTTCCCAGTCACGACC |
|
| ||
| Downstream Primer | ||
| FLAP1 | 20 | AGTAAAACGACGGCCAGTGC |
| FLAP5 | 24 | TCCAAGTAAAACGACGGCCAGTGC |
| FLAP15 | 34 | TTTTTTTTTTTCCAAGTAAAACGACGGCCAGTGC |
| FLAP26 | 45 | TTTTTTTTTTTTTTTTTTTTTTCCAAGTAAAACGACGGCCAGTGC |
| FLAP-Telo4 | 48 | TTAGGGTTAGGGTTAGGGTTAGGGTCCAAGTAAAACGACGGCCAGTGC |
| TFLP | 51 | TTTTTTTTTTTTTTTACAATCATCCTGACTCGAGACCGAGCTTGAGTGTAG |
RECQ1 unwinding assay
Reaction mixtures (20 μl) contained 20 mM Tris-HCl (pH 7.5), 10 mM KCl, 8 mM DTT, 5 mM MgCl2, 5 mM ATP, 10% glycerol, 80 μg/ml BSA, 0.5 nM DNA substrate, and the indicated concentrations of RECQ1 and/or FEN-1. Reactions were incubated for 15 min at 37°C, followed by addition of 20 μl of stop buffer (35 mM EDTA, 0.6% SDS, 25% glycerol, 0.04% bromophenol blue, and 0.04% xylene cyanol) with a 10-fold molar excess of unlabeled competitor oligonucleotide, and samples were loaded onto native 12% polyacrylamide gels (19:1 cross-linking ratio) and electrophoresed at 180 V for 2 h at 4°C using 1x TBE as the running buffer. The resolved radiolabeled species were visualized with a PhosphorImager and analyzed using ImageQuant software.
FEN-1 incision assays
Reactions (20 μl) contained 10 fmol of the indicated DNA substrate and the specified amounts of FEN-1, RECQ1 or RECQ1 deletion mutants in 30 mM HEPES (pH 7.6), 5% glycerol, 40 mM KCl, 0.1 mg/ml BSA, and 8 mM MgCl2. RECQ1 or RECQ1 variants were mixed with the substrate in assay buffer on ice prior to the addition of FEN-1 to start the incision reaction. Reactions were incubated at 37°C for 15 min, followed by proteinase K (2 mg/ml) treatment in the presence of 0.6% SDS at 37°C for 10 min. Reactions were terminated by the addition of 10 μl of formamide stop solution (80% formamide (v/v), 0.1% bromophenol blue and 0.1% xylene cyanol), and heated to 95°C for 5 min. Products were resolved on 16% polyacrylamide, 7 M urea denaturing gels. Detection and quantification of the reaction products were performed using a PhosphorImager and the ImageQuant software. Percent incision was calculated as described previously from the equation % incision = (P/ (S + P)) × 100, where P is the sum of the intensity of the bands representing incision products and S is the intensity of band representing the intact substrate [31]. Incision reactions for kinetic analysis, performed in triplicate, contained 0.3 nM FEN-1, 1 nM RECQ1, and the increasing amounts (0, 10, 20, 40, 80, 160 nM) of 15 nt 5’-flap DNA substrate. Kinetic parameters were obtained using Michaelis-Menten equation, v = Vmax[S]/ (Km + [S]), where v is the reaction rate and [S] is the concentration of substrate. The initial velocity was plotted against [S], and the values of Km and Vmax were calculated by nonlinear regression using PRISM3 (GraphPAD Software for Science). Data represent the mean of at least three independent experiments with SD shown by error bars.
Chromatin Immunoprecipitation (ChIP) and quantitative PCR (qPCR)
ChIP experiments were performed as described previously [9] and enrichment of telomeric chromatin was detected by a qPCR based method described by Cawthon [42] that has been used in other studies [43-46]. HeLa cells were cultured overnight at a density of 1 × 107 per 15 cm diameter dish and subjected to either no treatment or treatment with 2 mM HU or 15 μg/ml MMS for 16 h. ChIP experiments utilizing transient KD of FEN-1 were performed 48 h post siRNA transfection. Approximately 5 × 107 HeLa cells were washed with PBS and incubated with 1% formaldehyde for 10 min. After the reaction was quenched with 0.1 M glycine, the cells were sonicated into chromatin fragments with an average length of 400-1000 bp as determined by agarose gel. The chromatin solution was pre-cleared by incubation with protein-G-Sepharose/salmon sperm DNA beads (Millipore) at 4°C for 1 h, divided into aliquots, and incubated overnight at 4°C with 3 μg of rabbit antibodies specific for either RECQ1, FEN-1, Telomere repeat factor2 (TRF2) (all from Bethyl Lab) or phospho-histone H2AX (γH2AX) (Millipore); antibodies were confirmed for their IP specificity using Western blot. Antibody-chromatin complexes were pulled down by adding protein-G-sepharose/salmon sperm DNA beads and incubated for 2 h at 4°C. A reaction containing an equivalent amount of rabbit IgG was included as the background control. Immunoprecipitated pellets were washed, and DNA fragments were then recovered by phenol/chloroform extraction and ethanol precipitation and subjected to qPCR analysis using primers as below: 5’-CGGTTTGTTTGGGTTTGGGTTTGGGTTTGGGTTTGGGTT-3’ and 5’-GGCTTGCCTTACCCTTACCCTTACCCTTACCCTTACCCT-3’ for telomere repeats; 5’-TGTGCTGGCCCATCACTTTG-3’ and 5’-ACCAGCCACCACTTTCTGATAGG-3’ for HBG (a single copy gene as control is located on chromosome 11) [42, 44]. The enzyme was activated at 95°C for 3 min, followed by 40 cycles of 95°C for 15s, 54°C for 2 min, and 72°C for 30s. qPCR were performed using Taq Universal SYBR Green Supermix (Bio-Rad) with technical triplicates, and threshold cycle numbers (Ct) were determined with an iQ5 thermal cycler (BioRad). Fold enrichment of the telomeric sequences were calculated over IgG as: fold enrichment =2−(CtIP− CtIgG), where CtIP and CtIgG are mean threshold cycles of PCR done in triplicates on DNA samples immunoprecipitated with specified antibody and control IgG, respectively. All qPCR reactions were also checked by melt curve analyses and agarose gel electrophoresis to confirm the presence of smear ranging from 50-500 bp for telomeric sequence and a single band for HBG.
Measurement of Telomere Length
Genomic DNA was isolated from HeLa cells that have been stably transduced with control or RECQ1 shRNA by phenol-chloroform extraction and ethanol precipitation. Telomere length analysis was performed by qPCR [42]. 36B4 which encodes the acidic ribosomal phosphoprotein P0 was used as a single-copy gene control and similar cycling conditions as described above were used for amplification of 36B4 and telomere products. The absolute telomeric sequence in kilobases (kb) was calculated according to the method described by O’Callaghan et al. [47]. A one-tailed unpaired t-test was employed to determine whether the average telomere length in RECQ1-depleted cells is significantly shorter than the length in control cells.
RESULTS
RECQ1 interacts with FEN-1 in vivo and in vitro
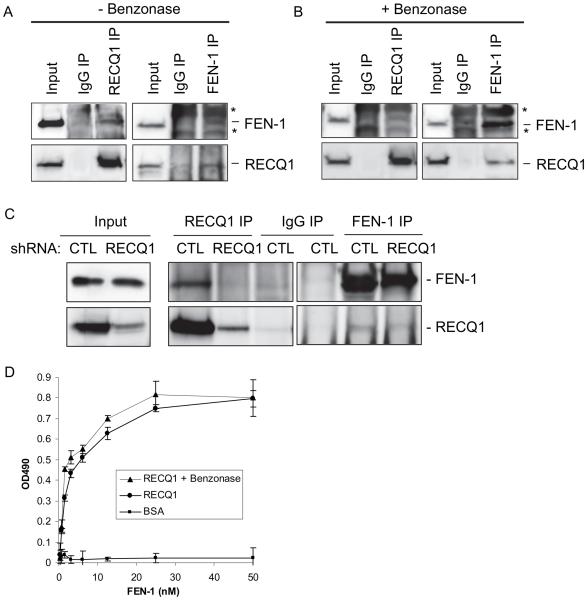
Given their overlapping roles in DNA repair processes, we characterized the putative physical interaction of RECQ1 with FEN-1 (Figure 1). To determine if RECQ1 interacts with FEN-1, we performed reciprocal IP from HeLa cell nuclear extracts using specific antibodies (Figure 1A). Western blot analyses showed RECQ1 antibody specifically co-precipitated FEN-1 and IP of FEN-1 resulted in co-precipitation of RECQ1 (Figure 1A). Similar IP using normal IgG failed to pull down RECQ1 or FEN-1 (Figure 1A). We note that the FEN-1 antibody also detected Ig heavy and light chains (indicated by asterisks) in IP-Western and interfered with FEN-1 signal (~ 42 kD). The presence of EtBr (data not shown) or the use of benzonase-treated extract in IP reaction did not abolish co-precipitation of FEN-1 and RECQ1, suggesting that the interaction is not mediated by DNA (Figure 1B). We next performed co-IP of RECQ1 and FEN-1 from HeLa cells depleted of RECQ1 by lentiviral-expressed RNA interference (RNAi) hairpins targeting RECQ1 (shRECQ1) or luciferase (negative control, shCTL) (Figure 1C). FEN-1 was specifically pulled down in RECQ1 IP from control cell extract. Similarly, FEN-1 IP from control cells contained RECQ1 and a relatively reduced RECQ1 was detected in FEN-1 IP from RECQ1-depleted cells. To determine whether RECQ1 and FEN-1 remain in a complex following DNA damage, we examined co-IP of RECQ1 and FEN-1 extracts prepared from HeLa cells that were untreated or treated with hydrogen peroxide, MMS, or mitomycin C. FEN-1 antibody co-immunoprecipitated a comparable amount of RECQ1 from the extracts of untreated or damage treated cells (Supplementary Figure 1A). These results suggest that endogenous RECQ1 coexists in a complex with FEN-1, and this interaction is unaffected following genotoxic exposure. Notably, these experiments were performed in the benzonase treated extracts to abolish DNA-mediated protein interactions. We examined localization of RECQ1 and FEN-1 in HeLa cells before and after treatment with MMS or HU. RECQ1 and FEN-1 proteins were detected in the nucleus of HeLa cells as reported previously, and this localization pattern was not significantly affected by DNA damage (Supplementary Figure 1B). To investigate if the interaction between RECQ1 and FEN-1 is direct, we performed ELISA using purified recombinant proteins (Figure 1D). FEN-1 bound to RECQ1 in a protein concentration-dependent manner, and a very low OD490 signal was detected in control experiments where BSA was substituted for RECQ1. Incubation with either benzonase or EtBr (not shown) during the binding did not affect the interaction appreciably, indicating that the interaction between FEN-1 and RECQ1 is not DNA-dependent (Figure 1D). Collectively, these data show a direct physical interaction between RECQ1 and FEN-1.
Figure 1. RECQ1 interacts with FEN-1 in vivo and in vitro.
A. Co-IP analysis of RECQ1 interaction with FEN-1 using HeLa nuclear extracts. Immunoprecipitations (IP) with antibodies specific for RECQ1, FEN-1 and preimmune IgG are indicated. Eluted proteins in immunoprecipitate were analyzed by Western blotting and are indicated. RECQ1 IP contained FEN-1. Reciprocal co-IPs of FEN-1 also contained RECQ1. FEN-1 antibody also detected Ig heavy and light chains (indicated by asterisks) in IP-Western and interfered with FEN-1 signal (~ 42 kD). B. Association of RECQ1 and FEN-1 is not mediated via DNA. RECQ1 antibody co-precipitated RECQ1 and FEN-1 using benzonase-treated extract in IP reaction. Reciprocal co-IPs of FEN-1 also contained RECQ1. C. Reciprocal co-IP of RECQ1 and FEN-1 from benzonase-treated extracts prepared from HeLa cells transduced with lentiviral-expressed RNA interference (RNAi) hairpins (shRNA) targeting RECQ1 or luciferase (negative control, CTL). D. Recombinant RECQ1 and FEN-1 proteins directly interact in vitro as shown by ELISA. Either BSA or purified recombinant RECQ1 was coated onto microtiter plates. Following blocking with 3% BSA, appropriate wells were incubated with the indicated concentrations of recombinant FEN-1 (0-50 nM) for 1 h at 30°C. Following washing, RECQ1-bound FEN-1 was detected by ELISA using anti-FEN-1 antibody. The values represent the mean of three independent experiments performed in duplicate with SD indicated by error bars.
FEN-1 binding activity of RECQ1 is contained within RQC and extreme C-terminal end
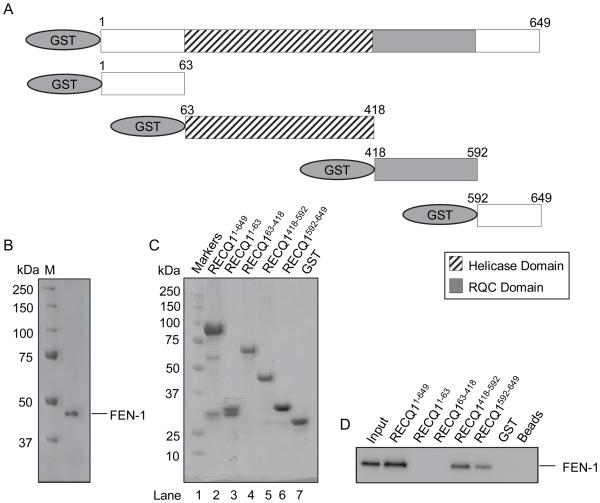
Having identified a direct interaction between the two proteins, we sought to map the FEN-1 interaction domain(s) within RECQ1 utilizing GST fusion proteins that encompass truncated versions of human RECQ1. These fusion proteins were expressed in bacterial cells and GST pull-down experiments were performed using purified recombinant FEN-1 followed by Western blot analysis (Figure 2). Recombinant FEN-1 efficiently bound full-length RECQ1 in a DNA-independent manner. A polypeptide fragment carryng the RQC domain (amino acid residues 418-592) of RECQ1 efficiently bound FEN-1. Moreover, C-terminus of RECQ1 (amino acid residues 592-649) displayed binding to FEN-1. In contrast, N-terminus of RECQ1 (amino acid residues 1-63) or the helicase domain (amino acid residues 63-418) failed to bind FEN-1. Altogether, these results demonstrate that RECQ1 forms a stable complex with FEN-1 and the DNA-independent direct protein-protein interaction between RECQ1-FEN-1 is mediated via the RQC domain with contribution from the C-terminus of RECQ1. The FEN-1 interacting domain of RECQ1 shares limited sequence homology with the WRN amino acid residues 949-1092 that was shown to mediate physical and functional interaction with FEN-1 (Supplementary Figure 2A).
Figure 2. FEN-1 binding activity of RECQ1 is contained within RQC and extreme C-terminal end.
A. Schematic representation of GST-RECQ1 recombinant fragments used for FEN-1 pull-down experiments. B. Coomassie-stained 10% SDS-PAGE gel showing purified FEN-1 used for binding assays. C. Ponceau S-stained membrane showing protein complexes bound to glutathione sepharose beads in pull-down assay. Beads were mixed with lysate from bacteria expressing GST fusion proteins containing human RECQ1 fragments or GST alone as indicated. D. Purified recombinant FEN-1 protein (200 ng) was added to the indicated GST-RECQ1 bound glutathione sepharose beads. After washing, protein complexes were eluted and resolved by SDS-PAGE. Bound FEN-1 was detected by Western blotting.
RECQ1 binding activity resides within amino acids 328-380 of FEN-1 encompassing the PCNA interacting domain
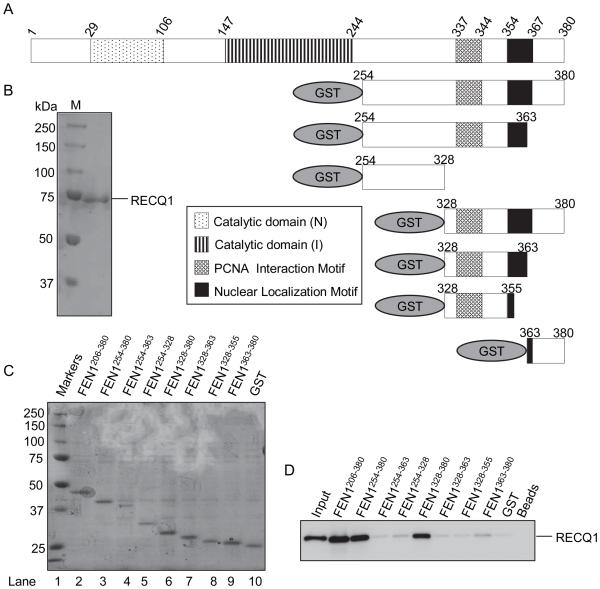
To map the RECQ1 interaction sites on FEN-1, we tested a series of GST fusion proteins that contain various regions of human FEN-1 for RECQ1 binding activity using our pull-down assay (Figure 3). PCNA, one of the best characterized FEN-1 interacting proteins, interacts with FEN-1 by a conserved PCNA binding box motif residing within residues 328-355 of FEN-1 [48]. Earlier study has shown that WRN or BLM binding activity is contained entirely within amino acids 363-380 of FEN-1 [39]. The presence of this 18 amino acid sequence was found to be critical, but not sufficient, for FEN-1 binding to RECQ1 (Figure 3). The recombinant FEN-1 protein fragments that contained the complete amino acid residues 328-380 displayed efficient RECQ1 binding activity; in contrast, deletion fragments of FEN-1 containing amino acid residues 328-363 or 363-380 failed to bind RECQ1 (Figure 3). These results suggest that the FEN-1 amino acids 328-380, spanning the WRN and BLM binding sequence and the PCNA binding motif, is essential for binding to RECQ1 (Supplementary Figure 2B).
Figure 3. RECQ1 binding activity of FEN-1 is contained within amino acids 328-380.
A. Schematic representation of GST-FEN-1 recombinant fragments used for pull-down experiments. B. Coomassie-stained 10% SDS-PAGE gel showing purified RECQ1. C. Ponceau S-stained membrane showing protein complexes bound to glutathione sepharose beads in pull-down assay. Beads were mixed with lysate from bacteria expressing GST fusion proteins containing human FEN-1 fragments or GST alone as indicated. D. Purified recombinant RECQ1 protein (200 ng) was added to the indicated GST-FEN-1 bound glutathione sepharose beads. After washing, protein complexes were eluted and resolved by SDS-PAGE. Bound RECQ1 was detected by Western blotting.
RECQ1 stimulates FEN-1 cleavage of 5’-flap DNA structures
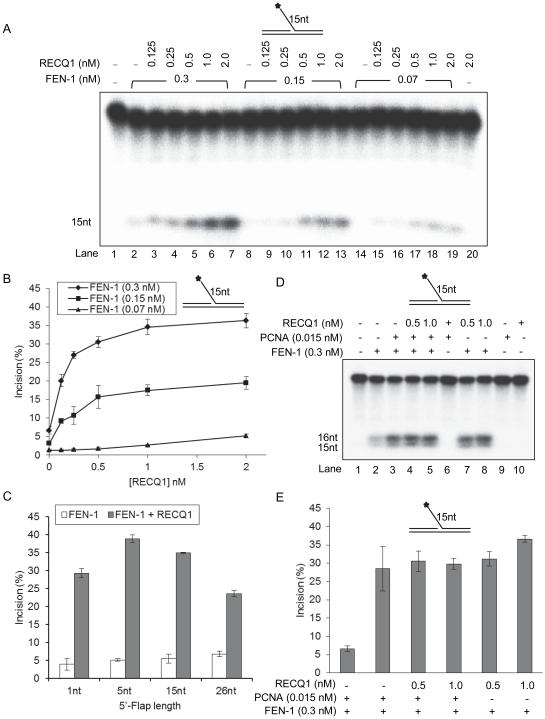
The finding that the RECQ1 protein interacts with FEN-1 prompted us to test whether these proteins exert any functional effect on their catalytic activities. To characterize the effect of RECQ1 on FEN-1 cleavage, we utilized a 19 bp duplex substrate with a 15 nt 5'-flap and analyzed FEN-1 cleavage as a function of RECQ1 concentration under standard reaction condition for FEN-1 incision (Figure 4). As shown previously, the 15 nt flap was susceptible to FEN-1 cleavage in a dose dependent manner [40]; and the presence of RECQ1 (0-2 nM) in the incision reaction resulted in stimulation of the cleavage reaction at all concentrations of FEN-1 tested (0.07, 0.15, and 0.3 nM) (Figure 4A, B). With 0.3 nM purified recombinant FEN-1 alone, approximately 5% of the substrate was incised (Figure 4A, lane 2; 4B) whereas FEN-1 (0.3 nM) incised 25% of the substrate in the presence of 0.25 nM RECQ1 (Figure 4A, lane 4; 4B) and 35% of the flap substrate was incised by FEN-1 (0.3 nM) in the presence of 2 nM RECQ1 (Figure 4A, lane 7; 4B). Thus, at nearly equimolar concentration, RECQ1 stimulated FEN-1 incision by 5-fold. Importantly, 2.0 nM RECQ1 alone did not catalyze cleavage of 5'-flap DNA substrate (Figure 4A, lane 20). Mechanistically, FEN-1 is suggested to slide from the single strand 5'-flap to the duplex junction to make incision [24]. Thus, we next tested the effect of RECQ1 on stimulating FEN-1 incision as a function of 5'-flap length. We examined the ability of RECQ1 to stimulate FEN-1 cleavage of a 1, 5, or 26 nt 5’-flap substrate (Figure 4C; Supplementary Figure 3). RECQ1 (0-2 nM) stimulated the FEN-1 cleavage of 5'-flap substrate with increasing flap length in a dose dependent manner at each concentration of FEN-1 tested (Supplementary Figure 3); and ~8-fold more 5 nt 5’-flap substrate was incised by FEN-1 (0.3 nM) in the presence of RECQ1 (2 nM) as compared to FEN-1 alone (Figure 4C).
Figure 4. RECQ1 stimulates FEN-1 cleavage of 5’-flap DNA substrate.
Reactions (20 μl) containing 10 fmol DNA substrate, indicted amounts of FEN-1 and increasing concentration of RECQ1 (0-2.0 nM) were incubated at 37°C for 15 min under conditions described in Experimental methods. Star indicates position of 32P label. A. Phosphorimage of a typical gel of FEN-1 incision activity on a 15 nt 5’-flap DNA substrate. B. Percent incision from the data shown in ‘A’, data points are the mean of three independent experiments with SDs indicated by error bars. C. RECQ1 stimulation of FEN-1 cleavage of 5’-flap substrates with increasing length of 5’-flap (1, 5, 15, and 26 nt). Percent incision by FEN-1 is shown as the mean of three independent experiments with SD indicated by error bars. D. FEN-1 stimulation by PCNA or RECQ1 is mutually exclusive. Incision reactions, performed as above, contained either FEN-1 alone, or the indicated amounts of PCNA or /and RECQ1. E. Percent incision from the representative experiment shown in ‘D’, data points are the mean of three independent experiments with SDs indicated by error bars.
Given our finding that RECQ1 interaction site on FEN-1 overlaps with the PCNA interaction site, we examined the effect of RECQ1 on PCNA stimulation of FEN-1 incision reaction (Figure 4D, E). We first determined the concentration of PCNA that stimulated FEN-1 activity to a level comparable with RECQ1 (data not shown). Approximately 5-fold greater 15 nt 5’-flap substrate was incised by FEN-1 (0.3 nM) in the presence of PCNA (0.015 nM) or RECQ1 (0.5 nM) (Figure 4D, lanes 2 vs 3 and 7; 4E). Thus, on a molar basis, PCNA was ~33-fold more effective than RECQ1 in stimulating FEN-1 cleavage of 15-nt flap substrate in specified reaction condition. However, the amount of FEN-1 incision product formed in the presence of PCNA or RECQ1 alone was not altered when both PCNA and RECQ1 were added to the FEN-1 reaction (Figure 4D, lanes 3 vs 4 and 5; 4E). This suggests that the stimulation of FEN-1 by PCNA or RECQ1 is mutually exclusive. In contrast, WRN doesn’t interfere with PCNA and coordinately act to stimulate FEN-1 [39].
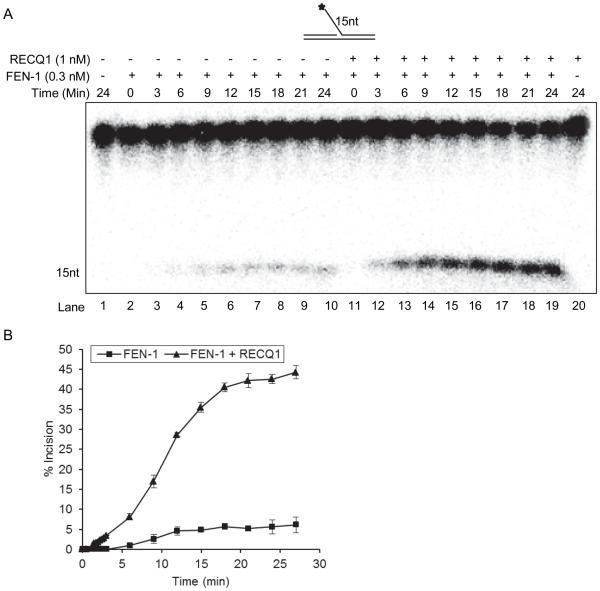
We next performed kinetic analysis of the FEN-1 catalyzed reaction on a 15 nt 5’-flap substrate in the presence or absence of RECQ1 (Figure 5A, B). These experiments utilized the minimum concentration of RECQ1 (1 nM) that was found to be sufficient to achieve maximum stimulation of FEN-1 in the incision experiments described above. In the absence of RECQ1, FEN-1 (0.3 nM) resulted in incision of ~5% of the 10 fmol of 5’-flap DNA substrate in a 15 min reaction, time point used for standard incision assays (Figure 5A, B). Stimulation of FEN-1 incision by RECQ1 was observed at time points as short as 3 min. FEN-1 cleavage in the absence of RECQ1 was less than 1%; however, in the presence of RECQ1, FEN-1 cleaved 7% of the DNA substrate (Figure 5A, lane 3 vs lane 12; 5B). FEN-1 cleavage in the presence or absence of RECQ1 was linear with respect to time from 0-9 min (Figure 5A, B). At 9-15 min, the FEN-1 cleavage reactions conducted in the absence of RECQ1 achieved a plateau of ~ 4-5% substrate incised and no significant increase was observed up to 25 min of reaction (Figure 5B). In contrast, FEN-1 reactions conducted in the presence of RECQ1 resulted in a progressively increased 5’-flap incision product up to 15 min (~ 40% incision), and ~ 44% of the substrate was incised by 25 min of reaction (Figure 5B). Next, we determined the reaction kinetic parameters, Km and Vmax, as described in “Experimental” section. The Vmax of the FEN-1 incision reaction for a 15 nt 5’-flap substrate was determined to be 1.4 ± 0.2 × 10−3 nM/s and 6.7 ± 0.1 × 10−3 nM/s in the absence or presence of RECQ, respectively. In contrast to the observed increase (>4-fold) in Vmax, the Km was determined to be 51.1 nM and 62.5 nM in the absence or presence of RECQ1, respectively. These results indicate that RECQ1 stimulates the rate of FEN-1 incision of a 5’-flap DNA substrate.
Figure 5. Kinetics of FEN-1 cleavage of the 15nt 5’-flap DNA substrate in the presence or absence of RECQ1.
Reactions (200 μl) containing 10 fmol of 15 nt 5’-flap DNA substrate and 0.3 nM of FEN-1 with or without RECQ1 (1 nM) were incubated at 37°C and 20 μl aliquots were removed at indicated time points. A. Phosphorimage of a typical gel from a time course experiment. Increasing times of incubation (0-24 min) for the FEN-1 cleavage reactions conducted in the absence of RECQ1 (lanes 2-10) or in the presence of RECQ1 (lanes 11-19) are indicated. No enzyme and RECQ1 alone reactions are indicated in lane 1 and 20, respectively. B. Percent incision from the data shown in ‘A’, data points are the mean of three independent experiments with SD indicated by error bars.
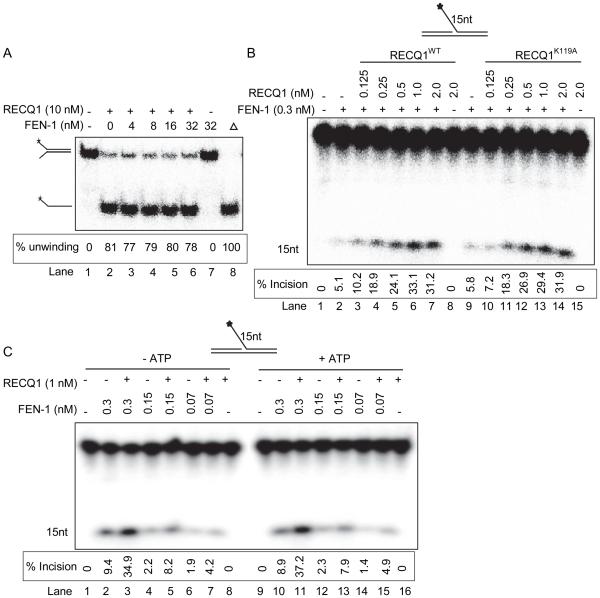
Helicase activity of RECQ1 is not necessary for stimulation of FEN-1 incision
Forked DNA substrates with either one or both of the arms in the double-stranded state are frequent intermediates of cellular processes such as DNA replication, repair, and recombination. RECQ1 unwinds forked duplex with noncomplementary 3’- and 5’-single-stranded DNA arms, as well as flap structures containing either a 3’- or 5’- single-stranded DNA in an ATP-dependent manner [15]. Presence of FEN-1 in a standard helicase assay did not modulate RECQ1 unwinding of a forked DNA duplex (Figure 6A). Thus we next tested whether the ATP-dependent helicase activity of RECQ1 plays a role in stimulation of FEN-1 cleavage of a 5’-flap substrate. First, we tested the effects of a previously characterized ATPase/helicase dead RECQ1 with a site-directed mutation, K119A, in the active site of its catalytic domain [49]. In a standard incision reaction, presence of the purified recombinant RECQ1-K119A mutant protein, devoid of ATPase or helicase activity [49], stimulated FEN-1 cleavage reaction (Figure 6B, lane 9 vs lanes 10-14) comparable to the wild-type RECQ1 in a dose dependent manner (Figure 6B; lane 2 vs lanes 3-7). No detectable incision products were obtained in control reaction conducted in the presence of RECQ1-K119A alone (Figure 6B, lane 14). DNA unwinding by RECQ1 is ATP-dependent, thus we next tested the effect of RECQ1 on FEN-1 cleavage reaction in the presence or absence of ATP (2 mM) (Figure 6C). RECQ1 (1 nM) similarly stimulated FEN-1 cleavage of 15 nt 5’-flap substrate irrespective of the presence or absence of ATP in the reaction (Figure 6C, lanes 2-7 vs. lanes 10-15). Altogether these results show that ATP hydrolysis and DNA unwinding are dispensable for RECQ1 stimulation of FEN-1 cleavage of a 15 nt 5’-flap DNA substrate, suggesting that the endonuclease activity enhancement produced by RECQ1 is not due to substrate modification.
Figure 6. Stimulation of FEN-1 cleavage of 5’-flap is independent of RECQ1 helicase activity and FEN-1 does not alter RECQ1 helicase activity.
A. FEN-1 does not alter RECQ1 helicase activity on a fork duplex. Phosphorimage of a typical helicase gel showing unwinding of a fork duplex by RECQ1 in the presence or absence of FEN-1; heat denatured substrate is indicated by triangle (lane 8). B. Reactions (20 μl) containing 10 fmol of 15nt 5’-flap DNA substrate and indicated concentrations of FEN-1 and/or RECQ1, wild-type (WT) or helicase dead mutant (K119A), proteins were incubated at 37°C for 15 min under standard conditions. Phosphorimage of a typical gel shows helicase-dead mutant of RECQ1 stimulates FEN-1 cleavage of 15nt 5’-flap DNA substrate. Lane 1, no enzyme; lane 2-7, FEN-1+ RECQ1 wild-type (0-2 nM); lane 8-13, FEN-1 + RECQ1 K119A mutant (0-2 nM). C. RECQ1 stimulates FEN-1 activity on 15nt 5’-flap DNA substrate in the absence of ATP. Phosphorimage of a typical gel shows FEN-1 incision products from reactions performed in the presence or absence of ATP.
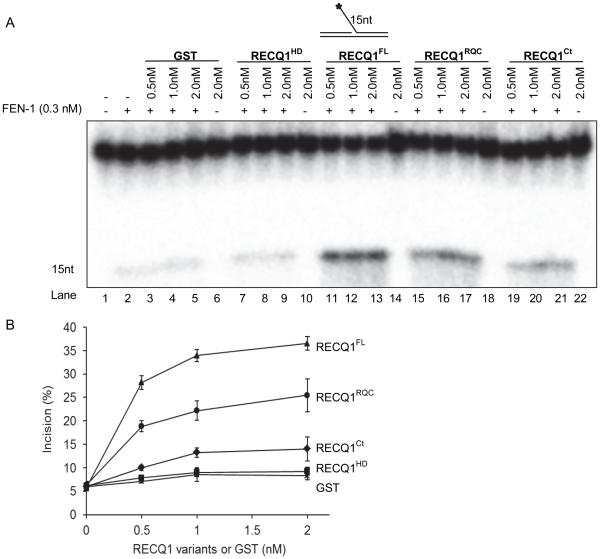
RQC and the C-terminus fragment of RECQ1 mediate functional stimulation of FEN-1
Having determined that the FEN-1 stimulation is independent of RECQ1 helicase activity, we asked whether the protein fragments of RECQ1 that were found to mediate physical interaction with FEN-1 could stimulate cleavage of 5’-flap substrate by FEN-1. Therefore, in addition to the wild-type full length RECQ1, we performed FEN-1 incision assays in the absence or presence of purified recombinant RECQ1 polypeptides GST-RECQ1418-592 (RQC domain), GST-RECQ1592-649 (C-terminus), GST-RECQ163-418 (helicase domain), or GST. No detectable incision of the 5’-flap substrate by RECQ1 polypeptides was observed in the absence of FEN-1 (Figure 7A). Full length RECQ1 was found to be most efficient in stimulating FEN-1 (0.3 nM) cleavage of 15 nt 5’-flap substrate (Figure 7A, lane 2 vs lanes 11-13; 7B). Stimulation in FEN-1 cleavage was observed in a dose dependent manner in the presence of the RQC domain (Figure 7A, lane 2 vs lanes 15-17) although to a significantly reduced extent than the wild-type full length RECQ1 (Figure 7B); and the C-terminal RECQ1 was found to be only slightly effective compared to the full length RECQ1 to stimulate FEN-1 cleavage (Figure 7A, lane 2 vs. lanes 19-21). In contrast, FEN-1 incision was not significantly altered in the presence of GST (Figure 7A, lane 2 vs. lanes 3-5; 7B) or helicase domain of RECQ1 that did not interact with FEN-1 in vitro (Figure 7A, lane 2 vs. lanes 7-9; 7B). These results indicate that both the RQC and C-terminal are essential to achieve optimal stimulation of FEN-1 activity by RECQ1 and the physical interaction between RECQ1-FEN-1 may be necessary.
Figure 7. Mapping of the FEN-1 interaction domains that mediate the functional interaction between RECQ1 and FEN-1.
Reactions (20 μl) containing 10 fmol of 15nt 5’-flap DNA substrate, 0.3 nM of FEN-1 and indicated concentrations of GST or GST-fused RECQ1, full-length (FL), helicase domain (HD), RecQ-C-terminus domain (RQC), or C-terminal (Ct), were incubated at 37°C for 15 min under standard conditions. A. Phosphorimage of a typical gel. Lane 1, no enzyme; B. Percent incision from the mean of three independent experiments is shown with SD indicated by error bars.
RECQ1 associates with telomere chromatin and stimulates FEN-1 cleavage of 5’-flap telomeric DNA substrates
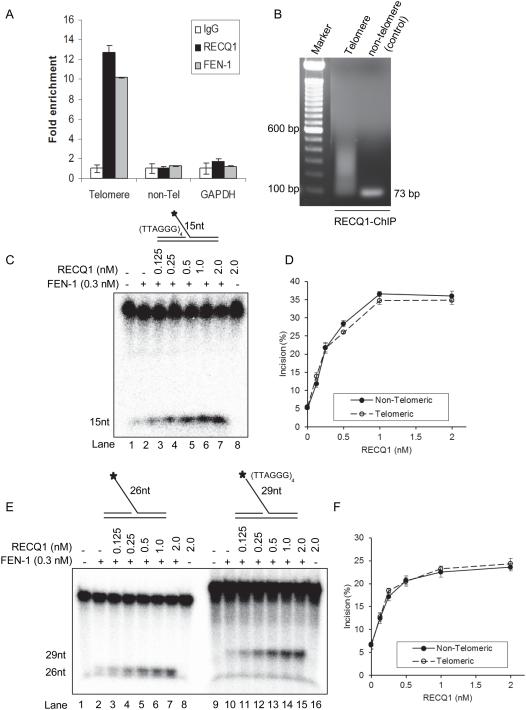
In addition to its essential roles in Okazaki fragment processing, FEN-1 is critical in telomeric lagging strand DNA synthesis [28-29]. Human telomeres consist of multiple tandem TTAGGG repeats which are highly susceptible to oxidative base damage due to their guanosine-rich nature [50]. Due to demonstrated roles of RECQ1 and FEN-1 in oxidative DNA damage and replication fork progression [7, 12-13, 23, 51-52], we investigated the intracellular association of RECQ1 with telomeric repeat DNA by ChIP followed by qPCR analysis using a previously established method that has been used for telomeric DNA detection in previous studies [42-46]
Cross-linked chromatin from asynchronously growing HeLa cells was immunoprecipitated with a control IgG or specific antibody against RECQ1. Following cross-link reversal, the immunoprecipitated chromatin was used in qPCR to determine the telomere repeat-containing DNA as well as HBG, a single copy gene located on chromosome 11 and used as control [42]. Telomere sequence-specific DNA was enriched nearly 13-fold in RECQ1-immunoprecipitate as compared to IgG in untreated HeLa cells (Figure 8A). Agarose gel electrophoresis of the PCR amplified products from the anti-RECQ1 ChIP DNA showed bands ranging from 50 to ~500 bp signifying enrichment of telomeric fragments (Figure 8B). As reported previously [29], FEN-1 associated with telomeres and ~11-fold enrichment of telomere-specific DNA was found in FEN-1 immunoprecipitate as compared to IgG (Figure 8A). In contrast, FEN-1 or RECQ1-immunoprecipitates were not enriched in DNA sequences corresponding to HBG or GAPDH, the negative control genomic loci (Figure 8A). Thus, our results indicate in vivo association of RECQ1 with telomeric DNA.
Figure 8. RECQ1 associates with telomere chromatin and stimulates FEN-1 cleavage of 5’-flap containing telomeric repeat sequence.
A. ChIP-qPCR of immunoprecipitated DNA with probes specific for telomeric region. HeLa cells were processed for ChIP using a RECQ1-specific antibody. FEN-1 antibody was used as a positive control for telomere enrichment and rabbit IgG served as negative control in ChIP experiments. Quantification of cross-linked telomere chromatin immunoprecipitated using the indicated antibodies is shown. Fold enrichment over IgG was determined and is shown for each primer pair for the ChIP. Relative occupancy at telomere versus a non-telomere negative control site (DNA containing HBG) and GAPDH shows preferential association of RECQ1 to telomeres. Results are expressed as means ± SEM for at least three independent experiments. B.A representative gel of the amplified telomere DNA immunoprecipitated with RECQ1 antibody. PCR amplified telomere fragments migrated as a smear (50 to ~500 bp). C. Phosphorimage of a typical gel of RECQ1 stimulation of FEN-1 incision of a 15 nt 5’-flap substrate containing TTAGGG repeats in duplex region upstream of 5’-flap. D. Percent incision of 15 nt 5’-flap substrate containing non-telomeric (solid line) or telomeric (dashed line) sequence. Data indicates mean of at least three independent experiments with SD shown as error bars. E. Phosphorimage of a typical gel of RECQ1 (0-2 nM) stimulation of FEN-1 incision of 5’-flap substrates containing non telomeric sequence or TTAGGG repeats in the 5’-flap. F. Percent incision of 5’-flap substrate containing non-telomeric (solid line) or telomeric (dashed line) sequence in the 5’-flap. Data indicates mean of at least three independent experiments with SD shown as error bars.
Consistent with our observation, Popuri et al [53] have recently reported that RECQ1 associates with telomeres in ALT cells and its interaction with TRF2 regulates its helicase activity on telomeric DNA in vitro. We used a 15 nt 5’-flap substrate containing (TTAGGG)4 sequence that has been previously reported to be bound by TRF2 [41] to ask whether RECQ1 can stimulate FEN-1 activity on telomeric DNA (Figure 8C, D). The addition of increasing concentrations of RECQ1 (0-2 nM) in FEN-1 (0.3 nM) reactions resulted in comparable stimulation of cleavage of 15 nt 5’-flap substrate with or without telomeric sequence in the duplex region 5’-to the flap (Figure 8D). We next tested whether the presence of telomeric repeats within the 5’-flap would affect cleavage by FEN-1 and its stimulation RECQ1. To address this, we compared the FEN-1 cleavage of 26 nt 5’-flap (used in Figure 4C) and a 29 nt 5’-flap substrate containing (TTAGGG)4 sequence in the 5’-flap region of the substrate in the presence or absence of RECQ1 (Figure 8E, F). Our results show RECQ1 stimulates FEN-1 cleavage of a 5’-flap substrate containing telomeric repeat sequence indicating a potential functional cooperation of RECQ1 and FEN-1 for DNA replication-repair pathways at telomeres.
RECQ1 regulates constitutive binding of FEN-1 to telomeres in vivo
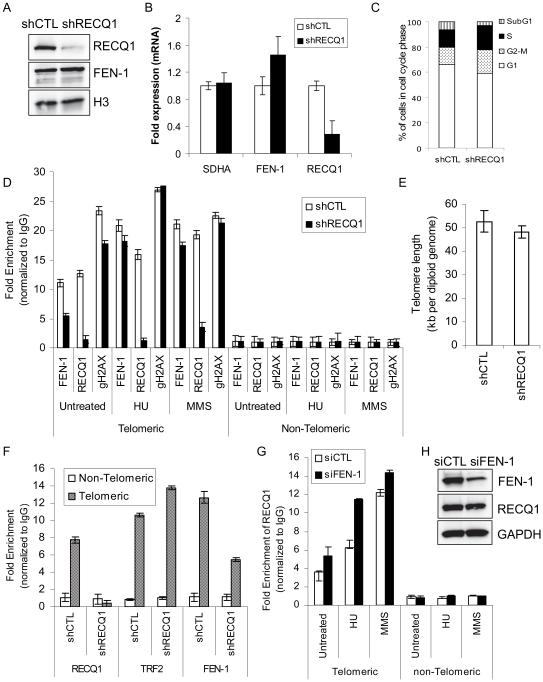
Precisely how FEN-1 nuclease acts at the chromosome ends is unknown, but FEN-1 deficiency affects telomere maintenance in both ALT and telomerase positive cell [29, 54]. Our finding that RECQ1 binds to the telomere DNA in telomerase-positive HeLa cells and its functional interaction with FEN-1 raised the possibility that RECQ1 might regulate FEN-1 at telomeres. To address this directly, effect of RECQ1 deficiency on the association of FEN-1 to the telomeric chromatin was determined by ChIP-qPCR from stable RECQ1-KD and control HeLa cells (Figure 9).
Figure 9. RECQ1 facilitates constitutive binding of FEN-1 at telomeres.
A. Total cell lysates were prepared from stable control-KD (shCTL) and RECQ1-KD (shRECQ1) cells and protein levels of RECQ1 and FEN-1 were measured by Western blotting. Histone H3 serves as loading control. B. RECQ1 silencing and mRNA expression of FEN-1 in control and RECQ1 KD cells was assessed by RT-qPCR normalized to GAPDH. SDHA was included as another housekeeping gene control. C. Cell cycle analysis of control-KD and RECQ1-KD HeLa cells used for ChIP assays. D. Stable control-KD or RECQ1-KD HeLa cells, untreated or treated with HU (2 mM, 16 h) or MMS (15 μg/ml, 16 h), were processed for ChIP using RECQ1, FEN-1, and γH2AX (gH2AX) antibodies. Quantification of telomere chromatin by qPCR of immunoprecipitated DNA with probes specific for telomeric region and a non-telomere negative control site is shown. Fold enrichment over IgG was determined and used to normalize the data to determine specific enrichment of telomere sequence in each case. Results are expressed as means ± SEM for at least three independent experiments. E. Telomere length in control-KD and RECQ1-KD HeLa cells. Results are expressed as means ± SEM for three independent experiments. F. Stable control-KD or RECQ1-KD HeLa cells (untreated) were processed for ChIP-qPCR using indicated antibodies. TRF2 antibody was used as a positive control for telomere enrichment and rabbit IgG served as negative control in ChIP experiments. GAPDH-normalized RT-qPCR quantification of cross-linked telomere chromatin immunoprecipitated using the indicated antibodies is shown. Fold enrichment over IgG was determined and is shown for each primer pair for the ChIP. Relative occupancy at telomere versus a non-telomere negative control site (DNA containing HBG) shows preferential association of RECQ1, FEN-1 and TRF2 to telomeres. As compared to control-KD cells, RECQ1-KD cells show reduced telomere binding of FEN-1 whereas TRF2 binding is comparable. Results are expressed as means ± SEM for at least three independent experiments. G. HeLa cells transfected with control (siCTL) or FEN-1-specific siRNA (siFEN-1), untreated or treated with HU (2 mM, 16 h) or MMS (15 μg/ml, 16 h), were processed for ChIP using RECQ1 and IgG antibodies. Fold enrichment over IgG was determined as described in “D”. Results are expressed as means ± SEM for at least three independent experiments. H. Protein levels of FEN-1 and RECQ1 in total lysates prepared from control or FEN-1 siRNA transfected cells were measured by Western blotting. GAPDH serves as loading control.
As expected, RECQ1-KD (shRECQ1) cells expressing anti-RECQ1 RNAi had dramatically decreased levels of RECQ1 protein and mRNA compared to control-KD (shCTL) cells (Figure 9A, B). FEN-1 mRNA and protein was not targeted by this treatment, and so was not decreased; on the contrary, FEN-1 levels were somewhat higher in the RECQ1-KD cells. FEN-1 binds to telomeres during S-phase [29] and cell cycle analyses revealed that 13.8% of control cells and 19.2% RECQ1-KD cells were in S-phase (Figure 9C; Supplementary Figure 4). With the efficacy of the RECQ1 depletion procedure confirmed, these cells were analyzed by ChIP-qPCR (Figure 9D). Telomere sequence-specific DNA was enriched 12-fold in RECQ1-immunoprecipitate as compared to IgG in untreated control-KD HeLa cells, showing the extent to which RECQ1 protein preferentially associates with telomeric chromatin. In untreated RECQ1-KD cells, RECQ1-immunoprecipitate showed only 1.5-fold enrichment of telomere sequence-specific DNA compared to IgG. Diminished telomeric signal upon shRNA-mediated depletion of RECQ1 further confirmed the specificity of RECQ1 antibody, because its effect compared to nonspecific IgG was antigen-dependent. FEN-1 is known to preferentially associate with telomeres [29], and we found that this association is, to a large degree, RECQ1-dependent. In FEN-1 immunoprecipitates, the yield of telomeric sequence-specific DNA was markedly lower in RECQ1-KD cells compared to control-KD cells (56% less than control, P< 0.05). FEN-1-immunoprecipitate from the control cells displayed nearly 11-fold enrichment of telomere sequence-specific DNA relative to non-telomeric DNA, whereas only ~5-fold enrichment of telomere sequence-specific DNA was observed in RECQ1-KD cells (Figure 9D). In order to compare these results to observations for a bona fide telomere-specific protein, an additional set of ChIP experiments also included specific antibody against TRF2 as positive control and normal IgG was used as negative control as usual. The immunoprecipitated DNAs were subjected to real time PCR amplification of telomere sequence-specific DNA and a non-telomeric DNA sequence (HBG). As expected, TRF2 specifically interacted with telomeres; an average of 10.6-fold and 13.6-fold enrichment of telomere-specific DNA was found in TRF2 immunoprecipitate in control-KD and RECQ1-KD cells, respectively (Figure 9F). The modest increase in telomere binding of TRF2 in RECQ1-KD cells may partly be due to increased S-phase population since telomeric association of TRF2 strongly increases in S-phase [55] (Figure 9C). FEN-1 immunoprecipitates showed 12.7-fold enrichment of telomere specific DNA in control-KD cells whereas only 5.5-fold enrichment of telomere sequence-specific DNA was observed in FEN-1 immunoprecipitates from RECQ1-KD cells (Figure 9F). This reduction in binding of FEN-1 to telomeric chromatin in RECQ1-KD cells cannot be due to reduced FEN-1 expression in RECQ1-depleted cells because FEN-1 expression remains robust in RECQ1-KD cells (Figure 9A, B). Depletion of FEN-1 in HeLa cells results in telomere shortening [54]. A qPCR based analysis of telomere length revealed that RECQ1-KD cells have shorter telomeres than shCTL HeLa cells (48.31 vs 52.82 kb, P= 0.038) (Figure 9E). This observation is qualitatively consistent with a recent report using Q-FISH analysis in RECQ1-depleted HeLa cells, however the magnitude of difference in telomere length in Popuri et al [53] is substantially greater and may be attributed to different assay methods used. To further characterize this system, γH2AX immunoprecipitates were analyzed. Even in untreated cells, telomeres are highly enriched in γH2AX [56]. We observed that γH2AX immunoprecipitates were enriched in telomere sequence-specific DNA in untreated control cells, and the extent of enrichment was somewhat reduced in RECQ1-KD cells.
We next examined the effect of DNA damage on the binding of RECQ1 and FEN-1 to telomeres in control and RECQ1-KD HeLa Cells (Figure 9D). ChIP experiments were performed using control or RECQ1-KD cells that were treated with HU, which induces replication stress, or MMS, an alkylating agent which generates lesions that are processed by BER [57]. For control cells, RECQ1-immunoprecipitates from HU- and MMS-treated cells showed even greater enrichment of telomere sequence-specific DNA (16-fold and 20-fold, respectively) than untreated cells (12-fold). Treatment with HU or MMS also increased FEN-1-bound telomere sequence-specific DNA, producing nearly 20-fold and 21-fold enrichment versus the 11-fold enrichment that had been observed in the untreated control cells. However, the fold enrichment values following treatment were only a little lower than this in the RECQ1-KD cells. Thus, while constitutive binding of FEN-1 to telomeres was RECQ1-dependent, this was not the case in the context of DNA damaging treatments. After HU or MMS treatment, FEN-1 was observed to be strongly associated with telomeres almost regardless of RECQ1 protein abundance. In contrast, as compared to control cells, telomeric association of RECQ1 was increased in FEN-1-depleted cells that were either unperturbed or exposed to DNA damage treatment (Figure 9G, H). FEN-1 is critical for re-initiation of stalled forks at telomeres [58], and increased telomere association of RECQ1 upon DNA damage, especially HU treatment, in FEN-1-depleted cells indicates that RECQ1 may be engaged in preventing replication fork collapse at telomeres (Figure 9G).
RECQ1-KD cells displayed a 1.7-fold increase in γH2AX-bound telomere sequence-specific DNA in response to HU treatment as compared to 1.2-fold in control cells, suggesting that RECQ1 depletion promotes DNA damage at telomeric sequences (Figure 9D). MMS treatment in control cells did not result in significant enrichment of γH2AX at telomeres but led to 1.4-fold greater γH2AX at telomeres in RECQ1-KD cells. Increased MMS-induced γH2AX at telomeres in RECQ1-depleted cells may indicate that replication stress in the absence of RECQ1 is a source of increased DNA damage at telomeres.
Collectively, our results indicate that RECQ1 contributes to the constitutive binding of FEN-1 to telomeric chromatin in HeLa cells but the DNA damage-induced enrichment of FEN-1 at telomeres is likely to be RECQ1-independent. Furthermore, results from γH2AX-ChIP experiments suggest that the role of RECQ1 at telomeres may be more important in the context of DNA damage.
DISCUSSION
Here, we report a direct protein interaction between RECQ1 and FEN-1, and demonstrate that RECQ1 stimulates FEN-1 cleavage of a 5’-flap DNA substrate independent of its helicase activity. Our study identifies a novel requirement of RECQ1 in facilitating constitutive, but not DNA damage-induced, residence of FEN-1 at telomeres in telomerase positive cycling cells. This study emphasizes role of RECQ1 in DNA replication and repair, and illustrates interaction between the RecQ helicase family proteins and the structure-specific nuclease FEN-1 as a conserved collaboration for genome maintenance in human cells.
Functions of FEN-1 in DNA transactions are analogous to those of RecQ helicases that play key roles by unwinding intermediate DNA structures in chromatin to regulate replication, recombination and repair. Brosh et al. [30] identified WRN as the first among the human RecQ homologs to be a FEN-1 interacting protein. FEN-1 can be specifically precipitated by the BLM 966-1417 fragment that shares homology with amino acid residues 949-1092 in the conserved RQC motif of WRN, essential for the physical and functional interaction with FEN-1 [31, 39, 59]. FEN-1 interaction has not been mapped on RECQ5β [36], however, RECQL4, which lacks RQC domain, also associates with FEN-1 in a common protein complex [35]. Our data indicate that interaction with FEN-1 involves the conserved RQC-domain and the C-terminus of RECQ1. The poorly conserved C-terminus of RECQ1 is also involved in interactions with PARP-1 [13, 51] and Ku70/80 [38] suggesting a critical role in mediating protein-protein interactions.
Stimulation of FEN-1 endonucleolytic cleavage of a 5’-flap substrate is independent of RECQ1 helicase activity in vitro, but our functional data using RECQ1 fragments suggests that full length protein, and consequently native conformation of RECQ1, is important to effect optimal FEN-1 stimulation. It is yet unknown whether RECQ1 modulates the gap endonuclease (GEN) activity of FEN-1 or process chicken foot structures [60-61], but the in vitro ability of RECQ1 to unwind intermediates of replication, repair and recombination [12, 15, 62] suggests that the coordinated activity of RECQ1 helicase and FEN-1 nuclease is important for replication restart similar to what has been proposed for WRN-FEN-1 [61]. Although the helicase activity of RECQ1 is unaffected by FEN-1, stimulation of FEN-1 may be an important role of RECQ1 for the endonucleolytic cleavage of 5’-flap structures during the processing of Okazaki fragments, rescue of stalled replication forks, and excision repair [59]. Similar to WRN, but unlike RECQL4 and RECQ5β, the 3’-5’ helicase activity of RECQ1 unwinds the leading strand of forked DNA duplex thereby unwinding the replication fork structure in the direction of the fork movement [12, 15]. Furthermore, RECQ1 preferentially catalyzes fork reversal and promotes resetting of replication forks in vitro; and this function of RECQ1 helicase is critical in preventing fork collapse upon replication stress in vivo [13].
Replication forks stall at fragile sites [63] and within telomeres containing G-rich hexameric TTAGGG repeats [64] as fork progression is inherently difficult at repetitive DNA sequences due to their propensity to form secondary structures [63]. Previously, we demonstrated that RECQ1 accumulates at common fragile sites where replication forks have stalled following stress [9]. We now show that RECQ1 is constitutively bound to telomeres consistent with a recent report from Bohr lab implicating RECQ1 in telomere maintenance [53]. Significantly reduced FEN-1 at telomeric chromatin observed in asynchronized RECQ1-KD HeLa cells suggests that RECQ1 is important for association of FEN-1 with telomeres in unperturbed, replicating cells. Stewart lab has established that FEN-1 nuclease activity is essential for its functions in maintaining telomere stability [29, 58]. Telomere dysfunction in RECQ1-depleted cells can, in part, be explained by reduced FEN-1 functions at lagging daughter telomeres [54]. In the absence of optimal FEN-1 levels, inappropriate Okazaki fragment processing might cause excessive dissociation or degradation of the last primer, leading to an increase in single stranded DNA of the template strand [28, 54] causing increased RPA phosphorylation foci at telomeric sites in RECQ1-KD cells [53]. Depletion of RECQ1- or FEN- 1 challenges replication fork progression genome-wide, though FEN-1 is especially important for the re-initiation of stalled replication forks at telomeres [29, 58]. It will be of interest, in the future, to determine whether RECQ1 and FEN1 depletion is epistatic for telomere dysfunction phenotype, or there is a synthetic telomere dysfunction phenotype attributable to interaction of RECQ1 and FEN1 when both are partially disabled. RECQ1 depletion did not increase γH2AX at telomeres in our analysis in telomerase positive HeLa cells, potentially indicating that the loss of FEN-1 functions at telomeres is compensated by telomerase in these cells [29]; however, insufficient FEN-1 may contribute to the accumulation of fragile telomeres reported in RECQ1-depleted cells since telomerase can not compensate for the loss of FEN-1 to prevent fragile telomeres [29].
Whether the RECQ1-FEN-1 interaction is essential for telomere maintenance remains to be determined but our data indicates RECQ1-dependence of FEN-1 binding to telomeres in unperturbed replicating cells. In contrast, damage-induced telomere-binding of FEN-1 in HeLa cells is independent of RECQ1 indicating that additional factors regulate FEN-1 recruitment and functions at telomeres. Nevertheless, both RECQ1 and FEN-1 are enriched at telomeres in HeLa cells treated with HU or MMS suggesting common functions in replication fork restart and repair at telomeres.
Interaction with RECQ1 is mediated through the PCNA interaction motif and the C-terminus of FEN-1. The interaction with PCNA that allows FEN-1 to associate with the replication machinery is not important for its functions at the telomere [58]. The C-terminus of FEN-1, and consequently interaction with WRN and TRF2, is essential for telomere functions as its deletion partially disables localization of FEN-1 to telomeres and fails to suppress telomere fragility and lagging strand sister telomere loss [29]. Given the interaction of RECQ1 [53] and WRN [65] with TRF2, it is conceivable that TRF2 might recruit the RecQ helicase-FEN-1 complex coordinately at the telomeres to resolve stalled replication forks and enable their efficient restart. However, the recruitment of RECQ1 [53] and FEN-1 [29] to telomeres is independent of TRF2.
TRF2, a component of Shelterin protein complex, binds to the double stranded telomeric repeat DNA resulting in a highly stable protein-DNA complex and is directly involved in inhibition of DNA damage signaling at telomeres and protection of telomeres [66-67]. Recent demonstration of RECQ1’s ability to dislodge TRF2 from telomeric substrate in an RPA-dependent manner presents a possible role in coordinating telomere protection by Shelterin proteins binding and DNA synthesis by the replication machinery [68]. While TRF2 promotes the helicase activity of WRN [65], it inhibits RECQ1 helicase on telomeric substrates [53]. Interestingly, TRF2 inhibition of RECQ1 helicase is overcome by the presence of oxidative lesion in the substrate[53] which also inhibits DNA binding of TRF2 [69]. However, RECQ1 can unwind DNA substrates containing oxidative base damage regardless of telomeric repeats [53, 70] and the sensitivity of RECQ1-deficient cells to oxidative stress [51] indicates a global role in the repair of oxidative DNA damage. Our observation that RECQ1 stimulates FEN-1 cleavage of a 5’-flap substrate containing telomeric repeat sequence comparable to that containing non-telomeric sequence further suggests that RECQ1 functions with FEN-1 to facilitate DNA replication and/or repair processes at telomeres and other genomic regions.
Including the present study, all five known human RecQ proteins have been shown to physically interact and stimulate FEN-1 catalytic activity. Conserved interaction of RecQ helicases with FEN-1 may signify functional compensation, by redundant and alternative mechanisms, essential for avoiding genomic instability since roles of FEN-1 are critically important in DNA replication and repair. Differential requirement of the concerted action of a specific RecQ homolog with FEN-1 is likely to be governed by distinct substrate specificity of individual RecQ helicases relevant to a given cellular and genomic context. For instance, unlike WRN and BLM, recombinant RECQ1 lacks the ability to resolve replication-impeding G4 DNA structures present at telomeres [71]. Our results emphasize a yet poorly addressed question that is what might be the specific physiological function of individual homolog of the five human RecQ helicases at specific genomic loci. Furthermore, the activities of individual RecQ homolog-FEN-1 complex may be assigned to lesion-specific pathways or sub-pathways of DNA repair. It has been suggested that FEN-1 forms dynamic protein complexes as necessary for the specific steps of the pathways in which it is participating [23]. Failure to detect a RECQ5β-FEN-1 complex supports the dynamic nature of RecQ-FEN-1 association in vivo [36]. Although FEN-1 C-terminus is commonly involved in mediating interactions with WRN, BLM, and RECQ1, physical interaction with RECQ1 and RECQ5β [36] spans the PCNA-interaction motif of FEN-1. WRN additively enhances FEN-1 endonuclease activity in the presence of PCNA [39] whereas RECQ1 stimulation of FEN-1 is not influenced by PCNA. PCNA mediates Okazaki fragment maturation through tight coordination of the activities of DNA polymerase δ, FEN-1 and DNA ligase I, and recent results support a mechanism of sequential switching of partners on the eukaryotic PCNA trimer during DNA replication and repair [72]. Given the critical roles of PCNA at the replication fork [73] and the unique association of RECQ1 (and RECQL4), but not other RecQ proteins, with replication initiation complex [8], interaction of FEN-1 with multiple RecQ proteins may ensure faithful replication and repair in cycling cells.
Collectively, our results provide several potential functional overlaps with FEN-1 [23] and other RecQ helicases [74], and support the emerging evidence suggesting that RECQ1 is involved in DNA replication, repair, and telomere maintenance. Genomic instability observed in RecQ-deficiency has been largely attributed to the inappropriate processing of DNA replication or repair intermediates that arise during conditions of replication stress [75]. In humans, RECQ1 is the most abundant RecQ helicase homolog and therefore it can be expected to play vital roles, either alone or in concert with protein partners such as FEN-1, in maintaining cellular homeostasis and genomic stability under basal and genotoxic stress conditions.
Supplementary Material
SUMMARY STATEMENT.
We report a physical and functional interaction between RECQ1 helicase and flap endonuclease-1, and identify a novel requirement of RECQ1 in facilitating constitutive, but not DNA damage-induced, binding of FEN-1 at telomeres in telomerase positive cycling cells.
ACKNOWLEDGEMENT
We thank Dr. Alessandro Vindigni (St. Louis University) for the shRNA constructs, Drs. Ashish Lal and Xiao Ling Li (NCI, NIH) for FACS analysis, and Dr. Robert Brosh and Joshua Sommers (NIA, NIH) for technical advice.
FUNDING
This research was supported by the National Institutes of General Medical Sciences [5SC1GM093999-05] to Sudha Sharma and National Institute on Minority Health and Health Disparities [G12MD007597] to Howard University.
REFERENCES
- 1.Sharma S, Doherty KM, Brosh RM., Jr. Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsen N, Hickson I. RecQ Helicases: Conserved Guardians of Genomic Integrity. In: Spies M, editor. In DNA Helicases and DNA Motor Proteins. Springer; New York: 2013. pp. 161–184. [DOI] [PubMed] [Google Scholar]
- 3.Brosh RM, Jr., Bohr VA. Human premature aging, DNA repair and RecQ helicases. Nucleic Acids Res. 2007;35:7527–7544. doi: 10.1093/nar/gkm1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brosh RM., Jr. DNA helicases involved in DNA repair and their roles in cancer. Nat. Rev. Cancer. 2013;13:542–558. doi: 10.1038/nrc3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma S, Brosh RM., Jr. Unique and important consequences of RECQ1 deficiency in mammalian cells. Cell Cycle. 2008;7:989–1000. doi: 10.4161/cc.7.8.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma S, Stumpo DJ, Balajee AS, Bock CB, Lansdorp PM, Brosh RM, Jr., Blackshear PJ. RECQL, a member of the RecQ family of DNA helicases, suppresses chromosomal instability. Mol. Cell. Biol. 2007;27:1784–1794. doi: 10.1128/MCB.01620-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sami F, Sharma S. Probing Genome Maintenance Functions of human RECQ1. Comput. Struct. Biotechnol. J. 2013;6:e201303014. doi: 10.5936/csbj.201303014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ, Jr., Falaschi A, Vindigni A. Human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol. Cell. Biol. 2010;30:1382–1396. doi: 10.1128/MCB.01290-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu X, Parvathaneni S, Hara T, Lal A, Sharma S. Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D. Mol. Cancer. 2013;12:29. doi: 10.1186/1476-4598-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharma S, Brosh RM., Jr. Human RECQ1 is a DNA damage responsive protein required for genotoxic stress resistance and suppression of sister chromatid exchanges. PLoS ONE. 2007;2:e1297. doi: 10.1371/journal.pone.0001297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeRoy G, Carroll R, Kyin S, Seki M, Cole MD. Identification of RecQL1 as a Holliday junction processing enzyme in human cell lines. Nucleic Acids Res. 2005;33:6251–6257. doi: 10.1093/nar/gki929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Popuri V, Croteau DL, Brosh RM, Jr., Bohr VA. RECQ1 is required for cellular resistance to replication stress and catalyzes strand exchange on stalled replication fork structures. Cell Cycle. 2012;11:4252–4265. doi: 10.4161/cc.22581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berti M, Chaudhuri AR, Thangavel S, Gomathinayagam S, Kenig S, Vujanovic M, Odreman F, Glatter T, Graziano S, Mendoza-Maldonado R, Marino F, Lucic B, Biasin V, Gstaiger M, Aebersold R, Sidorova JM, Monnat RJ, Lopes M, Vindigni A. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol. 2013;20:347–354. doi: 10.1038/nsmb.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui S, Arosio D, Doherty KM, Brosh RM, Jr., Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM., Jr. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 16.Watt PM, Louis EJ, Borts RH, Hickson ID. Sgs1: a eukaryotic homolog of E. coli RecQ that interacts with topoisomerase II in vivo and is required for faithful chromosome segregation. Cell. 1995;81:253–260. doi: 10.1016/0092-8674(95)90335-6. [DOI] [PubMed] [Google Scholar]
- 17.Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 18.Doherty KM, Sharma S, Uzdilla LA, Wilson TM, Cui S, Vindigni A, Brosh RM., Jr. RECQ1 helicase interacts with human mismatch repair factors that regulate genetic recombination. J. Biol. Chem. 2005;280:28085–28094. doi: 10.1074/jbc.M500265200. [DOI] [PubMed] [Google Scholar]
- 19.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat.Rev. Mol. Cell. Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 20.Harrington JJ, Lieber MR. Functional domains within FEN-1 and RAD2 define a family of structure-specific endonucleases: implications for nucleotide excision repair. Genes Dev. 1994;8:1344–1355. doi: 10.1101/gad.8.11.1344. [DOI] [PubMed] [Google Scholar]
- 21.Vallur AC, Maizels N. Complementary Roles for Exonuclease 1 and Flap Endonuclease 1 in Maintenance of Triplet Repeats. J. Biol. Chem. 2010;285:28514–28519. doi: 10.1074/jbc.M110.132738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vallur AC, Maizels N. Distinct Activities of Exonuclease 1 and Flap Endonuclease 1 at Telomeric G4 DNA. PLoS ONE. 2010;5:e8908. doi: 10.1371/journal.pone.0008908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Balakrishnan L, Bambara RA. Flap Endonuclease 1. Annu. Rev. Biochem. 2013;82:119–138. doi: 10.1146/annurev-biochem-072511-122603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finger LD, Atack J, Tsutakawa S, Classen S, Tainer J, Grasby J, Shen B. The Wonders of Flap Endonucleases: Structure, Function, Mechanism and Regulation. In: MacNeill S, editor. In The Eukaryotic Replisome: a Guide to Protein Structure and Function. Springer; Netherlands: 2012. pp. 301–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsutakawa, Susan E, Classen S, Chapados, Brian R, Arvai AS, Finger LD, Guenther G, Tomlinson, Christopher G, Thompson P, Sarker, Altaf H, Shen B, Cooper, Priscilla K, Grasby, Jane A, Tainer, John A. Human Flap Endonuclease Structures, DNA Double-Base Flipping, and a Unified Understanding of the FEN1 Superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Kao HI, Bambara RA. Flap endonuclease 1: a central component of DNA metabolism. Annu. Rev. Biochem. 2004;73:589–615. doi: 10.1146/annurev.biochem.73.012803.092453. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Prasad R, Beard WA, Hou EW, Horton JK, McMurray CT, Wilson SH. Coordination between polymerase beta and FEN1 can modulate CAG repeat expansion. J. Biol. Chem. 2009;284:28352–28366. doi: 10.1074/jbc.M109.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parenteau J, Wellinger RJ. Differential processing of leading-and lagging-strand ends at Saccharomyces cerevisiae telomeres revealed by the absence of Rad27p nuclease. Genetics. 2002;162:1583–1594. doi: 10.1093/genetics/162.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saharia A, Guittat L, Crocker S, Lim A, Steffen M, Kulkarni S, Stewart SA. Flap endonuclease 1 contributes to telomere stability. Curr. Biol. 2008;18:496–500. doi: 10.1016/j.cub.2008.02.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brosh RM, Jr., von Kobbe C, Sommers JA, Karmakar P, Opresko PL, Piotrowski J, Dianova I, Dianov GL, Bohr VA. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 2001;20:5791–5801. doi: 10.1093/emboj/20.20.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma S, Sommers JA, Wu L, Bohr VA, Hickson ID, Brosh RM., Jr. Stimulation of flap endonuclease-1 by the Bloom's syndrome protein. J. Biol. Chem. 2004;279:9847–9856. doi: 10.1074/jbc.M309898200. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Bambara RA. Human Bloom protein stimulates flap endonuclease 1 activity by resolving DNA secondary structure. J. Biol. Chem. 2005;280:5391–5399. doi: 10.1074/jbc.M412359200. [DOI] [PubMed] [Google Scholar]
- 33.Sharma S, Sommers JA, Driscoll HC, Uzdilla L, Wilson TM, Brosh RM., Jr. The exonucleolytic and endonucleolytic cleavage activities of human exonuclease 1 are stimulated by an interaction with the carboxyl-terminal region of the Werner syndrome protein. J. Biol. Chem. 2003;278:23487–23496. doi: 10.1074/jbc.M212798200. [DOI] [PubMed] [Google Scholar]
- 34.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. U. S. A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schurman SH, Hedayati M, Wang Z, Singh DK, Speina E, Zhang Y, Becker K, Macris M, Sung P, Wilson DM, 3rd, Croteau DL, Bohr VA. Direct and indirect roles of RECQL4 in modulating base excision repair capacity. Hum Mol. Genet. 2009;18:3470–3483. doi: 10.1093/hmg/ddp291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Speina E, Dawut L, Hedayati M, Wang Z, May A, Schwendener S, Janscak P, Croteau DL, Bohr VA. Human RECQL5beta stimulates flap endonuclease 1. Nucleic Acids Res. 2010;38:2904–2916. doi: 10.1093/nar/gkp1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pike AC, Shrestha B, Popuri V, Burgess-Brown N, Muzzolini L, Costantini S, Vindigni A, Gileadi O. Structure of the human RECQ1 helicase reveals a putative strand-separation pin. Proc.Natl. Acad. Sci. U. S. A. 2009;106:1039–1044. doi: 10.1073/pnas.0806908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parvathaneni S, Stortchevoi A, Sommers JA, Brosh RM, Jr., Sharma S. Human RECQ1 Interacts with Ku70/80 and Modulates DNA End-Joining of Double-Strand Breaks. PLoS ONE. 2013;8:e62481. doi: 10.1371/journal.pone.0062481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma S, Sommers JA, Gary RK, Friedrich-Heineken E, Hubscher U, Brosh RM., Jr. The interaction site of Flap Endonuclease-1 with WRN helicase suggests a coordination of WRN and PCNA. Nucleic Acids Res. 2005;33:6769–6781. doi: 10.1093/nar/gki1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brosh RM, Jr., Driscoll HC, Dianov GL, Sommers JA. Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry. 2002;41:12204–12216. doi: 10.1021/bi026031j. [DOI] [PubMed] [Google Scholar]
- 41.Muftuoglu M, Wong HK, Imam SZ, Wilson DM, 3rd, Bohr VA, Opresko PL. Telomere repeat binding factor 2 interacts with base excision repair proteins and stimulates DNA synthesis by DNA polymerase beta. Cancer Res. 2006;66:113–124. doi: 10.1158/0008-5472.CAN-05-2742. [DOI] [PubMed] [Google Scholar]
- 42.Cawthon RM. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Madlener S, Ströbel T, Vose S, Saydam O, Price BD, Demple B, Saydam N. Essential role for mammalian apurinic/apyrimidinic (AP) endonuclease Ape1/Ref-1 in telomere maintenance. Proc. Natl. Acad. Sci. U.S.A. 2013;110:17844–17849. doi: 10.1073/pnas.1304784110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kar A, Saha D, Purohit G, Singh A, Kumar P, Yadav VK, Kumar P, Thakur RK, Chowdhury S. Metastases suppressor NME2 associates with telomere ends and telomerase and reduces telomerase activity within cells. Nucleic Acids Res. 2012;40:2554–2565. doi: 10.1093/nar/gkr1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Dakic A, Zhang Y, Dai Y, Chen R, Schlegel R. HPV E6 protein interacts physically and functionally with the cellular telomerase complex. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18780–18785. doi: 10.1073/pnas.0906357106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hewitt G, Jurk D, Marques FDM, Correia-Melo C, Hardy T, Gackowska A, Anderson R, Taschuk M, Mann J, Passos JF. Telomeres are favoured targets of a persistent DNA damage response in ageing and stress-induced senescence. Nat. Commun. 2012;3:708. doi: 10.1038/ncomms1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.O'Callaghan N, Dhillon V, Thomas P, Fenech M. A quantitative real-time PCR method for absolute telomere length. Biotechniques. 2008;44:807–809. doi: 10.2144/000112761. [DOI] [PubMed] [Google Scholar]
- 48.Gary R, Ludwig DL, Cornelius HL, MacInnes MA, Park MS. The DNA Repair Endonuclease XPG Binds to Proliferating Cell Nuclear Antigen (PCNA) and Shares Sequence Elements with the PCNA-binding Regions of FEN-1 and Cyclin-dependent Kinase Inhibitor p21. J. Biol. Chem. 1997;272:24522–24529. doi: 10.1074/jbc.272.39.24522. [DOI] [PubMed] [Google Scholar]
- 49.Muzzolini L, Beuron F, Patwardhan A, Popuri V, Cui S, Niccolini B, Rappas M, Freemont PS, Vindigni A. Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 2007;5:e201. doi: 10.1371/journal.pbio.0050020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem. Sci. 2002;27:339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 51.Sharma S, Phatak P, Stortchevoi A, Jasin M, Larocque JR. RECQ1 plays a distinct role in cellular response to oxidative DNA damage. DNA Repair (Amst) 2012;11:537–549. doi: 10.1016/j.dnarep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen B, Singh P, Liu R, Qiu J, Zheng L, Finger LD, Alas S. Multiple but dissectible functions of FEN-1 nucleases in nucleic acid processing, genome stability and diseases. Bioessays. 2005;27:717–729. doi: 10.1002/bies.20255. [DOI] [PubMed] [Google Scholar]
- 53.Popuri V, Hsu J, Khadka P, Horvath K, Liu Y, Croteau DL, Bohr VA. Human RECQL1 participates in telomere maintenance. Nucleic Acids Res. 2014;42:5671–5688. doi: 10.1093/nar/gku200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sampathi S, Bhusari A, Shen B, Chai W. Human flap endonuclease I is in complex with telomerase and is required for telomerase-mediated telomere maintenance. J. Biol. Chem. 2009;284:3682–3690. doi: 10.1074/jbc.M805362200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li B, Jog SP, Reddy S, Comai L. WRN controls formation of extrachromosomal telomeric circles and is required for TRF2DeltaB-mediated telomere shortening. Mol. Cell. Biol. 2008;28:1892–1904. doi: 10.1128/MCB.01364-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaina B, Ochs K, Grosch S, Fritz G, Lips J, Tomicic M, Dunkern T, Christmann M. BER, MGMT, and MMR in defense against alkylation-induced genotoxicity and apoptosis. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:41–54. doi: 10.1016/s0079-6603(01)68088-7. [DOI] [PubMed] [Google Scholar]
- 58.Saharia A, Teasley DC, Duxin JP, Dao B, Chiappinelli KB, Stewart SA. FEN1 ensures telomere stability by facilitating replication fork re-initiation. J. Biol. Chem. 2010;285:27057–27066. doi: 10.1074/jbc.M110.112276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma S, Sommers JA, Brosh RM., Jr. Processing of DNA replication and repair intermediates by the concerted action of RecQ helicases and Rad2 structure-specific nucleases. Protein Pept. Lett. 2008;15:89–102. doi: 10.2174/092986608783330369. [DOI] [PubMed] [Google Scholar]
- 60.Zheng L, Zhou M, Chai Q, Parrish J, Xue D, Patrick SM, Turchi JJ, Yannone SM, Chen D, Shen B. Novel function of the flap endonuclease 1 complex in processing stalled DNA replication forks. EMBO Rep. 2005;6:83–89. doi: 10.1038/sj.embor.7400313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM., Jr. WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork. Mol. Biol. Cell. 2004;15:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bugreev DV, Brosh RM, Jr., Mazin AV. RECQ1 possesses DNA branch migration activity. J. Biol. Chem. 2008;283:20231–20242. doi: 10.1074/jbc.M801582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Durkin SG, Glover TW. Chromosome fragile sites. Annu. Rev. Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 64.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, de Lange T. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 66.van Steensel B, Smogorzewska A, de Lange T. TRF2 Protects Human Telomeres from End-to-End Fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 67.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JHJ, Lange T. d. The Telomeric Protein TRF2 Binds the ATM Kinase and Can Inhibit the ATM-Dependent DNA Damage Response. PLoS Biol. 2004;2:e240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sommers JA, Banerjee T, Hinds T, Wan B, Wold MS, Lei M, Brosh RM., Jr. Novel function of the Fanconi anemia group J or RECQ1 helicase to disrupt protein-DNA complexes in a replication protein A-stimulated manner. J. Biol. Chem. 2014;289:19928–19941. doi: 10.1074/jbc.M113.542456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Opresko PL, Fan J, Danzy S, Wilson DM, Bohr VA. Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucleic Acids Res. 2005;33:1230–1239. doi: 10.1093/nar/gki273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suhasini AN, Sommers JA, Mason AC, Voloshin ON, Camerini-Otero RD, Wold MS, Brosh RM., Jr. FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by replication protein A to unwind the damaged DNA substrate in a strand-specific manner. J. Biol. Chem. 2009;284:18458–18470. doi: 10.1074/jbc.M109.012229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Popuri V, Bachrati CZ, Muzzolini L, Mosedale G, Costantini S, Giacomini E, Hickson ID, Vindigni A. The Human RecQ helicases, BLM and RECQ1, display distinct DNA substrate specificities. J. Biol. Chem. 2008;283:17766–17776. doi: 10.1074/jbc.M709749200. [DOI] [PubMed] [Google Scholar]
- 72.Dovrat D, Stodola JL, Burgers PMJ, Aharoni A. Sequential switching of binding partners on PCNA during in vitro Okazaki fragment maturation. Proc. Natl. Acad. Sci. U.S.A. 2014;111:14118–14123. doi: 10.1073/pnas.1321349111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Moldovan G-L, Pfander B, Jentsch S. PCNA, the Maestro of the Replication Fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 74.Croteau DL, Popuri V, Opresko PL, Bohr VA. Human RecQ helicases in DNA repair, recombination, and replication. Annu. Rev. Biochem. 2014;83:519–552. doi: 10.1146/annurev-biochem-060713-035428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bachrati CZ, Hickson ID. RecQ helicases: guardian angels of the DNA replication fork. Chromosoma. 2008;117:219–233. doi: 10.1007/s00412-007-0142-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.