Abstract
Objective
We present a family with a mitochondrial DNA 3243A>G mutation resulting in MELAS, of which some members have hearing loss where a novel mutation in the P2RX2 gene was identified.
Methods
One hundred ninety-four (194) Japanese subjects from unrelated families were enrolled in the study. Targeted genomic enrichment and massively parallel sequencing of all known non-syndromic hearing loss genes were performed to identify the genetic causes of hearing loss.
Results
A novel mutation in the P2RX2 gene, that corresponded to c.601G>A (p.Asp201Tyr) was identified. Two patients carried the mutation, and had severe SNHL, while other members with MELAS (who did not carry the P2RX2 mutation) had normal hearing.
Conclusion
This is the first case report of a diagnosis of hearing loss caused by P2RX2 mutation in patients with MELAS. A potential explanation is that decreasing ATP production due to MELAS with mitochondrial 3243A>G mutation might suppress activation of P2X2 receptors. We also suggest that hearing loss caused by the P2RX2 mutation might be influenced by the decrease in ATP production due to MELAS, and that nuclear genetic factors may play a modifying role in mitochondrial dysfunction.
Keywords: Hearing loss, genetics, P2X2, MELAS, massively parallel sequencing
INTRODUCTION
Hearing loss affects over 300 million people worldwide (WHO data, http://www.who.int/pdb/deafness/estimates/en/) and is the most common sensory deficit. Genetic factors account for at least 50% of childhood sensorineural hearing loss (SNHL). The majority of genetic hearing loss was autosomal recessive (AR) inherited: about 75%, autosomal dominant (AD): about 20%, and X-linked was estimated to be 1–5 % of genetic causes.1 Genetic SNHL is mainly categorized into two forms, approximately 70% of non-syndromic SNHL, and 30% of syndromic SNHL accompanied by other specific manifestations.1
Among mitochondrial mutations, there have been many manifestations recognized as mitochondrial diseases involving various organs. SNHL is one of the most common manifestations in patients with mitochondrial diseases, and several mutations have been found to be maternally inherited SNHL2. A 3243A>G mutation in the mitochondrial DNA is associated with maternally inherited diabetes combined with mitochondrial myopathy, encephalopathy, lactic acidosis and stroke-like episodes (MELAS), and which also frequently involves SNHL.3
With regard to non-syndromic hearing loss, most of these cases are affected with severe and congenital prelingual deafness with AR inheritance. Meanwhile, AD SNHL is represented by a mostly late-onset, mild to moderate and progressive hearing loss with distinctive phenotypical audiological features correlating to causative genes.4 32 loci and 31 genes have been implicated in autosomal dominant SNHL (DFNA (Hereditary Hearing Loss Homepage, http://hereditaryhearingloss.org/)). DFNA 41 harbors the P2RX2 gene that encodes the P2X2 receptor expressed in the cochlear sensory epithelium and the spiral ganglion neurons.5–7 It comprises of a channel gated by extracellular ATP.8, 9 The P2RX2 gene has recently been identified as a cause of late-onset and progressive SNHL in two Chinese families and one Italian family.10, 11
Concerning modes of inheritance, it is difficult to distinguish between mitochondrial maternal inheritance and AD inheritance. Furthermore, some patients with mitochondrial diseases present variable symptoms, including different levels of hearing loss, as clinical expression may be altered by heteroplasmy. AD hearing loss may also have different levels at different ages due to its progressive nature. It can be difficult to recognize what gene would be a candidate, including mitochondrial gene mutation, and move on to the analysis using conventional DNA sequencing based on PCR. Recent advances in targeted genomic enrichment with massively parallel sequencing (TGE+MPS) have made the sequencing of all known causative genes simultaneously possible.12, 13
Here, we describe a family with a mitochondrial DNA 3243A>G mutation resulting in MELAS, of which some members have hearing loss where we identified a novel mutation in the P2RX2 gene. This is the first report of a diagnosis of hearing loss caused by P2RX2 in patients with MELAS and highlights the importance of comprehensive genetic testing for concomitant genomic and mitochondrial DNA mutations.
SUBJECTS and METHODS
Subjects
One hundred ninety-four (194) Japanese subjects (114 females) from unrelated and non-consanguineous families were ascertained through 33 otolaryngology clinics in 28 prefectures across Japan. All subjects had presumed non-syndromic SNHL. For each proband, informed consent was obtained to participate in this study, which was approved by the human subjects ethical committee associated with each clinic.
Clinical information and blood samples were obtained for each proband and for all consenting affected and unaffected relatives.
Targeted Genomic Enrichment and Massively Parallel Sequencing
Genomic DNA was assessed for quality by gel electrophoresis and spectrophotometry (Nanodrop 1000; Thermo Fisher Scientific, Waltham, MA; 260/280 ratio of 1.8–2.2) and quantity by fluorometry (Qubit 2.0 Fluorometer; Life Technologies, Carlsbad, CA). TGE of all exons of all genes implicated in non-syndromic SNHL, including non-syndromic SNHL mimics, was completed as described, targeting 89 genes as part of the OtoSCOPE® v5 platform. Libraries were prepared using a modification of the solution-based Agilent SureSelect target enrichment system (Agilent Technologies, Santa Clara, CA).14 Of the 198 samples, 58 samples were processed manually; the remainder was prepared robotically using the Sciclone NGS Workstation.
In brief, 3μg gDNA was randomly fragmented to an average size of 250 bp (Covaris Acoustic Solubilizer; Covaris Inc., Woburn, MA), fragment ends were repaired, A-tails were added, and sequencing adaptors were ligated before the first amplification. Solid phase reverse immobilization purifications were performed between each enzymatic reaction. Hybridization and capture with RNA baits was followed by a second amplification before pooling for sequencing. Minimal amplification was used – typically 8 cycles for the pre-hybridization PCR (range 8–10 cycles) using NEB Phusion HF Master Mix (New England BioLabs Inc, Ipswich, MA) and 14 cycles for the post-hybridization PCR (range 12–16 cycles) using Agilent Herculase II Fusion DNA Polymerase. All samples were barcoded and multiplexed before sequencing on either an Illumina MiSeq or HiSeq (Illumina Inc, San Diego, CA) in pools of 4–6 or 48, respectively, using 100-bp paired-end reads.
Bioinformatics Analysis
Data were analyzed as described using a local installation of the open-source Galaxy software (http://galaxyproject.org) and the following open-source tools: BWA 15 for read mapping, Picard for duplicate removal, GATK 16 for local re-alignment and variant calling and NGSRich 17 for enrichment statistics.13 We reported and annotated variants with custom software.
Variant Confirmation
All pathogenic variants were confirmed by Sanger sequencing and segregation analysis using exon-specific custom primers.
RESULTS
We indentified one family that had a causative mutation in P2RX2 in the cohort of this study (194 hearing loss patients).
Case Details
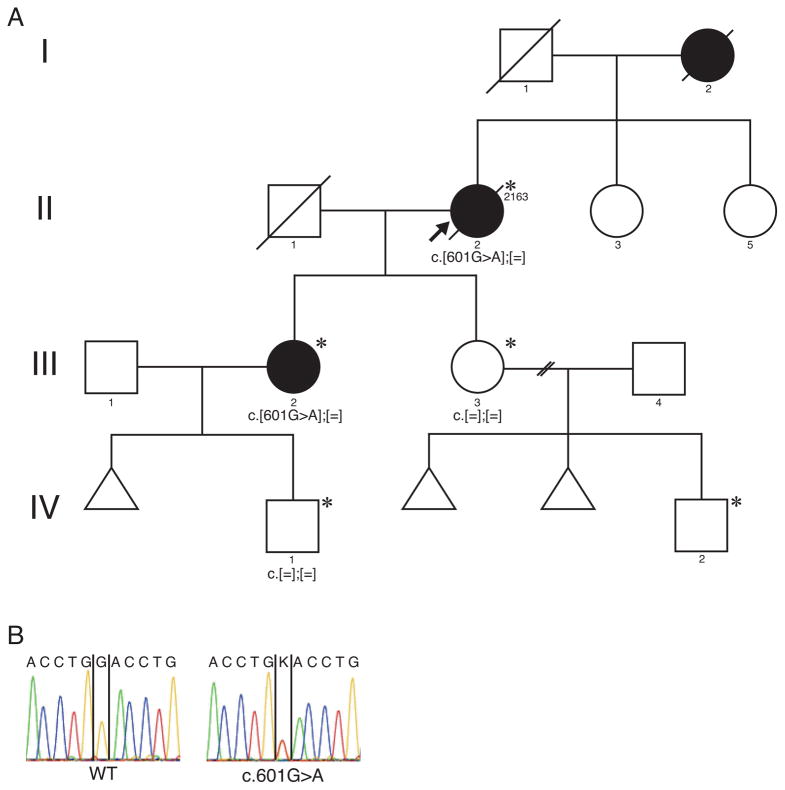
The proband (ID: 2163) is a 46-year-old female in a Japanese family spanning four generations (Figure 1). She was aware of hearing loss around the age of 12, and her hearing loss progressed gradually. She was diagnosed with diabetes mellitus (DM) and started insulin therapy. Furthermore, hypertrophic cardiomyopathy (HCM) was also pointed out at the age of 32. At the age of 46, she visited the Department of Otolaryngology at Shinshu University Hospital for a hearing examination. An audiogram showed severe SNHL by this time. She had a positive family history of DM, which was harbored in her mother and two daughters. Also her elder daughter (III-2) had bilateral hearing loss. At the age of 50, she and her two daughters were referred to the Department of Internal Medicine at Shinshu University Hospital for adequate control of their DM. A mitochondrial DNA 3243A>G mutation was identified at this time, and they were diagnosed with MELAS. The mother of the proband also suffered from DM and severe hearing loss, and died of cerebral embolism at age 68. The proband’s daughter (III-2) had an unstable status of DM, HCM, mild mental retardation and bilateral progressive SNHL. The other daughter (III-3) had the same manifestations except SNHL, and she had completely normal hearing over time. They were overweight and short stature. Unfortunately, the proband died of cardiac failure associated with HCM at the age of 57. Her grandson (IV-2) was also suspected to have MELAS, however, hearing assessment using auditory brainstem response (ABR) exhibited normal hearing bilaterally. More recently, her other grandson (IV-1) the son of (III-2) was identified as having a mitochondrial 3243A>G mutation, which was suspected MELAS due to fatigue and mild developmental delay, and he had normal hearing at the age of 8.
Figure 1.
Pedigree of Patient ID 2163. (A) Pedigree showed maternal or autosomal dominant inheritance. Target genome enrichment and massively parallel sequence was carried out for the patient II-2. (B) The electropherogram of the mutation in the P2RX2 gene. Asterisk (*) indicates individuals affected with MELAS.
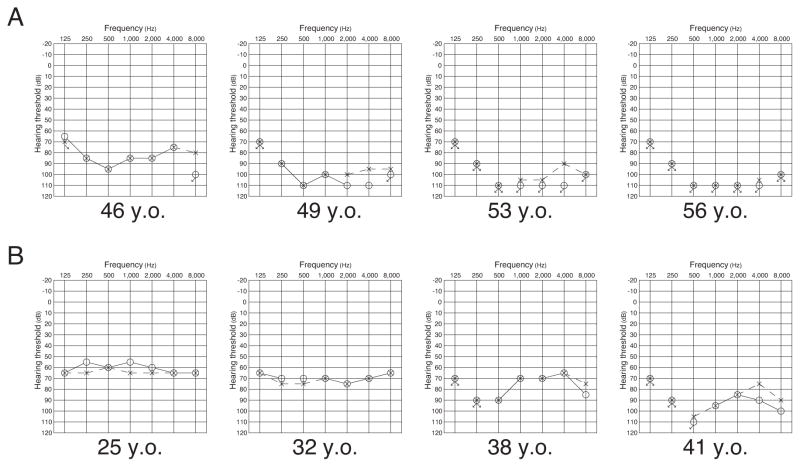
With respect to hearing loss status, the proband’s hearing loss rapidly deteriorated during three years (46 to 49 years old) and became to be undetectable. Her daughter’s (III-2) hearing loss also progressed over a period of 16 years (25 to 41 yeas old). Pedigree and serial audiograms are shown in Figure 1 and 2. Manifestations of each family member are shown in Table 1.
Figure 2.
Serial audiograms of affected patients II-2 and III-2. (A) Audiograms of the patient (II-2) over a period of 10 years. Hearing loss rapidly deteriorated during the period from 46 to 49 years old. (B) Audiograms of the patient (III-2) over a period of 16 years. Hearing loss was relatively stable around age of 30.
Table 1.
Summary of clinical findings of individuals with MELAS.
| Subject | Gender | Hearing | Diabetes mellitus | Cardiac disease | Mental retardation | Heteroplasmy of 3243AG |
|---|---|---|---|---|---|---|
| I-2 | F | Severe HL | Yes | na | na | na |
| II-2 | F | Severe HL | Yes | HCM | na | 1% |
| III-2 | F | Severe HL | Yes | HCM | Yes | 25% |
| III-3 | F | Normal | Yes | A-V block | Yes | 2% |
| IV-1 | M | Normal | No | No | Yes | 25% |
| IV-2 | M | Normal (ABR) | No | No | Yes | na |
HL: hearing loss, HCM: hypertrophic cardiomyopathy, na: not applicable
Mutation analysis
We performed comprehensive genetic testing using targeted genomic enrichment and massively parallel sequencing of all known non-syndromic hearing loss genes as well as non-syndromic mimic genes, as described previously.13 We identified a novel missense mutation in the P2RX2 gene, that corresponded to c.601G>A (p.Asp201Tyr, NM_012226). We employed in-silico pathogenicity prediction algorithms: PhyloP, SIFT, PolyPhen2, LRT, Mutation Taster, GERP, and all scores indicated “damaging” or “disease causing”. We also performed Sanger sequencing for a family segregation study, and confirmation of the variant in the proband. As shown in Figure 1, the Sanger sequencing results revealed that the proband and her elder daughter (III-2) had the mutation, although her younger daughter (III-3) did not. The P2RX2 mutation (p.Asp201Tyr) segregated with only the patients who had hearing loss in the family.
DISCUSSION
In this report, we identified a novel mutation in the P2RX2 gene in Japanese hearing loss patients. The P2RX2 gene is one of the latest identified as a cause of SNHL. There are only two previous reports on three families including their phenotypes of P2RX2 mutations. It seems extremely rare as there have only been two reported mutations: c.178G>T (p.Val60Leu) and c.1057G>C (p.Gly353Arg), from China and Italy respectively.10, 11
The molecular basis of the P2RX2 gene function, encodes the P2X2 receptor, which plays an essential role in the cochlea as an ATP-gated ion channel receptor through ATP-mediated regulation.7 P2X2 receptors are expressed in the epithelial cells surrounding the cochlear partition of the endolymphatic compartment which includes the organ of Corti. Sustained noise exposure induces an up-regulation of P2X2 transcripts in the surface of cells.18
ATP is thought to have a neurotransmission effect at the synapse of the hair cells, and contributes to regulation of the endocochlear potential. ATP is released into the endolymphatic compartment in which P2X2 receptors are expressed during the noise stress, and activates the P2X2 receptors, producing cation shunt conductance which reduces the endocochlear potential. This mechanism is beneficial for protective effects of the cochlea by reducing sound transduction and hair cell sensitivity during noise exposure.19, 20 Based on these, Yan et al. clearly reported that exacerbation of hearing loss occurred among families with p.Val60Leu heterozygous mutations in the P2RX2 gene, and was severer in the subjects having experience with noise exposure.10
ATP is derived from mitochondria, and mitochondrial disease is attributed to dysfunctions in the oxidative phosphorylation of the cell resulting in markedly reducing ATP production. Mit.3243A>G mutation generally causes MELAS, affects multiple biological aspects including ATP loss, increase of lactate and reactive oxygen species, and leads to systemic defect among various organs. In this family member presented with DM, short stature, stroke episodes, weakness, lactic acidosis and intellectual disability, as manifestations of MELAS. Nevertheless, there were only two patients in this family who were affected with severe progressive SNHL and they had relatively low heteroplasmy of Mit.3243A>G mutation (Table 1). It is noteworthy that only these two patients carried the mutation in the P2RX2 gene, and had significantly severe SNHL, while other members with MELAS who did not carry the P2RX2 mutation had normal hearing.
With regard to hearing levels and its progression, two affected patients (II-2, III-2) exhibited more severe hearing loss. Besides, hearing loss deteriorated more rapidly as compared with the result of progression of P2RX2 hearing loss, reported by Yan et al. (Figure 3). It is speculated that that hearing loss caused by the P2RX2 mutation might be influenced by the decrease in ATP production due to MELAS. Scuderi et al. have reported a similar case that had a nuclear gene DCX mutation coexisting with MELAS, a Mit.3243A>G mutation, and the manifestations of the DCX mutation were exacerbated by the mitochondrial dysfunction causing the MELAS.21 We also suggest that nuclear genetic factors may play a modifying role in the mitochondrial dysfunction.
Figure 3.
Progression of hearing loss in patients II-2 and III-2. Hearing loss rapidly deteriorated around age of 40–50.
Previously, genetic testing to identify mitochondrial mutation was carried out based on the clinical findings. If the corresponding mutation was identified, further testing was not deemed necessary. As such, even if patients with mitochondrial DNA mutation did not express different types of hearing loss, it could be interpreted as a variability of mitochondrial disease. In this family, the coexistence of the P2RX2 mutation and the Mit.3243A>G mutation might occur accidentally. TGE and MPS allowed us to identify the disease causing mutations, based upon all known hearing loss genes screened. This study supports the use of comprehensive genetic diagnosis for SNHL cases to provide the highest chance of diagnostic success. In addition, further studies are necessary for cases that identify multiple pathogenic mutations and investigations of these gene-gene interactions may have a potential to influence its phenotype.
Acknowledgments
This work was supported by NIDCD RO1s DC003544, DC002842 and DC012049 to RJHS. We thank Mr. David Callaghan for help in preparing the manuscript.
Footnotes
DECLARATION OF CONFLICTING INTERESTS
All authors have declared no competing financial interests.
References
- 1.Smith RJ, Bale JF, Jr, White KR. Sensorineural hearing loss in children. Lancet. 2005;365(9462):879–890. doi: 10.1016/S0140-6736(05)71047-3. [DOI] [PubMed] [Google Scholar]
- 2.Jacobs HT, Hutchin TP, Kappi T, et al. Mitochondrial DNA mutations in patients with postlingual, nonsyndromic hearing impairment. Eur J Hum Genet. 2005;13(1):26–33. doi: 10.1038/sj.ejhg.5201250. [DOI] [PubMed] [Google Scholar]
- 3.van den Ouweland JM, Lemkes HH, Ruitenbeek W, et al. Mutation in mitochondrial tRNA(Leu)(UUR) gene in a large pedigree with maternally transmitted type II diabetes mellitus and deafness. Nat Genet. 1992;1(5):368–371. doi: 10.1038/ng0892-368. [DOI] [PubMed] [Google Scholar]
- 4.Petersen MB. Non-syndromic autosomal-dominant deafness. Clin Genet. 2002;62(1):1–13. doi: 10.1034/j.1399-0004.2002.620101.x. [DOI] [PubMed] [Google Scholar]
- 5.Blanton SH, Liang CY, Cai MW, et al. A novel locus for autosomal dominant non-syndromic deafness (DFNA41) maps to chromosome 12q24-qter. J Med Genet. 2002;39(8):567–570. doi: 10.1136/jmg.39.8.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan D, Ouyang XM, Zhu X, Du LL, Chen ZY, Liu XZ. Refinement of the DFNA41 locus and candidate genes analysis. J Hum Genet. 2005;50(10):516–522. doi: 10.1007/s10038-005-0286-0. [DOI] [PubMed] [Google Scholar]
- 7.Housley GD, Kanjhan R, Raybould NP, et al. Expression of the P2X(2) receptor subunit of the ATP-gated ion channel in the cochlea: implications for sound transduction and auditory neurotransmission. J Neurosci. 1999;19(19):8377–8388. doi: 10.1523/JNEUROSCI.19-19-08377.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao HB, Yu N, Fleming CR. Gap junctional hemichannel-mediated ATP release and hearing controls in the inner ear. Proc Natl Acad Sci U S A. 2005;102(51):18724–18729. doi: 10.1073/pnas.0506481102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu Y, Zhao HB. ATP-mediated potassium recycling in the cochlear supporting cells. Purinergic Signal. 2010;6(2):221–229. doi: 10.1007/s11302-010-9184-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan D, Zhu Y, Walsh T, et al. Mutation of the ATP-gated P2X(2) receptor leads to progressive hearing loss and increased susceptibility to noise. Proc Natl Acad Sci U S A. 2013;110(6):2228–2233. doi: 10.1073/pnas.1222285110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faletra F, Girotto G, D’Adamo AP, Vozzi D, Morgan A, Gasparini P. A novel P2RX2 mutation in an Italian family affected by autosomal dominant nonsyndromic hearing loss. Gene. 2014;534(2):236–239. doi: 10.1016/j.gene.2013.10.052. [DOI] [PubMed] [Google Scholar]
- 12.Shearer AE, DeLuca AP, Hildebrand MS, et al. Comprehensive genetic testing for hereditary hearing loss using massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107(49):21104–21109. doi: 10.1073/pnas.1012989107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shearer AE, Black-Ziegelbein EA, Hildebrand MS, et al. Advancing genetic testing for deafness with genomic technology. J Med Genet. 2013;50(9):627–634. doi: 10.1136/jmedgenet-2013-101749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shearer AE, Hildebrand MS, Smith RJ. Solution-based targeted genomic enrichment for precious DNA samples. BMC Biotechnol. 2012;12(1):20. doi: 10.1186/1472-6750-12-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frommolt P, Abdallah AT, Altmuller J, et al. Assessing the enrichment performance in targeted resequencing experiments. Hum Mutat. 2012;33(4):635–641. doi: 10.1002/humu.22036. [DOI] [PubMed] [Google Scholar]
- 18.Housley GD, Bringmann A, Reichenbach A. Purinergic signaling in special senses. Trends Neurosci. 2009;32(3):128–141. doi: 10.1016/j.tins.2009.01.001. [DOI] [PubMed] [Google Scholar]
- 19.Telang RS, Paramananthasivam V, Vlajkovic SM, Munoz DJ, Housley GD, Thorne PR. Reduced P2x(2) receptor-mediated regulation of endocochlear potential in the ageing mouse cochlea. Purinergic Signal. 2010;6(2):263–272. doi: 10.1007/s11302-010-9195-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Housley GD, Morton-Jones R, Vlajkovic SM, et al. ATP-gated ion channels mediate adaptation to elevated sound levels. Proc Natl Acad Sci U S A. 2013;110(18):7494–7499. doi: 10.1073/pnas.1222295110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scuderi C, Borgione E, Castello F, et al. Coexistence of mitochondrial and nuclear DNA mutations in a woman with mitochondrial encephalomyopathy and double cortex. Mitochondrion. 2010;10(5):548–554. doi: 10.1016/j.mito.2010.04.004. [DOI] [PubMed] [Google Scholar]