Abstract
Nucleic acid-based logic devices were first introduced in 1994. Since then, science has seen the emergence of new logic systems for mimicking mathematical functions, diagnosing disease and even imitating biological systems. The unique features of nucleic acids, such as facile and high-throughput synthesis, Watson-Crick complementary base pairing, and predictable structures, together with the aid of programming design, have led to the widespread applications of nucleic acids (NA) for logic gating and computing in biotechnology and biomedicine. In this feature article, the development of in vitro NA logic systems will be discussed, as well as the expansion of such systems using various input molecules for potential cellular, or even in vivo, applications.
1. Introduction
Based on their ever-increasing computing and storage capability on a miniaturized scale, silicon semiconductor-based intelligent computing systems have impacted every aspect of daily life. At the same time, however, the capabilities of traditional silicon chips are becoming increasingly limited.1 This has only prompted researchers to explore alternatives to semiconductor-based computational systems, even at the molecular level. Molecules can be rationally designed, synthesized, and further integrated into Boolean operations, providing unprecedented potential for developing the basic components of molecular computing devices. More significantly, molecular assemblies that function as different modules, although not yet able to rival silicon-based computing devices, can, to some extent, emulate or mimic digital logic circuits.2
Several unique aspects characterize the design of molecular logic systems. First, logic device input and subsequent output to the next device must be considered. Second, a semiconductor device typically relies on electrical input and output signals transported through mechanical wires on the silicon chips. In contrast, the input/output of a molecular logic system is not limited to electrical circuitry. For example, the diffusion of molecules in solution can be used as the input and light emission as the output of a molecular logic device. Third, molecular computational devices do not have to be as durable as their semiconductor counterparts, especially when used in live biosystems that only require active function for hours to days.2 Finally, molecular computational devices, albeit relatively simple, can still be utilized in various fields, ranging from in vitro sensing and diagnosis to cellular drug delivery, and even gene expression regulation in vivo.
Nucleic acids (NA), as the significant carriers of genetic information, have recently displayed unprecedented potential in practical, high-capacity and low-maintenance digital information storage because of their predictable structures, high-throughput synthesis and sequencing techniques.3, 4 NA-based logic gate operation was first reported by Adleman and Lipton to solve the directed Hamiltonian path problem and “SAT” question in computer science with single-stranded DNA sequences and enzymes.5, 6 After that, researchers sought to design and create various DNA/RNA-based logic systems for feedback and cascading, mimicking mathematical function, diagnosing disease, and even imitating the neural network and memory system.7–14 Although NA-based logic and computing is still in its infancy, these attempts to substitute NA-based logic devices for conventional silicon chips and utilize them for cellular or in vivo applications have become the driving force behind a new generation of molecular logic systems.
This feature article first focuses on the in vitro design of NA-based logic and computational systems, followed by a review of recent progress in sensing and diagnosis with initial inputs of ions, small molecules and proteins. Finally, biological applications with NA-based logic platforms will be surveyed.
2. Rational design of nucleic acid molecular logic system
The basic construction modules of NA-based logic devices include input (DNA/RNA, ions, small molecules or proteins), computational “hardware” (double-stranded DNA/RNA and (DNA/RNA, or other readable signals). Thus, while it is easy to envision single-stranded DNA as input to double-stranded DNA computing “biohardware,” the output depends on triggering a readable signal, and in an NA-based logic network, this is typically achieved by a toehold-mediated displacement reaction.15 More specifically, the ssDNA input hybridizes to a specific domain (toehold) of the dsDNA, initiating the displacement process and releasing another ssDNA sequence as the output. Three basic steps are involved in this procedure: toehold binding, branch migration and strand dissociation (see Figure 1). It is worth noting that the length of toehold in a logic circuit quantitatively determines the rate constant of a strand-displacement reaction, which can vary by six orders of magnitude.16 Using this basic principle, Seelig et al. reported the implementation of DNA-based digital logic circuits to perform a series of functions, such as AND, OR and NOT gates.8 Moreover, this enthalpy-driven displacement reaction was also utilized in signal amplification, feedback, cascading and pattern transformation.17–21
Fig. 1.
Scheme illustrating the biomechanics of toehold-mediated strand displacement reaction (reprinted by permission from reference [16], Copyright 2011, Macmillan Publishers Ltd)
Since nucleic acids can be easily modified in the laboratory, either individual nucleotides or the entire sequence, unique properties and applications can be achieved. In addition, a double-stranded DNA (dsDNA) chain can provide long-range hole transport, which is caused by the ordered π-electron system of the bases, and different DNA pairs having distinct efficiency expression results. To build AND/OR logic gates, it is easy to modify the DNA bases and sequences with specific pairing, which can apparently affect the long-range hole transport efficiency. Thus, the Boolean results can be expressed by comparing the ratio of the measured long-range hole transport efficiencies of the normal and modified base pairs. Okamoto et al. reported a DNA logic gate system which employs MDA (methoxybenzodeazaadenine) paired to T base, instead of the natural A base, to deliver reasonable hole-transport efficiency (Gb/Ga = 0.39) provided by the MDA-T base pair.22 In this method, the GGG sites are used as an effective hole trap, and the result of hole transport is determined by the oxidative strand cleavage at GGG sites by PAGE (polyacrylamide gel electrophoresis) analysis upon hot piperidine treatment of reaction samples. Here, Gb and Ga represent oxidative strand cleavage at the distal GGG and the proximal GGG, respectively. The input strands also include photosensitizer, cyanobenzophenone-substituted uridine (S), for the hole injection to DNA. However, if MDA is replaced by C base, hole-transport efficiency is reduced significantly (Gb/Ga < 0.01). Therefore, the change of hole-transport efficiency can be used to distinguish the different logic gates. As shown in Figure 2, two logic gates are separated by two T/A base pairs, and they work independently such that only the input sequence with two T can provide a strong output signal, which provides the AND output. On the other hand, while the OR gate is designed according to the same principle, the logic gate is a combination of three different sequences such that the input can hybridize with any one of them to provide an output signal. By using parallel G-quadruplex structure to specifically enhance the fluorescence signal,23 Zhu et al. designed a logic circuit based on the three-way DNA junction driven strand displacement reaction, in which the output signal was enhanced by the G-quadruplex. Only when Stand R (input strand with toehold end), ABA (ATP-binding aptamer), and S (input strand which can draw G1 and G2 together to form G-quadruplex), are all input will the output fluorescence signal be significantly enhanced.24 Livshits et al. also employed guanine-quadruplex DNA molecules adsorbed on a mica substrate to develop DNA-based programmable circuits.25 Thus, while it is easy to envision single-stranded DNA as input to double-stranded DNA computing “hardware,” the output depends on triggering a readable signal, and in a NA-based logic network, this is achieved by a toehold-mediated displacement reaction.
Fig. 2.
Scheme of the DNA logic gate. Green line: logic gate strand. Orange line: Input strand; Xn: Logic bases; Yn: Input pyrimidines; S: Photosensitizer (reprinted by permission from reference [22], Copyright 2004, American Chemical Society).
The photon is one of the most useful and popular tools used in the construction of logic gates since light can be considered as the output signal. Saghatelian et al. used DNA-based photonic logic gates to provide AND, NAND and INHIBIT Boolean operations (Figure 3A).26 NAND gate (Negated AND or NOT AND) is a logic gate which produces an output that is false only if all its inputs are true; thus its output is complement to that of the AND gate. A LOW (0) output results only if both the inputs to the gate are HIGH (1); if one or both inputs are LOW (0), a HIGH (1) output results. It is made using transistors. The NAND gate is significant because any boolean function can be implemented by using a combination of NAND gates. This property is called functional completeness. Four possible input DNA strands (0,0; 1,0; 0,1; 1,1) are presented in the “AND” and “NAND” gates, in which the fluorescence signals are recognized by DNA strands labeled with different fluorophores based on fluorescence resonance energy transfer (FRET).26 For the AND gate, only having “1” at both inputs (1,1) can induce the FRET signal by two fluorophores on each oligonucleotide. This provides the maximum fluorescence intensity, in contrast to the low fluorescence signals for the other three input types. For the NAND gate, a (1,1) input results in a low fluorescence signal, while the other combinations provide strong signals, simply achieved by changing the fluorophore dyes and switching the excitation wavelength. Prokup et al. designed a photochemically controlled AND gate with the incorporation of caged thymidine nucleotides.27 As shown in Figure 3B, the input signals are lights (I1 and I2), and the output signal is the fluorescence signal from the TAMRA dye after the computing process. The AND output signal can be expressed only while applying two steps of the input light signals, because the decaging by A4 and excitation by I2 can be achieved only by introducing the lights, then the output signal can be detected.
Fig. 3.
(A) Photon-based DNA (top) AND and NAND (bottom) logic gates (reprinted by permission from reference [26], Copyright 2002, American Chemical Society). (B) Schematic of photocontrollable DNA computation (reprinted by permission from reference [27], Copyright 2012, American Chemical Society).
Nucleic acid hairpin-shaped structure has also been demonstrated as an effective tool in building molecular logic gates and automatons. For instance, the high selectivity of molecular beacons (MBs)28–30 is attributed to the specific opening of the hairpin loop by binding their complementary target sequences, resulting in the restoration of fluorescence at the end of the molecular beacon stem. The inactivated hairpin structure can quench fluorescence signal by FRET from the fluorophore to the quencher, while the fluorescence signal is restored when the hairpin structure is opened by the complementary sequence. Based on this working principle and specifically designed molecular beacons, Park and co-workers have developed simple and universal DNA logic gates.31 The input strands can open the loop part of the MB, and logic gate expression is achieved by specific recognition of various input sequences by the loop of the MB. Fluorescence intensity change then provides the output signal. Either one of the input sequences can open the MB loop to initiate an “OR” gate. However, if the opening process must be operated in the presence of both input sequences, it is an “AND” gate. Furthermore, multilevel circuits can be built by employing a combination of multiplex gates (Figure 4A). Genot et al. developed reversible logic circuits based on the AND logic gate by using the DNA hairpin (Hp).32 Input sequence 1 (I1) can open the domain 1 (d1); similarly, input 2 (I2) can open domain 2 (d2). The I2 is designed so that it is difficult to open d2; as a result, the AND gate can be opened only by using I1 and I2 sequentially. The final output signal, which can be measured by the reaction between I1 and I2 sequences with MB reporter, yields fluorescence signal as output. Furthermore, the logic circuit can be designed using more input sequences and several domains containing DNA hairpin structures.
Fig. 4.
DNA logic gates based on hairpin (Hp)-shaped structures. (A) Gate wiring in complex logic circuits based on molecular beacon probes (reprinted by permission from reference [31], reprinted with permission from WILEY-VCH Verlag GmbH & Co). (B) “YES” sensor gate with beacon module. i1: Input; BH2: Quencher; T: TAMRA. (C) Dual-input AND gate. (D) Tic-tac-toe game operated by deoxyribozyme-based automaton (reprinted by permission from reference [34], Copyright 2014, American Chemical Society).
Stojanovis et al. proposed the use of multiple catalytic nucleic acid hairpin structures (deoxyribozyme) in molecular computing design. As shown in Figure 4B, the YES sensor gate (The NOT gate is also called an inverting buffer, which stresses its ability to amplify. If we hook two NOT gates together we will now have a YES gate or non-inverting buffer) consists of an inactive deoxyribozyme module (ST) and another attached beacon module.33, 34 The output signal can be realized when the ST region is cleaved based on the following steps. First, the input sequence opens the loop of the beacon module and releases its stem. Second, the activated stem allows the ST module to form the active state by hybridizing with the substrate labeled with a fluorophore (TAMRA) and quencher (BH2). Finally, cleavage, which occurs in the active ST module, results in the restoration of fluorescence output signal. Because of the facile modification of the beacon module incorporated in the ST region, multiple input beacon modules can be precisely used with deoxyribozyme for different types of complex gates, such as “YES”, “AND” and “ANDAND” (Figure 4B and 4C).34–36 Furthermore, as shown in Figure 4D, the more complex game-playing automaton MAYA, was constructed and trained to play a symmetry-pruned game of tic-tac-toe using this strategy, which means that they can make the DNA strands “thinking” based on the logic gate they designed with the DNA moleculers.7, 34, 37, 38
3. Functional nucleic acid logic system involving metal ions and biomolecules
DNA not only forms a duplex structure, but it also interacts with ions, small molecules or proteins through a variety of mechanisms.39–43 Inspired by these properties, many researchers have designed logic gates by combining strand displacement with ion or molecule binding, thus largely expanding the scope of nucleic acid logic gate application. Moreover, some designs can not only detect the analyte, but also realize the control and manipulation of biological molecules in vitro with involvement of DNA logic gate.
3.1 NA logic systems mediated by metal ions and small molecules
Based on the interactions between DNA and metal ions, DNA logic gates can be mediated by the input of different ions that employ such mechanisms as mismatch base pairing by metal ions,44–47 ion-mediated catalytic deoxyribozyme48–52 and tetrad structure formation, such as G-quadruplexes and i-motifs.53–55 For example, mercury ions (Hg2+) can interact with thymine-thymine (T-T) mismatch in DNA double strands, thus increasing the stability of the duplex.41 A similar phenomenon also occurs between Ag+ ion and cytosine-cytosine (C-C) mismatch.56 Taking advantage of this property, Park et al. developed a logic gate in which the output is the PCR product of a double-stranded DNA.57 They deliberately put the T-T or C-C mismatches at the 3’ ends of both primers and templates, making elongation occur only when Hg2+ or Ag+ is present, respectively. They establish different gates systems using this property. Take AND gate as an example. T-T and C-C mismatches were deliberately inserted on the forward and reverse primers with the substrates, respectively (Figure 5A). With the mismatch at the end, the polymerase enzyme cannot work. Thus only with both the input ions (Ag+ and Hg2+) present can the 3’ end base pair with each other, which induce the polymerase reaction.
Fig. 5.
(A) AND gate for ion-induced pairing of mismatch bases utilizing PCR product as output. Only with both Ag+ and Hg2+ present can the PCR reaction occur (reprinted by permission from reference [57], Copyright 2010 WILEY-VCH Verlag GmbH & Co). (B) AND gate of electronic logic gate based on ion-mediated formation of oligonucleotide structural motifs. Only with Ag+ and Hg2+ presents can Faradaic current output produced. (reprinted by permission from reference [58], Copyright 2013 WILEY-VCH Verlag GmbH & Co).
While numerous techniques that have been developed for ion detection exploiting DNA logic gates, most of the output formats are fluorescence and colorimetric, which result in tedious management procedures, as well as difficulties in transferring the output signal into nonmolecular-based systems. Zhang et al tried to solve this problem by utilizing the change in Faradaic current as the output signal (Figure 5B). The T-/C-rich DNA strands on a modified gold electrode surface hybridize with T-/C-rich DNA strands with ferrocenecarboxylic acid (Fc) labeled at the end.58 For AND gate operation, T-T and C-C separately pair to form the duplex, but only with input of both Hg2+ and Ag+ ions. This brings Fc close to the surface of the gold electrode and produces a Faradaic current as the output. They also constructed NAND and NOR gates using similar strategies.
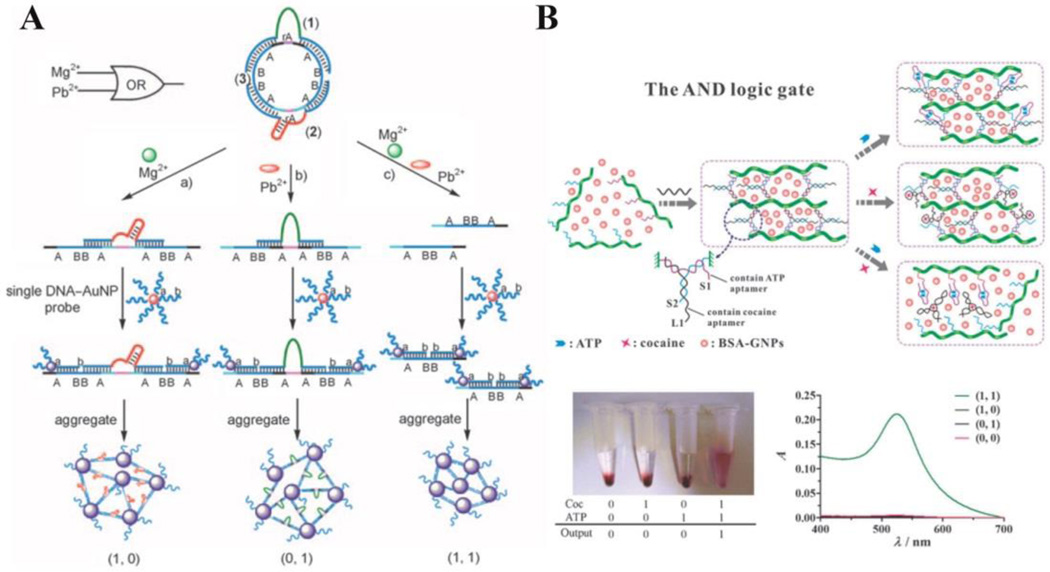
DNAzymes, which were first identified in 1994, are single DNA strands that show catalytic ability for certain chemical reactions.40 Often they need metal ions as cofactors.59 Taking advantage of this property, researchers have developed many strategies to detect ions. In addition, many DNA logic gate biosensing techniques have been based on DNAzymes, using ions as inputs and DNA/RNA strands produced by the catalytic reactions as reporters, which will produce fluorescence, electronic, colorimetric or other kinds of signals.48 Bi et al., for instance, developed a set of logic gates, including AND, OR, INHIBIT, XOR, XNOR, NOR, and NAND, with colorimetric output.60 Basically, they designed a series of supramolecular circular DNA strands containing certain patterns of repeated units, as well as the cleavage sites for DNAzymes. As shown in Figure 6A, by inputting the cofactor ions (Mg2+ and Pb2+), the two DNAzymes are activated, and the substrate is cleaved, resulting in overhung segments. When two hybridization overhangs are generated on one strand of DNA, gold nanoparticles modified with complementary oligonucleotides are brought close to each other upon hybridization. This aggregation induces a color change from red to purple as output. By rationally modifying the patterns of the circular DNA substrate, various logic gates can be realized.
Fig. 6.
(A) DNAzyme-based logic gates (AND, INHIBIT, and XOR) with specific design of substrate circular DNA. Cut of the substrate mediated by different ions induces nanoparticle aggregation, which result in colour change from red to purple. (reprinted by permission from reference [60], Copyright 2010 WILEY-VCH Verlag GmbH & Co). (B) AND logic gate of small molecule- (ATP and cocaine) mediated breakdown of aptamer-crosslinked hydrogel (reproduced from Ref. [71], with permission from The Royal Society of Chemistry).
Screened through an in vitro process called systematic evolution of ligands by exponential enrichment (SELEX), aptamers are single-stranded oligonucleotides which fold into unique secondary and tertiary structure with binding affinity to biological small molecules, proteins or even cells.39, 61–63 As an analog to antibodies, aptamers have many advantages, such as facile synthesis, good reproducibility, and thermal endurance. Most important, however, as DNA strands, aptamers can be incorporated into logic gate circuitry easily and effectively. Many logic gate systems have been designed utilizing the targets as inputs and binding-induced structural transformations of DNA strands as responses.24, 64–70 Yin et al., for example, developed a colorimetric logic gate system based on aptamer-crosslinked hydrogels.71 By modifying DNA molecules onto polymer chains, hybridization behavior with crosslinker strands induces hydrogel formation. Since the aptamer sequences of ATP and cocaine are incorporated into the DNA strands, the hydrogel dissociates upon target recognition. As output, the authors trapped BSA-modified gold nanoparticles in the hydrogel, and monitored particle release for detection. For the AND gate (Figure 6B), each strand partially hybridizes two others, forming a Y-shaped structure. Thus, one input can interrupt hybridization between two of them, but since the three strands remain linked together, no breakdown of the hydrogel occurs. For the OR gate, the crosslinker hybridizes with both DNA strands modified on polymer, while between the two strands there is no interaction. As such, either one of the molecules can decompose the hydrogel and release the gold nanoparticles.
3.2 Protein-mediated NA logic systems
Monitoring biological systems depends on the accurate detection of proteins. For that reason, many researchers have focused on making detection methods simple and effective including, for example, strip platforms,72 photoinduced electron transfer (PET),73 nanomaterials,74, 75 and electrochemiluminescent devices.76
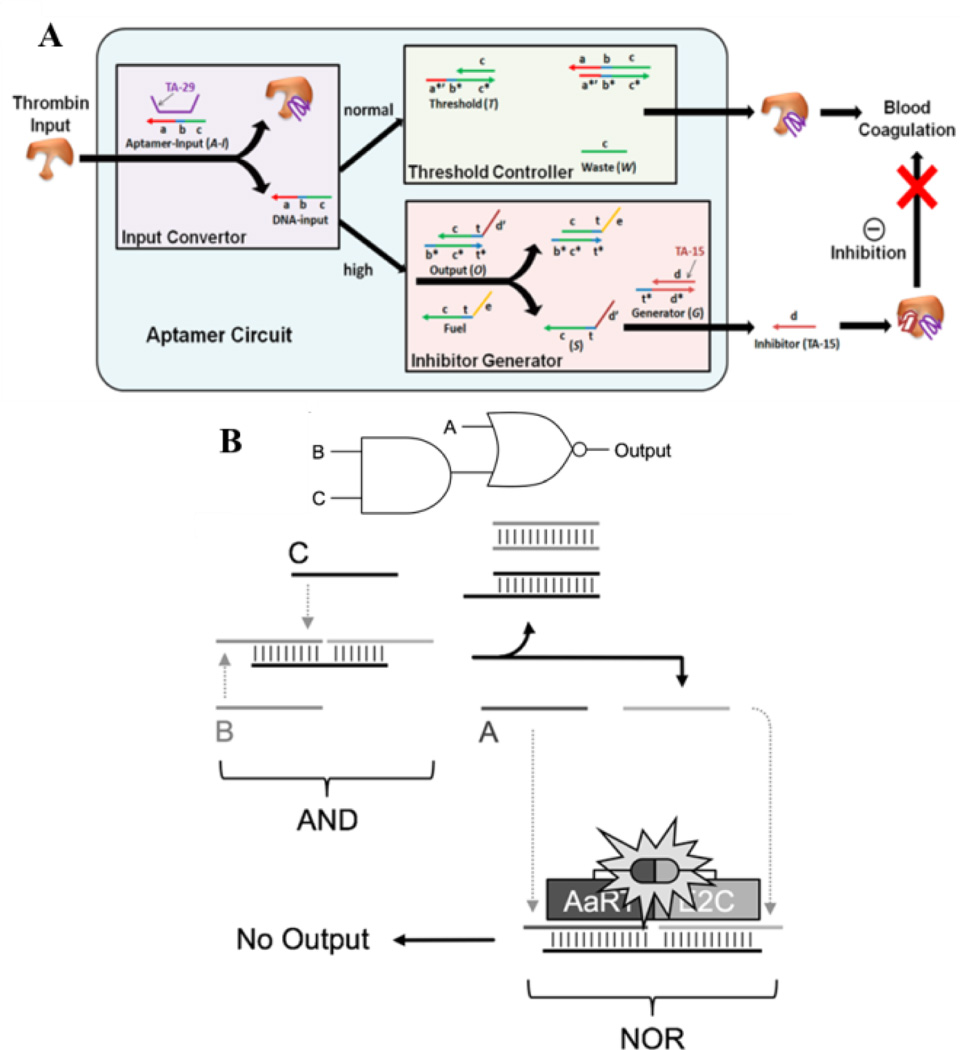
When compared with the most popular protein detection method, enzyme-linked immunosorbent assay (ELISA), which utilizes antibody and enzyme, aptamers need a less rigid environment. In addition, some aptamers can both recognize proteins and regulate protein functions upon recognition. For instance, thrombin aptamer TA-29 can target the protein without interference, while TA-15 that binds to the fibrinogen exosite of thrombin strongly inhibits blood coagulation.77 Taking this feature into consideration, Han et al. developed a logical molecular circuit that can realize programmable and autonomous manipulation of thrombin function via a set threshold.78 As shown in Figure 7A, the process includes three stages. In the Input Convertor, upon addition of thrombin, TA-29 binds to its target position and releases the partially complementary strand as DNA-input. In the normal input range, the process stops at the Threshold Controller step, and the DNA input hybridizes to one strand of the threshold duplex with the other strand as waste. However, if thrombin input exceeds the threshold, the Inhibitor Generator step is activated, and following strand displacement, inhibitor TA-15 is generated by recognizing thrombin, and blood coagulation is impeded.
Fig. 7.
(A) Logic circuit for thrombin detection and regulation. Three modules are involved: Input Convertor, Threshold Controller and Inhibitor Generator (reprinted by permission from reference [78], Copyright 2012, American Chemical Society). (B) Connection of AND and NOR logic gates using protein as output. The output strand of DNA gate become one of the input in NOR gate. (reprinted by permission from reference [79], Copyright 2014 WILEY-VCH Verlag GmbH & Co).
While previous methods have used proteins as inputs, Deiters et al. have recently engineered a set of DNA logic operations with proteins as outputs.79 Compared to typical outputs, such as oligonucleotides or fluorescent signals, direct control of protein activation enables the immediate triggering of enzyme functions. In this work, they exploited zinc-finger proteins AaRT and E2C, which are able to easily fuse with the split-protein component of luciferase, as well as bind to DNA with recognition ability. AaRT, which recognizes guanine-rich sequence, and E2C, which binds to adenine-rich group, are used as a proof-of-concept (Figure 7B). The two halves of the split-luciferase enzyme are separately modified with the two zinc finger proteins to generate luminescence readout. Integrated luciferase is formed and a signal produced only when the two zinc proteins are brought close together through binding to the DNA scaffold. Several gates, such as AND, OR and NOR, can be constructed. In addition to the single gates, more complex devices which integrate multiple logic gates can be fabricated. Figure 7B shows a combined AND/NOR gate. In this construct, the presence of both B and C strands activates the AND gate, and the output strand, as well as the A strand, acts as one of the inputs for the next NOR gate. Either input of the NOR gate releases the zinc protein binding strand from the scaffold, inducing luminescence quenching.
4. Nucleic acid logic systems for cancer theranostics
A key goal of NA-based logic systems is the ultimate translation of these devices for applications in vivo, for smart sensing, regulating, and even rewiring of biological systems.1 However, considering the complex environment of a biological system, a successful design in buffer system may have totally different results when applied in vivo, usually failing because of the unstable nature of nucleic acids, the lack of robustness of the designed hybridization reaction, or vulnerability of nucleic acids when exposed to enzymatic digestion.1 Scientists worldwide are trying to solve these problems. The effort includes those working on modifications of existing natural DNA, such as phosphorothioate replacing DNA,80 locked DNA (LNA),81, 82 2’-site-modified nucleic acids,83, 84 or enantiomers of natural DNA.85 Efforts to introduce artificial nucleotides into DNA will largely overcome the shortfalls of natural DNA and will endow nucleic acid logic systems with more building blocks and higher information density.86
Among all biological applications, cancer-related research undoubtedly spurs most attention. The images of building smart nucleic acid logic systems for cancer detection and therapy are indeed intriguing. Although this logic gate concept was proposed as early as in 1994, only recently has research in living cells advanced, and cancer-related research has followed.86, 87 Although reaching the goal of applying logic systems in vivo may still stretch far into the future, many excellent research results have already been published.
In 2007, the Benenson group constructed a molecular computing core implementing general Boolean logic using RNA interference (RNAi). In human kidney cells, this evaluator could directly evaluate any Boolean expression based on endogenous molecular inputs.88 In another work, this group built another RNAi-based logic circuit that could sense the expression levels of certain types of preinstalled endogenous microRNA (miRNA) and, as a consequence, classify cell type for a predetermined response. That is, whenever the expression level matches the default profile, the logic circuit correspondingly triggers a cellular response to induce apoptosis (Figure 8A).12 Deiters et al. reported the engineering of an AND gate to sense specific miRNA inputs in living cells with a fluorescence signal as output. This system could expand the potential of nucleic acid logic systems to monitor, image and respond to cell-specific markers.89 Very recently, Cai et al. designed a series of CRISPR-Cas9-based modular AND gate circuits that activate the output gene only if two promoter inputs are active in the tested cells. The designs exploit output genes, including luciferase reporter for bladder cancer cell detection and cellular functional genes to inhibit bladder cancer cell growth, and induce inducing apoptosis or decrease cell motility.90
Fig. 8.
(A) Scheme of integrated classifier for HeLa miRNA sensing and specific hBax protein regulation (from reference [12], reprinted with permission from AAAS). (B) Demonstration of automata assessing the existence of cell surface marker (from reference [91], reprinted by permission from Macmillan Publisher Ltd).
Besides successful work focusing on logic gate-based mRNA detection or gene regulation, other research has targeted cancer-related biomarkers. To achieve successful cancer detection and therapy, a biomarker makes it possible to distinguish cancer cells from normal cells or other types of cells and can largely enhance specificity and increase efficiency. As natural binding molecules, antibodies are the most useful binders in biomedicine today. Stojanovic et al. developed antibody-nucleic acid conjugates that can analyze cells by sensing surface biomarkers CD45 and CD20 (Figure 8B).91 These devices can detect the existence of two to three biomarkers in a logical pattern and give a corresponding fluorescence signal, thus avoiding the steps of transfecting oligonucleotides into cells. This capability largely extends the use of logic systems, thereby prompting operations directly on the surfaces of native cells.
As alternative binders, aptamers are excellent tools for incorporation with NA logic gates for cancer marker detection and targeted therapy. As noted above, SELEX technology was first developed in Tan group and has been utilized to generate over 300 aptamers targeting many kinds of cancer cells.63 Those aptamers are now being widely used in biomedical research as probes for cancer cell molecular recognition. In 2012, Douglas et al. reported pioneering work by developing a DNA nanorobot that could intelligently recognize cancer cells via biomarkers on cell membranes.13 They used DNA origami to build a hexagonal barrel-shaped nanostructure, with dimension of 35 nm × 35 nm × 45 nm. This structure can release its payload only when combinations of cell surface biomarkers match the aptamers fixed on the nanorobots, working much like combination locks where all the tumblers must fall into place. Amir et al. recently designed nanorobots capable of dynamically interacting with each other to generate logical outputs in live animals.92 Depending on the outputs, the molecular payloads could be switched either on or off. These origami robots have now been used in live cockroaches for controlling a molecule targeting their cells.
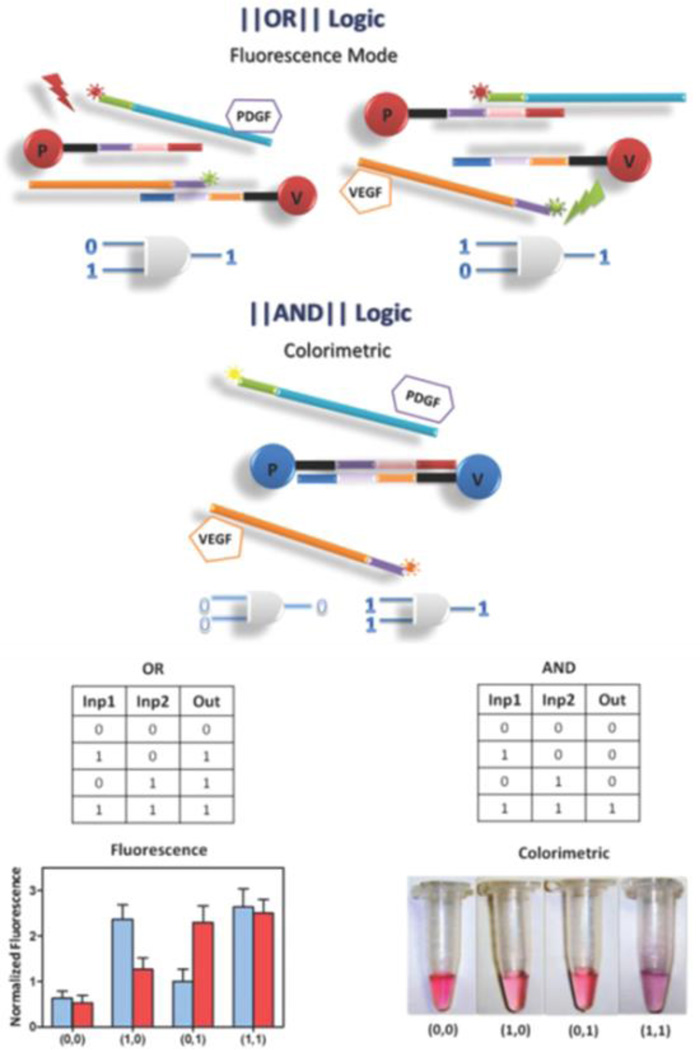
In Tan group, DNA-based logic systems have also been developed for cancer marker detection and targeted cancer therapy, using aptamers binding either to proteins or living cells. The first example utilized the special properties of gold nanoparticles (AuNP) to develop AND and OR logic gates for soluble biomarker detection (Figure 9).93 As a model to show the strategy, two important proangiogenic factors commonly used for cancer diagnosis and treatment, platelet-derived growth factor (PDGF) and vascular endothelial growth factor (VEGF), were tested. In the absence of both PDGF and VEGF, fluorescence from each probe is quenched by AuNP, and the AuNPs are monodispersed as surrounded by DNA probes, showing a red color in solution. However, when either one of these proteins is present in the solution, one type of duplex opens, restoring fluorescence, which is the output of the OR logic gate. When both targets are present, both types of duplexes open, and the AuNPs aggregate by complementary design of the connecting DNA. The solution becomes purple as the reporter output of the AND logic gate.
Fig. 9.
Schematic of aptamer-nanoparticle assembly-based logical detection. “OR” and “AND” gates for fluorescence and colorimetric output with inputs of PDGF and VEGF (reprinted by permission from reference [93], Copyright 2012, American Chemical Society).
Later, a logic circuit was engineered on the surface of the cells to achieve targeted and amplified photodynamic cancer therapy (PDT).94 In this work, a photosensitizer, Chlorin e6 (Ce6), was used as the killing module in the cell-surface sensing logic circuit (Figure 10A). In the absence of target cells, the two hairpin structures are stable. Upon introduction of the target cells, the aptamers selectively recognize them and bind specifically to the cell-surface targets. The overhanging catalyst sequence on the aptamer catalyzes the hybridization reaction between A1 and A2 to form A12, which further reacts with the R12 module to release the quencher-labeled strand and activate Ce6, which generates singlet oxygen to kill cancer cells by irradiation at 404 nm.
Fig. 10.
(A) Principle of specific cancer cell-surface aptamer circuit for enhanced photodynamic therapy (reprinted by permission from reference [94], Copyright 2013, American Chemical Society). (B) Construction of DNA-based “nano-claw”: two-input trivalent “Y”-shaped nano-claw and three-input tetravalent “X”-shaped nano-claw (reprinted by permission from reference [95], Copyright 2014, American Chemical Society). (C) Schematic of aptamer-based cell-surface “AND” logic gate for diagnosis and treatment of specific cancer cell type in the presence of two biomarkers (reprinted by permission from reference [96], Copyright 2014, American Chemical Society).
As aptamers become available against more cell types, programmable analysis of multiple biomarkers on the cell surface will be necessary for the clinician to establish a comprehensive disease profile and to conduct a more accurate intervention. To apply logic gates to cancer cell targeting and therapy, at least two aspects must be addressed. First, all modules need to be integrated into a single nanodevice to allow the use of logic systems as therapeutics. Second, logic gates more sophisticated than either AND or OR need to be developed to satisfy different biomedical criteria.
The Tan group has made progress towards meeting both of these requirements. To integrate all modules into one piece, a nanodevice termed “nano-claw” was designed for autonomous analysis of multiple cancer cell-membrane markers, yielding a diagnostic signal or targeted PDT response (Figure 10B).95 The structures were designed into either ‘X’ or ‘Y’ shapes, depending on the number of biomarkers to be tested. Either of these two types is composed of an oligonucleotide backbone as the scaffold, several modified switchable aptamers as “capture toes”, and a logic-gated DNA duplex as the “effector toe”. The “capture toes” recognize and bind to cell-surface targets, while, at the same time, generating “barcode strands”. The signal is released only if all “barcode strands” match the information stored at the “effector toe.” Using this strategy, both ON and INH gates were designed and tested successfully. Although this is only a preliminary attempt, it shows the potential of integrating logic gates into nanostructure assemblies.
Another effort involved the development of a programmable and universal platform to meet the requirement of screening for different complex conditions on cell membranes (Figure 10C).96 Again, either a fluorescence signal or PDT response is generated as output. With this platform, high-order multiple cell-surface marker identifications are possible. On the basis of basic Boolean operations (AND, OR and NOT), more complex and highly functional logic systems can be further programmed.
Concluding remarks
During the two decades since NA-based computational devices were first proposed, newly emerging logic systems have preliminarily demonstrated applications both in vitro and in vivo. The construction of NA-based logic devices can be integrated with DNA, RNA, small molecules, proteins, enzymes and even live cells with large variability, in contrast to conventional semiconductor-based computing systems. Also, the operation of NA-based logic devices is more flexible and diverse since they can be either static or oscillatory with various output generation modes. More significantly, the highly regulated and predictable properties of nucleic acids coupled with auxiliary programming software make molecular logic devices easy to control. All these unique features have advanced NA-based logic devices over the last ten years. However, many challenges must be overcome before these devices will see practical use. First, the relatively slow reaction kinetics between NA bases, or other interaction molecules, and the transport and output yield of scale-up NA logic devices could impede practical implementation. Second, the leak reaction and spurious binding between nucleic acid sequences may slow the desired reaction rates and decrease the effectiveness of NA logic devices.8, 10 Third, NA-based logic systems are more dependent on the diffusion among different nucleic acid gates, which may produce some unexpected spurious interactions.10 Moreover, current NA-based logic devices still cannot rival silicon chips in computational speed and capacity. These are critical issues in the future design of next-generation NA-based computing devices.
Nonetheless, the advantages of NA-based logic systems still offer tremendous opportunities for both in vitro and in vivo applications. From a fundamental point of view, NA logic circuitry must be scaled up to designs that can effectively substitute for semiconductor-based computing devices. For instance, the reversible strand displacement process was designed for DNA cascade reaction and introduced to construct a sophisticated biochemical circuit.10 In biomedicine, NA-based biological circuitry will facilitate personalized medicine by acquiring multiplexed physiological information inside the human body, then performing logical operations for analysis, and finally releasing loaded drugs, or even regulating the expression of specific disease genes.97–100 With further improvement of DNA/RNA nanotechnology and programming, more sophisticated artificial devices or systems based on NA and other biomolecules will be designed and created to satisfy growing needs in biotechnology and biomedicine.
Acknowledgements
C.W. acknowledges support from the American Chemical Society, Division of Analytical Chemistry Fellowship, sponsored by the Society for Analytical Chemists of Pittsburgh. This work is supported by grants awarded by the National Institutes of Health (GM079359 and CA133086). The authors thank Dr. K. R. Williams for manuscript review.
Notes and references
- 1.Han D, Kang H, Zhang T, Wu C, Zhou C, You M, Chen Z, Zhang X, Tan W. Chem. Eur. J. 2014;20:5866–5873. doi: 10.1002/chem.201304891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Silva AP, Uchiyama S. Nat Nano. 2007;2:399–410. doi: 10.1038/nnano.2007.188. [DOI] [PubMed] [Google Scholar]
- 3.Church GM, Gao Y, Kosuri S. Science. 2012;337:1628. doi: 10.1126/science.1226355. [DOI] [PubMed] [Google Scholar]
- 4.Goldman N, Bertone P, Chen S, Dessimoz C, LeProust EM, Sipos B, Birney E. Nature. 2013;494:77–80. doi: 10.1038/nature11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adleman L. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 6.Lipton R. Science. 1995;268:542–545. doi: 10.1126/science.7725098. [DOI] [PubMed] [Google Scholar]
- 7.Stojanovic MN, Stefanovic D. Nat Biotech. 2003;21:1069–1074. doi: 10.1038/nbt862. [DOI] [PubMed] [Google Scholar]
- 8.Seelig G, Soloveichik D, Zhang DY, Winfree E. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 9.Zhang DY, Turberfield AJ, Yurke B, Winfree E. Science. 2007;318:1121–1125. doi: 10.1126/science.1148532. [DOI] [PubMed] [Google Scholar]
- 10.Qian L, Winfree E. Science. 2011;332:1196–1201. doi: 10.1126/science.1200520. [DOI] [PubMed] [Google Scholar]
- 11.Qian L, Winfree E, Bruck J. Nature. 2011;475:368–372. doi: 10.1038/nature10262. [DOI] [PubMed] [Google Scholar]
- 12.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. [DOI] [PubMed] [Google Scholar]
- 13.Douglas SM, Bachelet I, Church GM. Science. 2012;335:831–834. doi: 10.1126/science.1214081. [DOI] [PubMed] [Google Scholar]
- 14.Siuti P, Yazbek J, Lu TK. Nat Biotech. 2013;31:448–452. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- 15.Yurke B, Turberfield AJ, Mills AP, Simmel FC, Neumann JL. Nature. 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 16.Zhang DY, Seelig G. Nat Chem. 2011;3:103–113. doi: 10.1038/nchem.957. [DOI] [PubMed] [Google Scholar]
- 17.Yin P, Choi HMT, Calvert CR, Pierce NA. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 18.Li B, Jiang Y, Chen X, Ellington AD. J. Am. Chem. Soc. 2012;134:13918–13921. doi: 10.1021/ja300984b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Briggs N, McLain JR, Ellington AD. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5386–5391. doi: 10.1073/pnas.1222807110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chirieleison SM, Allen PB, Simpson ZB, Ellington AD, Chen X. Nat Chem. 2013;5:1000–1005. doi: 10.1038/nchem.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang Y, Li B, Milligan JN, Bhadra S, Ellington AD. J. Am. Chem. Soc. 2013;135:7430–7433. doi: 10.1021/ja4023978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okamoto A, Tanaka K, Saito I. J. Am. Chem. Soc. 2004;126:9458–9463. doi: 10.1021/ja047628k. [DOI] [PubMed] [Google Scholar]
- 23.Li T, Wang E, Dong S. Anal. Chem. 2010;82:7576–7580. doi: 10.1021/ac1019446. [DOI] [PubMed] [Google Scholar]
- 24.Zhu J, Zhang L, Zhou Z, Dong S, Wang E. Anal. Chem. 2013;86:312–316. doi: 10.1021/ac403235y. [DOI] [PubMed] [Google Scholar]
- 25.Livshits GI, Stern A, Rotem D, Borovok N, Eidelshtein G, Migliore A, Penzo E, Wind SJ, Di Felice R, Skourtis SS, Cuevas JC, Gurevich L, Kotlyar AB, Porath D. Nat Nano. 2014;9:1040–1046. doi: 10.1038/nnano.2014.246. [DOI] [PubMed] [Google Scholar]
- 26.Saghatelian A, Völcker NH, Guckian KM, Lin VSY, Ghadiri MR. J. Am. Chem. Soc. 2002;125:346–347. doi: 10.1021/ja029009m. [DOI] [PubMed] [Google Scholar]
- 27.Prokup A, Hemphill J, Deiters A. J. Am. Chem. Soc. 2012;134:3810–3815. doi: 10.1021/ja210050s. [DOI] [PubMed] [Google Scholar]
- 28.Tyagi S, Kramer FR. Nat Biotech. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 29.Tan W, Wang K, Drake TJ. Curr. Opin. Chem. Biol. 2004;8:547–553. doi: 10.1016/j.cbpa.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Wang K, Tang Z, Yang CJ, Kim Y, Fang X, Li W, Wu Y, Medley CD, Cao Z, Li J, Colon P, Lin H, Tan W. Angew. Chem. Int. Ed. 2009;48:856–870. doi: 10.1002/anie.200800370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park KS, Seo MW, Jung C, Lee JY, Park HG. Small. 2012;8:2203–2212. doi: 10.1002/smll.201102758. [DOI] [PubMed] [Google Scholar]
- 32.Genot AJ, Bath J, Turberfield AJ. J. Am. Chem. Soc. 2011;133:20080–20083. doi: 10.1021/ja208497p. [DOI] [PubMed] [Google Scholar]
- 33.Stojanovic MN, de Prada P, Landry DW. ChemBioChem. 2001;2:411–415. doi: 10.1002/1439-7633(20010601)2:6<411::AID-CBIC411>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 34.Stojanovic MN, Stefanovic D, Rudchenko S. Acc. Chem. Res. 2014;47:1845–1852. doi: 10.1021/ar5000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stojanovic MN, Mitchell TE, Stefanovic D. J. Am. Chem. Soc. 2002;124:3555–3561. doi: 10.1021/ja016756v. [DOI] [PubMed] [Google Scholar]
- 36.Lederman H, Macdonald J, Stefanovic D, Stojanovic MN. Biochemistry. 2006;45:1194–1199. doi: 10.1021/bi051871u. [DOI] [PubMed] [Google Scholar]
- 37.Macdonald J, Li Y, Sutovic M, Lederman H, Pendri K, Lu W, Andrews BL, Stefanovic D, Stojanovic MN. Nano Lett. 2006;6:2598–2603. doi: 10.1021/nl0620684. [DOI] [PubMed] [Google Scholar]
- 38.Pei R, Matamoros E, Liu M, Stefanovic D, Stojanovic MN. Nat Nano. 2010;5:773–777. doi: 10.1038/nnano.2010.194. [DOI] [PubMed] [Google Scholar]
- 39.Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 40.Breaker RR, Joyce GF. Chemistry & Biology. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 41.Miyake Y, Togashi H, Tashiro M, Yamaguchi H, Oda S, Kudo M, Tanaka Y, Kondo Y, Sawa R, Fujimoto T, Machinami T, Ono A. J. Am. Chem. Soc. 2006;128:2172–2173. doi: 10.1021/ja056354d. [DOI] [PubMed] [Google Scholar]
- 42.Wu C, Yan L, Wang C, Lin H, Wang C, Chen X, Yang CJ. Biosensors and Bioelectronics. 2010;25:2232–2237. doi: 10.1016/j.bios.2010.02.030. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Z, Wu C, Liu H, Zou Y, Zhang X, Kang H, Yang CJ, Tan W. Angewandte Chemie International Edition. 2010;49:1052–1056. doi: 10.1002/anie.200905570. [DOI] [PubMed] [Google Scholar]
- 44.Zhu X, Xu H, Gao X, Li X, Liu Q, Lin Z, Qiu B, Chen G. Chem. Commun. 2011;47:9080–9082. doi: 10.1039/c1cc12734a. [DOI] [PubMed] [Google Scholar]
- 45.Park KS, Lee JY, Park HG. Chem. Commun. 2012;48:4549–4551. doi: 10.1039/c2cc17148a. [DOI] [PubMed] [Google Scholar]
- 46.Bi S, Ji B, Zhang Z, Zhu J-J. Chem. Sci. 2013;4:1858–1863. [Google Scholar]
- 47.Zhang L, Wang Z-X, Liang R-P, Qiu J-D. Langmuir. 2013;29:8929–8935. doi: 10.1021/la401887b. [DOI] [PubMed] [Google Scholar]
- 48.Moshe M, Elbaz J, Willner I. Nano Lett. 2009;9:1196–1200. doi: 10.1021/nl803887y. [DOI] [PubMed] [Google Scholar]
- 49.Orbach R, Mostinski L, Wang F, Willner I. Chem. Eur. J. 2012;18:14689–14694. doi: 10.1002/chem.201201995. [DOI] [PubMed] [Google Scholar]
- 50.Orbach R, Remacle F, Levine RD, Willner I. Proc. Natl. Acad. Sci. U.S.A. 2012;109:21228–21233. doi: 10.1073/pnas.1219672110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown CW, Lakin MR, Stefanovic D, Graves SW. ChemBioChem. 2014;15:950–954. doi: 10.1002/cbic.201400047. [DOI] [PubMed] [Google Scholar]
- 52.Orbach R, Wang F, Lioubashevski O, Levine RD, Remacle F, Willner I. Chem. Sci. 2014;5:3381–3387. [Google Scholar]
- 53.Li T, Wang E, Dong S. J. Am. Chem. Soc. 2009;131:15082–15083. doi: 10.1021/ja9051075. [DOI] [PubMed] [Google Scholar]
- 54.Li T, Zhang L, Ai J, Dong S, Wang E. ACS Nano. 2011;5:6334–6338. doi: 10.1021/nn201407h. [DOI] [PubMed] [Google Scholar]
- 55.Li T, Ackermann D, Hall AM, Famulok M. J. Am. Chem. Soc. 2012;134:3508–3516. doi: 10.1021/ja2108883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ono A, Cao S, Togashi H, Tashiro M, Fujimoto T, Machinami T, Oda S, Miyake Y, Okamoto I, Tanaka Y. Chem. Commun. 2008:4825–4827. doi: 10.1039/b808686a. [DOI] [PubMed] [Google Scholar]
- 57.Park KS, Jung C, Park HG. Angew. Chem. Int. Ed. 2010;49:9757–9760. doi: 10.1002/anie.201004406. [DOI] [PubMed] [Google Scholar]
- 58.Zhang Y-M, Zhang L, Liang R-P, Qiu J-D. Chem. Eur. J. 2013;19:6961–6965. doi: 10.1002/chem.201300625. [DOI] [PubMed] [Google Scholar]
- 59.Breaker RR, Joyce GF. Chemistry & Biology. 1995;2:655–660. doi: 10.1016/1074-5521(95)90028-4. [DOI] [PubMed] [Google Scholar]
- 60.Bi S, Yan Y, Hao S, Zhang S. Angew. Chem. Int. Ed. 2010;49:4438–4442. doi: 10.1002/anie.201000840. [DOI] [PubMed] [Google Scholar]
- 61.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 62.Shangguan D, Li Y, Tang Z, Cao ZC, Chen HW, Mallikaratchy P, Sefah K, Yang CJ, Tan W. Proc. Natl. Acad. Sci. U.S.A. 2006;103:11838–11843. doi: 10.1073/pnas.0602615103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan W, Donovan MJ, Jiang J. Chem. Rev. 2013;113:2842–2862. doi: 10.1021/cr300468w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elbaz J, Shlyahovsky B, Li D, Willner I. ChemBioChem. 2008;9:232–239. doi: 10.1002/cbic.200700436. [DOI] [PubMed] [Google Scholar]
- 65.Zhou M, Du Y, Chen C, Li B, Wen D, Dong S, Wang E. J. Am. Chem. Soc. 2010;132:2172–2174. doi: 10.1021/ja910634e. [DOI] [PubMed] [Google Scholar]
- 66.Lin Y, Tao Y, Pu F, Ren J, Qu X. Adv. Funct. Mater. 2011;21:4565–4572. [Google Scholar]
- 67.Wang L, Zhu J, Han L, Jin L, Zhu C, Wang E, Dong S. ACS Nano. 2012;6:6659–6666. doi: 10.1021/nn300997f. [DOI] [PubMed] [Google Scholar]
- 68.Zhou M, Dong S. Biomolecular Information Processing. Wiley-VCH Verlag GmbH & Co. KGaA; 2012. pp. 117–131. [Google Scholar]
- 69.Zhou M, Wang J. Electroanalysis. 2012;24:197–209. [Google Scholar]
- 70.Chen J, Zeng L. Biosens. Bioelectron. 2013;42:93–99. doi: 10.1016/j.bios.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 71.Yin B-C, Ye B-C, Wang H, Zhu Z, Tan W. Chem. Commun. 2012;48:1248–1250. doi: 10.1039/c1cc15639j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen J, Fang Z, Lie P, Zeng L. Anal. Chem. 2012;84:6321–6325. doi: 10.1021/ac301508b. [DOI] [PubMed] [Google Scholar]
- 73.Wang G, Zhu Y, Chen L, Zhang X. Biosens. Bioelectron. 2015;63:552–557. doi: 10.1016/j.bios.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 74.Huang Y, Wen W, Du D, Zhang X, Wang S, Lin Y. Biosens. Bioelectron. 2014;61:598–604. doi: 10.1016/j.bios.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 75.Lin B, Sun Q, Liu K, Lu D, Fu Y, Xu Z, Zhang W. Langmuir. 2014;30:2144–2151. doi: 10.1021/la4048769. [DOI] [PubMed] [Google Scholar]
- 76.Zhu X, Chen L, Lin Z, Qiu B, Chen G. Chem. Commun. 2010;46:3149–3151. doi: 10.1039/b926319e. [DOI] [PubMed] [Google Scholar]
- 77.Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 78.Han D, Zhu Z, Wu C, Peng L, Zhou L, Gulbakan B, Zhu G, Williams KR, Tan W. J. Am. Chem. Soc. 2012;134:20797–20804. doi: 10.1021/ja310428s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prokup A, Deiters A. Angew. Chem. Int. Ed. 2014;53:13192–13195. doi: 10.1002/anie.201406892. [DOI] [PubMed] [Google Scholar]
- 80.King DJ, Ventura DA, Brasier AR, Gorenstein DG. Biochemistry. 1998;37:16489–16493. doi: 10.1021/bi981780f. [DOI] [PubMed] [Google Scholar]
- 81.Koshkin AA, Rajwanshi VK, Wengel J. Tetrahedron Lett. 1998;39:4381–4384. [Google Scholar]
- 82.Koshkin AA, Singh SK, Nielsen P, Rajwanshi VK, Kumar R, Meldgaard M, Olsen CE, Wengel J. Tetrahedron. 1998;54:3607–3630. [Google Scholar]
- 83.Burmeister PE, Lewis SD, Silva RF, Preiss JR, Horwitz LR, Pendergrast PS, McCauley TG, Kurz JC, Epstein DM, Wilson C, Keefe AD. Chemistry & Biology. 2005;12:25–33. doi: 10.1016/j.chembiol.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 84.Burmeister PE, Wang C, Killough JR, Lewis SD, Horwitz LR, Ferguson A, Thompson KM, Pendergrast PS, McCauley TG, Kurz M, Diener J, Cload ST, Wilson C, Keefe AD. Oligonucleotides. 2006;16:337–351. doi: 10.1089/oli.2006.16.337. [DOI] [PubMed] [Google Scholar]
- 85.Eulberg D, Klussmann S. ChemBioChem. 2003;4:979–983. doi: 10.1002/cbic.200300663. [DOI] [PubMed] [Google Scholar]
- 86.Benenson Y. Curr. Opin. Biotechnol. 2009;20:471–478. doi: 10.1016/j.copbio.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hagiya M, Konagaya A, Kobayashi S, Saito H, Murata S. Acc. Chem. Res. 2014;47:1681–1690. doi: 10.1021/ar400318d. [DOI] [PubMed] [Google Scholar]
- 88.Rinaudo K, Bleris L, Maddamsetti R, Subramanian S, Weiss R, Benenson Y. Nat Biotech. 2007;25:795–801. doi: 10.1038/nbt1307. [DOI] [PubMed] [Google Scholar]
- 89.Hemphill J, Deiters A. J. Am. Chem. Soc. 2013;135:10512–10518. doi: 10.1021/ja404350s. [DOI] [PubMed] [Google Scholar]
- 90.Liu Y, Zeng Y, Liu L, Zhuang C, Fu X, Huang W, Cai Z. Nat Commun. 2014:5. doi: 10.1038/ncomms6393. [DOI] [PubMed] [Google Scholar]
- 91.Rudchenko M, Taylor S, Pallavi P, Dechkovskaia A, Khan S, Butler VP, Jr, Rudchenko S, Stojanovic MN. Nat Nano. 2013;8:580–586. doi: 10.1038/nnano.2013.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Amir Y, Ben-Ishay E, Levner D, Ittah S, Abu-Horowitz A, Bachelet I. Nat Nano. 2014;9:353–357. doi: 10.1038/nnano.2014.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shukoor MI, Altman MO, Han D, Bayrac AT, Ocsoy I, Zhu Z, Tan W. ACS Appl. Mater. Interfaces. 2012;4:3007–3011. doi: 10.1021/am300374q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Han D, Zhu G, Wu C, Zhu Z, Chen T, Zhang X, Tan W. ACS Nano. 2013;7:2312–2319. doi: 10.1021/nn305484p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.You M, Peng L, Shao N, Zhang L, Qiu L, Cui C, Tan W. J. Am. Chem. Soc. 2013;136:1256–1259. doi: 10.1021/ja4114903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.You M, Zhu G, Chen T, Donovan MJ, Tan W. J. Am. Chem. Soc. 2014 doi: 10.1021/ja509263k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Benenson Y, Gil B, Ben-Dor U, Adar R, Shapiro E. Nature. 2004;429:423–429. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- 98.Chen T, Wu CS, Jimenez E, Zhu Z, Dajac JG, You M, Han D, Zhang X, Tan W. Angewandte Chemie International Edition. 2013;52:2012–2016. doi: 10.1002/anie.201209440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wu C, Chen T, Han D, You M, Peng L, Cansiz S, Zhu G, Li C, Xiong X, Jimenez E, Yang CJ, Tan W. ACS Nano. 2013;7:5724–5731. doi: 10.1021/nn402517v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu C, Han D, Chen T, Peng L, Zhu G, You M, Qiu L, Sefah K, Zhang X, Tan W. J. Am. Chem. Soc. 2013;135:18644–18650. doi: 10.1021/ja4094617. [DOI] [PMC free article] [PubMed] [Google Scholar]