Abstract
Autophagy, an important catabolic pathway implicated in a broad spectrum of human diseases, begins by forming double membrane autophagosomes that engulf cytosolic cargo and ends by fusing autophagosomes with lysosomes for degradation1,2. Membrane fusion activity is required for early biogenesis of autophagosomes and late degradation in lysosomes3–7. However, the key regulatory mechanisms of autophagic membrane tethering and fusion remain largely unknown. Here we report that ATG14 (also known as beclin-1-associated autophagy-related key regulator (Barkor) or ATG14L), an essential autophagy-specific regulator of the class III phosphatidylinositol 3-kinase complex8–11, promotes membrane tethering of protein-free liposomes, and enhances hemifusion and full fusion of proteoliposomes reconstituted with the target (t)-SNAREs (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) syntaxin 17 (STX17) and SNAP29, and the vesicle (v)-SNARE VAMP8 (vesicle-associated membrane protein 8). ATG14 binds to the SNARE core domain of STX17 through its coiled-coil domain, and stabilizes the STX17–SNAP29 binary t-SNARE complex on autophagosomes. The STX17 binding, membrane tethering and fusion-enhancing activities of ATG14 require its homo-oligomerization by cysteine repeats. In ATG14 homo-oligomerization-defective cells, autophagosomes still efficiently form but their fusion with endolysosomes is blocked. Recombinant ATG14 homo-oligomerization mutants also completely lose their ability to promote membrane tethering and to enhance SNARE-mediated fusion in vitro. Taken together, our data suggest an autophagy-specific membrane fusion mechanism in which oligomeric ATG14 directly binds to STX17–SNAP29 binary t-SNARE complex on autophagosomes and primes it for VAMP8 interaction to promote autophagosome–endolysosome fusion.
Endoplasmic reticulum (ER)-derived STX17, SNAP29 and endolysosome-localized VAMP8 converge on the complete autophagosome and mediate autophagosome–lysosome fusion6,7. STX17 also recruits ATG14 to the ER–mitochondria contact site and promotes autophagosome formation and/or maturation12. However, a potential function of ATG14 in membrane tethering and fusion has not yet been explored.
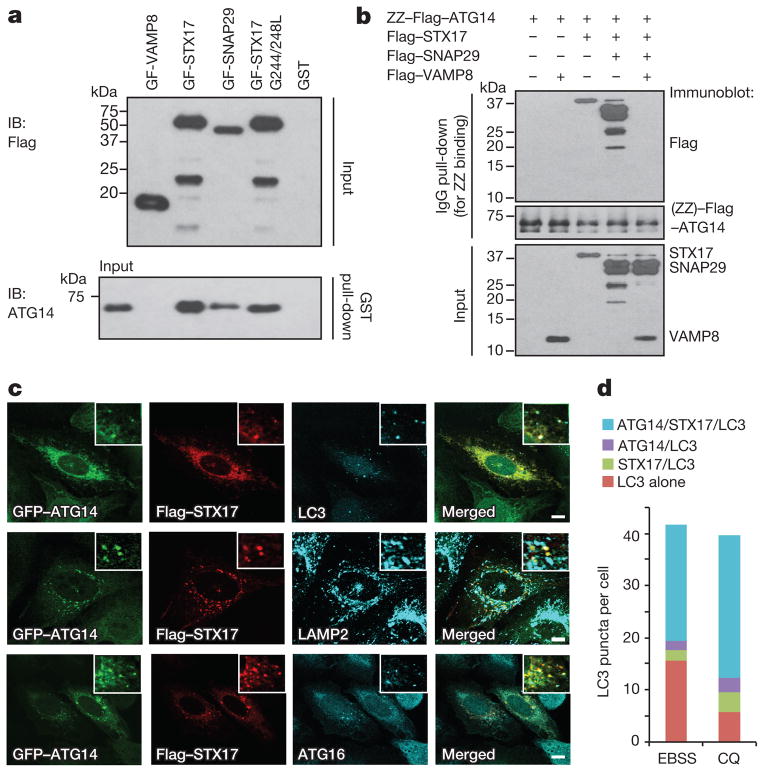
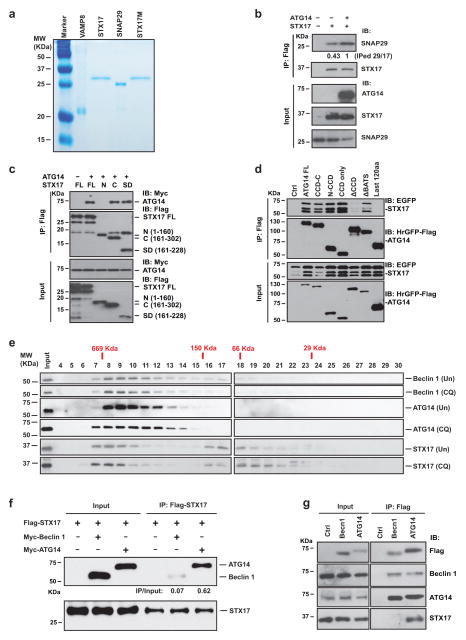
We first tested whether ATG14 physically interacts with autophagic SNAREs. In an in vitro glutathione S-transferase (GST) pull-down assay with recombinant full-length STX17, SNAP29 and VAMP8 proteins purified from Escherichia coli (Extended Data Fig. 1a), recombinant ATG14 bound to STX17 and SNAP29 but not toVAMP8 (Fig. 1a).STX17 G244/248L, an autophagosome targeting-deficient mutant6, still bound to ATG14 (Fig. 1a). Recombinant ATG14 bound to STX17 alone and the STX17–SNAP29 binary t-SNARE complex, but not to the STX17–SNAP29–VAMP8 ternary complex (Fig. 1b), suggesting that ATG14 binds before formation of trans-SNARE complex or that it can interact with partly folded trans-SNARE complex (but not fully folded cis-SNARE complex). Overexpression of ATG14 stabilized the STX17–SNAP29 binary complex in a co-immunoprecipitation assay (Extended Data Fig. 1b). Deletion mapping analysis showed that the coiled-coil domain (CCD) of ATG14 interacts with the SNARE core domain of STX17 (Extended Data Fig. 1c, d).
Figure 1. ATG14 interacts with STX17–SNAP29 on mature autophagosomes.
a, Interaction between purified recombinant ATG14 and GST–Flag-tagged (GF) autophagic SNAREs using an in vitro GST pull-down assay followed by western blot (IB, immunoblot). b, Recombinant ATG14 binds to STX17–SNAP29 binary t-SNARE complex but not STX17–SNAP29–VAMP8 ternary complex using an in vitro immunoglobulin-G (IgG) pull-down assay followed by western blot. c, Flag–STX17, GFP–ATG14, endogenous LC3, LAMP2 and ATG16 were detected by immunostaining in EBSS-starved U2OS cells (n =20). Scale bars, 5 μm. d, Corresponding statistical analysis of co-localization of ATG14, STX17 and LC3 upon treatment with EBSS or chloroquine (CQ).
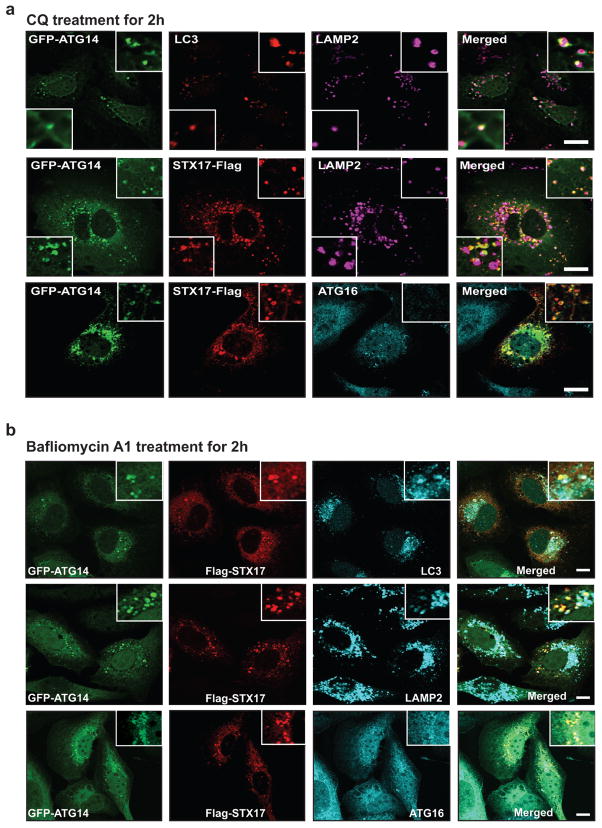
We also tested whether STX17 is a part of the beclin 1/class III phosphatidylinositol 3-kinase (PI3KC3) complex. ATG14 co-fractionated with STX17 and beclin 1 in a 669-kilodalton (kDa) complex separated by size-exclusion chromatography (SEC) (Extended Data Fig. 1e), and ATG14 interacted both with beclin 1 and with STX17. However, STX17 only bound to ATG14, but not beclin 1 and vice versa, in immunoprecipitation assays (Extended Data Fig. 1f, g). In cultured U2OS cells, ATG14 co-localized with STX17 and LC3, at least partly, on mature autophagosomes adjacent to or overlapping with LAMP2-labelled autolysosomes upon treatment with Earle’s balanced salt solution (EBSS) (Fig. 1c, d). ATG14 also localized to mature autophagosomes with STX17 but not with ATG16 upon treatments with chloroquine or bafilomycin A1 (Extended Data Fig. 2). Together, these results suggest that ATG14 physically and physiologically interacts with the STX17–SNAP29 binary t-SNARE complex on mature autophagosomes.
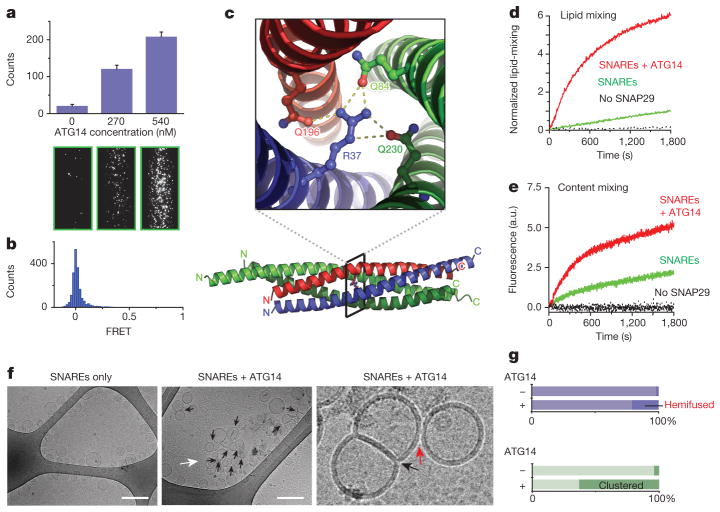
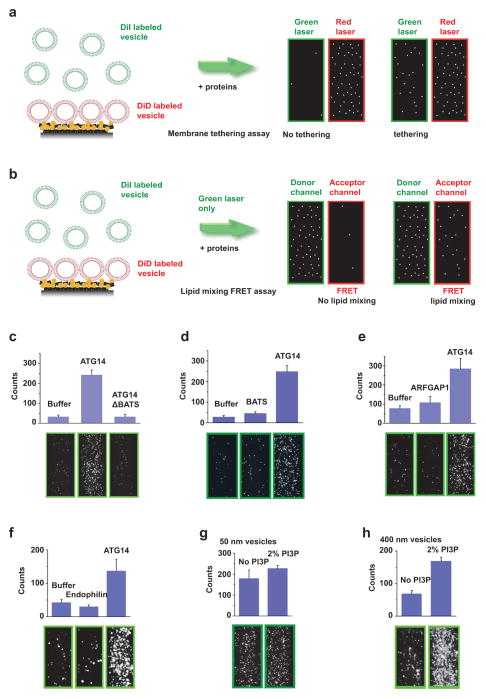
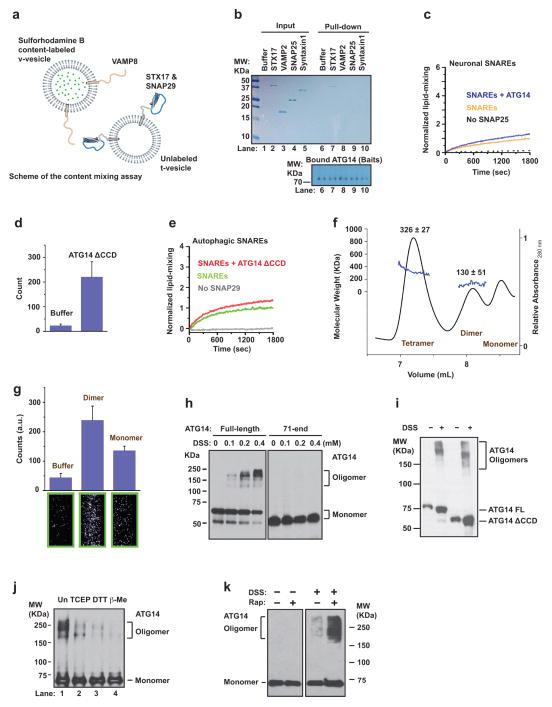
We then tested whether ATG14 participates directly in membrane tethering in a single-vesicle/liposome assay13,14 (Methods and Extended Data Fig. 3a). Recombinant ATG14 alone was sufficient to strongly promote protein-free liposome tethering (Fig. 2a). However, lipid-mixing of protein-free liposome membranes was not promoted by ATG14 (Extended Data Fig. 3b and Fig. 2b). The membrane-tethering activity of ATG14 required its membrane-binding Barkor/ATG14(L) autophagosome targeting sequence (BATS) domain15, but the BATS domain alone was insufficient for membrane tethering (Extended Data Fig. 3c, d). The membrane-tethering activity of ATG14 is not solely due to membrane curvature sensing, since no membrane-tethering activity was observed using another curvature sensing ALPS motif-containing ARFGAP1 protein16 or the membrane curvature-inducing protein BIF-1/endophilin B1 (ref. 17) (Extended Data Fig. 3e, f). ATG14 membrane-tethering activity was independent of phosphatidylinositol 3-phosphate (PI3P) for small (50 nm) liposomes, but increased in the presence of PI3P for large (400 nm) liposomes (Extended Data Fig. 3g, h). Taken together, ATG14 alone is a membrane tether.
Figure 2. ATG14 promotes membrane tethering and enhances autophagic SNARE-mediated fusion.
a, Purified recombinant ATG14 promotes protein-free single liposome tethering (Methods and Extended Data Fig. 3a). Top: mean number of tethered liposomes (±s.d.) (n =15) at random locations in the sample chamber; bottom: corresponding representative images (n =15). b, Fluorescence resonance energy transfer (FRET) efficiency profile between single donor/acceptor-dye liposome pairs upon addition of recombinant ATG14 (Methods and Extended Data Fig. 3b). c, The crystal structure of the autophagic SNARE complex is shown at the bottom and a close-up view of the ionic layer at the centre is shown at the top. d, ATG14 enhances ensemble lipid-mixing of proteoliposomes reconstituted with autophagic SNAREs (n =3). e, ATG14 enhances ensemble content-mixing of proteoliposomes reconstituted with autophagic SNAREs (n =3); a.u., arbitrary units. f, Representative cryo-electron micrographs (n =20) of proteoliposomes reconstituted with autophagic SNAREs. Left: without ATG14. Middle: with ATG14; black and white arrows indicate hemifusion diaphragms and proteoliposome clusters, respectively. Right: close-up view; black and red arrows indicate a hemifused diaphragm and a tethered proteoliposome pair, respectively. Scale bars, 200 nm. g, Percentage of hemifused or clustered proteoliposomes micrographs (n =20).
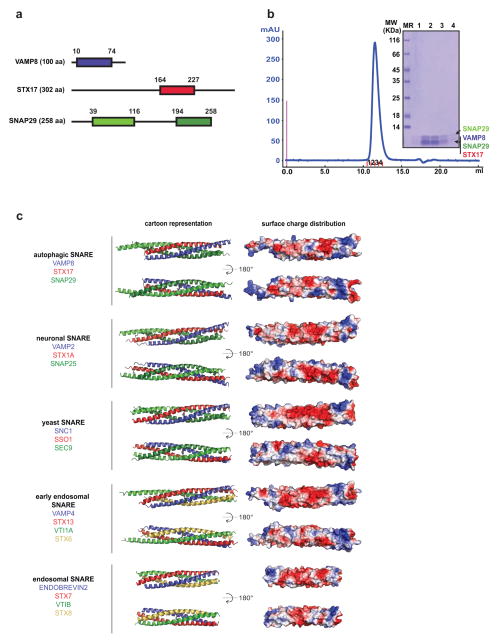
It is unclear whether autophagic SNAREs form a fusion-competent α-helical bundle and possess fusogenic activity. We co-expressed and co-purified the four SNARE core domains of VAMP8 (10–74), STX17 (164–227) and SNAP29 (39–116, 194–258), and determined the crystal structure of the complex at 1.4 Å resolution (Methods, Extended Data Fig. 4a–c and Extended Data Table 1). The autophagic SNARE complex forms a parallel four α-helix bundle (Fig. 2c). At the centre of the complex is a conserved ionic layer consisting of R37 from VAMP8, Q196 from STX17 and Q84/Q230 from SNAP29 that is characteristic for all SNARE complexes. The overall structure of the autophagic SNARE complex is similar to that of the neuronal, yeast and endosomal SNARE complexes determined so far (Extended Data Fig. 4c).
We tested the fusogenic activity of autophagic SNAREs using protein-reconstituted proteoliposome ensemble lipid- and content-mixing assays (Methods). Proteoliposomes reconstituted with autophagic SNAREs (mean diameter 50 nm) underwent lipid-mixing at a relatively low level, which was greatly enhanced by the addition of ATG14 (Fig. 2d). ATG14 also enhanced content-mixing in the presence of autophagic SNAREs (Fig. 2e), using an ensemble content-mixing assay with the content marker sulphorhodamine B that was used in previous studies18 (Methods and Extended Data Fig. 5a). Examination of membrane morphologies by cryo-electron microscopy revealed that hemifusion diaphragms were increased when ATG14 was added to proteoliposomes reconstituted with autophagic SNAREs (Fig. 2f, g). Thus, ATG14 enhances hemifusion and complete fusion of proteoliposomes reconstituted with autophagic SNAREs.
The ATG14 fusion-enhancing effect is specific to autophagic SNAREs, since ATG14 did not enhance membrane fusion mediated by neuronal SNAREs (Extended Data Fig. 5b, c). We tested whether the ATG14 fusion-enhancement is due to its tethering activity or its interaction with autophagic SNAREs. We observed that an STX17 binding-deficient mutant (CCD deletion) of ATG14 retained its membrane-tethering function but lost its fusion-enhancing activity (Extended Data Fig. 5d, e), suggesting that the interaction between ATG14 and STX17 (as part of the autophagic t-SNARE or trans-SNARE complex) contributes to the fusion enhancement.
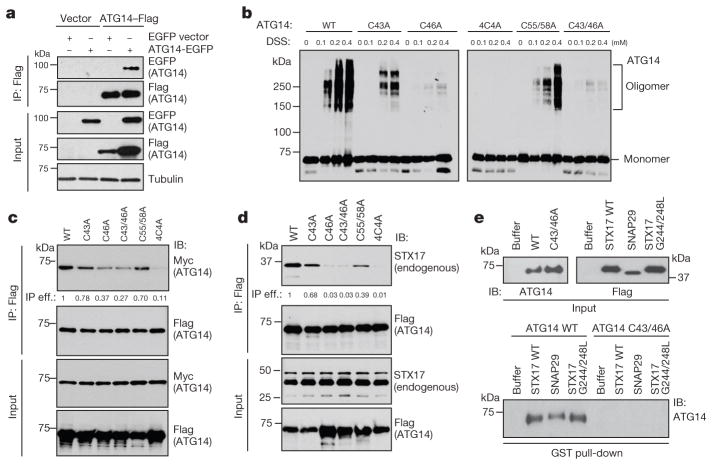
SEC coupled with multi-angle static light scattering (SEC–MALS) experiments indicated that recombinant ATG14 exists in monomeric, dimeric and tetrameric states (Extended Data Fig. 5f); upon separating different oligomeric forms, we found that the dimeric form of ATG14 is more active than the monomeric form in membrane tethering (Extended Data Fig. 5g). We confirmed ATG14 self-interaction in a co-immunoprecipitation assay (Fig. 3a). In the presence of the crosslinker disuccinimidyl suberate (DSS), we found that the amino terminus of ATG14 but not the CCD domain was required for homo-oligomerization (Extended Data Fig. 5h, i). Oligomeric forms of ATG14 could also be observed on native gels without crosslinking and were sensitive to reducing agents (Extended Data Fig. 5j). Upon rapamycin treatment, ATG14 oligomerization was increased compared with unstressed conditions (Extended Data Fig. 5k).
Figure 3. ATG14 homo-oligomerization is required for autophagic SNARE binding.
a, Immunoprecipitation (IP) of ATG14–eGFP by ATG14–Flag transfected into HEK293T cells. b, Mapping ATG14 oligomerization sites to its cysteine repeats by cross-linking with DSS and analysed by SDS–polyacrylamide gel electrophoresis (SDS–PAGE). c, Interaction between Flag-tagged wild-type (WT) ATG14 and mutants with Myc-ATG14 observed in a co-immunoprecipitation assay. IP eff., immunoprecipitation efficiency. d, Interaction between Flag-tagged WT ATG14 and mutants with endogenous STX17 observed in a co-immunoprecipitation assay. The immunoprecipitation efficiency (immunoprecipitation/input) in c and d was normalized by the ratio of immunoprecipitated Flag–ATG14 or interactants versus their inputs. e, In vitro GST pull-down of purified recombinant WT ATG14 or C43A/C46A mutant with GST-tagged autophagic SNAREs followed by western blot.
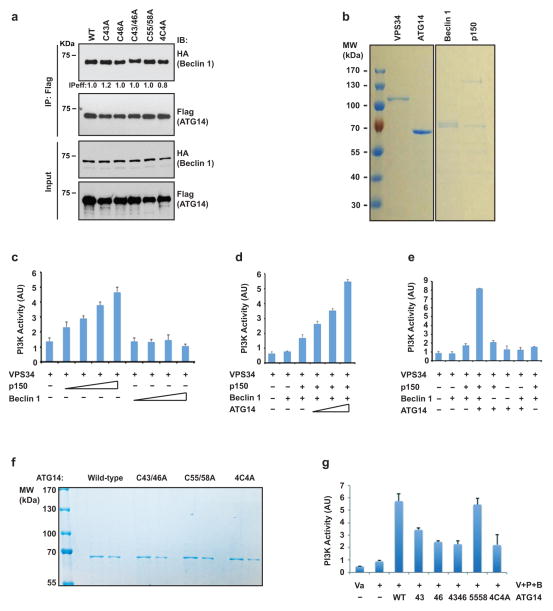
Four evolutionarily conserved cysteine repeats are localized to the N terminus of ATG14 and are essential for its ER localization and autophagy function19. We found that C46 is mainly responsible for ATG14 homo-oligomerization. C43 had a minor effect, while C55 and C58 had little impact on homo-oligomerization of ATG14 (Fig. 3b). The mutants C43A/C46A and C43A/C46A/C55A/C58A (abbreviated as 4C4A)showed less self-interaction in a co-immunoprecipitation assay (Fig. 3c); we refer to these mutants collectively as homo-oligomerization-deficient (HOD) mutants. Remarkably, ATG14 HOD mutants almost completely lost their ability to interact with the autophagic SNARE protein STX17 in a co-immunoprecipitation assay (Fig. 3d) and an in vitro pull-down assay (Fig. 3e). Thus, ATG14 homo-oligomerization is essential for its interaction with autophagic SNAREs.
The interaction between these ATG14 HOD mutants and beclin 1 remained intact (Extended Data Fig. 6a). In a reconstituted system purified in vitro, ATG14 enhanced the lipid kinase activity of the core VPS34–p150–beclin 1 complex (Extended Data Fig. 6b–e). Recombinant ATG14 HOD mutants could still enhance the lipid kinase activity catalysed by the VPS34–p150–beclin 1 core complex, but not as efficiently as wild-type (WT) ATG14 (Extended Data Fig. 6f, g). In addition, ATG14 HOD mutants could still localize to autophagosomes (Extended Data Fig. 7) and ER–mitochondria contact sites (Extended Data Fig. 8a), suggesting these membrane localization events are largely dispensable of ATG14 homo-oligomerization.
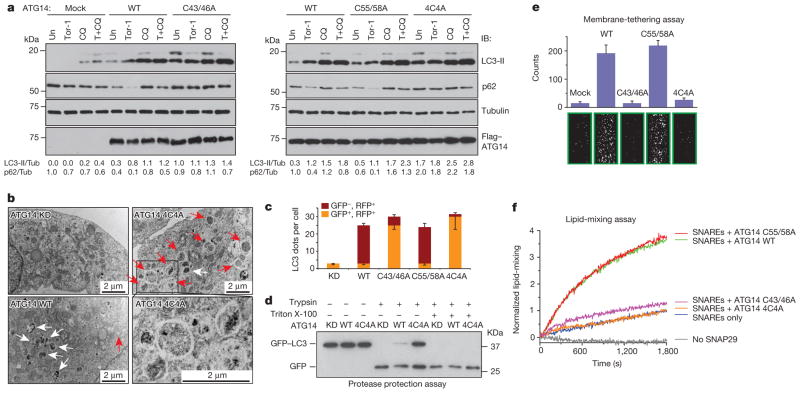
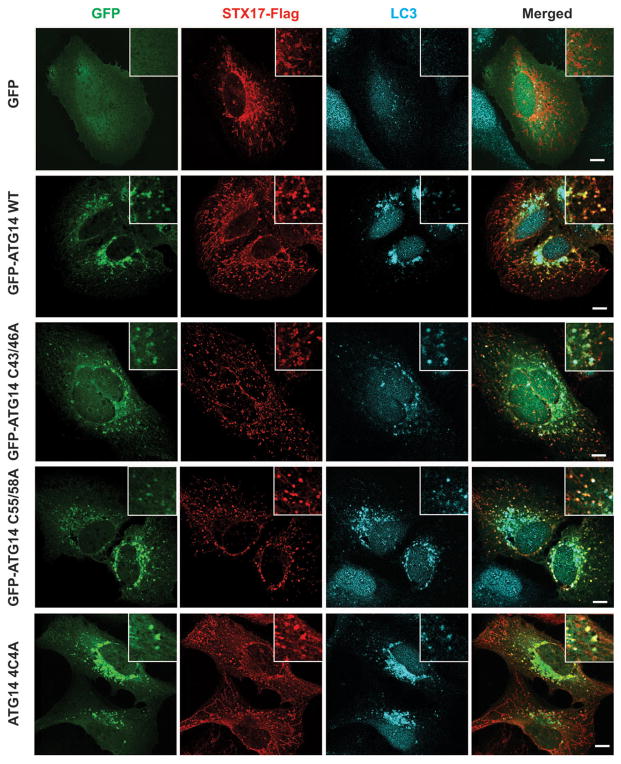
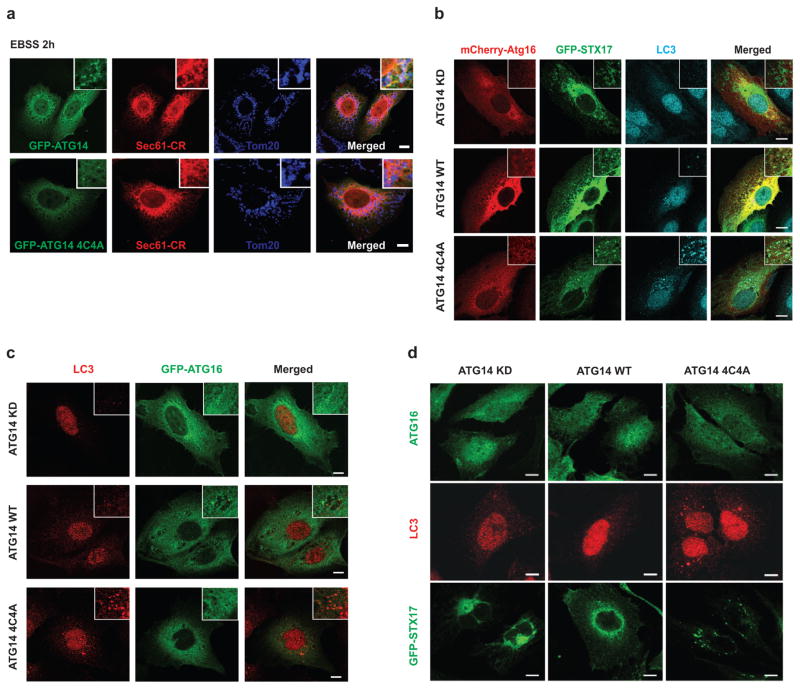
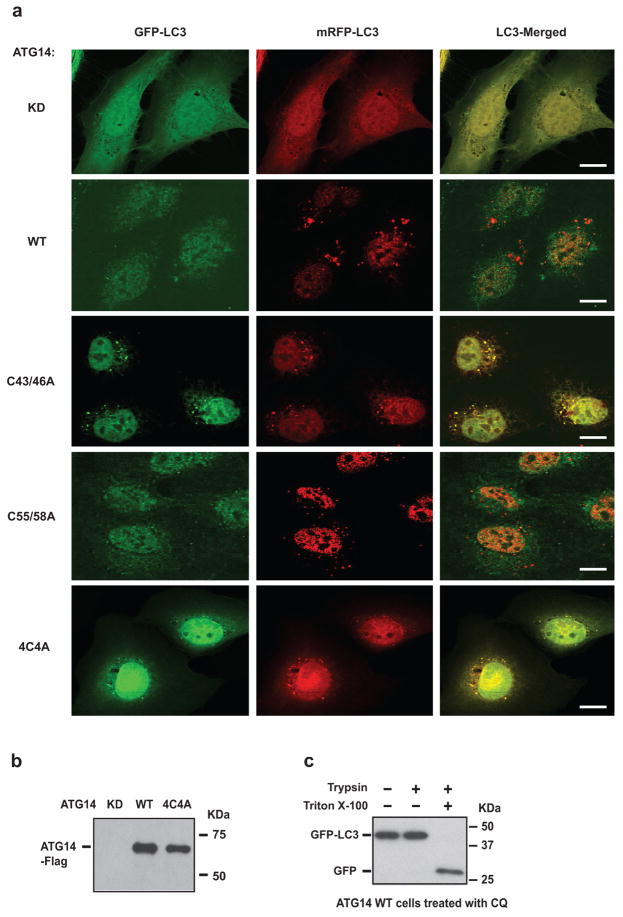
Furthermore, we investigated whether ATG14 oligomerization is important for autophagosomal fusion with endolysosome in vivo. We complemented ATG14-depleted U2OS cells8 with either WT ATG14 or ATG14 HOD mutants. The autophagy flux was blocked in cells expressing the ATG14 HOD mutants but not in cells expressing either WT ATG14 or the C55A/C58A mutant (Fig. 4a). Furthermore, in cells expressing ATG14 4C4A mutant, mature autophagosomes (LC3+ and STX17+) but not phagophores (Atg16+) accumulated (Extended Data Fig. 8b–d). Accumulation of sealed double-membrane autophagosomes in ATG14 4C4A mutant cells was detected by electron microscopy (Fig. 4b). In addition, we measured autophagosome maturation using LC3 tagged with acid-resistant monomeric red fluorescent protein (mRFP) and acid-sensitive green fluorescent protein (GFP)20. Autophagosome acidification/maturation was dramatically reduced in cells expressing ATG14 HOD mutants compared with WT ATG14 (Fig. 4c and Extended Data Fig. 9a). In a modified protease protection assay that was previously used in yeast studies21 (Extended Data Fig. 9b, c), more GFP–LC3 was protected from protease digestion in cells expressing ATG14 4C4A mutant than that in cells expressing WT ATG14 (Fig. 4d). Finally, we tested whether ATG14 homo-oligomerization is essential for its roles in membrane tethering and enhancement of SNARE-mediated fusion in vitro. Purified recombinant ATG14 HOD mutants lacked membrane-tethering activity (Fig. 4e) and lost their ability to enhance ensemble lipid mixing of proteoliposomes reconstituted with autophagic SNAREs (Fig. 4f).Together, all these results demonstrate a crucial role of ATG14 homo-oligomerization in regulating membrane fusion between autophagosome and endolysosomes.
Figure 4. ATG14 homo-oligomerization is required for autophagosomal fusion with endolysosomes in vivo and in vitro.
a, Autophagic flux analysis in ATG14-deficient U2OS cells reconstituted with WT ATG14 and mutants. Un, untreated; Tor-1, Torin 1. b, Accumulation of complete double-membrane autophagosome (red arrows) under TEM in ATG14 4C4A mutant but not in ATG14 WT or knockdown (KD) cells treated with rapamycin (n =15). White arrows denote autolysosomes. c, Quantitative analysis of acidified autophagosomes (GFP−RFP+) versus neutral autophagosomes (GFP+RFP+) per cell in rapamycin-treated ATG14 knockdown U2OS cells expressing WT or mutant ATG14 transfected with mRFP–GFP–LC3 (mean ±s.d.) (n =20). d, GFP–LC3 was protected from trypsin digestion in ATG14 4C4A mutant but not in ATG14 WT or knockdown cells. e, Tethering of protein-free liposomes by recombinant WT ATG14 and specified mutants using the single-vesicle membrane-tethering assay (mean number of tethered liposomes ±s.d.) (n =20)). f, Ensemble lipid-mixing activity of WT ATG14 and mutants between proteoliposomes reconstituted with autophagic SNAREs (n =3).
In this study, we observed that, in addition to its localization to phagophore8–11, ATG14 also localizes to mature autophagosomes. It is still not clear how ATG14 is recruited to mature autophagosomes. Most of ATG14 (or the autophagy-specific class III PI3K complex) dissociates from the phagophore membrane together with other ATGs during autophagosome formation22. It is possible that a small fraction remains on the membrane. Alternatively, after dissociation from the phagophore membrane, ATG14 (probably without other PI3KC3 components) is re-recruited to mature autophagosomes to facilitate the autophagosome–lysosome fusion. On autophagosomes, ATG14 promotes and stabilizes the STX17–SNAP29 binary t-SNARE complex assembly, and primes it for interaction with endolysosome-localized VAMP8 to promote membrane fusion between autophagosomes and endolysosomes. The membrane-tethering activity of ATG14 probably stabilizes fusion intermediates and facilitates fusion progression. This fusion process is probably tightly regulated and requires other protein factors23–25. Dissecting this regulated machinery will be essential to understand the molecular mechanisms of autophagy, and it will facilitate the development of therapeutics specifically to modulate autophagy in human diseases resulting from its dysregulation.
METHODS
Statistical analysis
In general, data shown in column graphs represent the mean ± s.d. or ± s.e.m., as indicated in the figure legends. Sample sizes were chosen after estimating effect size. To determine the group size necessary for adequate statistical power, power analysis was performed using preliminary data sets. Data were analysed for statistical significance after at least three repeated experiments. No data were excluded. The counts were averaged over the 20 images at random locations in each sample. Statistical analysis used Excel (Microscoft).
For the single-vesicle/liposome-tethering assay (Figs 2a and 4e and Extended Data Figs 3c–h and 5d, g), the counts were averaged from at least 15 images at random locations in each sample channel. No spots were excluded. For each set of comparisons between different conditions and/or mutants, the same protein and polyethylene glycol (PEG) surface preparations were used, and the experiments were run in separate channels on the same quartz slide with immobilized liposomes. Although there was some variation in absolute numbers of counts, the relative differences were statistically similar for different experiments. In all panels, error bars are standard deviations from at least 15 random imaging locations in the same sample channel.
Antibodies and cell lines
Antibodies used in this study included anti-ATG14 (#5504, Cell Signaling), anti-Flag M2 (F3165, Sigma), anti -HA (12CA5, Roche), anti-EGFP (GL-8, Clontech), anti-LC3 (7543, Sigma), anti-p62 (PM045, MBL), anti-ATG16 (PM040, M150-3, MBL), anti-STX17 (HPA001204, Sigma), anti-beclin 1 (sc-11427, Santa Cruz), anti-Tom20 (Ab78547, Abcam), anti-LAMP2 (sc-18822, Santa Cruz), anti-Myc (9E10, DSHB) and anti-β-tubulin (E7, DSHB). U2OS and HEK293T were described before25.
(Proteo-)liposome preparation
The lipids used in this study were phosphatidylcholine (POPC), phosphatidylethanolamine (POPE), biotinylated phosphatidylethanolamine (biotin-PE) and PI3P (Avanti Polar Lipids). Also, 2–4 mol% 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) or 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate (DiD) (Molecular Probes) were used as lipid dyes for the v- and t-proteoliposomes, respectively. The lipid mixtures POPE:POPC:DiI (molar ratio 20:78–76:2–4) or POPE:POPC:PI3P:DiI (molar ratio 20:76–74:2:2–4) were solubilized in chloroform and then dried to form a lipid film on the wall of a glass tube. The same procedures were applied to prepare another aliquot of lipid solution containing the same ratio of PE, PC, DiD, supplemented with 0.1 mol% biotinylated lipid (or with 2% PI3P). The dried lipid film was resuspended in proteoliposome buffer (20 mM HEPES, 100 mM KCl, pH 7.4). After five freeze–thaw cycles, unilamellar proteoliposomes were extruded through polycarbonate filters (50 nm pore size, Avanti Polar Lipids) at least 39 times.
Single vesicle/liposome-tethering assay
All surfaces were coated with PEG to eliminate non-specific binding of liposomes. A PEG-coated quartz slide was assembled into a flow chamber and coated with neutravidin (0.2 mg ml−1). A detailed protocol for this step has been previously described13. Briefly, the protein-free acceptor-dye DiD-labelled liposomes were immobilized on the PEG-coated surface during a 30-min incubation at ambient temperature (~25 °C). After two rounds of 200 μl buffer washing, protein-free donor-dye DiI-labelled liposomes (100–200 nM) with or without ATG14 (540 nM, unless specified otherwise) were injected into the flow chamber and placed in a 37 °C incubator for 30–120 min before buffer washing. The incubation time was long enough to reach equilibrium; no detectable increase of spot numbers was observed with a longer incubation time. For the single-vesicle/liposome-tethering assay (Extended Data Fig. 3a), the number of DiI–liposome interactions (tethering) was determined by counting the number of fluorescent spots from DiI dyes in the green detection channel upon excitation at 532 nm (ref. 13). The counts were averaged from at least 15 images at random locations in each sample channel. For each set of comparisons between different conditions and/or mutants, the same ATG14 and PEG surface preparations were used, and the experiments were run in separate channels on the same quartz slide with immobilized DiD-labelled liposomes. This assay was used in Figs 2a and 4e (concentration of ATG14 and mutants: 360 nM) and Extended Data Figs 3c–h and 5d, g.
Single vesicle/liposome FRET-based lipid-mixing assay
The setup and illumination scheme was similar to the single-vesicle/liposome-tethering assay described above, except that FRET to the acceptor (DiD) dyes from the excited donor dyes (DiI) was observed in the red channel (Extended Data Fig. 3b). The donor intensity bleed-through factor (BF) into the acceptor channel was measured as 15% for our setup. The FRET efficiency was then calculated as:
where is the ten-frame-averaged intensity value of acceptor dye emission upon excitation of the donor dye, and is the ten-frame-averaged intensity value of donor dye emission upon excitation of the donor dye13. This assay was used in Fig. 2b.
SNARE protein reconstitution
SNARE proteins were reconstituted by using the direct method described in ref. 13. Donor-dye and acceptor-dye proteoliposomes were reconstituted with autophagic t-SNAREs (STX17/SNAP29) and v-SNARE (VAMP8), respectively. SNAP29 and STX17 were mixed at a 1.5:1 molar ratio and incubated at 25 °C for 1 h to allow complex formation before reconstitution. The SNARE proteins and proteoliposomes were mixed together at the desired lipid to membrane-anchored protein (L/P) ratio of approximately 100–200. Then the mixture containing ~0.8 wt% octyl glucoside in the buffer was kept at 4 °C for 20 min. The mixture was diluted two times with proteoliposome buffer (20 mM HEPES, 100 mM KCl, pH 7.4), and dialysed against 2 l proteoliposome buffer at 4 °C for overnight. The same protocol was applied for proteoliposomes used in ensemble content-mixing experiments except that no lipid dyes were included and self-quenched sulphorhodamine B (50 mM) was encapsulated in v-SNARE (VAMP8) proteoliposmes18. For the experiments with neuronal SNAREs, proteoliposomes were reconstituted with t-SNAREs (syntaxin-1A/SNAP25) at an L/P ratio of 200 and v-SNARE (synaptobrevin-2/VAMP2) at an L/P ratio of 200, both at 0.1 mM lipid concentration.
Ensemble lipid/content-mixing assays
Protein-reconstituted t- and v-SNARE proteoliposomes were mixed at a molar ratio of 1:1. The ensemble lipid-mixing experiments were performed with DiI donor-dye and DiD acceptor-dye labelled t-SNARE and v-SNARE proteoliposomes, respectively, using the protocol described in ref. 26. Briefly, donor dyes were excited with 530 nm laser light. Emission fluorescence intensity was monitored in two channels, at 570 and 670 nm. Lipid mixing was measured as the fluorescence emission (670 nm) of DiD acceptor dyes arising from FRET upon excitation of DiI dyes with 530 nm light.
For the ensemble content-mixing assay, self-quenched sulphorhodamine B molecules encapsulated in v-SNARE proteoliposomes were used as a content indicator18. Content mixing was measured by an increase of fluorescence emission at 570 nm of the sulphorhodamine B dyes upon excitation with 530 nm laser light that results as the initially self-quenched dye is diluted upon complete fusion between labelled v-SNARE and unlabelled t-SNARE proteoliposomes.
Fluorescence emission was recorded with a Varian Cary Eclipse model fluorescence spectrophotometer using a quartz cell of 100 μl with a 5 mm path length. All lipid-mixing measurements were performed at 35 ±2 °C, whereas content-mixing measurements were performed at ambient temperature (~25 °C). The ATG14 concentrations used for the lipid- and content-mixing assays were 1 μM and 360 nM, respectively. The ensemble lipid-mixing assay was used in Figs 2d and 4f and Extended Data Fig. 5c, e. The lipid-mixing traces in these figures were normalized to the value at 1,800 s of the SNAREs-only trace. The ensemble content-mixing assay was used only in Fig. 2e.
Cryo-electron microscopy
Proteoliposomes reconstituted with autophagic SNARE proteins at an L/P ratio of 800 were incubated with or without Atg14 (54 nM) at 37 °C for 3 h. Samples were centrifuged at 800g. for 2 min to remove large aggregations. Frozen-hydrated samples were prepared using the procedures described previously18. Samples were imaged in low-dose conditions using a TF20 electron microscope (FEI) operating at 200 kV. Images of both conditions were collected at a nominal magnification of ×29,000 and an under-focus of 2.5–3.5 μm on a TemCam-F416 CMOS camera (TVIPS GmbH, 4,096 pixels ×4,096 pixels). Bar graphs of the percentage of hemifused or clustered proteoliposomes (Fig. 2g) were obtained by visual inspection of 20 micrographs, using 1,079 and 661 observed proteoliposomes without and with ATG14, respectively.
Insect cell recombinant protein purification
WT and mutant ATG14 proteins were purified from sf9 cells as described before15,27. Briefly, pFASTBACs containing the desired protein sequence were transformed into DH10α-competent cells to purify bacmid DNA. White colonies yielding bacmid DNA were selected from agar plates (50 μg ml−1 kanamycin, 7 μg ml−1 gentamicin, 10 μg ml−1 tetracycline, 100 μg ml−1 Blue-gal, 40 μg ml−1 IPTG). Baculovirus generated from cells containing bacmid DNA were then used to infect sf9 cells to reach an eventual culture volume of 1.0 l. Cells were harvested by centrifugation, resuspended in hypotonic buffer (20 mM Tris-HCl, pH 7.5, 5 mM KCl, 1 mM MgCl2, 1 mM DTT, protease inhibitors) on ice for 15 min, followed by lysis by using a dounce homogenizer with a pestle (Wheaton) 30 times. High-salt buffer (20 mM Tris-HCl, pH 7.5, 250 mM NaCl, 0.5 mM DTT, 20 mM Glycerol) was added at a 1:1 volume ratio to return the lysate to physiological salt levels. Cell lysate was first incubated with IgG beads to allow conjugation of the ZZ tag to beads. Bound protein was eluted using tobacco etch virus (TEV) protease cleavage. Protein purity levels were assessed with SDS–PAGE and Coomassie staining using standard protocols.
Full-length STX17, VAMP8 and SNAP29 expression and purification
All DNAs encoding human proteins were expressed and purified according to the detailed protocol previously reported28 with modifications. Briefly, WT STX17, VAMP8 and SNAP29 were expressed individually as N-terminal GST-fused proteins from pGEX-4T-1 (GE Healthcare) in BL21 (DE3) at 25 °C. After binding to Glutathione Sepharose 4 Fast Flow beads (GE Healthcare), the proteins were eluted by overnight cleavage with TEV protease. In a separate purification scheme, WT His-tagged SNAP29 was co-expressed with STX17 from pETDuet-1 (Novagen) in BL21 (DE3) at 25 °C, and His-tagged VAMP8 was purified in the system separately. His6-tagged SNAP29 and co-expressed STX17 were then extracted from membrane fractions in the presence of 110 mM octyl glucoside and purified using Ni2+-NTA agarose (Qiagen). His6-tag removal from purified SNAP29 was performed by TEV protease cleavage. All these three cleaved proteins (STX17, VAMP8 and SNAP29) were subjected to SEC using a Superdex 200 10/300 column (GE Healthcare) in 110 mM octyl glucoside, 20 mM HEPES, pH 7.4, 300 mM NaCl and 1 mM TCEP, respectively. For the in vitro binding assay for ATG14 and autophagic SNAREs, the STX17–SNAP29 binary t-SNARE complex or STX17–SNAP29–VAMP8 ternary complex was assembled in vitro and separated by SEC. Their binding to ZZ–Flag–ATG14 was then tested in an IgG pull-down experiment followed by a TEV cleavage assay.
Cloning, expression and purification of the autophagic SNARE complex used for crystallization
The SNARE domains of VAMP8 (10–74) and STX17 (164–227) were cloned into the pACYCDuet-1 vector, with the VAMP8 insert between BamHI and SalI restriction sites containing an engineered TEV protease cleavage site at the N terminus, and with the STX17 insert between NdeI and XhoI restriction sites, respectively. The SNARE domains of SNAP29 (39–116, 194–258) were cloned into the pETDuet-1 vector, with the former fragment inserted between NcoI and SalI restriction sites and the latter fragment inserted between NdeI and XhoI restriction sites, respectively. The two plasmids were co-transformed to E. coli BL21 (DE3) cells and expressed at 37 °C using auto-inducing LB medium28. After lysis and centrifugation, the cell lysate was loaded onto a 5-ml Ni-NTA agarose column (Qiagen), washed with 100 ml wash buffer (50 mM NaPi, pH 8.0, 300 mM NaCl, 20 mM imidazole), and eluted with elution buffer (wash buffer supplemented with 500 mM imidazole). Fractions containing the autophagic SNARE complex were pooled and concentrated using an Amicon Ultra-15 centrifugal filter with 10-kDa molecular mass cut off (Millipore) to reduce the volume. The complex was then purified on a Superdex 75 16/60 column (GE Healthcare) that was equilibrated with SEC buffer 1 (50 mM NaPi, pH 8.0, 100 mM NaCl, 0.5 mM TCEP). Fractions containing the autophagic SNARE complex were pooled, supplemented with TEV protease, and then dialysed at 4 °C overnight against anion exchange buffer A (50 mM HEPES, pH 7.5, 50 mM NaCl and 0.5 mM TCEP). The TEV protease digested complex was then loaded onto a MonoQ 10/10 anion exchange column (GE Healthcare) and eluted with a linear NaCl gradient up to 500 mM to remove TEV protease. Fractions containing autophagic SNARE complex were pooled and purified on a Superdex 75 10/300 column (GE Healthcare) that was equilibrated with SEC buffer 2 (50 mM HEPES, pH7.5, 150 mM NaCl and 0.5 mM TCEP) for a final purification. The purified autophagic SNARE complex was concentrated to 0.83 mg ml−1 on the basis of 280 nm ultraviolet absorption and an extinction coefficient of 22460 M−1 cm−1.
Crystallization, data collection and structure determination of the autophagic SNARE complex
Crystals were obtained at 22 °C by the hanging-drop vapour diffusion method in a mother liquor containing 0.1 M Bicine, pH 8.5 and 55% 2-methyl-2,4-pentanediol (MPD). The crystals were allowed to grow for 2 weeks before flash freezing in liquid nitrogen. X-ray diffraction data were collected at the Northeastern Collaborative Access Team beamline 24-ID-C at Advanced Photon Source (Extended Data Table 1) using a beam corresponding to a wavelength of 0.9792 Å. The diffraction data were integrated and scaled using the XDS/XSCALE program packages29. The structure was determined by molecular replacement using the program PHASER30 and a poly-alanine search model derived from the crystal structure of the neuronal SNARE complex (Protein Data Bank accession number 1SFC). The model was built using COOT31 and was refined using PHENIX32. In the final model, all residues (100%) fall into the favoured regions of Ramachandran plot. Coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under accession number 4WY4. In Extended Data Fig. 4, neuronal (1SFC)33, yeast (3B5N)34, early endosomal (2NPS)35 and endosomal (1GL2)36 SNARE structures and electrostatic potential surfaces are shown in two different orientations, and compared with the autophagic SNARE complex structure.
Expression and purification of full-length neuronal SNAREs
Full-length rat syntaxin-1A and VAMP2 were prepared as previously described37. The proteins were expressed separately in E. coli with an N-terminal, TEV protease-cleavable, hexa-histidine tag fused to syntaxin using plasmid pJEXPRESS414, and to VAMP2 using plasmid pTEV5 (ref. 38). Protein was expressed overnight at 25 °C in auto-inducing media28 in E. coli strain C43 (ref. 39). Cell pellets from 8 l of culture were suspended in 500 ml of 1× phosphate-buffered saline, 5 mM EDTA and 1 mM phenylmethylsulphonyl fluoride (PMSF) supplemented with Complete Protease Inhibitor Cocktail tablets (Roche), and broken by three passes through a M-110-EH microfluidizer (Mircrofluidics) at 15,000 p.s.i. Inclusion bodies were removed by two consecutive 10 min spins at 15,344g. in a JA-14 rotor (Beckman Coulter), and the membrane fraction collected by centrifugation at 125,000g. for 2 h in a Ti-45 (Beckman Coulter) rotor. Membranes containing syntaxin-1A were further washed with a buffer containing 10 mM Tris-H2SO4, pH 7.5, 10 mM EDTA, 10% glycerol (w/v), centrifuged at 125,000g. for 1 h in a Ti-45 rotor. Membrane pellets were resuspended in 20 mM HEPES, pH 7.5, 500 mM NaCl, and 1 mM TCEP, and 10% glycerol (w/v), and centrifuged for an additional 1 h in the same rotor. Membranes were suspended to a concentration of 5 mg of protein per millitre in 20 mM HEPES, pH 7.5, 500 mM NaCl, 1 mM TCEP, 10 mM imidazole, and 10% glycerol (w/v), 1 mM PMSF and EDTA-free Complete Protease Inhibitor Cocktail. Dodecylmaltoside (Anatrace) was added to 2%, and after incubation at 4 °C for 1 h, the sample was centrifuged for 35 min at 125,000g in a Ti-45 (Beckman Coulter) rotor, and the supernatant loaded onto a 1 ml column of Nickel-NTA agarose (Qiagen). The column was washed with 20 mM HEPES, pH 7.5, 300 mM NaCl, 1 mM TCEP, 20 mM imidazole, 110 mM octyl glucoside and 10% glycerol (w/v), and the proteins were eluted in the same buffer containing 450 mM imidazole and 1 M NaCl. EDTA (1 mM) was immediately added to the pooled fraction, and loaded onto a Superdex 200 HR 10/300 GL (GE Healthcare) that was equilibrated with 20 mM HEPES, pH 7.5, 300 mM NaCl, 1 mM TCEP, 110 mM octyl glucoside, and 10% glycerol (w/v). Protein fractions were pooled, and digested with 0.2 mg ml−1 TEV protease for 30 min at ambient temperature, after which the TEV protease had precipitated. TEV was removed by centrifugation at 4,500g for 10 min in an Eppendorf model 5804 R tabletop centrifuge (Eppendorf North America). The sample in the supernatant was again digested with 100 mg ml−1 TEV protease for 30 min at ambient temperature, after which the reaction was complete and the TEV protease had precipitated. TEV was removed by centrifugation at 4,500g for 10 min in the same rotor. Cysteine-free SNAP25 was expressed with an N-terminal TEV protease cleavable hexa-histidine tag from plasmid pTEV542. Proteins were expressed overnight at 25 °C in autoinducing media32 in E. coli strain BL21(DE3) at 30 °C. Cells from 4 l of culture were resuspended in 200 ml of 50 mM sodium phosphate pH 8.0, 300 mM NaCl, 20 mM imidazole (bufferA) supplemented with 1 mM PMSF and four EDTA-free protease inhibitor cocktail tablets (Roche). Cells were lysed by three passes through a Emulsiflex C5 homogenizer (Avestin) at 15,000 p.s.i. The lysate was clarified by centrifugation in the Ti45 rotor (Beckman Coulter) for 1.5 h at 125,000g The supernatant was bound to a 5 ml Nickel NTA column (Qiagen) by flowing the lysate on to the column at 1 ml min−1 using the Akta prime (GE Healthcare). The column was washed with 150 ml of buffer A supplemented with 50 mM imidazole and eluted with buffer A supplemented with 350 mM imidazole. Protein-containing fractions were combined, DTT was added to 5 mM, EDTA was added to 1 mM and 150 μg of TEV protease was added to remove the hexa-histidine tag. This mixture was dialysed against buffer B (20 mM HEPES pH 7.5, 100 mM NaCl, 4 mM DTT) overnight at 4 °C. The TEV-cleaved SNAP25 was concentrated in a 15 ml Amicon Ultra centrifugal concentrator with a 10,000 molecular mass cutoff membrane (Millipore) to 5 ml and injected on the Superdex 200 (16/60) column (GE Healthcare) equilibrated in buffer B. Protein-containing fractions were combined, the concentration of SNAP25 measured by absorbance at 280 nm and aliquots were frozen in liquid nitrogen.
Cell culture, cell transfection and cell lysate preparation
293T and U2OS cells were cultured in DMEM (Gibco) supplemented with 10% FBS (HyClone) and 1% Penicillin-Streptomycin Solution (Gibco). Tet-approved FBS (Clontech) was used for tetracycline-inducible short hairpin RNA cells. Cell transfection was performed using Lipofectin 2000 (Invitrogen) or PEI (Polysciences) according to protocols provided by manufacturers. Whole-cell lysates used for immunoprecipitation and immunoblotting were prepared in tandem affinity purification buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% Igepal CA-360, 1 mM NaF, 1 mM Na3VO4, 1 mM EDTA and a protease inhibitor mixture) on ice. Unless used for further immunoprecipitation, lysates were mixed with SDS loading buffer (200 mM Tris-HCl, pH 6.8, 400 mM DTT, 16% β-mercaptoethanol, 8% SDS, 2× loading dye base (Amresco) and 40% glycerol), boiled for 10 min and subjected to SDS–PAGE and western blotting according to standard protocols.
Immunoprecipitation assays
Cells were lysed using tandem affinity purification buffer as described above. Whole-cell lysates (input) were collected after pelleting cellular debris using centrifugation. Lysate was then incubated with a 20:1 ratio of lysate to anti-Flag M2 Affinity Gel (Sigma) for 16 h at 4 °C. Beads were washed three times with tandem affinity purification buffer and then eluted with a 1:10 ratio of 200 μg ml−1 Flag peptide to lysate and subjected to SDS–PAGE. Immunoblotting was performed following standard procedures.
Immunofluorescence staining
Cells were transfected according to the protocol described above with noted plasmids or treated with doxycycline either to knock down or to overexpress inducible constructs. Twenty-four hours after transfection, cells were trypsinized and transferred to six-well dishes containing coverslips. After another 24 h, cells were fixed using ice-cold methanol for 4 min at 4 °C. Cells were then washed three times with PBS and blocked with blocking buffer (2.5% BSA +0.1% Triton X-100 in PBS) at 25 °C for 2 h. Cells were incubated with primary antibodies at 4 °C overnight, washed with PBS buffer and then incubated with appropriate secondary antibodies for 2 h at 25 °C. Slides were examined using under a laser scanning confocal microscope (Zeiss LSM 510 META ultraviolet–visible).
Autophagy analysis
Autophagy was induced under starvation or rapamycin treatment. For starvation, cells were washed with PBS three times and incubated with EBSS (Gibco) for 1 h at 37 °C. For rapamycin (LC Laboratories) treatment, cells were incubated with 500 nM rapamycin in complete medium for 16 h at 37 °C. To block autophagy flux, 400 nm bafilomycin A1 (LC Laboratories) was added to complete medium and incubated for 4 h at 37 °C. Autophagy activity was assessed using two approaches: LC3 punctae per cell and LC3-II formation. The amounts of LC3-positive punctae per cell were quantitated manually and averaged. Results are representative of three independent experiments. For LC3-II formation detection, cell lysates were prepared as described above and subjected to standard western blotting protocols using 1:10,000 dilution of the antibody against LC3 (Sigma). Western blotting quantitation was conducted using ImageJ band analysis (NIH), relative LC3-II peak values were normalized to β-tubulin loading control peak values.
Protease protection assay in mammalian cells
Two million human U2OS cells were infected with lentivirus expressing GFP–LC3 for 24 h and treated with chloroquine for 2 h before harvest. The cell pellet was collected by centrifugation at 600g for 5 min and resuspended in 1 ml buffer Q (400 mM sucrose, 200 mM Hepes pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA and 1 mM DTT). Resuspended cells were forced to go through a needle of a 25-gauge syringe 30 times. The resulting solution was centrifuged twice at 500g for 10 min to remove the unbroken cells and cells debris. The supernatant fraction was aliquoted into 300 μl per tube. Trypsin was added to each tube to the final concentration at 10 μg ml−1, along with 0.4% Triton X-100 if desired. The reaction mix was incubated at 30 °C for 25 min, and SDS loading dye was immediately added to stop the reaction. The sample was then boiled for 10 min.
SEC–MALS analysis for ATG14 homo-oligomerization
SEC–MALS was performed by injecting 25 μg of purified recombinant ATG14 on a WTC-030S5 column (Wyatt Technology) that was previously equilibrated in 10 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, 1 mM EDTA, pH 8.0 at a flow rate of 0.5 ml min−1. The eluted sample was monitored by ultraviolet absorption at 280 nm (Jasco UV-975 ultraviolet–visible system), light scattering at 658 nm (HELEOS system, Wyatt Technology) and differential refractometry (Optilab system, Wyatt Technology). The data analyses used ASTRA 6.0 software (Wyatt Technology). The protein absolute molecular mass was calculated in ASTRA 6.0 (Wyatt Technology) assuming a dn/dc value of 0.185 ml g−1 and a theoretical ultraviolet extinction coefficient value of 1.0 ml mg−1 cm−1.
Extended Data
Extended Data Figure 1. Interaction among ATG14, STX17 and beclin 1.
a, Coomassie-stained SDS gel of recombinantly expressed and purified full-length Flag-tagged STX17, SNAP29 and VAMP8. b, Overexpression of Myc-ATG14 stabilizes the STX17–SNAP29 binary t-SNARE complex in a co-immunoprecipitation assay. c, ATG14 interacts with the SNARE core domain of STX17 in a co-immunoprecipitation assay. d, The CCD domain of ATG14 interacts with STX17 in an co-immunoprecipitation assay. e, Fractionation of beclin 1, ATG14, STX17 with or without chloroquine treatment by Superdex 200. f, STX17 does not associate with beclin 1. Flag-tagged STX17 was co-transfected with either Myc-ATG14 or Myc-beclin 1 in HEK293T cells. ATG14 but not beclin 1 co-immunoprecipitated with STX17. The immunoprecipitation efficiency (immunoprecipitation/input) was normalized by the ratio of immunoprecipitated ATG14 or beclin 1 versus their inputs. g, STX17 interacts with ATG14 in a complex that is distinct from the beclin 1/PI3KC3 complex. Cell lysates from U2OS cells stably expressing Flag–beclin 1 or ATG14 were immunoprecipitated with anti-Flag M2 beads, and endogenous beclin 1, ATG14 and STX17 were detected in the immunoprecipitates.
Extended Data Figure 2. Co-localization of ATG14 and STX17 on autophagosomes upon chloroquine or bafilomycin A1 treatments.
a, b, GFP–ATG14 was co-transfected with STX17–Flag in chloroquine- (a) or bafilomycin-A1-treated (b) U2OS cells, and detected by GFP fluorescence and anti-Flag antibody in immunostaining. Endogenous LC3, LAMP1 or Atg16 were detected by anti-LC3, LAMP2 or Atg16 antibodies in immunostaining (n =20). Scale bars, 5 μm.
Extended Data Figure 3. Characterization of ATG14-mediated membrane tethering.
a, Scheme of the single-vesicle/liposome-tethering assay13,14. DiD-labelled liposomes were attached to the imaging surface through the interaction between biotin/NeutrAvidin; surface binding was assessed by red laser excitation (633 nm). Tethered DiI-labelled liposomes were detected by green laser excitation (532 nm). This assay was used in Figs 2a and 4e and Extended Data Figs 3c–h, 5d and 5g. b, Scheme of the FRET-based single-vesicle/liposome lipid-mixing assay13. Tethered DiI-labelled liposomes were excited by illumination with a green laser (532 nm). Detection of emission in both green and red spectral regions was performed simultaneously by using a dichroic beam-splitter. The total number of tethered DiI-labelled liposomes were counted in the green fluorescence channel, and FRET to the DiD acceptor dyes was observed in the red fluorescence channel. This assay was only used in Fig. 2b. The field of view is 45 μm ×90 μm. See Methods for more details. c, The BATS domain deletion mutant of ATG14 (900 nM) does not promote liposome tethering. Top panels: quantitation. Bottom panels: representative fluorescence images of tethered liposomes (n =15). d, The BATS domain alone does not promote liposome tethering. Top panels: quantitation. Bottom panels: representative fluorescence images of tethered liposomes (n =15). e, Purified recombinant ADP-ribosylation factor GTPase-activating protein 1 (ARFGAP1, 870 nM) does not promote liposome tethering. Top panels: quantitation. Bottom panels: representative fluorescence images of tethered liposomes (n =15). f, Purified recombinant endophilin 1/Bif 1 (2 μM) does not promote liposome tethering. Top panels: quantitation. Bottom panels: representative fluorescence images of tethered liposomes (n =15). g, Incorporation of 2% PI3P into small liposomes (with a 50 nm diameter) failed to enhance the liposome-tethering activity by ATG14. Top panels: quantitation. Bottom panels: representative fluorescence images of tethered liposomes (n =15). h, Incorporation of 2% PI3P into large liposomes (with a 400 nm diameter) enhances the liposome-tethering activity by ATG14. Top panels: quantitation. Bottom panels: representative fluorescence images of tethered liposomes (n =15). Results in c–h are presented as the mean (±s.d.) of random imaging locations (n =15) in the same sample channel.
Extended Data Figure 4. Structure of the autophagic SNARE complex.
a, Boundaries of SNARE domains in VAMP8, STX17 and SNAP29. b, Purification of the autophagic SNARE complex. The four SNARE core domains of VAMP8, STX17 and SNAP29 (with two SNARE core domains) were cloned, expressed in E. coli and co-purified (see Methods). The four fragments form a complex that appears as a single peak by SEC. The SDS–PAGE gel of the indicated four eluted fractions shows individual bands corresponding to the four SNARE core domains. Note that VAMP8 (10–74), STX17 (164–277) and SNAP29 (194–258) migrate at approximately the same position. The leftmost lane is a protein ladder with the molecular masses labelled. c, Comparison of SNARE structures. Shown are cartoon representation and surface charge distribution of the autophagic, neuronal, yeast, early endosomal and endosomal SNARE complex structures in two different orientations. The structures were placed such that the carboxy termini of the SNARE complexes are facing to the right. The surface charge distribution was generated using default ‘vacuum electrostatics’ in PyMOL for qualitative
Extended Data Figure 5. Determining the specificity of ATG14 to autophagic SNAREs-mediated membrane fusion and characterizing recombinant ATG14 homo-oligomerization.
a, Scheme of the ensemble content-mixing assay using autophagic SNARE-reconstituted proteoliposomes (v- and t-proteoliposomes). b, Purified recombinant Flag–STX17, His-VAMP2, His-SNAP25 and His-Syntaxin1 were incubated with IgG Sepharose associated with recombinant ZZ–Flag–ATG14, and the ATG14 binding proteins were cleaved by TEV protease and detected by Coomassie blue staining (upper panel). ZZ–Flag–ATG14 bound to IgG Sepahrose was detected by western blotting in the bottom panel. ATG14 binds to STX17 but not to neuronal SNAREs. c, ATG14 had no detectable effect on promoting ensemble lipid-mixing of proteoliposomes reconstituted with neuronal SNAREs (n =3). d, The membrane-tethering activity of ATG14 CCD deletion mutant is largely intact. Shown is the mean number of tethered vesicles (±s.d.) (n =15) in the same sample channel. e, The ATG14 CCD deletion mutant fails to enhance ensemble lipid mixing of proteoliposomes reconstituted with autophagic SNAREs (n =3). f, The oligomeric states of recombinant ATG14 were determined by SEC–MALS. g, Membrane-tethering activities of ATG14 monomer and dimer measured by the single protein-free vesicle/liposome membrane-tethering assay. Shown are the mean numbers of tethered vesicles (±s.d.) (n =15) in the same sample channel. Representative images are shown in the bottom panels (n =15). h, Mapping ATG14 oligomerization sites to its N terminus. Flag-tagged full-length ATG14 or an ATG14 truncating mutant (lacking the first 70 residues) were transfected into HEK293T cells and treated with cross-linking agent DSS (0, 0.1, 0.2, 0.4 mM) for 30 min, then subjected to SDS–PAGE analysis. ATG14 was probed by anti-Flag antibody. i, ATG14 CCD deletion mutant still forms oligomer in the DSS cross-linking assay. j, Purified recombinant ATG14 (36 nM) was boiled in non-reducing SDS sample buffer with 12.5 mM TCEP (lane 2), 25 mM DTT (lane 3) or 2.5% β-mercaptoethanol (lane 4), or mock-treated (lane 1), and the samples were loaded on non-reducing SDS–PAGE and probed for anti-Flag antibody for ATG14. k, Flag–ATG14 was transfected into HEK293T cells and treated with rapamycin (500 nM for 12 h) and/or cross-linking agent DSS (0.2 μM) for 30 min, then subjected to SDS–PAGE analysis. ATG14 was probed by anti-Flag antibody.
Extended Data Figure 6. ATG14 homo-oligomerization is not essential for beclin 1 interaction and PI3KC3 activation.
a, Interaction between Flag-tagged WT ATG14 and mutants with HA-tagged beclin 1 observed in a co-immunoprecipitation assay. b, Purification of Flag-tagged PI3KC3 complex subunits from insect cells. c, Graph showing p150 but not beclin 1 stimulated Vps34 lipid kinase activity using in vitro TLC kinase assays. Shown are the mean intensity values of radioactive PI3P spots (±s.d.) (n =3). AU, arbitrary unit. d, ATG14 enhanced the lipid kinase activity of Vps34–p150–beclin 1. Shown are the mean intensity values of radioactive PI3P spots (±s.d.) (n =3). e, The requirement of beclin 1 and p150 to the promotion effect of ATG14. Shown are the mean intensity values of radioactive PI3P spots (±s.d.) (n =3). f, Purification of ATG14 WT and mutants from insect cells. Two doses of each recombinant protein were loaded. g, Effect of ATG14 WT and mutants on Vps34(V)–p150(P)–beclin 1(B) lipid kinase activity. Shown are the mean intensity values of radioactive PI3P spots (±s.d.) (n =3).
Extended Data Figure 7.
Autophagosome targeting of ATG14 homo-oligomerization-deficient mutants. GFP, GFP–ATG14 WT, GFP–ATG14 C43/46A, GFP–ATG14 C55/58A and GFP–ATG14 4C4A were co-transfected with STX17–Flag in U2OS cells and detected by immunostaining. Endogenous LC3 was detected by anti-LC3 antibody in immunostaining (n =20). Scale bars, 5 μm.
Extended Data Figure 8. Characterization of the subcellular localization of ATG14 HOD mutants.
a, U2OS cells transfected with GFP–ATG14 WT or ATG14 4C4A and mCherry-Sec61β (ER marker) were treated with EBSS for 2 h and co-stained with endogenous Tom20 (mitochondria marker). Both GFP–ATG14 and GFP–ATG14 4C4A were found at the junction of ER (Sec61β) and mitochondria (Tom20) (n =20). Scale bars, 5 μm. b, Accumulation of endogenous LC3 and GFP–STX17, but not mCherry-Atg16, in ATG14 4C4A mutant but not in ATG14 WT or knockdown (KD) cells (n =20). Scale bars, 5 μm. c, ATG14 knockdown U2OS cells stably complemented with ATG14 WT or 4C4A mutant ATG14 were transfected with GFP–ATG16, and endogenous LC3 staining and ATG16 fluorescence were imaged. LC3 but not ATG16 puncta were accumulated in ATG14 4C4A cells (n =20). Scale bars, 5 μm. d, ATG14 knockdown U2OS cells stably complemented with ATG14 WT or 4C4A mutant ATG14 were transfected with GFP–STX17, and endogenous ATG16 staining and STX17 fluorescence were imaged. STX17 but not ATG16 puncta were accumulated in ATG14 4C4A cells (n =20). Scale bars, 5 μm.
Extended Data Figure 9. ATG14 homo-oligomerization is required for autophagosome maturation.
a, mRFP–GFP–LC3, with WT or mutant ATG14, were expressed in U2OS cells, and LC3 puncta (overlapped green and red fluorescence) in ATG14-expressing cells were imaged (n =20). Scale bars, 5 μm. Yellow LC3 signals represent phagophores or autophagosomes before fusion. Red-only LC3 signals represent acidified mature autophagosomes. Un, untreated. Rap, rapamycin. The corresponding quantitative statistical analysis is shown in Fig. 4c. b, Stable expression of Flag–ATG14 or Flag–ATG14 4C4A in ATG14 knockdown U2OS cells. c, Protease protection assay in U2OS ATG14 WT cells treated with chloroquine (2 h). U2OS ATG14 WT cells were infected with lentivirus expressing GFP–LC3 for 24 h and treated with chloroquine 2 h before harvest. The autophagosome-enriched fraction from these cells was isolated by centrifugation, and incubated with trypsin (10 μg ml−1) with and without 0.4% Triton X-100 for 25 min at 30 °C. GFP or GFP–LC3 were detected by western blotting.
Extended Data Table 1.
Statistics of X-ray data collection and refinement for the autophagic SNARE complex
| Autophagic SNARE complex | |
|---|---|
| Data collection | |
| Space group | C2 |
| Cell dimensions | |
| a, b, c (Å) | 82.85, 35.03, 104.2 |
| α, β, γ (°) | 90, 94.09, 90 |
| Resolution (Å) | 51.97-1.40(1.44-1.40)* |
| Rsym or Rmerge | 6.3 (67.1) |
| I/σI | 14.3 (2.5) |
| Completeness (%) | 98.7 (98.1) |
| Redundancy | 6.6(6.8) |
| Refinement | |
| Resolution (Å) | 51.97-1.40 |
| No. reflections | 58386 |
| Rwork/Rfree | 13.6/17.7 |
| No. atoms | |
| Protein | 2317 |
| Water | 299 |
| B-factors | |
| Protein | 26.8 |
| Water | 45.1 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.011 |
| Bond angles (°) | 1.17 |
Values in parentheses are of the highest-resolution shell.
Acknowledgments
We thank Q. Sun, W. Fan, M. Padolina and R. Bellerose for technical assistance, Y. Cheng and S. Wu for help in the cryo-electron microscopy experiments, A. Liang at OCS Microscopy Core of New York University Langone Medical Center for electron microscope analysis, the Northeastern Collaborative Access Team (supported by National Institutes of Health (NIH) P41 GM103403) at the Advanced Photon Source for X-ray data collection, and B. Levine and R. Sumpter for reading the manuscript. The work was supported by grants to Q.Z. from the Welch Foundation (I-1864), the Cancer Prevention & Research Institute of Texas (RP140320), an American Cancer Society Research Scholar Grant (RSG-11-274-01-CCG) and NIH R01 (CA133228), and NIH R01 (R37-MH63105) to A.T.B. The work was partly supported by China Scholarship Council to R.L. This work was also supported by the National Cancer Institute of the NIH under award number 5P30CA142543.
Footnotes
Online Content Methods, along with any additional Extended Data display items and Source Data, are available in the online version of the paper; references unique to these sections appear only in the online paper.
Author Contributions R.L., Y.R., J.Z., L.M.W. and J.L. performed the biological and biochemical experiments characterizing ATG14 function. J.D., Y.L. and R.A.P. performed the in vitro membrane tethering and fusion experiments; M.Z. and Q.Z. determined the crystal structure of the autophagic SNARE complex and performed the cryo-electron microscopy experiments; S.V. performed the SEC–MALS experiments, J.D., R.L., A.T.B. and Q.Z. conceived the project, designed the experiments, analysed the data and wrote the manuscript with the help of all authors.
Author Information Coordinates and structure factor amplitudes have been deposited in the RCSB Protein Data Bank (http://www.rcsb.org) under accession code 4WY4
The authors declare no competing financial interests. Readers are welcome to comment on the online version of the paper.
References
- 1.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 2.Huang WP, Klionsky DJ. Autophagy in yeast: a review of the molecular machinery. Cell Struct Funct. 2002;27:409–420. doi: 10.1247/csf.27.409. [DOI] [PubMed] [Google Scholar]
- 3.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nair U, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Takats S, et al. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013;201:531–539. doi: 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun Q, et al. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci USA. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsunaga K, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nature Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- 11.Zhong Y, et al. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nature Cell Biol. 2009;11:468–476. doi: 10.1038/ncb1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamasaki M, et al. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 13.Diao J, et al. A single vesicle–vesicle fusion assay for in vitro studies of SNAREs and accessory proteins. Nature Protocols. 2012;7:921–934. doi: 10.1038/nprot.2012.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Diao J, et al. Native alpha-synuclein induces clustering of synaptic-vesicle mimics via binding to phospholipids and synaptobrevin-2/VAMP2. eLife. 2013;2:e00592. doi: 10.7554/eLife.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc Natl Acad Sci USA. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. EMBO J. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Farsad K, et al. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J Cell Biol. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyoung M, et al. In vitro system capable of differentiating fast Ca2+-triggered content mixing from lipid exchange for mechanistic studies of neurotransmitter release. Proc Natl Acad Sci USA. 2011;108:E304–E313. doi: 10.1073/pnas.1107900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsunaga K, et al. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimura S, Fujita N, Noda T, Yoshimori T. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol. 2009;452:1–12. doi: 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- 21.Nair U, Thumm M, Klionsky DJ, Krick R. GFP-Atg8 protease protection as a tool to monitor autophagosome biogenesis. Autophagy. 2011;7:1546–1550. doi: 10.4161/auto.7.12.18424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koyama-Honda I, Itakura E, Fujiwara TK, Mizushima N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy. 2013;9:1491–1499. doi: 10.4161/auto.25529. [DOI] [PubMed] [Google Scholar]
- 23.Takats S, et al. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014;25:1338–1354. doi: 10.1091/mbc.E13-08-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 25.Chen D, et al. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell. 2012;45:629–641. doi: 10.1016/j.molcel.2011.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su Z, Ishitsuka Y, Ha T, Shin YK. The SNARE complex from yeast is partially unstructured on the membrane. Structure. 2008;16:1138–1146. doi: 10.1016/j.str.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun Q, et al. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. J Biol Chem. 2011;286:185–191. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr. 1993;26:795–800. [Google Scholar]
- 30.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sutton RB, Fasshauer D, Jahn R, Brunger AT. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution. Nature. 1998;395:347–353. doi: 10.1038/26412. [DOI] [PubMed] [Google Scholar]
- 34.Strop P, Kaiser SE, Vrljic M, Brunger AT. The structure of the yeast plasma membrane SNARE complex reveals destabilizing water-filled cavities. J Biol Chem. 2008;283:1113–1119. doi: 10.1074/jbc.M707912200. [DOI] [PubMed] [Google Scholar]
- 35.Zwilling D, et al. Early endosomal SNAREs form a structurally conserved SNARE complex and fuse liposomes with multiple topologies. EMBO J. 2007;26:9–18. doi: 10.1038/sj.emboj.7601467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Antonin W, et al. A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 2000;19:6453–6464. doi: 10.1093/emboj/19.23.6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Diao J, et al. Synaptic proteins promote calcium-triggered fast transition from point contact to full fusion. eLife. 2012;1:e00109. doi: 10.7554/eLife.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rocco CJ, Dennison KL, Klenchin VA, Rayment I, Escalante-Semerena JC. Construction and use of new cloning vectors for the rapid isolation of recombinant proteins from Escherichia coli. Plasmid. 2008;59:231–237. doi: 10.1016/j.plasmid.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]