Abstract
Background: Hepatic lipase (HL) plays a pivotal role in the metabolism of HDL and LDL. Recent genome-wide association studies have identified common variants in the HL gene (LIPC) associated with HDL cholesterol.
Objective: We tested the effect of a common variant in LIPC on changes in blood lipids in response to weight-loss diets in the Preventing Overweight Using Novel Dietary Strategies Trial.
Methods: We genotyped LIPC rs2070895 in 743 overweight or obese adults aged 30–70 y (61% women) who were assigned to high-fat (40% energy) or low-fat (20% energy) diets for 2 y. We measured serum concentrations of total cholesterol (TC), triglycerides, LDL cholesterol, and HDL cholesterol at baseline and 2 y of intervention.
Results: At 2 y of intervention, dietary fat modified effects of the variant on changes in serum TC, LDL cholesterol, and HDL cholesterol (P-interaction: 0.0008, 0.004, and 0.03, respectively). In the low-fat group, as compared to the G allele, the A allele tended to be related to the decrease in TC and LDL cholesterol concentrations [TC (β ± SE): −5.5 ± 3.0, P = 0.07; LDL cholesterol: −4.8 ± 2.5, P = 0.06] and a lower increase in HDL cholesterol concentrations (β ± SE: −1.37 ± 0.69, P = 0.048), whereas an opposite effect in the high-fat diet group was evident [TC (β ± SE): 7.3 ± 2.7, P = 0.008; LDL cholesterol: 4.1 ± 2.3, P = 0.07], and there was no genetic effect on changes in HDL cholesterol concentrations (P = 0.54).
Conclusion: Dietary fat intake modifies the effect of a common variant in LIPC on changes in serum lipids during a long-term weight-loss intervention in overweight or obese adults. This trial was registered at clinicaltrials.gov as NCT00072995.
Keywords: hepatic lipase, LIPC, high-fat diet, gene-diet interaction, weight-loss intervention
Introduction
Dyslipidemia, such as low HDL cholesterol, and high LDL cholesterol and TGs, has been well associated with increased cardiovascular risk. Hepatic lipase (HL)11, which is expressed in the liver, plays a pivotal role in the metabolism of lipoproteins as a lipolytic enzyme that hydrolyzes TGs and phospholipids in chylomicron remnants, intermediate-density lipoprotein, and HDL. HL has a multifunctional effect on the development of atherosclerosis. Previous studies support proatherogenic and antiatherogenic functions for HL (1).Candidate gene studies have reported that variation in the HL gene (LIPC) affected the effectiveness of the therapy in reducing the risk of cardiovascular disease (2, 3). Recent genome-wide association studies have identified and confirmed a common variant rs2070895 in LIPC that is associated with HDL cholesterol concentrations (4, 5), probably through affecting the activity of HL. A family study showed that the total allelic variations at LIPC might account for up to 25% of the total variance in plasma HDL cholesterol concentrations (6).
Several previous studies found significant interaction between LIPC genetic variants and dietary fat intake on the blood concentrations of lipids such as HDL cholesterol (7–11). However, those studies were largely observational in nature and might be biased by reverse causation and confounding. To our knowledge, no study has assessed the long-term effect of LIPC variants on changes in lipid profile and the potential interactions with dietary fat intake in randomized clinical trials.
In the present study, we tested the effects of the common variant rs2070895 in LIPC on changes in blood lipid concentrations in response to a 2-y diet intervention in a randomized clinical trial, the Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) Trial, and particularly examined the gene-diet interactions.
Methods
Participants.
The POUNDS LOST Trial (NCT00072995) was designed to compare the effects of energy-reduced diets with different compositions of macronutrients on body weight during 2 y of follow-up. The study design has been described in detail elsewhere (12, 13). Briefly, 811 overweight and obese subjects (25 ≤ BMI < 40 kg/m2) aged 30–70 y with no diabetes or unstable cardiovascular disease, no use of medications affecting body weight, and insufficient motivation to take part in the trial, were randomly assigned to 4 diets; the target percentages of energy derived from fat, protein, and carbohydrate in the 4 diets were respectively 20%, 15%, and 65%; 20%, 25%, and 55%; 40%, 15%, and 45%; and 40%, 25%, and 35%. Thus, 2 diets were low fat (20%) and 2 diets were high fat (40%), or 2 diets were average protein (15%) and 2 diets were high protein (25%), which constituted a 2-by-2 factorial design. The nutrient goals of the 4 diets were to be low in saturated fat and cholesterol and high in fiber. In detail, the diets included 8% saturated fat for each group; 6% monounsaturated fat for low-fat and 22% for high-fat groups; 6% polyunsaturated fat for low-fat and 10% for high-fat groups; at least 20 g dietary fiber/d; and ≤150 mg cholesterol/1000 kcal. Carbohydrate-rich foods with a lower glycemic index were used. Each participant’s diet prescription represented a 750-kcal/d deficit from baseline, which was calculated from the person’s resting energy expenditure and activity level. After 2 y, 80% of the participants (n = 645) completed the trial. The study was approved by the human subjects committee at the Harvard School of Public Health and Brigham and Women’s Hospital and the Pennington Biomedical Research Center of the Louisiana State University System and by a data and safety monitoring board appointed by the National Heart, Lung, and Blood Institute. All participants gave written informed consent.
The present study reported the secondary outcomes of the POUNDS LOST Trial. We included 743 participants’ genotype data available at baseline in the current study. There was no significant difference in the baseline characteristics between the participants with and without genotype data.
Assessment of outcomes and covariates.
Body height was measured at baseline. Body weight and waist circumference were measured in the morning before breakfast at each intervention visit. Blood pressure was measured on 2 d, at baseline, 6 mo, and 2 y, by using an automated device (HEM-907XL; Omron). Dietary intake was assessed in a random sample of 50% of the participants by a review of 5-d diet record at baseline and by 24-h recall during a telephone interview on 3 nonconsecutive days at 6 mo and 2 y. Respiratory quotient was measured at baseline and during follow-up at 6 mo and 2 y as a biomarker of adherence to the assigned diet, i.e., specific carbohydrate, protein, and fat content. Fasting blood samples, 24-h urine samples, and measurement of resting metabolic rate were obtained on 1 d. Concentrations of fasting serum glucose, TG, total cholesterol (TC), HDL cholesterol, and LDL cholesterol and 24-h urinary nitrogen excretion were measured at the clinical laboratory at the Pennington Biomedical Research Center. TG, TC, LDL cholesterol, and HDL cholesterol were measured on the Synchron CX7 (Beckman Coulter). LDL was calculated for each participant according to the following formula: LDL cholesterol = TC − HDL cholesterol − TG / 5 (14), except when TG concentration exceeded 400 mg/dL, in which case LDL cholesterol was measured directly on all samples of the participant. BMI was calculated as weight in kilograms divided by height in squared meters.
Genotyping.
DNA was extracted from the buffy coat fraction of centrifuged blood by using the QIAmp Blood Kit (Qiagen). We genotyped single nucleotide polymorphism (SNP) rs2070895 near LIPC that was associated with the serum lipid concentrations in recent genome-wide association studies (4). Genotyping was performed with use of the OpenArray SNP Genotyping System (BioTrove), and the genotyping success rate was 99%. Replicated quality control samples (10%) were included in every genotyping plate with >99% concordance (15).
Statistical analysis.
We used SAS version 9.1 (SAS Institute, Inc.) to perform the data analysis. Because we aimed to examine the long-term changes in blood lipids during the intervention, the primary outcomes of the analysis were 2-y changes in blood lipids. Baseline data are presented as means ± SDs or medians [IQRs] for the continuous variables and n (%) for the categorical variables. Variables with skewed distribution (serum TG) were log transformed before analysis. The Hardy-Weinberg equilibrium of rs2070895 genotypes and comparison of categorical variables at baseline were accessed by χ2 test. We used ANCOVA to test the differences in continuous variables at baseline, with adjustment for age, sex, and ethnicity groups. The primary outcomes were changes (i.e., the concentrations at 2 y of follow-up time minus the baseline concentrations) in fasting serum TC, TG, HDL cholesterol, and LDL cholesterol over the time the participant remained in the trial. Multivariate general linear models were used to test the main genetic effects and the potential interactions between the genetic variation and diet intervention (high vs. low fat) on changes in weight and lipid profile (TC, TG, HDL cholesterol, and LDL cholesterol). The significance of gene-diet intervention interactions was tested by including the genotype-by-diet interaction multiplicative terms in the models. The covariates included age, sex, ethnicity, baseline BMI, and weight loss, and values for the respective outcome were used in the analysis. Linear mixed models were used to test genetic associations with the trajectory of changes in weight or lipids according to diet groups. Time was treated as a repeated measurement factor, with which genotype-time interaction terms were included in the mixed models. An additive genetic model was analyzed. Because the majority of study participants were white (80%), we also performed sensitivity analysis in white participants. All P values were two-sided. A P value of 0.05 was considered statistically significant.
Results
The mean age of the total participants was 51 y and mean BMI was 32.7 kg/m2. Among the participants, 61% were women, 80% were white, 15% were black, 3% were Hispanic, and 2% were Asian or other ethnic groups. The minor allele (A) frequency of LIPC rs2070895 was 26.4% in total participants, and the genotype distribution fits the Hardy-Weinberg equilibrium.
Table 1 shows baseline characteristics of participants according to the LIPC rs2070895 genotypes. Genotype frequencies were similar between men and women and across diet groups. Significant difference of the genotype distribution was observed among the ethnic groups (P < 0.0001). The A allele of SNP rs2070895 was related to higher concentrations of HDL cholesterol. No other differences in baseline characteristics across the genotype were observed (all P ≥ 0.05). The results were similar in the white participants (Supplemental Table 1). In addition, there were no significant differences in nutrient intakes and biomarkers of adherence to the assigned diet at 2 y across the LIPC rs2070895 genotype in low- and high-fat diet groups.
TABLE 1.
Baseline characteristics of the participants according to LIPC rs2070895 genotypes1
| AA (n = 62) | AG (n = 268) | GG (n = 413) | P2 | |
| Age, y | 47.9 ± 8.3 | 50.5 ± 9.1 | 51.9 ± 9.4 | 0.06 |
| Female | 21 (33.9) | 103 (38.4) | 166 (40.2) | 0.62 |
| Dietary fat group | 0.83 | |||
| Low fat | 30 (8.1) | 138 (37.1) | 204 (54.8) | |
| High fat | 32 (8.6) | 130 (35.0) | 209 (56.3) | |
| Race or ethnic group | <0.0001 | |||
| White | 31 (5.2) | 198 (33.3) | 366 (61.5) | |
| Black | 27 (24.1) | 53 (47.3) | 32 (28.6) | |
| Hispanic | 3 (12.0) | 13 (52.0) | 9 (36.0) | |
| Asian or other | 1 (9.0) | 4 (36.4) | 6 (54.6) | |
| Height, cm | 167 ± 8.5 | 169 ± 9.2 | 169 ± 8.5 | 0.09 |
| Weight, kg | 91.8 ± 14.2 | 93.5 ± 15.2 | 93.3 ± 16.0 | 0.22 |
| Waist circumference, cm | 103 ± 12.5 | 103 ± 12.9 | 104 ± 13.3 | 0.75 |
| BMI, kg/m2 | 33.0 ± 3.8 | 32.7 ± 3.8 | 32.6 ± 3.9 | 0.75 |
| Blood pressure, mm Hg | ||||
| Systolic | 122 ± 16 | 120 ± 13 | 119 ± 13 | 0.05 |
| Diastolic | 77 ± 11 | 76 ± 9 | 75 ± 9 | 0.20 |
| Fasting serum glucose, mg/dL | 93 ± 16 | 91 ± 11 | 92 ± 12 | 0.88 |
| Fasting serum lipid profile, mg/dL | ||||
| TGs | 98.5 [70.0–168] | 121 [80.0–165] | 124 [90.0–193] | 0.17 |
| TC | 205 ± 41.3 | 201 ± 37.1 | 203 ± 36.1 | 0.47 |
| HDL cholesterol | 51.0 ± 14.3 | 49.6 ± 15.6 | 47.9 ± 13.0 | 0.002 |
| LDL cholesterol | 129 ± 34.2 | 125 ± 31.3 | 126 ± 32.2 | 0.99 |
| Dietary intake per day | ||||
| Carbohydrate, % of energy | 45.9 ± 7.5 | 45.2 ± 8.0 | 44.1 ± 7.4 | 0.28 |
| Fat, % of energy | 35.8 ± 6.9 | 37.0 ± 6.3 | 37.1 ± 5.7 | 0.48 |
| Protein, % of energy | 18.3 ± 3.2 | 17.7 ± 3.3 | 18.4 ± 3.4 | 0.17 |
| Energy, kcal | 1881 ± 572 | 2036 ± 539 | 1936 ± 569 | 0.33 |
| Urinary nitrogen, g/d | 11.3 ± 4.6 | 12.2 ± 4.5 | 12.4 ± 4.3 | 0.54 |
| Respiratory quotient | 0.84 ± 0.05 | 0.84 ± 0.04 | 0.84 ± 0.04 | 0.72 |
Values are means ± SDs, medians [IQRs], or n (%). LIPC, hepatic lipase gene; TC, total cholesterol.
P values were calculated by χ2 test for categorical variables and ANCOVA for continuous variables after adjusting for age, sex, and ethnicity.
The weight losses (means ± SDs) in the high- and low-fat diet groups were 4.0 ± 7.3 kg and 4.2 ± 7.6 kg at 2 y, respectively. The corresponding weight losses among the 3 genotypes of LIPC rs2070895 variant were 2.9 ± 4.9 kg in the AA, 3.8 ± 7.6 kg in the GA, and 4.4 ± 7.6 kg in the GG genotype at 2 y, respectively. No main genetic effect or gene-diet interaction was found on weight loss during 2 y of intervention. Genotype did not affect the changes in serum TC, LDL cholesterol, or HDL cholesterol concentrations during 2 y of intervention (all P ≥ 0.35).
We then examined the interaction of dietary fat intake and the genetic variation on changes in serum lipids. At 2 y of intervention, we observed that dietary fat significantly modified the genetic effects of rs2070895 on changes in serum TC, LDL cholesterol, and HDL cholesterol (P-interaction: 0.0008, 0.004, and 0.03, respectively), with adjustment for age, sex, ethnicity, baseline BMI, weight loss, and baseline measurements for the respective outcomes (Figure 1, Table 2). In the low-fat diet group, the A allele was associated with a decrease of TC and LDL cholesterol concentrations [TC (β ± SE): −5.5 ± 3.0, P = 0.07; LDL cholesterol: −4.8 ± 2.5, P = 0.06], whereas an opposite genetic effect was found in the high-fat diet group [TC (β ± SE): 7.3 ± 2.7, P = 0.008; LDL cholesterol: 4.1 ± 2.3, P = 0.07]. Then we tested the independent associations between the genetic variation and lipids. We did not find a significant difference in changes of TC after further adjustment for changes in LDL cholesterol at 2 y in the low-fat or high-fat groups. We found opposite genetic effects on changes in HDL cholesterol concentrations. The A allele was significantly associated with less increase of HDL cholesterol concentrations in the low-fat group (β ± SE: −1.37 ± 0.69, P = 0.048), whereas no association was observed in the high-fat group (β ± SE: 0.44 ± 0.71, P = 0.54). We did not detect any interactions between the genetic variation and dietary fat intake on changes in serum TG at 2 y of intervention (P-interaction: 0.62). Similar interactions were observed when the analysis was restricted to the white participants (Supplemental Table 2).
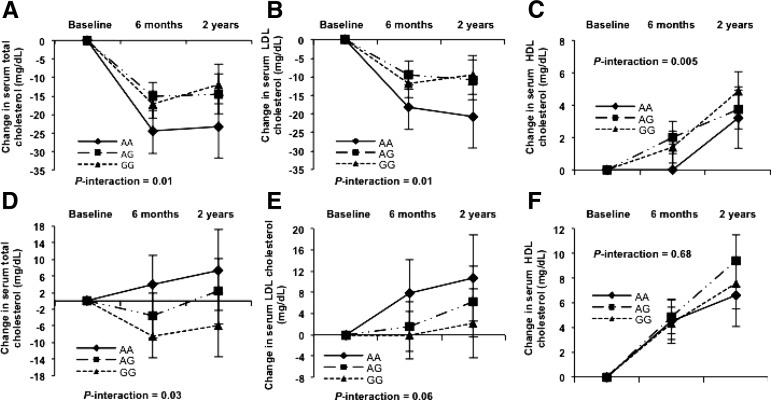
FIGURE 1.
Trajectory analysis on 2-y changes in serum total cholesterol (A, D), LDL cholesterol (B, E), and HDL cholesterol (C, F) by LIPC rs2070895 genotype of participants that consumed the low-fat (upper panels) or high-fat (lower panels) diets. The number of participants in the low-fat diet group (A–C), for AA: n = 30 at baseline, n = 22 at 6 mo, and n = 20 at 2 y; AG: n = 138 at baseline, n = 118 at 6 mo, and n = 101 at 2 y; and GG: n = 204 at baseline, n = 182 at 6 mo, and n = 155 at 2 y. The corresponding numbers in the high-fat diet group (D–F), AA: n = 32, 24, and 20, at baseline, 6 mo, and 2 y, respectively; AG: n = 130, 110, and 98, at baseline, 6 mo, and 2 y, respectively; and GG: n = 209,180, and 149, at baseline, 6 mo, and 2 y, respectively. Values are adjusted means ± SEs after adjusting for age, sex, ethnicity, baseline BMI, weight loss, and baseline values for respective outcomes. P-interactions were derived from the linear mixed models, in which intervention time was treated as a repeated measurement factor, and genotype time as the interaction terms. LIPC, hepatic lipase gene.
TABLE 2.
Dietary fat intakes modulated the genotype effect of the LIPC rs2070895 variant on the changes in lipids in response to the dietary fat intervention at 2 y1
| Low-fat diet | High-fat diet | P-interaction | |
| ΔTotal cholesterol,2 mg/dL | |||
| AA | 20 (−24.7 ± 8.2) | 20 (7.4 ± 9.5) | |
| AG | 101 (−15.0 ± 5.2) | 98 (1.3 ± 7.8) | |
| GG | 155 (−11.2 ± 5.2) | 149 (−6.3 ± 7.6) | |
| P-trend3 | 0.07 | 0.008 | 0.0008 |
| ΔLDL cholesterol,2 mg/dL | |||
| AA | 20 (−21.8 ± 6.8) | 20 (10.7 ± 8.1) | |
| AG | 101 (−11.2 ± 4.3) | 98 (6.3 ± 6.7) | |
| GG | 155 (−8.7 ± 4.3) | 149 (2.1 ± 6.5) | |
| P-trend3 | 0.06 | 0.07 | 0.004 |
| ΔHDL cholesterol,2 mg/dL | |||
| AA | 20 (2.73 ± 1.85) | 20 (5.67 ± 2.42) | |
| AG | 101 (3.32 ± 1.16) | 98 (9.02 ± 1.99) | |
| GG | 155 (4.81 ± 1.16) | 149 (7.09 ± 1.95) | |
| P-trend3 | 0.048 | 0.54 | 0.03 |
Values are n (means ± SEs). LIPC, hepatic lipase gene.
The Δ variables indicate the change of respective outcomes.
P-trend was for general linear regression in which an additive genetic model was analyzed for the rs2070895 variant. The adjustment was including age, sex, ethnicity, baseline BMI, weight loss, and baseline values for respective outcomes.
We then examined the trajectory changes in lipids according to the LIPC rs2070895 genotypes by dietary fat intake over the 2-y intervention period. We observed significant genotype-time interactions on changes in TC, LDL cholesterol, and HDL cholesterol in the low-fat diet group (P-interactions: 0.01, 0.01, and 0.005, respectively). The LIPC genetic effects on TC and LDL cholesterol displayed a cumulative pattern throughout the intervention and reached the maximum values at 2 y (all P-trend values were ≤0.002 for AA and AG genotype, but there were no significant trends for GG genotype), whereas continued cumulative increasing trends were found in HDL cholesterol concentrations (both P-trend values were ≤0.0001 for GG and AG genotype, but there were no significant trends for GG genotype) (Figure 1A–C). In the high-fat diet group, we observed a significant genotype-time interaction on changes in TC (P-interaction: 0.03), a marginal interaction on changes in LDL cholesterol (P-interaction: 0.06), and no significant interaction on changes in HDL cholesterol (P-interaction: 0.68) (Figure 1D–F).
Discussion
In this 2-y randomized weight-loss intervention trial, we observed a significant interaction between the LIPC SNP rs2070895 and dietary fat intake on changes in serum TC, LDL cholesterol, and HDL cholesterol concentrations. The A allele was related to a decrease in TC and LDL cholesterol and a small increase in HDL cholesterol in response to the low-fat diet, whereas an opposite genetic effect was observed when participants were assigned a high-fat diet. The genetic effect on changes of blood cholesterol concentrations showed a long-term, cumulative pattern throughout the intervention.
Our findings are in line with several previous studies. These studies have reported that the effects of −514C/T (rs1800588) of LIPC, which is in high linkage disequilibrium with rs2070895 (r2 = 0.97), on HDL cholesterol concentrations were modified by dietary fat intake (7, 10, 11). Like rs2070895, rs1800588 is located in the promoter region and associated with decreased plasma HL activity and increased HDL cholesterol concentrations (3, 16, 17). In the Framingham Study, the T allele (in linkage disequilibrium with allele A in rs2070895) was associated with significantly higher HDL cholesterol concentrations and large particle size only in subjects consuming 30% of energy from fat (10). The study indicated an intraindividual difference in the plasma lipid response to the dietary fat. Similar results were found in African Americans (16) and subjects of Indian origin living in Singapore (11).
However, few studies have assessed interactions between LIPC genotype and dietary factors in intervention trials (18). In the Finnish Diabetes Prevention Study (18), the A allele was associated with a low conversion rate to type 2 diabetes in the lifestyle intervention group, which was assigned to a reduced fat intake of <30% of energy consumed and reduced intake of saturated fat to <10% of energy consumed. In addition, a greater increase in HDL cholesterol concentration from baseline to 3 y was found in subjects carrying the A allele than those with the G allele. In the current study, the minor allele A was associated with less of an increase in HDL cholesterol in response to a low-fat diet. Our data suggest that LIPC may partly account for the interindividual heterogeneity in HDL cholesterol changes induced by different dietary fat intake, although the underlying mechanisms remain to be determined.
In addition to HDL cholesterol, we also found that dietary fat intake significantly modified the genetic effect of LIPC on changes in TC and LDL cholesterol concentrations during the weight-loss trial. Our results were consistent with previous investigation (19). Lindi et al. (19) reported that individuals with the AA genotype were responsive to the multiunsaturated FA–enriched diet, and their serum LDL cholesterol concentrations decreased more than in subjects with other genotypes. Interestingly, we found that the genetic effect on changes in TC and LDL cholesterol in response to a low-fat diet showed a cumulative pattern throughout the intervention and reached the maximum values at 2 y. The gene-time interactions were less significant in the high-fat diet. These findings suggest that the gene-diet interactions on the changes of cholesterol concentrations might be persistent.
HL plays a pivotal role in lipid metabolism by affecting both reverse cholesterol transport and remodeling of TG-rich lipoprotein particles and, thereby, the formation of atherogenic small dense LDLs (1). Increased HL is associated with smaller and denser LDL and HDL particles that are proatherogenic, whereas decreased HL is associated with larger and more buoyant LDL and HDL particles (20). Previous animal studies showed that the activity of HL in the liver could be depressed by high-fat feeding (21). Dysfunctional HL may associate with elevated blood HDL cholesterol concentrations with varying size of HDL particles and paradoxically increased atherosclerosis risk. In the present study, we observed a greater decrease in TC and LDL cholesterol and a small increase in HDL cholesterol in the A allele carriers assigned to the low-fat diet, which may give suggestive implications in preventive medicine and public health.
To our knowledge, this is the first report about the interactions of the genetic effect of the HL activity determinant gene and dietary fat intake on changes of serum lipids, especially cholesterols, in a long-term, randomized weight-loss intervention trial. Our results indicate that the effect of genetic variation of LIPC on changes of lipid concentrations was modified by dietary fat intake. In contrast to observational studies, the study conditions in randomized clinical trials are controlled directly by the investigators, including the specifically defined dietary intakes. This control minimizes the possibility of bias and increases the level of causality (22).
However, some limitations should be acknowledged. We did not measure the HL activity, which did not allow assessing the direct genetic effect of LIPC on HL. However, a genetic marker could be a surrogate for the biomarker in yielding causal relation according to the Mendelian randomization principle (23, 24). Moreover, previous studies have shown that the minor alleles of these promoter variants (rs2070895 and rs1800588) have been associated with diminished transcriptional activity in vitro (25) and a 15–45% decrease in plasma HL activity in vivo (26). Also, because the participants are all overweight or obese, our results should be cautiously generalized to the general population with normal body weight. Finally, given that 80% of the participants are white, future studies are warranted to verify the gene-diet interactions in other ethnicity groups.
In conclusion, we observed that dietary fat intake significantly modified the LIPC rs2070895 genetic effect on changes in serum TC, LDL cholesterol, and HDL cholesterol concentrations during a 2-y randomized weight-loss intervention trial. Our data suggest that the carriers of A alleles might benefit more in terms of improved lipid profiles by eating a low-fat (20%), high-carbohydrate (55–65%) diet.
Acknowledgments
MX and LQ designed the research; MX and SSN conducted the research and analyzed the data; GAB, DHR, FMS, and GN provided essential materials; MX and LQ wrote the paper; and LQ had primary responsibility for final content. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: HL, hepatic lipase; LIPC, hepatic lipase gene; POUNDS LOST, Preventing Overweight Using Novel Dietary Strategies; SNP, single nucleotide polymorphism; TC, total cholesterol.
References
- 1.Santamarina-Fojo S, Haudenschild C, Amar M. The role of hepatic lipase in lipoprotein metabolism and atherosclerosis. Curr Opin Lipidol 1998;9:211–9. [DOI] [PubMed] [Google Scholar]
- 2.Peters BJ, Pett H, Klungel OH, Stricker BH, Psaty BM, Glazer NL, Wiggins KL, Bis JC, de Boer A, Maitland-van der Zee AH. Genetic variability within the cholesterol lowering pathway and the effectiveness of statins in reducing the risk of MI. Atherosclerosis 2011;217:458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zambon A, Deeb SS, Pauletto P, Crepaldi G, Brunzell JD. Hepatic lipase: a marker for cardiovascular disease risk and response to therapy. Curr Opin Lipidol 2003;14:179–89. [DOI] [PubMed] [Google Scholar]
- 4.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. . Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010;466:707–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraja AT, Vaidya D, Pankow JS, Goodarzi MO, Assimes TL, Kullo IJ, Sovio U, Mathias RA, Sun YV, Franceschini N, et al. . A bivariate genome-wide approach to metabolic syndrome: STAMPEED Consortium. Diabetes 2011;60:1329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen JC, Wang Z, Grundy SM, Stoesz MR, Guerra R. Variation at the hepatic lipase and apolipoprotein AI/CIII/AIV loci is a major cause of genetically determined variation in plasma HDL cholesterol levels. J Clin Invest 1994;94:2377–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Lopez-Ridaura R, Rimm EB, Rifai N, Hunter DJ, Hu FB. Interactions between the −514C→T polymorphism of the hepatic lipase gene and lifestyle factors in relation to HDL concentrations among US diabetic men. Am J Clin Nutr 2005;81:1429–35. [DOI] [PubMed] [Google Scholar]
- 8.Bos G, Dekker JM, Feskens EJ, Ocke MC, Nijpels G, Stehouwer CD, Bouter LM, Heine RJ, Jansen H. Interactions of dietary fat intake and the hepatic lipase –480C→T polymorphism in determining hepatic lipase activity: the Hoorn Study. Am J Clin Nutr 2005;81:911–5. [DOI] [PubMed] [Google Scholar]
- 9.Deeb SS, Zambon A, Carr MC, Ayyobi AF, Brunzell JD. Hepatic lipase and dyslipidemia: interactions among genetic variants, obesity, gender, and diet. J Lipid Res 2003;44:1279–86. [DOI] [PubMed] [Google Scholar]
- 10.Ordovas JM, Corella D, Demissie S, Cupples LA, Couture P, Coltell O, Wilson PW, Schaefer EJ, Tucker KL. Dietary fat intake determines the effect of a common polymorphism in the hepatic lipase gene promoter on high-density lipoprotein metabolism: evidence of a strong dose effect in this gene-nutrient interaction in the Framingham Study. Circulation 2002;106:2315–21. [DOI] [PubMed] [Google Scholar]
- 11.Tai ES, Corella D, Deurenberg-Yap M, Cutter J, Chew SK, Tan CE, Ordovas JM; Singapore National Health Survey. Dietary fat interacts with the -514C>T polymorphism in the hepatic lipase gene promoter on plasma lipid profiles in a multiethnic Asian population: the 1998 Singapore National Health Survey. J Nutr 2003;133:3399–408. [DOI] [PubMed] [Google Scholar]
- 12.Sacks FM, Bray GA, Carey VJ, Smith SR, Ryan DH, Anton SD, McManus K, Champagne CM, Bishop LM, Laranjo N, et al. . Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N Engl J Med 2009;360:859–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi Q, Bray GA, Smith SR, Hu FB, Sacks FM, Qi L. Insulin receptor substrate 1 gene variation modifies insulin resistance response to weight-loss diets in a 2-year randomized trial: the Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) Trial. Circulation 2011;124:563–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972;18:499–502. [PubMed] [Google Scholar]
- 15.Xu M, Qi Q, Liang J, Bray GA, Hu FB, Sacks FM, Qi L. Genetic determinant for amino acid metabolites and changes in body weight and insulin resistance in response to weight-loss diets: the Preventing Overweight Using Novel Dietary Strategies (POUNDS LOST) Trial. Circulation 2013;127:1283–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nettleton JA, Steffen LM, Ballantyne CM, Boerwinkle E, Folsom AR. Associations between HDL-cholesterol and polymorphisms in hepatic lipase and lipoprotein lipase genes are modified by dietary fat intake in African American and White adults. Atherosclerosis 2007;194:e131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isaacs A, Sayed-Tabatabaei FA, Njajou OT, Witteman JC, van Duijn CM. The -514 C->T hepatic lipase promoter region polymorphism and plasma lipids: a meta-analysis. J Clin Endocrinol Metab 2004;89:3858–63. [DOI] [PubMed] [Google Scholar]
- 18.Todorova B, Kubaszek A, Pihlajamäki J, Lindström J, Eriksson J, Valle TT, Hämäläinen H, Ilanne-Parikka P, Keinänen-Kiukaanniemi S, Tuomilehto J, et al. ; Finnish Diabetes Prevention Study. The G-250A promoter polymorphism of the hepatic lipase gene predicts the conversion from impaired glucose tolerance to type 2 diabetes mellitus: the Finnish Diabetes Prevention Study. J Clin Endocrinol Metab 2004;89:2019–23. [DOI] [PubMed] [Google Scholar]
- 19.Lindi V, Schwab U, Louheranta A, Vessby B, Hermansen K, Tapsell L, Riccardi G, Rivellese AA, Laakso M, Uusitupa MI; KANWU Study Group. The G-250A polymorphism in the hepatic lipase gene promoter is associated with changes in hepatic lipase activity and LDL cholesterol: the KANWU Study. Nutr Metab Cardiovasc Dis 2008;18:88–95. [DOI] [PubMed] [Google Scholar]
- 20.Brunzell JD, Zambon A, Deeb SS. The effect of hepatic lipase on coronary artery disease in humans is influenced by the underlying lipoprotein phenotype. Biochim Biophys Acta 2012;1821:365–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cahova M, Dankova H, Palenickova E, Papackova Z, Kazdova L. The opposite effects of high-sucrose and high-fat diet on fatty acid oxidation and very low density lipoprotein secretion in rat model of metabolic syndrome. J Nutr Metab 2012;2012:757205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qi L, Cho YA. Gene-environment interaction and obesity. Nutr Rev 2008;66:684–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi L. Mendelian randomization in nutritional epidemiology. Nutr Rev 2009;67:439–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Voight BF, Peloso GM, Orho-Melander M, Frikke-Schmidt R, Barbalic M, Jensen MK, Hindy G, Hólm H, Ding EL, Johnson T, et al. . Plasma HDL cholesterol and risk of myocardial infarction: a Mendelian randomisation study. Lancet 2012;380:572–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deeb SS, Peng R. The C-514T polymorphism in the human hepatic lipase gene promoter diminishes its activity. J Lipid Res 2000;41:155–8. [PubMed] [Google Scholar]
- 26.Tahvanainen E, Syvanne M, Frick MH, Murtomaki-Repo S, Antikainen M, Kesaniemi YA, Kauma H, Pasternak A, Taskinen MR, Ehnholm C. Association of variation in hepatic lipase activity with promoter variation in the hepatic lipase gene. The LOCAT Study Investigators. J Clin Invest 1998;101:956–60. [DOI] [PMC free article] [PubMed] [Google Scholar]