Abstract
The mammalian heart regenerates poorly, and damage commonly leads to heart failure. Hippo signaling is an evolutionarily conserved kinase cascade that regulates organ size during development and prevents adult mammalian cardiomyocyte regeneration by inhibiting the transcriptional coactivator Yap, which also responds to mechanical signaling in cultured cells to promote cell proliferation. To identify Yap target genes that are activated during cardiomyocyte renewal and regeneration, we performed Yap chromatin immunoprecipitation sequencing (ChIP-Seq) and mRNA expression profiling in Hippo signaling-deficient mouse hearts. We found that Yap directly regulated genes encoding cell cycle progression proteins, as well as genes encoding proteins that promote F-actin polymerization and that link the actin cytoskeleton to the extracellular matrix. Included in the latter group were components of the dystrophin glycoprotein complex (DGC), a large molecular complex that, when defective, results in muscular dystrophy in humans. Cardiomyocytes near scar tissue of injured Hippo signaling-deficient mouse hearts showed cellular protrusions suggestive of cytoskeletal remodeling. The hearts of mdx mutant mice, which lack functional dystrophin and are a model for muscular dystrophy, showed impaired regeneration and cytoskeleton remodeling, but normal cardiomyocyte proliferation after injury. Our data showed that, in addition to genes encoding cell cycle progression proteins, Yap regulated genes that enhance cytoskeletal remodeling Thus, blocking the Hippo pathway input to Yap may tip the balance so that Yap responds to the mechanical changes associated with heart injury to promote repair.
INTRODUCTION
Although some vertebrates, such as zebrafish, can regenerate the heart, heart regeneration in mammals is limited (1, 2). Rather than regenerate, human cardiomyocytes undergo a maladaptive stress response commonly termed “pathologic remodeling,” including fibrosis and scarring, that leads to heart failure, a leading killer worldwide (3, 4). The mammalian heart has a transient regenerative capacity that terminates by postnatal day 7 (P7) in mice (5). This observation has led to the idea that manipulating relevant genetic pathways can therapeutically enhance cardiomyocyte regeneration.
The Hippo signaling pathway is a kinase cascade that links changes in cellular density or mechanical stress to changes in cell proliferation (6). In mammals, Hippo signaling limits heart size and inhibits cardiomyocyte proliferation during development and adult cardiac regeneration (7, 8). The downstream Hippo effector Yes-associated protein (Yap) is a transcriptional cofactor that interacts with transcription factors such as Tead. When Hippo activity is high, Yap is phosphorylated by Lats and is excluded from the nucleus. When Hippo activity is low, such as during early heart development, Yap shuttles into the nucleus where it promotes cardiomyocyte proliferation (6). Yap activity is not only regulated by Hippo kinases but also by mechanical signaling. In cells subjected to high amounts of mechanical stress, Yap is preferentially localized in the nucleus and promotes proliferation (9, 10).
Hippo signaling inhibits adult cardiomyocyte regeneration through Yap. Hippo deficiency due to loss of function of the adaptor protein Salvador (Salv) promotes regeneration in mouse models of apex resection, neonatal myocardial infarction, and adult myocardial infarction (8). Furthermore, overexpression of a constitutively active form of Yap in the myocardium also enhances regeneration in adult cardiomyocytes (11,12).
In this study, we investigated Yap target genes in heart regeneration by performing ChIP-sequencing (ChIP-Seq) analysis and mRNA expression profiling in Hippo-deficient hearts. Our data indicated that Yap directly regulates genes involved in cell cycle progression, genes that promote F-actin polymerization, and genes linking the actin cytoskeleton to the extracellular matrix.
RESULTS
Yap directly regulates genes encoding proteins that control cytoskeletal dynamics and cell proliferation
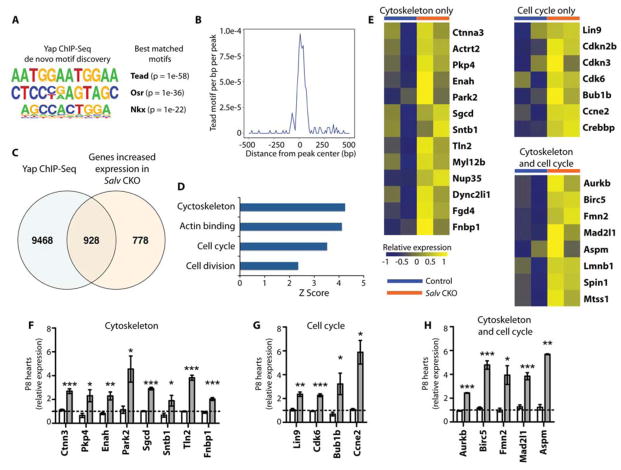
To identify direct transcriptional targets of Yap, we performed ChIP-Seq experiments in Salv conditional knockout (CKO) mutant mouse hearts at P8. We chose P8 because it is a non-regenerative stage in wild-type mouse hearts but is a regenerative stage in Hippo-deficient mouse hearts (8). We predicted that in P8 Hippo-deficient hearts, Yap binding would be enriched for genes that are directly involved in cardiac regeneration. We performed ChIP-Seq experiments with an anti-Yap antibody in dissected Salv CKO mouse hearts and generated libraries that were sequenced by using an Ion Torrent sequencer (13). A total of 25 million Yap ChIP-Seq reads were evaluated with Homer (14). Motif analysis comparing Yap ChIP-Seq reads indicated that Tead binding elements were among the most enriched peaks, which validated the specificity of the ChIP-Seq experiment (Fig. 1, A and B). In addition, we performed mRNA expression profiling of P8 Salv CKO mouse hearts to analyze changes in gene expression in Hippo-deficient hearts (fig. S1). We then compared differentially expressed genes to those in the Yap ChIP-Seq datasets to identify direct transcriptional targets of Yap. Overlay of the ChIP-Seq and mRNA expression profiling datasets revealed that Yap bound to 928 genes that showed increased expression (Fig. 1C). From these data, we generated a list of Yap target genes that included 3 categories: cell cycle progression, cytoskeleton, and both cell cycle and cytoskeleton (Fig. 1D–H).
Figure 1. Integrated genomic analysis for identifying Yap target genes.
(A) Motif analysis for enriched Yap ChIP-Seq peaks (total number = 35,412 from 2 independent biological replicates). De novo motifs and their best matches are shown. (B) The density of Tead binding motifs within a 500-bp distance of the Yap ChIP-Seq peaks is shown. (C) Overlay of genes with increased expression in Salv CKO mouse hearts (total number = 1,706 from 2 mice) and genes annotated from Yap ChIP-Seq peaks (total number = 10,396). (D) Gene ontology analysis of genes with increased expression in Salv CKO mouse hearts and with Yap binding peaks (total number = 928). Enriched terms were calculated by using over-representation statistics and measured by using Z-scores. (E) Heat map of Yap target genes identified by the overlay of microarray and Yap ChIP-Seq genes in the labeled categories. Heat maps show relative expression between Salv CKO and control mouse hearts. (F–H) qRTPCR validation of Yap target genes in P8 control (unshaded bar) and P8 Salv CKO (shaded bar) mouse hearts. N=3 independent biological replicates. *P<0.05; **P<0.01; ***P<0.001.
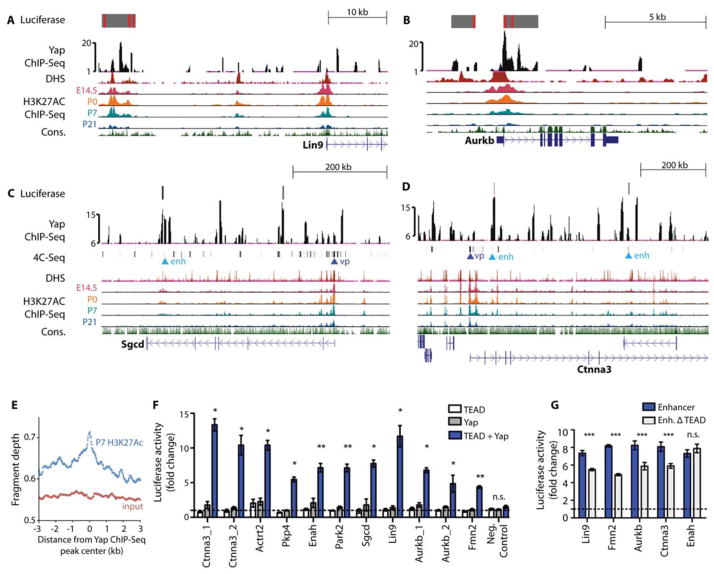
To further characterize productive Yap binding sites, we compared conserved Tead sites in our ChIP-Seq data to available DNAase hypersensitivity (DHS) and H3K27Ac datasets that mark enhancers (15–17). Many Yap peaks from Hippo-deficient hearts were enriched in putative enhancer regions (15) in the cell cycle genes Lin9 and Aurkb and in genes encoding proteins that are involved in both cytoskeletal remodeling and that link the cytoskeleton to the extracellular matrix (ECM), such as Sgcd and Ctnna3 (Fig. 2A–E). Furthermore, comparison of a mouse heart H3K27Ac ChIP-Seq dataset that marks enhancers with the Yap ChIP-Seq dataset showed that Yap peaks from Hippo-deficient hearts were globally enriched in putative enhancer regions (15) (Fig. 2E). We also performed chromosome conformation capture (4C) analysis to define enhancer contact maps for Sgcd and Ctnna3 and compared those maps to the Yap ChIP-Seq data (Fig. 2, C and D). Chromosome conformation capture provides 3-dimensional information about enhancer-promoter interactions and can help identify previously unknown enhancers (18, 19). We noted that enhancer-promoter contact points for both genes included Yap ChIP-Seq peaks, further supporting the conclusion that Yap peaks were found in enhancer regions.
Figure 2. Preferential expression of Yap target genes in the fetal heart.
(A–D) Genome browser view of Yap ChIP-Seq enriched peaks for the labeled genes. Alignments are shown for stage-specific H3K27Ac ChIP-Seq, DHS, and 4C anchor points (for Ctnna3 and Sgcd). For 4C, “vp” denotes viewpoint and “enh” denotes enhancer. Grey blocks show regulatory regions used as luciferase reporters with red lines indicating consensus Tead binding motif. The y-axes of Yap ChIP peaks show the normalized read number. (E) Quantification of P7 heart H3K27Ac ChIP-Seq reads in the 6-kb range around Yap ChIP-Seq peaks. N=2 biologic replicates. (F) Luciferase reporter assay data showing that Yap/Tead co-activated target gene expression through regulatory regions identified in Yap ChIP-Seq. N=3 independent transfection experiments. *P<0.05; **P<0.01; ***P<0.001. (G) Luciferase reporter assay data of Yap enhancers with or without TEAD motifs. N=3 independent transfection experiments. All luciferase constructs were co-transfected with Yap and TEAD expression vectors. ***p<0.001 (Mann-Whitney), n.s. not significant. Error bars are standard deviations. Activity was normalized to that of cells expressing the pGL3 vector.
We used luciferase assays to validate the transcriptional activation of the genes mentioned above, as well as additional genes encoding cytoskeletal proteins (Actrt2, Pkp4, Enah, and Fmn2). A luciferase reporter gene was fused to amplified genomic regions containing conserved Yap ChIP-Seq peaks that aligned with DHS peaks and/or H3K27Ac peaks that also contained Tead binding elements (15, 16) or that were also 4C contact points (Fig. 2, A–D). These reporter constructs were activated in cells by Yap and Tead2 co-expression (Fig. 2F,G). Moreover, deletion of the Tead elements resulted in loss of enhancer activity in all genes with the exception of Enah. It is possible that Enah has a poorly conserved Tead element that we failed to identify or that Yap may bind Enah by cooperating with a different DNA binding partner (Fig. 2, F and G). Together, these data support the hypothesis that Yap directs the transcription of genes promoting cell cycle progression, genes that link the actin cytoskeleton to the ECM, and genes that remodel the actin cytoskeleton.
Yap target genes are preferentially expressed in the fetal heart
Analysis of human RNA-Seq data revealed that orthologs of most of our Yap target genes were more highly expressed in human fetal hearts than in adult hearts (17) (fig. S2, A and B). Comparison of the human fetal RNA-Seq data with genes that we found had increased expression in Salv CKO hearts indicated an enrichment for genes that encode cell cycle and cytoskeletal proteins (fig. S2C). In addition, genes encoding proteins typical of the adult heart, including lipid metabolism and muscle contraction proteins, had reduced expression in Salv CKO mouse hearts (fig. S2D). This is consistent with findings that developing and early postnatal hearts rely on glycolytic metabolism rather than lipid oxidation and have an immature contractile apparatus (20). We also compared our Yap target gene set to mRNA expression data from P4 rat cardiomyocytes overexpressing Yap. Almost all Yap targets we identified had higher mRNA expression in the Yap gain of function rat cardiomyocytes (fig. S2E) (21).
Analysis of DHS datasets from human fetal and adult hearts further supported the hypothesis that Yap binding sites are enriched in fetal heart genes (22, 23). Motif analysis of the human fetal and human adult DHS data indicated that Tead elements were highly enriched in human fetal heart DHS peaks, along with recognition elements for other important cardiac development transcription factors such as Tbx and Mef2 (fig. S3A). Although Tead elements were enriched in human fetal heart DHS peaks, this was not the case for DHS peaks from human embryonic stem cells (fig. S3B). Tead elements were also found in DHS peaks from tissues such as the kidney and lung indicating that Tead elements are not cardiac specific (fig. S3C). Tead elements were enriched in fetal heart DHS peaks even for genes whose physical location was not conserved between humans and mice (Fgd4, Crebbp, and Mad2I1) (Fig. S3, D–F). These data support the hypothesis that Yap and Tead enhance the expression of fetal cardiac genes.
Hippo-deficient adult cardiomyocytes are proliferative
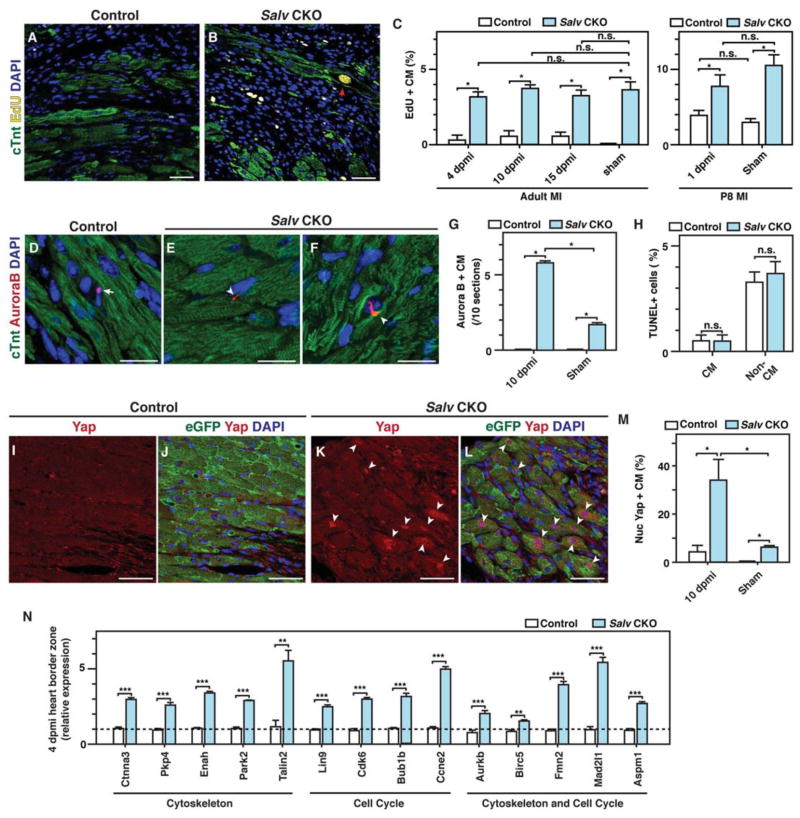
We evaluated proliferation and apoptosis in adult and neonatal control and Salv CKO mutant mouse hearts after inducing myocardial infarction by left anterior descending artery occlusion (LADO; Fig. 3). Compared to controls, Salv CKO mutant mouse hearts showed an increased number of EdU-positive cardiomyocytes at 4, 10, and 15 days post myocardial infarction for adults (Fig. 3, A–C) and 1 day post myocardial infarction for P8 neonatal hearts (Fig. 3C). Notably, S-phase entry was induced by Hippo deficiency and not by injury, as shown by equivalent amounts of EdU incorporation in Hippo-deficient sham hearts and Hippo-deficient injured hearts (Fig. 3C). The number of M-phase cardiomyocytes, as indicated by Aurora B immunostaining, was higher in Hippo-deficient injured hearts than in Hippo-deficient sham hearts (Fig. 3, D–G). These results suggest that Hippo-deficient hearts respond to injury by progressing through cytokinesis more efficiently than do Hippo-deficient sham hearts.
Figure 3. DNA synthesis and Yap localization in border zone cardiomyocytes during adult heart regeneration.
LAD occlusion was performed on Myh6-CreErt; mTmG (control) and Myh6-CreErt; Salvfx/fx; mTmG (Salv CKO) mice, and hearts were collected at 1, 4, 10, and 15 days post myocardial infarction (dpmi). (A, B) De novo DNA synthesis was detected by measuring EdU incorporation in control (A) and Salv CKO (B) mouse hearts at 10 dpmi. Red arrowhead shows EdU-stained nucleus. Bars=50 μm. (C) Quantification of de novo DNA synthesis in the border zone at 1, 4, 10, and 15 dpmi or in sham mice (N=3 mice for each genotype and time point). *P<0.05. (D–F) Aurora B kinase immunostaining served as a proxy of cytokinesis in control (D) and Salv CKO (E, F) mouse hearts at 10 dpmi. Arrow shows staining in a non-cardiomyocyte cell, and arrowheads show staining in cardiomyocytes. Bars=20 μm. (G, H): Quantification of Aurora B kinase (G) and TUNEL immunostaining (H) (N=3 mice for each genotype and treatment). **P<0.01. (I–M) Yap localization in border zone of control (I, J) and Salv CKO mouse hearts (K, L) at 10 dpmi. Arrowheads show nuclear localized Yap. Bars=50 μm. Quantification (M) of nuclear Yap in border zone cardiomyocytes at 10 dpmi (N=3 mice for each genotype and treatment). *P<0.05; remaining column comparisons were nonsignificant. (N) Gene expression of Yap downstream target genes were quantified with qPCR in border zones from heart tissues after myocardial infarction (N=3 biologic replicates). **P<0.01, ***P<0.001
The number of apoptotic cells in the myocardium was similar between control and Salv CKO mice after LADO (Fig. 3H). Moreover, the frequency of cell fusion events was similar in control and Salv CKO mutant mouse hearts (fig. S4). We also examined cell size at multiple stages after myocardial infarction and found that Salv CKO mouse cardiomyocytes were smaller than those of controls (fig. S5). We conclude that Salv CKO mouse cardiomyocytes showed increased EdU incorporation for 15 days post myocardial infarction and that after injury, Salv CKO mouse cardiomyocytes progress through M-phase more efficiently than do cardiomyocytes of sham mouse hearts.
In the border zone (tissue adjacent to the scar) of Salv CKO mouse hearts, nuclear localized Yap was increased (Fig. 3, I–M), suggesting that the expression of Yap-regulated genes may also be increased in this region in the regenerating heart. To examine this possibility, we isolated RNA from dissected border zones of Salv CKO mouse hearts and control hearts 4 days after inducing myocardial infarction. Quantitative real-time PCR (qRT-PCR) analysis revealed that Yap target genes had increased expression in the border zone of Salv CKO mouse hearts as compared to that of controls (Fig. 3N).
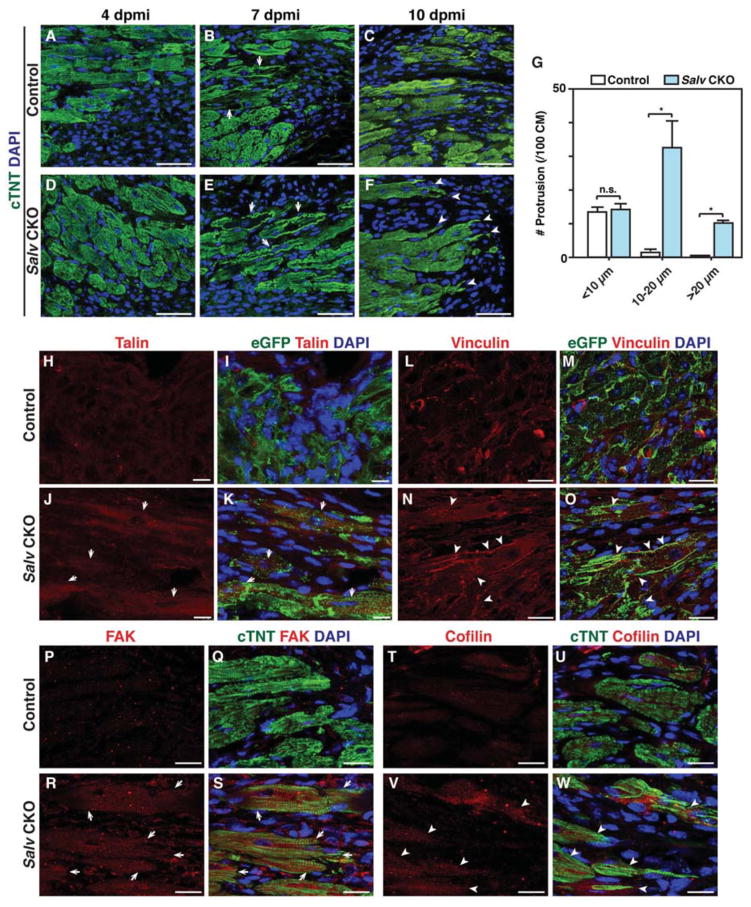
Hippo-deficient cardiomyocytes extend sarcomere-filled protrusions
Our finding that Yap-regulated genes encode proteins that regulate the actin cytoskeleton suggests that Hippo-deficient cardiomyocytes may have different cytoskeletal characteristics than control cardiomyocytes. We examined cardiomyocytes at 4, 7, and 10 days after myocardial infarction in adult control and Salv CKO mouse hearts. No differences in cellular morphology were observed between control and Salv CKO mouse hearts until 7 days post myocardial infarction, when the Salv CKO mouse cardiomyocytes showed more prevalent sarcomere breakdown in the border zone (Fig. 4, A–G). At 10 days post myocardial infarction, adult Salv CKO mouse cardiomyocytes, but not those from control mice, showed extended protrusions into the scar (Fig. 4C, F, and G). Immunofluoresence experiments performed by using anti-talin and anti-vinculin antibodies that recognize costameres linking the ECM to the actin cytoskeleton through the integrin-vinculin-talin complex (24) revealed that the distribution of both talin and vinculin was markedly expanded in the cardiomyocytes of adult Salv CKO mice at 10 days post myocardial infarction (Fig. 4H–O). Similarly, we noted that the distribution of focal adhesion kinase (FAK) and cofilin, both of which have been previously implicated in cell migration, was expanded in Salv CKO mutant cardiomyocytes (Fig. 4P–W) (25,26). Protrusions were commonly seen on multiple sections surrounding the infarct in adult Salv CKO mouse cardiomyocytes but not in those of control mice (fig. S6, A–F).
Figure 4. Cardiomyocyte morphology and cytoskeleton rearrangement during adult heart regeneration.
(A to F) LAD occlusion was performed on the hearts of Myh6-CreErt; mTmG (control) and Myh6-CreErt; Salvfx/fx; mTmG (Salv CKO) mice. Control (A–C) and Salv CKO (D–F) mouse hearts were stained for the cardiomyocyte marker cTNT to visualize morphology of the cardiomyocytes in the border zone at 4, 7, and 10 dpmi (N=3 mice for each genotype and time point). More images from different hearts of 10 dpmi hearts are shown in Supplemental fig. S6A-F. Arrows show cardiomyocytes with sarcomere disassembly. Arrowheads show cardiomyocyte protrusion. Bars=50 μm. (G) Quantification of protrusions. Cardiomyocytes adjacent to the scar were analyzed for length and number of protrusions at 10 dpmi. One hundred cardiomyocytes from each mouse were analyzed (N=3 mice per genotype). *P<0.05. (H to O) Control (H, I) and Salv CKO (J, K) mouse heart sections at 10 dpmi were stained for Talin (N=2 mice per genotype). Arrows show increased Talin staining in border zone cardiomyocytes. Bars=25 μm. Control (L, M) and Salv CKO (N, O) mouse heart sections at 10 dpmi were stained for the focal adhesion molecule vinculin. Arrowheads show the rearrangement of vinculin in the protruding front of the cardiomyocytes (N=3 mice per genotype). Bars=25 μm. (P to W) Control (P, Q) and Salv CKO (R, S) mouse heart sections at 10 dpmi were stained for FAK (N=2 mice per genotype). Arrows show increased FAK staining in border zone cardiomyocytes. Bars=20 μm. Control (T, U) and Salv CKO (V, W) mouse heart sections at 10 dpmi were stained for FAK (N=2 mice per genotype). Arrowheads show increased Cofilin staining in border zone cardiomyocytes. Bars=20 μm.
In the apex resection model, a defined region of cardiac apex is removed, allowing for the accurate characterization of renewing cardiomyocytes (5). In resected Salv CKO hearts, cells derived from the cardiomyocyte lineage [as denoted by cardiac troponin T (cTnt) staining] were observed in the resected zone that contained a large number of non-cardiomyocyte cells (fig. S6, J). Similar to what we observed in adult mouse hearts after myocardial infarction, Hippo-deficient cardiomyocyte lineage–derived cells extended cellular protrusions and showed evidence of remodeling of vinculin-positive focal adhesions (fig. S6, K and L). In control mouse hearts, border-zone GFP-positive cells did not infiltrate the resected region of the heart or remodel vinculin (fig. S6, G, H and I).
Hippo-deficient cardiomyocytes mobilize into collagen gels
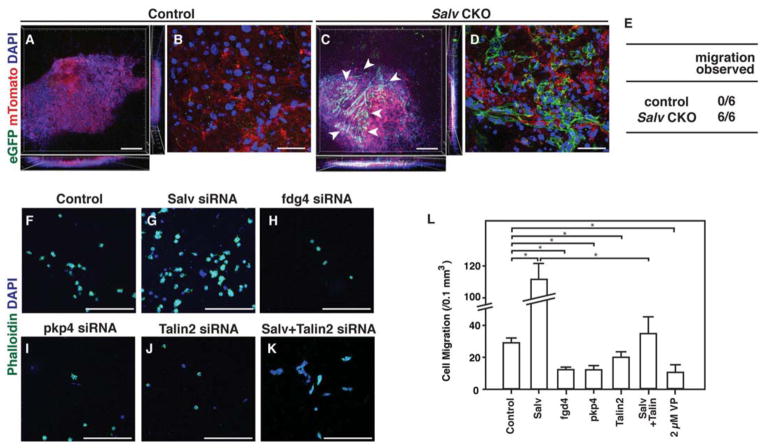
Our ChIP-Seq and immunofluorescence data support the notion that Salv CKO mouse cardiomyocytes display extensive cytoskeletal remodeling with protrusive activity. Because cellular protrusions are characteristic of migrating cells (27), we determined whether Salv CKO mouse cardiomyocytes could mobilize into a collagen gel. Cardiomyocytes from cardiac explants from Salv CKO mice, but not those from control mice, efficiently migrated into collagen gels (Fig. 5, A–E). Therefore, our findings suggest that Salv CKO mouse cardiomyocytes extend protrusions that could enable cellular movement into a collagen gel.
Figure 5. Cardiomyocyte migration through collagen.
(A to E) Collagen migration assays were performed with P8 cardiomyocytes from control (Myh6-CreErt; mTmG )(A, B) or Salv CKO (Myh6-CreErt; Salvfx/fx; mTmG) mouse hearts (C, D). Cardiomyocyte lineage was visualized with eGFP and non-cardiomyocytes were visualized with mTomato. Whole gel view (A, C). Arrowheads show migrated cardiomyocytes. Bars=250 μm. High-magnification images of the area with eGFP positive cells (B, D). Bars=50 μm. Quantification (E) of the number of hearts in which migration was observed (N=6 hearts per genotype). P=0.002, control compared to Salv CKO mice. (F–K) P19 cell migration in collagen gel after siRNA treatment with the labeled siRNA. Bars=250 μm. (L) Quantification of migrated cells after each treatment. Cells were treated with either siRNA or Yap inhibitor verteporfin (VP). n=3 biological replicates for all groups.
We also used the collage invasion assay to test whether the small interfering RNA (siRNA)-mediated knockdown of Salv promotes cell migration into the gel. siRNA knockdown experiments in P19 embryonal carcinoma cells revealed that the siRNA-mediated knockdown of Salv resulted in increased cell invasion into the gel that was not due to increased cell proliferation (Fig. 5, F–L; fig. S7). In addition, the siRNA-mediated knockdown of the Yap target genes Fgd4, Pkp4, and Talin2 resulted in decreased cell migration into collagen gels. Furthermore, the simultaneous siRNA-mediated knockdown of both Salv and Talin2 showed that the knockdown of Talin2 suppressed the Salv knockdown phenotype. Moreover, treating P19 cells with verteporfin, a small molecule that disrupts Yap-Tead interaction (28), blocked the migration of P19 cells into the gel (Fig. 5L).
The dystrophin glycoprotein complex is required for cardiac regeneration
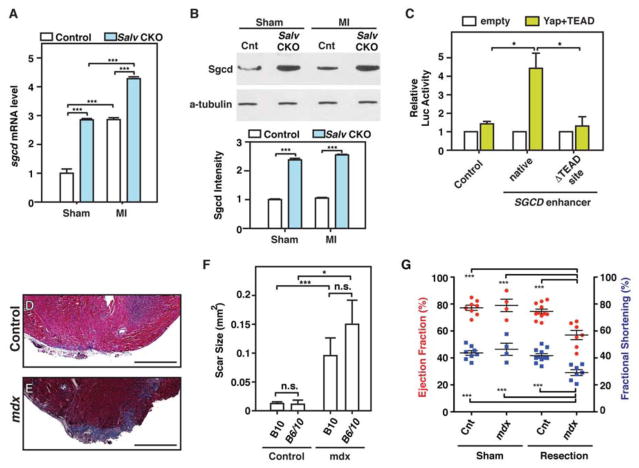
Many Yap target genes encode proteins that link the actin cytoskeleton to the sarcolemma and ECM. Sarcoglycan delta (Sgcd) and Syntrophin B1 (Sntb1) are both DGC components that connect the actin cytoskeleton to the extracellular matrix and transmit force from muscle cells to the extracellular matrix (29, 30). Sgcd is mutated in human patients with limb-girdle muscular dystrophy and is thought to stabilize the plasma membrane in response to mechanical stress (31). qRT-PCR and Western blot analyses showed that the abundance of Sgcd mRNA and Sgcd protein was increased in Salv CKO mouse hearts when compared with control mouse hearts (Fig. 6, A and B). After myocardial infarction, Sgcd mRNA expression was increased in both control and Salv CKO mouse hearts. The abundance of Sgcd protein was not further increased after myocardial infarction, suggesting the posttranscriptional regulation of Sgcd (Fig. 6, A and B; fig. S8A). Western blot analysis indicated that Sntb1 protein abundance was unchanged between control and Salv CKO mouse hearts (fig. S8B). Luciferase assays using a luciferase reporter gene fused to the region of the Sgcd gene containing the Yap/Tead binding element revealed that Yap and Tead2 transactivated the Sgcd reporter gene but not a Sgcd reporter with a mutated Tead element (Fig. 6C).
Figure 6. The dystrophic complex is downstream of the Hippo pathway and is required for cardiac regeneration.
(A, B) LADO and sham surgery were performed in control and Salv CKO mouse hearts at P8, and heart samples were collected at 4 dpmi. Delta-sarcoglycan (Sgcd) mRNA was detected in control and Salv CKO mouse hearts by using qRT-PCR and was normalized to Gapdh (N=3 hearts for all groups) (A). Sgcd protein was detected by using Western blot analysis (N=3 hearts per genotype and treatment) (B). Sgcd band intensities were quantified and normalized to those of alpha-tubulin. ***P<0.001, remaining column comparisons were non-significant (n.s.) (C) Luciferase assays were performed with P19 embryonal carcinoma cells. Cells were transfected with either the control luciferase reporter, a reporter containing the Sgcd enhancer, or a reporter containing the Sgcd enhancer but lacking the Tead site. Three independent experiments with technical triplicates were performed. *P<0.05. (D–G) Dystophin glycoprotein complex (DGC) is required for endogenous cardiac regeneration. Representative images of trichrome-stained heart sections from B10 control (D) and Mdx-B10 (E) mice subjected to resection of the cardiac apex. Images of 2 additional control and mutant apexes are shown in fig. S9. Bars=500 μm. Quantification (F) of the scar size at 21 dpr in B10, (N=11), B6/10 (N=4), Mdx-B10 (N=7), and Mdx-B6/10 (N=6) mouse hearts.*P<0.05, ***P<0.001. Echocardiography analysis (G) of control sham (N=3), control apex resection (n=7), Mdx sham (N=4), and Mdx apex resection (N=7) mouse hearts 21 days after surgery. ***P<0.001, remaining column comparisons were n.s.
Yap is required for mammalian heart regeneration during the postnatal regenerative period (11). Therefore, we examined whether the Yap target gene Sgcd and its functional activity in the DGC are also required for cardiac regeneration. As a model for impaired DGC, we used Mdx mutant mice that harbor a null dystrophin mutation that disrupts the DGC. Cardiac apexes from Mdx and control mice were amputated at P1, when regeneration is possible (5). Whereas control mouse hearts had a robust regenerative capacity, all Mdx mouse hearts formed a large scar (Fig. 6, D–F; fig. S9). Furthermore, cardiac function as measured by ejection fraction and fractional shortening was compromised in resected Mdx mouse hearts (Fig. 6G).
The dystrophin glycoprotein complex is required for cellular protrusions
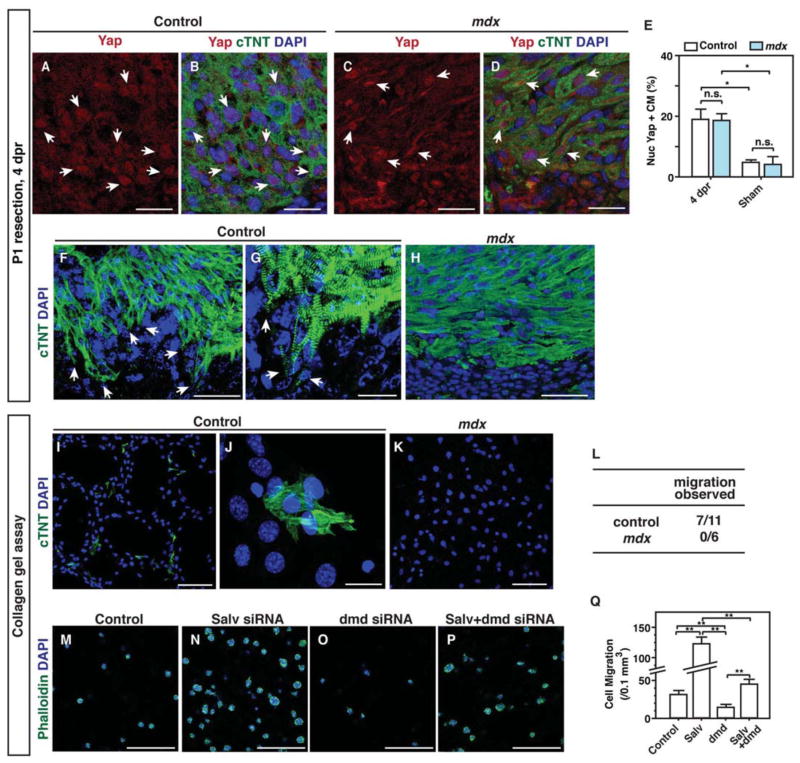
We examined the border zone of control and Mdx mouse hearts 4 days after resection. No difference was observed in the amount of nuclear localized Yap between Mdx and control mouse hearts, suggesting that DGC function is dispensable for the shuttling of Yap from the nucleus to the cytoplasm during regenerative stages (Fig. 7, A–E). Whereas cardiomyocytes of control mice exhibited extensive cytoskeletal remodeling with protrusion formation, Mdx mouse cardiomyocytes failed to remodel their cytoskeleton and extend protrusions (Fig. 7, F–H; fig. S10, A and B). Using collagen gel assays to functionally evaluate protrusive activity, we found that explanted myocardial cells from control mice, but not those from Mdx mice, moved through a filter into a lower collagen gel (Fig. 7, I–L). We also performed EdU incorporation and AurkB immunofluorescence assays to determine whether Mdx mouse cardiomyocytes had a proliferative defect that was responsible for failed regeneration. We found that EdU incorporation and AurkB immunoreactivity were equivalent between cardiomyocytes of Mdx and control mice (fig. S10). Proliferation was increased equally in both control and Mdx mouse hearts after injury, supporting the notion that Mdx mice failed to regenerate the heart because of a defect in myocardial protrusive activity rather than because of a proliferative defect (fig. S10). To determine whether dystrophin depletion could suppress the increased migration of Salv knockdown cells, we evaluated the migration of p19 cells with concomitant knock down of Salv and Dmd. Indeed, knocking down both Dmd and Salv in p19 cells suppressed the increased cell migration observed in Salv knockdown cells (Fig. 7, M–Q).
Figure 7. Regulation of cardiomyocyte protrusion by the dystrophin complex.
(A–D) Yap localization in border zone cardiomyocytes of B6/10 control (A, B) and Mdx-B6/10 (C, D) mouse hearts at 4 days post resection (dpr). Arrows show nuclear localized Yap. Bars=25 μm. (E) Quantification of nuclear Yap in border zone cardiomyocytes at 4 dpr (N=3 mice per genotype). *P<0.05. (F–H) DGC is required for cardiomyocyte protrusion. Sections of the border zone of B6/10 control (F, G) and Mdx-B6/10 (H) mouse hearts were stained with the cardiac marker cTnt at 4 dpr. Arrows show protruding cells at the border zone. (G) is a higher magnification image of (F). For (F) and (H), bars=50 μm; for (G), bars=25 μm. (I–K) Collagen gel assay for P1 B10 (I, J) and Mdx-B10 (K) hearts. (J) is a higher magnification image of (I). For (I) and (K), bars=100 μm; for (J), bars=20 μm. (L) Quantification of cardiomyocyte migration from B10 control (N=11) and Mdx-B10 (N=6) mouse hearts. P=0.035, control compared to Mdx. (M–P) P19 cell migration in collagen gel after transfection with the indicated siRNA. Negative control (M), Salv (N), dystrophin (dmd) (O), and Salv and dmd combined (P). Bars=250 μm. (Q) Quantification of migrated cells after each treatment. N=3 biological replicates for all groups. *P<0.01.
DISCUSSION
Mobilization of endogenous cardiomyocyte regenerative capacity may be a viable option to treat heart failure. Hippo pathway loss of function or Yap gain of function promotes cardiomyocyte regeneration after myocardial infarction (8, 11, 12). In this study, we identified direct Yap transcriptional target genes and discovered the importance of cytoskeletal remodeling with cellular protrusions in heart regeneration. Yap target genes directly control cell shape and protrusive activity.
Yap regulates genes encoding proteins that link the ECM to the actin cytoskeleton
Cardiomyocytes constantly sense and interpret local microenvironment rigidity (32). Yap-regulated genes, including Ctnna3, Sgcd, and Tln2, are important for linking the actin cytoskeleton to the ECM. Our data, along with data the regulation of Yap subcellular localization by matrix rigidity, support the idea that Yap interprets and responds to local ECM mechanical characteristics (10, 33). Yap-regulated genes are components of 2 plasmalemmal complexes that link the actin cytoskeleton to the ECM: the DGC and the talin-vinculin-integrin complex (34). These genes include Sgcd, which encodes a DGC component and Talin2, which encodes an actin-binding protein that links the actin cytoskeleton to integrins and ECM. Other Yap-regulated genes such as Ctnna3, a gene that encodes a cadherin-associated protein localized to the intercalated disc, connect the actin cytoskeleton to both the intercalated disc and ECM and most likely function to sense tension between cardiomyocytes (35).
Linking complexes are critical for cardiac muscle homeostasis. The DGC is disrupted in multiple types of muscular dystrophy with dilated cardiomyopathy (DCM). Mutations in Vinculin also cause DCM in humans (36). Likewise, loss of function mutations in Ctnna3 result in DCM, revealing a critical requirement of this protein in cardiomyocyte homeostasis (35). Our molecular findings suggesting that the myocardium is linked to the ECM more efficiently in Salv CKO mouse hearts are also supported by the observation that Lats1/2 loss-of-function myocardium can transmit tissue deformations more effectively, as determined by using optical coherence tomography (37).
Actin cytoskeleton remodeling with cellular protrusion is required for heart regeneration
In the border zone of Salv CKO mouse hearts, we observed changes in cellular phenotypes, including the appearance of sarcomere-filled protrusions surrounding the wound in apex resection and adult myocardial infarction models. Our data also showed that Salv CKO mouse cardiomyocytes can invade a collagen gel. Cardiomyocytes in hearts subjected to apex resection or LADO showed protrusive activity.
Our genomic analyses indicated that Yap regulates genes promoting lamellipodia formation and actin cytoskeleton protrusion. Certain Yap target genes encode proteins that localize to lamellipodia and filopodia such as Enah, which encodes an Ena/VASP actin regulator that causes cardiac dysfunction when disrupted in mice (38). Fgd4, another Yap target gene, is mutated in Charcot-Marie-Tooth type 4H disease. Fgd4 encodes an F-actin binding protein with RhoGEF activity specific for Cdc42, which is crucial to filopodia and lamellipodia formation and is essential for cytokinesis (39).
The Yap target gene Mtss1, also named missing in metastasis (MIM), encodes a protein that localizes to adherens junctions where it promotes actin filament assembly. Mtss1 induces the formation of lamellipodial microspikes and membrane ruffles that contribute to protrusions and may be involved in the sensing of fluid shear stress in the kidney (40). Other findings indicate that Mtss1 is required for neural tube closure (41). Intriguingly, Yap is regulated by polymerized F actin, which promotes Yap nuclear localization, suggesting a positive feedback loop that maintains nuclear Yap localization and cellular protrusions.
Our data indicate that in Salv CKO mutants, there are more cell protrusions in border zone cardiomyocytes supporting the hypothesis that active cell movement is required for effective heart regeneration. Further experiments performed in vivo are required to determine if cardiomyocytes move individually or collectively as coherent groups of cells. Approaches such as vital imaging on cultured heart slices will be necessary to directly observe cell migration in Salv CKO hearts.
It is conceivable that collective cell migration in the border zone is initiated by a subpopulation of cardiomyocytes that can migrate while other cardiomyocytes move passively to fill in the tissue defect (42). Consistent with this hypothesis, we found that Cofilin and FAK are highly abundant in Salv CKO border zone cardiomyocytes. Cofilin promotes asymmetric actin polymerization and is found in locomotory and invasive protrusions such as lamellipodia and invadopodia. Likewise, FAK is a component of integrin adhesion complexes and promotes tissue polarity and collective cell migration (25, 26). In the context of heart regeneration, such polarized cell movement may be required for a structurally well-organized myocardium that has adequate pumping function.
Yap directly regulates genes encoding proteins that promote cell cycle progression and cytokinesis
Our findings indicated that apoptosis is unaffected in Salv CKO mice after myocardial infarction. Similar findings have been reported for Yap gain-of-function mice (12). Our Yap target gene dataset indicated that Yap directly regulates multiple genes that enhance progression through multiple cell cycle phases and cytokinesis. Yap directly activated genes (Aurkb and Birc5) which encode cytokinesis factor. Another direct gene target of Yap, Lin9, encodes a MuvB complex component that enhances mitotic gene expression (43). Considerable cross-talk has been reported between the MuvB and Lats2 in senescent cells, which warrants further investigation (44). Other direct gene targets of Yap identified in our study included Spin1, a cytokinesis-associated gene expressed in cardiomyocytes, and Aspm, which shows increased expression in cardiac hypertrophy.
Aspm is required for proper spindle orientation and cytokinesis, and promotes Wnt signaling in neurogenesis (45). Yap target genes identified in our study also included Bub1b and Mad2l, components of the spindle assembly checkpoint that ensure mitotic fidelity. Bub1b gain of function extends lifespan and inhibits aneuploidy and chromosome instability. In addition, other Yap target genes identified in this study were the cell cycle genes Ccne2 and Cdk6, which play important roles in G1 progression. Moreover, the genes that encode 2 inhibitors of cell cycle progression, Cdkn2b and Cdkn3, were identified as Yap targets that likely function in a negative feedback mechanism. In some contexts, Cdkn3 may act to promote proliferation through poorly understood mechanism(s) (46).
Yap promotes the expression of its target genes that are more highly expressed in the fetal heart: evidence for dedifferentiation
Yap promoted the expression of its target genes in the postnatal heart, revealing a molecular mechanism for reversion to the fetal gene program that occurs after injury but poorly understood mechanistically. Consistent with our molecular data, border zone cardiomyocytes of Hippo-deficient mouse hearts displayed more sarcomere disassembly than did border zone cardiomyocytes of control mouse hearts, suggesting that Hippo-deficient cardiomyocytes dedifferentiate after injury and acquire an immature phenotype typical of developing cardiomyocytes (47). Cardiomyocyte dedifferentiation also occurs in zebrafish heart regeneration, implying that there are evolutionarily conserved mechanisms between fish and mammals (48). The mechanisms for sarcomere disassembly in the context of regeneration are poorly understood.
MATERIALS AND METHODS
Mice
Mouse strains used in this study were as follows: Myh6-CreErt; mTmG, Myh6-CreErt; Salvfx/fx; mTmG (8), C57BL/10J (B10), B10 crossed with C57BL/6J (B6/10), C57BL/10ScSn-Dmdmdx/J (Mdx-B10), and Mdx crossed with B6 (Mdx-B6/10). B10, B6, and Mdx-10 mice were obtained from Jackson Laboratory.
ChIP-seq analysis
Immunoprecipitation was performed with an anti-Yap antibody (Novus) in whole-heart lysates to generate libraries that were then sequenced by using an Ion Proton sequencer (Life Technologies). Two biological replicates were performed, and the reproducibility between samples was assessed by using Pearson’s correlation coefficient. About 24 million Yap binding reads were evaluated by using a Homer software package.
Microarray and ChIP-seq analysis
Microarray analysis was performed on a Phalanx Mouse OneArray v2 platform. Two biological replicates, each with 3 technical replicates, were collected for each genotype. The value distribution of raw intensity from a total of 4 samples was graphically viewed in box plots. Linear correlation coefficients between biological replicates were measured by using R software function var.
Differentially expressed genes were detected by using the R bioconductor package ‘Limma,’ version 3.16.8. Briefly, log transformation was performed to the raw intensity values; a Student t test was performed to identify differences in mRNA expression level between control and Salv CKO mouse hearts for each probe. Differentially expressed genes were called by using the cutoff p-value ≤ 0.05 and fold change ≥ 1.5. Activated Yap1 targets were extracted from dataset GSE33019 by using the same criteria.
Ion Torrent Proton reads were mapped to the mm9 assembly (NCBI Build 37) using Torrent Suite (2.0.1) aligner Tmap (0.2.3) (Life Technologies). Two biological replicates were performed. Only uniquely mapped reads were kept. A total of 11 million Yap ChIP-Seq reads were generated from replicate 1 and 13 million from replicate 2. The reproducibility between the 2 runs was measured by using Pearson’s correlation coefficient to compare signal intensities in 5-kbp bins across the genome (Fig. S1G, r=0.69). ChIP-enriched peaks were identified from combined reads with the Homer v4.2 parameters “findPeaks -style factor -o -i input -tagThreshold 15”. The peak size was calculated automatically as 162 bp. The cutoffs for calling the peaks were read number enriched over input control values ≤ 4 fold, minimum read number of 15 which limited Poisson p-value to less than 1e-10. The clonal peaks were filtered out. This process produced 35,412 enriched peaks. Nearest genes associated to the enriched peaks were annotated with the “annotatePeaks” function, and de novo motif discovery was performed with the “findMotifsGenome” function in Homer.
1,706 genes had increased expression in Salv CKO mouse hearts and were overlaid with 10,396 Yap ChIP-Seq binding genes. Overlaid genes were subjected to over-represented gene ontology analysis by using GO Elite with 5000 permutations. The false discovery rate was calculated with the Benjamini-Hochberg correction. Gene ontology terms were plotted by using the Z score. Only terms with a false discovery rate <0.10 were reported.
Human heart RNA-seq data analysis
Human RNA-Seq data were obtained from Gene Expression Omnibus (GEO) and DNA Data Bank of Japan (DDBJ). The sample IDs of fetal hearts were SRR643778, SRR643779; of adult hearts: GSM1010964, ERR030894. The reads were aligned to reference genome hg19 by using Tophat. The splicing junctions were detected by using known annotations. Uniquely mapped reads within exons were counted with HTSeq-count. Differentially expressed genes were detected by using R package DESeq with a threshold false discovery rate (FDR)≤0.1. Gene ontology analysis of 700 genes with increased expression and 419 genes with reduced expression was performed in GO Elite.
Human DHS-seq data analysis
Mapped human DHS-Seq data were extracted from GEO. We used data for 11 fetal heart samples (GSM530654, GSM530661, GSM665811, GSM665814, GSM665817, GSM665824, GSM665830, GSM665831, GSM774203, GSM817220, GSM878630, bed file) and 2 adult heart samples (GSM1008559, bam file); tissue-specific motif calling was performed on a mouse DNaseI digital genomic footprinting dataset GSE40869. Peaks enriched in fetal hearts were detected against adult DHS data by using Homer with the following script: “findPeaks fetal_dhs -style dnase -o auto -i adult_dhs”. A total of 73,949 peaks were called. De novo motif enrichment was performed with the Homer function “findMotifsGenome.” Hierarchical clustering of enriched motifs in each tissue was computed and visualized on the basis of -log(p-value) by using R package pheatmap.
Mouse heart H3K27Ac ChIP-seq data analysis
Mouse heart H3K27Ac ChIP-Seq data were available in GEO (GSE52386). BedGraph files were generated from mapped bam files by using the Homer “makeUCSCfile” function and were visualized in the UCSC genome browser. The total read number of individual samples was normalized to 10 million. P7 H3K27Ac ChIP-Seq data were used to plot genome-wide density profiling within a 6-kb range of Yap binding peaks.
Left anterior descending coronary artery occlusion and apex resection
Left anterior descending coronary artery occlusion (LADO) was performed as previously described on P8 or adult (8–10 week old) mice (8). In experiments using (P8) 1 dpmi mice, tamoxifen (0.5 mg) was injected in Myh6-CreErt; mTmG (control) and Myh6-CreErt; Salvfx/fx; mTmG (Salv CKO) mice 1 day before and 5 min after surgery. For 8–10 week old mice, tamoxifen was injected at 1.5 mg 7 and 6 days before surgery and at 2 mg within 2 hours after surgery. Hearts were collected at 4, 7, and 15 days after LADO, and EdU was injected 4 times starting at 3 days before collection of the heart.
Resection of the heart apex was performed on P1 mice as previously described (8) by using B6/10 or B10 (control) and Mdx (with B6/10 or B10 background) mice. Hearts were collected at either 4 or 21 days after resection. At 21 days after resection before collection, echocardiography was performed in resected and sham animals.
Echocardiographic measurements were performed in the Mouse Phenotyping Core at Baylor College of Medicine. One-way ANOVA with Bonferroni’s multiple comparison test was performed for statistical analysis.
After echocardiography, hearts were collected, embedded in paraffin, sectioned (7-μm thickness), and stained by using Masson’s trichrome staining. After images were documented, scar size was measured by using ImageJ. Every other 3 slides were documented, and the sizes of the sections with the largest scars were recorded. The Mann-Whitney U test was performed for statistical analysis.
Edu staining, TUNEL assay, and immunohistochemistry
For the adult mouse study, EdU (5′ethynyl-2′-deoxyuridine) was injected into mice for 4 consecutive days starting 3 days before the heart was harvested. For experiments using P8 mice, hearts were collected at 1 day post myocardial infarction, and EdU was injected within 5 min after surgery and at 4 h before the heart was harvested. For experiments using neonatal mice, hearts were collected at 4 days post resection, and EdU was injected 4 h before tissue collection. Hearts were embedded in paraffin and sectioned at a thickness of 7 μm. EdU staining was performed by using the Click-iT EdU Alexa Fluor 647 Imaging kit (Life Technologies). Immunohistochemistry with eGFP or cTNT was performed to label cardiomyocytes after EdU staining.
The TUNEL assay was performed by using the DeadEnd fluorometric TUNEL assay (Promega). To mark cardiomyocytes, immunohistochemistry with cTNT was performed after TUNEL staining.
Immunohistochemistry was performed by using standard protocols. Antibodies used in this study were as follows: rabbit anti-GFP (1:400 dilution, Abcam), rabbit anti-AuroraB kinase (1:200 dilution, Abcam), mouse anti-GFP (1:500 dilution, Clontech), rabbit anti-RFP (1:200 dilution, Clontech), rabbit anti-Yap (1:200 dilution, Novus), mouse anti-cTNT (1:200 dilution, Thermo), rabbit anti-talin (1:200 dilution, Abcam), and mouse anti-vinculin (1:200 dilution, Thermo). Fluorescence images were documented by using a Leica TCS SP5 confocal microscope with Leica LAS AF software. All images presented in the manuscript were prepared by using Adobe Photoshop CS5.1 (Adobe Systems). Border zone cardiomyocytes were defined as cells within 300 μm from the scar.
For the quantification of positive cells, 3 biological replicates were collected per genotype and time points. For each stain, 3 images were captured for each biological replicate. Cell counting was performed blindly. A total of 100–250 cardiomyocytes were counted in each image for quantification. For the quantification of Aurora B kinase staining, a total of 10 sections were counted per biological replicate. For quantitation of protrusions, 100 cells adjacent to the scar were examined for the number of protrusions (less than 10μm, 10–20 μm, and greater than 20 μm). The Mann-Whitney U test was performed for statistical analysis.
Collagen gel assay with heart tissue
For the study of P10 hearts, Myh6-CreErt; mTmG (control) and Myh6-CreErt; Salvfx/fx; mTmG (Salv CKO) mice were used. Tamoxifen (0.6 mg) was injected twice at P8 and P9, and hearts were dissected at P10. Apexes were cut and placed on collagen gel (Millipore). Tissues were mounted by overlaying collagen gel and were cultured in DMEM with 10% bovine calf serum (BCS) at 37°C. After 5 days, collagen gels were collected and stained with eGFP. Each gel was analyzed for the presence of cardiomyocytes that migrated to the bottom gel. If any cardiomyocytes were found in the bottom gel, it was counted as “migration observed.” The Fisher’s exact test was performed for statistical analysis.
For the study of P1 hearts, B10 (control) and Mdx-B10 mice were used. Hearts were harvested at P1, and apexes were collected. Nylon mesh (100 μm) was placed on the collagen gel, and the tissues were placed on top of the mesh. Tissues were mounted by overlaying collagen gel and were cultured in DMEM media with 10% BCS at 373C. After 5 days, the top gel and nylon mesh were removed, and the bottom collagen gel was stained with cTNT. Quantification was performed as described above.
Luciferase assay and transfection experiments
Each enhancer element was amplified and cloned into the pGL3-promoter plasmid (Promega). The primers used are listed in Supplemental Table 1. The Tead motif for each element was deleted by using the Quick Change XL Site-directed mutagenesis kit (Agilent Technologies) for Sgcd or Gibson assembly kit (NEB) for Aurkb, Ctnna3, Enah, Fmn2, and Lin9. The locations and sequences of TEAD motifs are listed in Table S1. Plasmids were co-transfected with Yap and Tead expression plasmids (7) into P19 embryonal carcinoma cells. Luciferase activity was analyzed by using the Dual Glo luciferase assay system (Promega). Three independent transfection experiments were performed with triplicate wells. Mann-Whitney test was used for statistical analysis.
Quantitative RT-PCR
Total RNA was extracted from dissected heart tissues by using the RNeasy Micro Kit (Qiagen), and the concentration was determined by using an Infinite M200PRO Nanodrop spectrophotometer (Tecan). First-strand cDNA synthesis was performed by using the iScript™ cDNA synthesis kit (Invitrogen) using 1 μg of total RNA input. For qRT-PCR, 1 μl of cDNA (1:10 dilution) was added to SYBR® Green Jumpstart™ Taq Ready Mix (Sigma). Each sample was analyzed in triplicate and run on a StepOnePlus Real-Time PCR System (Life Technologies). Mean dCt values for each Yap target gene were normalized against those of GAPDH mRNA levels, and corresponding ddCt values were log2-transformed to obtain fold-change values. The standard deviations of control and mutant samples were calculated within groups. The non-parametric Mann-Whitney test was performed to determine statistical significance of the differences between control and mutant groups for each target gene (P<0.05 was considered significant). Sequences of primers used in qPCR experiments are available upon request.
P19 migration assay
To examine the influence of Yap target genes on cell motility, we performed siRNA experiments in P19 embryonal carcinoma cells. siRNA oligos were obtained from Integrated DNA Technologies (Coralville, Iowa). The methods used for siRNA knockdown experiments have been described previously (8). After 24 hours of siRNA treatment, trypsinized cells were suspended in collagen gel mix (Millipore). Nylon mesh (20 μm) was applied to solidified collagen gel spots in 4-well plates, and cell/collagen mixes were dispensed to meshes. Plates were incubated at 37°C (5% CO2) for 1 h and α-MEM culture media with 10% fetal bovine serum (FBS) was subsequently added. After 24 h of culture, the top gel and nylon mesh were removed, and the bottom gel was immmunostained accordingly. We examined the statistical significance of the differences between groups by using Mann-Whitney U tests. P<0.05 was considered significant.
Western blotting
Methods for Western Blot quantitation were previously described (7). Primary antibodies used were anti-α-tubulin (1:8000 dilution; Sigma) and anti-SGCD (1:500 dilution; Santa Cruz Biotechnology). Differences between groups were evaluated for statistical significance by using two-way ANOVA with the post-hoc Bonferroni correction.
Cell surface area measurements
Immunofluorescence staining for GFP and DAPI was performed as described previously (8). For visualization of cell membranes, samples were incubated with wheat germ agglutin Alexa Fluor® 647 conjugate (1:200 dilution; Life Technologies) for 30 min at room temperature before blocking and primary antibody application. Cardiomyocyte cell surface area measurements were obtained from multiple transverse sections in series, and averages were obtained by using ImageJ software (http://rsbweb.nih.gov/ij/). For 4 and 10 dpmi hearts, we used Mhccre-Ert; mTmG (control) and Mhccre-Ert; mTmG; Salvf/f (Salv CKO) mice. For 21 dpmi and 21 dpmi sham hearts, we used Mhccre-Ert (control) and Mhccre-Ert; Salvf/f (Salv CKO) mice. Differences between groups were examined for statistical significance by using The Mann-Whitney U tests. P<0.05 was considered significant.
4C sequencing and analysis
The 4C templates were generated from P8 Salv CKO mouse hearts (2 biological replicates), as previously described (49, 50). HindIII and DpnII were used as first and second restriction enzymes, respectively. Promoter sequences from Ctnna3 and Sgcd were used to design viewpoint amplification primers (Ctnna3 DpnII: 5′ ACTGGCTGTGGAGGTTTTGAATTAC; HindIII: 5′ CCCTCTGTATTGACTTTCTATCCC; Sgcd DpnII: 5′ GAAACCCTACTCTGCCTCTACTTC; HindIII: 5′ CAGAGGGACTGGGTGCTTCT). Libraries were amplified from 100 ng of template DNA by using the following PCR protocol: 94°C 2 min, 30 cycles of 94°C 15 s, 56 °C 1 min, 68 °C 1 min. The libraries were then prepared for Ion Proton Sequencer. Ion Proton reads were separated by PCR primers. Over 700,000 reads were generated for each viewpoint library. For extracting unknown contact point sequences, only reads were kept that contained a recognition restriction enzyme site jointed to the PCR primer and, at most, one secondary restriction site. After trimming the primer sequences, unknown sequences with length ≥ 40 bp were kept for aligning against mouse genome mm9. Significant contact points in the proximal range (500 kb) were detected by using a previously described algorithm (49, 51) and were visualized in the UCSC genome browser.
Acknowledgments
We thank Richard Behringer for comments on the manuscript and Nicole Stancel for expert editing.
Funding: This work was supported by grants from the National Institutes of Health (NIH)[1U01HL087365 to J.T.W.; 5T32HL007676-23 to J.L., HL 118761 to J.F.M., NIH R01HG007538 to W.L.], CPRIT RP150292 [W.L.] the Vivian L. Smith Foundation [J.F.M.], and the American Heart Association (AHA) [AHA10POST4140029 and AHA12POST11760019 to T.H.; AHA NCRP SDG 0930240N to Y.M.; AHA 13POST17040027 to G. T.; AHA 12PRE11720003 to Y. B.]. Also supported by Baylor College of Medicine IDDRC Grant Number 1 U54 HD083092 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development and the Mouse Phenotyping Core at Baylor College of Medicine with funding from the NIH (U54 HG006348).
Footnotes
Author contributions: Y.M. and M.Z. designed and performed experiments and analyzed data; Y.B., T.H, J.L., G.T., and Y.X. performed experiments and analyzed data; W.L. analyzed data; J.T.W. provided reagents; J.F.M. designed and supervised experiments and analyzed data; Y.M., M.Z., and J.F.M. wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data availability: Microarray and ChIP-Seq data are available through the NCBI Gene Expression Omnibus (GEO) data repository under accession GSE44103.
REFERENCES AND NOTES
- 1.Kikuchi K, Poss KD. Cardiac regenerative capacity and mechanisms. Annual review of cell and developmental biology. 2012;28:719–741. doi: 10.1146/annurev-cellbio-101011-155739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabe Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisen J. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Berlo JH, Maillet M, Molkentin JD. Signaling effectors underlying pathologic growth and remodeling of the heart. J Clin Invest. 2013;123:37–45. doi: 10.1172/JCI62839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy SL, Xu J, Kochanek KD. Deaths: final data for 2010. National vital statistics reports : from the Centers for Disease Control and Prevention, National Center for Health Statistics, National Vital Statistics System. 2013;61:1–117. [PubMed] [Google Scholar]
- 5.Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu FX, Guan KL. The Hippo pathway: regulators and regulations. Genes Dev. 2013;27:355–371. doi: 10.1101/gad.210773.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heallen T, Zhang M, Wang J, Bonilla Claudio M, Klysik E, Johnson RL, Martin JF. Hippo pathway inhibits Wnt signaling to restrain cardiomyocyte proliferation and heart size. Science. 2011;332:458–461. doi: 10.1126/science.1199010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heallen T, Morikawa Y, Leach J, Tao G, Willerson JT, Johnson RL, Martin JF. Hippo signaling impedes adult heart regeneration. Development. 2013;140:4683–4690. doi: 10.1242/dev.102798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mosqueira D, Pagliari S, Uto K, Ebara M, Romanazzo S, Escobedo Lucea C, Nakanishi J, Taniguchi A, Franzese O, Di Nardo P, Goumans MJ, Traversa E, Pinto do OP, Aoyagi T, Forte G. Hippo Pathway Effectors Control Cardiac Progenitor Cell Fate by Acting as Dynamic Sensors of Substrate Mechanics and Nanostructure. ACS nano. 2014 doi: 10.1021/nn4058984. [DOI] [PubMed] [Google Scholar]
- 10.Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- 11.Xin M, Kim Y, Sutherland LB, Murakami M, Qi X, McAnally J, Porrello ER, Mahmoud AI, Tan W, Shelton JM, Richardson JA, Sadek HA, Bassel Duby R, Olson EN. Hippo pathway effector Yap promotes cardiac regeneration. Proc Natl Acad Sci U S A. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Z, von Gise A, Zhou P, Gu F, Ma Q, Jiang J, Yau AL, Buck JN, Gouin KA, van Gorp PR, Zhou B, Chen J, Seidman JG, Wang DZ, Pu WT. Cardiac Specific YAP Activation Improves Cardiac Function and Survival in an Experimental Murine Myocardial Infarction Model. Circ Res. 2014 doi: 10.1161/CIRCRESAHA.115.303632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J. An integrated semiconductor device enabling non optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 14.Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage determining transcription factors prime cis regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer Frick I, Shoukry M, Afzal V, Rubenstein JL, Rubin EM, Pennacchio LA, Visel A. Rapid and pervasive changes in genome wide enhancer usage during mammalian development. Cell. 2013;155:1521–1531. doi: 10.1016/j.cell.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neph S, Vierstra J, Stergachis AB, Reynolds AP, Haugen E, Vernot B, Thurman RE, John S, Sandstrom R, Johnson AK, Maurano MT, Humbert R, Rynes E, Wang H, Vong S, Lee K, Bates D, Diegel M, Roach V, Dunn D, Neri J, Schafer A, Hansen RS, Kutyavin T, Giste E, Weaver M, Canfield T, Sabo P, Zhang M, Balasundaram G, Byron R, MacCoss MJ, Akey JM, Bender MA, Groudine M, Kaul R, Stamatoyannopoulos JA. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, Farnham PJ, Hirst M, Lander ES, Mikkelsen TS, Thomson JA. The NIH Roadmap Epigenomics Mapping Consortium. Nature biotechnology. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghavi Helm Y, Klein FA, Pakozdi T, Ciglar L, Noordermeer D, Huber W, Furlong EE. Enhancer loops appear stable during development and are associated with paused polymerase. Nature. 2014;512:96–100. doi: 10.1038/nature13417. [DOI] [PubMed] [Google Scholar]
- 19.de Laat W, Duboule D. Topology of mammalian developmental enhancers and their regulatory landscapes. Nature. 2013;502:499–506. doi: 10.1038/nature12753. [DOI] [PubMed] [Google Scholar]
- 20.Lopaschuk GD, Jaswal JS. Energy metabolic phenotype of the cardiomyocyte during development, differentiation, and postnatal maturation. Journal of cardiovascular pharmacology. 2010;56:130–140. doi: 10.1097/FJC.0b013e3181e74a14. [DOI] [PubMed] [Google Scholar]
- 21.von Gise A, Lin Z, Schlegelmilch K, Honor LB, Pan GM, Buck JN, Ma Q, Ishiwata T, Zhou B, Camargo FD, Pu WT. YAP1, the nuclear target of Hippo signaling, stimulates heart growth through cardiomyocyte proliferation but not hypertrophy. Proc Natl Acad Sci U S A. 2012;109:2394–2399. doi: 10.1073/pnas.1116136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercer TR, Edwards SL, Clark MB, Neph SJ, Wang H, Stergachis AB, John S, Sandstrom R, Li G, Sandhu KS, Ruan Y, Nielsen LK, Mattick JS, Stamatoyannopoulos JA. DNase I hypersensitive exons colocalize with promoters and distal regulatory elements. Nat Genet. 2013;45:852–859. doi: 10.1038/ng.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zemljic Harpf A, Manso AM, Ross RS. Vinculin and talin: focus on the myocardium. Journal of investigative medicine : the official publication of the American Federation for Clinical Research. 2009;57:849–855. doi: 10.231/JIM.0b013e3181c5e074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bravo Cordero JJ, Magalhaes MA, Eddy RJ, Hodgson L, Condeelis J. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Biol. 2013;14:405–415. doi: 10.1038/nrm3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bjerke MA, Dzamba BJ, Wang C, DeSimone DW. FAK is required for tension dependent organization of collective cell movements in Xenopus mesendoderm. Dev Biol. 2014;394:340–356. doi: 10.1016/j.ydbio.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 28.Liu Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. Genetic and pharmacological disruption of the TEAD YAP complex suppresses the oncogenic activity of YAP. Genes Dev. 2012;26:1300–1305. doi: 10.1101/gad.192856.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barton ER. Impact of sarcoglycan complex on mechanical signal transduction in murine skeletal muscle. American journal of physiology Cell physiology. 2006;290:C411–419. doi: 10.1152/ajpcell.00192.2005. [DOI] [PubMed] [Google Scholar]
- 30.Goyenvalle A, Seto JT, Davies KE, Chamberlain J. Therapeutic approaches to muscular dystrophy. Hum Mol Genet. 2011;20:R69–78. doi: 10.1093/hmg/ddr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace GQ, McNally EM. Mechanisms of muscle degeneration, regeneration, and repair in the muscular dystrophies. Annual review of physiology. 2009;71:37–57. doi: 10.1146/annurev.physiol.010908.163216. [DOI] [PubMed] [Google Scholar]
- 32.Kshitiz, Hubbi ME, Ahn EH, Downey J, Afzal J, Kim DH, Rey S, Chang C, Kundu A, Semenza GL, Abraham RM, Levchenko A. Matrix rigidity controls endothelial differentiation and morphogenesis of cardiac precursors. Science signaling. 2012;5:ra41. doi: 10.1126/scisignal.2003002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- 34.Anastasi G, Cutroneo G, Gaeta R, Di Mauro D, Arco A, Consolo A, Santoro G, Trimarchi F, Favaloro A. Dystrophin glycoprotein complex and vinculin talin integrin system in human adult cardiac muscle. International journal of molecular medicine. 2009;23:149–159. [PubMed] [Google Scholar]
- 35.Li J, Goossens S, van Hengel J, Gao E, Cheng L, Tyberghein K, Shang X, De Rycke R, van Roy F, Radice GL. Loss of alphaT catenin alters the hybrid adhering junctions in the heart and leads to dilated cardiomyopathy and ventricular arrhythmia following acute ischemia. Journal of cell science. 2012;125:1058–1067. doi: 10.1242/jcs.098640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNally EM, Golbus JR, Puckelwartz MJ. Genetic mutations and mechanisms in dilated cardiomyopathy. J Clin Invest. 2013;123:19–26. doi: 10.1172/JCI62862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, Lopez AL, Morikawa Y, Tao G, Li J, Larina IV, Martin JF, Larin KV. Noncontact quantitative biomechanical characterization of cardiac muscle using shear wave imaging optical coherence tomography. Biomedical Optics Express. 2014;5:1980–1992. doi: 10.1364/BOE.5.001980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell. 2004;118:363–373. doi: 10.1016/j.cell.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 39.Wang ZB, Jiang ZZ, Zhang QH, Hu MW, Huang L, Ou XH, Guo L, Ouyang YC, Hou Y, Brakebusch C, Schatten H, Sun QY. Specific deletion of Cdc42 does not affect meiotic spindle organization/migration and homologous chromosome segregation but disrupts polarity establishment and cytokinesis in mouse oocytes. Mol Biol Cell. 2013;24:3832–3841. doi: 10.1091/mbc.E13-03-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saarikangas J, Mattila PK, Varjosalo M, Bovellan M, Hakanen J, Calzada Wack J, Tost M, Jennen L, Rathkolb B, Hans W, Horsch M, Hyvonen ME, Perala N, Fuchs H, Gailus Durner V, Esposito I, Wolf E, de Angelis MH, Frilander MJ, Savilahti H, Sariola H, Sainio K, Lehtonen S, Taipale J, Salminen M, Lappalainen P. Missing in metastasis MIM/MTSS1 promotes actin assembly at intercellular junctions and is required for integrity of kidney epithelia. Journal of cell science. 2011;124:1245–1255. doi: 10.1242/jcs.082610. [DOI] [PubMed] [Google Scholar]
- 41.Liu W, Komiya Y, Mezzacappa C, Khadka DK, Runnels L, Habas R. MIM regulates vertebrate neural tube closure. Development. 2011;138:2035–2047. doi: 10.1242/dev.058800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 43.Sadasivam S, Duan S, DeCaprio JA. The MuvB complex sequentially recruits B Myb and FoxM1 to promote mitotic gene expression. Genes Dev. 2012;26:474–489. doi: 10.1101/gad.181933.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tschop K, Conery AR, Litovchick L, Decaprio JA, Settleman J, Harlow E, Dyson N. A kinase shRNA screen links LATS2 and the pRB tumor suppressor. Genes Dev. 2011;25:814–830. doi: 10.1101/gad.2000211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lindsey ML, Goshorn DK, Comte Walters S, Hendrick JW, Hapke E, Zile MR, Schey K. A multidimensional proteomic approach to identify hypertrophy associated proteins. Proteomics. 2006;6:2225–2235. doi: 10.1002/pmic.200500013. [DOI] [PubMed] [Google Scholar]
- 46.Xing C, Xie H, Zhou L, Zhou W, Zhang W, Ding S, Wei B, Yu X, Su R, Zheng S. Cyclin dependent kinase inhibitor 3 is overexpressed in hepatocellular carcinoma and promotes tumor cell proliferation. Biochemical and biophysical research communications. 2012;420:29–35. doi: 10.1016/j.bbrc.2012.02.107. [DOI] [PubMed] [Google Scholar]
- 47.Ahuja P, Perriard E, Perriard JC, Ehler E. Sequential myofibrillar breakdown accompanies mitotic division of mammalian cardiomyocytes. Journal of cell science. 2004;117:3295–3306. doi: 10.1242/jcs.01159. [DOI] [PubMed] [Google Scholar]
- 48.Jopling C, Sleep E, Raya M, Marti M, Raya A, Izpisua Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Splinter E, de Wit E, van de Werken HJ, Klous P, de Laat W. Determining long range chromatin interactions for selected genomic sites using 4C seq technology: from fixation to computation. Methods (San Diego, Calif) 2012;58:221–230. doi: 10.1016/j.ymeth.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 50.Stadhouders R, Kolovos P, Brouwer R, Zuin J, van den Heuvel A, Kockx C, Palstra RJ, Wendt KS, Grosveld F, van Ijcken W, Soler E. Multiplexed chromosome conformation capture sequencing for rapid genome scale high resolution detection of long range chromatin interactions. Nature protocols. 2013;8:509–524. doi: 10.1038/nprot.2013.018. [DOI] [PubMed] [Google Scholar]
- 51.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, Lee JH, Puviindran V, Tam D, Shen M, Son JE, Vakili NA, Sung HK, Naranjo S, Acemel RD, Manzanares M, Nagy A, Cox NJ, Hui CC, Gomez Skarmeta JL, Nobrega MA. Obesity associated variants within FTO form long range functional connections with IRX3. Nature. 2014;507:371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]