Abstract
The individual human genome and epigenome are being defined at unprecedented resolution by current advances in sequencing technologies with important implications for human disease. This review uses examples relevant to clinical practice to illustrate the functional consequences of genetic and epigenetic variation. The insights gained from genome-wide association studies are described together with current efforts to understand the role of rare variants in common disease, set in the context of recent successes in Mendelian traits through the application of whole exome sequencing. The application of functional genomics to interrogate the genome and epigenome, build up an integrated picture of the regulatory genomic landscape and inform disease association studies is discussed, together with the role of expression quantitative trait mapping and analysis of allele-specific gene expression.
Keywords: common disease, genetic variation, genome-wide association, sequencing
Introduction
We live in remarkable times in the field of human genetics. Our ability to define variation at the sequence and structural level in the human genome is unprecedented, with whole genome sequencing now being performed on thousands of individuals [1]. Indeed, the pace of technological advances in sequencing is such that the target of the $1000 genome [2] now appears likely to be achieved within 3–5 years. For medicine, the opportunities afforded by genomic science are wide-ranging and potentially paradigm shifting, but amid the scientific optimism there is some justifiable concern from clinicians and patients about whether ‘genomic medicine’ will deliver. The investments from public and private funding in this field of research over the last 20 years have been enormous, and deliverables in terms of tangible change or benefit in the clinic are yet to be widely appreciated. In the current economic climate, the importance of translational research output is increasingly sought and should be more actively considered by those engaged in genomic research. This will be facilitated by the growing involvement of other professionals in this field, notably from public health and the pharmaceutical industry. The potential is, however, undoubtedly present: enormous strides have been made in our understanding of the heritable component of common multifactorial disease, while research into the genetics of rare diseases showing a Mendelian pattern of inheritance is undergoing a renaissance as new sequencing technologies offer the opportunity to tackle diseases where linkage and other approaches had previously been unsuccessful [3].
In this review, I will briefly outline the historical context of current progress in genomic research as applied to human disease, illustrating how much of this has been made possible by radical technological advances. This has allowed us to resolve sequence level and structural genomic variation and apply this in a high-throughput manner to study thousands of individuals, for example, using genotyping arrays comprising hundreds of thousands of common biallelic single nucleotide substitutions [4] and more recently massively parallel ‘next generation’ high-throughput DNA sequencing [5, 6]. I will discuss the successful application of genome-wide association studies (GWAs) to common disease, the insights this has provided but also the challenges that lie ahead as we seek to define the substantial proportion of the estimated heritable risk remaining unexplained in multifactorial traits. Current efforts to define the role of rarer variants, structural genomic variants, gene–gene and gene–environment interactions, epigenetic and other factors are being actively pursued to try and address this problem.
A fundamental question that remains unanswered for the majority of disease associations in common traits, as well as many Mendelian diseases, is the identity and functional consequence of the causal genetic variants for gene expression, the nature and function of the encoded protein, and disease pathogenesis [7]. As we understand more about the remarkably complex processes by which gene expression is regulated, proteins synthesized and cellular systems operate, this challenge to define functional variants appears increasingly daunting. However, advances in functional genomics, notably taking advantage of new sequencing technologies to allow genome-wide resolution of transcription and the regulatory chromatin landscape, offer exciting new opportunities to do so, particularly when combined with proteomic, metabolomic and other approaches. This review discusses some of the approaches that can be taken to define functional variants, current limitations and future directions.
Linkage, GWAs and rare variants
For Mendelian disorders, the application of linkage-based approaches has been extremely successful with thousands of gene loci and specific mutations identified over the last 30 years [8]. By contrast, progress in common multifactorial disease without a clear Mendelian pattern of inheritance was slow [9]. Linkage did yield some notable successes such as the role of NOD2 in Crohn’s disease [10] but was not a tractable approach in the vast majority of cases. Similarly, candidate gene analysis, while being fruitful in some instances such as APOE e4 in Alzheimer’s disease [11, 12] or factor V Leidin in venous thrombosis [13], in most cases was often unsuccessful or yielded associations that failed to replicate [14]. The common disease: common variant hypothesis became tractable to test with the advent of affordable high-throughput genotyping, and growing insights into the nature and coinheritance of genetic variation across different populations through large collaborative studies such as the International HapMap Project [15]. This set the scene for GWAs in which informative common biallelic genetic markers could be genotyped in thousands of cases and controls to look for the evidence of association [9]. One minor note in terms of terminology: single nucleotide variants (SNVs) include single nucleotide substitutions, which when present in the human population with a frequency of both alleles of >1% are also referred to as single nucleotide polymorphisms (SNPs). Rare SNVs may be defined based on a minor allele frequency (MAF) <1% but of note, GWAs typically involve genotyping SNPs with a MAF >5% which means both rare and less common variants (MAF 1–5%) are not well captured.
Genome-wide association studies
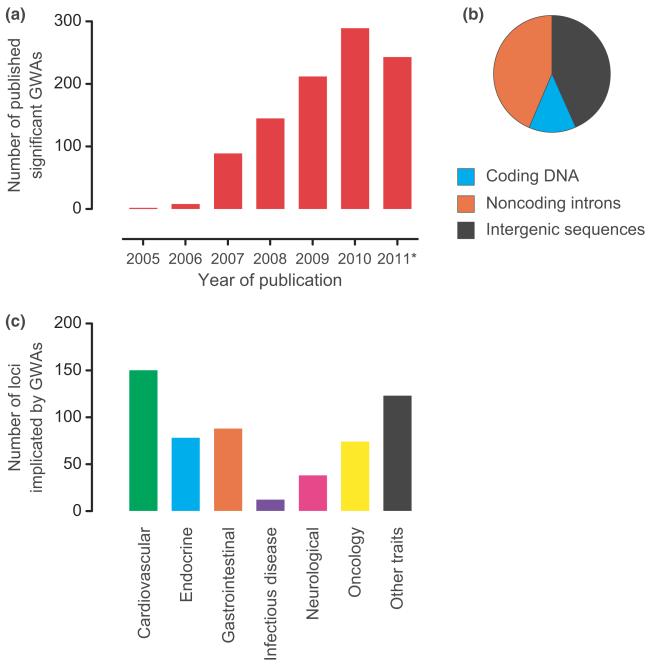
By June 2011, 1449 genome-wide associations have been reported at a P value of <5 × 10−8 for 237 traits (http://www.genome.gov/gwastudies/) (Fig. 1). The results have been striking in terms of the strength of association, with many variants implicated with considerable statistical confidence for diseases ranging from type I diabetes [16, 17] to leprosy [18, 19] and cancer [20]. However, the magnitude of effect of individual disease-associated variants was in almost all cases very modest, typically 1.2-fold, and the proportion of the estimated heritability explained by such variants was relatively low, for example, ranging from 5% to 10% in type II diabetes [21] to 25% in Crohn’s disease [22]. It would be wrong, however, to interpret this as meaning that GWAs have been unsuccessful: for the first time, we have a substantial number of robustly replicated associations with common genetic markers which are starting to be translated into risk modelling and prediction of clinical utility, notably in cancer, [23, 24]. More evident are the new insights into disease pathogenesis which GWAs are providing, ranging from the role of complement factor H in age-related macular degeneration [25] to Crohn’s disease where the significance of autophagy [26–29] and IL23 signalling [28, 30] has been highlighted and is providing new targets for therapeutic intervention [31, 32].
Fig. 1.
Genome-wide association studies. (a) Number of published genome-wide association studies (GWAs) reporting at least one significant single-nucleotide polymorphisms (SNP) trait association by year to October 2011 catalogued by the National Human Genome Research Institute (NHGRI) GWAS Catalog [146] [Hindorff LA, MacArthur J, Wise A, Junkins HA, Hall PN, Klemm AK, and Manolio TA. A Catalog of Published Genome-Wide Association Studies. Available at: http://www.genome.gov/gwastudies. Accessed 10/30/2011]; (b) Schematic showing location of associated marker SNPs from GWAs by frequency [147]; (c) Reported GWAs loci by disease classification [147].
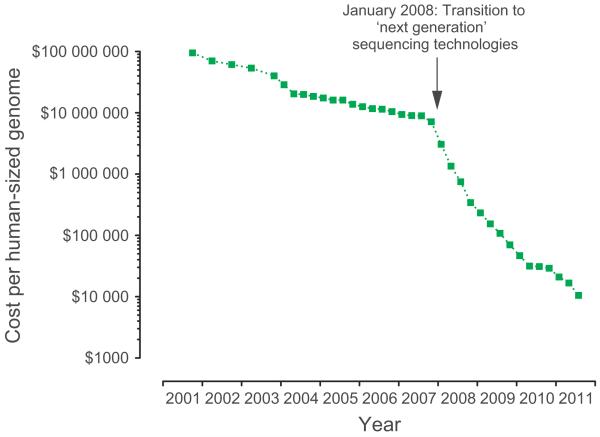
The ‘missing heritability’ in common disease following GWAs has been the subject of much debate [33–35]. Dubbed by some investigators as the ‘dark matter’, which underlined the elusive nature of resolving the basis of this heritable risk, current views highlight the potential role of rarer variants with moderate or high magnitude of effect. GWAs have not interrogated such variants to date, and their analysis has become achievable through the increasing application of massively parallel sequencing as costs continue to fall (Fig. 2). Anticipated results over the next 12 months for ongoing studies involving in large numbers of cases will be highly informative in the context of common disease. There may also be much potentially useful information within GWAs data sets involving associated variants just below the selected thresholds for statistical significance: mining such data and sifting the wheat from the chaff is challenging and may be facilitated by increasing sample sizes (with a note of caution in terms of the cost benefit of doing so) and using functional genomics and other approaches to try and inform this process. There are many other potentially relevant contributors to this phenomenon of unexplained heritability: the estimations of heritable risk may be overinflated, and epigenetic factors are increasingly recognized to be significant contributors to heritable risk (including parent of origin effects and environmental modulators), while gene–gene and gene–environment interactions have not yet been well characterized.
Fig. 2.
DNA sequencing cost of the human genome. The dramatic fall in sequencing costs is illustrated by data arising from sequencing centres funded by the NHGRI [Wetterstrand KA. DNA Sequencing Costs: Data from the NHGRI Large-Scale Genome Sequencing Program Available at: http://www.genome.gov/sequencingcosts. Accessed 10/30/2011].
Rare variants
The relentless pace of technological advances in our ability to detect and quantify genetic variation in a high-throughput manner now makes the analysis of rarer variants a feasible option at the whole exome and increasingly the whole genome level. While for common disease, the jury remains out on the relative importance of such variants, there is growing optimism that for rare ‘orphan’ diseases with very robust phenotypes such as primary immunodeficiencies and metabolic disorders, the potential is very great, while for Mendelian diseases, considerable success has already been reported [36–40].
The potential of whole exome sequencing was underlined in 2009 by data from sequencing four unrelated individuals with the rare autosomal dominant disorder Freeman Sheldon syndrome that resolved the known causal gene [41]. Whole exome sequencing has since been successfully used to determine the genetic basis of a number of unresolved Mendelian disorders. This includes autosomal dominant traits where, for example, sequencing 10 unrelated pro-bands resolved mutations of MLL2 as a major cause of Kabuki syndrome [38], while for Schinzel–Giedion syndrome, SETB1 was implicated following whole exome sequencing of four unrelated individuals which revealed de novo mutations involving this gene [42]. For autosomal recessive diseases, success has also been achieved as illustrated by work involving whole exome sequencing of four individuals from three families with Miller syndrome which defined DHODH as the disease gene [39], and for hyperphosphatasia mental retardation syndrome where mutations in PIGV were resolved following whole exome sequencing of three siblings with validation in additional families [43]. This latter work also highlighted the power of filtering regions based on identity by descent.
The value of whole exome sequencing using a family-based approach for sporadic disease was illustrated for 10 cases of unexplained severe mental retardation where case parent trios were sequenced and de novo likely pathogenic nonsynonymous SNVs identified in seven of the affected individuals [44]. For very specific phenotypes where extensive biochemical and functional data and validation are possible, whole exome sequencing of a single individual may be informative as illustrated for a mitochondrial respiratory chain disorder where a mutation involving ACAD9 (encoding acyl-CoA dehydrogenase 9) was identified and causally implicated in disease, with other mutations in the same gene identified in further cases [45].
The utility of whole exome sequencing for clinical diagnosis is also increasingly recognized. This is illustrated by a patient referred with renal disease in whom Bartter syndrome was suspected: sequencing revealed a mutation in SLC26A3, leading to the diagnosis of congenital chloride diarrhoea [36]. A further case involving a young patient with intractable and atypical inflammatory bowel disease illustrates how the therapeutic implications can be significant. In this instance, whole exome sequencing revealed a mutation in XIAP, knowledge of which contributed to a clinical decision to carry out stem cell transplantation [46, 47].
The optimal strategic approach to apply whole exome and whole genome sequencing in Mendelian disease is still in the process of being resolved for different scenarios, while for common multifactorial traits, how to apply high-throughput sequencing approaches is a source of considerable debate. If families can be identified, then sequencing distantly related individuals within the pedigree, looking for cosegregation and testing specific implicated variants in large cohorts may be fruitful, while for other traits, studying individuals in the extreme tails of the phenotype distribution, particularly where there is younger age of onset, is advocated [48]. As costs continue to fall, however, whole genome sequencing of hundreds or thousands of cases and controls to identify all variants will be carried out.
The bioinformatic and analytical challenges such data sets represent should not be underestimated [49]. The mapping and accurate calling of sequence and structural variants remain a very active area of development and research, in which further progress is being made and urgently needed [50–54]. The amounts of data involved are prodigious and on a scale more commonly encountered in astrophysics. Accepting that these challenges can be overcome, the subsequent analysis to narrow down the lists of potentially deleterious variants causing disease is challenging enough in Mendelian traits. Recent data from the 1000 Genomes Project have highlighted how on average, each of us has 250–300 loss-of-function variants in annotated genes and 50–100 variants previously associated with inherited disease [1].
There are several examples of common multifactorial traits where rare variants have been shown to play a significant role. Resequencing candidate genes identified through GWAs has been productive, with rare variants with large effects resolved in hypertriglyceridaemia [55], Crohn’s disease [56] and type I diabetes [57]. The latter highlighted rare variants in IFIH1, a gene encoding interferon induced with helicase domain 1, which is important in the recognition of RNA from picornaviruses, and may be highly relevant given the link between enteroviruses and development of diabetes [57]. Other candidate genes resolved through animal studies such as SIAE (encoding the enzyme sialic acid acetyl transferase) have revealed several functionally important rare variants associated with autoimmune disease [58]. Further examples from autoimmune disease include the association of rare variants in the DNA exonuclease gene TREX1 with systemic lupus erythematosus [59].
For common traits, we can anticipate that if rare variants play a role, lessons should be learned from Mendelian diseases such that analysing association with rare variants present at a given gene or locus may be of value with many different mutations resulting in a common phenotype. Various analytical strategies are being advocated and have been reviewed elsewhere [49]. A blurring of the distinction between common and Mendelian disease is apparent as we also appreciate the role of modifier genetic variants and the environment in observed penetrance and phenotypic heterogeneity in Mendelian disease, such that conditions such as sickle cell disease are viewed as complex multigenic disorders rather than monogenic disease [60]. The role of modifier variants is highlighted by recent work in cystic fibrosis, where genome-wide association and linkage analysis have highlighted variation at chromosome 11p13 and 20q13.2, respectively, in modulating observed variation in the severity of lung disease among patients with two copies of loss-of-function CFTR alleles [61].
Functional genomics and epigenomics of the individual
Advances in our ability to sequence DNA have had important ramifications beyond the identification and screening of genetic and genomic variants. Application of the technologies for high-throughput sequencing to analyse RNA (RNA-seq) represents a significant advance on microarray-based approaches in terms of the dynamic range that can be achieved with single-base-pair resolution [62]. Being able to interrogate the transcriptome at this level of resolution using increasingly small amounts of input RNA is revolutionizing our ability to understand the function of the genome and more particularly in the context of this review, to appreciate how genetic variation may modulate the critical processes of alternative splicing [63–65] and the generation of noncoding RNAs which have profound implications for gene regulation [66–68].
In parallel, sequencing technologies are radically advancing our understanding of the broader transcriptional landscape at genome-wide resolution, for example, in terms DNA methylation, chromatin accessibility, specific histone modifications and transcription factor binding (Fig. 3). Such data, notably through international collaborative studies such as the ENCODE (ENCyclopedia Of DNA Elements) Project [69], are publically available for a range of cell lines allowing investigators to interrogate specific loci or integrate data sets using genome browsers [70] such that markers from disease GWAs may be over-laid onto RNA-seq and ChIP-seq data, together with other information on sequence conservation and putative regulatory elements, to help generate hypotheses and prioritize disease-associated variants. It is important, however, to consider the disease-relevant context for such analyses, as any effects of specific variants are increasingly recognized to be cell or tissue type specific [71–73]. This means that we need to continue to expand such data sets for a range of cell and tissue types, including primary cells from healthy individuals as well as patients with disease.
Fig. 3.
Resolving the transcriptional landscape of allele-specific gene expression. Allele-specific differences in gene expression may arise because of sequence variation, for example, in distant enhancer regions. Such sites may be indicated by allelic differences in chromatin accessibility, specific histone modifications and DNA methylation, as well as recruitment of specific transcription factors.
We are also beginning to understand the three-dimensional structure of the transcribing genome, in particular how different genomic regions interact, through experimental approaches based on chromosome conformation capture which can now be performed at genome-wide resolution. These take advantage of new sequencing technologies to sequence the products of proximity-based ligation (Hi-C) [74] and chromatin interaction analysis by paired end tag sequencing (ChIA-PET) [75]. Other approaches and analyses can also be highly informative, for example, based on systems biology to define interactions and biological pathways. By using information from many different sources now available at genome-wide resolution, we can adopt an integrated approach to understanding the functional genomic context of genetic and epigenetic variation [76] and do so in a disease-relevant manner.
Such integrative approaches should facilitate the generation of specific hypotheses regarding the mechanism of action and location of putative functional variants in a more systematic and high-throughput manner than has previously been possible. In some instances, this will relate to structural changes in the encoded protein with consequences for function, which may be profound; in other cases, it may involve variants modulating levels of gene expression in many different ways [7]. Infectious disease has provided striking examples of such events, notably malaria. Here, the protective role of sickle cell trait was established as arising from a glutamic acid to valine substitution that converts normal adult haemoglobin (HbA) to haemoglobin S (Hb S, sickle variant haemoglobin) [77] and arises because of an A to T single nucleotide substitution in the HBB gene [78] that protects against the development of severe cerebral malaria because of Plasmodium falciparum. The molecular mechanisms underlying this include the induction of heme oxygenase-1, suppression of circulating free heme by carbon monoxide and independent immunoregulatory effects on pathogenic CD8+ T cells [79]. By contrast, a G to A single nucleotide substitution in the promoter region of the DARC gene (Duffy blood group, chemokine receptor) was found to modulate transcription factor binding by GATA-1, dramatically reducing the levels of gene expression in a cell-type-specific manner and rendering red blood cells resistant to invasion by Plasmodium vivax [80]. Structural variants may also be highly significant, as noted for a 32-bp deletion in the CCR5 gene encoding the major host coreceptor for HIV-1, the CC chemokine receptor CCR5. The deletion resulted in a frameshift, prematurely terminating translation and truncating the protein and rendering cells resistant to invasion by HIV-1 when present in the homozygous state [81–83]. Copy number variation also plays a critical role. Indeed, copy number of a segmental duplication spanning CCL3L1, encoding chemokine (C-C motif) ligand 3-like 1, which is the most significant ligand for CCR5 and is a potent HIV-1 suppressive chemokine, varies by individual with most people having 1–6 copies: when present at lower than the population average, copy number of CCL3L1 was associated with increased HIV-1/AIDS susceptibility [84].
Expression quantitative trait mapping
A powerful approach aiding the interpretation of GWAs is based on mapping gene expression as a quantitative trait [85, 86]. Gene expression is recognized to vary widely between and within populations and to be heritable [87]. The application of ‘genetical genomic’ approaches in model systems and humans has highlighted that expression quantitative trait loci (eQTL) and more specifically in the context of GWAs, expression-associated SNVs, are common and informative [88, 89]. Many such studies have been carried out in EBV-transformed lymphoblastoid cell lines (LCLs), while more recent eQTL analyses in humans are being carried out in specific cell types and tissues [72, 90–96]. This is important, as association with differential gene expression is recognized to be context specific – for example, over 50% of cis-eQTL defined in LCLs or T cells were cell type specific [72].
An elegant early example of integrating GWAs and eQTL data was provided by the work of Cookson and colleagues [97] who found that SNVs associated with childhood onset asthma at chromosome 17q21 were also significantly associated with expression of the neighbouring gene ORMDL3 in LCLs established from children in the asthma family panel. This cis association was subsequently also noted in peripheral blood leucocytes [98], and the locus is of broad interest given significant association with other autoimmune diseases including type 1 diabetes [99], Crohn’s disease [100] and primary biliary cirrhosis [101]. ORMDL3 encodes a protein involved in regulating endoplasmic reticulum-mediated calcium signalling, in turn modulating the unfolded protein response which is proposed to provide a link with inflammation [102]. This situation is complex with a number of variants implicated and likely several genes involved, with evidence of allele-specific chromatin remodelling in the region and involvement of the insulator factor CTCF [103]. More recently, other investigators have shown the value of eQTL mapping in interpreting GWAs for a range of traits including coeliac disease [104], body mass index [105] and psoriasis [106].
When considering eQTL data, it is important to note that the vast majority of genome-wide data sets to date have been generated using expression microarrays and that observed associations for specific oligonucleotide probes may be confounded in many cases by sequence variation falling within the sequence bound by the probe – this may lead to spurious results apparently suggesting an eQTL is present [107].
Recently published work involving type II diabetes and metabolic traits further highlights the informativeness of an integrated approach to following up GWAs signals [108]. A striking example was noted at chromosome 7q32.3 of significant association with type II diabetes [21] and HDL cholesterol [109], with a parent of origin effect involving the maternal allele and differential expression of the KLF14 gene [110]. A local likely cis-acting eQTL was defined in adipose tissue for the expression of KLF14, a gene that encodes the transcription factor Kruppel Factor 14. Strikingly, the same associated variants show strong trans-eQTL with at least 10 genes in adipose tissue which were found to be enriched for KLF binding sites and whose expression correlates with metabolic phenotypes and themselves harbour metabolic trait-associated variants [108]. This data underlines the complexity of how particular variants may modulate function in a given tissue, dependent on epigenetic mechanisms and in turn helping resolve further disease associations within existing GWAs data sets as well as providing new insights into disease process.
Transcript profiling using RNA-seq
RNA-seq has been successfully applied to eQTL mapping. This is exciting given the inherent advantages of the technology, which does not rely on hybridization but directly sequences the transcripts. Data for LCLs established from individuals of European and African ancestry have highlighted the increased resolution that can be achieved, notably including associations with the expression of alternatively spliced isoforms and long noncoding RNAs, as well as highlighting effects on transcriptional termination [95, 96]. With greater read length and coverage, analysis arising from RNA-seq will be increasingly informative while falling reagent costs and opportunities for multiplexing should ensure broad application across the field [111]. There are, however, significant bioinformatic challenges remaining in the analysis of RNA-seq data, not least relating to read mapping, quantification of expression at transcript isoform resolution and differential expression [112]. Potential bias introduced by the reference genome used for mapping is a particular issue for allele-specific quantification using RNA-seq as discussed later in this review.
Transcription landscape and epigenetics
The analysis of chromatin immunoprecipitation experiments using high-throughput sequencing (ChIP-seq) is providing genome-wide resolution of binding by specific transcription factors in a variety of contexts, as well as the presence of specific histone modifications allowing interrogation of epigenetic mechanisms. ChIP-seq analysis of 10 LCLs for NF-κB and RNA polymerase II binding underlined how often any two individuals differ in observed binding regions (7.5% and 25%, respectively), while human/chimp comparison indicated differences in 32% of sites [113]. Strong correlation with SNVs and structural variants was noted.
Understanding the regulatory transcriptional landscape is highly informative when considering GWAs, as illustrated by our work mapping binding by the ligand-activated vitamin D receptor (VDR) to DNA which demonstrated significant enrichment in GWAs intervals for a variety of autoimmune diseases as well as cancer and other specific traits related to vitamin D [114]. This provides a route-map for GWAs signals that may relate to genes modulated by vitamin D and provides a link with growing epidemiological evidence implicating vitamin D in autoimmune disease susceptibility. Other data for specific loci such as HLA-DRB1 illustrate how disease risk haplotypes may be associated with allele-specific recruitment of VDR, providing a potential link between genetic and environmental risk factors for disease [115].
Specific histone marks such as H3K4me1 are often associated with more distant enhancer elements [116]. Such sites may also be characterized by open chromatin as revealed by DNase I hyper-sensitivity (DHS) mapping. While conventionally analysed by Southern blotting, DHS experiments can also be analysed using high-throughput sequencing (DNase-seq) [117]. Active chromatin sites based on DNase-seq and ChIP-seq for the transcription factor CTCF showed evidence of heritability when analysed in LCLs from family pedigrees with 10% of sites individual specific [118].
A further, experimentally more straightforward method to assay open chromatin is FAIRE (Formaldehyde-Assisted Isolation of Regulatory DNA Elements) [119], which proved very informative in understanding GWAs signals for type II diabetes [120]. FAIRE-seq analysis in pancreatic islet cells resolved that a variant associated with type II diabetes in the TCF7L2 gene (encoding the transcription factor 7-like 2) was not only located in a region of open chromatin but showed allelic differences in accessibility and enhancer activity with evidence of tissue specificity [121]. Such work underlines the future importance of characterizing the function of disease-associated variants in the most disease-relevant cell/tissue type and context, making use of a variety of approaches to resolve associations. The recent generation of publically available ChIP-seq, DNase-seq and FAIRE-seq data sets for a number of different cell types [122, 123] is an important next step in such work.
The power of analysis defining chromatin accessibility and modifications in combination with gene expression is illustrated by the work of Higgs and colleagues in the α globin locus. Careful study of this region in the context of thalassaemia has been highly informative in understanding fundamental processes in gene regulation and the impact of particular mutations [124, 125]. These can be dramatic, as found for a gain-of-function intergenic regulatory variant identified in individuals with α-thalassaemia which was associated with allele-specific histone acetylation, recruitment of transcription factors and Pol II binding resulting in a new transcriptionally active region, ‘stealing’ transcriptional activity from downstream α globin genes whose expression was significantly reduced [126].
DNA methylation is a critical epigenetic alteration modulating gene expression that involves addition of a methyl group to the 5 position of the pyrimidine ring of cytosine residues in CpG dinucleotides [127]. DNA methylation is known to be dependent on cell type, developmental stage and environmental factors. Recently, it was found that allele-specific differences in DNA methylation at nonimprinted loci are common across the genome [128] and that CpG methylation can be mapped as a quantitative trait [129]. A number of techniques are available for analysing DNA methylation at genome-wide resolution, based primarily on restriction enzyme digestion as seen with Methyl-seq, or affinity enrichment, for example, with antibodies specific for methylated cytosines (Me-DIP-seq) [130]. Whole genome bisulphite sequencing remains technically challenging but would offer significant advantages including single-base resolution.
High-throughout sequencing is also dramatically advancing our understanding of the role of microRNAs in gene expression acting through posttranscriptional mechanisms, a process thought to be critical in 30% of genes [131]. A striking example of how underlying sequence variation may modulate this process was seen for HLA-C where an insertion deletion polymorphism in the 3′UTR affected binding by miR-148 [132]. This provided a mechanism for the observed association of a linked SNP 35 kb upstream of HLA-C that was previously strongly associated with HIV control [133].
Allele-specific gene expression
The analysis of allele-specific gene expression has proved a powerful approach to try and resolve functional variants. When present in the heterozygous state in an individual, a biallelic genetic variant such as a single nucleotide substitution can provide a useful marker of the allelic origin of a transcript and allele-specific expression. For example, when located in coding sequence, transcript abundance specific to each allele can be quantified based on the presence of the variant in the transcribed RNA [134]. For genes without transcribed genetic markers, relative allelic expression can be assessed by haplotype-specific chromatin immunoprecipitation (haploChIP) for RNA polymerase II using antibodies specific for the phosphorylated serine residues in the C-terminal domain characteristic of actively transcribing RNA polymerase II [135]. This approach can also be used to resolve allele-specific recruitment of specific transcription factors such as activated B cell factor-1 which we demonstrated was recruited to the LTA (encoding lymphotoxin alpha) gene [136] in the presence of a genetic variant subsequently associated with susceptibility to leprosy [137].
For a small number of imprinted genes, monoallelic expression is seen dependent on the parental origin of alleles [138]. Genome-wide surveys have shown that smaller differences in allelic expression are common, involving an estimated 20% of autosomal genes with typically differences in relative allelic expression of 1.5-fold [139–141]. Genome-wide allele-specific discrimination is now possible to high resolution using RNA-seq [95, 96], but there is potential bias dependent on the reference sequence to which reads are mapped which may or may not be a match for the particular read from a given individual [142]. Analysis of allele-specific gene expression is highly complementary to eQTL data and can be integrated to facilitate mapping of likely regulatory variants [95, 96, 143].
The analysis of individuals homozygous for particular genomic regions can also be informative. We recently analysed gene expression for LCLs established from individuals homozygous for autoimmune disease risk haplotypes spanning 3.5 Mb of the classical Major Histocompatibility Complex (MHC) on chromosome 6p21 and demonstrated that allelic differences were common and often involved alternative splicing [144]. This was made possible by using a custom microarray that combined a strand-specific tiling path probe set with probes specific to known and predicted splice junctions and included alternate allele probes for sequence variants identified by resequencing as part of the MHC Haplotype Project [145].
Conclusions
There is no doubt that recent advances in genomics, currently driven by new high-throughput sequencing techniques, are taking us to remarkable new levels in our understanding of the human genome, and the genetic and epigenetic variation that exists, with important implications for our understanding of human disease. As this knowledge grows, our appreciation of the complexity with which we are faced is also underlined. For common multifactorial traits, GWAs have been very informative but leave much heritable risk unresolved. Rarer variants may prove important but in general, more integrated approaches are needed in which environmental risk factors are considered and combined with functional genomic analyses. Moreover, we need to derive functional genomic data in a disease-relevant setting as the consequences of underlying genetic and epigenetic diversity are increasingly recognized to be highly context specific.
Current technologies that can interrogate the whole genome carry with them significant caveats: these tools are new, and successful application to important biological problems requires careful experimental design and consideration of the limitations inherent in such approaches. The data sets involved are highly complex, and analysis remains extremely challenging with significant risks of false positive and negative results until the field matures. High-throughput sequencing is not a panacea but a critical tool in current genomics. Used wisely, it is resolving the individual genome and epigenome, at a structural and functional level, and will radically advance our understanding of disease. For Mendelian traits, the impact is already being felt. For common multifactorial diseases, this may take a little longer.
Acknowledgements
The work of the author is funded by the Wellcome Trust [grant number 074318 /075491/Z/04], the Medical Research Council [grant ID 98082] and the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013) / ERC Grant agreement number 281824.
Footnotes
Conflicts of interest statement
No conflicts of interest to declare.
References
- 1.Durbin RM, Abecasis GR, Altshuler DL, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mardis ER. Anticipating the 1,000 dollar genome. Genome Biol. 2006;7:112. doi: 10.1186/gb-2006-7-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ku CS, Naidoo N, Pawitan Y. Revisiting Mendelian disorders through exome sequencing. Hum Genet. 2011;129:351–70. doi: 10.1007/s00439-011-0964-2. [DOI] [PubMed] [Google Scholar]
- 4.Wang DG, Fan JB, Siao CJ, et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science. 1998;280:1077–82. doi: 10.1126/science.280.5366.1077. [DOI] [PubMed] [Google Scholar]
- 5.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 6.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–45. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 7.Knight JC. Human Genetic Diversity. Functional Consequences for Health and Disease. 1st edn Oxford University Press; Oxford: 2009. [Google Scholar]
- 8.Botstein D, Risch N. Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet. 2003;33:228–37. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 9.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–8. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugot JP, Laurent-Puig P, Gower-Rousseau C, et al. Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature. 1996;379:821–3. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 11.Saunders AM, Strittmatter WJ, Schmechel D, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–72. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 12.Strittmatter WJ, Saunders AM, Schmechel D, et al. Apolipoprotein E: high-avidity binding to beta-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:1977–81. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bertina RM, Koeleman BP, Koster T, et al. Mutation in blood coagulation factor V associated with resistance to activated protein C. Nature. 1994;369:64–7. doi: 10.1038/369064a0. [DOI] [PubMed] [Google Scholar]
- 14.Hirschhorn JN, Lohmueller K, Byrne E, Hirschhorn K. A comprehensive review of genetic association studies. Genet Med. 2002;4:45–61. doi: 10.1097/00125817-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Altshuler D, Brooks LD, Chakravarti A, Collins FS, Daly MJ, Donnelly P. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Polychronakos C, Li Q. Understanding type 1 diabetes through genetics: advances and prospects. Nat Rev Genet. 2011;12:781–92. doi: 10.1038/nrg3069. [DOI] [PubMed] [Google Scholar]
- 17.Bradfield JP, Qu HQ, Wang K, et al. A genome-wide meta-analysis of six type 1 diabetes cohorts identifies multiple associated Loci. PLoS Genet. 2011;7:e1002293. doi: 10.1371/journal.pgen.1002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang FR, Huang W, Chen SM, et al. Genomewide association study of leprosy. N Engl J Med. 2009;361:2609–18. doi: 10.1056/NEJMoa0903753. [DOI] [PubMed] [Google Scholar]
- 19.Zhang F, Liu H, Chen S, et al. Identification of two new loci at IL23R and RAB32 that influence susceptibility to leprosy. Nat Genet. 2011;43:1247–51. doi: 10.1038/ng.973. [DOI] [PubMed] [Google Scholar]
- 20.Chung CC, Chanock SJ. Current status of genome-wide association studies in cancer. Hum Genet. 2011;130:59–78. doi: 10.1007/s00439-011-1030-9. [DOI] [PubMed] [Google Scholar]
- 21.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J, Kader AK, Hsu FC, et al. Inherited genetic markers discovered to date are able to identify a significant number of men at considerably elevated risk for prostate cancer. Prostate. 2011;71:421–30. doi: 10.1002/pros.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jostins L, Barrett JC. Genetic risk prediction in complex disease. Hum Mol Genet. 2011;20:R182–8. doi: 10.1093/hmg/ddr378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–9. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hampe J, Franke A, Rosenstiel P, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–11. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 27.Parkes M, Barrett JC, Prescott NJ, et al. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn’s disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rioux JD, Xavier RJ, Taylor KD, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.WTCCC Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duerr RH, Taylor KD, Brant SR, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mathew CG. New links to the pathogenesis of Crohn disease provided by genome-wide association scans. Nat Rev Genet. 2008;9:9–14. doi: 10.1038/nrg2203. [DOI] [PubMed] [Google Scholar]
- 32.Cho JH, Brant SR. Recent insights into the genetics of inflammatory bowel disease. Gastroenterology. 2011;140:1704–12. doi: 10.1053/j.gastro.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eichler EE, Flint J, Gibson G, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11:446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spencer C, Hechter E, Vukcevic D, Donnelly P. Quantifying the underestimation of relative risks from genome-wide association studies. PLoS Genet. 2011;7:e1001337. doi: 10.1371/journal.pgen.1001337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi M, Scholl UI, Ji W, et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci U S A. 2009;106:19096–101. doi: 10.1073/pnas.0910672106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Musunuru K, Pirruccello JP, Do R, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–7. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–3. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42:30–5. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sobreira NL, Cirulli ET, Avramopoulos D, et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet. 2010;6:e1000991. doi: 10.1371/journal.pgen.1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hoischen A, van Bon BW, Gilissen C, et al. De novo mutations of SETBP1 cause Schinzel–Giedion syndrome. Nat Genet. 2010;42:483–5. doi: 10.1038/ng.581. [DOI] [PubMed] [Google Scholar]
- 43.Krawitz PM, Schweiger MR, Rodelsperger C, et al. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat Genet. 2010;42:827–9. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- 44.Vissers LE, de Ligt J, Gilissen C, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–12. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 45.Haack TB, Danhauser K, Haberberger B, et al. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat Genet. 2010;42:1131–4. doi: 10.1038/ng.706. [DOI] [PubMed] [Google Scholar]
- 46.Manolio TA, Green ED. Genomics reaches the clinic: from basic discoveries to clinical impact. Cell. 2011;147:14–6. doi: 10.1016/j.cell.2011.09.012. [DOI] [PubMed] [Google Scholar]
- 47.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–62. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 48.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–25. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 49.Bansal V, Libiger O, Torkamani A, Schork NJ. Statistical analysis strategies for association studies involving rare variants. Nat Rev Genet. 2010;11:773–85. doi: 10.1038/nrg2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lunter G, Goodson M. Stampy: a statistical algorithm for sensitive and fast mapping of Illumina sequence reads. Genome Res. 2010;21:936–9. doi: 10.1101/gr.111120.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin Y, Li J, Shen H, Zhang L, Papasian CJ, Deng HW. Comparative studies of de novo assembly tools for next-generation sequencing technologies. Bioinformatics. 2011;27:2031–7. doi: 10.1093/bioinformatics/btr319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen Y, Wan Z, Coarfa C, et al. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Res. 2010;20:273–80. doi: 10.1101/gr.096388.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Handsaker RE, Korn JM, Nemesh J, McCarroll SA. Discovery and genotyping of genome structural polymorphism by sequencing on a population scale. Nat Genet. 2011;43:269–76. doi: 10.1038/ng.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johansen CT, Wang J, Lanktree MB, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42:684–7. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rivas MA, Beaudoin M, Gardet A, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat Genet. 2011;43:1066–73. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type1 diabetes. Science. 2009;324:387–9. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Surolia I, Pirnie SP, Chellappa V, et al. Functionally defective germline variants of sialic acid acetylesterase in autoimmunity. Nature. 2010;466:243–7. doi: 10.1038/nature09115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Namjou B, Kothari PH, Kelly JA, et al. Evaluation of the TREX1 gene in a large multi-ancestral lupus cohort. Genes Immun. 2011;12:270–9. doi: 10.1038/gene.2010.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Higgs DR, Wood WG. Genetic complexity in sickle cell disease. Proc Natl Acad Sci U S A. 2008;105:11595–6. doi: 10.1073/pnas.0806633105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wright FA, Strug LJ, Doshi VK, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at11p13 and 20q13.2. Nat Genet. 2011;43:539–46. doi: 10.1038/ng.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hull J, Campino S, Rowlands K, et al. Identification of common genetic variation that modulates alternative splicing. PLoS Genet. 2007;3:e99. doi: 10.1371/journal.pgen.0030099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nembaware V, Lupindo B, Schouest K, Spillane C, Scheffler K, Seoighe C. Genome-wide survey of allele-specific splicing in humans. BMC Genomics. 2008;9:265. doi: 10.1186/1471-2164-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang GS, Cooper TA. Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–61. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 66.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24:489–97. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 67.Duan S, Mi S, Zhang W, Dolan ME. Comprehensive analysis of the impact of SNPs and CNVs on human microRNAs and their regulatory genes. RNA Biol. 2009;6:412–25. doi: 10.4161/rna.6.4.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barenboim M, Zoltick BJ, Guo Y, Weinberger DR. MicroSNiPer: a web tool for prediction of SNP effects on putative microRNA targets. Hum Mutat. 2010;31:1223–32. doi: 10.1002/humu.21349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Raney BJ, Cline MS, Rosenbloom KR, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–5. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kwan T, Grundberg E, Koka V, et al. Tissue effect on genetic control of transcript isoform variation. PLoS Genet. 2009;5:e1000608. doi: 10.1371/journal.pgen.1000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dimas AS, Deutsch S, Stranger BE, et al. Common regulatory variation impacts gene expression in a cell type-dependent manner. Science. 2009;325:1246–50. doi: 10.1126/science.1174148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nica AC, Parts L, Glass D, et al. The architecture of gene regulatory variation across multiple human tissues: the MuTHER study. PLoS Genet. 2011;7:e1002003. doi: 10.1371/journal.pgen.1002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li G, Fullwood MJ, Xu H, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11:476–86. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ingram VM. Gene mutations in human haemoglobin: the chemical difference between normal and sickle cell haemoglobin. Nature. 1957;180:326–8. doi: 10.1038/180326a0. [DOI] [PubMed] [Google Scholar]
- 78.Kan YW, Dozy AM. Polymorphism of DNA sequence adjacent to human beta-globin structural gene: relationship to sickle mutation. Proc Natl Acad Sci U S A. 1978;75:5631–5. doi: 10.1073/pnas.75.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ferreira A, Marguti I, Bechmann I, et al. Sickle hemoglobin confers tolerance to Plasmodium infection. Cell. 2011;145:398–409. doi: 10.1016/j.cell.2011.03.049. [DOI] [PubMed] [Google Scholar]
- 80.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–8. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 81.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–62. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 82.Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–77. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 83.Samson M, Libert F, Doranz BJ, et al. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–5. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 84.Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–40. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 85.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10:184–94. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Montgomery SB, Dermitzakis ET. From expression QTLs to personalized transcriptomics. Nat Rev Genet. 2011;12:277–82. doi: 10.1038/nrg2969. [DOI] [PubMed] [Google Scholar]
- 87.Schadt EE, Monks SA, Drake TA, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 88.Nica AC, Montgomery SB, Dimas AS, et al. Candidate causal regulatory effects by integration of expression QTLs with complex trait genetic associations. PLoS Genet. 2010;6:e1000895. doi: 10.1371/journal.pgen.1000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheung VG, Conlin LK, Weber TM, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–5. doi: 10.1038/ng1094. [DOI] [PubMed] [Google Scholar]
- 91.Emilsson V, Thorleifsson G, Zhang B, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–8. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- 92.Monks SA, Leonardson A, Zhu H, et al. Genetic inheritance of gene expression in human cell lines. Am J Hum Genet. 2004;75:1094–105. doi: 10.1086/426461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stranger BE, Nica AC, Forrest MS, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–24. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goring HH, Curran JE, Johnson MP, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39:1208–16. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 95.Montgomery SB, Sammeth M, Gutierrez-Arcelus M, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature. 2010;464:773–7. doi: 10.1038/nature08903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pickrell JK, Marioni JC, Pai AA, et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–72. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–3. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 98.Halapi E, Gudbjartsson DF, Jonsdottir GM, et al. A sequence variant on 17q21 is associated with age at onset and severity of asthma. Eur J Hum Genet. 2010;18:902–8. doi: 10.1038/ejhg.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Barrett JC, Clayton DG, Concannon P, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type1 diabetes. Nat Genet. 2009;41:703–7. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–62. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hirschfield GM, Liu X, Xu C, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. N Engl J Med. 2009;360:2544–55. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cantero-Recasens G, Fandos C, Rubio-Moscardo F, Valverde MA, Vicente R. The asthma-associated ORMDL3 gene product regulates endoplasmic reticulum-mediated calcium signaling and cellular stress. Hum Mol Genet. 2010;19:111–21. doi: 10.1093/hmg/ddp471. [DOI] [PubMed] [Google Scholar]
- 103.Verlaan DJ, Berlivet S, Hunninghake GM, et al. Allele-specific chromatin remodeling in the ZPBP2/GSDMB/ORMDL3 locus associated with the risk of asthma and autoimmune disease. Am J Hum Genet. 2009;85:377–93. doi: 10.1016/j.ajhg.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dubois PC, Trynka G, Franke L, et al. Multiple common variants for celiac disease influencing immune gene expression. Nat Genet. 2010;42:295–302. doi: 10.1038/ng.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stuart PE, Nair RP, Ellinghaus E, et al. Genome-wide association analysis identifies three psoriasis susceptibility loci. Nat Genet. 2010;42:1000–4. doi: 10.1038/ng.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Alberts R, Terpstra P, Li Y, Breitling R, Nap JP, Jansen RC. Sequence polymorphisms cause many false cis eQTLs. PLoS ONE. 2007;2:e622. doi: 10.1371/journal.pone.0000622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Small KS, Hedman AK, Grundberg E, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43:561–4. doi: 10.1038/ng.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kong A, Steinthorsdottir V, Masson G, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–74. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Majewski J, Pastinen T. The study of eQTL variations by RNA-seq: from SNPs to phenotypes. Trends Genet. 2010;27:72–9. doi: 10.1016/j.tig.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 112.Garber M, Grabherr MG, Guttman M, Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8:469–77. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 113.Kasowski M, Grubert F, Heffelfinger C, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–5. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ramagopalan SV, Heger A, Berlanga AJ, et al. A ChIP-seq defined genome-wide map of vitamin D receptor binding: associations with disease and evolution. Genome Res. 2010;20:1352–60. doi: 10.1101/gr.107920.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ramagopalan SV, Maugeri NJ, Handunnetthi L, et al. Expression of the multiple sclerosis-associated MHC class II Allele HLA-DRB1*1501 is regulated by vitamin D. PLoS Genet. 2009;5:e1000369. doi: 10.1371/journal.pgen.1000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancersin the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 117.Crawford GE, Holt IE, Whittle J, et al. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS) Genome Res. 2006;16:123–31. doi: 10.1101/gr.4074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.McDaniell R, Lee BK, Song L, et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–9. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (Form-aldehyde-Assisted Isolation of Regulatory Elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–85. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Helgason A, Palsson S, Thorleifsson G, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39:218–25. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 121.Gaulton KJ, Nammo T, Pasquali L, et al. A map of open chromatin in human pancreatic islets. Nat Genet. 2010;42:255–9. doi: 10.1038/ng.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Boyle AP, Song L, Lee BK, et al. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011;21:456–64. doi: 10.1101/gr.112656.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Song L, Zhang Z, Grasfeder LL, et al. Open chromatin defined by DNaseI and FAIRE identifies regulatory elements that shape cell-type identity. Genome Res. 2011;21:1757–67. doi: 10.1101/gr.121541.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Higgs DR. Gene regulation in hematopoiesis: new lessons from thalassemia. Hematology Am Soc Hematol Educ Program. 2004;1:1–13. doi: 10.1182/asheducation-2004.1.1. [DOI] [PubMed] [Google Scholar]
- 125.Higgs DR, Wood WG. Long-range regulation of alpha globin gene expression during erythropoiesis. Curr Opin Hematol. 2008;15:176–83. doi: 10.1097/MOH.0b013e3282f734c4. [DOI] [PubMed] [Google Scholar]
- 126.De Gobbi M, Viprakasit V, Hughes JR, et al. A regulatory SNP causes a human genetic disease by creating a new transcriptional promoter. Science. 2006;312:1215–7. doi: 10.1126/science.1126431. [DOI] [PubMed] [Google Scholar]
- 127.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 128.Schalkwyk LC, Meaburn EL, Smith R, et al. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gibbs JR, van der Brug MP, Hernandez DG, et al. Abundant quantitative trait Loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Laird PW. Principles and challenges of genomewide DNA methylation analysis. Nat Rev Genet. 2010;11:191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 131.Buermans HP, Ariyurek Y, van Ommen G, den Dunnen JT, t Hoen PA. New methods for next generation sequencing based microRNA expression profiling. BMC Genomics. 2010;11:716. doi: 10.1186/1471-2164-11-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kulkarni S, Savan R, Qi Y, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–8. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Thomas R, Apps R, Qi Y, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet. 2009;41:1290–4. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 135.Knight JC, Keating BJ, Rockett KA, Kwiatkowski DP. In vivo characterization of regulatory polymorphisms by allele-specific quantification of RNA polymerase loading. Nat Genet. 2003;33:469–75. doi: 10.1038/ng1124. [DOI] [PubMed] [Google Scholar]
- 136.Knight JC, Keating BJ, Kwiatkowski DP. Allele-specific repression of lymphotoxin-alpha by activated B cell factor-1. Nat Genet. 2004;36:394–9. doi: 10.1038/ng1331. [DOI] [PubMed] [Google Scholar]
- 137.Alcais A, Alter A, Antoni G, et al. Stepwise replication identifies a low-producing lymphotoxin-alpha allele as a major risk factor for early-onset leprosy. Nat Genet. 2007;39:517–22. doi: 10.1038/ng2000. [DOI] [PubMed] [Google Scholar]
- 138.Hirasawa R, Feil R. Genomic imprinting and human disease. Essays Biochem. 2010;48:187–200. doi: 10.1042/bse0480187. [DOI] [PubMed] [Google Scholar]
- 139.Pant PV, Tao H, Beilharz EJ, Ballinger DG, Cox DR, Frazer KA. Analysis of allelic differential expression in human white blood cells. Genome Res. 2006;16:331–9. doi: 10.1101/gr.4559106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Serre D, Gurd S, Ge B, et al. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Zhang K, Li JB, Gao Y, et al. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat Methods. 2009;6:613–8. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Degner JF, Marioni JC, Pai AA, et al. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–12. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ge B, Pokholok DK, Kwan T, et al. Global patterns of cis variation in human cells revealed by high-density allelic expression analysis. Nat Genet. 2009;41:1216–22. doi: 10.1038/ng.473. [DOI] [PubMed] [Google Scholar]
- 144.Vandiedonck C, Taylor M, Lockstone H, et al. Pervasive haplotypic variation in the spliceo-transcriptome of the human major histocompatibility complex. Genome Res. 2011;21:1042–54. doi: 10.1101/gr.116681.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Horton R, Gibson R, Coggill P, et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics. 2008;60:1–18. doi: 10.1007/s00251-007-0262-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010;363:166–76. doi: 10.1056/NEJMra0905980. [DOI] [PubMed] [Google Scholar]