ABSTRACT
Since May 2013, outbreaks of porcine epidemic diarrhea have devastated the U.S. swine industry, causing immense economic losses. Two different swine enteric coronaviruses (porcine epidemic diarrhea virus and Delta coronavirus) have been isolated from the affected swine population. The disease has been reported from at least 32 states of the United States and other countries, including Mexico, Peru, Dominican Republic, Canada, Columbia, Ecuador, and Ukraine, with repeated outbreaks in previously infected herds. Here we report the isolation and characterization of a novel mammalian orthoreovirus 3 (MRV3) from diarrheic feces of piglets from these outbreaks in three states and ring-dried swine blood meal from multiple sources. MRV3 could not be isolated from healthy or pigs that had recovered from epidemic diarrhea from four states. Several MRV3 isolates were obtained from chloroform-extracted pig feces or blood meal in cell cultures or developing chicken embryos. Biological characterization of two representative isolates revealed trypsin resistance and thermostability at 90°C. NextGen sequencing of ultrapurified viruses indicated a strong homology of the S1 segment to mammalian and bat MRV3. Neonatal piglets experimentally infected with these viruses or a chloroform extract of swine blood meal developed severe diarrhea and acute gastroenteritis with 100% mortality within 3 days postinfection. Therefore, the novel porcine MRV3 may contribute to enteric disease along with other swine enteric viruses. The role of MRV3 in the current outbreaks of porcine epidemic diarrhea in the United States remains to be determined, but the pathogenic nature of the virus warrants further investigations on its epidemiology and prevalence.
IMPORTANCE
Porcine orthoreoviruses causing diarrhea have been reported in China and Korea but not in the United States. We have isolated and characterized two pathogenic reassortant MRV3 isolates from swine fecal samples from porcine epidemic diarrhea outbreaks and ring-dried swine blood meal in the United States. These fecal and blood meal isolates or a chloroform extract of blood meal induced severe diarrhea and mortality in experimentally infected neonatal pigs. Genetic and phylogenetic analyses of two MRV3 isolates revealed that they are identical but differed significantly from nonpathogenic mammalian orthoreoviruses circulating in the United States. The present study provides a platform for immediate development of suitable vaccines and diagnostics to prevent and control porcine orthoreovirus diarrhea.
INTRODUCTION
In May 2013, a devastating outbreak of epidemic diarrhea in young piglets commenced in swine farms of the United States, causing immense economic concerns. The mortality can reach up to 100% in piglets less than 10 days of age, with a recorded loss of at least 8 million neonatal pigs since 2013 (1, 2). Enteric viruses, such as swine enteric coronaviruses (SECoVs), porcine epidemic diarrhea virus (PEDV), and porcine deltacoronavirus (PDCoV), were isolated from these outbreaks (3, 4) and characterized (5). However, despite intensive biosecurity measures adopted to prevent the spread of SECoV in many farms and the use of two U.S. Department of Agriculture (USDA) conditionally licensed vaccines against PEDV, the outbreaks continue and have now spread to many other countries, including Mexico, Peru, Dominican Republic, Canada, Columbia, and Ecuador in the Americas (6) and Ukraine (7). Repeated outbreaks have also been reported on the same farms that were previously infected with PEDV. In June 2014, the USDA issued a federal order to report, monitor, and control swine enteric coronavirus disease (SECD) (8). In our efforts to understand the seemingly uncontrollable porcine epidemic diarrhea outbreaks, we discovered a novel mammalian orthoreovirus type 3 (MRV3) in feces of pigs from these outbreaks and ring-dried swine blood meal (RDSB). We have also reproduced severe diarrhea and acute gastroenteritis in neonatal pigs experimentally infected with purified MRV3 strains.
The family Reoviridae comprises 15 genera of double-stranded RNA (dsRNA) viruses (9). Orthoreoviruses with 10 discrete RNA segments have been isolated from a wide variety of animal species, including bats, civet cats, birds, reptiles, pigs, and humans (10, 11). Most orthoreoviruses are recognized to cause respiratory infections, gastroenteritis, hepatitis, myocarditis, and central nervous system disease in humans, animals, and birds (11); orthoreovirus genomes are prone to genetic reassortment and intragenic rearrangement (11, 12). The exchange of RNA segments between viruses could lead to molecular diversity and evolution of viruses with increased virulence and host range (13, 14). MRV serotypes 1 to 3 were associated with enteritis, pneumonia, or encephalitis in swine around the world, including China and South Korea (15–18). The zoonotic potential of MRV3 has been reported recently (19–21). However, porcine orthoreovirus infection of pigs was unknown previously in the United States.
RESULTS
Isolation of a novel MRV3 from diarrheic feces of pigs and ring-dried swine blood meal.
Nine out of 11 ring-dried swine blood meal (RDSB) samples from different manufacturing sources (82%) and 18 out of 48 fecal samples (37%) from neonatal pigs from farms with epidemic diarrhea outbreaks in North Carolina, Minnesota, and Iowa amplified a 326-bp S1 fragment with orthoreovirus group-specific primers. Among the 18 orthoreovirus-positive fecal samples, 11 samples were further sequence verified using MRV3-S1 gene-specific primers amplifying a 424-bp fragment. Characteristic cytopathic effects (CPE), including syncytium formation and rounding of individual cells, were evident at 48 h postinfection (hpi) in BHK-21 cells inoculated with chloroform-extracted samples of feces and blood meal (Fig. 1A to C). The infected cell monolayers were completely detached by 72 to 96 hpi. Developing chicken embryos died 2 to 5 days postinoculation (dpi) after inoculation by the chorioallantoic membrane (CAM) route. Infected chicken embryos showed hemorrhages (“cherry red appearance”) on the body and/or stunted growth (“dwarfing”) (Fig. 1D). MRV3 antigen was detected in infected BHK-21 cells using monoclonal antibody clone 2Q2048 against σ1 protein (Fig. 1E to G). The virus isolates from infected BHK-21 cells or chicken embryos were further confirmed as MRV3 by reverse transcription-PCR (RT-PCR) and sequencing. Eight virus isolates were obtained, and two representative isolates (T3/Swine/FS03/USA/2014 and T3/Swine/BM100/USA/2014) were used for further studies.
FIG 1 .
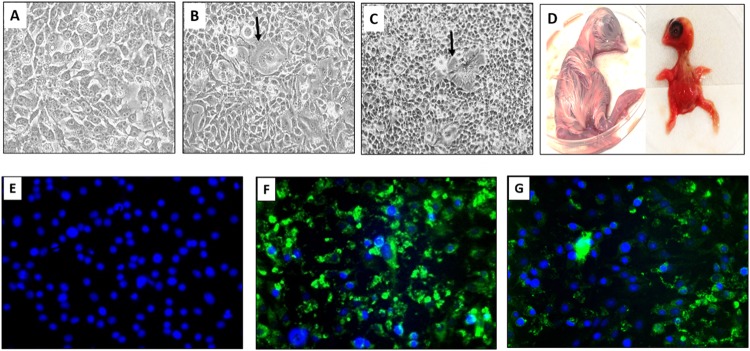
U.S. porcine orthoreovirus induces syncytia in BHK-21 cells and dwarfing in developing chicken embryos. (A) Mock-infected BHK-21 cells. (B) BHK-21 cells infected with T3/Swine/FS03/USA/2014 (FS03) virus showing syncytia (arrows) at 48 hpi. (C) BHK-21 cells infected with T3/Swine/BM100/USA/2014 (BM100) virus showing CPE with cell clumping and syncytia at 48 hpi. (D) Developing chicken embryos. An infected embryo has a cherry red appearance and dwarfing (right); a mock-infected embryo is shown to the left. (E to G) MRV3-specific antigen in infected BHK-21 cells 72 hpi. Mock-infected cells (E), cells infected with FS03 (F), and cells infected with BM100 (G) are shown. Green fluorescence represents localization of mammalian orthoreovirus 3 σ1 antigen within infected cells, and nuclei are stained (blue) with DAPI (4′,6-diamidino-2-phenylindole).
To determine whether normal, healthy pigs harbor orthoreoviruses, we obtained 36 samples of feces and matched samples of plasma from different states (Indiana, Ohio, Iowa, and Illinois) from farms with or without a PEDV outbreak. Six samples of feces and plasma each were obtained from uninfected farms in Indiana and Ohio, 12 samples of feces and plasma each were obtained from a farm in Illinois collected 6 weeks postepidemic diarrhea, and 12 samples of feces and plasma each were obtained from a farm in Iowa collected 6-month post-epidemic diarrhea. None of these samples was found to be positive for orthoreovirus by RT-PCR. Furthermore, chloroform extracts of feces from a few randomly selected MRV3-negative samples were blindly passaged twice on BHK-21 cells, and no CPE was observed.
The novel porcine orthoreovirus is unique in morphology and biological characteristics.
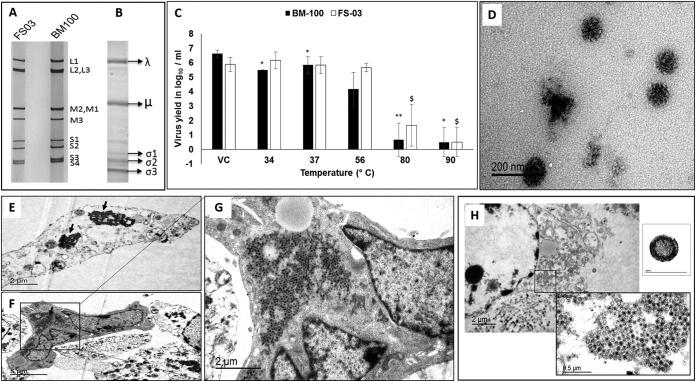
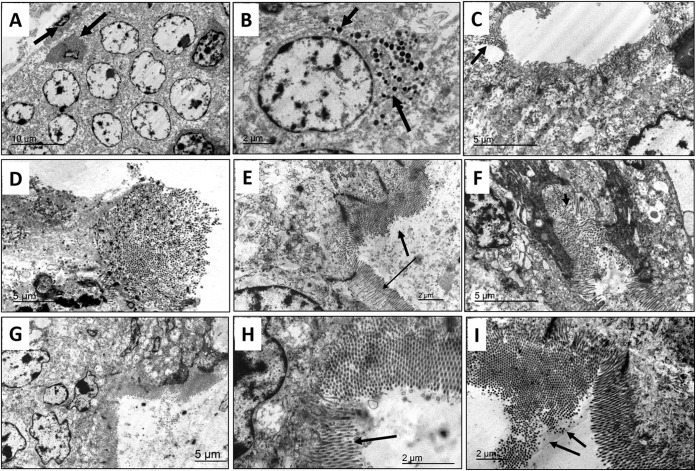
Genomic RNA from sucrose density gradient-purified virions was resistant to S1 nuclease treatment, confirming the double-stranded nature of the viral genome. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) indicated that the viral genome consists of 10 segments (Fig. 2A). The protein profile of the viruses was consistent with λ, μ, and σ proteins and their subclasses (Fig. 2B). The virions were stable at 56°C without significant loss of infectivity and remained viable after exposure to 80 or 90°C for 1 h (Fig. 2C). Transmission electron microscopy (TEM) analysis of negatively stained virions revealed icosahedral, nonenveloped, double-layered uniform-sized particles reminiscent of members of the family Reoviridae (Fig. 2D). In infected Vero cells, the presence of paracrystalline arrays of virus particles free of organelles (Fig. 2E to G) and viral factories in the cytoplasm (Fig. 2H) was evident. The mean diameter of the virus particles was 82 nm (Fig. 2H, inset), with particle sizes ranging from 80 to 85 nm. The MRV3 isolates (FS03 and BM100) replicated efficiently in BHK-21 cells, with a mean tissue culture infective dose (TCID50) of 6.7 log10/ml. Virus infectivity to BHK-21 cells increased after treatment with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) trypsin (6.7 to 7.7 log10/ml), suggesting trypsin resistance. The U.S. porcine MRV3 strains were able to hemagglutinate swine erythrocytes, and this property could be specifically inhibited with MRV3 anti-σ1 monoclonal antibody.
FIG 2 .
Characteristics of U.S. porcine orthoreovirus. (A) RNA profile of FS03 and BM100 on 7.5% SDS-PAGE gel. (B) Protein profile of FS03 purified virus on 7.5% SDS-PAGE gel. (C) Temperature sensitivity of FS03 and BM100. The TCID50 virus titers (mean values ± standard deviation) after treatment at different temperatures (34, 37, 56, 80, and 90°C) are plotted along with that of the untreated virus control (VC). Differences in the titers were evaluated by two-tailed t test, and statistically significant (P < 0.05) titers of FS03 ($) and BM100 (*) are indicated. (D) Negative-stained, purified FS03 virions, reminiscent of members of Reoviridae, shown as 80- to 85-nm particles with the characteristic double-shelled, icosahedral morphology. (E and F) FS03-infected Vero cells showing a paracrystalline array of virus particles free of organelles (black arrows). (G) Magnified view of a single Vero cell infected with FS03 with a paracrystalline array. (H) Viral factory within the cytoplasm of an FS03-infected Vero cell and a single virus particle in an enlarged view to show the double-layered virus structure and size (inset).
The U.S. porcine orthoreovirus isolates possess virulence-associated mutations.
Deep sequencing (MiSeq) of purified viral RNAs from two selected MRV3 isolates (FS03 from a pig fecal sample and BM100 from a porcine blood meal) confirmed their genomic identity with MRV3. No other contaminating viral sequences were detected in the deep sequence data. The high level of sequence identity between FS03 and BM100 sequences validated our immunofluorescence, gel electrophoresis and virus protein profile data. The total length of the porcine orthoreovirus genome is 23,561 nucleotides (nt). The two porcine isolates have consensus genome termini at the 5′ and 3′ ends similar to other MRVs. The 5′ untranslated region (UTR) ranged in length from 12 to 31 nt, and the 3′ UTR ranged in length from 32 to 80 nt, with variations from prototype MRV3 T3D (Table 1). The 5′ UTRs of both FS03 and BM100 have a 6-nt deletion in L1 and a 1-nt deletion in each of the L2 and S4 segments. In addition, a deletion of 3 nt in the M2 segment open reading frame (ORF) was noticed. The genome of these novel viruses contains reassorted gene segments from other MRVs.
TABLE 1 .
U.S. porcine orthoreovirus strains show altered UTRs
| Segment | Size (bp) | 5′ end |
ORF/protein |
3′ end |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Terminal sequencea | UTR (bp) | Region | Size (aa) | Class | Protein functionb | UTR (bp) | Terminal sequencea | ||
| L1 | 3,854 | GCUACA | 18 | 19–3822 | 1,267 | λ3 | RNA-dependent RNA polymerase | 32 | ACUCAUC |
| L2 | 3,915 | GCUAUU | 12 | 13–3882 | 1,289 | λ2 | Guanyltransferase, methyltransferase | 33 | AUUCAUC |
| L3 | 3,901 | GCUAAU | 13 | 14–3841 | 1,275 | λ1 | RNA binding, NTPase, helicase, RNA triphosphatase | 60 | AUUCAUC |
| M1 | 2,304 | GCUAUU | 13 | 14–2224 | 736 | μ2 | Binds RNA NTPase | 80 | CUUCAUC |
| M2 | 2,205 | UGCUAAU | 30 | 31–2157 | 708 | μ1 | Cell penetration, transcriptase activation | 48 | AUCAUCA |
| M3 | 2,241 | GCUAAA | 18 | 19–2184 | 721 | μNS | Unknown | 57 | AUUCAUC |
| S1 | 1,416 | UGCUAUU | 14 | 15–1382, 73–435 | 455, 120 | σ1, σ1s | Cell attachment | 34 | CACUUAA |
| S2 | 1,331 | GCUAUU | 18 | 19–1275 | 418 | σ2 | Binds dsRNA | 56 | ACUGACC |
| S3 | 1,198 | GCUAAA | 27 | 28–1128 | 366 | σNS | Inclusion formation, binds ssRNA | 70 | AAUCAUC |
| S4 | 1,196 | GCUAUU | 31 | 32–1129 | 365 | σ3, σ3a, σ3b | Binds dsRNA | 67 | AUUCAUC |
The 5′ and 3′ untranslated regions (UTRs) of U.S. porcine strains FS03 and BM100 show mutations on the M2, S1, and S2 segments. The conserved terminal sequences are shown in boldface, and mutations are italicized.
Predicted functions of different proteins encoded by the 10 segments analogous to known members of the Orthoreovirus genus are also indicated.
The deduced amino acid sequences of FS03 and BM100 are homologous except for σ1 protein, with 1 amino acid (aa) change between them. The percentage of homology of each of the different proteins coded by these two viruses with prototype MRV1 to -4 is provided in Table S1 in the supplemental material. On comparison of the deduced amino acids, it appears that with proteins of the L class segment, λ2 protein was homologous to MRV1, while the λ1 and λ3 proteins were highly similar to the MRV1 and -3 prototypes, T1-Lang (T1L) and T3/Dearing (T3D), respectively. In M class proteins, only μNS was identical to T3D, while μ1 and μ2 were identical to T1L. The sequence alignment of μ1 protein indicated 6 amino acid substitutions that were unique to these isolates in comparison to the T3/Bat/Germany, T3D, T1L, and T2J isolates (see Fig. S1 in the supplemental material). The μ2 protein alignment revealed 15 unique amino acid substitutions compared to the T3/Bat/Germany, T3D, T1L, and T2J sequences (see Fig. S2 in the supplemental material) and possessed the S208P mutation compared to T3D. In the S class proteins, all of them appear to originate from European bat (MRV3) viruses, with 88% to 98% identity at amino acid level (see Table S1 in the supplemental material). The highest diversity among all proteins was observed for the σ1 protein, with close homology to T3/Bat/Germany virus (91%). Deduced amino acid sequence analysis of σ1 protein revealed that the sialic acid binding domain (NLAIRLP), and protease resistance (249I) and neurotropism (340 D and 419E) residues were conserved in the U.S. porcine orthoreovirus strains. The novel viruses possessed 31 and 11 unique amino acid substitutions in the σ1 and σ1s proteins in comparison to T3/Bat/Germany and other MRV prototypes, respectively (see Fig. S3 in the supplemental material).
The novel U.S. porcine orthoreovirus is evolutionarily related to MRV3.
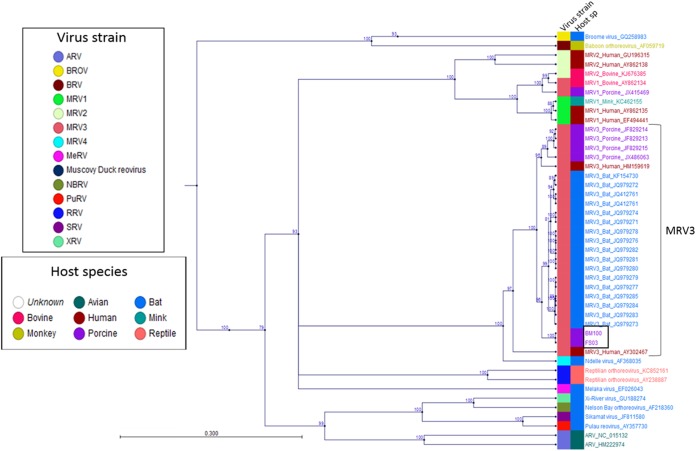
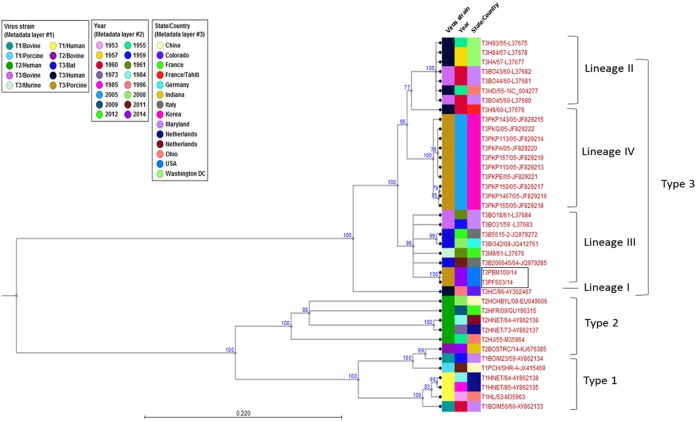
Phylogenetic analysis of the FS03 and BM100 isolates revealed a strong evolutionary relationship with MRV3 strains. The ORFs of the nucleotide sequences of the L1, S1, S2, S3, and S4 segments (see Table S2 and S3 in the supplemental material) were used to construct the phylogenetic trees. Based on S1 phylogeny, both isolates were monophyletic with MRV3 of bat origin (Fig. 3) and formed a distinct lineage together with the bat strains under lineage 3 (Fig. 4). Phylogenetic analysis of segment S2 indicated that the novel MRV3 isolates were monophyletic with the human T3D, T1L, and Chinese porcine T1 strains (Fig. 5A). The S3 phylogeny indicated that U.S. MRV3 strains were closely related to T1L and Chinese pig and European bat MRV3 strains (Fig. 5B). The topologies of S4 segment phylogenetic trees revealed that the U.S. porcine MRV3 isolates were closely related to Chinese T1 and T3 pig isolates (Fig. 5C). The L1 segment phylogeny revealed a close relationship to Chinese porcine T3 strains (Fig. 5D).
FIG 3 .
U.S. porcine orthoreoviruses are evolutionarily related to MRV3. Phylogenetic evolutionary analysis based on the complete S1 segment open reading frames of U.S. porcine isolates, all MRV serotypes, and closely related reoviruses was performed by the maximum likelihood method using the Jukes-Cantor evolution model in CLC workbench 7.0.4. Branches of FS03 and BM100 in lineage III are boxed. The metadata layers, including the virus strain and host species, are color coded. Branches with less than 75% bootstrap support were collapsed. The scale bar shows the evolutionary distance of 0.3 substitution per site. The tree calculations were unrooted.
FIG 4 .
Phylogenetic analysis of novel orthoreoviruses. Shown is evolutionary analysis for tree topologies of the complete MRV3 S1 segment open reading frames constructed through the maximum likelihood method using the Jukes-Cantor evolutionary model in CLC workbench 7.0.4. Branches with less than 75% bootstrap support were collapsed. All MRV3 strains are grouped into lineages I to IV. Branches of FS03 and BM100 within lineage III are boxed. The metadata layers, including virus strain, isolation year, and place of origin, are color coded. The scale bar shows the evolutionary distance of 0.220 substitution per site. The tree calculations were unrooted.
FIG 5 .
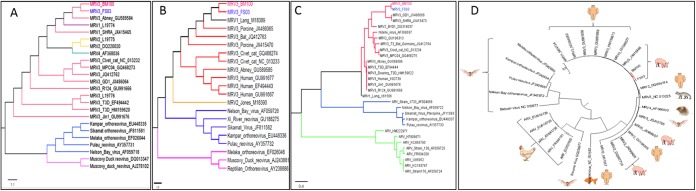
Phylogenetic analysis of S class and L1 nucleotide sequences of U.S. porcine orthoreoviruses. Unrooted maximum likelihood trees based on complete S2, S3, and S4 nucleotide sequences and L1 open reading frame nucleotide sequences (A to D) were constructed for U.S. porcine orthoreoviruses (FS03 and BM100) using the JTT w/freq model with 1,000 bootstrap replicates in MEGA 6.06. Shown are maximum likelihood trees of the S2 segment (A), S3 segment (B), and S4 segment (C). The branch tips of fecal sample and blood meal isolates are shown in blue and red, respectively. The scale bar indicates the nucleotide substitutions per site. The circular unrooted L1 phylogenetic tree represents the evolutionary relatedness of U.S. porcine strains (red branch) with that of other viruses and their respective host species (D). The scale bar shows the evolutionary distance of nucleotide substitutions per position. The branches with less than 75% bootstrap support were collapsed. The final trees were drawn using FigTree v1.4.2. GenBank accession numbers for each sequence are given next to the virus or strain name.
The U.S. porcine orthoreovirus is highly pathogenic to pigs.
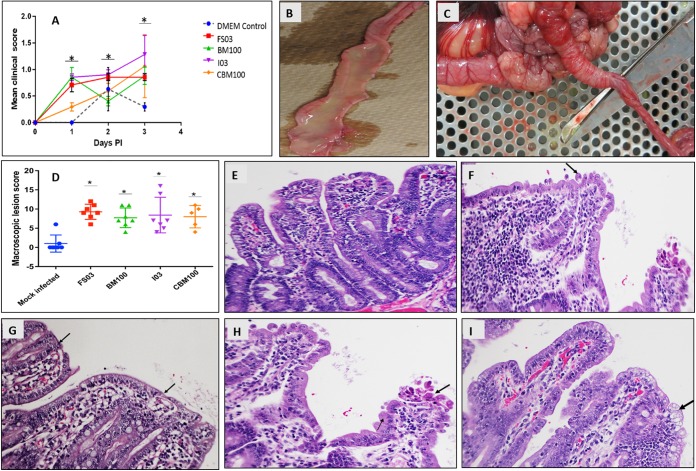
Experimental neonatal pigs were screened for swine deltacoronavirus, PEDV, Kobuvirus, swine transmissible gastroenteritis virus (TGEV), rotavirus, and orthoreoviruses by RT-PCR and found to be negative, except for three pigs that were positive for Kobuvirus, whose pathogenicity is yet to be established. Neonatal pigs orally inoculated with purified viruses FS03, BM100, T3/Swine/I03/USA/2014 (I03), or a chloroform extract of blood meal 100 (CBM100) developed clinical illness in all infected animals (100%), with loss of physical activity, severe diarrhea, and decrease in body weight. Infected animals had significantly high mean clinical scores compared to the mock-infected group (P < 0.01). Piglets infected with FS03 and I03 had the highest clinical scores as early as 1 dpi, which peaked at 3 dpi (Fig. 6A). Three pigs in the mock-infected group had a slow recovery from parenteral anesthetics, with elevated mean clinical scores for the first 2 days but returned to normal later. Gross lesions, such as catarrhal enteritis (Fig. 6B) and intussusception (Fig. 6C), were observed in all of the infected animals. The cumulative macroscopic lesion scores of FS03 and I03 were higher than those of other groups on day 4 dpi (Fig. 6D). Compared to mock-infected pigs (Fig. 6E), small intestines of the virus-infected pigs showed mild to severe villous blunting and fusion (crypt/villous ratios of 1:1 to 1:4) (Fig. 6F), occasional villous epithelial syncytial cells (Fig. 6G), swollen epithelial cells with granular cytoplasm and multifocal necrosis of mucosal epithelium (Fig. 6H), and round to oval vacuoles in the intestinal epithelial cells (Fig. 6I). In a few pigs, protein casts in renal tubules, minimal to mild hepatic lipidosis and hepatocellular vacuolar changes, and mild to moderate suppurative bronchopneumonia were also seen.
FIG 6 .
Pathogenicity of U.S. porcine orthoreovirus isolates and chloroform extract of blood meal. Neonatal pigs were infected with the FS03, BM100, and I03 virus strains and chloroform extract of blood meal 100 (CBM100). (A) Mean clinical scores of neonatal pigs from 1 to 3 days postinfection (dpi). The clinical signs were scored from 0 to 3, and mean values ± standard deviation (SD) are shown. (B and C) Gross lesions in virus-infected pigs, including catarrhal enteritis (B) and intussusception (C). (D) Mean macroscopic lesion scores of experimentally infected piglets. Lesions from different organs were scored on a scale of 0 to 6, and the cumulative mean score ± SD is shown. (E to I) Histology of small intestine in mock-infected (E) and virus-infected (F to I) pigs. Shown are villous blunting and fusion (F), villous epithelial syncytium (G), granular cytoplasm (thin arrow) and multifocal necrosis of mucous epithelium (thick arrow) (H), and vacuolation (I).
Ultrastructural examination revealed multinucleated cells with apoptotic nuclei (Fig. 7A), and in some cells, dark granular bodies resembling stress granules were seen (Fig. 7B). Viral particles were localized in regions of the cytoplasm that lacked typical cytoplasmic organelles (Fig. 7C). Large numbers of viral particles egressed by cell lysis (Fig. 7D) or as a string of beads through microvilli from infected villous epithelial cells (Fig. 7E) into the lumen of the intestine (Fig. 7F). Multinucleated cells with virions egressing through microvilli were evident (Fig. 7G). Virions disrupt microvilli before release and were still surrounded by the cell membrane of microvilli (Fig. 7H), and after release were devoid of membranes in the lumen of the intestine (Fig. 7I).
FIG 7 .
U.S. porcine orthoreoviruses egress from infected intestinal villous epithelial cells by lysis and also exit through microvilli. Ultrathin sections of intestine from experimentally infected pigs. (A) Multinucleated villous epithelial cell with apoptotic nuclei (arrows). (B) Villous epithelial cells showing stress granules (arrows). (C) Virus particles in the cytoplasm devoid of organelles. (D) Virus egress through cell lysis. (E) Virions egressing through microvilli as a string of beads (thick arrow) in comparison with normal microvilli (thin arrow). (F) Virus particles released from villi in the lumen of intestine (arrow). (G) Multinucleated cells with virions egressing through microvilli. (H) Virus particles encased in microvillous membrane (arrow). (I) Virus particles free of membranes in the intestinal lumen (arrows).
Virus replication in the intestines and fecal virus shedding in infected pigs were also confirmed by virus isolation in cell culture and by S1-segment-specific RT-PCR. The intestinal contents had MRV3 virus in 80% of the infected piglets through RT-PCR, suggesting the virus replication in the intestine is consistent with electron microscopic findings of virus replication within the enterocytes.
DISCUSSION
The ongoing outbreaks of epidemic piglet diarrhea have devastated the swine industry in the United States, with significant economic losses. Since a federal mandate to report the SECD cases on 5 June 2014, a total of 731 swine premises had been confirmed positive until 22 January 2015 in the United States (22). Strict and intensive biosecurity measures have been adopted on many farms to control SECD, but the disease spread is unabated, and at least 32 states are now affected. Repeated outbreaks in previously infected herds were also reported, suggesting that the SECoVs may not be the sole agents responsible for this unprecedented epidemic diarrhea outbreak in the United States and other countries, including Mexico, Japan, Canada, Colombia, Dominican Republic, Ecuador (6), and Ukraine (7).
Here, we report the isolation and characterization of a novel MRV3 from SECD fecal samples and swine blood meal, which is a by-product of the slaughtering industry and is used as a protein source in the diets of livestock. We also provide evidence that these novel MRV3 strains are pathogenic to neonatal pigs, leading to lethal enteric disease. More than 80% of ring-dried blood meal feed supplements and 37.5% of the fecal samples from epidemic diarrheic pigs tested in this study were found positive for the novel MRV3. It remains to be determined whether porcine MRV3 is responsible, either primarily or in combination with SECoV, for the current uncontrollable epidemic piglet diarrhea. There have been conflicting reports on the presence of live pathogenic SECoVs in porcine blood products (23, 24). The World Organization for Animal Health Office International des Epizooties (OIE) ad hoc group on PEDV recently concluded that contaminated pig blood products, including spray-dried plasma are not a likely source of infectious PEDV (2, 25) as spray-drying typically inactivates enveloped coronaviruses. Therefore, the presence of chloroform-resistant, infectious MRV3 in RDSB as identified in this study is of great significance considering the current difficulties in arresting the spread of this economically important disease in the U.S. and Canadian swine industries.
Our results showed the U.S. porcine MRV3 isolates are thermostable and trypsin resistant, kill developing chicken embryos, and produce syncytium in BHK-21 cells but not in Vero cells. Fusogenic MRVs carry a FAST protein (26), but the U.S. porcine MRV3 strains lack this protein but produce syncytium in infected BHK-21 cells or intestinal epithelium. Additional studies are needed to understand this feature. The virions were double layered with a mean diameter of 82 nm, in concordance with the reported size for MRVs (11) but larger than the reported sizes of 70 to 72 nm for bat orthoreoviruses (20, 27). Size differences in MRV particle forms, such as virions, intermediate subvirion particles (ISVPs), and core particles, have been reported (11). Viral factories with paracrystalline arrays of virions in infected Vero cells are an important characteristic of these strains, unlike the tubular viral factories seen in T3D type strains (11). Interestingly, in infected intestinal villous epithelial cells of young pigs, the virions fuse cells and egress by lysis or by using microvilli. Reovirus egress from infected cells is not completely understood (11). Our results suggest that MRV3 may use intestinal microvilli to release complete virions as arrays in addition to cell lysis.
Deep sequencing analysis of the purified cell culture or developing chicken embryo isolates revealed novel MRV3 sequence. The sequencing data from two selected porcine MRV3 isolates (one each from feces and blood meal) revealed a high sequence homology, thus strongly suggesting that blood meal could be a possible mode of transmission along with other undetermined modes. The thermostability of these MRV3 strains at 56, 80, and 90°C for 1 h lends further credence to this notion. Ring drying of blood meal entails coagulation of blood by heating to 90°C, which may not be sufficient to inactivate these heat-resistant MRV3 strains. The current European Union regulation for pig blood products for use in pig feeds (EU 483/2014)—that they must have been treated at 80°C and kept in storage for 2 weeks at room temperature to inactivate PEDV—may not be sufficient to inactivate porcine MRV3. The genome sequences of the 10 segments of these strains revealed interesting features in combination like never before, making them unique and novel. For example, they carry specific mutations in σ1 protein that would impart trypsin resistance (28) and neurotropism (29, 30), in μ2 protein for interferon antagonism (31), and possessed multiple basic residues in σ1s protein for hematogenous dissemination (11). The observed nine unique amino acid substitutions on the μ1 protein may have a role in conferring thermostability to these strains as specific amino acid residues in μ1 have been associated with thermostability in T3-type strains (32).
Phylogenetic analysis of “S” class and “L” class segments of the novel U.S. porcine MRV3 strains indicates that these are T3 divergents. The S1 segment is bicistronic and highly similar to that of bat MRV3. The novel porcine MRV strains fall into lineage III (18), along with the human, bovine, murine, and bat strains with close evolutionary distance to German and Italian bat MRV3 S1 sequences (20). The sequence diversity of S2, S3, and S4 segments does not correlate with host species, geographic location, or year of isolation, suggesting their origin from different evolutionarily distinct strains from humans, pigs, and bats and reaffirming that MRVs reassort in nature (13, 33–35).
Even though MRVs are not common in causing severe disease outbreaks in livestock, several strains of porcine MRVs have been isolated from diarrheic pigs in China and Korea (18, 36). Similarly, MRV3 strains have been reported from bats in Europe suffering from clinical disease (20) and in children with bat origin nonfusogenic MRV3 in Europe (37). All of these studies and our results confirm that porcine MRV3 strains are pathogenic.
At necropsy, all infected piglets had accumulation of fluid in the intestine. The reproducibility of severe diarrhea and clinical disease with mortality in experimentally infected piglets with isolated MRV3 confirms the pathogenic nature of these strains. Villous blunting is a consistent feature of piglets affected by neonatal diarrhea syndrome (3, 38). The observed protein casts in the renal tubules and mild hepatic lipidosis could be attributed to the metabolic disorder. The presence of isoleucine at position 249 probably prevented the cleavage of σ1 protein by intestinal luminal proteases, enabling efficient viral growth and migration to other tissues (10) compared to the trypsin-sensitive σ1 protein (threonine at 249) in endemic T3D type strains with attenuated virulence (39). Previous studies with mice also indicated that reovirus virulence correlates strongly with the age of the host (40). The naive immune system in the newborn piglets could also contribute to this increased pathogenicity.
The chloroform extract of blood meal and a virus derived from the same sample caused similar disease in experimental pigs, suggesting blood meal as a source of infection. There is also an additional possibility that the virus could be mechanically transmitted through insect vectors and other fomites, although there is no evidence from this study for this type of transmission. If proven, this would be the first MRV to be transmitted by vectors. Such a possibility does exist as related seadornaviruses infect pigs and are vector borne (41).
The PEDV strain or strains currently circulating in the United States possibly originated from bats in China (42). It is tempting to speculate that PEDV and porcine MRV3 were introduced together into the domestic swine industry based on the cocirculation of these viruses and the genetic distance of the bat origin MRV3 to circulating strains in the United States. However, with the limited number of bat MRV3 and porcine orthoreovirus sequences available to compare, the epidemiological link remains to be identified. Similarly, a large-scale epidemiological study on the prevalence of this virus in U.S. swine farms with or without diarrhea is warranted to definitively incriminate MRV3 as either the primary causative agent or coagent of the current porcine epidemic diarrhea outbreaks. Continued surveillance and development of preventive and control measures against MRV3 would ease the economic burden of diarrheal disease in the swine industry. The availability of cell culture and the developing chicken embryo for isolation and propagation of these viruses would greatly enhance the development of vaccines and diagnostics for MRV3-induced disease in the United States.
MATERIALS AND METHODS
RT-PCR for porcine MRV3.
Viral RNA was isolated from fecal and ring-dried swine blood meal samples using the QIAmp RNA kit (Qiagen, United States), and reverse transcription-PCR (RT-PCR) was performed using MRV3-S1 gene-specific primers (primer sequences available upon request). The amplified PCR products were analyzed by electrophoresis on a 1.5% (wt/vol) agarose gel, and the PCR products were purified and directly sequenced.
Virus isolation.
Virus isolation was performed on RT-PCR-positive fecal and blood meal samples. The chloroform extracts of a 20% fecal suspension and 10% ring-dried blood meal samples were filtered through 0.2-µm-pore membrane filters (Millipore, United States) and inoculated into 9- to 11-day-old, specific-pathogen-free (SPF), developing chicken embryos (via the chorioallantoic membrane [CAM] route) and BHK-21 cells. Embryos and cells were incubated at 37°C for 5 days and monitored daily for mortality and cytopathic effects (CPE), respectively. At 5 days postinfection (dpi), allantoic fluid and CAM were harvested from eggs, and the cell culture supernatant was collected from BHK-21 cultures, chloroform extracted, and further passaged in SPF chicken embryos or BHK-21 cells, respectively. Viral RNA was detected by RT-PCR using MRV3 S1 segment-specific primers. Amplified MRV3-S1 PCR products were sequenced to confirm the viral genome. The virus isolates obtained from BHK-21 cells were further confirmed using an indirect immunofluorescence assay (IFA), employing a mouse monoclonal antibody directed against type 3 orthoreovirus σ1 protein (clone 2Q2048; Abcam, United States,).
Virus purification.
BHK-21 cell monolayers grown in T-175 flasks were infected with the MRV3 isolates at a multiplicity of infection (MOI) of 0.1 in Dulbecco’s modified Eagle’s medium (DMEM) containing 1% fetal calf serum (FCS). The cells were harvested at 3 dpi and subjected to three freeze-thaw cycles. The cellular debris was clarified by centrifugation at 3,700 × g at 4°C. Crude virus was pelleted from the clarified supernatant by ultracentrifugation at 66,000 × g for 2 h using an SW-28 rotor (Beckman Coulter, United States). The virus pellet was resuspended in 1 ml TN buffer (20 mM Tris, 400 mM NaCl, 0.01% N-lauryl sarcosine [pH 7.4]). The virus suspension was then layered onto a 15 to 45% (wt/vol) discontinuous sucrose gradient and centrifuged at 92,300 × g for 2 h at 4°C using an SW-41 Ti swing-out rotor (Beckman Coulter, United States). The virus band at the interface was collected and used for characterization and genomic studies.
Virus characterization.
Hemagglutination (HA) and hemagglutination inhibition (HI) assays were performed as previously described (43), with modifications. Briefly, the viruses were serially diluted in 50 µl of phosphate-buffered saline (PBS [pH 7.4]) in 96-well V-bottom microtiter plates (Corning-Costar, United States) followed by 50 µl of 1% pig erythrocytes (Lampire Biological Laboratories, United States). The plates were incubated for 2 h at 37°C to record the HA titer. The HI assay was performed using mouse monoclonal antibody directed against type 3 orthoreovirus σ1 protein (clone 2Q2048; Abcam, United States) and 4 HA units of the virus. The HI assay plates were incubated initially at 37°C for 1 h and then at 4°C overnight before scoring.
For electron microscopy, ultrathin sections of virus-infected BHK-21 cells (3 dpi), intestines of experimentally infected pigs, or purified virions were placed on Formvar-carbon-coated electron microscope grids and negatively stained with 2% (wt/vol) uranyl acetate or 1% sodium phosphotungstic acid for 30 s. The specimens were then examined in a JEOL 1400 transmission electron microscope (JEOL, United States) at an accelerating voltage of 80 kVA.
To determine the temperature sensitivity, the virus strains were subjected to five different temperature treatments at 34, 37, 56, 80, and 90°C for 1 h. Serial dilution of the virus was then made in DMEM, which was then titrated for infectivity in BHK-21 cells. For trypsin sensitivity, virus was incubated with 1 µg/ml tosyl phenylalanyl chloromethyl ketone (TPCK) trypsin in DMEM for 1 h at 37°C and titrated for infectivity in BHK-21cells. To demonstrate the double-stranded nature of the viral genome, total RNA extracted from purified virions was subjected to S1 nuclease digestion and 7.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and silver nitrate staining. For protein profiling, the purified virus was denatured in protein sample buffer and analyzed by standard 7.5% SDS-PAGE and Coomassie blue staining.
Deep sequencing.
The double-stranded RNA (dsRNA) isolated from two purified viruses—FS03 isolated from fecal samples and BM100 isolated from swine ring-dried blood meal—were subjected to NextGen genome sequencing. The NEBNext Ultr directional RNA library prep kit for Illumina (catalog no. e74205; NEB) was used to prepare the RNA library with some modifications. Using a standard protocol, 100 ng of viral RNA was fragmented to 250 nucleotides at 94°C for 10 min. After adapter ligation, 350- to 375-bp libraries (250- to 275-bp insert) were selected using Pippin Prep (Sage Science, United States). The template molecules with the adapters were enriched by 12 cycles of PCR to create the final library. The generated library was validated using the Agilent 2100 bioanalyzer and quantitated using the Quant-iT dsDNA H.S. kit (Invitrogen) and quantitative PCR (qPCR). Two individually barcoded libraries (FS03 virus with A006-GCCAAT, and BM100 virus with A012-CTTGTA) were pooled and sequenced on Illumina MiSeq. Briefly, the individual libraries were pooled in equimolar amounts, denatured, and loaded onto MiSeq. The pooled library was spiked with 5% phiX and sequenced to 2 × 250 paired-end reads (PE) on the MiSeq using the MiSeq reagent kit V2 at 500 cycles (MS-102-2003) to generate 24 million PE.
Genome assembly.
Reference-based mapping and de novo assembly methods were applied to the raw data for assembly into viral genomes. Reference-based mapping was performed against the mammalian orthoreovirus genome by using the CLC Genomics Workbench software (version 7.0.4; CLC Bio, Denmark). The de novo assembly was performed with the following overlap settings: mismatch cost of 2, insert cost of 3, minimum contig length of 1,000 bp, a similarity of 0.8, and a trimming quality score of 0.05. This assembly yielded 3,444 contigs that were annotated according to Gene Ontology terms with the Blast2Go program (44), which was executed as a plugin of CLC by mapping against the UniprotKB/Swiss-Prot database with a cutoff E value of 1e−05. Furthermore, to determine putative gene descriptions, homology searches were carried out through querying the NCBI database using the tBLASTx algorithm. The de novo-assembled sequences were used to confirm the validity of the reference-based sequence assembly. Both de novo assembly and the reference-based mapping produced identical sequences.
Phylogenetic analysis.
The nucleotide and deduced amino acid sequences of L1 and S class segments (S1, S2, S3, and S4) were compared with those of other closely related orthoreoviruses using the BioEdit sequence alignment editor software (version 7.0.0; BioEdit, Ibis Biosciences, Carlsbad, CA). The phylogenetic evolutionary histories for the virus strains were inferred using the maximum likelihood method based on either JTT w/freq model (45) for the S2, S3, S4, and L1 segments in Mega 6.06 (46) or the Jukes-Cantor evolution model (47) with “WAG” (i.e., Whelan and Goldman model) protein substitution for S1 segment in CLC workbench 7.0.4 after testing for their appropriateness to be the best fit. The bootstrap consensus tree inferred from 1,000 replicates was taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 75% bootstrap replicates were collapsed.
Pathogenicity study in neonatal pigs.
All animal studies were performed as approved by the Institutional Animal Care and Use Committee of Virginia Tech (IACUC no. 14-105-CVM, 5 June 2014). Thirty-five 2-day-old piglets, purchased from the Virginia Tech Swine Center, were housed as 7 animals/group in HEPA-filtered level 2 biosecurity facility. The experimental design is provided in Table S4 in the supplemental material. Prior to the start of the experiment, pigs were tested for most common enteric RNA viruses, such as rotavirus, PEDV, swine deltacoronavirus, Kobuvirus, and TGEV, by RT-PCR of the fecal samples using specific primers (primer sequences available upon request). The amplified PCR products were analyzed by electrophoresis on 1.5% (wt/vol) agarose gel.
After acclimatizing for a day, the animals were anesthetized, and 2 ml of 5 × 105 TCID50/ml of each virus strain or chloroform extract of 10% blood meal suspension (2.5 g ring-dried blood meal) was homogenized in 12.5 ml DMEM to get a 20% solution that was extracted with an equal volume of chloroform. The upper aqueous phase obtained was diluted with an equal volume of DMEM to get a final concentration of 10%, and the piglet was orally inoculated using a 5-ml syringe. Mock-infected animals received 2 ml DMEM orally. The animals were monitored two times a day: rectal temperature, body weight, and clinical scores based on physical appearance, activity, respiratory, gastrointestinal, and systemic signs were recorded on a scale of 0 to 3. Fecal swabs were collected daily and suspended in 1 ml of DMEM containing 10× antibiotic solution (HyClone, United States), mixed vigorously, incubated for 1 h, and stored at −80°C until tested. At 4 dpi, or when they reached the clinical endpoint, all animals were euthanized. Gross and microscopic lesions were scored by a board-certified veterinary pathologist blind to the experimental groups (48). The S1 gene-specific RT-PCR was performed to confirm the production of orthoreovirus in the intestine using the intestinal contents of the experimentally infected piglets.
Statistical analysis.
Summary statistics were calculated to assess the overall quality of the data. Analysis of variance (ANOVA) was used for assessment of the mean clinical score and microscopic lesion scores. The significance level was set for a P value of <0.01 and a 95% confidence interval. Statistical analysis was performed using GraphPad Prism software (version 6.0; Graph Pad Software, Inc., San Diego, CA).
Nucleotide sequence accession numbers.
The complete genome sequences of both viruses FS03 and BM100 have been deposited in GenBank under accession no. KM820744 to KM820763.
SUPPLEMENTAL MATERIAL
The μ1 protein of U.S. porcine orthoreovirus isolates carries unique mutations. Shown is deduced amino acid sequence comparison of the μ1 protein of U.S. porcine orthoreovirus isolates (FS03 and BM100) with those of the T1L, T2J, and T3D prototypes and T3/Bat/Germany sequences. Amino acid residues that are identical to T3D sequence are indicated by dashes. Observed mutations with respect to T3D are boxed. Download
The μ2 protein of U.S. porcine orthoreovirus isolates carries unique mutations. Deduced amino acid sequence comparison of the μ2 protein of U.S. porcine orthoreovirus isolates (FS03 and BM100) with those of the T1L, T2J, T3D prototype and T3/Bat/Germany sequences. Amino acid residues that are identical to those in the T3D sequence are indicated by dashes. Observed mutations with respect to T3D are boxed. Download
Sequence comparison of σ1 and σ1s proteins of U.S. porcine orthoreovirus isolates with closely related MRV3 sequences. (A) Alignments of deduced σ1 protein sequences of U.S. porcine orthoreovirus isolates (FS03 and BM100) were compared with those of T3/D, T3/Bat/Germany, and T3/porcine/Korea sequences. Positions of the amino acids are numbered above the sequences. The residues boxed in red represent the predicted NLAIRLP sialic acid-binding domain, trypsin resistance residue (249I), and residues involved in neurotropism (340D and 419E). (B) Deduced amino acid comparison of σ1s protein of U.S. porcine orthoreovirus isolates (FS03 and BM100) with those of the T1L, T2J, and T3D prototype and T3/Bat/Germany virus sequences. Amino acid residues that are identical to those in the T3D sequence are indicated by dashes. Observed mutations with respect to T3D are boxed. Download
Proteins encoded by U.S. porcine orthoreovirus isolates are homologous to T1 and T3 serotypes.
List of accession numbers of viral strains used for phylogenetic analysis in this study.
GenBank accession numbers of U.S. porcine orthoreovirus isolates and prototype sequences used in this study.
Experimental design.
ACKNOWLEDGMENT
We thank Renukaradhya Gourapura for providing swine fecal and plasma samples from multiple farms.
Footnotes
Citation Thimmasandra Narayanappa A, Sooryanarain H, Deventhiran J, Cao D, Ammayappan Venkatachalam B, Kambiranda D, LeRoith T, Heffron CL, Lindstrom N, Hall K, Jobst P, Sexton C, Meng X-J, Elankumaran S. 2015. A novel pathogenic mammalian orthoreovirus from diarrheic pigs and swine blood meal in the United States. mBio 6(3):e00593-15. doi:10.1128/mBio.00593-15.
REFERENCES
- 1.Mitchell D. 2014. Porcine epidemic diarrhea virus is nasty—and it could get worse. http://modernfarmer.com/2014/03/piglet-attacking-virus-get-worse/. Accessed 9 November 2014.
- 2.Kareim R. 2014. Virus kills millions of American pigs, pushing up pork prices. http://news.nationalgeographic.com/news/2014/05/140501-pigs-virus-meat-prices-food-science-health/ Accessed 9 November 2014.
- 3.Stevenson GW, Hoang H, Schwartz KJ, Burrough ER, Sun D, Madson D, Cooper VL, Pillatzki A, Gauger P, Schmitt BJ, Koster LG, Killian ML, Yoon KJ. 2013. Emergence of porcine epidemic diarrhea virus in the United States: clinical signs, lesions, and viral genomic sequences. J Vet Diagn Invest 25:649–654. doi: 10.1177/1040638713501675. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Byrum B, Zhang Y. 2014. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg Infect Dis 20:1227–1230. doi: 10.3201/eid2007.140296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vlasova AN, Marthaler D, Wang Q, Culhane MR, Rossow KD, Rovira A, Collins J, Saif LJ. 2014. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg Infect Dis 20:1620–1628. doi: 10.3201/eid2010.140491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EFSA 2014. Scientific opinion on porcine epidemic diarrhea and emerging porcine deltacoronavirus. EFSA J 12:3877. doi: 10.2903/j.efsa.2014.3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.USDA 2014. Gain-report. 2014. Multiple PED outbreaks likely to impact pork production. http://gain.fas.usda.gov/Recent%20GAIN%20Publications/Multiple%20PED%20Outbreaks%20Likely%20to%20Impact%20Pork%20Production%20%20_Kiev_Ukraine_12-10-2014.pdf Accessed 30 January 2015.
- 8.USDA 2014. Reporting, herd monitoring and management of novel swine enteric coronavirus diseases, June 5 2014. wwwaphisusdagov/newsroom/2014/06/pdf/secd_federal_orderpdf. Accessed 9 November 2014.
- 9.Attoui H, Mertens PPC, Becnel J, Belaganahalli S, Bergoin M, Brussaard CP, Chappell JD, Ciarlet M, del Vas M, Dermody TS, Dormitzer PR, Duncan R, Fang Q, Graham R, Guglielmi KM, Harding RM, Hillman B, Makkay A, Marzachì C, Matthijnssens J, Milne RG, Mohd Jaafar F, Mori H, Noordeloos AA, Omura T, Patton JT, Rao S, Maan M, Stoltz D, Suzuki N, Upadhyaya NM, Wei C, Zhou H. 2011. Orthoreovirus, Reoviridae, p 546–554. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy classification and nomenclature of viruses: ninth report of the International Committee on the Taxonomy of Viruses. Elsevier Academic Press, London, United Kingdom. [Google Scholar]
- 10.Tyler KL, Barton ES, Ibach ML, Robinson C, Campbell JA, O’Donnell SM, Valyi-Nagy T, Clarke P, Wetzel JD, Dermody TS. 2004. Isolation and molecular characterization of a novel type 3 reovirus from a child with meningitis. J Infect Dis. 189:1664–1675. doi: 10.1086/383129. [DOI] [PubMed] [Google Scholar]
- 11.Dermody TS, Parker JSL, Sherry B. 2013. Orthoreoviruses, p 1304–1346. In Knipe DM, Howley PM (ed), Fields virology, 6th ed. Lippincott Williams, Philadelphia, PA. [Google Scholar]
- 12.Duncan R. 1999. Extensive sequence divergence and phylogenetic relationships between the fusogenic and nonfusogenic orthoreoviruses: a species proposal. Virology 260:316–328. doi: 10.1006/viro.1999.9832. [DOI] [PubMed] [Google Scholar]
- 13.Chapell JD, Goral MI, Rodgers SE, dePamphilis CW, Dermody TS. 1994. Sequence diversity within the reovirus S2 gene: reovirus genes reassort in nature, and their termini are predicted to form a panhandle motif. J Virol 68:750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ouattara LA, Barin F, Barthez MA, Bonnaud B, Roingeard P, Goudeau A, Castelnau P, Vernet G, Paranhos-Baccalà G, Komurian-Pradel F. 2011. Novel human reovirus isolated from children with acute necrotizing encephalopathy. Emerg Infect Dis 17:1436–1444. doi: 10.3201/eid1708.101528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baskerville A, McFerran JB, Connor T. 1971. The pathology of experimental infection of pigs with type I reovirus of porcine origin. Res Vet Sci 12:172–174. [PubMed] [Google Scholar]
- 16.Kasza L. 1970. Isolation and characterisation of a reovirus from pigs. Vet Rec 87:681–686. doi: 10.1136/vr.87.22.681. [DOI] [PubMed] [Google Scholar]
- 17.Zhang C, Liu L, Wang P, Liu S, Lin W, Hu F, Wu W, Chen W, Cui S. 2011. A potentially novel reovirus isolated from swine in northeastern China in 2007. Virus Genes 43:342–349. doi: 10.1007/s11262-011-0642-4. [DOI] [PubMed] [Google Scholar]
- 18.Kwon HJ, Kim HH, Kim HJ, Park JG, Son KY, Jung J, Lee WS, Cho KO, Park SJ, Kang MI. 2012. Detection and molecular characterization of porcine type 3 orthoreoviruses circulating in South Korea. Vet Microbiol 157:456–463. doi: 10.1016/j.vetmic.2011.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chua KB, Voon K, Yu M, Keniscope C, Abdul Rasid K, Wang LF. 2011. Investigation of a potential zoonotic transmission of orthoreovirus associated with acute influenza-like illness in an adult patient. PLoS One 6:e25434. doi: 10.1371/journal.pone.0025434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohl C, Lesnik R, Brinkmann A, Ebinger A, Radonić A, Nitsche A, Mühldorfer K, Wibbelt G, Kurth A. 2012. Isolation and characterization of three mammalian orthoreoviruses from European bats. PLoS One 7:e43106. doi: 10.1371/journal.pone.0043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steyer A, Gutiérrez-Aguire I, Kolenc M, Koren S, Kutnjak D, Pokorn M, Poljšak-Prijatelj M, Racki N, Ravnikar M, Sagadin M, Fratnik Steyer A, Toplak N. 2013. High similarity of novel orthoreovirus detected in a child hospitalized with acute gastroenteritis to mammalian orthoreoviruses found in bats in Europe. J Clin Microbiol 51:3818–3825. doi: 10.1128/JCM.01531-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.USDA-APHIS. 2014. Swine enteric coronavirus disease. SECD: situation report—October 30, 2014. USDA-Animal and Plant Health Inspection Service, Washington, DC: www.aphis.usda.gov/animal-health/secd Accessed 9 November 2014. [Google Scholar]
- 23.Pasick J, Berhane Y, Ojkic D, Maxie G, Embury-Hyatt C, Swekla K, Handel K, Fairles J, Alexandersen S. 2014. Investigation into the role of potentially contaminated feed as a source of the first-detected outbreaks of porcine epidemic diarrhea in Canada. Transbound Emerg Dis 61:397–410. doi: 10.1111/tbed.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Opriessnig T, Xiao CT, Gerber PF, Zhang J, Halbur PG. 2014. Porcine epidemic diarrhea virus RNA present in commercial spray-dried porcine plasma is not infectious to naive pigs. PLoS One 9:e104766. doi: 10.1371/journal.pone.0104766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.OIE 2014. Technical-Facsheet. Infection with porcine epidemic diarrhoea virus. World Organization for Animal Health (OIE), Paris, France: http://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/pdf/A_factsheet_PEDV.pdf. Accessed 10 November 2014. [Google Scholar]
- 26.Wong AH, Cheng PK, Lai MY, Leung PC, Wong KK, Lee WY, Lim WW. 2012. Virulence potential of fusogenic orthoreoviruses. Emerg Infect Dis 18:944–948. doi: 10.3201/eid1806.111688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lelli D, Moreno A, Lavazza A, Bresaola M, Canelli E, Boniotti MB, Cordioli P. 2013. Identification of mammalian orthoreovirus type 3 in Italian bats. Zoonoses Public Health 60:84–92. doi: 10.1111/zph.12001. [DOI] [PubMed] [Google Scholar]
- 28.Chappell JD, Barton ES, Smith TH, Baer GS, Duong DT, Nibert ML, Dermody TS. 1998. Cleavage susceptibility of reovirus attachment protein sigma1 during proteolytic disassembly of virions is determined by a sequence polymorphism in the sigma1 neck. J Virol 72:8205–8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bassel-Duby R, Spriggs DR, Tyler KL, Fields BN. 1986. Identification of attenuating mutations on the reovirus type 3 S1 double-stranded RNA segment with a rapid sequencing technique. J Virol 60:64–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaye KM, Spriggs DR, Bassel-Duby R, Fields BN, Tyler KL. 1986. Genetic basis for altered pathogenesis of an immune-selected antigenic variant of reovirus type 3 (Dearing). J Virol 59:90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irvin SC, Zurney J, Ooms LS, Chappell JD, Dermody TS, Sherry B. 2012. A single-amino-acid polymorphism in reovirus protein mu2 determines repression of interferon signaling and modulates myocarditis. J Virol 86:2302–2311. doi: 10.1128/JVI.06236-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Middleton JK, Agosto MA, Severson TF, Yin J, Nibert ML. 2007. Thermostabilizing mutations in reovirus outer-capsid protein mu1 selected by heat inactivation of infectious subvirion particles. Virology 361:412–425. doi: 10.1016/j.virol.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dermody TS, Nibert ML, Bassel-Duby R, Fields BN. 1990. Sequence diversity in S1 genes and S1 translation products of 11 serotype 3 reovirus strains. J Virol 64:4842–4850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goral MI, Mochow-Grundy M, Dermody TS. 1996. Sequence diversity within the reovirus S3 gene: reoviruses evolve independently of host species, geographic locale, and date of isolation. Virology 216:265–271. doi: 10.1006/viro.1996.0059. [DOI] [PubMed] [Google Scholar]
- 35.Kedl R, Schmechel S, Schiff L. 1995. Comparative sequence analysis of the reovirus S4 genes from 13 serotype 1 and serotype 3 field isolates. J Virol 69:552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai Y, Zhou Q, Zhang C, Song Y, Tian X, Zhang X, Xue C, Xu S, Bi Y, Cao Y. 2012. Complete genome sequence of a porcine orthoreovirus from southern China. J Virol 86:12456. doi: 10.1128/JVI.02254-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chua KB, Voon K, Crameri G, Tan HS, Rosli J, McEachern JA, Suluraju S, Yu M, Wang LF. 2008. Identification and characterization of a new orthoreovirus from patients with acute respiratory infections. PLoS One 3:e3803. doi: 10.1371/journal.pone.0003803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kongsted H, Jonach B, Haugegaard S, Angen Ø, Jorsal SE, Kokotovic B, Larsen LE, Jensen TK, Nielsen JP. 2013. Microbiological, pathological and histological findings in four Danish pig herds affected by a new neonatal diarrhoea syndrome. BMC Vet Res 9:206. doi: 10.1186/1746-6148-9-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clarke P, Meintzer SM, Spalding AC, Johnson GL, Tyler KL. 2001. Caspase 8-dependent sensitization of cancer cells to TRAIL-induced apoptosis following reovirus-infection. Oncogene 20:6910–6919. doi: 10.1038/sj.onc.1204842. [DOI] [PubMed] [Google Scholar]
- 40.Tardieu M, Powers ML, Weiner HL. 1983. Age dependent susceptibility to reovirus type 3 encephalitis: role of viral and host factors. Ann Neurol 13:602–607. doi: 10.1002/ana.410130604. [DOI] [PubMed] [Google Scholar]
- 41.Attoui H, Mohd Jaafar F, de Micco P, de Lamballerie X. 2005. Coltiviruses and seadornaviruses in North America, Europe, and Asia. Emerg Infect Dis 11:1673–1679. doi: 10.3201/eid1111.050868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang YW, Dickerman AW, Piñeyro P, Li L, Fang L, Kiehne R, Opriessnig T, Meng XJ. 2013. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 4(5):e00737-13. doi: 10.1128/mBio.00737-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dermody TS, Nibert ML, Bassel-Duby R, Fields BN. 1990. A sigma 1 region important for hemagglutination by serotype 3 reovirus strains. J Virol 64:5173–5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 45.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jukes TH, Cantor CR. 1969. Evolution of protein molecules, p 21–132. In Munro HM (ed), Mammalian protein metabolism. Academic Press, Waltham, MA. [Google Scholar]
- 48.Elazhary MA, Morin M, Derbyshire JB, Lagacé A, Berthiaume L, Corbeil M. 1978. The experimental infection of piglets with a porcine reovirus. Res Vet Sci 25:16–20. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The μ1 protein of U.S. porcine orthoreovirus isolates carries unique mutations. Shown is deduced amino acid sequence comparison of the μ1 protein of U.S. porcine orthoreovirus isolates (FS03 and BM100) with those of the T1L, T2J, and T3D prototypes and T3/Bat/Germany sequences. Amino acid residues that are identical to T3D sequence are indicated by dashes. Observed mutations with respect to T3D are boxed. Download
The μ2 protein of U.S. porcine orthoreovirus isolates carries unique mutations. Deduced amino acid sequence comparison of the μ2 protein of U.S. porcine orthoreovirus isolates (FS03 and BM100) with those of the T1L, T2J, T3D prototype and T3/Bat/Germany sequences. Amino acid residues that are identical to those in the T3D sequence are indicated by dashes. Observed mutations with respect to T3D are boxed. Download
Sequence comparison of σ1 and σ1s proteins of U.S. porcine orthoreovirus isolates with closely related MRV3 sequences. (A) Alignments of deduced σ1 protein sequences of U.S. porcine orthoreovirus isolates (FS03 and BM100) were compared with those of T3/D, T3/Bat/Germany, and T3/porcine/Korea sequences. Positions of the amino acids are numbered above the sequences. The residues boxed in red represent the predicted NLAIRLP sialic acid-binding domain, trypsin resistance residue (249I), and residues involved in neurotropism (340D and 419E). (B) Deduced amino acid comparison of σ1s protein of U.S. porcine orthoreovirus isolates (FS03 and BM100) with those of the T1L, T2J, and T3D prototype and T3/Bat/Germany virus sequences. Amino acid residues that are identical to those in the T3D sequence are indicated by dashes. Observed mutations with respect to T3D are boxed. Download
Proteins encoded by U.S. porcine orthoreovirus isolates are homologous to T1 and T3 serotypes.
List of accession numbers of viral strains used for phylogenetic analysis in this study.
GenBank accession numbers of U.S. porcine orthoreovirus isolates and prototype sequences used in this study.
Experimental design.