ABSTRACT
Listeria monocytogenes enters nonphagocytic cells by a receptor-mediated mechanism that is dependent on a clathrin-based molecular machinery and actin rearrangements. Bacterial intra- and intercellular movements are also actin dependent and rely on the actin nucleating Arp2/3 complex, which is activated by host-derived nucleation-promoting factors downstream of the cell receptor Met during entry and by the bacterial nucleation-promoting factor ActA during comet tail formation. By genome-wide small interfering RNA (siRNA) screening for host factors involved in bacterial infection, we identified diverse cellular signaling networks and protein complexes that support or limit these processes. In addition, we could precise previously described molecular pathways involved in Listeria invasion. In particular our results show that the requirements for actin nucleators during Listeria entry and actin comet tail formation are different. Knockdown of several actin nucleators, including SPIRE2, reduced bacterial invasion while not affecting the generation of comet tails. Most interestingly, we observed that in contrast to our expectations, not all of the seven subunits of the Arp2/3 complex are required for Listeria entry into cells or actin tail formation and that the subunit requirements for each of these processes differ, highlighting a previously unsuspected versatility in Arp2/3 complex composition and function.
IMPORTANCE
Listeria is a bacterial pathogen that induces its internalization within the cytoplasm of human cells and has been used for decades as a major molecular tool to manipulate cells in order to explore and discover cellular functions. We have inactivated individually, for the first time in epithelial cells, all the genes of the human genome to investigate whether each gene modifies positively or negatively the Listeria infectious process. We identified novel signaling cascades that have never been associated with Listeria infection. We have also revisited the role of the molecular complex Arp2/3 involved in the polymerization of the actin cytoskeleton, which was shown previously to be required for Listeria entry and movement inside host cells, and we demonstrate that contrary to the general dogma, some subunits of the complex are dispensable for both Listeria entry and bacterial movement.
INTRODUCTION
The Gram-positive bacterium Listeria monocytogenes has emerged as a model for the study of intracellular parasitism (1, 2). Listeria is able to enter mammalian cells, to disrupt its internalization vacuole, and to replicate in the cytoplasm of target cells, avoiding host cytoplasmic surveillance mechanisms. In addition, actin nucleation at the bacterial surface and formation of actin tails propel Listeria through the host cytoplasm and into neighboring cells (3).
Cellular invasion is achieved by engagement of either the adherens junction molecule E-cadherin or the receptor tyrosine kinase Met (4, 5) by the bacterial surface proteins internalin A (InlA) and InlB, respectively. Phosphorylation and ubiquitination of receptors upon ligand binding lead to the recruitment of a clathrin-associated complex containing the adaptor molecules Dab2, Hip1R, and myosin 6, which connect clathrin to the actin cytoskeleton (6, 7). Clathrin recruitment is required for the initial actin reorganization at the bacterial entry site (8, 9). Further actin rearrangements are induced by the small GTPases Rac1 and Cdc42, which activate WAVE and N-WASP, respectively, and promote actin nucleation via the Arp2/3 complex (10, 11). The Arp2/3 complex, whose function was initially characterized in the Listeria actin comet tail system (12), is a seven-protein complex that consists of two actin-related proteins, Arp2 and Arp3, and five additional components, p16 (ArpC5), p20 (ArpC4), p21 (ArpC3), p34 (ArpC2), and p41 (ArpC1). In humans, two isoforms of subunit p41 are encoded by the genes arpc1A and arpc1B (13). By nucleating a new daughter filament on preexisting actin filaments, the Arp2/3 complex generates branched actin networks (14). Other classes of actin nucleators, including VASP (15), formins, and WH2-containing members of the Spire and Leiomodin families, generate and elongate linear actin filaments (reviewed in reference 16).
Here we first report the development of a high-throughput human genome-wide small interfering RNA (siRNA) screening protocol to identify new host factors involved in different steps of the Listeria infection process and to obtain new insights concerning its regulation. Imaging of the bacterial secreted protein InlC in the host cell cytoplasm was used to quantify infection levels (17), and actin labeling was used to detect actin tails associated with bacteria. Our siRNA screens allowed us to identify several cellular processes that regulate Listeria infection. In particular, several actin nucleators could be identified that, in addition to the Arp2/3 complex, contribute to early steps of InlB/Met-dependent Listeria infection but are dispensable for actin comet tail formation. Strikingly, subunits of the Arp2/3 complex are not equally required for bacterial entry and actin tail formation: while the subunits Arp2, Arp3, p21, p34, and p41 are required for entry, with p16 and p20 being dispensable for this step, all of the subunits except p16 are necessary for actin tail formation. Our results highlight the novel concept that different biological processes may use alternative Arp2/3 complexes.
RESULTS
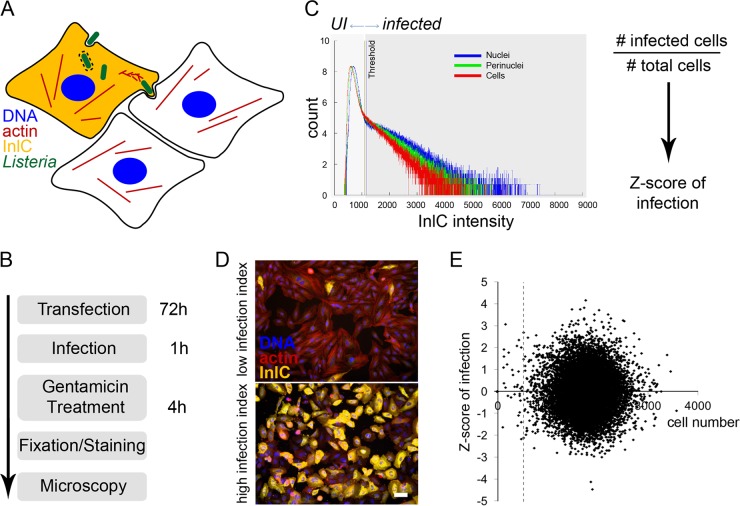
In order to obtain a more comprehensive understanding of global signaling cascades governing host cell infection by Listeria, we performed a high-throughput siRNA screen in HeLa cells using a commercial human genome-wide siRNA library (ON-TARGETplus SMARTpool; Dharmacon) that contains pools of 4 different RNA oligonucleotides targeting the same transcript. To monitor infection, we developed an image-based assay relying on the detection of the bacterial protein InlC, which is actively secreted by Listeria once it reaches the cytoplasm of infected cells; immunofluorescence staining for this protein therefore allows for unambiguous discrimination between infected versus noninfected cells (Fig. 1A) (17). To perform our screen, at 72 h posttransfection with the genome-wide siRNA library, HeLa cells were infected for 1 h with enhanced green fluorescent protein (EGFP)-expressing Listeria and were incubated for an additional 4 h in the presence of the non-cell-permeable antibiotic gentamicin in order to allow internalized bacteria to reach the cytoplasm and secrete InlC, while preventing further bacteria from entering cells and saturating the infection. The HeLa cells were then fixed and stained for InlC, for actin (using fluorescent phalloidin), and for DNA (using 4[prime],6-diamidino-2-phenylindole [DAPI]) and were imaged using automated microscopy (Fig. 1B). After segmentation of nuclei and cell bodies using DAPI and actin staining, respectively, the InlC signal was quantified for each nucleus, cell, and perinuclear region; thresholding of the InlC intensities of these cellular objects allowed for binary quantification of infected versus noninfected cells and calculation of a normalized Z score (Fig. 1C). Indeed, if in most cases the InlC signal was evenly distributed across the cell and the nucleus, upon knockdown of some cellular proteins and depending on the cell shape, the InlC distribution could change. Manual quality control showed that infection could be reliably detected if the InlC intensities in all three aforementioned cellular regions were taken into account. A cell was considered infected if the InlC intensity threshold was reached in at least one of the analyzed regions. Representative examples of HeLa cells displaying either low (where siRNA treatment decreased infection) or high (where siRNA treatment increased infection) Z score indexes are shown (Fig. 1D). Knockdown of certain cellular factors influences cell viability or proliferation. In order to test for a general effect of the cell number on Listeria infection, the infection Z scores from genome-wide siRNA screens were plotted against the respective cell numbers (Fig. 1E). Even though the cell counts per well varied between 0 and more than 3,000, a correlation with the infection level was not observed. Nevertheless, knockdowns that reduced the cell number to fewer than 500 were not considered for further analyses or specifically flagged in order to filter out possible random effects due to the limited sample size.
FIG 1 .
Screening strategy for an image-based siRNA screening of Listeria infection. (A) Cellular infection by Listeria was assessed by immunofluorescent detection of the protein InlC, which is secreted by intracellular bacteria and accumulates in the cytoplasm of infected cells. Bacteria were detected by expression of EGFP, the actin cytoskeleton was detected using fluorescent phalloidin, and nuclei were stained with DAPI. (B) The infection screening protocol included 72 h of HeLa cell reverse siRNA transfection, 1 h of infection with overnight-grown EGFP-expressing Listeria, gentamicin treatment for 4 h to kill extracellular bacteria, sample fixation and staining with antibodies and fluorescent dyes, and automated microscopy. (C) A binary quantification of infection (UI, uninfected versus infected cells) was established by thresholding the InlC intensities in nuclei, the perinuclear region, and in the cytoplasm of HeLa cells. The Z score of infection for a given gene was calculated as z = (x − µ)/σ, in which x is the infection index (no.of infected cells divided by the total no. of cells in a well of a 384-well plate), μ is the infection index mean per plate, and σ is the standard deviation (SD) of the infection index per plate. (D) Representative examples are shown of cells displaying low (up) or high (down) infection indexes according to the proportion of InlC-positive cells (yellow signal). Bar, 50 µm. (E) Scatter plot showing the cell number on the horizontal axis and Listeria infection levels on the vertical axis. Values are averages of 2 genome-wide screens. The dashed line shows the threshold of 500 imaged cells per well.
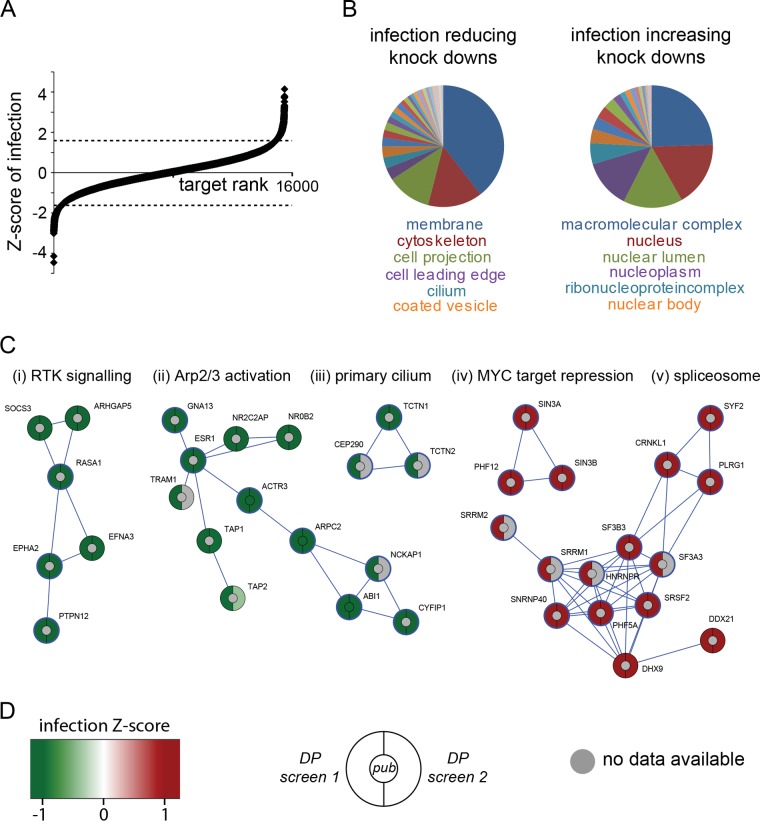
Figure 2A shows the distribution of all siRNA targets from a genome-wide screen performed in duplicate and ranked according to the normalized infection level, microscopically quantified based on InlC secretion: a negative Z score indicates that cell infection by Listeria was impaired upon knockdown of the target gene of interest, while a positive value is associated with knockdowns resulting in a higher fraction of infected cells. (Genome-wide results are presented in Table S1 in the supplemental material.) Further analysis of the strongest 500 infection up- and downregulating hits using the “cellular components enrichment” tool of the STRING database (http://www.string-db.org) revealed substantial differences between these two classes of targets: while infection-limiting targets (the knockdown of which increases infection) are predominantly nuclear proteins, those hits whose knockdown limited infection are mostly associated with peripheral cellular structures related to cell membrane and cytoskeleton (Fig. 2B). Among the strongest 500 infection-limiting targets, we detected a cluster of spliceosome-related proteins: the heterogeneous nuclear ribonucleoprotein R (HNRNPR) as well as the 40-kDa small nuclear ribonucleoprotein (SNRNP40), together with the nuclear matrix component SRRM1 and the pre-mRNA splicing factors SF3A3 and SRSF2 (Fig. 2C). Another cluster of infection-limiting factors is made of components that are involved in transcriptional repression, for example, of MYC target genes (Fig. 2C). Interestingly, knockdown of MYC itself showed the opposite effect on Listeria infection (Fig. 3A), indicating that MYC-dependent gene expression plays an important role in cellular susceptibility to Listeria infection. We identified, comprised within the strongest 500 infection-promoting targets, a cluster that includes the Arp2/3 complex subunits Arp3 and p34 (encoded by the genes actr3 and arpc2, respectively) together with the estrogen receptor 1 (ESR1) and several components of the WAVE complex (NCKAP1, ABI1 and CYFIP1) (Fig. 2C). Other relevant downregulating clusters include, for example, components related to the primary cilium (TCTN1, TCTN2, and CEP290) or factors involved in tyrosine kinase signal transduction, including the ephrin receptor EPHA2 and the protein tyrosine phosphatase PTPN12 (Fig. 2C).
FIG 2 .
Results from two repetitions of the genome-wide siRNA screen for Listeria infection. (A) Distribution of the Z scores of infection for roughly 16,000 genes, with negative values associated with infection-reducing knockdowns and positive values associated with infection-increasing knockdowns. (Dotted lines indicate thresholds for the strongest 500 infection up- and downregulating hits.) (B) Cellular functions associated with infection-reducing (left) and infection-increasing (right) knockdowns using the “cellular components enrichment” tool from the STRING database; infection-reducing hits are associated with cortical cell functions, while infection-increasing hits are related to nuclear functions. (C) Gene clusters (STRING stringency, 0.7) of infection-reducing (green) and infection-increasing (red) knockdowns: (i) cluster related to receptor tyrosine kinase signaling, (ii) Arp2/3 complex activation-related cluster, (iii) cluster related to the primary cilium, (iv) cluster related to transcriptional repression of MYC-responsive genes, and (v) spliceosome-related cluster. Hits with a blue circle belong to the cellular components of cell projections (i and ii), cilia (iii), nucleoplasm (iv), or spliceosomal complex (v). (D) Infection Z score color code and diagram depicting the expectation according to published results where available (center) as well as results from the two different genome-wide screens using a Dharmacon pooled small interfering RNA (siRNA) library.
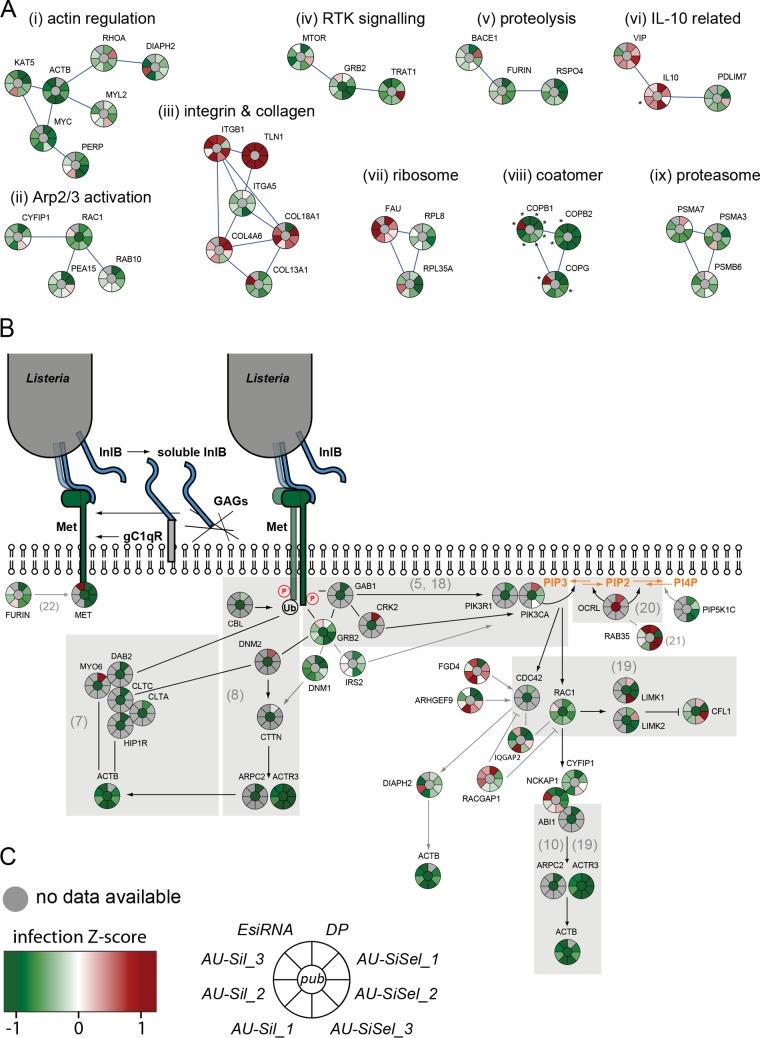
FIG 3 .
Results from validation screens for Listeria infection. (A) Gene clusters (STRING stringency, 0.7) of functional groups obtained with all hits that gave consistent results for infection with no more than one outlier out of 7 or 8 individual siRNA sequences or pools: (i) action-related cluster, (ii) RAC1 and Arp2/3 activation-related cluster, (iii) integrin/collagen-related cluster, (iv) receptor tyrosine kinase (RTK) signaling and GRB2-related cluster, (v) proteolytic cleavage, (vi) genes connected to interleukin-10 (IL-10), (vii) ribosome-related cluster, (viii) coatomer-related cluster, and (ix) proteasome-related cluster. Knockdowns that resulted in cell numbers lower than 500 are marked with asterisks. (B) Validation results for genes that have been shown to be involved in Met/InlB-dependent Listeria invasion (gray background) or that can be associated with the Met/InlB pathway (STRING stringencies of 0.235 for furin/MET and 0.4 for other connections). References are indicated in parentheses. (C) Infection Z score color code and diagram depicting the expectation according to published results (center) as well as results obtained with the Dharmacon pooled genome-wide libraries (DP), with the Ambion silencer select unpooled siRNAs (AU-SiSel_1 to -3), the Ambion silencer unpooled siRNAs (AU-Sil_1 to -3), and the Sigma esiRNA pooled siRNAS (EsiRNA).
A subset of candidate genes was selected for further validation using siRNAs from different commercial providers (see Table S2 in the supplemental material). Figure 3A depicts Z-scored infection results for nine molecular clusters, which include mainly effectors not previously associated to signaling cascades affecting Listeria invasion. Among these, COPB1/COPB2 and COPG form a cluster of coat molecules which highly decreases infection (but also reduces cell viability) upon inactivation (Fig. 3A). An integrin-related cluster includes molecules like integrin β1, talin, and 18A1/4A6 collagens, which highly increase infection upon inactivation; however, in the same cluster, inactivation of integrin α5 and collagen 13A1 reduces infection (Fig. 3A). A Rac1-related cluster of infection-reducing targets includes the small GTPase RAB10, which was previously identified in a proteomic screen of molecules associated with Listeria internalization vacuoles (J. Pizarro-Cerdá, J. Garin, and P. Cossart, unpublished data) (Fig. 3A). An actin-related cluster also includes the lysine acetyl-transferase KAT5 (Fig. 3A).
We then focused our analysis on the InlB/Met invasion signaling cascade. As shown in Fig. 3B, our screens confirmed the requirement of a core signal transduction pathway for Listeria invasion in HeLa cells that includes the InlB receptor Met and the adaptor molecules GAB1 and GRB2 (5, 18), as well as two molecular routes dependent on clathrin or RAC1 that lead to actin rearrangements and nucleation by the Arp2/3 complex (7, 8, 10, 19) (Fig. 3B). We confirmed the inhibitory role of the phosphoinositide phosphatase OCRL on bacterial invasion (20) and identified the small GTPase RAB35 as a putative upstream activator of OCRL during infection, similarly to what has been shown for cytokinesis (21) (Fig. 3B). In addition, by STRING analysis, we identified novel cellular factors that influenced Listeria infection in siRNA screens and, according to their molecular function and reported role in related processes, could be involved in the regulation of the InlB-dependent invasion pathway. These include the insulin receptor substrate 2 (IRS2), the phosphoinositide kinase PIP5K1C, the Met precursor protease furin (22), and the formin DIAPH2 (Fig. 3B).
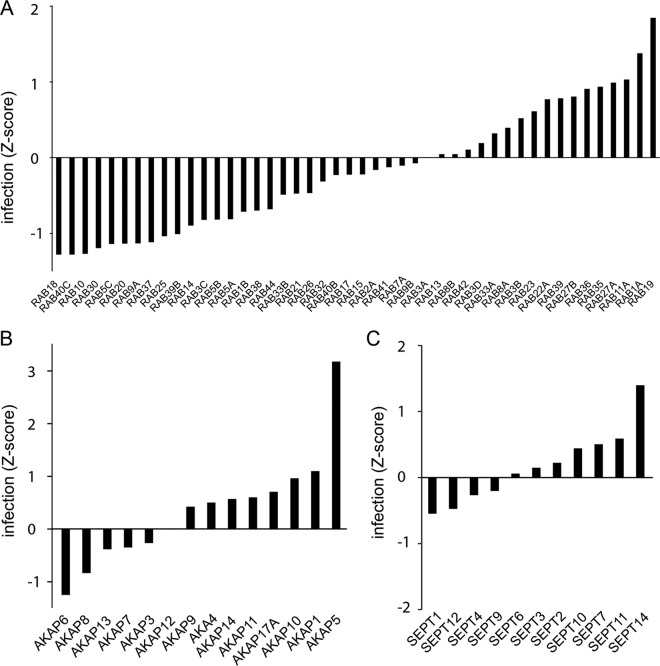
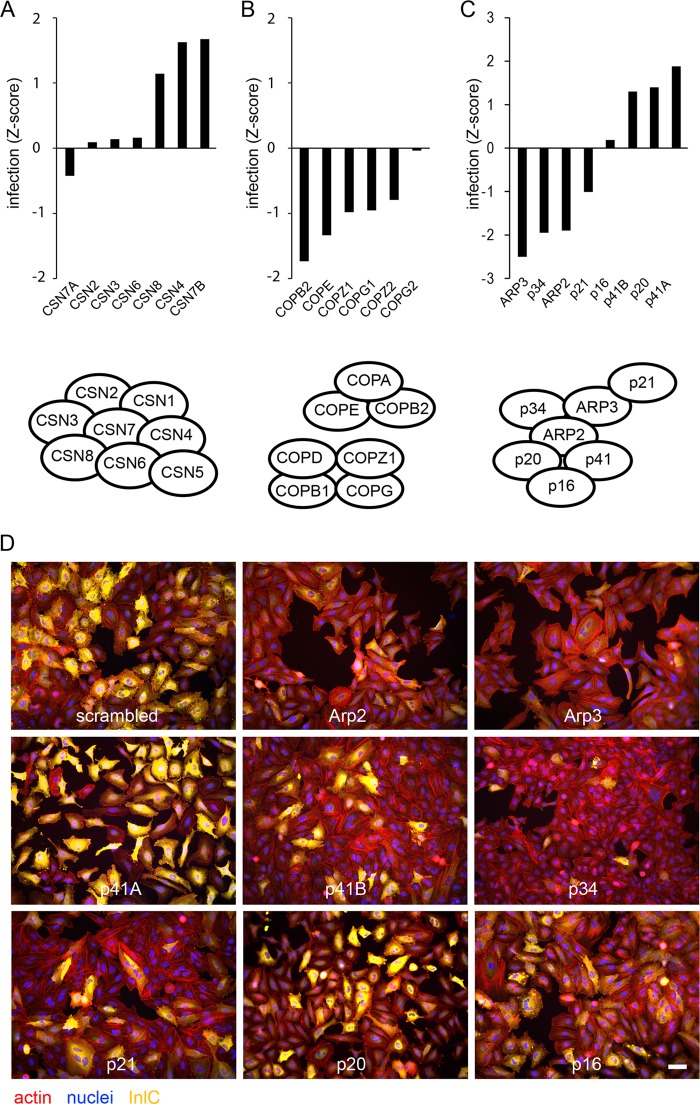
In macrophages, the contribution of members of the Rab family of GTPases to Listeria infection has been previously investigated, and RAB5A has been described as fostering the maturation of Listeria phagosomes toward lysosomes, while no role has been attributed to RAB5C (23, 24). In our screen, the three isoforms of RAB5 (RAB5A, RAB5B, and RAB5C) are found to be required for Listeria infection in epithelial cells, together with RAB20, a GTPase that has been recently described as a novel partner of phosphatidylinositol 3-kinase (PI 3-kinase) and RAB5 in delaying phagosomal maturation (25), and RAB10, as mentioned above. Interestingly, the most pronounced reduction in infection was observed upon inactivation of RAB18 and RAB40C, two GTPases that have been implicated in the formation of lipid droplets (26, 27). On the other hand, inactivation of recycling endosome-related Rab-GTPases RAB11 and RAB35, as well as RAB27A- and -B, promotes Listeria infection (Fig. 4A). Other examples of protein families that showed diverse results in our screens are the A kinase anchor proteins (AKAPs) and septins (Fig. 4B and C) (28). As mentioned above (Fig. 3A), a cluster of subunits of the coatomer was shown to play a role in Listeria infection. Similar results were observed for all the other subunits of the complex, with only COPG2 displaying no effect on infection (Fig. 5B). Another complex example is the COP9/signalosome, which has been implicated in a broad range of functions, including proteolysis, cell cycle, and DNA repair (29): with the exception of CSN7A, inactivation of all of the subunits of the complex appeared to promote infection, at least mildly (Fig. 5A).
FIG 4 .
Infection of cells depleted of proteins belonging to selected protein families. Shown are infection Z scores of cells infected with Listeria for 5 h after depletion of members of the Rab-GTPase family (A), the A-kinase anchor protein family (B), or the septin family (C). Bars represent the mean of results from two genome-wide siRNA screens.
FIG 5 .
Infection of cells depleted of proteins belonging to selected protein complexes. Infection Z score of cells infected with Listeria for 5 h after depletion of subunits of the COP9/signalosome complex (A), the coatomer complex (B), or the Arp2/3 complex (C). Protein complex schemes were adapted from references 29, 30, and 37. Bars represent the means of results from two genome-wide siRNA screens. (D) Examples of images from the primary genome-wide siRNA screen for InlC staining upon inactivation of each subunit of the Arp2/3 complex. Bar, 50 µm.
A particular and surprising situation was observed for the Arp2/3 complex (Fig. 5C and D). This actin nucleating complex has previously been shown to participate in the InlB-dependent invasion process by Listeria: in particular, the subunit Arp3 has been localized by fluorescence microscopy to the entry site of bacteria or InlB-coated latex beads, and sequestration of the Arp2/3 complex by overexpression of the Scar1 C-terminal domain blocks bacterial entry into host cells (19). In our screens, we demonstrated a role for the Arp2/3 complex subunits Arp2, Arp3, p34, and p21 in cellular infection by Listeria (Fig. 3 and 5C and D), but surprisingly, no role for p16 was observed, and we even detected positive infection scores upon inactivation of p41A, p41B, and p20 (Fig. 5C and D). We verified the effect of siRNA transfection on each Arp2/3 complex subunit on expression of the other subunits, and while siRNA-mediated knockdown of the targeted Arp2/3 complex subunits was efficient, expression of the respective other subunits was not significantly altered (see Text S1 and Fig. S1 in the supplemental material). Together, our observations strongly suggest that an Arp2/3 complex containing only a subset of subunits is required for cellular infection by Listeria or that only some subunits of the complex are functional in this context.
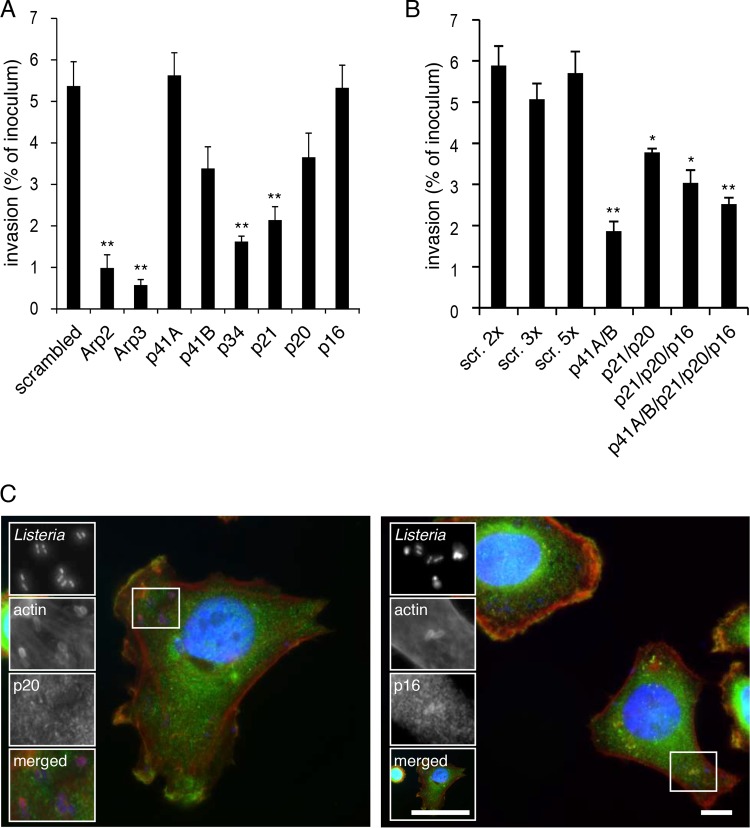
Since our InlC infection readout is performed after 5 h of infection, in order to specifically analyze the early stages of bacterial internalization, we performed gentamicin invasion assays for 2 h and quantitation of Listeria colony-forming units (CFU) after knockdown of the individual Arp2/3 complex subunits. Here again, inactivation of Arp2, Arp3, p34, and p21 resulted in the strongest inhibition of Listeria entry into HeLa cells, while p16 did not affect bacterial invasion at all, and only a moderate effect on entry was observed upon inactivation of p20 (Fig. 6A). Inactivation of p41A and p41B did not inhibit invasion or only moderately affected this process, respectively (Fig. 6A); we tested whether these p41 isoforms have overlapping functions during Listeria invasion, and as shown in Fig. 6B, simultaneous knockdown of both molecules has a strong effect on entry, demonstrating that the subunit p41 is important for the function of the Arp2/3 complex during Listeria entry into cells. All tested combined depletions of p41, p21, p20, and p16 never did block Listeria invasion as efficiently as the single knockdowns of Arp2 or Arp3 (Fig. 6B). These results therefore suggest on one hand that Arp2 and Arp3 are the key components of the complex, but also demonstrate that the subunits p20 and p16 are dispensable for the internalization of Listeria within host cells.
FIG 6 .
Contributions of the different subunits of the Arp2/3 complex to Listeria invasion. Subunits of the Arp2/3 complex were knocked down by siRNA transfection of HeLa cells either individually (A) or in combination (B). One hour after infection with Listeria, cells were washed with gentamicin in order to kill extracellular bacteria, and Listeria entry was measured by counting CFU. Bars represent the mean and standard error of the mean (SEM) from 3 experiments. Significant differences (Student’s t test) from the scramble treatments are indicated as ** (P < 0.01) and * (P < 0.05). (C) HeLa cells infected with Listeria for 10 min were stained for actin (red) and the Arp2/3 complex subunit p20 (left panel, green) or p16 (right panel, green). Bars, 5 µm.
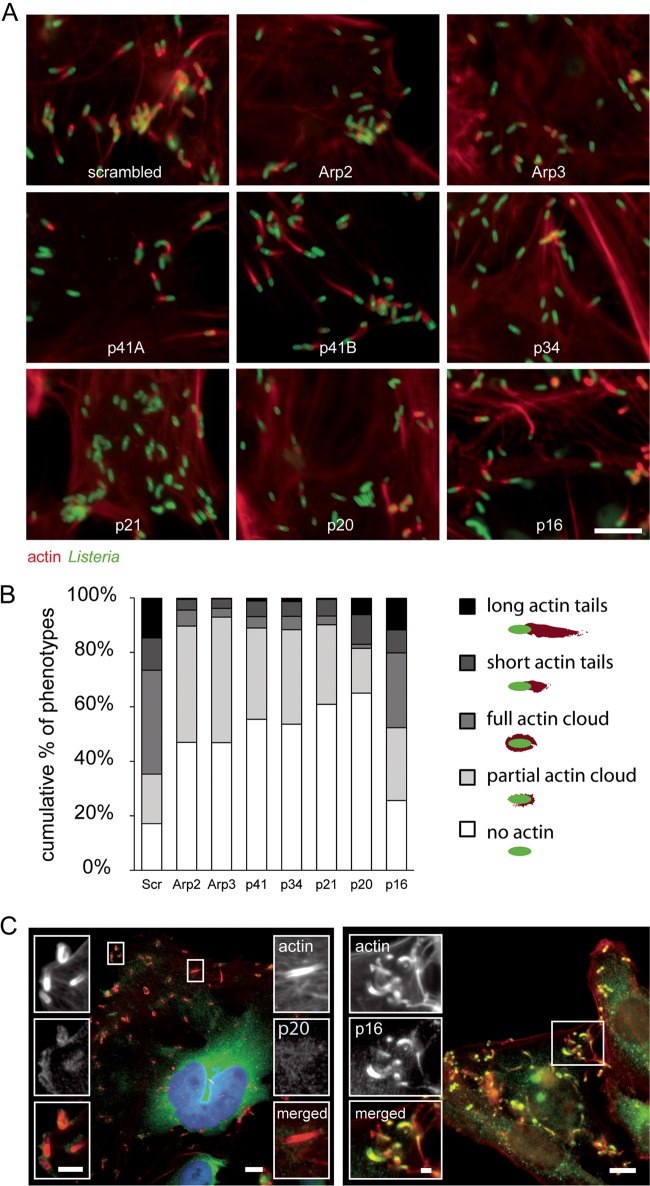
We next analyzed whether actin tail formation by cytoplasmic Listeria, a well-characterized Arp2/3-dependent mechanism (12), may take place in the absence of specific complex subunits. Qualitative analysis of actin tails in images from validation experiments indicated that inactivation of Arp2, Arp3, p34, and p21 abrogates actin comet tail formation by cytoplasmic Listeria completely, and knockdown of p20 reduces the number of actin-positive bacteria, even though the fraction of bacteria presenting elongated tails is less strongly altered. Simultaneous knockdown of the two p41 isoforms blocked actin polymerization, while inactivation of either p41A, p41B, or p16 led to partial inhibition of actin tail formation (Fig. 7A). Quantitative morphometric analysis of actin polymerization in cells inactivated for each subunit of the Arp2/3 complex demonstrates that the subunits Arp2, Arp3, p41, p34, p21, and p20 are required for tail formation, but p16 is dispensable (Fig. 7B). Interestingly, while knockdown of p16 did not block Listeria entry and actin comet tail formation, it can still be detected at bacterial entry sites and actin comet tails by fluorescence staining (Fig. 6C, right panel, and 7C, right panel). In contrast, p20 could be detected neither at the bacterial entry site (Fig. 6C, left panel) nor at actin comet tails (Fig. 7C, left panel). However, closer examination of p20 localization in cells infected with Listeria revealed that this Arp2/3 complex subunit localizes to the bacterial surface at early stages of actin comet tail formation (Fig. 7C, left panel). p20 may therefore be mainly beneficial during initial actin polymerization at the surface of cytoplasmic Listeria, while it seems to be dispensable for subsequent comet tail elongation. This is in agreement with the observation that p20 knockdown strongly reduced the number of bacteria with partial or complete actin clouds at the surface, while the number of bacteria with short or long actin tails was altered only relatively mildly (Fig. 7B). Altogether, our results clearly demonstrate that the subunit p16 is required neither for Listeria entry into host cells nor for actin tail formation and that p20 is required for actin tail formation, probably at initial stages, but not for cell invasion.
FIG 7 .
Contributions of the different subunits of the Arp2/3 complex to actin comet tail formation by Listeria. (A) Examples of images from validation experiments for actin and Listeria upon inactivation of each subunit of the Arp2/3 complex. Bar, 5 µm (B) Percentage of Listeria cells displaying long actin tails, short tails, full actin clouds, partial clouds, or no actin association upon inactivation of Arp2, Arp3, p41 (both subunits simultaneously), p34, p21, p20, and p16. Bars represent the means from 4 experiments. (C) HeLa cells infected with Listeria for 6 h and stained for actin (red) and for the Arp2/3 complex subunit p20 (left panel, green) or p16 (right panel, green). Bars, 5 µm; insert bar left, 2 µm; insert bar right, 1 µm.
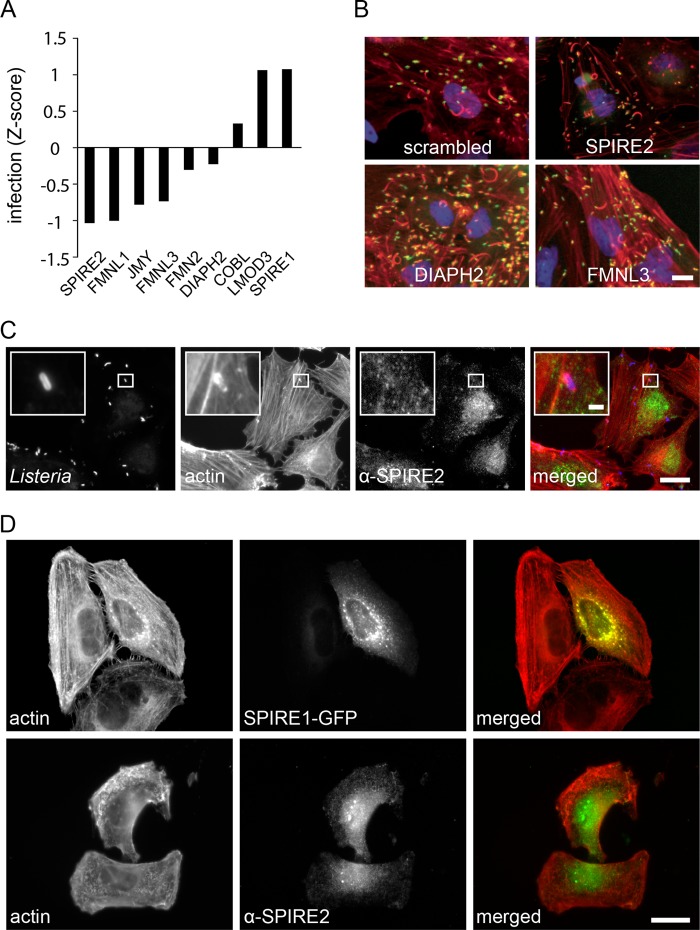
Broader analysis of screening results for described actin nucleators indicates that, besides the Arp2/3 complex, other actin nucleators may also participate in the Listeria infection process (Fig. 8A). For SPIRE2 and the formin homology 2 (FH2) domain-containing proteins FMNL3 and DIAPH2, a contribution to Listeria infection was confirmed by validation experiments (Fig. 2A; see Fig. S2A and Text S1 in the supplemental material). Interestingly, SPIRE and DIAPH2 were previously implicated in the interplay between endosomal compartments and the actin cytoskeleton (31, 32), which raises the question of whether these actin nucleators play similar roles during Listeria invasion. SPIRE2, however, could not be detected in the vicinity of invading Listeria (Fig. 8C). While SPIRE2 knockdown reduced bacterial invasion, knockdown of the closely related SPIRE1 did not. Indeed, the subcellular localizations of both proteins were not identical. SPIRE2 could be found throughout the cell body; SPIRE1 instead was concentrated in perinuclear structures probably belonging to the exocytic pathway (Fig. 8D). This is in agreement with the previously described subcellular localization of this protein (33). Remarkably, the phenotype of cells visualized by actin staining was altered after depletion of SPIRE2. These cells were more circular, with altered stress fiber orientation and distinct cell borders. Depletion of SPIRE1 did not affect the cell shape and general actin phenotype (see Text S1 and Fig. S2B in the supplemental material). A phenotype similar to the one observed in SPIRE2-depleted cells was also seen upon knockdown of a few other genes that reduced Listeria infection (not shown), strongly supporting that macroscopic actin structures like stress fibers and general cell shape influence the bacterial invasion process. While SPIRE2, DIAPH2, or FMNL3 seem to be required for successful Listeria invasion of host cells, actin comet tails can still form in their absence (Fig. 8B), indicating that they are not required for this process.
FIG 8 .
Involvement of other described and potential actin nucleators in Listeria infection. (A) Infection Z score of alternative described and potential actin nucleators from 2 genome-wide siRNA screens. (B) Example of images from genome-wide siRNA screens showing actin comet tails after knockdown of different alternative actin nucleators. Bar, 10 µm. (C) HeLa cells infected for 10 min with Listeria and stained for SPIRE2. Bar, 10 µm; insert bar, 1 µm. (D) Images showing cellular localization of SPIRE1 expressed as a GFP fusion protein and fluorescently stained SPIRE2 in HeLa cells. Bar, 10 µm.
DISCUSSION
Here we report a genome-wide siRNA screen for factors critical for Listeria infection in epithelial human cells. Two genome-wide siRNA screens have been previously performed in phagocytic Drosophila S2 cells (34, 35). The screen that we present in this article is therefore the first genome-wide siRNA screen in a cellular system in which Listeria actively triggers its own internalization, using the InlB/Met-dependent signaling cascade, as HeLa CCL2 cells were our infection model. Even though the entry pathways in these different screens are different, we found common hits related to vesicle trafficking (including VPS28, COPB2, ATP6V1C1, and MON2) and to regulation of actin dynamics by the Arp2/3 complex (for example, NCKAP1, ABI1, ARPC2, and CFL2), highlighting therefore that these two cellular processes are essential for Listeria invasion irrespective of the host species and cell-specific entry pathway.
Our genome-wide siRNA screen not only confirmed and refined previously described signaling pathways involved in this process but also shed new light on the contribution of diverse cellular functions like RNA splicing, vesicle trafficking mechanisms, and protein complexes like the coatomer and the COP9/signalosome to cellular infection by Listeria. Interesting results were obtained for factors involved in actin polymerization: the actin nucleators SPIRE2, DIAPH2, and FMNL3 were found to contribute to early stages of infection but are dispensable for actin comet tail formation by cytosolic Listeria. Particularly surprising results were obtained for the involvement of the Arp2/3 complex in both steps of infection. It has been previously shown that for successful invasion of host cells by Listeria, local actin rearrangements by the Arp2/3 complex at bacterial entry sites are essential (19). By performing human genome-wide siRNA screens and selected validations, here we confirm that inactivation of central components of cytoskeletal rearrangement pathways dependent on the Arp2/3-complex results in significantly reduced cellular infection by Listeria. More importantly, we also demonstrate, contrary to the prevalent view in the field, that not all of the subunits of the Arp2/3 complex are equally required for Listeria cell entry and that alternative complexes displaying only the Arp2, Arp3, p41, p34, and p21 subunits, in the absence p20 and p16, appear sufficient to allow invasion. Moreover, we also show that the subunit p16 is dispensable for Listeria actin comet tail formation. The function of the Arp2/3 complex as one major actin nucleator in eukaryotic cells was initially identified and characterized in the Listeria actin-based motility system, in which the bacterial protein ActA directly recruits and activates Arp2/3 at the Listeria surface to promote cell-to-cell spread (12). Detailed analysis of the contribution of each Arp2/3 subunit to actin polymerization using ActA as a nucleation-promoting factor in a baculovirus expression system within insect cells indicated that all subunits are important for the ActA-dependent Arp2/3 function, even though complexes lacking p16, p21, or p41 still showed basal activity (36). Arp3 was shown to be particularly crucial for actin nucleation, while p20 and p34 were proposed to form a structural core that is essential for complex formation (36). Our present results indicate that in the context of mammalian cell infection by Listeria, Arp2, Arp3, p41, and p34 are the most critical subunits for successful cell invasion, with p21 partially contributing to this process and p20 and p16 being dispensable. Inactivation of both p41A and p41B strongly affects Listeria cellular invasion, but knockdown of p41A has no effect at all on bacterial entry in HeLa cells, suggesting that p41B is the predominant isoform in this context. Consistently, Arp2, Arp3, p34, and p41B were all detected at the bacterial entry site (see Fig. S3 and Text S1 in the supplemental material). Interestingly, inactivation of p41A, but not p41B, has a distinct effect on actin tail formation (Fig. 7A), suggesting that p41A is the predominant isoform in the Arp2/3 complex recruited by ActA. Altogether, these results show that unexpectedly not all of the Arp2/3 complex subunits are necessary for efficient Listeria entry into host cells and actin-based motility. The fact that p16 can nevertheless be detected at both bacterial entry sites and actin comet tails indicates that Arp2/3 complexes containing this subunit, while not required, may still be recruited during both processes. It is possible that Arp2/3 complexes of different compositions have overlapping functions during Listeria infection, but it is also possible that the precise composition of different Arp2/3 complex types plays a role in fine-tuning actin rearrangements in both instances. This is supported by the observations that p20 can be found predominantly early during actin comet tail formation and that knockdown of p20 affects the initial actin polymerization at the bacterial surface rather than actin tail elongation, indicating that different Arp2/3 complexes may be required in a sequential manner.
Interestingly, during the preparation of this article, a study was published by Chorev and coworkers showing that hybrid complexes consisting of an Arp2/3 core complex (Arp2, Arp3, and p34), α-actinin, and the focal adhesion molecule vinculin or Arp2, Arp3, p21, p34, and vinculin are involved in focal adhesion formation (37). Vinculin had been previously shown to modulate Arp2/3 function at focal adhesions (38). The study by Chorev et al. (37) supports the notion that alternative Arp2/3 complexes that do not consist of the 7 classical subunits are involved in specific cellular processes. Notably, these alternative complexes contain vinculin, which allows for recruitment to focal adhesions and competes with p41; knockdown of p41, therefore, has a positive effect on focal adhesion and stress fiber formation, as the equilibrium is shifted toward Arp2/3-vinculin hybrid complexes (37); double knockdown of both p41 isoforms strongly decreases bacterial entry in gentamicin invasion assays, indicating that p41 function is absolutely required for actin polymerization by Arp2/3 at Listeria entry foci. Interestingly, in our siRNA screens, we found that vinculin knockdown positively affects cell infection by Listeria, indicating that alternative complexes formed by the Arp2/3 core Arp2, Arp3, and p34, as well as additional factors other than vinculin, are implicated in Listeria entry. Determination of the conditions that promote the contribution of alternative Arp2/3 complexes to specific cellular functions should provide important information on the signaling pathways involved. In addition, this work strongly provides a framework for an integrative view of bacterial infection: future efforts will aim at deciphering the specific contribution of novel identified signaling networks to the Listeria infection process, in vitro as well as in vivo.
MATERIALS AND METHODS
Cells, bacteria, and plasmids.
HeLa CCL-2 cells from the American Type Culture Collection (ATCC) were cultured at 37°C in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal calf serum (FCS) in a humidified 5 or 10% CO2 atmosphere. Listeria monocytogenes strains EGDe.PrfA* (BUG3057) and EGDe.PrfA*.GFP (BUG3357) were cultured in brain heart infusion (BHI) medium. The plasmid pEGFP-C3-SPIRE1 was obtained from Isabelle Tardieux (Institut Cochin, Paris, France) and was described previously (33).
siRNA transfection.
For genome-wide siRNA screens, a commercially available library of pooled siRNA sequences (human ON-TARGETplus SMARTpool; Dharmacon) was used in a 384-well-plate format. Validation screens were done using up to 6 individual commercially available siRNA sequences (Silencer and Silencer Select; Ambion) and endoribonuclease-prepared siRNA (esiRNA) pools (Sigma). Secondary validation experiments were performed using siRNA ON-TARGETplus SMARTpools of 4 siRNA sequences, custom-made pools of 2 to 3 ON-TARGETplus siRNA sequences, or individual ON-TARGETplus siRNAs (Dharmacon). HeLa cells were transfected in DMEM containing 0.1 to 0.125% Lipofectamine RNAiMAX (Life Technologies) and a total siRNA concentration of 20 nM and cultured for 72 h in the presence of 10% FCS. Each 384-well screening plate contained controls for siRNA transfection (Kif11 siRNA) as well as negative controls (mock and nontargeting siRNA) and a control for Listeria infection (Met siRNA). For siRNA screens, transfections were done using a Multidrop 384 device (Thermo Electron Corporation).
Listeria infection and siRNA screening.
For infection experiments, L. monocytogenes strain EGDe.PrfA* and for siRNA screens EGDe.PrfA*.GFP overnight liquid cultures in BHI medium were washed 3 times with phosphate-buffered saline (PBS). The bacterial concentration was quantified by measuring the optical density at 600 nm, and bacteria were diluted in DMEM supplemented with 1% FCS. For siRNA screens, cells were infected at a multiplicity of infection (MOI) of 25 bacteria per cell in 30 µl infection medium calculated for 2,000 cells per well. The infection process was synchronized by centrifugation for 5 min at 1,000 rpm, and the plates were then transferred to a 37°C incubator with a 5% CO2 atmosphere for 1 h. Extracellular bacteria were then killed by exchanging the medium with fresh DMEM supplemented with 10% FCS and 40 µg/ml gentamicin (Gibco) and incubated for another 4 h at 37°C in a humidified 5%-CO2-containing atmosphere. Infection and medium replacement were carried out using a plate washer (ELx50-16; BioTek). Cells were fixed for 15 min by adding 30 µl 8% paraformaldehyde in PBS to each well using a Multidrop 384 device and washed with 500 µl PBS per well using a Power Washer 384 (Tecan). Nuclei were stained with DAPI (Roche), the actin cytoskeleton was labeled with Dy-647–phalloidin (Dyomics), and InlC was stained with a homemade rabbit-derived anti-InlC serum (39) and a secondary Alexa Fluor 546-coupled anti-rabbit antibody (Invitrogen). Antibodies and probes were diluted in PBS supplemented with 0.2% saponin, and the staining procedure was carried out with a Freedom Evo robot (Tecan). After staining, cells were kept in 40 µl PBS, sealed, and stored at 4°C until microscopy. Nine images per well were taken automatically with an ImageExpress microscope (Molecular Devices) equipped with a 10× S Fluor objective. HeLa cells plated on glass coverslips and transfected with siRNA targeting the indicated subunits of the Arp2/3 complex were infected with L. monocytogenes strain EGDe.PrfA*. After 1 h, the medium was replaced with 40 µg/ml gentamicin-containing medium in order to kill extracellular bacteria. After another 4 h, cells were fixed, permeabilized with 0.05% saponin or 0.1% Triton X-100, and stained for actin using fluorescent phalloidin (Molecular Probes) and for total Listeria (R11, in-house produced rabbit polyclonal antibody). For staining at bacterial entry sites of individual Arp2/3 subunits (rabbit polyclonal anti-p20 antibody [Santa Cruz sc-68394] or mouse monoclonal anti-p16 antibody [Santa Cruz sc-166760]) or for SPIRE2 (rabbit polyclonal AP54024PU-N; Acris GmbH), cells were infected at an MOI of 25 and fixed 10 min postinfection; for staining of Arp2/3 subunits at actin comet tails, cells were infected at an MOI of 5 and fixed 6 h postinfection. Images were taken using an inverted wide-field fluorescent Axiovert 200-M microscope (Carl Zeiss) equipped with an EMCCD Neo camera (Andor).
Image analysis.
Images from high-throughput screens were scaled to an intensity range between 0 and 1 on a full assay plate. After shading correction (flat-field correction and vignetting correction), objects were detected using CellProfiler (cellprofiler.org). First, labeled nucleus objects were segmented in the DAPI channel. Second, nucleus objects were extended by 8 pixels and the nuclear area was removed from this extended nuclear area in order to construct a perinuclear ring object. Finally, cell body objects were segmented in the actin channel using the “propagation” method with the nucleus objects as seed objects. Actin comet tails from high-magnification images were classified and quantified manually.
Infection detection and measurement for high-throughput screens.
Listeria infection, visualized by InlC staining, appears as dispersed Cy3 signals of various intensities across the cell body. The strength of this signal is dependent on the amount of bacteria inside the cell. While high infection leads to a very strong fluorescence intensity, very low or no infection results in low background intensity. To quantify infection, Cy3 intensity, which is highest in the perinuclear area, was measured in the objects “nuclei,” “perinuclei,” and “cells” using a custom CellProfiler pipeline. Infection was quantified by setting a threshold of InlC signal intensity that corresponds to uninfected cells. In order to obtain comparable results, Z scoring was used to normalize variations between individual plates and screens.
Gentamicin invasion assay.
Gentamicin invasion assays were performed as described previously (40). Briefly, cells were plated and siRNA transfected in 96-well plates 72 h before infection. After infection for 1 h, cells were washed once with DMEM supplemented with 10% FCS and 40 µg/ml gentamicin and incubated in the same medium for 1 h at 37°C in a humidified 10% CO2-containing atmosphere. To quantify entry, cells were lysed with distilled water, serial dilutions of the lysates were plated on BHI agar plates, and colonies were counted the day after. Values of invasion are given as a percentage of the inoculum.
SUPPLEMENTAL MATERIAL
Supplemental methods. Download
Western blots to validate the inactivation of the different subunits of the Arp2/3 complex. (A) Cell lysates were loaded on polyacrylamide-SDS gels, transferred to polyvinylidene difluoride (PVDF) membranes, and blotted using antibodies against the different subunits of the Arp2/3 complex; the TopoIIα protein (170 kDa) or histone H3 (15 kDa) was used as a loading control. (B) Relative expression levels of all 7 Arp2/3 complex subunits after knockdown of individual subunits. mRNA concentrations 72 h post-siRNA transfection were quantified by quantitative reverse transcription-PCR (qRT-PCR). One representative experiment of a biological triplicate is shown. Bars represent the means of two technical replicates. Download
Validation experiments for alternative actin nucleators. (A) Infection Z score of cells transfected with siRNAs targeting different actin nucleators and infected with Listeria for 5 h. Results for 3 different silencer select siRNAs for each target are shown. Bars represent the means of duplicate screens. Results of primary screens for SPIRE2, FMNL3, FMN2, and DIAPH2 could be confirmed with 2 or 3 additional siRNAs. (B) Examples of images from genome-wide screen depicting cellular actin phenotypes of cells transfected with nontargeting control siRNA or siRNA targeting SPIRE1 or SPIRE2 and infected with Listeria. Bar, 50 µm. Download
Colocalization of Arp2/3 subunits with Listeria at bacterial entry sites. Cells infected with Listeria for 10 min were stained for Arp2, Arp3, p34, or p41B. Bar, 5 µm. Download
Genome-wide infection Z scores upon gene inactivation by siRNA pools.
Validation screen Z scores upon gene inactivation by single siRNAs. DP, Dharmacon pooled primary screening results (average from 2 screens); AU SiSel, Ambion unpooled silencer select siRNA; AU sel, Ambion unpooled silencer siRNA; SI esiRNA, Sigma esiRNA pools. Negative infection Z scores are highlighted with green, and positive infection Z scores are highlighted with red. Genes are ranked according to the median infection Z score.
ACKNOWLEDGMENTS
This work received financial support from the Institut Pasteur, Institut National de la Santé et de la Recherche Médicale (Unité 604), Institut National de la Recherche Agronomique (Unité Sous Contrat 2020), Fondation Louis Jeantet, European Research Council Advanced (grant 233348 MODELIST). Support was also provided by the Swiss National Science Foundation and grant 51RT 0_126008 for the Research and Technology Development (RTD) project InfectX in the frame of SystemsX.ch, the Swiss Initiative for Systems Biology. P.C. is a Howard Hughes Medical Institute Senior International Research Scholar. A.K. is a recipient of a scholarship from the Pasteur-Paris University International Doctoral Program/Institut Carnot Maladies Infectieuses.
We thank Alice Lebreton and Juan-José Quereda-Torres for assistance on quantitative reverse transcription-PCR (qRT-PCR) experiments and Isabelle Tardieux for providing the plasmid expressing a SPIRE1-GFP fusion protein.
Footnotes
Citation Kühbacher A, Emmenlauer M, Rämo P, Kafai N, Dehio C, Cossart P, Pizarro-Cerdá J. 2015. Genome-wide siRNA screen identifies complementary signaling pathways involved in Listeria infection and reveals different actin nucleation mechanisms during Listeria cell invasion and actin comet tail formation. mBio 6(3):e00598-15. doi:10.1128/mBio.00598-15.
REFERENCES
- 1.Cossart P. 2011. Illuminating the landscape of host-pathogen interactions with the bacterium Listeria monocytogenes. Proc Natl Acad Sci USA 108:19484–19491. doi: 10.1073/pnas.1112371108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pizarro-Cerdá J, Kühbacher A, Cossart P. 2012. Entry of Listeria monocytogenes in mammalian epithelial cells: an updated view. Cold Spring Harb Perspect Med 2:a010009. doi: 10.1101/cshperspect.a010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gouin E, Welch MD, Cossart P. 2005. Actin-based motility of intracellular pathogens. Curr Opin Microbiol 8:35–45. doi: 10.1016/j.mib.2004.12.013. [DOI] [PubMed] [Google Scholar]
- 4.Mengaud J, Ohayon H, Gounon P, Mege RM, Cossart P. 1996. E-cadherin is the receptor for internalin, a surface protein required for entry of L. monocytogenes into epithelial cells. Cell 84:923–932. doi: 10.1016/S0092-8674(00)81070-3. [DOI] [PubMed] [Google Scholar]
- 5.Shen Y, Naujokas M, Park M, Ireton K. 2000. InIB-dependent internalization of Listeria is mediated by the Met receptor tyrosine kinase. Cell 103:501–510. doi: 10.1016/S0092-8674(00)00141-0. [DOI] [PubMed] [Google Scholar]
- 6.Bonazzi M, Veiga E, Pizarro-Cerdá J, Cossart P. 2008. Successive post-translational modifications of E-cadherin are required for InlA-mediated internalization of Listeria monocytogenes. Cell Microbiol 10:2208–2222. doi: 10.1111/j.1462-5822.2008.01200.x. [DOI] [PubMed] [Google Scholar]
- 7.Bonazzi M, Vasudevan L, Mallet A, Sachse M, Sartori A, Prevost MC, Roberts A, Taner SB, Wilbur JD, Brodsky FM, Cossart P. 2011. Clathrin phosphorylation is required for actin recruitment at sites of bacterial adhesion and internalization. J Cell Biol 195:525–536. doi: 10.1083/jcb.201105152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Veiga E, Cossart P. 2005. Listeria hijacks the clathrin-dependent endocytic machinery to invade mammalian cells. Nat Cell Biol 7:894–900. doi: 10.1038/ncb1292. [DOI] [PubMed] [Google Scholar]
- 9.Veiga E, Guttman JA, Bonazzi M, Boucrot E, Toledo-Arana A, Lin AE, Enninga J, Pizarro-Cerdá J, Finlay BB, Kirchhausen T, Cossart P. 2007. Invasive and adherent bacterial pathogens co-opt host clathrin for infection. Cell Host Microbe 2:340–351. doi: 10.1016/j.chom.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bierne H, Miki H, Innocenti M, Scita G, Gertler FB, Takenawa T, Cossart P. 2005. WASP-related proteins, Abi1 and Ena/Vasp are required for Listeria invasion induced by the Met receptor. J Cell Sci 118:1537–1547. doi: 10.1242/jcs.02285. [DOI] [PubMed] [Google Scholar]
- 11.Sousa S, Cabanes D, Bougnères L, Lecuit M, Sansonetti P, Tran-Van-Nhieu G, Cossart P. 2007. Src, cortactin and Arp2/3 complex are required for E-cadherin-mediated internalization of Listeria into cells. Cell Microbiol 9:2629–2643. doi: 10.1111/j.1462-5822.2007.00984.x. [DOI] [PubMed] [Google Scholar]
- 12.Welch MD, Iwamatsu A, Mitchison TJ. 1997. Actin polymerization is induced by Arp2/3 protein complex at the surface of Listeria monocytogenes. Nature 385:265–269. doi: 10.1038/385265a0. [DOI] [PubMed] [Google Scholar]
- 13.Goley ED, Welch MD. 2006. The Arp2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol 7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- 14.Svitkina TM, Borisy GG. 1999. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol 145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winkelman JD, Bilancia CG, Peifer M, Kovar DR. 2014. Ena/Vasp enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundle F-actin networks with fascin. Proc Natl Acad Sci U S A 111:4121–4126. doi: 10.1073/pnas.1322093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Campellone KG, Welch MD. 2010. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol 11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kühbacher A, Gouin E, Mercer J, Emmenlauer M, Dehio C, Cossart P, Pizarro-Cerdá J. 2013. Imaging InlC secretion to investigate cellular infection by the bacterial pathogen Listeria monocytogenes. J Vis Exp 79:e51043. doi: 10.3791/51043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ireton K, Payrastre B, Cossart P. 1999. The Listeria monocytogenes protein InlB is an agonist of mammalian phosphoinositide 3-kinase. J Biol Chem 274:17025–17032. doi: 10.1074/jbc.274.24.17025. [DOI] [PubMed] [Google Scholar]
- 19.Bierne H, Gouin E, Roux P, Caroni P, Yin HL, Cossart P. 2001. A role for cofilin and LIM kinase in Listeria-induced phagocytosis. J Cell Biol 155:101–112. doi: 10.1083/jcb.200104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kühbacher A, Dambournet D, Echard A, Cossart P, Pizarro-Cerdá J. 2012. Phosphatidylinositol 5-phosphatase oculocerebrorenal syndrome of Lowe protein (OCRL) controls actin dynamics during early steps of Listeria monocytogenes infection. J Biol Chem 287:13128–13136. doi: 10.1074/jbc.M111.315788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dambournet D, Machicoane M, Chesneau L, Sachse M, Rocancourt M, El Marjou A, Formstecher E, Salomon R, Goud B, Echard A. 2011. Rab35 GTPase and OCRL phosphatase remodel lipids and F-actin for successful cytokinesis. Nat Cell Biol 13:981–988. doi: 10.1038/ncb2279. [DOI] [PubMed] [Google Scholar]
- 22.Komada M, Hatsuzawa K, Shibamoto S, Ito F, Nakayama K, Kitamura N. 1993. Proteolytic processing of the hepatocyte growth factor/scatter factor receptor by furin. FEBS Lett 328:25–29. doi: 10.1016/0014-5793(93)80958-W. [DOI] [PubMed] [Google Scholar]
- 23.Alvarez-Dominguez C, Madrazo-Toca F, Fernandez-Prieto L, Vandekerckhove J, Pareja E, Tobes R, Gomez-Lopez MT, Del Cerro-Vadillo E, Fresno M, Leyva-Cobián F, Carrasco-Marín E. 2008. Characterization of a Listeria monocytogenes protein interfering with Rab5a. Traffic 9:325–337. doi: 10.1111/j.1600-0854.2007.00683.x. [DOI] [PubMed] [Google Scholar]
- 24.Alvarez-Dominguez C, Stahl PD. 1999. Increased expression of Rab5a correlates directly with accelerated maturation of Listeria monocytogenes phagosomes. J Biol Chem 274:11459–11462. doi: 10.1074/jbc.274.17.11459. [DOI] [PubMed] [Google Scholar]
- 25.Segawa T, Hazeki K, Nigorikawa K, Morioka S, Guo Y, Takasuga S, Asanuma K, Hazeki O. 2014. Inpp5e increases the Rab5 association and phosphatidylinositol 3-phosphate accumulation at the phagosome through an interaction with Rab20. Biochem J 464:365–375. doi: 10.1042/BJ20140916. [DOI] [PubMed] [Google Scholar]
- 26.Tan R, Wang W, Wang S, Wang Z, Sun L, He W, Fan R, Zhou Y, Xu X, Hong W, Wang T. 2013. Small GTPase Rab40c associates with lipid droplets and modulates the biogenesis of lipid droplets. PLoS One 8:e63213. doi: 10.1371/journal.pone.0063213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin S, Driessen K, Nixon SJ, Zerial M, Parton RG. 2005. Regulated localization of Rab18 to lipid droplets: effects of lipolytic stimulation and inhibition of lipid droplet catabolism. J Biol Chem 280:42325–42335. doi: 10.1074/jbc.M506651200. [DOI] [PubMed] [Google Scholar]
- 28.Greenwald EC, Saucerman JJ. 2011. Bigger, better, faster: principles and models of AKAP anchoring protein signaling. J Cardiovasc Pharmacol 58:462–469. doi: 10.1097/FJC.0b013e31822001e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wei N, Deng XW. 2003. The COP9 signalosome. Annu Rev Cell Dev Biol 19:261–286. doi: 10.1146/annurev.cellbio.19.111301.112449. [DOI] [PubMed] [Google Scholar]
- 30.von Arnim AG, Schwechheimer C. 2006. Life is degrading—thanks to some zomes. Mol Cell 23:621–629. doi: 10.1016/j.molcel.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 31.Kerkhoff E, Simpson JC, Leberfinger CB, Otto IM, Doerks T, Bork P, Rapp UR, Raabe T, Pepperkok R. 2001. The Spir actin organizers are involved in vesicle transport processes. Curr Biol 11:1963–1968. doi: 10.1016/S0960-9822(01)00602-9. [DOI] [PubMed] [Google Scholar]
- 32.Gasman S, Kalaidzidis Y, Zerial M. 2003. RhoD regulates endosome dynamics through diaphanous-related formin and Src tyrosine kinase. Nat Cell Biol 5:195–204. doi: 10.1038/ncb935. [DOI] [PubMed] [Google Scholar]
- 33.Lagal V, Abrivard M, Gonzalez V, Perazzi A, Popli S, Verzeroli E, Tardieux I. 2014. Spire-1 contributes to the invadosome and its associated invasive properties. J Cell Sci 127:328–340. doi: 10.1242/jcs.130161. [DOI] [PubMed] [Google Scholar]
- 34.Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. 2005. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science 309:1248–1251. [DOI] [PubMed] [Google Scholar]
- 35.Cheng LW, Viala JP, Stuurman N, Wiedemann U, Vale RD, Portnoy DA. 2005. Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proc Natl Acad Sci U S A 102:13646–13651. doi: 10.1073/pnas.0506461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gournier H, Goley ED, Niederstrasser H, Trinh T, Welch MD. 2001. Reconstitution of human Arp2/3 complex reveals critical roles of individual subunits in complex structure and activity. Mol Cell 8:1041–1052. doi: 10.1016/S1097-2765(01)00393-8. [DOI] [PubMed] [Google Scholar]
- 37.Chorev DS, Moscovitz O, Geiger B, Sharon M. 2014. Regulation of focal adhesion formation by a vinculin-Arp2/3 hybrid complex. Nat Commun 5:3758. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- 38.DeMali KA, Barlow CA, Burridge K. 2002. Recruitment of the Arp2/3 complex to vinculin: coupling membrane protrusion to matrix adhesion. J Cell Biol 159:881–891. doi: 10.1083/jcb.200206043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gouin E, Adib-Conquy M, Balestrino D, Nahori MA, Villiers V, Colland F, Dramsi S, Dussurget O, Cossart P. 2010. The Listeria monocytogenes InlC protein interferes with innate immune responses by targeting the IκB kinase subunit IKKα. Proc Natl Acad Sci USA 107:17333–17338.doi: 10.1073/pnas.1007765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kühbacher A, Cossart P, Pizarro-Cerdá J. 2014. Internalization assays for Listeria monocytogenes. Methods Mol Biol 1157:167–178. doi: 10.1007/978-1-4939-0703-8_14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods. Download
Western blots to validate the inactivation of the different subunits of the Arp2/3 complex. (A) Cell lysates were loaded on polyacrylamide-SDS gels, transferred to polyvinylidene difluoride (PVDF) membranes, and blotted using antibodies against the different subunits of the Arp2/3 complex; the TopoIIα protein (170 kDa) or histone H3 (15 kDa) was used as a loading control. (B) Relative expression levels of all 7 Arp2/3 complex subunits after knockdown of individual subunits. mRNA concentrations 72 h post-siRNA transfection were quantified by quantitative reverse transcription-PCR (qRT-PCR). One representative experiment of a biological triplicate is shown. Bars represent the means of two technical replicates. Download
Validation experiments for alternative actin nucleators. (A) Infection Z score of cells transfected with siRNAs targeting different actin nucleators and infected with Listeria for 5 h. Results for 3 different silencer select siRNAs for each target are shown. Bars represent the means of duplicate screens. Results of primary screens for SPIRE2, FMNL3, FMN2, and DIAPH2 could be confirmed with 2 or 3 additional siRNAs. (B) Examples of images from genome-wide screen depicting cellular actin phenotypes of cells transfected with nontargeting control siRNA or siRNA targeting SPIRE1 or SPIRE2 and infected with Listeria. Bar, 50 µm. Download
Colocalization of Arp2/3 subunits with Listeria at bacterial entry sites. Cells infected with Listeria for 10 min were stained for Arp2, Arp3, p34, or p41B. Bar, 5 µm. Download
Genome-wide infection Z scores upon gene inactivation by siRNA pools.
Validation screen Z scores upon gene inactivation by single siRNAs. DP, Dharmacon pooled primary screening results (average from 2 screens); AU SiSel, Ambion unpooled silencer select siRNA; AU sel, Ambion unpooled silencer siRNA; SI esiRNA, Sigma esiRNA pools. Negative infection Z scores are highlighted with green, and positive infection Z scores are highlighted with red. Genes are ranked according to the median infection Z score.