ABSTRACT
Clostridium clariflavum is an anaerobic, cellulosome-forming thermophile, containing in its genome genes for a large number of cellulosomal enzyme and a complex scaffoldin system. Previously, we described the major cohesin-dockerin interactions of the cellulosome components, and on this basis a model of diverse cellulosome assemblies was derived. In this work, we cultivated C. clariflavum on cellobiose-, microcrystalline cellulose-, and switchgrass-containing media and isolated cell-free cellulosome complexes from each culture. Gel filtration separation of the cellulosome samples revealed two major fractions, which were analyzed by label-free liquid chromatography-tandem mass spectrometry (LC-MS/MS) in order to identify the key players of the cellulosome assemblies therein. From the 13 scaffoldins present in the C. clariflavum genome, 11 were identified, and a variety of enzymes from different glycoside hydrolase and carbohydrate esterase families were identified, including the glycoside hydrolase families GH48, GH9, GH5, GH30, GH11, and GH10. The expression level of the cellulosomal proteins varied as a function of the carbon source used for cultivation of the bacterium. In addition, the catalytic activity of each cellulosome was examined on different cellulosic substrates, xylan and switchgrass. The cellulosome isolated from the microcrystalline cellulose-containing medium was the most active of all the cellulosomes that were tested. The results suggest that the expression of the cellulosome proteins is regulated by the type of substrate in the growth medium. Moreover, both cell-free and cell-bound cellulosome complexes were produced which together may degrade the substrate in a synergistic manner. These observations are compatible with our previously published model of cellulosome assemblies in this bacterium.
IMPORTANCE
Because the reservoir of unsustainable fossil fuels, such as coal, petroleum, and natural gas, is overutilized and continues to contribute to environmental pollution and CO2 emission, the need for appropriate alternative energy sources becomes more crucial. Bioethanol produced from dedicated crops and cellulosic waste can provide a partial answer, yet a cost-effective production method must be developed. The cellulosome system of the anaerobic thermophile C. clariflavum comprises a large number of cellulolytic and hemicellulolytic enzymes, which self-assemble in a number of different cellulosome architectures for enhanced cellulosic biomass degradation. Identification of the major cellulosomal components expressed during growth of the bacterium and their influence on its catalytic capabilities provide insight into the performance of the remarkable cellulosome of this intriguing bacterium. The findings, together with the thermophilic characteristics of the proteins, render C. clariflavum of great interest for future use in industrial cellulose conversion processes.
INTRODUCTION
Our modern lifestyle relies heavily on technology, which is continuously and rapidly developing. The energetic cost for this kind of lifestyle is high and still growing and will eventually lead to the exhaustion of fossil fuels, thus raising the demand for alternative, sustainable energy sources. Biofuels are a proper alternative, since they are produced from monosaccharides, derived from degradation of plant-derived cellulosic biomass, that can be fermented to different useful chemicals, such as bioethanol (1–3). The rate-limiting step in the process is the deconstruction of biomass from complex polysaccharides to sugar monomers, in order to make the energy sources accessible for fermenting microorganisms (1). The main component of the plant cell wall is cellulose, a crystalline-structured, recalcitrant polysaccharide, which is the most abundant source of carbon on earth (4–6). An efficient way to degrade cellulose will promote the production and utilization of biofuels worldwide.
A wide range of microorganisms are capable of plant cell wall and cellulose degradation (6). Cellulose degradation can be carried out by individually secreted enzymes or by highly efficient multienzymatic machinery called the cellulosome. The cellulosome is produced by anaerobic bacteria and is either attached to or secreted from the cell, in order to degrade the cellulose and hemicellulose into soluble sugars for subsequent assimilation by the various microbes in the immediate environment (6–9). The cellulosome is constructed from structural subunits called scaffoldins, which are responsible for the intricate and multimodular structure of the cellulosome. The scaffoldins bear modules called cohesins that interact with their modular counterparts, called dockerins, usually conjugated to enzymatic subunits or other scaffoldins (7–11). In addition, the scaffoldin may contain a carbohydrate-binding module (CBM) that guides the complex and its intricate set of component enzymes to the surface of the cellulosic substrate (12–16). The cohesin-dockerin interactions define the high-molecular-weight cellulosome complex, whose enzymes are arranged in a contiguous fashion, thus promoting their heightened synergistic action on cellulosic and hemicellulosic substrates (11). The cellulosome was first discovered in the thermophilic anaerobe Clostridium thermocellum, which serves as the prototype of cellulosome-forming bacteria (17–19).
Clostridium clariflavum is a recently discovered Gram-positive, thermophilic, cellulolytic bacterium, isolated from an anaerobic sewage sludge (20, 21). Its genome was sequenced and annotated, and an abundance of genes encoding putative polysaccharide-degrading enzymes and cellulosomal proteins were found (22). The thermophilic characteristics of C. clariflavum are of great interest to the biomass degradation research community, since it is one of the few thermophilic cellulosome-producing bacterial species known today. In addition, C. clariflavum is capable of utilizing cellulose, cellobiose, and natural substrates, such as switchgrass. Recently, a new strain of C. clariflavum was isolated which has the ability to utilize xylan and xylose as sole sources of carbon (23). The bacterium also demonstrates high similarity to C. thermocellum, the most studied cellulosomal bacterium, which exhibits the most efficient cellulosome machinery to date (11). Moreover, the cellulosome system of C. clariflavum closely resembles the cellulosome system of Acetivibrio cellulolyticus, a mesophilic anaerobe which possesses a particularly elaborate cellulosome, containing 16 scaffoldin subunits (24, 25).
In our previous work, we further characterized the cellulosomal system of C. clariflavum by bioinformatic analysis that resulted in determination of the scaffoldin architectures and evaluation of the cellulosomal enzyme contents (26). From this analysis, 13 scaffoldins and 75 type I dockerin-containing proteins were found, 48 of which are dockerin-containing enzymes. In addition, we examined biochemically the interaction patterns among a variety of recombinant cohesin and dockerin modules, and a model of the possible cellulosome assemblies was established. Based on the biochemical data, we suggested that the major cellulosome assembly is based on a primary scaffoldin, ScaA, which contains eight type I cohesin modules (interacting with enzymes bearing type I dockerins), a family 3 carbohydrate binding module (CBM3) and a type II dockerin module conjugated to an X module. This noncatalytic ScaA subunit is incorporated into the type II cohesins of the adaptor scaffoldin ScaB via a characteristic type II cohesin-dockerin interaction, which, in turn, is incorporated into unique type I cohesin modules of the anchoring scaffoldin ScaC, via a divergent type I cohesin-dockerin interaction. Several other cellulosomes may also be assembled (26), thereby comprising additional types of cell-bound and cell-free cellulosomes. To date, detailed characterization of the catalytic activity of the C. clariflavum cellulosome and its breadth of plant cell wall-degrading enzyme activity has not been performed.
Recently, different proteomics-based methods have been applied to cellulosome research. Cellulosomes of several bacterial species were investigated, the proteins constituting the complexes were characterized, and differences in expression of cellulosomal proteins were identified, based on the substrate upon which the bacteria were grown. The cellulosomes of Clostridium cellulolyticum (27, 28), Clostridium cellulovorans (29, 30), and C. thermocellum were widely studied by proteomics methods (34, 66). The cellulosomal systems of these bacterial species are based on a primary scaffoldin, containing various numbers of type I cohesin modules (usually 8 or 9), which function as dockerin-binding modules for incorporation of the parent protein (notably enzyme) into the complex (6, 12, 35, 36). In the C. thermocellum cellulosome, there are also several cell-anchoring scaffoldins, which attach the primary scaffoldin to the cell wall. However, the C. clariflavum cellulosome system was shown to be much more elaborate than that of C. thermocellum and appears to be more similar to the cellulosome system of A. cellulolyticus, both of which are believed to form cellulosome complexes that are diverse in both architecture and enzyme contents.
In the present study, we cultivated C. clariflavum on three different sources of carbon: cellobiose, microcrystalline cellulose, and acid-pretreated switchgrass. The cellulosomes were isolated into two major fractions, the composition of each was characterized by label-free liquid chromatography-tandem mass spectrometry (LC-MS/MS), and their catalytic activities on different substrates were examined. The findings revealed the identity of the expressed cellulosomal proteins that are necessary for polysaccharide degradation as a function of the carbon source in the cultivation medium and provided insight into the possible types of cellulosome assemblies in vivo.
RESULTS
Fermentation and isolation of cellulosomes.
In order to characterize the secreted cellulosome complexes of C. clariflavum, cells were grown on three different carbon sources: cellobiose (CB), microcrystalline cellulose (MCC), and acid-pretreated switchgrass (SG) in three biological replicates. Growth was assessed by monitoring NaOH consumption during fermentation for pH stabilization, since the pH decreases during the fermentation. In addition, CMCase activity of the supernatant fluids was examined during fermentation, in order to assess the growth phases and increasing levels of cellulase production in the medium (data not shown). Cells grown on cellobiose and pretreated switchgrass reached late stationary phase after 30 h, while cells grown on microcrystalline cellulose reached late stationary phase after 50 h. Samples of spent growth media were collected, and high-molecular-mass proteins (500-kDa cutoff) were concentrated. Concentrated proteins were loaded onto a Sephacryl S-500 gel filtration column with a separation range of 40 kDa to 20 MDa, in order to separate high-molecular-weight cellulosome complexes. We observed two major peaks for each of the samples, with estimated molecular masses of ~1,200 and ~400 kDa, respectively (gel filtration elution profiles are found in Fig. S1 in the supplemental material). SDS-PAGE was employed to distinguish between the different complexes (Fig. 1): fraction I (the first peak eluted from the column) and the smaller fraction II from the cellobiose medium, designated CB I and CB II; MCC I and MCC II from the microcrystalline cellulose medium; and SG I and SG II from the switchgrass medium. Cellulolytic and xylanolytic activities of selected fractions were measured on CMC and xylan substrates (data not shown), and the active fractions of each complex were pooled.
FIG 1 .
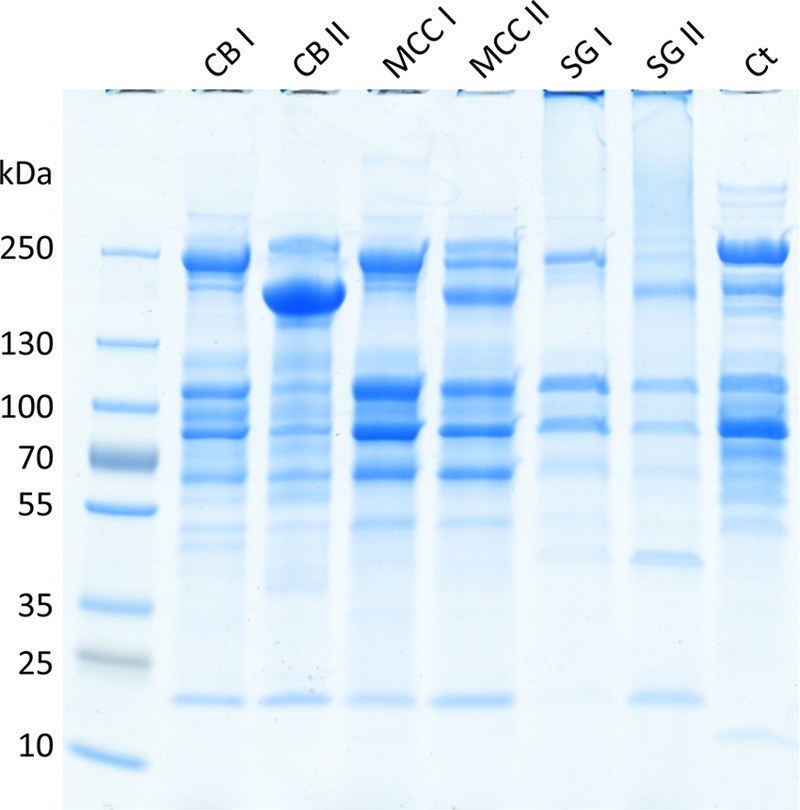
SDS-PAGE analysis of the cellulosome fractions. The C. clariflavum cellulosomes produced in cellobiose, microcrystalline cellulose, and switchgrass growth media and the purified C. thermocellum cellulosome were concentrated and isolated by gel filtration chromatography. For each growth medium, two peaks of cellulosome proteins were identified (I and II). The resultant cellulosomes (20 µg) were subjected to 4-to-15% gradient SDS-PAGE. CB, cellobiose; MCC, microcrystalline cellulose; SG, switchgrass; Ct, C. thermocellum.
Mass spectrometry analysis.
Cellulosome fractions (two fractions isolated from each of the three growth conditions and three biological replicates for each fraction) were analyzed by label-free LC-MS/MS. The MS data were compared with the C. clariflavum protein data from the UniProt and NCBI-nr databases for protein identification. Proteins were annotated more accurately using the CAZY database (http://www.cazy.org) (37), and mean values for each of the three biological replicates were calculated. Proteins were quantified by two values: (i) intensity-based absolute quantification (iBAQ), calculated by the sum of peak intensities of all the peptides belonging to a specific protein divided by the number of theoretically observable peptides (38), allowing comparison of the quantities of the proteins within the same sample, and (ii) the protein abundance value, label-free quantification (LFQ), computed by a label-free algorithm that takes the maximum number of identified peptides between any two samples and compares the intensity of these peptides for determining the ratio between them, after which protein abundance is calculated using the mean values of all peptide ratios of the given protein (39, 40). This value allows comparison of protein amounts between samples. The iBAQ values for the identified cellulosomal proteins were normalized relative to the ScaA protein in each sample, in order to demonstrate the abundance of each protein corresponding to the primary cellulosomal scaffoldin, which binds the highest number of enzymes, thus comprising the main component of the cellulosome (26, 31–33). LFQ values were normalized according to the ScaA values in fraction I, isolated from the cellobiose growth condition, to facilitate comparison of the protein quantities among the different complexes. Both values are presented in Table 1 (the complete data are found in Table S1 in the supplemental material, supported by statistical analysis of the data in Table S2). In general, the data reveal that the largest amount of cellulosomal proteins was produced by cells grown on microcrystalline cellulose, while cells grown on switchgrass produced the smallest amount (Table 1, LFQ values). It is thus clear from the table that the LFQ values for both scaffoldins and enzyme components are consistently higher in the cellulose-grown cells than in cells grown on cellobiose or, especially, switchgrass.
TABLE 1 .
Label-free LC-MS/MS analysis of cellulosome fractionsa
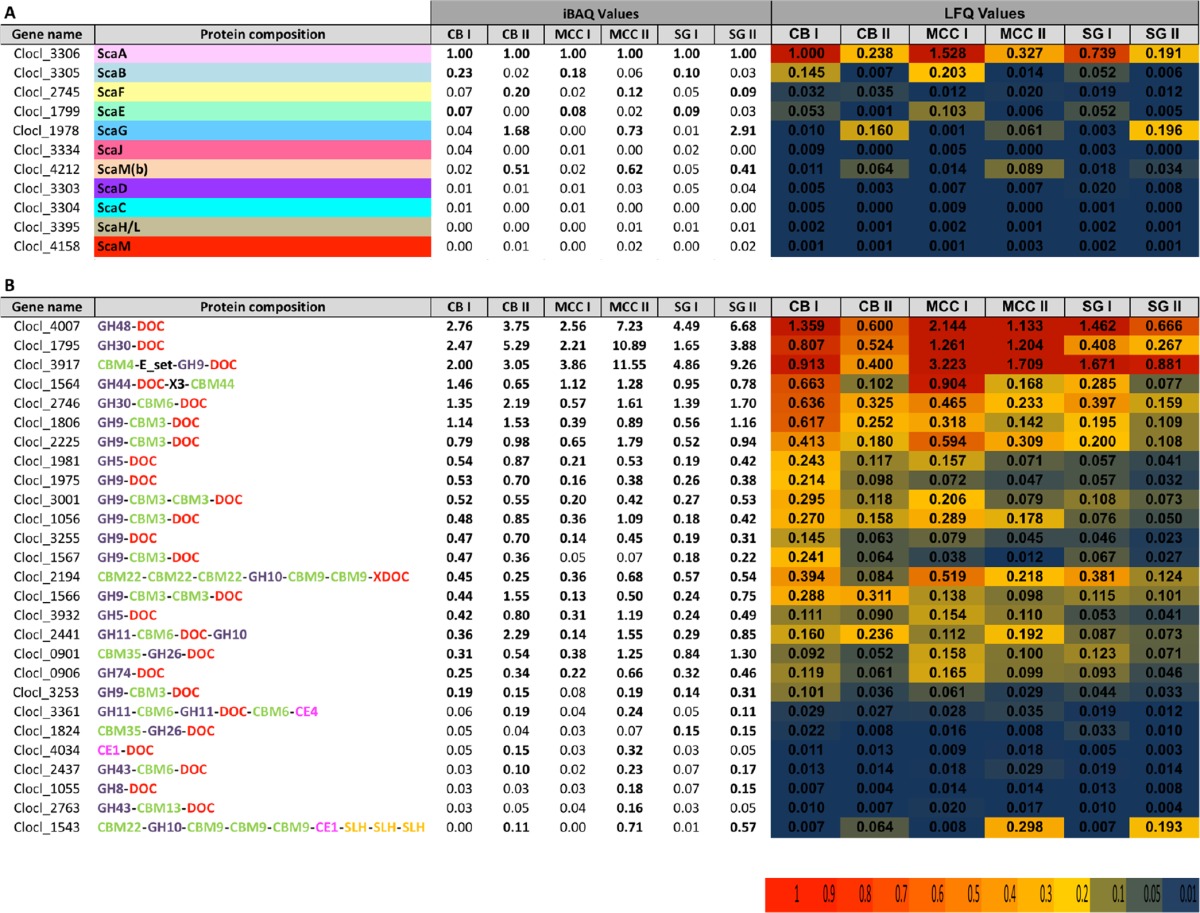
The quantity and abundance of each protein in the six cellulosome fractions were assessed by iBAQ and LFQ, which are described in detail in Results. All iBAQ values were normalized by dividing each value by that for ScaA in each sample. The LFQ values were normalized by dividing all values by the ScaA value of CB I. The top portion shows scaffoldins that were identified in each fraction, and the bottom portion shows the most abundant dockerin-containing proteins that were identified in each fraction. CB, cellobiose; MCC, microcrystalline cellulose; SG, switchgrass; DOC, dockerin; GH, glycoside hydrolase; CBM, carbohydrate-binding module; CE, carbohydrate esterase; SLH, S-layer homology domain. Significant iBAQ values (≥0.10) are shown in bold. The range of LFQ values is color coded according to the scale shown at the bottom.
Scaffoldins.
The genome of C. clariflavum bears 13 open-reading frames predicted to encode putative cellulosomal structural proteins (scaffoldins) containing type I and type II cohesin and dockerin modules, carbohydrate-binding modules (CBMs), and cell wall-anchoring elements. Proteomics analysis revealed the presence of 11 scaffoldins: ScaA, ScaB, ScaC, ScaD, ScaE, ScaF, ScaG, ScaH/L, ScaJ, ScaM and ScaM(b); scaffoldins ScaM(a) and ScaO, both of which lack a signal peptide sequence (26), were not found in the different fractions. The expressed scaffoldins differ between fraction I and fraction II in their distribution and abundance for all three growth conditions. However, the ratios of the scaffoldin subunits in CB I, MCC I, and SG I are very similar, as are those in CB II, MCC II, and SG II. ScaA is the primary scaffoldin of the cellulosome, with eight type I cohesin modules and a CBM3 module, and it was found to be the most abundant scaffoldin in CB I, MCC I, and SG I. It is also very abundant in CB II, MCC II, and SG II. Quantitative proteomics revealed that ScaA is produced at the highest level by cells grown on microcrystalline cellulose and is most abundant in the MCC I complex. The lowest level of ScaA was detected in the switchgrass growth medium (Table 1, top).
ScaB is an adaptor scaffoldin of the cellulosome, bearing five type II cohesin modules which bind the type II ScaA dockerin, and its unique type I dockerin binds to the type I cohesins of the anchoring ScaC as described earlier (26). ScaB was detected in fractions CB I and MCC I (~5-fold less compared to ScaA). This finding is compatible with the proposed C. clariflavum cellulosome assembly, in which five ScaA subunits would integrate 40 enzymes into a single ScaB subunit by virtue of its five type II cohesins. In SG I fraction, however, ScaB is found in 10-fold-smaller amounts than ScaA, which may be explained by the complexity of the switchgrass composition, which may cause a lag in expression of cellulosomal genes and protein production. This observation was shown before for C. thermocellum by Raman et al. (31). However, the representation of ScaB in fractions CB II, MCC II, and SG II was significantly lower, presumably reflecting the characteristic lower-molecular-mass fraction (Table 1, top).
Interestingly, the quantity of ScaC, a putative cell-anchoring scaffoldin, was substantially lower than expected considering the four type I cohesin modules which would interact specifically with the type I dockerin of ScaB in all the analyzed complexes (less than 1% compared to ScaA). This finding is surprising, since the cellulosomes of C. thermocellum are reported to be released from the bacterium cell wall to the medium during the late stationary phase (17, 31, 41). Levels of other anchoring scaffoldins, including ScaD, ScaF, and ScaJ, which bear an S-layer homology (SLH) module, were also much lower than expected. ScaJ is the only other scaffoldin in the C. clariflavum cellulosome system that contains the same unique type I cohesin as ScaC, with the capacity to interact with the ScaB dockerin. It is thus probable that most of the anchoring scaffoldins are not released from the cell wall to the medium and remain attached to the cell, while ScaA and ScaB are detached from them and are present in the growth medium. Scaffoldins ScaM and ScaH/L were also at low levels for all the complexes (<2% versus ScaA).
ScaE is a cell-free scaffoldin comprising seven type II cohesins interacting with the type II dockerin of ScaA. ScaE was detected in relatively large amounts in CB I, MCC I, and SG I (7, 8, and 9% of the amount of ScaA in the respective fractions) and in lower quantities in CB II, MCC II, and SG II (<3% compared to ScaA) (Table 1, top, iBAQ values). Quantitative proteomics showed that the MCC I complex contained the largest quantity of ScaE (Table 1, top, LFQ values). The interaction between the cohesin modules of ScaE and the dockerin module of ScaA creates a large complex that can contain up to 7 ScaA subunits and 56 enzymes, which explains the presence of ScaE in the high-molecular-mass fraction I for all growth conditions.
ScaG is a scaffoldin that bears a single type I cohesin and a cell surface-binding module (CSBM). The CSBM is different from the SLH module of the other anchoring scaffoldins mentioned above. ScaG was identified as the most abundant scaffoldin in fractions CB II and SG II, in quantities 1.68-fold and 2.91-fold higher than that of ScaA, respectively. ScaG was also found at high levels in MCC II, 73% of the quantity of ScaA (Table 1, top, iBAQ values). This is the only scaffoldin subunit found to be more abundant than ScaA in any of the fractions, which suggests that ScaG has a significant role in cellulosome activity, especially when the bacterium grows on cellobiose or natural substrates like switchgrass. ScaG demonstrates high similarity to the C. thermocellum scaffoldin OlpC, also an abundant protein on the bacterial cell surface (31, 42). Pinheiro et al. (42) suggested that OlpC may bind cellulosomal enzymes temporarily before these enzymes are recruited to the higher-structured cellulosome complexes. A similar role was suggested for the membrane-bound, cohesin-containing OrfXp in C. cellulolyticum (36). This assumption is reasonable, due to the high diversity and great number of C. clariflavum cellulosomal enzymes revealed from proteomic analysis, all recruited to degrade complicated polysaccharide substrates. Incorporation of these essential enzymes into the cellulosomes could thus be assisted by ScaG.
Another relatively abundant scaffoldin present in CB II, MCC II, and SG II is ScaM(b) (51%, 62%, and 41% in comparison to ScaA, respectively). ScaM(b) holds a CBM2 module and six type I cohesin modules. Its presence in the isolated cellulosome fractions indicates the existence of a cell-free cellulosome with a CBM module, which would presumably target the hexavalent complex to the substrate.
Cellulosomal enzymes.
Proteomics analysis of the isolated fractions revealed a variety of cellulosomal (dockerin-bearing) enzymes which participate in cellulose and biomass degradation. Intriguingly, the molar amount of dockerin-containing enzymes was much higher than the amount of available cohesin modules in the scaffoldins in the low-molecular-weight fractions, indicating the presence of free uncomplexed enzymes. The most abundant protein in the C. clariflavum CB I cellulosome fraction and one of the most abundant proteins in the other fractions is the putative exoglucanase GH48 (Clocl_4007), which shows high similarity to the C. thermocellum exoglucanase Cel48S (22). Cel48S was shown early on (43–46) to be the most abundant enzyme in the cellulosome of C. thermocellum, and this was confirmed in several proteomics studies (31, 32). A family 48 enzyme is a major component of all known cellulosomal systems and also comprises a major enzyme in some noncellulosomal (free and bifunctional enzyme) bacterial systems. In C. clariflavum, GH48 is found in all of the analyzed complexes, in greater quantities for CB I, MCC I, and SG I than for CB II, MCC II, and SG II (Table 1, bottom, LFQ values). Compared to ScaA, the quantity of GH48 in each complex is 2- to 7-fold higher, and the GH48/ScaA ratio is even greater in fractions CB II, MCC II, and SG II. The highest ratio was demonstrated for MCC II and SG II (7.23 and 6.68, respectively).
GH30, a putative xylanase (Clocl_1795), is the second most prominent enzyme in the CB I fraction and is one of the most abundant enzymes in all of the fractions. This enzyme shows high similarity to GH30 from C. josui (a moderately thermophilic bacterium) and C. cellulolyticum (a mesophilic bacterium). Interestingly, the C. clariflavum GH30s were notably distinct from those of A. cellulolyticus, despite the remarkable similarities of their cellulosomal systems. The GH30/ScaA ratios vary between cellulosomes, and the highest ratio was found for the MCC II fractions, where the level of GH30 is almost 11-fold higher than that of ScaA. In general, this enzyme is more abundant than ScaA in the second peak for each growth condition.
Another highly abundant putative xylanase is an enzyme bearing a GH30 module, a CBM6 module, and a dockerin module (Clocl_2746), also in a higher GH30/ScaA ratio in CB II, MCC II, and SG II than in CB I, MCC I, and SG I. Interestingly, this particular GH30 enzyme did not appear to be one of the abundant cellulosomal enzymes in the C. thermocellum cellulosome (31, 32), while in our study, the GH30 enzymes are highly abundant in all the cellulosome fractions examined. In order to determine the activity of the GH30-family enzymes, we cloned and expressed them recombinantly and tested their activity on phosphoric acid-swollen cellulose (PASC) and xylan. Both Clocl_1795 and Clocl_2746 demonstrated xylanolytic activity (data not shown). This may suggest that the GH30-family xylanases have a pivotal role in the C. clariflavum cellulosome as hemicellulases.
Family 9 enzymes (GH9s) are very common in cellulosomes. The C. clariflavum genome contains 13 GH9-encoding genes, and we detected the 12 cellulosomal enzymes in significant quantities in the proteomics analysis. A GH9 enzyme bearing a CBM4 module (Clocl_3917) had the highest expression level of all GH9s, and it shares great similarity with Cel9K from C. thermocellum, characterized as an exoglucanase (47). The Cel9K-like enzyme was the most abundant enzyme produced when cells were grown on microcrystalline cellulose and switchgrass. The lowest level was found in cellulosomes produced on cellobiose growth medium, but even here, Cel9K was still the third most highly expressed enzyme in fractions CB I and CB II (Table 1, bottom, iBAQ values). From a quantitative standpoint, Cel9K-like expression levels were decreased significantly in cellobiose-containing growth media in comparison to those in media containing cellulose and switchgrass (Table 1, bottom, LFQ values). This expression pattern of the Cel9K-like enzyme was also observed for C. thermocellum Cel9K, which is expressed in lower levels when cells are grown on cellobiose than on complex substrates (31, 32). Other putative GH9 endoglucanases that were produced are Cel9Q-like (Clocl_2225), two GH9s similar to Cel9M from C. cellulolyticum (Clocl_1975 and Clocl_3255), Cel9U-like enzymes (Clocl_3001 and Clocl_1864), which are also highly similar to Cel9B from A. cellulolyticus, a Cel9F-like enzyme (Clocl_1056), a GH9 highly similar to Cthe_2761 (Clocl_1567), a Cel9V-like enzyme (Clocl_1566), a Cel9N-like enzyme which is also similar to a Cel9 enzyme from A. cellulolyticus (Clocl_3253), a Cel9J-like enzyme (Clocl_3029), and a Cel9T-like enzyme (Clocl_1806). In general, the distribution of GH9 endoglucanases is higher in CB II, MCC II, and SG II than in CB I, MCC I, and SG I. GH9 endoglucanases were expressed at higher levels when cells were grown on cellobiose and microcrystalline cellulose than on switchgrass. Growth on cellobiose showed a slight increase in the production of GH9 endoglucanases, findings which contradict previous reports which indicated a decrease in the levels of GH9 endoglucanases when C. thermocellum was grown on cellobiose compared to cultivation on insoluble cellulose-containing carbon sources (31, 32, 48).
Intriguingly, C. clariflavum, A. cellulolyticus, and C. thermocellum produce a small number of noncellulosomal enzymes in the free form. One of the most interesting ones is the bifunctional cellulase, Clocl_3038, which exhibits a GH48-GH9-CMB3-CBM3 modular arrangement (22). However, the levels of this enzyme were virtually undetectable in the CB-grown cells and high-molecular-weight fractions of the MCC- and SG-grown cells. The levels of this protein in the low-molecular-weight fractions of the latter cells were detectable, but at levels about a thousandfold lower than those of the dockerin-bearing cellulosomal GH48. A homologous gene encoding a cell-free, noncellulosomal enzyme with an identical modular architecture and very similar overall sequence is present in the genome of A. cellulolyticus. A similar bifunctional enzyme is not present in C. thermocellum; however, two separate CBM-containing noncellulosomal enzymes, Cel48Y and Cel9I, together appear to complement Clocl_3038 and its A. cellulolyticus homolog.
The C. thermocellum enzyme CelJ is a particularly intricate, bifunctional, cellulosomal enzyme that contains two catalytic modules (GH9 and GH44) and two CBM modules (CBM44 and CBM30). In C. thermocellum, the GH9 module was shown previously to have CMCase activity, while the GH44 module was active on CMC, xylan, and xyloglucan. The CBM30 module is a cellulose- and xyloglucan-binding module, and the CBM44 module binds to cellulose, xylan, and xyloglucan (49–52). Interestingly, the status of the GH9/44 pair between C. clariflavum and C. thermocellum is the reverse of that described above for the GH48/9 pair. Thus, the genome of C. clariflavum contains two discrete polypeptide chains containing the same catalytic modules and CBMs as Cel9/44J, with high sequence similarity to the cellulosomal C. thermocellum CelJ: a CBM30-GH9-Doc protein (Clocl_3029) and a GH44-Doc-X3-CBM44 protein (Clocl_1564). Proteomics analysis revealed that the GH44 enzyme is produced in high quantity, and is one of the most abundant proteins in all the different fractions. However, the CelJ-like GH9 enzyme is found in very low quantities compared to ScaA (<2%). This may suggest that the GH44 enzymatic unit and the CBM44 conjugated to it play an important role in the cellulose and hemicellulose degradation, while the GH9 homolog of CelJ plays a relatively minor role. Our results correspond with the findings of Raman et al., which show that for C. thermocellum CelJ was one of the highly abundant enzymes in its cellulosome (31).
The presence of GH5-family enzymes in the C. clariflavum genome is relatively low (4 genes) in comparison to C. thermocellum (10 genes). All four GH5 enzymes were detected in cellulosomes and are similar in their sequences to C. thermocellum GH5 enzymes. According to their sequences, the enzymes are predicted to act as endoglucanases. The two highly abundant GH5s are similar to Cel5B (Clocl_1981) and Cel5G (Clocl_3932) endoglucanases. The two other enzymes are produced at significantly lower levels and resemble Cel5E (Clocl_0350) and Cel5L (Clocl_1122). Compared to the quantity of ScaA in each complex, the Cel5B-like and Cel5G-like enzymes are both more abundant in complexes CB II, MCC II, and SG II. Quantitatively, expression of both Cel5B and Cel5G expression is elevated in the cellulose- and cellobiose-containing growth media compared to expression in the switchgrass-containing growth medium.
Endoglucanase Cel8A (GH8 enzyme) was reported to be one of the most highly expressed enzymes in the C. thermocellum cellulosome, thus playing a pivotal role in the biomass degradation (31, 32). Surprisingly, its GH8 homolog in the C. clariflavum genome (Clocl_1055) was not expressed in significant levels compared to other endoglucanases produced during the growth of C. clariflavum on different carbon sources.
In addition to the cellulases that were discovered in the cellulosome complexes, various hemicellulases were also expressed and detected in the complexes. Putative β-mannanases belonging to the GH26 enzymes (Clocl_0901, Clocl_1824, and Clocl_3132) were produced, and one of them (Clocl_0901) was expressed in increased levels compared to the other two.
Expression of this enzyme was elevated in cells grown on cellulose and switchgrass (Table 1, bottom, LFQ values), and its levels were 1.25- and 1.30-fold higher than ScaA in fractions MCC II and SG II, respectively (Table 1, iBAQ values). A putative chitinase, GH18, was also identified, but in relatively small amounts. In addition to the high abundance of GH30 xylanases, additional predicted xylanases were identified, such as those from GH11 and GH10, which may also contain CE1, CE2, CE4, CE6, and CE12 catalytic modules, as well as GH74 and GH43-family enzymes. The bifunctional enzyme GH11-CBM6-Doc-GH10 (Clocl_2441) occurred at the most increased levels in the cellobiose growth medium, particularly in the CB II fraction. In general, the enzyme was detected at high levels in the CB II, MCC II, and SG II fractions. An unusually large GH10-family enzyme (Clocl_2194), CBM22-CBM22-CBM22-GH10-CBM9-CBM9-XDoc, was identified in large quantities when cells were grown on all three substrates. A homolog of this enzyme exists in the genome of A. cellulolyticus but not in that of C. thermocellum. This enzyme is exceptional due to its X-Doc module, which is common in scaffoldins but a unique component in enzymes. Its presence in this enzyme would imply that it would bind to scaffoldins which bear type II cohesins, such as ScaB, ScaD, ScaE, and ScaF. Another putative high-molecular-weight hemicellulase is an enzyme containing a CBM22 module, a GH10 and a CE1 module, three CBM9 modules, and an SLH (S-layer homology) module at the C terminus of the protein. This protein is highly abundant in fractions MCC II and SG II, although it is predicted to be a cell-bound enzyme. This may indicate a higher level of expression of the protein on cellulose- and switchgrass-containing growth media, in comparison to expression in cells grown on cellobiose, resulting in release of the enzyme from the cell wall in larger amounts. Interestingly, similar genes for SLH-containing GH10 enzymes exist in the genomes of A. cellulolyticus and C. thermocellum.
Catalytic activity of the cellulosomes.
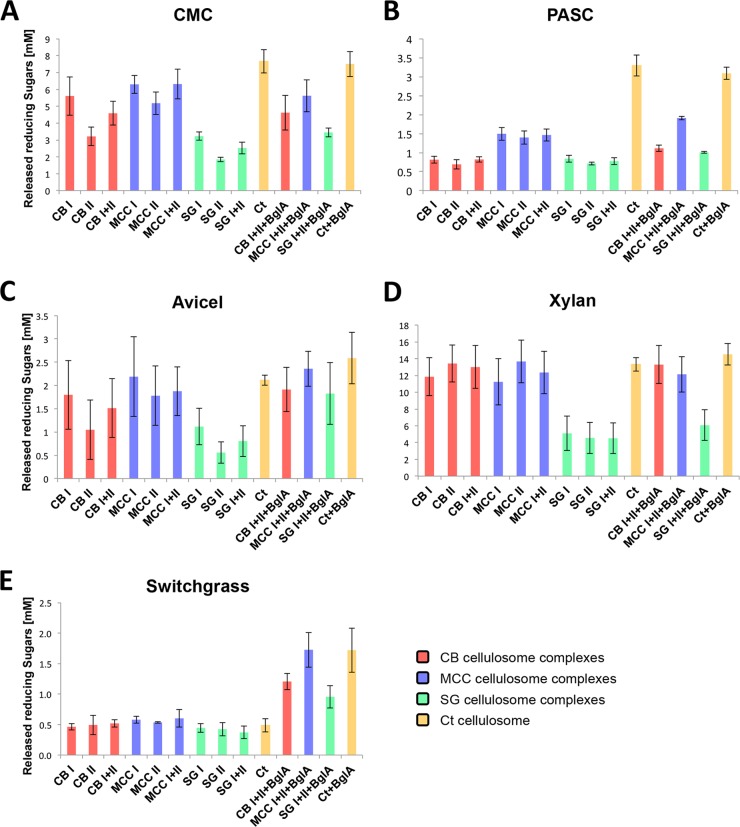
The isolated cellulosome complexes were examined for their catalytic activity on five substrates: carboxymethyl cellulose (CMC), phosphoric acid-swollen cellulose (PASC), microcrystalline cellulose (Avicel), beechwood xylan, and pretreated switchgrass. The catalytic activity was tested for each fraction separately and also, for each substrate, a combination of fractions I and II (CB I and CB II together, MCC I and MCC II together, and SG I and SG II together) in a 1:1 ratio (totaling 25 µg/ml) (Fig. 2). The combination of the two fractions was examined in order to test for synergy between them. For calculating synergism, the sum of the activity of each individual fraction was compared to the value of released reducing sugars received for the combination of the fractions multiplied by 2 (since the two complexes were combined together at the same loads used in individual assays). Furthermore, the recombinant β-glucosidase BglA, originated from C. thermocellum, was added to the combined complexes from each carbon source medium in order to reduce inhibition of activity of the cellulosomal enzymes by cellobiose, which is the main product of cellulose degradation. It was shown previously that addition of β-glucosidase results in enhancement of the catalytic activity (53–55). The purified C. thermocellum cellulosome was used as a reference (11).
FIG 2 .
Comparative hydrolysis of cellulosic substrates, beechwood xylan, and switchgrass by cellulosome preparations. The two complexes isolated from each growth medium were tested for their catalytic activity on (A) CMC (carboxymethyl cellulose), (B) PASC (phosphoric acid-swollen cellulose), (C) microcrystalline cellulose (Avicel), (D) beechwood xylan, and (E) switchgrass. Each complex was tested individually or combined with the second complex from the same growth medium. To avoid product inhibition, a recombinant β-glucosidase (BglA) from C. thermocellum was added to the combined complexes. The C. thermocellum cellulosome was also tested for catalytic activity with or without the addition of BglA, to serve as a reference for the catalytic activity of the C. clariflavum cellulosomes. CB, cellobiose; MCC, microcrystalline cellulose; SG, switchgrass; Ct, C. thermocellum.
The isolated cellulosome fractions displayed varied efficiencies on the different substrates, depending both on the carbon source on which the bacterium was cultivated and the substrate that was degraded. The MCC-grown cellulosome was found to be the most active cellulosome on all substrates. Degradation of cellulosic substrates was evaluated by using three substrates—soluble CMC for determining endoglucanase activity, PASC (amorphous cellulose), and microcrystalline cellulose. In general, complexes CB I, MCC I, and SG I demonstrated higher activity on all the cellulosic substrates than CB II, MCC II, and SG II, and the MCC-grown cellulosome fractions demonstrated the highest cellulolytic activity.
For degradation of CMC (Fig. 2A), the combination of MCC I and MCC II and the individual MCC I activities were highest of all C. clariflavum cellulosome fractions. SG cellulosome fractions exhibited the lowest levels of endoglucanase activity, and the C. thermocellum cellulosome was the most active complex on CMC. Addition of BglA to the reactions did not influence CMC degradation levels. The combination of MCC I and MCC II together displayed a very minor synergistic effect compared to the activity of each complex alone.
For PASC (Fig. 2B), the degradation activity by C. thermocellum cellulosomes was significantly higher than that of the C. clariflavum cellulosome fractions, and the MCC-derived cellulosome fractions were the most active among the C. clariflavum cellulosome fractions. The enzyme BglA enhanced the cellulolytic activity of the combined C. clariflavum cellulosome fractions, especially the activity of the combined MCC I and MCC II.
For microcrystalline cellulose (Fig. 2C), the MCC I fraction exhibited the highest levels of catalytic activity, which was equivalent to that of the purified C. thermocellum cellulosome. Addition of BglA to the reactions slightly enhanced the cellulose degradation for all of the combined cellulosome fractions tested and for the C. thermocellum cellulosome.
Xylan hydrolysis, however, resulted in a different picture: fractions CB II and MCC II demonstrated higher degradation activities than CB I and MCC I (Fig. 2D), which may suggest that fractions II contain higher concentrations of xylanases than fractions I. The CB, MCC, and C. thermocellum cellulosomes showed very similar degradation levels, whereas the SG cellulosomes were significantly less active. This finding is surprising, since we had expected to see elevated degradation levels of hemicellulose by the cellulosomes isolated from cells grown on a natural substrate.
C. clariflavum grown on switchgrass exhibited the lowest degradation levels for all cellulosome fractions tested (Fig. 2E). When BglA was added to the three combined cellulosome pairs, the release of reducing sugars was increased. The highest level of degradation was achieved by the combined activity of MCC-derived cellulosomes and the purified C. thermocellum cellulosome, with addition of BglA to both. The SG cellulosome again failed to reach the degradation levels of the MCC-grown C. clariflavum and the purified C. thermocellum cellulosomes and showed the lowest levels of activity.
DISCUSSION
C. clariflavum is a cellulolytic bacterium possessing in its genome an extensive set of cellulosomal genes with great potential to orchestrate a highly efficient plant cell wall-degrading machinery. In our recent study (26), we expressed representative recombinant cohesin and dockerin modules and demonstrated specific interaction patterns among them, verifying that the different components of the cellulosome interact to form several different intricate complexes. In this work, we show by proteomics analysis the natural organization of the cellulosomes and their enzyme composition, depending on the carbon source upon which the bacterium was cultivated.
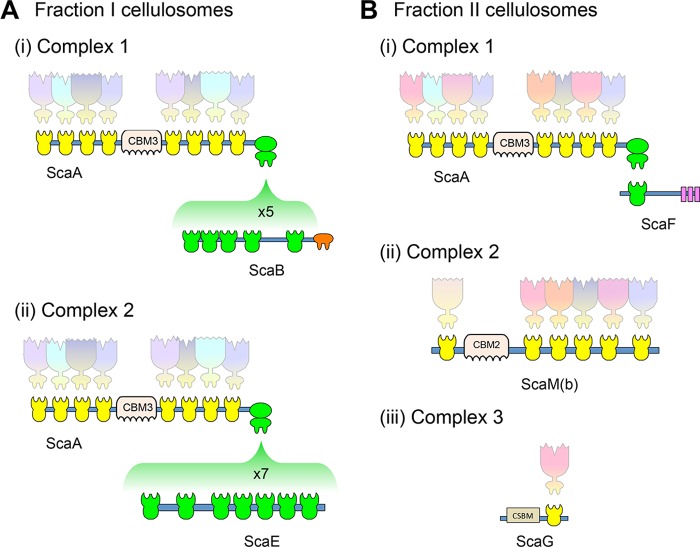
The separation of the cellulosome complexes from the spent growth medium by gel filtration resulted in an elution profile of two broad peaks, containing two discrete groups of cellulosome fractions. Mass spectrometry analysis showed that each peak contains cellulosomal proteins that differ in content and ratios of their component parts. The results allowed us to deduce the existence of several different types of cellulosome assemblies, shown in Fig. 3. The higher-molecular-mass first peak is mainly composed of two main complexes: (i) the ScaB adaptor scaffoldin, which carries five molecules of the primary scaffoldin ScaA, each of which bears eight enzymatic subunits, yielding a complex of 40 enzymatic subunits (assuming full occupancy), and (ii) the cell-free ScaE, which carries seven ScaA molecules, resulting in complexes containing up to 56 enzymes (Fig. 3A). Considering the abundance of ScaB and ScaE in fraction I, the ScaB-based complex is the most abundant. In contrast, the second peak includes a different set of complex compositions of lower molecular mass: (i) the monovalent cell wall ScaF interacts with a single ScaA molecule and its eight enzymes; (ii) ScaM(b), a scaffoldin bearing six type I cohesins, interacts with six type I dockerins conjugated to a variety of enzymes; and (iii) the single type I cohesin module of ScaG interacts with a type I dockerin of a cellulosomal enzyme (Fig. 3B). The variety of expressed cellulosomes discovered in this work reveal two complementary mechanisms of action employed by the bacterium in order to degrade the plant cell wall efficiently: cell-bound cellulosomes and cell-free cellulosomes, both cooperating to achieve effective deconstruction of plant cell wall polysaccharides. The lack of the anchoring scaffoldins ScaC and ScaD in the two fractions was surprising, since we would have expected to find them at levels that would fit the amounts of ScaA and ScaB according to the molar number of ScaC and ScaD cohesins versus that of the ScaA and ScaB dockerins. This may suggest that (unlike ScaF and ScaG) ScaC and ScaD remain bound to the cell wall by a particularly strong interaction and are not easily released to the medium.
FIG 3 .
Major cellulosomes produced by C. clariflavum. Gel filtration separation of the spent growth medium resulted in two fractions which contain five major types of cellulosome complexes. (A) Two very large cellulosomes are present in fraction I. (i) Complex 1 is composed of five subunits of the octavalent ScaA, 40 enzymes, and the pentavalent ScaB. (ii) Complex II contains 7 subunits of ScaA, 56 enzymes, and the heptavalent ScaE. (B) Fraction II contains three cellulosomes. (i) Complex 1 contains a single ScaA subunit, 8 enzymes, and a monovalent ScaF. (ii) Complex 2 contains the hexavalent ScaM(b) subunit and 6 enzymes. (iii) Complex 3 contains ScaG, which binds a single enzyme via its type I cohesin module.
The mass spectrometry data revealed that the cellulosomal enzymes are found in the two fractions in distributions and ratios different from those of ScaA. In all of the cellulosome types, fraction II contained enzyme complexes in significantly higher enzyme/ScaA ratios. This corresponds to the relatively high expression levels of ScaM(b), which contains 6 additional type I cohesin modules that would be expected to bind enzyme-borne dockerin modules, thus increasing the number of enzymes in fraction II. The hemicellulase/ScaA ratios were notably higher in these complexes than in those of fraction I, in particular the GH30-Doc/ScaA ratio (10.89 [Table 1, iBAQ values]). These data fit the results for the xylanase activity in fraction II, which was consistently higher than that in fraction I or the combination of fractions I and II in cellulosomes derived from the CB- and MCC-grown cells.
The MCC cellulosome demonstrated the highest cellulolytic activity of all three cellulosomes isolated. The reason for this observation may be the level of expression of ScaA, which was most elevated when cells were grown on microcrystalline cellulose. ScaA is the primary scaffoldin of the cellulosome, containing eight type I cohesins and a CBM3 module that targets the cellulosome for more efficient degradation. Previous studies have shown that mutations of CipA (the ScaA equivalent in C. thermocellum) caused a significant decrease in cellulolytic activity (56), indicating that CipA is the key player in the C. thermocellum cellulosome, and its expression is crucial to cellulolytic activity. The elevated levels of cellulosomal ScaA derived from the MCC-grown culture may explain the difference we observed for the cellulolytic activity for the MCC cellulosome.
Our study provides in vivo insight into the intricate, elaborate cellulosome system of C. clariflavum, in which the number of assembled cellulosomal complexes is revealed, and sheds light on the key players in biomass degradation by this bacterium. Although the abundance of each protein in the cellulosome was determined, the potency of catalytic activity of each enzyme and the individual contribution of each enzyme remain a mystery. Characterization of individual cellulosomal enzymes is thus required for better understanding of the plant cell wall deconstruction mechanisms. To date, the cellulosomal system of C. clariflavum is the most complicated one that has been explored by proteomics approaches so far. Several bacterial cellulosomal systems were characterized earlier by proteomics, such as C. cellulolyticum, C. cellulovorans, and C. thermocellum, but all these systems were based on a single major scaffoldin subunit and its appended enzymes (27–33), whereas the C. clariflavum system is much more complicated. This system is highly similar to the mesophilic A. cellulolyticus system, in terms of architecture and sequence, which was investigated previously only by in vitro assays (25, 58–60). Further investigation of the A. cellulolyticus cellulosomal components participating in cellulose degradation by proteomics or transcriptomics analyses may be of interest in order to compare the cellulosome assemblies and the cellulosomal catalytic activity between the thermophilic and mesophilic cellulosomes.
In this work, we display the complexity of the C. clariflavum cellulosomes and show the potency of the biomass and cellulose degradation activity, which approaches the remarkable degradation capabilities of the C. thermocellum system. The thermostability of the enzymes and structural proteins is critical for industrial applications, and the multiplicity of its cellulosome system substantially broadens our current resources of thermophilic enzymes. These characteristics render C. clariflavum an appropriate candidate for development of highly efficient biomass degradation systems, by utilizing its cellulosomal components for deconstruction of plant cell wall polysaccharides into their monomers, which can be used as substrates for production of biofuels and other alternative energy sources.
MATERIALS AND METHODS
Bacterial strains.
C. clariflavum DSM 19732 and C. thermocellum DSM 1313 were purchased from the Leibniz Institute DSMZ (German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany).
Fermentation.
C. clariflavum was grown on GS-2 medium containing (per liter) 0.5 g K2HPO4, 0.5 g MgCl2·6H2O, 0.5 g KH2PO4, 1.3 g NH4SO4, 10.5 g morpholinopropane sulfonic acid (MOPS), 5 g yeast extract, 2 mg of resazurin (pH 7.2) (17), and mineral solution as described previously (20). The bacterium was grown at 55°C and pH 7.2 on the following carbon sources (added to the medium) in 10-liter glass fermentors: 8 g/liter cellobiose (MP Biomedicals, Illkirch, France), 2 g/liter microcrystalline cellulose (Avicel; Sigma-Aldrich), and 2 g/liter switchgrass [Nott Farms (Ont.) Ltd. and Switch Energy Corp., Ontario, Canada]. The chemical composition of the initial switchgrass was 37% cellulose, 28% hemicellulose, 18% lignin, and 17% miscellaneous materials (waxes, proteins, salts, etc.). The switchgrass was subjected to acid pretreatment as follows: 100 g of the biomass was placed in a beaker, and 700 ml of 5% (wt/vol) sulfuric acid was added. The beaker containing the biomass and reagent was heated to boiling with stirring and treated for 1 h. The pretreated biomass was washed using a vacuum glass filter until the effluent reached neutral pH and compressed to a final solids content of 20 to 30% (wt/wt).
For each carbon source, two additional cultures were cultivated in 1-liter fermentors for biological repeats. C. thermocellum was grown at 60°C and pH 7.2 on GS-2 medium containing 2 g/liter microcrystalline cellulose as carbon source. Fermentors were inoculated with 5% (vol/vol) of a culture that was grown on cellobiose and were sparged during the whole process with nitrogen. During the fermentation process, samples were taken for evaluation of the growth phase, and the CMCase activity of each sample was measured on carboxymethyl cellulose (CMC; VWR International Ltd., Poole, England) by the dinitrosalicylic acid (DNS) assay (61) in order to determine the commencement of the stationary phase. Cells were harvested and removed, and the supernatant fluids were filtered with sterile plastic filter units (Thermo, Fisher Scientific, Waltham, MA, USA) and stored for further analysis.
Isolation of high-molecular-weight complexes.
The growth medium supernatant fluids were concentrated 100-fold using a peristaltic pump (MasterFlex l/S pump system, Easy-Load II pump head [Cole-Parmer, Vernon Hills, IL]) and a 500-kDa-cutoff Pellicon 2 membrane (Millipore, Darmstadt, Germany). The concentration of proteins in the concentrated and unconcentrated fractions was measured by a Bradford assay, and a CMCase activity assay (see below) was performed to confirm the presence of cellulolytic complexes. The concentrated fraction showed high CMCase activity, and high-molecular-weight complexes were isolated by gel filtration chromatography using a preparative chromatography system for laboratory-scale protein purification (Äkta start; GE Healthcare, Uppsala, Sweden). The samples were loaded onto a HiPrep 26/60 Sephacryl S-500 HR gel filtration column (GE Healthcare) with Tris-buffered saline as the running buffer (TBS; 137 mM NaCl, 2.7 mM KCl, 25 mM Tris-HCl [pH 7.4]) for separation of the different complexes. Fractions of each peak were pooled and concentrated with a Vivaspin concentrator (100-kDa cutoff; Sartorius Stedim Biotech GmbH, Göttingen, Germany). Protein concentrations were measured by the bicinchoninic acid (BCA) assay. The isolated high-molecular-weight complexes were boiled for 5 min at 100°C and subjected to 4-to-15% gradient SDS-PAGE (Bio-Rad, Hercules, CA). The results revealed a wide range of proteins for each complex, indicative of cellulosome complexes.
Label-free LC-MS/MS analysis. (i) Proteolysis.
Protein pellets were dissolved in a solution containing 8 M urea in 100 mM ammonium bicarbonate. The proteins were reduced with 2.8 mM dithiothreitol (DTT) at 60°C for 30 min, modified with 8.8 mM iodoacetamide in 100 mM ammonium bicarbonate (room temperature for 30 min in the dark), and digested in 2 M urea in 25 mM ammonium bicarbonate with modified trypsin (Promega, Fitchburg, WI) at a 1:50 enzyme-to-substrate ratio.
(ii) Mass spectrometry analysis.
The peptides were desalted using C18 stage tips (homemade from membrane; 3M Maplewood, MN, USA) dried and resuspended in 0.1% formic acid. The peptides were resolved by reverse-phase chromatography on 0.075- by 180-mm fused silica capillaries (J&W) packed with Reprosil reversed-phase material (Dr. Maisch GmbH, Germany). The peptides were eluted with a linear 120-min gradient of 5 to 28% acetonitrile with 0.1% formic acid, a 5-min gradient of 28% to 95%, and a 15-min step at 95% acetonitrile with 0.1% formic acid in water at flow rates of 150 nl/min. Mass spectrometry was performed with a Q Exactive Plus mass spectrometer (Thermo) in positive mode using a repetitively full MS scan followed by collision-induced dissociation (CID) of the 10 most dominant ions selected from the first MS scan. The mass spectrometry data were analyzed using MaxQuant v1.5 software (62, 63) versus the C. clariflavum section of the UniProt database (http://www.uniprot.org/) or of the NCBI-nr database (http://www.ncbi.nlm.nih.gov) with a 1% false discovery rate (FDR). Statistical analysis was done using Perseus v1.4.1.3 (64).
β-Glucosidase expression and purification.
A pET28a cassette containing the His-tagged wild-type (WT) bglA gene from the C. thermocellum genome was obtained from Designer Energy, Ltd., Rehovot, Israel (53). The plasmid was transformed into Escherichia coli BL21 (DE3), and the cells were grown in 1 liter of Luria-Bertani broth (LB), containing 50 µg/ml kanamycin and 2 mM CaCl2, for 2 h at 37°C to an A600 of ~0.8. Isopropyl-1-thio-β-d-galactoside (IPTG; 0.2 mM) (Fermentas UAB, Vilnius, Lithuania) was added to induce protein expression, and cells were incubated for an additional 3 h at 37°C. Cells were harvested (5,000 rpm, 15 min) and sonicated, and the sonicate was heated for 30 min at 50°C and centrifuged (15,000 rpm, 30 min). The protein was purified on nickel-nitrilotriacetic acid (Ni-NTA) beads in a batch purification system as described previously (65). The purity of the protein was verified by subjecting it to SDS-PAGE (10%). Protein concentration was determined by absorbance at 280 nm and evaluated based on the extinction coefficient calculated using the Expasy ProtParam tool (http://web.expasy.org/protparam/). The protein was stored in 50% (vol/vol) glycerol at −20°C.
Activity assays.
Activity assays were performed in a total volume of 200 µl, containing 50 mM acetate buffer (pH 5.5), 12 mM CaCl2, 2 mM EDTA, and 25 µg/ml of each cellulosome complex, or combined complexes. When β-glucosidase (BglA) was added, its concentration was 8.33 µg/ml. Endoglucanase activity was assayed using a final concentration of 1% CMC, for 1 h at 60°C. Degradation of phosphoric acid-swollen cellulose (PASC) was assayed at a final concentration of 5.6 mg/ml PASC, for 3 h at 60°C. Avicel degradation was assayed at a final concentration of 7.5 mg/ml Avicel, for 24 h at 60°C. Xylan degradation was assayed at a final concentration of 1% beechwood xylan (Sigma Aldrich, Rehovot, Israel), for 1 h at 60°C. Switchgrass degradation was assayed at a final concentration of 5 mg/ml acid-pretreated switchgrass, for 24 h at 60°C. For assays containing the BglA enzyme, citrate buffer (pH 6) was used instead of acetate buffer. All assays were performed with C. clariflavum cellulosome complexes containing 50 mM acetate buffer (pH 5.5) and were incubated at 60°C, according to the predetermined optimal conditions for C. clariflavum cellulosome activity. Assays performed for the C. thermocellum cellulosome were incubated at 70°C (the optimal temperature for C. thermocellum cellulosome activity).
Experiments were performed three times in duplicate samples in 1.5-ml tubes. Tubes were incubated with shaking, and the reaction was terminated by flash-cooling the tubes on ice. The tubes were centrifuged (14,000 rpm, 5 min), and 100 µl was transferred into 150 µl dinitrosalicylic acid (DNS) solution. The tubes were boiled for 10 min at 100°C, and absorbance was measured at 540 nm in a plate reader. The enzymatic activity was evaluated by measuring the concentration (millimolar) of released reducing sugars of each reaction and by using a glucose standard curve for determining the amount of reducing sugars.
SUPPLEMENTAL MATERIAL
Gel filtration elution profile of cellulosome fractions. The supernatant fractions of the cellobiose, microcrystalline cellulose, and switchgrass growth media were concentrated and loaded onto a HiPrep 26/60 Sephacryl S-500 HR gel filtration column. The first eluted peak did not contain any detectable proteins and is therefore called F0. The second peak (including any higher molecular-mass shoulder) is fraction I, and the third is lower-molecular-mass fraction II, as described in Results. The fraction I from cellobiose-grown cells eluted as two connected peaks (described as A and B), but SDS-PAGE analysis showed that the protein contents were indistinguishable, so A and B were pooled and considered one fraction. Download
Complete LC-MS/MS data. The full list of proteins identified in the various fractions of the 18 samples analyzed by LC-MS/MS is given. The proteins are annotated according to the CAZy and BLAST databases.
Statistical analysis of LC-MS/MS data. In order to determine significant differences or similarities among the different samples, statistical analysis of the LC-MS/MS was performed. t tests and analysis of variance (ANOVA) were performed for the following sets of samples: t test of complex (fraction) I versus complex (fraction) II for each of the growth media (blue, proteins which are more abundant in fraction II; green, proteins which are more abundant in fraction I [P < 0.0015; difference >│1│]); t test of CB (cellobiose) versus MCC (microcrystalline cellulose) samples (orange, proteins which are more abundant in MCC; purple, proteins which are more abundant in CB I [P < 0.0024; difference > │1.3│]); t test of MCC versus SG (switchgrass) (orange, proteins which are more abundant in MCC; pink, proteins which are more abundant in SG [P < 0.00058; difference > │1.8│]); t test of CB versus SG (orange, proteins which are more abundant in MCC; pink, proteins which are more abundant in SG; purple, proteins which are more abundant in CB [P < 0.0006; difference > │1.4│]); t test of complex (fraction) I versus complex (fraction) II from all the samples for proteins with at least two peptides (blue, proteins which are more abundant in fraction II; green, proteins which are more abundant in fraction I [P < 0.0015; difference >│1│]); t test between the CB fractions (P < 0.0074; difference >│1│); t test between the MCC fractions (P < 0.008; difference > │1.15│); t test between the SG fractions (P < 0.0.0115; difference > │1.38│); ANOVA test between CB, MCC, and SG; two-way ANOVA between the group of all growth media and the subgroup pf the complexes (fractions); blue letters, proteins which are more abundant in fraction II for one (or two) of the growth media; green letters, proteins which are more abundant in fraction I for one (or two) of the growth media.
ACKNOWLEDGMENTS
We are grateful to Tamar Ziv and Keren Bendalak (Smoler Protein Research Center, Technion, Israel) for performing the LC-MS/MS and their assistance with the data analysis and to Ghil Jona and Amnon Naziri (Bacteriology Unit, Weizmann Institute of Science) for their conscientious help with the cultivation of the bacteria.
We appreciate the support of the European Union, Area NMP.2013.1.1-2: Self-assembly of naturally occurring nanosystems: CellulosomePlus Project no. 604530 and an ERA-IB Consortium (EIB.12.022), acronym FiberFuel. In addition, E.A.B. is grateful for a grant from the F. Warren Hellman Grant for Alternative Energy Research in Israel in support of alternative energy research in Israel administered by the Israel Strategic Alternative Energy Foundation (I-SAEF). This research was also supported by a grant (no. 1349) to E.A.B. also from the Israel Science Foundation (ISF) and a grant (no. 24/11) issued to R.L. by The Sidney E. Frank Foundation also through the ISF. Additional support was obtained from the establishment of an Israeli Center of Research Excellence (I-CORE Center no. 152/11) managed by the ISF, Jerusalem, Israel, by the Weizmann Institute of Science Alternative Energy Research Initiative (AERI), and by the Helmsley Foundation. E.A.B. is the incumbent of the Maynard I. and Elaine Wishner Chair of Bio-organic Chemistry.
Footnotes
Citation Artzi L, Morag E, Barak Y, Lamed R, Bayer EA. 2015. Clostridium clariflavum: key cellulosome players are revealed by proteomic analysis. mBio 6(3):e00411-15. doi:10.1128/mBio.00411-15.
REFERENCES
- 1.Demain AL, Newcomb M, Wu JH. 2005. Cellulase, clostridia, and ethanol. Microbiol Mol Biol Rev 69:124–154. doi: 10.1128/MMBR.69.1.124-154.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Himmel ME, Ding S-Y, Johnson DK, Adney WS, Nimlos MR, Brady JW, Foust TD. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 3.Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ, Hallett JP, Leak DJ, Liotta CL, Mielenz JR, Murphy R, Templer R, Tschaplinski T. 2006. The path forward for biofuels and biomaterials. Science 311:484–489. doi: 10.1126/science.1114736. [DOI] [PubMed] [Google Scholar]
- 4.O’Sullivan AC. 1997. Cellulose: the structure slowly unravels. Cellulose 4:173–207. doi: 10.1023/A:1018431705579. [DOI] [Google Scholar]
- 5.Gilbert HJ. 2010. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol 153:444–455. doi: 10.1104/pp.110.156646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bayer EA, Shoham Y, Lamed R. 2013. Lignocellulose-decomposing bacteria and their enzyme systems, p 216–266. In Rosenberg E. (ed), The prokaryotes, 4th ed. Springer Verlag, Berlin, Germany. [Google Scholar]
- 7.Bayer EA, Morag E, Lamed R. 1994. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol 12:379–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 8.Bayer EA, Chanzy H, Lamed R, Shoham Y. 1998. Cellulose, cellulases and cellulosomes. Curr Opin Struct Biol 8:548–557. doi: 10.1016/S0959-440X(98)80143-7. [DOI] [PubMed] [Google Scholar]
- 9.Bayer EA, Belaich J-P, Shoham Y, Lamed R. 2004. The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Annu Rev Microbiol 58:521–554. doi: 10.1146/annurev.micro.57.030502.091022. [DOI] [PubMed] [Google Scholar]
- 10.Doi RH, Kosugi A. 2004. Cellulosomes: plant-cell-wall-degrading enzyme complexes. Nat Rev Microbiol 2:541–551. doi: 10.1038/nrmicro925. [DOI] [PubMed] [Google Scholar]
- 11.Fontes CM, Gilbert HJ. 2010. Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu Rev Biochem 79:655–681. doi: 10.1146/annurev-biochem-091208-085603. [DOI] [PubMed] [Google Scholar]
- 12.Shoseyov O, Takagi M, Goldstein MA, Doi RH. 1992. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc Natl Acad Sci U S A 89:3483–3487. doi: 10.1073/pnas.89.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poole DM, Morag E, Lamed R, Bayer EA, Hazlewood GP, Gilbert HJ. 1992. Identification of the cellulose-binding domain of the cellulosome subunit S1 from Clostridium thermocellum YS. FEMS Microbiol Lett 78:181–186. doi: 10.1016/0378-1097(92)90022-G. [DOI] [PubMed] [Google Scholar]
- 14.Morag E, Lapidot A, Govorko D, Lamed R, Wilchek M, Bayer EA, Shoham Y. 1995. Expression, purification, and characterization of the cellulose-binding domain of the scaffoldin subunit from the cellulosome of Clostridium thermocellum. Appl Environ Microbiol 61:1980–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shoseyov O, Shani Z, Levy I. 2006. Carbohydrate binding modules: biochemical properties and novel applications. Microbiol Mol Biol Rev 70:283–295. doi: 10.1128/MMBR.00028-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382:769–781. doi: 10.1042/BJ20040892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bayer EA, Kenig R, Lamed R. 1983. Adherence of Clostridium thermocellum to cellulose. J Bacteriol 156:818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lamed R, Setter E, Bayer EA. 1983. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J Bacteriol 156:828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lamed R, Setter E, Kenig R, Bayer EA. 1983. The cellulosome—a discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulose-binding and various cellulolytic activities. Biotechnol Bioeng Symp 13:163–181. [Google Scholar]
- 20.Shiratori H, Sasaya K, Ohiwa H, Ikeno H, Ayame S, Kataoka N, Miya A, Beppu T, Ueda K. 2009. Clostridium clariflavum sp. nov. and Clostridium caenicola sp. nov., moderately thermophilic, cellulose-/cellobiose-digesting bacteria isolated from methanogenic sludge. Int J Syst Evol Microbiol 59:1764–1770. doi: 10.1099/ijs.0.003483-0. [DOI] [PubMed] [Google Scholar]
- 21.Shiratori H, Ikeno H, Ayame S, Kataoka N, Miya A, Hosono K, Beppu T, Ueda K. 2006. Isolation and characterization of a new Clostridium sp. that performs effective cellulosic waste digestion in a thermophilic methanogenic bioreactor. Appl Environ Microbiol 72:3702–3709. doi: 10.1128/AEM.72.5.3702-3709.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izquierdo JA, Goodwin L, Davenport KW, Teshima H, Bruce D, Detter C, Tapia R, Han S, Land M, Hauser L, Jeffries CD, Han J, Pitluck S, Nolan M, Chen A, Huntemann M, Mavromatis K, Mikhailova N, Liolios K, Woyke T, Lynd LR. 2012. Complete genome sequence of Clostridium clariflavum DSM 19732. Stand Genomic Sci 6:104–115. doi: 10.4056/sigs.2535732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Izquierdo JA, Pattathil S, Guseva A, Hahn MG, Lynd LR. 2014. Comparative analysis of the ability of Clostridium clariflavum strains and Clostridium thermocellum to utilize hemicellulose and unpretreated plant material. Biotechnol Biofuels 7:136. doi: 10.1186/s13068-014-0136-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dassa B, Borovok I, Lamed R, Henrissat B, Coutinho P, Hemme CL, Huang Y, Zhou J, Bayer EA. 2012. Genome-wide analysis of Acetivibrio cellulolyticus provides a blueprint of an elaborate cellulosome system. BMC Genomics 13:210. doi: 10.1186/1471-2164-13-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamberg Y, Ruimy-Israeli V, Dassa B, Barak Y, Lamed R, Cameron K, Fontes CM, Bayer EA, Fried DB. 2014. Elaborate cellulosome architecture of Acetivibrio cellulolyticus revealed by selective screening of cohesin–dockerin interactions. PeerJ 2:e636. doi: 10.7717/peerj.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Artzi L, Dassa B, Borovok I, Shamshoum M, Lamed R, Bayer EA. 2014. Cellulosomics of the cellulolytic thermophile Clostridium clariflavum. Biotechnol Biofuels 7:100. doi: 10.1186/1754-6834-7-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fendri I, Tardif C, Fierobe H-P, Lignon S, Valette O, Pagès S, Perret S. 2009. The cellulosomes from Clostridium cellulolyticum: identification of new components and synergies between complexes. FEBS J 276:3076–3086. doi: 10.1111/j.1742-4658.2009.07025.x. [DOI] [PubMed] [Google Scholar]
- 28.Blouzard J-C, Coutinho PM, Fierobe H-P, Henrissat B, Lignon S, Tardif C, Pagès S, de Philip P. 2010. Modulation of cellulosome composition in Clostridium cellulolyticum: adaptation to the polysaccharide environment revealed by proteomic and carbohydrate-active enzyme analyses. Proteomics 10:541–554. doi: 10.1002/pmic.200900311. [DOI] [PubMed] [Google Scholar]
- 29.Morisaka H, Matsui K, Tatsukami Y, Kuroda K, Miyake H, Tamaru Y, Ueda M. 2012. Profile of native cellulosomal proteins of Clostridium cellulovorans adapted to various carbon sources. AMB Express 2:37. doi: 10.1186/2191-0855-2-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han SO, Yukawa H, Inui M, Doi RH. 2005. Effect of carbon source on the cellulosomal subpopulations of Clostridium cellulovorans. Microbiology 151:1491–1497. doi: 10.1099/mic.0.27605-0. [DOI] [PubMed] [Google Scholar]
- 31.Raman B, Pan C, Hurst GB, Rodriguez M, McKeown CK, Lankford PK, Samatova NF, Mielenz JR. 2009. Impact of pretreated switchgrass and biomass carbohydrates on Clostridium thermocellum ATCC 27405 cellulosome composition: a quantitative proteomic analysis. PLoS One 4:e5271. doi: 10.1371/journal.pone.0005271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gold ND, Martin VJ. 2007. Global view of the Clostridium thermocellum cellulosome revealed by quantitative proteomic analysis. J Bacteriol 189:6787–6795. doi: 10.1128/JB.00882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zverlov VV, Kellermann J, Schwarz WH. 2005. Functional subgenomics of Clostridium thermocellum cellulosomal genes: identification of the major catalytic components in the extracellular complex and detection of three new enzymes. Proteomics 5:3646–3653. doi: 10.1002/pmic.200401199. [DOI] [PubMed] [Google Scholar]
- 34.Dykstra AB, St Brice L, Rodriguez M, Raman B, Izquierdo J, Cook KD, Lynd LR, Hettich RL. 2014. Development of a multipoint quantitation method to simultaneously measure enzymatic and structural components of the Clostridium thermocellum cellulosome protein complex. J Proteome Res 13:692–701. doi: 10.1021/pr400788e. [DOI] [PubMed] [Google Scholar]
- 35.Gerngross UT, Romaniec MP, Kobayashi T, Huskisson NS, Demain AL. 1993. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal S L-protein reveals an unusual degree of internal homology. Mol Microbiol 8:325–334. doi: 10.1111/j.1365-2958.1993.tb01576.x. [DOI] [PubMed] [Google Scholar]
- 36.Pagès S, Bélaïch A, Fierobe HP, Tardif C, Gaudin C, Bélaïch JP. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J Bacteriol 181:1801–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 39.Liu NQ, Braakman RB, Stingl C, Luider TM, Martens JW, Foekens JA, Umar A. 2012. Proteomics pipeline for biomarker discovery of laser capture microdissected breast cancer tissue. J Mammary Gland Biol Neoplasia 17:155–164. doi: 10.1007/s10911-012-9252-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buschow SI, Figdor CG. 2010. Dendritic cell subsets digested: RNA sensing makes the difference! Immunity 32:149–151. doi: 10.1016/j.immuni.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 41.Lemaire M, Ohayon H, Gounon P, Fujino T, Béguin P. 1995. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J Bacteriol 177:2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinheiro BA, Gilbert HJ, Sakka K, Sakka K, Fernandes VO, Prates JA, Alves VD, Bolam DN, Ferreira LM. 2009. Functional insights into the role of novel type I cohesin and dockerin domains from Clostridium thermocellum. Biochem J 424:375–384. doi: 10.1042/BJ20091152. [DOI] [PubMed] [Google Scholar]
- 43.Bayer EA, Setter E, Lamed R. 1985. Organization and distribution of the cellulosome in Clostridium thermocellum. J Bacteriol 163:552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morag E, Halevy I, Bayer EA, Lamed R. 1991. Isolation and properties of a major cellobiohydrolase from the cellulosome of Clostridium thermocellum. J Bacteriol 173:4155–4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dror TW, Morag E, Rolider A, Bayer EA, Lamed R, Shoham Y. 2003. Regulation of the cellulosomal celS (cel48A) gene of Clostridium thermocellum is growth rate dependent. J Bacteriol 185:3042–3048. doi: 10.1128/JB.185.10.3042-3048.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang WK, Kruus K, Wu JH. 1994. Cloning and expression of the Clostridium thermocellum celS gene in Escherichia coli. Appl Microbiol Biotechnol 42:346–352. doi: 10.1007/BF00902740. [DOI] [PubMed] [Google Scholar]
- 47.Kataeva I, Li XL, Chen H, Choi SK, Ljungdahl LG. 1999. Cloning and sequence analysis of a new cellulase gene encoding CelK, a major cellulosome component of Clostridium thermocellum: evidence for gene duplication and recombination. J Bacteriol 181:5288–5295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dror TW, Rolider A, Bayer EA, Lamed R, Shoham Y. 2005. Regulation of major cellulosomal endoglucanases of Clostridium thermocellum differs from that of a prominent cellulosomal xylanase. J Bacteriol 187:2261–2266. doi: 10.1128/JB.187.7.2261-2266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirano N, Hasegawa H, Nihei S, Haruki M. 2013. Cell-free protein synthesis and substrate specificity of full-length endoglucanase CelJ (Cel9D-Cel44A), the largest multi-enzyme subunit of the Clostridium thermocellum cellulosome. FEMS Microbiol Lett 344:25–30. doi: 10.1111/1574-6968.12149. [DOI] [PubMed] [Google Scholar]
- 50.Ahsan MM, Kimura T, Karita S, Sakka K, Ohmiya K. 1996. Cloning, DNA sequencing, and expression of the gene encoding Clostridium thermocellum cellulase CelJ, the largest catalytic component of the cellulosome. J Bacteriol 178:5732–5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arai T, Araki R, Tanaka A, Karita S, Kimura T, Sakka K, Ohmiya K. 2003. Characterization of a cellulase containing a family 30 carbohydrate-binding module (CBM) derived from Clostridium thermocellum CelJ: importance of the CBM to cellulose hydrolysis. J Bacteriol 185:504–512. doi: 10.1128/JB.185.2.504-512.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Najmudin S, Guerreiro CI, Carvalho AL, Prates JA, Correia MA, Alves VD, Ferreira LM, Romão MJ, Gilbert HJ, Bolam DN, Fontes CM. 2006. Xyloglucan is recognized by carbohydrate-binding modules that interact with beta-glucan chains. J Biol Chem 281:8815–8828. doi: 10.1074/jbc.M510559200. [DOI] [PubMed] [Google Scholar]
- 53.Gefen G, Anbar M, Morag E, Lamed R, Bayer EA. 2012. Enhanced cellulose degradation by targeted integration of a cohesin-fused β-glucosidase into the Clostridium thermocellum cellulosome. Proc Natl Acad Sci U S A 109:10298–10303. doi: 10.1073/pnas.1202747109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kadam SK, Demain AL. 1989. Addition of cloned beta-glucosidase enhances the degradation of crystalline cellulose by the Clostridium thermocellum cellulose complex. Biochem Biophys Res Commun 161:706–711. doi: 10.1016/0006-291X(89)92657-0. [DOI] [PubMed] [Google Scholar]
- 55.Lamed R, Kenig R, Morgenstern E, Calzada JF, De Micheo F, Bayer Ea. 1991. Efficient cellulose solubilization by a combined cellulosome-β-glucosidase system. Appl Biochem Biotechnol 27:173–183. doi: 10.1007/BF02921525. [DOI] [Google Scholar]
- 56.Zverlov VV, Klupp M, Krauss J, Schwarz WH. 2008. Mutations in the scaffoldin gene, cipA, of Clostridium thermocellum with impaired cellulosome formation and cellulose hydrolysis: insertions of a new transposable element, IS1447, and implications for cellulase synergism on crystalline cellu. J Bacteriol 190:4321–4327. doi: 10.1128/JB.00097-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reference deleted.
- 58.Xu Q, Gao W, Ding S-Y, Kenig R, Shoham Y, Bayer EA, Lamed R. 2003. The cellulosome system of Acetivibrio cellulolyticus includes a novel type of adaptor protein and a cell surface anchoring protein. J Bacteriol 185:4548–4557. doi: 10.1128/JB.185.15.4548-4557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ding SY, Bayer EA, Steiner D, Shoham Y, Lamed R. 1999. A novel cellulosomal scaffoldin from Acetivibrio cellulolyticus that contains a family 9 glycosyl hydrolase. J Bacteriol 181:6720–6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Q, Barak Y, Kenig R, Shoham Y, Bayer EA, Lamed R. 2004. A novel Acetivibrio cellulolyticus anchoring scaffoldin that bears divergent cohesins. J Bacteriol 186:5782–5789. doi: 10.1128/JB.186.17.5782-5789.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miller GL. 1959. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. doi: 10.1021/ac60147a030. [DOI] [Google Scholar]
- 62.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 63.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. 2011. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 64.Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M. 2010. Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol 189:739–754. doi: 10.1083/jcb.200911091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vazana Y, Barak Y, Unger T, Peleg Y, Shamshoum M, Ben-Yehezkel T, Mazor Y, Shapiro E, Lamed R, Bayer EA. 2013. A synthetic biology approach for evaluating the functional contribution of designer cellulosome components to deconstruction of cellulosic substrates. Biotechnol Biofuels 6:182. doi: 10.1186/1754-6834-6-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zverlov VV, Schwarz WH. 2008. Bacterial cellulose hydrolysis in anaerobic environmental subsystems—Clostridium thermocellum and Clostridium stercorarium, thermophilic plant-fiber degraders. Ann N Y Acad Sci 1125:298–307. doi: 10.1196/annals.1419.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gel filtration elution profile of cellulosome fractions. The supernatant fractions of the cellobiose, microcrystalline cellulose, and switchgrass growth media were concentrated and loaded onto a HiPrep 26/60 Sephacryl S-500 HR gel filtration column. The first eluted peak did not contain any detectable proteins and is therefore called F0. The second peak (including any higher molecular-mass shoulder) is fraction I, and the third is lower-molecular-mass fraction II, as described in Results. The fraction I from cellobiose-grown cells eluted as two connected peaks (described as A and B), but SDS-PAGE analysis showed that the protein contents were indistinguishable, so A and B were pooled and considered one fraction. Download
Complete LC-MS/MS data. The full list of proteins identified in the various fractions of the 18 samples analyzed by LC-MS/MS is given. The proteins are annotated according to the CAZy and BLAST databases.
Statistical analysis of LC-MS/MS data. In order to determine significant differences or similarities among the different samples, statistical analysis of the LC-MS/MS was performed. t tests and analysis of variance (ANOVA) were performed for the following sets of samples: t test of complex (fraction) I versus complex (fraction) II for each of the growth media (blue, proteins which are more abundant in fraction II; green, proteins which are more abundant in fraction I [P < 0.0015; difference >│1│]); t test of CB (cellobiose) versus MCC (microcrystalline cellulose) samples (orange, proteins which are more abundant in MCC; purple, proteins which are more abundant in CB I [P < 0.0024; difference > │1.3│]); t test of MCC versus SG (switchgrass) (orange, proteins which are more abundant in MCC; pink, proteins which are more abundant in SG [P < 0.00058; difference > │1.8│]); t test of CB versus SG (orange, proteins which are more abundant in MCC; pink, proteins which are more abundant in SG; purple, proteins which are more abundant in CB [P < 0.0006; difference > │1.4│]); t test of complex (fraction) I versus complex (fraction) II from all the samples for proteins with at least two peptides (blue, proteins which are more abundant in fraction II; green, proteins which are more abundant in fraction I [P < 0.0015; difference >│1│]); t test between the CB fractions (P < 0.0074; difference >│1│); t test between the MCC fractions (P < 0.008; difference > │1.15│); t test between the SG fractions (P < 0.0.0115; difference > │1.38│); ANOVA test between CB, MCC, and SG; two-way ANOVA between the group of all growth media and the subgroup pf the complexes (fractions); blue letters, proteins which are more abundant in fraction II for one (or two) of the growth media; green letters, proteins which are more abundant in fraction I for one (or two) of the growth media.