ABSTRACT
Acinetobacter baumannii is an emerging Gram-negative pathogen found in hospitals and intensive care units. In order to persist in hospital environments, A. baumannii withstands desiccative conditions and can rapidly develop multidrug resistance to conventional antibiotics. Cationic antimicrobial peptides (CAMPs) have served as therapeutic alternatives because they target the conserved lipid A component of the Gram-negative outer membrane to lyse the bacterial cell. However, many Gram-negative pathogenic bacteria, including A. baumannii, fortify their outer membrane with hepta-acylated lipid A to protect the cell from CAMP-dependent cell lysis. Whereas in Escherichia coli and Salmonella, increased production of the outer membrane acyltransferase PagP results in formation of protective hepta-acylated lipid A, which reinforces the lipopolysaccharide portion of the outer membrane barrier, A. baumannii does not carry a gene that encodes a PagP homolog. Instead, A. baumannii has evolved a PagP-independent mechanism to synthesize protective hepta-acylated lipid A. Taking advantage of a recently adapted A. baumannii genetic recombineering system, we characterized two putative acyltransferases in A. baumannii designated LpxLAb (A. baumannii LpxL) and LpxMAb (A. baumannii LpxM), which transfer one and two lauroyl (C12:0) acyl chains, respectively, during lipid A biosynthesis. Hepta-acylation of A. baumannii lipid A promoted resistance to vertebrate and polymyxin CAMPs, which are prescribed as last-resort treatment options. Intriguingly, our analysis also showed that LpxMAb-dependent acylation of lipid A is essential for A. baumannii desiccation survival, a key resistance mechanism for survival in hospital environments. Compounds that inhibit LpxMAb-dependent hepta-acylation of lipid A could act synergistically with CAMPs to provide innovative transmission prevention strategies and treat multidrug-resistant infections.
IMPORTANCE
Acinetobacter baumannii infections can be life threatening, and disease can progress in a variety of host tissues. Current antibiotic regimen and disinfectant strategies have failed to limit nosocomial A. baumannii infections. Instead, the rate of A. baumannii infection among health care communities has skyrocketed due to the bacterium’s adaptability. Its aptitude for survival over extended periods on inanimate objects, such as catheters, respirators, and surfaces in intensive care units, or on the hands of health care workers and its ability to rapidly develop antibiotic resistance make A. baumannii a threat to health care communities. Emergence of multidrug- and extremely drug-resistant A. baumannii illustrates the ineffectiveness of current prevention and treatment options. Our analysis to understand how A. baumannii resists cationic antimicrobial peptide (CAMP)-mediated and desiccative killing revealed two lipid A acyltransferases that produce protective hepta-acylated lipid A. Our work suggests that inhibiting lipid A biosynthesis by targeting the acyltransferase LpxMAb (A. baumannii LpxM) could provide a novel target to combat this pathogen.
INTRODUCTION
The outer membrane of Gram-negative bacteria is a highly conserved barrier consisting of an inner monolayer of glycerophospholipids and a surface-exposed monolayer of lipopolysaccharide (LPS). The amphipathic properties of LPS and phospholipids allow spontaneous formation of a membrane bilayer where the hydrophobic lipid moieties are sandwiched between the hydrophilic groups. The biophysical membrane properties restrict diffusion of toxic molecules (e.g., antibiotics) across the membrane into the cell. LPS is a biologically distinct glycolipid that contains three domains: the bioactive membrane anchor called lipid A, core sugars that extend from lipid A, and a core-ligated O-antigen carbohydrate repeat (1). Many mucosal pathogens, including Acinetobacter baumannii synthesize lipooligosaccharide (LOS), which includes only core and lipid A. Whereas the O-antigen domain is dispensable, lipid A and core are required for bacterial survival in a host. In fact, inhibitors that target essential enzymatic steps in lipid A biosynthesis have provided promising antimicrobial chemotherapeutics (2–4).
In the well-defined Gram-negative bacterium Escherichia coli K-12, LPS/LOS biosynthesis initiates with formation of Kdo2-lipid A (Kdo stands for 3-deoxy-d-manno-octulosonic acid) or endotoxin. Nine conserved enzymes, termed the Raetz pathway (1), coordinately synthesize hexa-acylated lipid A. While the first seven enzymes assemble the precursor Kdo2-lipid IVA, the last two biosynthetic steps are completed by LpxL and LpxM. LpxL first catalyzes transfer of laurate (C12:0) followed by LpxM-dependent myristate (C14:0) addition in a stepwise manner to complete synthesis of the hexa-acylated and bis-phosphorylated Kdo2-lipid A molecule (Fig. 1) (5–7).
FIG 1 .
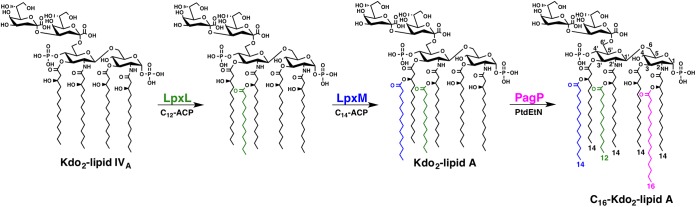
Synthesis of hexa- and hepta-acylated lipid A in E. coli K-12. In E. coli, LpxL catalyzes transfer of a laurate (C12:0) group from an acyl carrier protein (ACP) onto the R-2′-hydroxymyristate acyl chain of Kdo2-lipid IVA. Subsequently, LpxM-dependent addition of myristate (C14:0) onto the R-3′-hydroxymysristate residue completes synthesis of hexa-acylated Kdo2-lipid A. Upon sensing specific environmental cues, PagP activation results in palmitoylation (C16:0) of the R-2-hydroxymyristate primary acyl chain, altering membrane fluidity and increasing resistance to antimicrobial peptides. PtdEtN, phosphatidylethanolamine.
Despite E. coli K-12 providing a basic model to understand lipid A biosynthesis in Gram-negative bacteria, many pathogens remodel the hexa-acylated molecule into diverse lipid A-based structures. Lipid A modifications directly affect the ability of a pathogen to survive in its host by altering outer membrane permeability, by camouflaging the pathogen from host immune detection, and by promoting resistance to antimicrobial peptides (1, 8). While pathogens exploit diverse strategies to survive in a host, a detailed understanding of the molecular mechanisms that mediate bacterial survival is key for the development of new and more-effective antimicrobial treatments.
Gram-negative bacteria can alter their lipid A structure by incorporating additional chemical moieties or by altering the lipid A phosphate or acyl chain groups (8, 9). Well-defined transcriptional and posttranscriptional regulatory systems tightly control the lipid A enzymatic modification machinery, illustrating the importance of lipid A on bacterial survival (8, 10, 11). One well-characterized lipid A modification involves palmitate addition to form hepta-acylated lipid A, which promotes bacterial resistance to cationic antimicrobial peptides (CAMPs), fortifies the outer membrane, and reduces host immune recognition (). In a number of pathogens, hepta-acylation is a protective response to a specific environmental stressor. In the presence of the signal, the bacterium initiates a regulatory phosphorylation cascade that culminates in increased expression of the outer membrane protein PagP (12). In the outer membrane, PagP transfers a palmitoyl (C16:0) group to lipid A with phosphatidylethanolamine serving as the acyl donor (13).
Unlike most Gram-negative pathogens, which synthesize hexa-acylated lipid A under standard growth conditions, the nosocomial pathogen Acinetobacter baumannii produces a predominant hepta-acylated lipid A molecule in a PagP-independent fashion (15, 16). In this study, we investigate the biosynthetic mechanism utilized for production of hepta-acylated lipid A in A. baumannii and characterize how it impacts bacterial fitness. A. baumannii is a resilient Gram-negative pathogen that is well adapted to survive in hospital or intensive care unit environments. Over the past decade, the rate of A. baumannii transmission and infection in health care communities has significantly increased patient morbidity and mortality (17). A. baumannii infections are diverse, and treatment options have become extremely limited due to a propensity for A. baumannii to develop resistance to conventional antibiotics. The bacterium develops antibiotic resistance through acquisition of resistance genes, upregulation of efflux pumps, and through modification of outer membrane structures (18). Oftentimes acquisition of resistance mechanisms results in reduced bacterial fitness (19–21). Emergence of multidrug-resistant (MDR) and extensively drug-resistant strains has required increased prescription of last-resort antibiotics (17, 22–25). Currently, last-line treatments include a CAMP called colistin which targets lipid A to perturb the membrane and lyse the bacterium (26). Despite success of last-resort CAMPs like colistin, recent studies have shown that A. baumannii can develop resistance under selective pressure (25). Therefore, a detailed understanding of the bacterial factors essential for CAMP resistance in A. baumannii is necessary to direct innovative chemotherapeutic strategies.
Another important factor contributing to the spread of A. baumannii is its propensity to withstand desiccation. Uniquely, A. baumannii survives on inanimate objects without nutrients or water for extended periods, which promotes transmission throughout health care environments (27). The molecular factors that mediate desiccation survival are not well understood, but identification of desiccation fitness determinants could result in more effective eradication strategies.
In this study, we characterized two genes designated lpxLAb (A. baumannii lpxL) and lpxMAb (A. baumannii lpxM) that are required for biosynthesis of hepta-acylated lipid A in A. baumannii. Unlike lipid A biosynthesis in most other Gram-negative pathogens, A. baumannii encodes a dual acyltransferase designated LpxMAb (A. baumannii LpxM), which transfers two lauroyl groups onto lipid A to synthesize hepta-acylated lipid A via a PagP-independent mechanism. Our analysis demonstrated that hepta-acylated lipid A is the dominant glycolipid on the surface of A. baumannii and activates human Toll-like receptor 4 (TLR-4). In contrast, LOS isolated from lpxLAb and lpxMAb mutants failed to stimulate the human TLR-4 innate immune pathway. Similar to studies performed in Salmonella, hepta-acylated lipid A promoted bacterial resistance to vertebrate CAMPs (10); however, hepta-acylated lipid A in A. baumannii also conferred resistance to polymyxin CAMPs such as the last-resort antimicrobial agent colistin. In addition, we determined that hepta-acylated lipid A is important for bacterial desiccation survival, indicating that loss of outer membrane fatty acids is detrimental to A. baumannii persistence on inanimate objects in the hospital.
RESULTS
A. baumannii encodes LpxL and LpxM lipid A acyltransferase homologs.
Previous studies to characterize the biosynthesis of lipid A, the bioactive moiety that anchors LPS/LOS to the outer membranes of Gram-negative bacteria, established that two secondary acyltransferases catalyze the final two enzymatic steps. In E. coli, LpxL functions first by transferring a laurate group onto Kdo2-lipid IVA followed by myristate addition via LpxM to complete biosynthesis of hexa-acylated Kdo2-lipid A (Fig. 1) (5–7). The two secondary acylation steps in lipid A biosynthesis are generally conserved among Gram-negative bacteria. However, lipid A secondary acyltransferases that recognize noncanonical substrates in an epsilonproteobacterium have been reported (28).
Instead of synthesizing hexa-acylated lipid A, like many Gram-negative bacterial pathogens, A. baumannii synthesizes a predominant hepta-acylated lipid A species under normal growth conditions (15, 16). While the mechanism that leads to A. baumannii lipid A hepta-acylation has not been characterized, analysis of E. coli and Salmonella determined that lipid A hepta-acylation is dependent on the outer membrane protein PagP (Fig. 1) (10, 13, 29). PagP expression increases in response to specific stimuli that perturb the outer membrane, leading to migration of phospholipids in the outer leaflet (30). Mechanistically, PagP transfers a palmitate acyl chain (C16:0) from phosphatidylethanolamine to the lipid A R-2-hydroxymyristate to generate hepta-acylated lipid A (Fig. 1) (13). In E. coli and Salmonella, palmitate addition protects the cell from vertebrate CAMPs by reducing the permeability of the outer membrane barrier (10). However, the biological role of hepta-acylated lipid A in A. baumannii has not been examined. Intriguingly, A. baumannii does not carry a gene that encodes a PagP homolog, suggesting that it has evolved a PagP-independent acylation mechanism.
In order to identify putative lipid A acyltransferases in A. baumannii, E. coli LpxL and LpxM amino acid sequences were analyzed by Basic Local Alignment Search Tool (BLAST). Bioinformatics analysis identified two A. baumannii genes that encode putative lipid A acyltransferases, designated in strain ATCC 17978 as A1S_0431 and A1S_2609. E. coli LpxL shared 39% identity with the A1S_0431 gene product, designated LpxLAb, while LpxM shared 23% identity to the A1S_2609 gene product, which we designated LpxMAb. LpxLAb and LpxMAb shared 29% identity to each other.
Targeted removal of each putative acyltransferase gene was performed by genetic recombineering, which was recently adapted for use in A. baumannii (31). Strains harboring nonpolar markerless deletions of lpxLAb and lpxMAb were isolated. While lpxMAb is not transcribed in an operon, lpxLAb could potentially be cotranscribed with A1S_0430, encoding LpsB, and A1S_0432, which encodes a putative transport protein. LpsB is a glycosyltransferase required for core oligosaccharide attachment to lipid A (32), which is important for A. baumannii virulence, CAMP resistance, and polymyxin resistance (33). For complementation of each acyltransferase, each respective coding sequence was expressed from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter on a replicon in the mutant strain (see Table S1 in the supplemental material). Growth curve analysis of the mutants and complemented strains did not reveal any major growth defect compared with the wild-type strain (see Fig. S1 in the supplemental material).
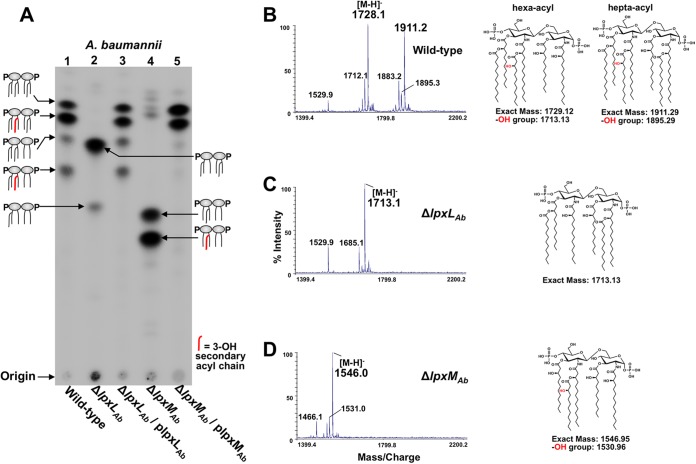
To analyze the effect on lipid A produced by each A. baumannii strain, 32P-radiolabeled lipid A was isolated and chromatographically separated based on hydrophobicity. Quantitative thin-layer chromatography (TLC) showed that wild-type A. baumannii produced a predominant hepta-acylated lipid A species with relatively minor amounts of a hexa-acylated species (Fig. 2A, lane 1). Strains with deletions in the LpxLAb and LpxMAb genetic coding sequences exhibited reduced hydrophobic migration patterns indicative of decreased acylation relative to the wild-type strain (Fig. 2A, lanes 2 and 4). Complementation restored production of wild-type hepta-acylated lipid A in each mutant strain (Fig. 2A, lanes 3 and 5). Unlike the well-defined E. coli lipid A biosynthetic pathway, these initial results also indicate that secondary acylation in A. baumannii is not strictly ordered, since both lpxLAb and lpxMAb mutants retain secondary acyl chains.
FIG 2 .
Structural characterization of the A. baumannii secondary lipid A acyltransferases. Characterization of lipid A isolated from A. baumannii strains. (A) 32P-radiolabeled lipid A from A. baumannii was isolated and separated based on hydrophobicity using TLC. Fatty acyl chains bearing a hydroxyl group at position 3 are indicated in red. (B) MALDI-TOF MS and structures of wild-type A. baumannii lipid A with m/z ratios of 1,729.12 and 1,911.29, corresponding to the hexa- and hepta-acylated species, respectively. Addition of a hydroxyl group (red) corresponds to the m/z ratios 1,713.13 and 1,895.16. (C) MALDI-TOF MS of the ΔlpxLAb mutant indicating a major species at m/z 1,713.1. (D) MALDI-TOF MS of the ΔlpxMAb mutant indicating a major species at m/z 1,546.0. Addition of the hydroxyl group to lipid A (red) results in a lipid A at m/z 1,530.96.
To determine the mutant acylation pattern, matrix-assisted laser desorption ionization−time of flight (MALDI-TOF) mass spectrometry (MS) was performed on isolated lipid A. Spectra collected from wild-type and complemented A. baumannii strains indicated major molecular ions at m/z 1,728.1 and 1,911.2 (Fig. 2B; see Fig. S2 in the supplemental material), while the ΔlpxLAb strain revealed one major ion at m/z 1,713.1 (Fig. 2C) and the ΔlpxMAb strain revealed one major ion species at m/z 1,546.0 (Fig. 2D). On the basis of our observed m/z ratios, we proposed chemical structures to fit the exact theoretical mass where wild-type A. baumannii synthesizes two lipid A species, including a minor hexa-acylated lipid A and a predominant hepta-acylated lipid A (Fig. 2B). Based on our chemical structures in the mutant strains, the ΔlpxLAb strain lacked a hydroxylauric acid, while the ΔlpxMAb strain resulted in production of penta-acylated lipid A lacking two lauroyl groups (Fig. 2C and D).
To verify the A. baumannii lipid A secondary acyl groups, higher-energy collisional dissociation (HCD) and UV photodissociation (UVPD) fragmentation spectra were collected for the previously indicated molecular ions. HCD and UVPD are high-energy ion activation methods that produce rich fragmentation patterns and thus enable a detailed structural analysis (34, 35). Unique fragments in the HCD and UVPD spectra at m/z 1,911.2, corresponding to wild-type hepta-acylated lipid A, confirmed the presence and length of all seven acyl chains (see Fig. S3 in the supplemental material). Specifically, our analysis revealed four acyl chains attached to the distal side and three on the proximal side of the lipid A molecule as shown in Fig. 3C.
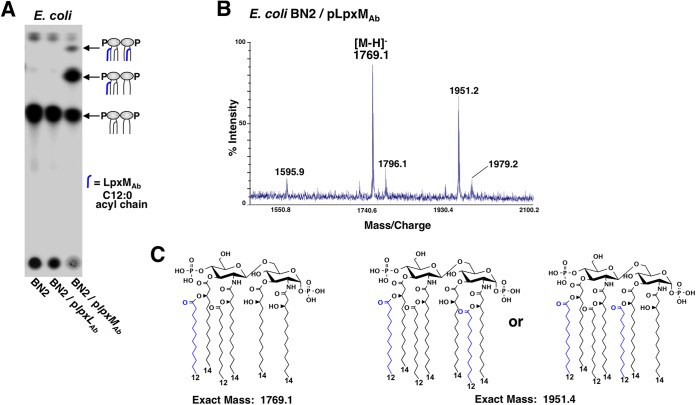
FIG 3 .
LpxMAb is a dual acyltransferase. Characterization of LpxMAb revealed a unique dual acyltransferase activity. (A) 32P-radiolabeled lipid A from E. coli BN2 was isolated and separated based on hydrophobicity using TLC. The blue acyl chains in all lipid A structures correspond to lauroyl groups derived from LpxMAb. (B) MALDI-TOF MS analysis of lipid A isolated from a PagP- and LpxM-deficient strain, BN2, expressing LpxMAb. The m/z at 1,769.1 and 1,951.2 correspond to LpxMAb-dependent acylation. (C) Proposed chemical structures of LpxMAb-dependent acylation in E. coli.
Heterologous expression of LpxMAb in E. coli revealed dual transferase activity.
Previously characterized LpxL and LpxM acyltransferases exclusively add one fatty acid to lipid A (5). However, our data indicated that A. baumannii ΔlpxMAb strains lacked two lauroyl groups. To determine whether LpxMAb is sufficient to catalyze transfer of two laurate groups onto A. baumannii lipid A, we expressed the lpxLAb and lpxMAb genetic coding sequences in BN2, an E. coli strain with deletions in both lpxM and pagP. E. coli strain BN2 synthesizes penta-acyl lipid A with one laurate group attached at position 2′ (Fig. 3A, leftmost lane) (9). Expression of LpxLAb in strain BN2 did not alter the lipid A structure, suggesting that it is not active in E. coli or that it functions like E. coli LpxL and only adds a secondary acyl chain at position 2′ (Fig. 3A, middle lane, and Fig. 1) (36). Importantly, expression of LpxMAb in strain BN2 resulted in formation of both hexa- and hepta-acylated lipid species (Fig. 3A, rightmost lane). Thus, LpxMAb is sufficient to attach lauroyl groups at two distinct locations of the lipid A molecule, a characteristic unique to LpxMAb. We confirmed attachment of the acyl chains by mass spectrometry where spectra showed two major species at m/z 1,768.2 and 1,951.2, which indicate two separate acylation sites (Fig. 3B and C).
Determining LpxMAb acyl chain placement and hydroxylation.
Importantly, the palmitate acyl chain added by PagP at the R-2-hydroxymyristate position of lipid A in E. coli and Salmonella was shown to be important for resistance against host CAMPs (Fig. 1) (10). Therefore, we sought to determine whether placement of the secondary acyl chain in A. baumannii lipid A occurs at an equivalent position.
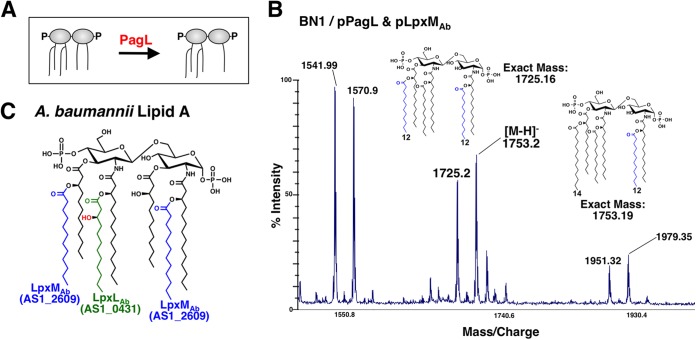
Formation of hepta-acylated lipid A through addition of an acyl chain onto A. baumannii lipid A at the same site that PagP functions in E. coli and Salmonella could be important for survival during host infection. To investigate LpxMAb-dependent acyl chain placement, we used a genetic approach to confirm attachment of a lauroyl group to the R-2-hydroxymyristate in A. baumannii, which is the equivalent position that PagP-dependent myristate addition occurs in E. coli (37). To do this, LpxMAb was coexpressed with the lipid A deacylase PagL in an E. coli strain. PagL efficiently removes the 3-O-linked acyl chain of lipid A following transport of LPS/LOS to the outer membrane (Fig. 4A) (38). Lipid A isolated from cells coexpressing LpxMAb and PagL was analyzed by MALDI-TOF mass spectrometry, and major species at m/z 1,725.2 and 1,753.2 were present in the spectra (Fig. 4B). These data indicate that deacylation at position 3 by PagL, which takes place in the outer membrane, is not inhibited by inner membrane-associated LpxMAb-dependent acylation (Fig. 4B). Furthermore, HCD and UVPD fragmentation spectra of m/z 1,712.12, corresponding to the ΔlpxLAb mutant, also confirmed that the secondary laurate group is attached at position 2 (see Fig. S4 in the supplemental material). On the basis of our genetic data and high-resolution HCD and UVPD data, we confirmed that LpxMAb and PagP add a lauroyl group and a palmitoyl group, respectively, at the R-2-hydroxymyristate position of lipid A (Fig. 1 and 4C). In summary, LpxMAb-dependent lauroyl group addition occurs at the R-3′- and R-2-hydroxymyrsitate positions of lipid A (Fig. 4C).
FIG 4 .
Determining LpxMAb-dependent acyl chain placement. (A) PagL removes the 3-O-linked acyl chain of lipid A. (B) MALDI-TOF MS analysis of lipid A from the PagP-deficient E. coli strain BN1 expressing PagL and LpxMAb. The m/z at 1,725.16 and 1,753.19 correspond to lipid A structures that have a secondary laurate group (blue) at positions 3′ and 2 and only at position 2, respectively. (C) Chemical structure of wild-type A. baumannii lipid A that is based on both genetic studies and chemical analysis. LpxLAb catalyzes addition of a secondary lauroyl group to position 2′ (green), which can be hydroxylated (red). LpxMAb catalyzes transfer of secondary acyl chain at both positions 2 and 3′ (blue).
In addition, lipid A from A. baumannii strains was analyzed by TLC; lipid A from the wild-type strain, lpxMAb mutant, and all complemented A. baumannii strains migrated as doublets, indicating the presence of a modified fatty acid in all strains except the lpxLAb mutant (Fig. 2A). Mass spectrometry indicated a mass difference of 16 in lipid A, which is indicative of hydroxylation modification (Fig. 2B and D). Therefore, we concluded that the lpxLAb-dependent acyl chain is hydroxylated. Hydroxylation of lipid A fatty acids is common in Gram-negative bacteria where LpxO hydroxylates fatty acids. Furthermore, BLAST analysis revealed that gene A1S_0308 encodes a putative A. baumannii LpxO homolog with 59% identity to Salmonella enterica serovar Typhimurium LT-2 LpxO.
A. baumannii lipid A differentially stimulates human TLR-4/MD-2.
Microbes continuously challenge the host immune system, but mammals have evolved receptors like TLR-4/MD-2 to detect invading bacterial cells via pathogen-associated molecular patterns (39). Specifically, the lipid A portion of Gram-negative bacterial LPS/LOS is a pathogen-associated molecular pattern that is bound by the TLR-4/MD-2 complex with high affinity (40). The interaction between lipid A and TLR-4/MD-2 strongly activates MyD88- and TRIF-dependent innate immune pathways to result in inflammation and clearance of the infection via innate immune recruitment (41). Coevolution of the TLR-4/MD-2 receptor and lipid A ligand has resulted in strong immune activation when hexa-acylated lipid A is detected. However, many pathogens modify the lipid A chemical structure to avoid TLR-4-dependent immune detection (8). Therefore, we analyzed human TLR-4/MD-2 activation using LOS isolated from the A. baumannii acyltransferase mutants.
To analyze immune activation by A. baumannii LOS, we used a human embryonic kidney reporter cell line (HEK-blue; InvivoGen) that expressed the human TLR-4, MD-2, and CD-14 receptor complex along with the NF-κB and activator protein-1-dependent reporter secreted embryonic alkaline phosphatase (SEAP) (9). LOS was isolated and purified from wild-type, ΔlpxLAb, ΔlpxMAb, and complemented strains (see Fig. S5 in the supplemental material). Increasing concentrations of LOS were incubated with HEK-blue cells to activate potential signaling pathways. The wild type and the complemented strains activated reporter expression in a dose-dependent manner indicating that A. baumannii hepta-acylated lipid A activates the human TLR-4/MD-2 innate immune pathway (Fig. 5). In contrast, LOS stimulation by lpxLAb and lpxMAb was dramatically reduced (Fig. 5). Similar to findings in E. coli, the pattern of TLR-4/MD-2 reporter activation indicates that acylation of A. baumannii lipid A is important for immune recognition by the host immune system (8, 37).
FIG 5 .
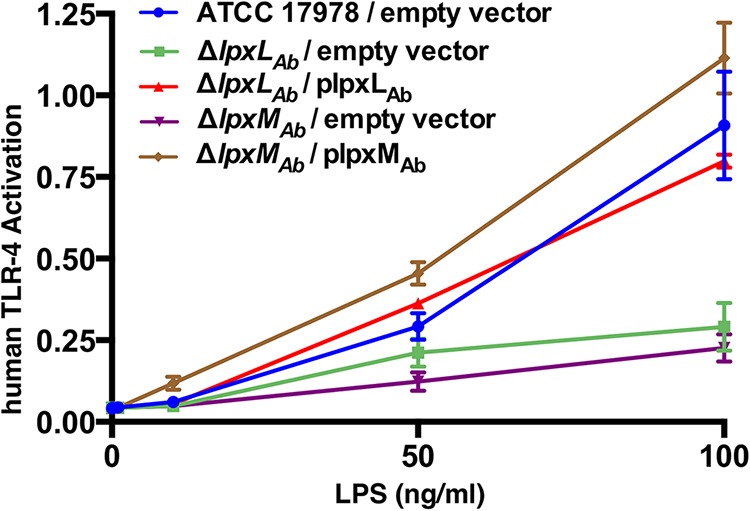
Stimulation of the human TLR-4 immune signaling pathway by A. baumannii LOS. Stimulation of human TLR-4 following incubation with increasing concentrations (0, 0.1, 1.0, 10, 50, and 100 ng/ml) of isolated LOS with HEK-blue cells expressing TLR-4, MD-2, and CD-14 is depicted. Detection of a secreted reporter indicates differential activation of TLR-4 in response to LOS.
LpxMAb protects A. baumannii from CAMPs.
The innate immune system produces CAMPs that bind to and lyse Gram-negative bacteria through disruption of the outer membrane. CAMPs are directed to LPS/LOS on the bacterial surface by electrostatic interactions (42). Presumably PagP-dependent lipid A palmitoylation increases the hydrophobic van der Waals forces of the lipid bilayer to prevent CAMP insertion and membrane disruption. In Salmonella, hepta-acylation of lipid A protected bacterial cells from vertebrate CAMPs such as C18G, an α-helical peptide found at the C terminus of the secreted cytokine human platelet factor IV (10, 43).
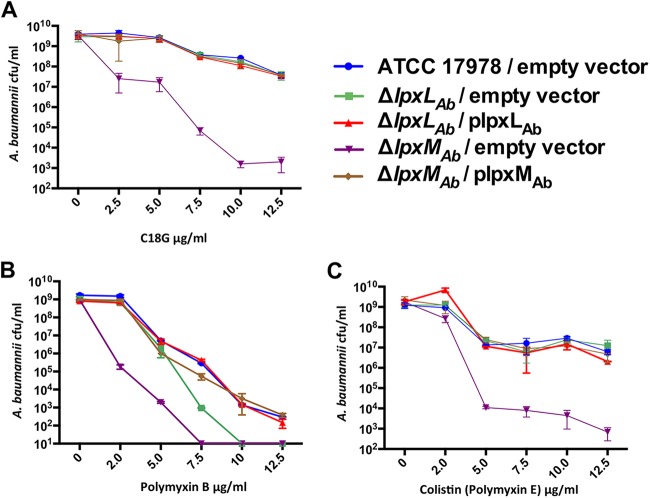
Based on quantitative TLC analysis of wild-type A. baumannii lipid A, hepta-acylated lipid A dominated the cell surface (Fig. 2A, leftmost lane). Therefore, we first determined whether hepta-acylated lipid A in A. baumannii conferred resistance to vertebrate CAMPs. Increasing concentrations of the C18G CAMP facilitated concentration-dependent killing in all A. baumannii strains. Importantly, the lpxMAb mutant displayed increased sensitivity to C18G, where a 1,000-fold decrease in the number of CFU per milliliter was recovered at a concentration of 10 µg/ml compared to the wild-type strain. Complementation fully restored resistance of the lpxMAb mutation back to wild-type levels, indicating that LpxMAb-mediated hepta-acylation confers protection against vertebrate CAMPs in A. baumannii (Fig. 6A).
FIG 6 .
LpxMAb contributes to vertebrate and polymyxin CAMP resistance in A. baumannii. A. baumannii CAMP survival assays. A. baumannii strains were incubated with the indicated concentrations of the mammalian CAMP, C18G (A), polymyxin A (B), or polymyxin E (C) for 2 h. Bacterial cells were serially diluted and plated to recover viable bacteria. Colony counts, reported as CFU per milliliter, determined cell survival.
Another class of CAMPs called polymyxins includes polymyxin B and E, which are lipopeptides synthesized by Gram-positive soil bacilli. These cyclic peptides contain a hydrophobic tail that inserts into the membrane to kill by forming pores in the Gram-negative outer membrane (44). Polymyxin B is currently used in topical solutions such as neosporin, while polymyxin E (colistin) is a last-line antimicrobial used to treat Gram-negative multidrug-resistant bacterial infections. We observed increased killing by polymyxin B (Fig. 6B) and colistin (Fig. 6C), indicating that these drugs could kill A. baumannii more effectively when LpxMAb-dependent acylation was inhibited. The enhanced killing that we observed was fully complemented when each gene was expressed in trans. Therefore, LpxMAb-dependent acylation in A. baumannii promotes resistance not only to vertebrate CAMPs but also to medically relevant polymyxin antibiotics.
LpxMAb is required for virulence in Galleria mellonella.
A well-established model to determine virulence of A. baumannii strains is the Galleria mellonella infection model (45–47). Importantly, a strong correlation between the virulence of several bacteria, including A. baumannii, in G. mellonella and mammalian virulence models has been established (47–49). Furthermore, G. mellonella carries a gene that encodes a humoral immune response, which is activated when large bacterial loads are injected. While invertebrates do not mount an antibody response, within 3 h of challenge, antimicrobial peptides are synthesized and released into the hemolymph to neutralize the bacterial infection (50, 51).
To determine the susceptibility of killing in the A. baumannii acyltransferase mutants, we injected equivalent numbers of CFU from wild-type, mutant, and complemented strains into G. mellonella and monitored bacterial killing. After 72 h, all wax moth larvae injected with the wild-type and complemented A. baumannii strains died, whereas all worms that were challenged with phosphate-buffered saline (PBS) and the A. baumannii ΔlpxMAb strain survived. In contrast, only one of the larvae injected with the A. baumannii LpxLAb mutant survived (Fig. 7). These data suggest that LpxMAb-dependent lipid A acylation is important for A. baumannii survival and virulence in invertebrates.
FIG 7 .
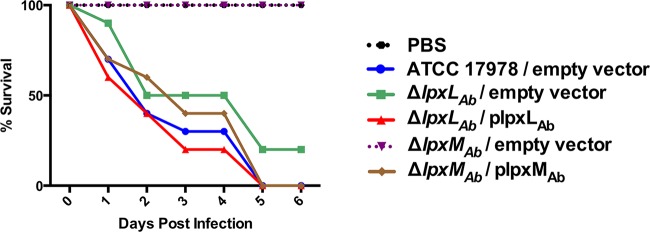
Galleria mellonella virulence model. G. mellonella survival is dependent on LpxMAb. G. mellonella larvae were injected with approximately 1 × 106 CFU and incubated at 37°C. Killing was monitored every 24 h for 6 days.
LpxMAb-dependent acylation of A. baumannii lipid A contributes to desiccation survival.
A. baumannii is a nosocomial pathogen that persists on inanimate objects in hospitals for months and even years (52). The organism’s unique ability to survive under desiccative conditions results in increased infections and transmission rates. The molecular mechanisms that contribute to desiccation tolerance in A. baumannii have not been characterized and are not well understood. In other Gram-negative bacteria, proteins involved in exopolysaccharide biosynthesis, in maintaining protein stability, and in detoxification of reactive oxygen species were identified as important to desiccation resistance (53, 54). However, the protective role of the outer membrane barrier has not been explored.
In order to further characterize our lipid A acyltransferase mutants, we subjected the wild type, ΔlpxLAb, ΔlpxMAb, and the complemented strains to a desiccation survival assay. Strains were spotted on agar in a 10-fold dilution series before and after desiccation on polystyrene (Fig. 8; see Fig. S6 in the supplemental material). Following desiccation, bacterial CFUs were recovered for all strains except for the A. baumannii ΔlpxMAb strain, for which no viable cells were recovered at either cell density tested, indicating a >1,000-fold decrease in CFU (Fig. 8A and Fig. S6A). In addition, we performed desiccation assays where we resuspended A. baumannii cells in human serum before desiccation. Following desiccation, cells were recovered in all groups and spotted (Fig. 8B and Fig. S6B). However, we recovered 100-fold less of the lpxMAb mutant strain following desiccation (Fig. 8B and Fig. S6B). Under each condition, in trans expression of lpxMAb fully complemented the mutant phenotype and restored bacterial survival to the wild-type level. These results suggest that the lipid components in the outer membrane contribute to desiccation survival in A. baumannii.
FIG 8 .
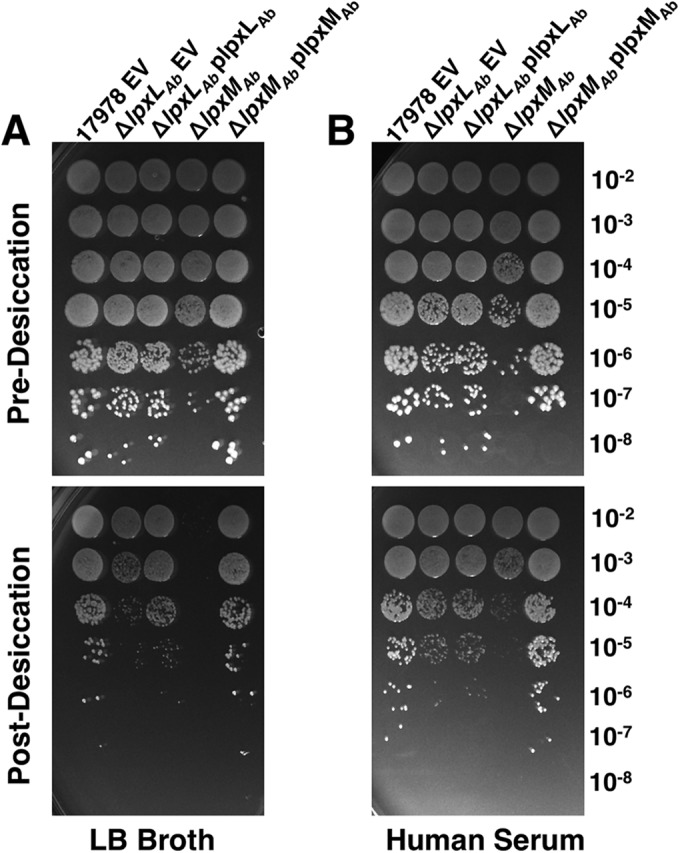
LpxMAb contributes to A. baumannii desiccative survival. An A. baumannii desiccation survival assay was performed. A. baumannii strains, including ATCC 17978 carrying an empty vector (EV), at an OD600 of 0.1 were serially diluted and spotted onto LB plates before and after desiccation on polystyrene. Cells were washed in LB broth (A) or human serum (B) before desiccation.
DISCUSSION
The lipid A domain of LPS/LOS provides Gram-negative bacteria with a formidable barrier to protect the cell from its environment. The work described in this study suggests that hepta-acylation of lipid A fortifies the outer membrane to protect A. baumannii from CAMPs and desiccation. While many pathogens synthesize hexa-acylated lipid A under standard growth conditions, PagP-dependent hepta-acylation of lipid A occurs upon exposure to stressful factors (10). Hepta-acylation reinforces the outer membrane by packing the hydrophobic membrane with palmitate added at the R-2-hydroxymyristate position of lipid A (37). PagP-dependent acylation of lipid A protects Gram-negative bacteria from antimicrobial peptides in vertebrate mucosal secretions (e.g., CAMPs) (10). Whereas most pathogens regulate synthesis of hepta-acylated lipid A by regulating PagP expression, A. baumannii does not carry a gene that encodes a PagP homolog and constitutively synthesizes hepta-acylated lipid A under standard growth conditions. The PagP-independent mechanism utilized by A. baumannii illustrates an additional strategy for the addition of a secondary acyl chain at position 2 on lipid A. Intriguingly, A. baumannii could have evolved its PagP-independent mechanism to continuously produce hepta-acylated lipid A on its surface due to the selective pressures encountered in its environmental niche. Since A. baumannii is primarily found in health care settings, assault by hospital-like environmental factors may suggest why A. baumannii encodes a LpxM dual acyltransferase homolog to continually reinforce its outer membrane.
The previously characterized LpxL and LpxM acyltransferases of E. coli add acyl chains to a specific site of the lipid A molecule in an ordered fashion (5–7). In contrast, our results suggest that the A. baumannii homologs LpxLAb and LpxMAb catalyze lipid A acylation independently of the other. Furthermore, the dual acyltransferase activity of LpxMAb, illustrated in a heterologous E. coli system, involved a unique transferase activity where two lauroyl groups are attached to lipid A. While beyond the scope of this study, a more detailed examination of the LpxMAb molecular structure could reveal the requirements that mediate the relaxed acyltransferase fidelity. Intriguingly, Moraxella catarrhalis, which is responsible for otitis media in infants and is closely related to Acinetobacter, also carries a gene that encodes an enzyme with potential dual acyltransferase activity. While A. baumannii adds two C12:0 groups, M. catarrhalis adds two C10:0 groups to lipid A, both of which are essential for its survival in the mouse lung and human serum (55).
While the host immune system exploits a number of immune mechanisms to clear bacterial infections, a common lipid A structure, including the number, length, and arrangement of fatty acyl chains and exposed phosphates, is key (40). The human TLR-4/MD-2 receptor complex has evolved to bind hexa-acylated bis-phosphorylated lipid A with high affinity to result in strong immune activation. TLR-4-dependent pathways include MyD88- and TRIF-dependent signaling cascades, which trigger an immune response at the site of infection (56). While we cannot directly compare TLR-4/MD-2 activation by A. baumannii LOS to hepta-acylated E. coli LPS because of the different lipid A acyl chain lengths, hepta-acylated LOS extracted from wild-type A. baumannii activated a human TLR-4 reporter cell line (Fig. 5). In contrast, hexa-acylated lpxLAb and penta-acylated lpxMAb mutants failed to stimulate our reporter cell line, suggesting that the acyl chains added via these enzymes are important for human TLR-4/MD-2 recognition of Acinetobacter LOS. The inability of the lpxMAb and lpxMAb mutants to stimulate human TLR-4 illustrates the importance of acyl chain placement in human immune recognition of LPS/LOS.
In addition to immune surveillance systems, the mammalian innate immune system secretes CAMPs to deter bacterial invasion into host tissues. In Salmonella, PagP-dependent hepta-acylation of lipid A increased resistance to vertebrate CAMPs, such as C18G CAMP, which is a cytokine secreted by host cells to inhibit bacterial growth and prevent infection (57). However, hepta-acylation did not result in increased resistance to polymyxin CAMPs (10). Similar to Salmonella pagP mutants, A. baumannii lpxMAb mutants exhibited sensitivity to C18G (Fig. 6A). However, removal of lpxMAb also sensitized A. baumannii to the bactericidal effects of polymyxin CAMPs (Fig. 6B and C). Whereas pagP mutants produce hexa-acylated lipid A, our analysis showed that the A. baumannii lpxMAb mutant synthesizes penta-acylated lipid A (Fig. 2D). Therefore, we suspect that the observed polymyxin killing sensitivity results from the loss of two laurate acyl chains from A. baumannii lipid A. Importantly, LpxMAb inhibition effectively sensitizes A. baumannii cells to a wide range of host and polymyxin CAMPs, suggesting potential for enhanced drug delivery as a synergistic antimicrobial agent.
To demonstrate the essentiality of LpxMAb-dependent survival in a host, we used an invertebrate model to assess CAMP-dependent killing. Galleria mellonella provides an optimal model, because it carries a gene(s) encoding a humoral immune response that synthesizes and releases antimicrobial peptides into the hemolymph upon sensing bacterial assault (50). Our results demonstrate that deletion of lpxMAb resulted in attenuated virulence (Fig. 7). Production of antimicrobial peptides likely plays a large role in neutralizing A. baumannii infections (58). The inability of the lpxMAb mutant A. baumannii to kill wax moth larvae is likely because the humoral antimicrobial peptide response can rapidly lyse the penta-acylated lipid A lpxMAb mutant, but not cells that synthesize wild-type lipid A.
Furthermore, CAMPs that target the outer membrane such as magainin and cecropin are potential alternatives to currently prescribed antibiotics. It was previously demonstrated that magainin disorders the outer membrane acyl chains of Gram-negative bacteria, whereas cecropin disrupts lipid moieties in biological membranes to form pores (59–61). These CAMPs and countless others could act synergistically with an LpxMAb inhibitor to effectively lyse multidrug-resistant A. baumannii cells, providing an innovative antimicrobial strategy.
Unexpectedly, our analysis of lipids in the outer membrane determined that the fatty acid lipid content of lipid A is important for A. baumannii survival during desiccation (Fig. 8; see Fig. S6 in the supplemental material). While a detailed understanding of the factors required for desiccation survival in any bacterium is lacking, surface structures such as carbohydrates and oxidative damage have been hypothesized to play a role. We suspect that the penta-acylated lipid A outer membrane synthesized by the lpxMAb mutant is more fluid than the wild-type hepta-acylated membrane. Increased membrane fluidity would likely permit leakage of water and hydrophilic nutrients out of the cell. The resultant loss of essential growth factors would potentially be deadly to the bacterial cell. In this study, the penta-acylated A. baumannii bacteria could not survive for even 48 h in a desiccative environment, while the wild-type A. baumannii cells could persist under desiccative conditions for weeks and even months. Identification of outer membrane lipid species as important factors for desiccation survival may aid in the development of new antiseptics.
MATERIALS AND METHODS
Bacterial strains and growth.
All strains and plasmids used in this study are listed in Table S1 in the supplemental material. A. baumannii ATCC 17978 strains were initially grown from freezer stocks on Luria-Bertani (LB) agar at 37°C. Isolated colonies were used to inoculate LB broth at 37°C. Carbenicillin was used at 150 µg/ml, and kanamycin was used at 25 or 7.5 µg/ml for selection.
Construction of mutant and complementation A. baumannii strains.
Primers used in this study are listed in Table S2 in the supplemental material. All A. baumannii genetic mutants were engineered as previously described (31). In brief, A. baumannii ATCC 17978 carrying plasmid pMMB67EH containing the RECAb coding sequences was diluted from a culture grown overnight into LB broth at an optical density at 600 nm (OD600) of 0.05 and grown for 45 min. Expression of RECAb (pAT03) was induced by the addition of 2 mM IPTG, and cells were grown at 37°C until they were in mid-log growth phase (OD600 of 0.4). After the cells were washed in ice-cold 10% glycerol, 1010 cells were electroporated in a 2-mm cuvette at 1.8 mV with 5 µg of a recombineering linear PCR product. The cells were immediately grown for 4 h in 4 ml of LB broth with 2 mM IPTG and plated on LB agar containing 7.5 µg/ml or 25 µg/ml of kanamycin. All mutants were verified by PCR and sequenced.
To cure isolated mutants of the pMMB67EH::RECAb Tetr plasmid following mutant isolation, strains were streaked for isolated colonies on 2 mM NiCl2 to select for cells that have lost the tetracycline cassette (62). Cured mutants were electroporated with pMMBR67EH carrying the FLP recombinase (pAT04). Cells were recovered for 1 h in 4 ml of LB broth and plated on LB agar containing 2 mM IPTG to induce expression of the FLP recombinase. Excision of the kanamycin cassette was confirmed by PCR and sequenced.
To complement A. baumannii mutants, the coding sequences from A1S_0431 (lpxLAb) and A1S_2609 (lpxMAb) were cloned into the KpnI and SalI sites in pMMB67EH. The plasmids were expressed in the respective mutants, and all strains were grown in 0.05 mM IPTG for expression.
Isolation of lipid A.
Isolation of lipid A for TLC analysis involved 32P radiolabeling of whole cells as previously described (63). In brief, 14 ml of A. baumannii was grown at 37°C to an OD600 of 1.0. Bacteria were harvested by centrifugation at 10,000 × g for 10 min. Lipid A extraction was carried out by mild-acid hydrolysis as previously described (64).
Isolation and quantification of LOS.
LOS was isolated as previously described (63). In brief, A. baumannii (1 liter) was grown at 37°C to an OD600 of 1.0. Cells were harvested by centrifugation and lyophilized overnight. The phenol-water method was used to isolate LOS (65). Quantification of LOS was performed by using the 3-deoxy-3-manno-octulosonic acid (Kdo) colorimetric assay as previously described (66).
Mass spectrometry.
For mass spectrometry, lipid A was analyzed using a MALDI-TOF/TOF (ABI 4700 proteom ics analyzer) mass spectrometer in the negative ion mode as previously described (63). All UVPD and HCD spectra were collected in the negative ion mode on a Thermo Scientific Orbitrap Elite mass spectrometer (Bremen, Germany) modified with a Coherent ExciStar XS ArF excimer laser (Santa Clara, CA), as previously described (34, 35). HCD was performed with the normalized collision energy (NCE) of 55% with a cell pressure of approximately 0.13 × 102 Pa. UVPD was performed with the laser emitting 193-nm photons at 4 mJ per laser pulse with four pulses per scan. The laser pulse repetition rate was 500 Hz. The instrument was operated at 120,000 resolving power with a precursor isolation window of 5 m/z. All samples were dissolved in 50:50 methanol (MeOH)-CHCl3 and directly infused into the mass spectrometer at a rate of 3 µl/min with a spray voltage of 4 kV. The presented HCD and UVPD spectra are composed of 104to 330 averages.
TLR-4 signaling assays.
The HEK-blue human TLR-4 (hTLR-4) cell line was maintained according to the manufacturer’s specifications (InvivoGen). LOS samples were serially diluted for assays as previously described (9, 63). At least two biological replicates were each done in triplicate, and one representative set was shown.
Bactericidal assays.
Overnight cultures of A. baumannii strains were diluted to an OD600 of 0.15, and 100 µl was added to three replicate wells on a 96-well microtiter plate. CAMPs were added to each well at the desired concentration, and the plate was incubated at 37°C for 2 h (10). Serial dilutions were plated on LB agar and incubated overnight at 37°C. Colony counts were reported as CFU per milliliter. Two biological replicates were performed in triplicate for each strain, and all groups were included in the reported data.
Desiccation assays.
Wild-type and mutant A. baumannii strains were diluted to OD600s of 0.1 and 0.01 in 10-µl and 100-µl volumes, respectively, and diluted in LB broth or human serum. Predesiccated samples were serially diluted in 10-fold increments and spotted. Samples were desiccated on polystyrene at 25°C and 40% humidity for 48 h. Following desiccation, samples were resuspended in LB broth, serially diluted in 10-fold increments, and spotted. Survival of each strain was assessed by the relative loss of CFU compared to the wild type.
Galleria mellonella virulence assay.
The Galleria mellonella virulence assay was performed as previously described (45). Larvae (Vanderhorst, Inc., St. Marys, OH) were stored in the dark and used within 7 days of receipt. Ten randomly chosen caterpillars of similar weight were used for each group in the experiment. Following bacterial injection, caterpillars were stored at 37°C and monitored every 12 h for death. Caterpillars were considered dead when they became nonresponsive to touch. The experiment was performed in triplicate with data from one representative group reported.
SUPPLEMENTAL MATERIAL
Growth curve of A. baumannii strains. A. baumannii strains were inoculated into fresh LB broth at an OD600 of 0.1 in triplicate in a microtiter plate for 16 h at 37°C. Absorbance was read every 0.5 h. Download
MALDI-TOF MS of complemented strains of A. baumannii. Lipid A was isolated from complemented strains of A. baumannii and analyzed via MALDI-TOF MS as described in the text. Download
HCD and UVPD MS of wild-type A. baumannii lipid A. (A and B) HCD (A) and UVPD (B) spectra for m/z 1,911.29 with the corresponding fragmentation map and fragment list. The numbers next to each m/z value in the fragment lists correspond to all cleavage sites or combination cleavage sites on the fragmentation map that result in that m/z. For the sake of simplicity, only informative/nonredundant ions are labeled. Download
HCD and UVPD MS of ΔlpxLAb lipid A. (A and B) HCD (A) and UVPD (B) spectra for m/z 1,712.12 with the corresponding fragmentation map and fragment list. The numbers next to each m/z value in the fragment lists correspond to all cleavage sites or combination cleavage sites on the fragmentation map that result in that m/z. For the sake of simplicity, only informative/nonredundant ions are labeled. Download
LOS isolated from A. baumannii strains. LOS isolated from wild-type, mutant, and complemented A. baumannii strains was separated by SDS-PAGE and stained using Pro-Q emerald 300 lipopolysaccharide gel stain kit (Life Technologies). Download
LpxMAb contributes to A. baumannii desiccative survival. An A. baumannii desiccation survival assay was performed. A. baumannii strains at an OD600 of 0.01 were serially diluted and spotted onto LB plates before and after desiccation on polystyrene. (A and B) Cells were washed in LB (A) or human serum (B) before desiccation. Download
Strains and plasmids used in this study.
Primers used in this study.
ACKNOWLEDGMENTS
Funding from NIH (grants AI064184 and AI076322 to M.S.T., grant F32GM113488 to J.M.B. and grant GM103655 to J.S.B.), the Welch Foundation (grant F-1155 to J.S.B.), and the Army Research Office (grant W911NF-12-1-0390 to M.S.T.) is gratefully acknowledged.
Footnotes
Citation Boll JM, Tucker AT, Klein DR, Beltran AM, Brodbelt JS, Davies BW, Trent MS. 2015. Reinforcing lipid A acylation on the cell surface of Acinetobacter baumannii promotes cationic antimicrobial peptide resistance and desiccation survival. mBio 6(3):e00478-15. doi:10.1128/mBio.00478-15.
REFERENCES
- 1.Whitfield C, Trent MS. 2014. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem 83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 2.Coggins BE, Li X, McClerren AL, Hindsgaul O, Raetz CRH, Zhou P. 2003. Structure of the LpxC deacetylase with a bound substrate-analog inhibitor. Nat Struct Biol 10:645–651. doi: 10.1038/nsb948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McClerren AL, Endsley S, Bowman JL, Andersen NH, Guan Z, Rudolph J, Raetz CRH. 2005. A slow, tight-binding inhibitor of the zinc-dependent deacetylase LpxC of lipid A biosynthesis with antibiotic activity comparable to ciprofloxacin. Biochemistry 44:16574–16583. doi: 10.1021/bi0518186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng D, Zhao J, Chung HS, Guan Z, Raetz CRH, Zhou P. 2013. Mutants resistant to LpxC inhibitors by rebalancing cellular homeostasis. J Biol Chem 288:5475–5486. doi: 10.1074/jbc.M112.447607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brozek KA, Raetz CR. 1990. Biosynthesis of lipid A in Escherichia coli. Acyl carrier protein-dependent incorporation of laurate and myristate. J Biol Chem 265:15410–15417. [PubMed] [Google Scholar]
- 6.Clementz T, Bednarski JJ, Raetz CR. 1996. Function of the htrB high temperature requirement gene of Escherichia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J Biol Chem 271:12095–12102. doi: 10.1074/jbc.271.20.12095. [DOI] [PubMed] [Google Scholar]
- 7.Clementz T, Zhou Z, Raetz CR. 1997. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A. Acylation by MsbB follows laurate incorporation by HtrB. J Biol Chem 272:10353–10360. doi: 10.1074/jbc.272.16.10353. [DOI] [PubMed] [Google Scholar]
- 8.Needham BD, Trent MS. 2013. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11:467–481. doi: 10.1038/nrmicro3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Needham BD, Carroll SM, Giles DK, Georgiou G, Whiteley M, Trent MS. 2013. Modulating the innate immune response by combinatorial engineering of endotoxin. Proc Natl Acad Sci U S A 110:1464–1469. doi: 10.1073/pnas.1218080110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen HD, Groisman EA. 2013. The biology of the PmrA/PmrB two-component system: the major regulator of lipopolysaccharide modifications. Annu Rev Microbiol 67:83–112. doi: 10.1146/annurev-micro-092412-155751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trent MS, Stead CM, Tran AX, Hankins JV. 2006. Diversity of endotoxin and its impact on pathogenesis. J Endotoxin Res 12:205–223. doi: 10.1179/096805106X118825. [DOI] [PubMed] [Google Scholar]
- 12.Guo L, Lim KB, Poduje CM, Daniel M, Gunn JS, Hackett M, Miller SI. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189–198. doi: 10.1016/S0092-8674(00)81750-X. [DOI] [PubMed] [Google Scholar]
- 13.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, Raetz CR. 2000. Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J 19:5071–5080. doi: 10.1093/emboj/19.19.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fukuoka S, Howe J, Andrä J, Gutsmann T, Rössle M, Brandenburg K. 2008. Physico-chemical and biophysical study of the interaction of hexa- and heptaacyl lipid A from Erwinia carotovora with magainin 2-derived antimicrobial peptides. Biochim Biophys Acta 1778:2051–2057. doi: 10.1016/j.bbamem.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 15.Beceiro A, Llobet E, Aranda J, Bengoechea JA, Doumith M, Hornsey M, Dhanji H, Chart H, Bou G, Livermore DM, Woodford N. 2011. Phosphoethanolamine modification of lipid A in colistin-resistant variants of Acinetobacter baumannii mediated by the pmrAB two-component regulatory system. Antimicrob Agents Chemother 55:3370–3379. doi: 10.1128/AAC.00079-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arroyo LA, Herrera CM, Fernandez L, Hankins JV, Trent MS, Hancock REW. 2011. The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 55:3743–3751. doi: 10.1128/AAC.00256-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falagas ME, Bliziotis IA. 2007. Pandrug-resistant Gram-negative bacteria: the dawn of the post-antibiotic era? Int J Antimicrob Agents 29:630–636. doi: 10.1016/j.ijantimicag.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Manchanda V, Sanchaita S, Singh N. 2010. Multidrug resistant Acinetobacter. J Glob Infect Dis 2:291–304. doi: 10.4103/0974-777X.68538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang Y-S, Park W. 2010. Trade-off between antibiotic resistance and biological fitness in Acinetobacter sp. strain DR1. Environ Microbiol 12:1304–1318. doi: 10.1111/j.1462-2920.2010.02175.x. [DOI] [PubMed] [Google Scholar]
- 20.López-Rojas R, Domínguez-Herrera J, McConnell MJ, Docobo-Peréz F, Smani Y, Fernández-Reyes M, Rivas L, Pachón J. 2011. Impaired virulence and in vivo fitness of colistin-resistant Acinetobacter baumannii. J Infect Dis 203:545–548. doi: 10.1093/infdis/jiq086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beceiro A, Moreno A, Fernández N, Vallejo JA, Aranda J, Adler B, Harper M, Boyce JD, Bou G. 2014. Biological cost of different mechanisms of colistin resistance and their impact on virulence in Acinetobacter baumannii. Antimicrob Agents Chemother 58:518–526. doi: 10.1128/AAC.01597-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan P-C, Huang L-M, Lin H-C, Chang L-Y, Chen M-L, Lu C-Y, Lee P-I, Chen J-M, Lee C-Y, Pan H-J, Wang J-T, Chang S-C, Chen Y-C. 2007. Control of an outbreak of pandrug-resistant Acinetobacter baumannii colonization and infection in a neonatal intensive care unit. Infect Control Hosp Epidemiol 28:423–429. doi: 10.1086/513120. [DOI] [PubMed] [Google Scholar]
- 23.Falagas ME, Karveli EA, Siempos II, Vardakas KZ. 2008. Acinetobacter infections: a growing threat for critically ill patients. Epidemiol Infect 136:1009–1019. doi: 10.1017/S0950268807009478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Souli M, Galani I, Giamarellou H. 2008. Emergence of extensively drug-resistant and pandrug-resistant Gram-negative bacilli in Europe. Euro Surveill 13(47):pii=19045 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=19045. [PubMed] [Google Scholar]
- 25.Moffatt JH, Harper M, Harrison P, Hale JDF, Vinogradov E, Seemann T, Henry R, Crane B, St Michael F, Cox AD, Adler B, Nation RL, Li J, Boyce JD. 2010. Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob Agents Chemother 54:4971–4977. doi: 10.1128/AAC.00834-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas S, Brunel J-M, Dubus J-C, Reynaud-Gaubert M, Rolain J-M. 2012. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther 10:917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 27.Jawad A, Seifert H, Snelling AM, Heritage J, Hawkey PM. 1998. Survival of Acinetobacter baumannii on dry surfaces: comparison of outbreak and sporadic isolates. J Clin Microbiol 36:1938–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rubin EJ, O’Brien JP, Ivanov PL, Brodbelt JS, Trent MS. 2014. Identification of a broad family of lipid A late acyltransferases with non-canonical substrate specificity. Mol Microbiol 91:887–899. doi: 10.1111/mmi.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bishop RE, Kim S-H, El Zoeiby A. 2005. Role of lipid A palmitoylation in bacterial pathogenesis. J Endotoxin Res 11:174–180. doi: 10.1179/096805105X35242. [DOI] [PubMed] [Google Scholar]
- 30.Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A 86:5054–5058. doi: 10.1073/pnas.86.13.5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tucker AT, Nowicki EM, Boll JM, Knauf GA, Burdis NC, Trent MS, Davies BW. 2014. Defining gene-phenotype relationships in Acinetobacter baumannii through one-step chromosomal gene inactivation. mBio 5(4):e01313-14. doi: 10.1128/mBio.01313-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luke NR, Sauberan SL, Russo TA, Beanan JM, Olson R, Loehfelm TW, Cox AD, St Michael F, Vinogradov EV, Campagnari AA. 2010. Identification and characterization of a glycosyltransferase involved in Acinetobacter baumannii lipopolysaccharide core biosynthesis. Infect Immun 78:2017–2023. doi: 10.1128/IAI.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hood MI, Becker KW, Roux CM, Dunman PM, Skaar EP. 2013. Genetic determinants of intrinsic colistin tolerance in Acinetobacter baumannii. Infect Immun 81:542–551. doi: 10.1128/IAI.00704-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw JB, Li W, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS. 2013. Complete protein characterization using top-down mass spectrometry and ultraviolet photodissociation. J Am Chem Soc 135:12646–12651. doi: 10.1021/ja4029654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Brien JP, Needham BD, Henderson JC, Nowicki EM, Trent MS, Brodbelt JS. 2014. 193 nm ultraviolet photodissociation mass spectrometry for the structural elucidation of lipid A compounds in complex mixtures. Anal Chem 86:2138–2145. doi: 10.1021/ac403796n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raetz CRH. 1990. Biochemistry of endotoxins. Annu Rev Biochem 59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 37.Bishop RE. 2005. The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol Microbiol 57:900–912. doi: 10.1111/j.1365-2958.2005.04711.x. [DOI] [PubMed] [Google Scholar]
- 38.Trent MS, Pabich W, Raetz CRH, Miller SI. 2001. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J Biol Chem 276:9083–9092. doi: 10.1074/jbc.M010730200. [DOI] [PubMed] [Google Scholar]
- 39.Takeda K, Kaisho T, Akira S. 2003. Toll-like receptors. Annu Rev Immunol 21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 40.Park BS, Song DH, Kim HM, Choi B-S, Lee H, Lee J-O. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 41.Kawai T, Akira S. 2005. Pathogen recognition with Toll-like receptors. Curr Opin Immunol 17:338–344. doi: 10.1016/j.coi.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 42.Hancock RE, Falla T, Brown M. 1995. Cationic bactericidal peptides. Adv Microb Physiol 37:135–175. doi: 10.1016/S0065-2911(08)60145-9. [DOI] [PubMed] [Google Scholar]
- 43.Guina T, Yi EC, Wang H, Hackett M, Miller SI. 2000. A PhoP-regulated outer membrane protease of Salmonella enterica serovar typhimurium promotes resistance to alpha-helical antimicrobial peptides. J Bacteriol 182:4077–4086. doi: 10.1128/JB.182.14.4077-4086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carr C, Morrison DC. 1985. Mechanism of polymyxin B-mediated lysis of lipopolysaccharide-treated erythrocytes. Infect Immun 49:84–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peleg AY, Jara S, Monga D, Eliopoulos GM, Moellering RC, Mylonakis E. 2009. Galleria mellonella as a model system to study Acinetobacter baumannii pathogenesis and therapeutics. Antimicrob Agents Chemother 53:2605–2609. doi: 10.1128/AAC.01533-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antunes LC, Imperi F, Carattoli A, Visca P. 2011. Deciphering the multifactorial nature of Acinetobacter baumannii pathogenicity. PLoS One 6:e22674. doi: 10.1371/journal.pone.0022674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Iwashkiw JA, Seper A, Weber BS, Scott NE, Vinogradov E, Stratilo C, Reiz B, Cordwell SJ, Whittal R, Schild S, Feldman MF. 2012. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog 8:e1002758. doi: 10.1371/journal.ppat.1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, Calderwood SB, Ausubel FM, Diener A. 2005. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun 73:3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jander G, Rahme LG, Ausubel FM. 2000. Positive correlation between virulence of Pseudomonas aeruginosa mutants in mice and insects. J Bacteriol 182:3843–3845. doi: 10.1128/JB.182.13.3843-3845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ratcliffe NA. 1985. Invertebrate immunity−a primer for the non-specialist. Immunol Lett 10:253–270. doi: 10.1016/0165-2478(85)90100-2. [DOI] [PubMed] [Google Scholar]
- 51.Cytryńska M, Mak P, Zdybicka-Barabas A, Suder P, Jakubowicz T. 2007. Purification and characterization of eight peptides from Galleria mellonella immune hemolymph. Peptides 28:533–546. doi: 10.1016/j.peptides.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 52.Peleg AY, Seifert H, Paterson DL. 2008. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev 21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roberson EB, Firestone MK. 1992. Relationship between desiccation and exopolysaccharide production in a soil Pseudomonas sp. Appl Environ Microbiol 58:1284–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Potts M. 1994. Desiccation tolerance of prokaryotes. Microbiol Rev 58:755–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gao S, Peng D, Zhang W, Muszyński A, Carlson RW, Gu X-X. 2008. Identification of two late acyltransferase genes responsible for lipid A biosynthesis in Moraxella catarrhalis. FEBS J 275:5201–5214. doi: 10.1111/j.1742-4658.2008.06651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kagan JC, Medzhitov R. 2006. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell 125:943–955. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 57.Darveau RP, Blake J, Seachord CL, Cosand WL, Cunningham MD, Cassiano-Clough L, Maloney G. 1992. Peptides related to the carboxyl terminus of human platelet factor IV with antibacterial activity. J Clin Invest 90:447–455. doi: 10.1172/JCI115880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogel H, Altincicek B, Glöckner G, Vilcinskas A. 2011. A comprehensive transcriptome and immune-gene repertoire of the lepidopteran model host Galleria mellonella. BMC Genomics 12:308. doi: 10.1186/1471-2164-12-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rana FR, Sultany CM, Blazyk J. 1990. Interactions between Salmonella typhimurium lipopolysaccharide and the antimicrobial peptide, magainin 2 amide. FEBS Lett 261:464–467. doi: 10.1016/0014-5793(90)80616-Q. [DOI] [PubMed] [Google Scholar]
- 60.Silvestro L, Gupta K, Weiser JN, Axelsen PH. 1997. The concentration-dependent membrane activity of cecropin A. Biochemistry 36:11452–11460. doi: 10.1021/bi9630826. [DOI] [PubMed] [Google Scholar]
- 61.Silvestro L, Axelsen PH. 2000. Membrane-induced folding of cecropin A. Biophys J 79:1465–1477. doi: 10.1016/S0006-3495(00)76398-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Podolsky T, Fong ST, Lee BT. 1996. Direct selection of tetracycline-sensitive Escherichia coli cells using nickel salts. Plasmid 36:112–115. doi: 10.1006/plas.1996.0038. [DOI] [PubMed] [Google Scholar]
- 63.Hankins JV, Madsen JA, Giles DK, Childers BM, Klose KE, Brodbelt JS, Trent MS. 2011. Elucidation of a novel Vibrio cholerae lipid A secondary hydroxy-acyltransferase and its role in innate immune recognition. Mol Microbiol 81:1313–1329. doi: 10.1111/j.1365-2958.2011.07765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou Z, Lin S, Cotter RJ, Raetz CR. 1999. Lipid A modifications characteristic of Salmonella typhimurium are induced by NH4VO3 in Escherichia coli K12. Detection of 4-amino-4-deoxy-l-arabinose, phosphoethanolamine and palmitate. J Biol Chem 274:18503–18514. doi: 10.1074/jbc.274.26.18503. [DOI] [PubMed] [Google Scholar]
- 65.Jann K, Jann B, Orskov F, Orskov I, Westphal O. 1965. Immunochemical studies of K antigens from Escherichia coli. II. K antigen from E. coli 08:K42(A):H-. Biochem Z 342:1–22. (In German.) [PubMed] [Google Scholar]
- 66.Marolda CL, Lahiry P, Vinés E, Saldías S, Valvano MA. 2006. Micromethods for the characterization of lipid A-core and O-antigen lipopolysaccharide. Methods Mol Biol 347:237–252. doi: 10.1385/1-59745-167-3:237. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth curve of A. baumannii strains. A. baumannii strains were inoculated into fresh LB broth at an OD600 of 0.1 in triplicate in a microtiter plate for 16 h at 37°C. Absorbance was read every 0.5 h. Download
MALDI-TOF MS of complemented strains of A. baumannii. Lipid A was isolated from complemented strains of A. baumannii and analyzed via MALDI-TOF MS as described in the text. Download
HCD and UVPD MS of wild-type A. baumannii lipid A. (A and B) HCD (A) and UVPD (B) spectra for m/z 1,911.29 with the corresponding fragmentation map and fragment list. The numbers next to each m/z value in the fragment lists correspond to all cleavage sites or combination cleavage sites on the fragmentation map that result in that m/z. For the sake of simplicity, only informative/nonredundant ions are labeled. Download
HCD and UVPD MS of ΔlpxLAb lipid A. (A and B) HCD (A) and UVPD (B) spectra for m/z 1,712.12 with the corresponding fragmentation map and fragment list. The numbers next to each m/z value in the fragment lists correspond to all cleavage sites or combination cleavage sites on the fragmentation map that result in that m/z. For the sake of simplicity, only informative/nonredundant ions are labeled. Download
LOS isolated from A. baumannii strains. LOS isolated from wild-type, mutant, and complemented A. baumannii strains was separated by SDS-PAGE and stained using Pro-Q emerald 300 lipopolysaccharide gel stain kit (Life Technologies). Download
LpxMAb contributes to A. baumannii desiccative survival. An A. baumannii desiccation survival assay was performed. A. baumannii strains at an OD600 of 0.01 were serially diluted and spotted onto LB plates before and after desiccation on polystyrene. (A and B) Cells were washed in LB (A) or human serum (B) before desiccation. Download
Strains and plasmids used in this study.
Primers used in this study.