ABSTRACT
Influenza A and B viruses are human pathogens that are regarded to cause almost equally significant disease burdens. Neuraminidase (NA) inhibitors (NAIs) are the only class of drugs available to treat influenza A and B virus infections, so the development of NAI-resistant viruses with superior fitness is a public health concern. The fitness of NAI-resistant influenza B viruses has not been widely studied. Here we examined the replicative capacity and relative fitness in normal human bronchial epithelial (NHBE) cells of recombinant influenza B/Yamanashi/166/1998 viruses containing a single amino acid substitution in NA generated by reverse genetics (rg) that is associated with NAI resistance. The replication in NHBE cells of viruses with reduced inhibition by oseltamivir (recombinant virus with the E119A mutation generated by reverse genetics [rg-E119A], rg-D198E, rg-I222T, rg-H274Y, rg-N294S, and rg-R371K, N2 numbering) or zanamivir (rg-E119A and rg-R371K) failed to be inhibited by the presence of the respective NAI. In a fluorescence-based assay, detection of rg-E119A was easily masked by the presence of NAI-susceptible virus. We coinfected NHBE cells with NAI-susceptible and -resistant viruses and used next-generation deep sequencing to reveal the order of relative fitness compared to that of recombinant wild-type (WT) virus generated by reverse genetics (rg-WT): rg-H274Y > rg-WT > rg-I222T > rg-N294S > rg-D198E > rg-E119A ≫ rg-R371K. Based on the lack of attenuated replication of rg-E119A in NHBE cells in the presence of oseltamivir or zanamivir and the fitness advantage of rg-H274Y over rg-WT, we emphasize the importance of these substitutions in the NA glycoprotein. Human infections with influenza B viruses carrying the E119A or H274Y substitution could limit the therapeutic options for those infected; the emergence of such viruses should be closely monitored.
IMPORTANCE Influenza B viruses are important human respiratory pathogens contributing to a significant portion of seasonal influenza virus infections worldwide. The development of resistance to a single class of available antivirals, the neuraminidase (NA) inhibitors (NAIs), is a public health concern. Amino acid substitutions in the NA glycoprotein of influenza B virus not only can confer antiviral resistance but also can alter viral fitness. Here we used normal human bronchial epithelial (NHBE) cells, a model of the human upper respiratory tract, to examine the replicative capacities and fitness of NAI-resistant influenza B viruses. We show that virus with an E119A NA substitution can replicate efficiently in NHBE cells in the presence of oseltamivir or zanamivir and that virus with the H274Y NA substitution has a relative fitness greater than that of the wild-type NAI-susceptible virus. This study is the first to use NHBE cells to determine the fitness of NAI-resistant influenza B viruses.
INTRODUCTION
Influenza B viruses are important human respiratory pathogens causing a significant disease burden. Although the consequences of influenza B virus infections on human influenza disease in epidemic seasons were frequently discounted in the past, they are now viewed as being almost equal to those of influenza A virus infections (1, 2). From the 2004-2005 to the 2013-2014 influenza seasons, influenza B viruses comprised, on average, 21.8% of the influenza viruses circulating in the United States (peak, 35.7% in the 2012-2013 influenza season), with the percentage of influenza-associated pediatric deaths being attributable to influenza B virus averaging 26.9% (peak, 51.9% in the 2012-2013 influenza season). Clinical reports also suggest that links exist between influenza B virus and lethal secondary bacterial infections and myocardial or neurological complications (3–7).
Annual vaccination is an effective method for controlling influenza disease. The current FDA-approved quadrivalent seasonal influenza vaccine includes both antigenically distinct hemagglutinin (HA) lineages of influenza B virus (i.e., Yamagata and Victoria) (8, 9). Antiviral treatment is another option for the control of influenza, and the neuraminidase (NA) inhibitors (NAIs) are currently the only class of antivirals approved for prophylaxis and treatment of influenza B virus infections. NAIs limit influenza disease by competitively binding the NA active site, inhibiting NA-mediated cleavage of cell surface and virus-associated sialic acids, and preventing the release and spread of influenza virus. The FDA-approved NAIs in the United States are oral oseltamivir, inhaled zanamivir, and intravenous peramivir (10).
For influenza A and B viruses, the development of NAI resistance is associated with amino acid substitutions in NA, typically at 1 of 19 highly conserved residues in or near the NA active site (11, 12). These amino acids are principally responsible for the sialidase activity of the NA enzyme, as they either directly contact the terminal sialic acid (catalytic residues R118, D151, R152, R224, E276, R292, R371, and Y406; the N2 numbering is used here and throughout the text) or support the NA enzymatic binding pocket (framework residues E119, R156, W178, S179, D198, I222, E227, H274, E277, N294, and E425). Substitutions at these conserved residues disrupt NAI inhibition, while at least some NA sialidase activity is maintained (13, 14). Occasionally, substitutions affecting NAI inhibition are identified elsewhere in the NA protein of influenza B virus; these may reduce inhibition by altering NA glycosylation (G142R and N146K) or NA tetramer stabilization (E105K) (12, 15, 16). The World Health Organization's (WHO's) Global Influenza Surveillance and Response System recommends monitoring influenza B viruses for 6 single amino acid substitutions in NA (R152K, D198E, D198N, I222T, N294S, and G402S) that reduce inhibition by NAIs (17).
Importantly, such substitutions can affect influenza virus fitness, i.e., the summation of parameters that quantify the degree of virus adaptation to a given environment (18). Influenza virus populations naturally exist as quasispecies, or pools of individuals with diverse genetic variations. Influenza virus variants compete against one another, and those best fit to their environment come to dominate the population during infection. The capacity of the NAI-resistant influenza virus to efficiently transmit among humans can be an important indicator of fitness advantages (19). To date, only sporadic transmission of NAI-resistant influenza B viruses has been reported, with small clusters of viruses containing either the D198N, I222T, or I222V NA substitution being identified from Japan, China, or the United States, respectively (20–23). Additionally, limited information about the consequence of NAI resistance-associated substitutions on the fitness of influenza B viruses is available from animal models. In ferrets, the “gold standard” animal model for evaluating influenza A virus pathogenesis and transmission, influenza B virus infections are generally mild and are not known to be transmissible (24–26). In ferret coinfection models, the catalytic site substitution R152K was found to be deleterious to influenza B virus replication, and a virus carrying the framework substitution D198N was shown to have replication kinetics similar to those of an NAI-susceptible wild-type (WT) virus (27, 28). Hartley strain guinea pigs have recently been shown to be a suitable model for the transmission of influenza B virus but fail to develop pathogenic infections (29). Infection of standard mouse models with influenza B virus is generally not informative, as it produces subclinical infections; lethal infections require mouse-adapted virus strains or gene-knockout mice (30, 31).
In this study, we addressed the question of whether NAI-resistant influenza B viruses may retain fitness comparable to that of a susceptible counterpart. Using 6 recombinant NAI-resistant B/Yamagata/166/1998 influenza viruses, each containing a single substitution in NA generated by reverse genetics (rg), we applied a competitive coinfection model in differentiated normal human bronchial epithelial (NHBE) cells. The proportions of individual variants present in the resultant mixed populations were determined by next-generation deep sequencing. Differentiated NHBE cells contain a mixture of epithelial cell types, including ciliated, nonciliated, and goblet cells. These cultures recapitulate many features of the human upper respiratory tract, which is the primary site of influenza virus infection and a major source of respiratory droplets that mediate transmission (32–34). The outcomes achieved with this highly relevant model are expected to be predictive of the outcomes of human infection by NAI-susceptible and -resistant influenza B viruses. Our results reveal that while the recombinant WT virus generated by reverse genetics (rg-WT) was able to outcompete five of the NAI-resistant viruses (recombinant virus with the E119A mutation generated by reverse genetics [rg-E119A], rg-D198E, rg-I222T, rg-N294S, and rg-R371K), the rg-H274Y NAI-resistant variant was more fit than the susceptible rg-WT strain.
MATERIALS AND METHODS
Cells.
Madin-Darby canine kidney (MDCK) cells were obtained from the American Type Culture Collection and were maintained as previously described (35). NHBE cells (lot number 0000105104; Lonza) were plated on collagen-coated Transwell inserts with 0.4-μm pores (Corning), brought to the air-liquid interface, and differentiated over a period of 4 to 6 weeks (35–37). Differentiated cells were maintained for 1 month before infection, and mucus was removed by washing the inserts every 2 to 3 days with Hanks' balanced salt solution (HBSS).
Compounds.
The NAIs oseltamivir carboxylate [oseltamivir, ethyl (3R,4R,5S)-4-acetamido-5-amino-3-(1-ethylpropoxy)-1-cyclohexene-1-carboxylate] and zanamivir (2,4-dideoxy-2,3-didehydro-4-guanidineosialic acid) were provided by Hoffmann-La Roche. The compounds were dissolved in sterile distilled water and stored in aliquots at −20°C until use.
Generation of recombinant influenza B viruses.
Experimental protocols were approved by the Institutional Biosafety Committee of St. Jude Children's Research Hospital. Recombinant B/Yamanashi/166/1998 (Yamagata lineage) WT (rg-WT) or single substitution-containing influenza viruses (rg-E119A, rg-D198E, rg-I222T, rg-H274Y, rg-N294S, rg-R371K) were rescued by using pAD3000 plasmid vectors (38), and their NA sequences were confirmed as previously described (35).
Virus infectivity.
The infectivity of the virus stocks was determined by performing plaque assays in MDCK cells. Briefly, confluent monolayers of MDCK cells in 6-well plates were infected with one-half-log serial dilutions of virus stock, rinsed with phosphate-buffered saline (PBS), overlaid with medium containing 0.4% agarose (MP Biochemicals) and 1 μg/ml l-tosylamido-2-phenylmethyl chloromethyl ketone (TPCK)-treated trypsin (Worthington), incubated at 33°C for 72 h, and stained with 1% crystal violet in 10% formaldehyde.
Infectivity of virus released from NHBE cells was determined by performing 50% tissue culture infective dose (TCID50) assays in MDCK cells. Briefly, 10-fold serial dilutions of virus collected in medium containing 1 μg/ml TPCK-treated trypsin were used to infect confluent monolayers of MDCK cells in a 96-well plate. After incubation at 33°C for 72 h, virus replication was detected by hemagglutination assay with 0.5% turkey red blood cells (tRBCs), and results were calculated by using the method of Reed and Muench (39).
NA enzyme inhibition assay.
The NA inhibition (NI) assay used was a modified version of the technique of Potier and colleagues (40) and was described previously (35, 41). Mixtures of NAI-susceptible (rg-WT) and -resistant recombinant B/Yamanashi/166/1998 influenza viruses containing 0, 10, 20, 50, 80, or 100% resistant virus were prepared. The percentage of NAI-susceptible and -resistant viruses was determined on the basis of the number of infectious virus particles, as determined by performing plaque assays in MDCK cells (35). Twofold dilutions of each virus mixture at each ratio were made in MES (4-morpholineethanesulfonic acid sodium salt) buffer (32.5 mM MES, 4 mM CaCl2 [Sigma-Aldrich], pH 6.5) in a black 96-well plate (Costar). The MUNANA (2′-4-methylumbelliferyl-α-d-N-acetylneuraminic acid, sodium salt hydrate; Sigma-Aldrich) substrate was added to a final concentration of 100 μM, and the mixture was incubated at 37°C for 30 min before the reaction was terminated with stop solution (0.1 M glycine in 25% ethanol, pH 10.7). Fluorescence was measured on a BioTek Synergy 2 microplate reader (BioTek) with 360-nm/460-nm excitation/emission filters. Mixtures of virus were then standardized in MES buffer to an NA enzyme activity equivalent to the fluorescence of 10 μM 4-methylumbelliferone (4-MU; Sigma-Aldrich). In a new reaction, NA activity-standardized virus mixtures were preincubated with 10-fold dilutions of an NAI (oseltamivir or zanamivir) at 37°C for 30 min. The MUNANA substrate was added, and the assay was performed as described above. Drug concentrations inhibiting NA activity by 50% (IC50s) were calculated by using the variable-slope, four-parameter dose-response curve in GraphPad Prism (v5.04) software.
Kinetics of virus replication.
The replication kinetics of recombinant influenza B viruses were assessed by the use of multistep growth curves in NHBE cells with and without NAIs. Prior to infection, NHBE cells were rinsed 3 times with HBSS and equilibrated in bronchial epithelial growth medium (BEGM; Lonza) plus 0.5% bovine serum albumin (BSA; Sigma-Aldrich) for 30 min at 37°C. Infections with virus at a multiplicity of infection (MOI) of 0.01 PFU/cell were performed in duplicate with an inoculum containing oseltamivir (10 μM) or zanamivir (10 μM). After adsorption for 1 h at 33°C, the inoculum was removed; then, all inserts were rinsed once with 0.9% saline (pH 2.2) to remove residual virus and twice with HBSS to restore the pH to 7.2. Oseltamivir or zanamivir was then added to the respective basal chamber at a 10 μM final concentration and left for the duration of the experiment. For each recombinant virus, duplicate control infections were performed in the absence of drug. Virus released by NHBE cells was collected by adding 300 μl BEGM plus 0.5% BSA to the apical surface and incubating the plate at 33°C for 30 min. Samples were collected at 24, 48, and 72 h postinfection (p.i.) and stored at −80°C until use.
Competitive fitness experiment.
After competitive coinfection, the growth of recombinant NAI-susceptible (rg-WT) and -resistant influenza B viruses was assessed in multistep growth curves. NAI-susceptible (rg-WT) and -resistant viruses were mixed in an equal ratio (50:50, determined by PFU) at an MOI of 0.01 PFU/cell per virus strain. NHBE cells were infected with the mixed inoculum at a final MOI of 0.02 PFU/cell. Infections were performed in triplicate in the presence of oseltamivir (1 or 10 μM) or zanamivir (1 or 10 μM) as described above. Infections of control wells containing each virus mixture in the absence of drug were performed in triplicate. Samples were collected at 24, 48, and 72 h p.i. and stored at −80°C until use.
RNA extractions and deep sequencing.
RNA was extracted from the virus-containing supernatants of NHBE cells or from individual plaques of MDCK cells by using a MagMAX 96 AI/ND viral RNA isolation kit (Life Technologies) and a KingFisher Flex magnetic particle processor (Thermo Fisher Scientific) and then stored at −80°C until use.
Quantification of mixed populations.
Two independent assays were used to quantify the amount of NAI-susceptible and -resistant viruses released from coinfected NHBE cells: (i) next-generation deep sequencing of the NA gene via the Illumina platform and (ii) high-resolution melt (HRM) analysis of the single-nucleotide substitution in the NA of plaque-purified viruses (42, 43). In the first assay, RNA was extracted from virus collected from NHBE cells, and the NA gene was amplified by reverse transcription-PCR (RT-PCR) using a SuperScript III One-Step RT-PCR system with Platinum Taq (Life Technologies) (forward primer, 5′-GCACTCCTAATTAGCCCTCATAGA-3′; reverse primer, 5′-TAAGGACAATTGTTCAAAC-3′). PCR amplicons were extracted from agarose gels by using a QIAquick 96 PCR purification kit (Qiagen), prepared for the library by using a 96-plex bar-coding scheme, and sequenced on the Illumina MiSeq platform (Illumina) using paired-end sequencing technology. After the indices were demultiplexed, the CLC Genomics Workbench (v6.5.1; CLCbio) was used to remove short and poor-quality reads. The remaining reads with a quality score greater than 30 were then mapped to the B/Yamanashi/166/1998 WT NA gene, and the percentage of NAI-resistant viruses present in each mixture was determined by using the quality-based variant detection algorithm, examining only the single-nucleotide variant of interest. The average read depth was 3,300, and the average quality score was 37.01. Deep sequencing was performed on the following samples of competitive mixtures at 24, 48, and 72 h p.i.: 3 replicates of the virus mixture grown without drug pressure and 2 replicates of the virus mixture grown with drug pressure (oseltamivir or zanamivir at 1 and 10 μM). Library preparation, sample quality control, flow cell preparation, and deep sequencing were performed by the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital.
For high-resolution melt analysis, viral supernatants collected from NHBE cells were serially diluted and used to infect monolayers of MDCK cells in a plaque assay. For each mixture, all 6 wells of a 6-well plate were infected with a specific virus dilution necessary to produce ∼10 to 20 plaques per well, and the plate was incubated at 33°C. Fourteen well-separated, individual plaques from each mixture were picked with wide-bore, low-binding P200 tips (Axygen) after 3 days (mixtures of rg-WT and rg-H274Y) or 4 days (to ensure that the plaque size was large enough for efficient RNA recovery; mixtures of rg-WT and rg-E119A or rg-R371K) after infection and stored in PBS at −80°C. RNA from each plaque was extracted and subjected to reverse transcription using Maxima reverse transcriptase (Thermo Fisher Scientific) with random hexamers (final concentration, 2.5 μM; Roche Applied Science). The resulting cDNA was diluted 1:4 with sterile water, and 8 μl was mixed with 2 μl NA substitution-specific primers (final concentration, 0.4 μM; primer sequences are available upon request) and 10 μl 2× PrecisionMelt Supermix (Bio-Rad) in thin-walled, white, 96-well plates (Bio-Rad) before PCR amplification in a CFX96 real-time PCR system (Bio-Rad). Melt curves of the resulting PCR amplicons were generated by measuring the fluorescence of the mixture at 0.2°C increments between 67°C and 90°C. The melt curves from the experimental samples were compared to the melt curves of plasmid templates by using Precision Melt Analysis software (v1.2; Bio-Rad).
Fitness calculations.
Calculations of fitness coefficients (fitness difference) and relative viral fitness were performed as described elsewhere (44, 45). Each fitness coefficient was calculated as the slope of a linear regression of the log ratio of resistant/susceptible virus populations over 4 time points (0, 24, 48, and 72 h p.i.). Negative values indicate a fitness advantage for the NAI-susceptible rg-WT; positive values indicate a fitness advantage for the NAI-resistant virus. For relative fitness, the selection coefficient (s) was first calculated by using the following formula:
The percentages of NAI-susceptible (WT) or -resistant (RES) influenza viruses were determined by variant call analysis of deep sequencing data as described above, where the total population of WT virus (total WT; in numbers of TCID50s/ml) was calculated from the percentage of rg-WT in the mixture and the viral titer, T is the difference in time (2 days) between the start and the end of the experiment, and δ is the estimated death rate (0.7) of NHBE cells infected with influenza B virus (46). Relative fitness was then calculated as 1 + s (44).
Statistical analyses.
Differences in virus yield in NHBE cells with and without drug pressure were tested by two-way analysis of variance (ANOVA) followed by the Bonferroni multiple-comparison posttest. Changes in the total area under the curve (AUC) of virus yield were tested by one-way ANOVA followed by Dunnett's multiple-comparison test. The correlation between the deep sequencing data and HRM analysis was tested by performing linear regression. All analyses were performed by using GraphPad Prism software (v5.04).
RESULTS
Inhibitory activity of NAIs on mixed populations of NAI-susceptible and -resistant influenza B viruses.
NAI-resistant influenza viruses often represent a subpopulation within a clinical specimen, which can complicate their detection. To gauge the severity of this problem for the influenza B viruses generated here, we determined the amount of NAI-resistant virus needed to overcome concealment by competing nonresistant virus in the same sample. To this end, WHO Antiviral Working Group criteria were used to evaluate whether samples containing different proportions of NAI-susceptible and -resistant influenza B viruses had reduced inhibition by NAIs (47–49).
For oseltamivir, mean IC50s 5-fold greater than those for rg-WT were observed for 3 mixtures in which the resistant virus comprised at least 10% (rg-D198E and rg-N294S) or 20% (rg-I222T) of the mixture (Table 1). Mixtures of rg-WT and either rg-H274Y or rg-R371K had increased mean IC50s only when the NAI-resistant population comprised at least 50% of the mixture. The oseltamivir-resistant phenotype was the most difficult to detect for rg-E119A, for which an increase in the mean IC50 was detected only when it comprised at least 80% of the viral mixture.
TABLE 1.
Inhibitory activity of oseltamivir and zanamivir on mixed populations of NAI-susceptible and -resistant recombinant influenza B viruses with single-nucleotide substitutions in NA
| NAI | Resistant virus (%)a | Mean IC50 (nM) ± SD for influenza B viruses with single NA substitutionsb |
|||||
|---|---|---|---|---|---|---|---|
| E119A | D198E | I222T | H274Y | N294S | R371K | ||
| Oseltamivir | 0 | 5.9 (1) | 5.9 (1) | 5.9 (1) | 5.9 (1) | 5.9 (1) | 5.9 (1) |
| 10 | 8.3 ± 1.7 (1) | 43.2 ± 0.1 (7) | 22.6 ± 5.0 (4) | 14.2 ± 1.2 (2) | 29.5 ± 5.6 (5) | 9.1 ± 0.7 (2) | |
| 20 | 6.9 ± 0.6 (1) | 60.2 ± 0.2 (10) | 29.3 ± 2.1 (5) | 16.0 ± 0.5 (3) | 62.0 ± 8.7 (11) | 9.3 ± 0.7 (2) | |
| 50 | 8.1 ± 0.7 (1) | 69.5 ± 0.3 (12) | 54.8 ± 6.4 (9) | 32.9 ± 1.4 (6) | 112.9 ± 11.3 (19) | 35.3 ± 2.2 (6) | |
| 80 | 51.7 ± 8.4 (9) | 57.0 ± 5.9 (10) | 77.5 ± 7.1 (13) | 46.8 ± 0.2 (8) | 149.9 ± 1.5 (25) | 199.6 ± 32.8 (34) | |
| 100 | 12,532.5 ± 750.6 (2124) | 54.4 ± 0.5 (9) | 75.2 ± 7.2 (13) | 43.5 ± 5.6 (7) | 167.2 ± 13.1 (28) | 385.7 ± 4.9 (65) | |
| Zanamivir | 0 | 1.1 (1) | 1.1 (1) | 1.1 (1) | 1.1 (1) | 1.1 (1) | 1.1 (1) |
| 10 | 1.0 ± 0.1 (1) | 1.3 ± 0.1 (1) | 1.2 ± 0.0 (1) | 0.9 ± 0.0 (1) | 0.7 ± 0.6 (1) | 1.0 ± 0.2 (1) | |
| 20 | 1.0 ± 0.0 (1) | 1.7 ± 0.3 (2) | 1.4 ± 0.1 (1) | 0.8 ± 0.0 (1) | 2.2 ± 0.1 (2) | 1.3 ± 0.2 (1) | |
| 50 | 1.3 ± 0.2 (1) | 2.0 ± 0.2 (2) | 1.7 ± 0.1 (2) | 0.7 ± 0.1 (1) | 3.0 ± 0.3 (3) | 4.3 ± 0.4 (4) | |
| 80 | 3.5 ± 0.1 (3) | 2.6 ± 0.3 (2) | 2.0 ± 0.2 (2) | 0.6 ± 0.1 (1) | 3.5 ± 0.2 (3) | 37.3 ± 0.0 (34) | |
| 100 | 6,260.0 ± 1,026.6 (5,690) | 2.6 ± 0.5 (2) | 1.4 ± 0.1 (1) | 0.5 ± 0.1 (1) | 3.9 ± 0.9 (4) | 65.7 ± 7.0 (60) | |
NAI-susceptible (rg-WT) and -resistant influenza B viruses were mixed in different ratios (on the basis of PFU).
Data were collected in fluorescence-based assays; the values shown were calculated from two independent experiments. Substitutions are identified by N2 numbering. Data in parentheses represent the fold increase over the value for the WT.
Four of the studied viruses were susceptible to zanamivir and, thus, did not have increased mean IC50s when they were present at any proportion. Mean IC50s in the presence of zanamivir were higher when rg-R371K comprised at least 80% of the mixed population. Increased IC50s were not observed with any mixture of rg-WT and rg-E119A; elevated IC50s were observed only in a homogeneous (100%) population of the NAI-resistant virus.
Thus, using a phenotypic assay, only rg-D198E, rg-I222T, and rg-N294S could be detected in a mixed population with NAI-susceptible virus when they comprised less than 50% of the viral population. Importantly, influenza B viruses carrying the E119A NA substitution could go undetected in a mixture with NAI-susceptible virus, emphasizing that both phenotypic and genotypic assays are required for its identification.
Replication kinetics of influenza B viruses in NHBE cells under oseltamivir and zanamivir pressure.
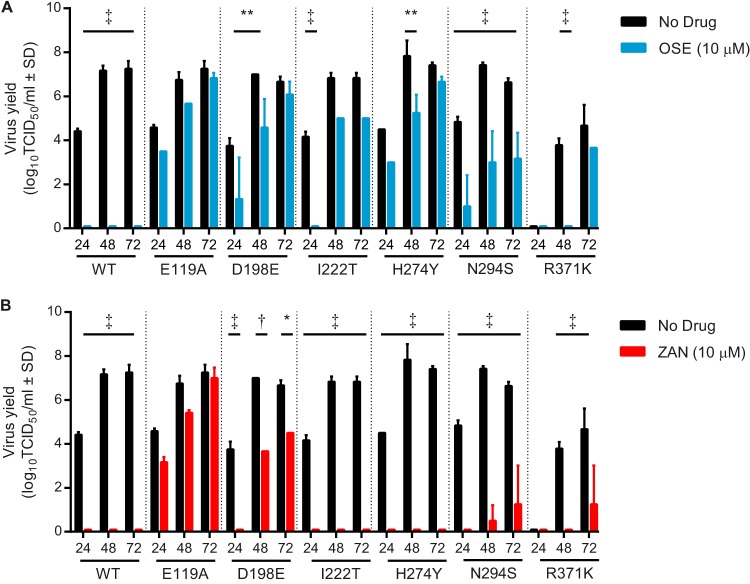
We next examined the ability of NAIs to inhibit the replication of NAI-susceptible and -resistant influenza B viruses in NHBE cells by determining the kinetics of virus growth in the absence or presence of oseltamivir (Fig. 1A) or zanamivir (Fig. 1B). In the absence of oseltamivir, all tested viruses except rg-R371K replicated productively, with virus yields being similar to those of rg-WT (Fig. 1A). The replication of rg-R371K in NHBE cells was delayed. In the presence of oseltamivir, the level of replication of rg-WT was below the limit of detection, but the replication of all 6 oseltamivir-resistant viruses was detected in NHBE cells (Fig. 1A). The yields of rg-D198E, rg-H274Y, and rg-N294S were significantly lower and the replication of rg-I222T and rg-R371K was impaired, with their detection being delayed until 48 and 72 h p.i., respectively. Importantly, the yields of rg-E119A were not impaired by oseltamivir at any time point studied.
FIG 1.
Effect of NAI treatment on replication kinetics of recombinant B/Yamanashi/166/1998 influenza viruses in NHBE cells. (A and B) Multicycle growth curves after infection of NHBE cells with recombinant B/Yamanashi/166/1998 influenza virus at an MOI of 0.01 PFU/cell grown either without drug pressure (A and B), with 10 μM oseltamivir (A), or with 10 μM zanamivir (B). Virus yield in MDCK cells was determined by the TCID50 assay at 33°C and expressed as log10 TCID50/ml. Each data point represents the mean ± SD from two independent experiments. The limit of detection was 0.75 log10 TCID50/ml. *, P < 0.05; **, P < 0.01; †, P < 0.001; ‡, P < 0.0001. Abbreviations: OSE, oseltamivir; ZAN, zanamivir.
Zanamivir inhibited or delayed the replication of the rg-WT and 4 zanamivir-susceptible viruses (Fig. 1B). Among the 2 viruses that were resistant to zanamivir (rg-E119A and rg-R371K), detection of virus replication was delayed until 72 h p.i. for rg-R371K. The replication of rg-E119A in the presence of zanamivir was not delayed, nor was the virus yield reduced at any time point studied.
Overall, all tested NAI-resistant viruses were capable of replicating in NHBE cells under oseltamivir pressure. Zanamivir inhibited or delayed the replication of viruses susceptible to the NAI. Importantly, rg-E119A had reduced inhibition by oseltamivir and zanamivir in the phenotypic assays but replicated without a significant reduction of virus yield in the presence of either NAI.
Yields of NAI-susceptible and -resistant influenza B viruses following coinfection of NHBE cells.
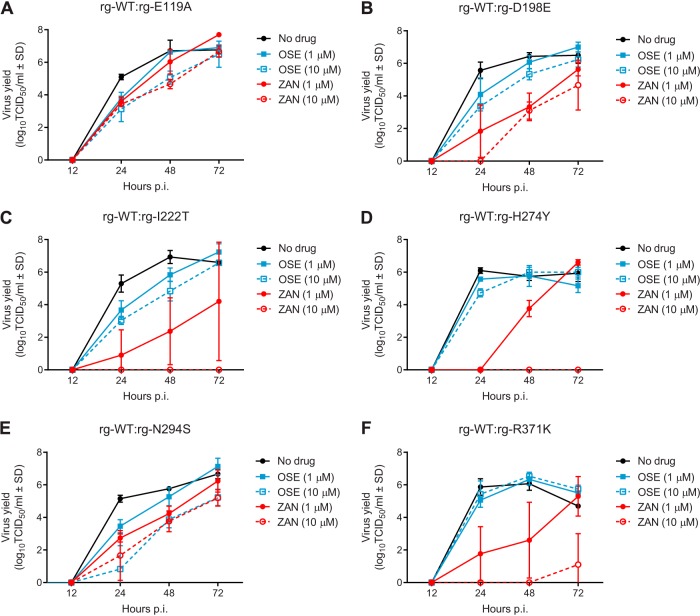
To model the competition between viral populations in the human upper respiratory tract, we coinfected NHBE cells with equal proportions of NAI-susceptible (rg-WT) and -resistant viruses and examined their replicative capacities in the absence and presence of oseltamivir (1 or 10 μM) or zanamivir (1 or 10 μM) (Fig. 2). We evaluated the efficacy of NAI at reducing the total viral load by performing area under the curve (AUC) analysis because the composition of the mixed populations was not known a priori. In the absence of NAI pressure, each pair of virus mixtures had similar total viral loads in NHBE cells (Table 2; Fig. 2A to F).
FIG 2.
Yield of NAI-susceptible and -resistant influenza B virus populations following coinfection of NHBE cells. Multicycle growth curves after coinfection of NHBE cells with mixtures of NAI-susceptible and -resistant influenza B viruses grown either without drug pressure, with oseltamivir (1 or 10 μM), or with zanamivir (1 or 10 μM) are shown. NAI-susceptible rg-WT and rg-E119A (A), rg-D198E (B), rg-I222T (C), rg-H274Y (D), rg-N294S (E), and rg-R371K (F) at an MOI of 0.01 PFU/cell were mixed 50:50 and then used to infect NHBE cells at a final MOI of 0.02 PFU/cell. Virus yield in MDCK cells was determined by the TCID50 assay at 33°C and expressed as log10 TCID50/ml. Total virus yield was determined by performing AUC analysis of the viral load at 24, 48, and 72 h p.i. Each data point represents the mean ± SD from two or three independent experiments. The limit of detection was 0.75 log10 TCID50/ml. Abbreviations: OSE, oseltamivir; ZAN, zanamivir.
TABLE 2.
Total yield of NAI-susceptible and -resistant recombinant influenza B viruses following coinfection of NHBE cells in the absence or presence of NAIs
| Virus in mixturea | Mean AUC (degree of restriction after treatment with NAI)b |
||||
|---|---|---|---|---|---|
| No Drug | Oseltamivir |
Zanamivir |
|||
| 1 μM | 10 μM | 1 μM | 10 μM | ||
| rg-E119A | 303.2 | 287.6 (15.6) | 237.2 (66.0)** | 280.4 (22.8) | 233.2 (70.0)** |
| rg-D198E | 299.2 | 278.8 (20.4) | 249.8 (49.4) | 169.6 (129.6)† | 130.4 (168.8)† |
| rg-I222T | 304.8 | 270.8 (34.0) | 231.2 (73.6) | 118 (186.8)** | 0 (304.8)† |
| rg-H274Y | 282.0 | 267.2 (14.8) | 272.8 (9.2) | 169.6 (112.4)† | 0 (282.0)† |
| rg-N294S | 276.8 | 253.6 (23.2) | 165.6 (111.2)† | 209.2 (67.6)* | 172.8 (104.0)† |
| rg-R371K | 272.4 | 278.8 (−6.4) | 290.8 (−18.4) | 147.2 (125.2)* | 13.2 (259.2)† |
Mixture of rg-WT and a recombinant influenza B virus carrying the indicated single substitution in NA.
The mean AUC indicates the total virus load at 24, 48, and 72 h p.i. The degree of restriction indicates the difference in the mean AUC between the control group (no drug) and the group treated with NAIs. *, P < 0.05; **, P < 0.01; †, P < 0.001 compared to the group not treated with drug by one-way ANOVA followed by Dunnett's multiple-comparison test.
With the addition of oseltamivir, a dose-dependent decrease in total virus yield occurred in 4 virus mixtures (rg-WT and rg-E119A, rg-D198E, rg-I222T, or rg-N294S; Fig. 2A to C and E), but significant reductions were observed only with 10 μM oseltamivir in the rg-WT–rg-E119A (P < 0.05) and rg-WT–rg-N294S mixtures (P < 0.01). The yield of 2 other virus mixtures (rg-WT and either rg-H274Y or rg-R371K) was unchanged by either concentration of oseltamivir tested (Fig. 2D and F).
The addition of zanamivir during coinfection of NHBE cells was more effective at reducing the total virus yield than was the addition of oseltamivir. Addition of 1 μM zanamivir significantly decreased the total virus yields for 4 virus mixtures susceptible to zanamivir (rg-WT and rg-D198E, rg-I222T, rg-H274Y, or rg-N294S; P < 0.05 to 0.001), and 10 μM zanamivir significantly decreased the yield of all 6 mixtures (Fig. 2; Table 2). The rg-WT–rg-E119A mixture was the least sensitive to zanamivir at the concentrations tested, as only the highest concentration (10 μM) significantly restricted the virus yield (P < 0.05). Thus, for the 2 viruses that possessed cross-resistance to both NAIs (i.e., rg-E119A and rg-R371K), zanamivir was more effective than oseltamivir at inhibiting growth.
Relative proportions of NAI-susceptible and -resistant influenza B viruses following coinfection of NHBE cells.
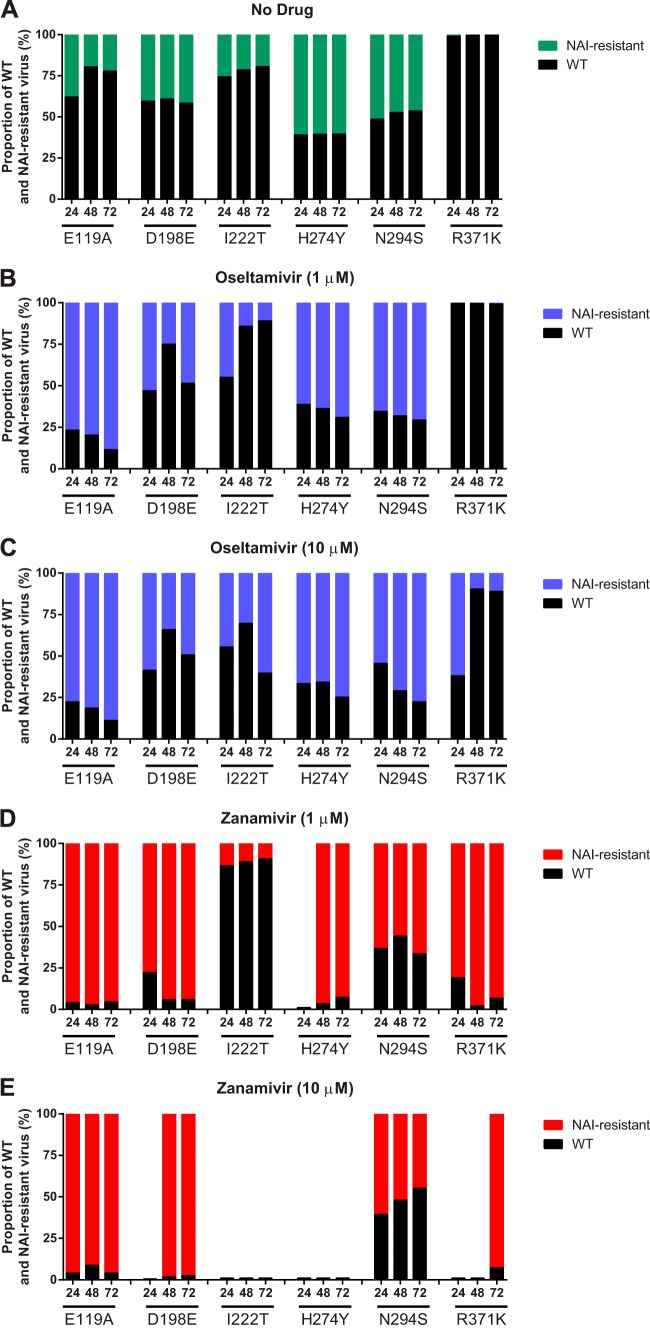
We next quantified the proportions of NAI-susceptible (rg-WT) and -resistant viruses released after coinfection of NHBE cells in the presence or absence of either oseltamivir or zanamivir by using next-generation deep sequencing. In the absence of oseltamivir, rg-WT was the dominant virus in 5 mixed populations (rg-WT and either rg-E119A, rg-D198E, rg-I222T, rg-N294S, or rg-R371K) (Fig. 3A). The proportion of rg-WT remained constant when it was used in combination with rg-D198E but increased over the course of infection for mixtures with rg-E119A, rg-I222T, or rg-N294S. Virus with the R371K substitution was detected at very low proportions (<0.5%) in the absence of NAI. Interestingly, the yield of rg-WT was reduced to a minority of the population after coinfection with rg-H274Y.
FIG 3.
Relative proportions of NAI-susceptible and -resistant influenza B virus populations following coinfection of NHBE cells. The cells were infected as described in the Fig. 2 legend. The proportions of NAI-susceptible and -resistant populations were determined by deep sequencing analysis of the NA gene and are expressed as a percentage of the total population after cells were grown either without drug (A), with oseltamivir at 1 μM (B) or 10 μM (C), or with zanamivir at 1 μM (D) or 10 μM (E). Each data point represents the mean from two or three independent experiments.
Oseltamivir was effective at reducing the proportion of rg-WT in each mixture after coinfection of NHBE cells. Additionally, rg-WT was reduced to a minority of the population, and its proportion diminished over the duration of coinfection with rg-E119A, rg-H274Y, or rg-N294S (Fig. 3B). Only 10 μM oseltamivir was effective at reducing the proportion of rg-WT in the rg-WT–rg-I222T population (Fig. 3C). In a mixture with rg-D198E, rg-WT started as a minority population but became the dominant virus by 72 h p.i. rg-R371K was again detected at a very low frequency (<1.0%) after addition of 1 μM oseltamivir. However, with addition of 10 μM oseltamivir, rg-WT was reduced to a minority of the population at 24 h p.i. but was the major viral population at 48 and 72 h p.i. (Fig. 3C).
With the addition of either concentration of zanamivir, the proportion of rg-WT in the viral population in mixtures with rg-E119A or rg-R371K was an average of 5% over the course of infection (Fig. 3D and E), indicating the superior fitness of the zanamivir-resistant viruses in the presence of drug. Both rg-D198E and rg-H274Y were susceptible to zanamivir, as indicated by reductions in total virus yield following infection and the NAI susceptibility assay, but the significant reduction in rg-WT following infection may suggest a slight difference in functional resistance to zanamivir between rg-WT and the 2 viruses containing NA substitutions. Although the composition of the rg-WT–rg-I222T and rg-WT–rg-N294S mixtures was not markedly different after addition of zanamivir, the total virus yield of these mixtures was reduced, as expected on the basis of their NAI susceptibility (Table 2; Fig. 3D and E).
Overall, we found that, except for severely compromised rg-R371K, influenza B viruses carrying any of the other 5 single NA substitutions contributed to a biologically relevant portion of the mixed viral population in the absence of oseltamivir or zanamivir pressure.
Independent quantification of mixed populations by high-resolution melt analysis.
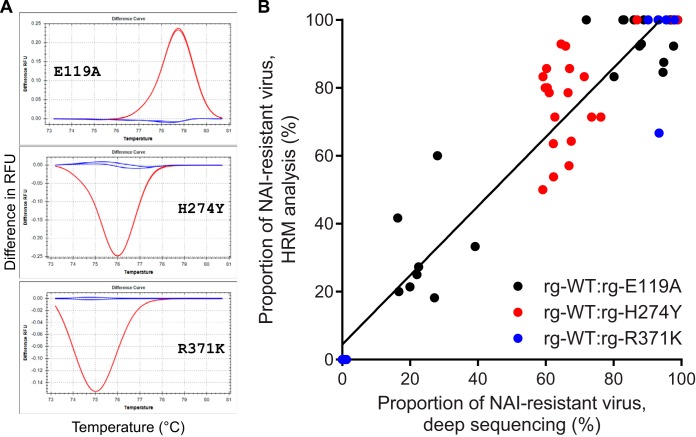
Defective interfering (DI) or other noninfectious influenza virus particles can be produced during the course of infection and can be detected by deep sequencing (50–52). To independently quantify the proportions of NAI-susceptible and -resistant influenza B viruses after coinfection of NHBE cells and to confirm the results of deep sequencing, we performed high-resolution melt (HRM) analysis (42) on individually purified plaques from 3 pairs of mixtures (rg-WT and either rg-E119A, rg-H274Y, or rg-R371K; example melt curves are shown in Fig. 4A). This analysis is based on the single-nucleotide difference in the NA gene of NAI-susceptible and -resistant infectious virus particles and, thus, overcomes possible limitations of whole-supernatant deep sequencing in determining the proportion of a particular virus in the mixture. The proportions of NAI-resistant virus in the viral population determined by HRM analysis and next-generation deep sequencing were correlated, with an overall R2 value of 0.9025 (Fig. 4B). There was a difference in the correlation between the rg-E119A- and rg-R371K-containing mixtures (R2 = 0.8815 and 0.9715, respectively) and the mixture containing rg-H274Y (R2 = 0.3864). This outcome is likely because we picked insufficient numbers of plaques (n = 14) to adequately sample the rg-WT–rg-H274Y population. The probability of selecting a representative viral population from 14 individual plaques increases as the composition of a mixed population becomes more homogeneous, as exemplified by the samples from the rg-WT–rg-E119A or rg-WT–rg-R371K mixture. Indeed, across all samples, as the population became less heterogeneous, the correlation between deep sequencing and HRM analysis increased (for 30 to 70% NAI-resistant virus, R2 = 0.3257; for 70 to 85% or 15 to 30% NAI-resistant virus, R2 = 0.8453; for >85% or <15% NAI-resistant virus, R2 = 0.9762). The correlation between the 2 analysis methods was similar among all tested time points (24 h p.i., R2 = 0.8334; 48 h p.i., R2 = 0.8087; 72 h p.i., R2 = 0.7798). Overall, our independent HRM analysis confirms the next-generation deep sequencing data for the mixed viral populations and the lack of bias by DI or other types of noninfectious influenza virus particles.
FIG 4.
Correlation of deep sequencing and high-resolution melt analysis in determining the proportions of NAI-susceptible and -resistant influenza B viruses following coinfection of NHBE cells. (A) Examples of the different melt curves used for the analysis by HRM of individually purified plaques derived from mixed viral populations are shown. Each graph shows the difference between the number of relative fluorescence units (RFUs) of the WT (blue lines) and those of each of the 3 single-nucleotide substitution-containing templates (red lines) over increasing temperatures (°C). Analyses were performed by using Precision Melt Analysis software (v1.2; Bio-Rad). (B) The percentage of NAI-resistant viruses determined by the results of deep sequencing variant call analysis is plotted against the percentage of NAI-resistant viruses determined by HRM analysis of individually purified plaques for three pairs of virus mixtures: rg-WT and rg-E119A, rg-WT and rg-H274Y, and rg-WT and rg-R371K. Data obtained from all time points (24, 48, 72 h p.i.) and with all NAI concentrations (0, 1, and 10 μM oseltamivir and 1 and 10 μM zanamivir) are shown for each of the three mixtures. The result of linear regression analysis of all samples is depicted by a solid black line.
Relative fitness of NAI-susceptible and -resistant influenza B viruses following coinfection of NHBE cells.
We next quantified both the fitness difference and the relative fitness of each recombinant influenza B virus. The fitness difference between 2 viruses was calculated as the change in the population ratios (WT/mutant) over time (45), where a negative value indicates an absolute fitness advantage for rg-WT. We also calculated the relative fitness of each NAI-resistant virus from the selection coefficient (s), taking into account the various replication rates for each virus, where values less than 1 indicate that the relative fitness of an NAI-resistant virus is less than that of rg-WT (44). In the absence of drug treatment, all viruses except rg-H274Y had a diminished fitness compared to that of rg-WT (Table 3). rg-H274Y had a minor fitness advantage over rg-WT, having a positive slope and relative fitness greater than 1. Fitness could not be calculated for rg-R371K, and it was clearly the least fit virus among all viruses tested. These results demonstrate that the order of relative fitness for the tested NAI-susceptible and -resistant influenza B viruses in the absence of drug is as follows: rg-H274Y > rg-WT > rg-I222T > rg-N294S > rg-D198E > rg-E119A ≫ rg-R371K.
TABLE 3.
Fitness of NAI-susceptible and -resistant recombinant influenza B viruses following coinfection in NHBE cells
| Experimental treatment group (dose) | Virus in mixturea | Fitness parameter valueb |
Qualitative fitnessc | |
|---|---|---|---|---|
| Fitness coefficient | Relative fitness | |||
| No drug | rg-E119A | −0.009 ± 0.002 | 0.860 ± 0.019 | Diminished |
| rg-D198E | −0.002 ± 0.002 | 0.956 ± 0.035 | Diminished | |
| rg-I222T | −0.007 ± 0.003 | 0.981 ± 0.072 | Diminished | |
| rg-H274Y | 0.004 ± 0.002 | 1.098 ± 0.122 | Superior | |
| rg-N294S | −0.003 ± 0.001 | 0.976 ± 0.009 | Diminished | |
| rg-R371K | —d | — | — | |
| Oseltamivir (1 μM) | rg-E119A | 0.097 ± 0.025 | 1.123 ± 0.041 | Superior |
| rg-D198E | −0.007 ± 0.008 | 0.933 ± 0.089 | Diminished | |
| rg-I222T | −0.014 ± 0.003 | 0.785 ± 0.032 | Diminished | |
| rg-H274Y | 0.016 ± 0.004 | 1.758 ± 1.733 | Superior | |
| rg-N294S | 0.017 ± 0.004 | 1.028 ± 0.024 | Superior | |
| rg-R371K | — | — | — | |
| Oseltamivir (10 μM) | rg-E119A | 0.135 ± 0.067 | 1.079 ± 0.142 | Superior |
| rg-D198E | 0.002 ± 0.009 | 0.999 ± 0.045 | Comparable | |
| rg-I222T | 0.005 ± 0.01 | 1.047 ± 0.041 | Superior | |
| rg-H274Y | 0.024 ± 0.005 | 1.088 ± 0.098 | Superior | |
| rg-N294S | 0.039 ± 0.007 | 1.06 ± 0.065 | Superior | |
| rg-R371K | −0.017 ± 0.008 | 0.104 ± 0.277 | Diminished | |
| Zanamivir (1 μM) | rg-E119A | 0.002 ± 0.498 | 1.048 ± 0.025 | Superior |
| rg-D198E | 0.203 ± 0.160 | 1.213 ± 0.01 | Superior | |
| rg-I222T | −0.01 ± 0.004 | 0.966 ± 0.012 | Diminished | |
| rg-H274Y | −0.629 ± 0.57 | — | - | |
| rg-N294S | −0.009 ± 0.007 | 1.104 ± 0.234 | Comparable | |
| rg-R371K | 0.359 ± 0.364 | 1.107 ± 0.147 | Superior | |
| Zanamivir (10 μM) | rg-E119A | 0.174 ± 0.188 | 1.134 ± 0.197 | Superior |
| rg-D198E | 0.621 ± 0.451 | — | Superior | |
| rg-I222T | — | — | — | |
| rg-H274Y | — | — | — | |
| rg-N294S | −0.004 ± 0.006 | 0.914 ± 0.01 | Comparable | |
| rg-R371K | 0.198 ± 0.147 | — | — | |
Mixture of rg-WT and a recombinant influenza B virus carrying the indicated single substitution in NA.
See the Materials and Methods section for the calculation.
Qualitative fitness compared to that of rg-WT.
—, fitness calculations not applicable.
The addition of oseltamivir during coinfection of NHBE cells resulted in the superior fitness of influenza B viruses containing the E119A, H274Y, or N294S NA substitution compared to that of rg-WT. rg-D198E or rg-I222T exhibited fitness either comparable to or superior to that of rg-WT only in the presence of 10 μM oseltamivir. Neither of the concentrations of oseltamivir tested provided a fitness advantage to rg-R371K, again confirming the poor fitness conferred by the catalytic site NA substitution.
The addition of zanamivir during coinfection of NHBE cells was also effective at inhibiting the proportions of rg-WT in the virus mixtures, up to the point of inhibiting or delaying the virus replication of the entire mixture. At 1 μM, zanamivir increased the fitness advantage compared to that of rg-WT of 3 viruses (rg-E119A, rg-D198E, and rg-R371K). Consistent with a small reduction in virus yield, rg-E119A was more fit than rg-WT in the presence of both concentrations of zanamivir tested. Our calculations indicate a comparable qualitative fitness of rg-N294S at 1 and 10 μM zanamivir.
Together, these data indicate that in the absence of drug pressure, the fitness in NHBE cells of 5 of the 6 NAI-resistant influenza B viruses studied was diminished compared to that of rg-WT. The only NA substitution to increase the fitness of influenza B virus was H274Y. Inclusion of oseltamivir during infection of NHBE cells provided an advantage to all NAI-resistant virus populations except the severely compromised rg-R371K. While zanamivir also provided an increased fitness advantage to the NAI-resistant viruses, it was more effective than oseltamivir at inhibiting their replication. Finally, our data indicate that although rg-E119A can replicate to high viral titers in the presence of oseltamivir or zanamivir, it is less fit than rg-WT.
DISCUSSION
To date, antiviral surveillance studies conducted during different seasons and in different geographic areas reported a low frequency (0.1 to 0.8%) of circulation of NAI-resistant influenza B viruses (53–59). Monitoring of the NAI susceptibility of influenza B viruses is limited by a lack of well-established genetic markers of drug resistance and a lack of understanding of the fitness advantages and disadvantages conferred by mutations in mutant viruses compared to the fitness of an NAI-susceptible virus. In this study, using a competitive coinfection model in NHBE cells, we examined the replicative capacity and relative fitness of 6 influenza B/Yamanashi/166/1998 viruses with different single NA substitutions associated with NAI resistance. Among all viruses studied, we identified that the viruses with the E119A or H274Y substitution in NA deserve close and continued monitoring. This conclusion is based on our findings that virus containing the E119A NA substitution was less fit than the NAI-susceptible WT virus but could productively replicate in the presence of both oseltamivir and zanamivir and that virus with the H274Y NA substitution was more fit than the NAI-susceptible virus in the absence or presence of oseltamivir or zanamivir drug pressure.
Although several studies using recombinant influenza B virus passage in cell culture under NAI pressure have shown that amino acid substitutions at position 119 (E119A/D/G/V) in the NA protein are associated with highly reduced inhibition by oseltamivir and zanamivir (60–63), only one report described the isolation of a virus with the E119A substitution from a patient sample (64). Here we demonstrated that detection of influenza B viruses with the E119A NA substitution by NA inhibition assays is easily masked by the presence of NAI-susceptible virus and thus can be missed if only the phenotypic assay is used for identification. Genotypic assays alone are not sufficient for antiviral surveillance studies, and a phenotypic assay is still needed to identify novel substitutions that reduce inhibition or to identify the impact of a substitution in a specific genetic background. Different substitutions in NA result in different levels of NA activity in buffers of different pHs (65, 66). It might be possible to reduce the masking effect through simple modifications of the NI assay, although more studies are necessary to examine this effect on the breadth of NA substitutions in influenza B viruses. The NA substitutions E119D and E119V were also shown to reduce the growth kinetics of influenza B viruses in vitro, but E119G conferred no change in growth kinetics (60). An influenza B virus that acquired the E119A substitution after passages in MDCK cells in the presence of zanamivir grew to higher titers than the parental WT virus in cell culture but also contained N145S and N150S substitutions in HA (influenza B virus numbering) (61). These HA substitutions move a glycosylation site within the 150-loop antigenic region, potentially altering the sialic acid-binding properties of HA (61, 67).
The side chains of oseltamivir and zanamivir were specifically designed to contact the E119 residue to increase the binding affinity with the NA protein (68). Although the E119 residue does not directly contact the host-derived sialic acid, the E119A NA substitution reduces NA activity but not NA enzyme kinetics (35). Thus, although the mechanism of resistance caused by the E119A NA substitution in influenza B viruses is not fully understood, it is most likely due to changes allowing a reduced affinity for the NAI and maintaining biologically sufficient NA activity.
The emergence of influenza B viruses with the H274Y substitution was reported in a patient with no known previous antiviral treatment (69) and from surveillance studies (57). Our results show that H274Y-containing influenza B virus is more fit than the parental WT virus in NHBE cells. For influenza A viruses (H5N1 influenza A viruses, H1N1 2009 pandemic [H1N1pdm09] influenza A viruses, and seasonal H1N1 influenza A viruses prior to the 2008-2009 season), the H274Y NA substitution was generally found to reduce in vitro and in vivo viral fitness compared to that of its NAI-susceptible counterparts (70–72). Only by the addition of secondary permissive NA substitutions was viral fitness restored for seasonal H1N1 influenza A viruses, leading to their spread worldwide (73–75). It was reported that the viruses involved in outbreaks of H274Y-containing H1N1pdm09 influenza A viruses possessed permissive secondary NA substitutions that also enhanced viral fitness (76, 77). Our previous work demonstrated that for the B/Yamanashi/166/1998 influenza virus, the H274Y NA substitution alone increased NA protein expression, similar to the effect of the permissive substitutions in seasonal H1N1 and H1N1pdm09 influenza A viruses (35, 76, 77). Thus, it is possible that the genetic background of the B/Yamanashi/166/1998 influenza virus already contains the requirements permissive for increased fitness with acquisition of the H274Y NA substitution.
Researchers recently found that the H274Y NA substitution in H1N1pdm09 influenza A viruses reduces viral fitness after infection in ferrets, potentially by increasing the eclipse phase of viral growth (i.e., the time between infection and production of infectious virus) (78). In a genetic background without permissive mutations, reduced cell surface NA protein expression could increase the length of time necessary for infectious virus to bud and be released from an infected cell. Accordingly, elevated cell surface NA protein expression would decrease this length of time. Although our and other models of influenza virus fitness do not take into account the eclipse phase of viral growth, it is evident that variations in NA protein expression impact viral fitness for multiple subtypes of influenza viruses, and an altered eclipse phase is one potential mechanism of such changes (79). Further studies in the NHBE cell model will be required to properly determine these kinetic parameters of influenza B virus replication.
Notably, the fitness advantages and disadvantages conferred by 5 of the 6 NA substitutions in influenza B viruses presented here were not studied previously, and 2 different amino acid changes were reported earlier for residue 198. We observed a small fitness deficit for rg-D198E in vitro, but another group observed no fitness difference in vivo for an influenza B virus containing the D198N NA substitution (28). An influenza B virus with a D198Y substitution was found during antiviral surveillance studies (53). The aspartic acid residue at position 198 of the influenza B virus NA protein interacts with arginine at neighboring residue 152, and thus, there is likely an NA substitution-specific effect on virus fitness for residue 198 (13). Consistent with our results for the R371K NA substitution, results from 2 previous studies show that other catalytic site NA substitutions are also deleterious to the fitness (R152K) or replication (R292K) of influenza B viruses (27, 60). A cluster of influenza B viruses with an I222V NA substitution was identified during a surveillance study, representing the first report of the identification of this specific substitution in influenza B viruses (22). Pyrosequencing of the clinical specimens indicated mixed populations of NAI-resistant (V222) and -susceptible (I222) viruses, possibly indicating an increased fitness of the NAI-resistant virus. Thus, for influenza B viruses, in addition to substitutions at position 198, changes at position 222 can confer resistance and alter fitness relative to that of the NAI-susceptible parent.
Advantages of our study include the use of NHBE cells as a model system and the use of next-generation deep sequencing to determine the proportions of mixed viral populations. Influenza B viruses are strict human pathogens: the NHBE cell model best approximates the human upper respiratory tract, which is both the primary site of infection and the source of influenza virus-containing respiratory droplets. NHBE cells also produce mucus—which is absent from other in vitro cell-based systems—in which NA sialidase activity plays a critical role during the infectious cycle (80). Although many methods for the discrimination of viral populations on the basis of genetic differences are available, our use of next-generation deep sequencing provides a precise quantification of the composition of the virus mixtures. As costs continue to decrease, next-generation deep sequencing will likely become a more attractive analysis method for quantification of any type of mixed viral population. One limitation of this technology is the inability to distinguish individual virus populations. Through the course of analysis, we identified a minor percentage of additional secondary substitutions in NA (data not shown). However, because our population contained mixed NAI-susceptible and -resistant viruses, we were unable to determine which virus population contained the additional substitutions. Our study utilized a single ratio of WT and NAI-resistant viruses (50:50) to examine viral fitness. Further experiments with different virus ratios could provide an additional depth to the analysis of the fitness characteristics but were unfeasible due to experimental considerations.
In conclusion, we identified influenza B viruses containing either the E119A or H274Y NA substitution to be of potential public health concern. Although virus with the E119A NA substitution possessed diminished fitness in the absence of drug pressure, addition of oseltamivir or zanamivir increased the fitness relative to that of rg-WT. We also found that influenza B virus with the H274Y NA substitution had fitness superior to that of rg-WT, even in the absence of oseltamivir or zanamivir drug pressure. Oseltamivir was largely ineffective at inhibiting the replication of the H274Y variant and further increased its fitness relative to that of the NAI-susceptible rg-WT. Although NHBE cells are a simplified model system which does not allow direct evaluation of viral transmission between hosts, these data suggest that an influenza B virus carrying an H274Y NA substitution could have the potential to spread widely in the human population even in the absence of widespread oseltamivir use. Importantly, rg-H274Y remained susceptible to zanamivir. Finally, as influenza viruses are constantly evolving human pathogens, surveillance studies are necessary to continually monitor the antiviral susceptibility of currently circulating viruses, and these studies can soon be extended to include novel anti-influenza virus drugs that are undergoing preclinical and clinical evaluation.
ACKNOWLEDGMENTS
This work was partially supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, under Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract numbers HHSN266200700005C, HHSN266200700006C, and HHSN272201400006C, under R01 AI099000 (to A.C.L.), and by ALSAC.
We are grateful to Bindumadhav M. Marathe for technical support, Cherise Guess for excellent editing of the manuscript, and the Hartwell Center for Bioinformatics and Biotechnology at St. Jude Children's Research Hospital for its support with deep sequencing.
REFERENCES
- 1.McCullers JA, Hayden FG. 2012. Fatal influenza B infections: time to reexamine influenza research priorities. J Infect Dis 205:870–872. doi: 10.1093/infdis/jir865. [DOI] [PubMed] [Google Scholar]
- 2.Gutierrez-Pizarraya A, Perez-Romero P, Alvarez R, Aydillo TA, Osorio-Gomez G, Milara-Ibanez C, Sanchez M, Pachon J, Cordero E. 2012. Unexpected severity of cases of influenza B infection in patients that required hospitalization during the first postpandemic wave. J Infect 65:423–430. doi: 10.1016/j.jinf.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Paddock CD, Liu L, Denison AM, Bartlett JH, Holman RC, Deleon-Carnes M, Emery SL, Drew CP, Shieh WJ, Uyeki TM, Zaki SR. 2012. Myocardial injury and bacterial pneumonia contribute to the pathogenesis of fatal influenza B virus infection. J Infect Dis 205:895–905. doi: 10.1093/infdis/jir861. [DOI] [PubMed] [Google Scholar]
- 4.Aebi T, Weisser M, Bucher E, Hirsch HH, Marsch S, Siegemund M. 2010. Co-infection of influenza B and streptococci causing severe pneumonia and septic shock in healthy women. BMC Infect Dis 10:308. doi: 10.1186/1471-2334-10-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen SH, Huang IA, Wu CT, Hsia SH, Hung PC, Chiu CH. 2011. Complicated features in a young child with influenza B virus pneumonia and co-infection with Stenotrophomonas maltophilia. Ann Trop Paediatr 31:159–162. doi: 10.1179/1465328111Y.0000000012. [DOI] [PubMed] [Google Scholar]
- 6.Yusuf K, Soraisham AS, Fonseca K. 2007. Fatal influenza B virus pneumonia in a preterm neonate: case report and review of the literature. J Perinatol 27:623–625. doi: 10.1038/sj.jp.7211802. [DOI] [PubMed] [Google Scholar]
- 7.Goenka A, Michael BD, Ledger E, Hart IJ, Absoud M, Chow G, Lilleker J, Lunn M, McKee D, Peake D, Pysden K, Roberts M, Carrol ED, Lim M, Avula S, Solomon T, Kneen R. 2014. Neurological manifestations of influenza infection in children and adults: results of a national British surveillance study. Clin Infect Dis 58:775–784. doi: 10.1093/cid/cit922. [DOI] [PubMed] [Google Scholar]
- 8.Barr IG, Jelley LL. 2012. The coming era of quadrivalent human influenza vaccines: who will benefit? Drugs 72:2177–2185. doi: 10.2165/11641110-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 9.Ambrose CS, Levin MJ. 2012. The rationale for quadrivalent influenza vaccines. Hum Vaccin Immunother 8:81–88. doi: 10.4161/hv.8.1.17623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.FDA. 2014. FDA approves rapivab to treat flu infection. FDA, Rockville, MD: http://www.fda.gov/NewsEvents/Newsroom/PressAnnouncements/ucm427755.htm Accessed 28 December 2014. [Google Scholar]
- 11.Nguyen HT, Fry AM, Gubareva LV. 2012. Neuraminidase inhibitor resistance in influenza viruses and laboratory testing methods. Antivir Ther 17:159–173. doi: 10.3851/IMP2067. [DOI] [PubMed] [Google Scholar]
- 12.Burnham AJ, Baranovich T, Govorkova EA. 2013. Neuraminidase inhibitors for influenza B virus infection: efficacy and resistance. Antiviral Res 100:520–534. doi: 10.1016/j.antiviral.2013.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oakley AJ, Barrett S, Peat TS, Newman J, Streltsov VA, Waddington L, Saito T, Tashiro M, McKimm-Breschkin JL. 2010. Structural and functional basis of resistance to neuraminidase inhibitors of influenza B viruses. J Med Chem 53:6421–6431. doi: 10.1021/jm100621s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKimm-Breschkin JL. 2000. Resistance of influenza viruses to neuraminidase inhibitors—a review. Antiviral Res 47:1–17. doi: 10.1016/S0166-3542(00)00103-0. [DOI] [PubMed] [Google Scholar]
- 15.Okomo-Adhiambo M, Sleeman K, Lysen C, Nguyen HT, Xu X, Li Y, Klimov AI, Gubareva LV. 2013. Neuraminidase inhibitor susceptibility surveillance of influenza viruses circulating worldwide during the 2011 Southern Hemisphere season. Influenza Other Respir Viruses 7:645–658. doi: 10.1111/irv.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujisaki S, Takashita E, Yokoyama M, Taniwaki T, Xu H, Kishida N, Sato H, Tashiro M, Imai M, Odagiri T. 2012. A single E105K mutation far from the active site of influenza B virus neuraminidase contributes to reduced susceptibility to multiple neuraminidase-inhibitor drugs. Biochem Biophys Res Commun 429:51–56. doi: 10.1016/j.bbrc.2012.10.095. [DOI] [PubMed] [Google Scholar]
- 17.WHO. 2012. WHO information for molecular diagnosis of influenza virus in humans—update. WHO, Geneva, Switzerland: http://www.who.int/influenza/gisrs_laboratory/molecular_diagnosis/en/ Accessed 28 December 2014. [Google Scholar]
- 18.Domingo E, Menendez-Arias L, Holland JJ. 1997. RNA virus fitness. Rev Med Virol 7:87–96. doi:. [DOI] [PubMed] [Google Scholar]
- 19.Kelso A, Hurt AC. 2012. The ongoing battle against influenza: drug-resistant influenza viruses: why fitness matters. Nat Med 18:1470–1471. doi: 10.1038/nm.2954. [DOI] [PubMed] [Google Scholar]
- 20.Garg S, Moore Z, Lee N, McKenna J, Bishop A, Fleischauer A, Springs CB, Nguyen HT, Sheu TG, Sleeman K, Finelli L, Gubareva L, Fry AM. 2013. A cluster of patients infected with I221V influenza B virus variants with reduced oseltamivir susceptibility—North Carolina and South Carolina, 2010-2011. J Infect Dis 207:966–973. doi: 10.1093/infdis/jis776. [DOI] [PubMed] [Google Scholar]
- 21.Hatakeyama S, Sugaya N, Ito M, Yamazaki M, Ichikawa M, Kimura K, Kiso M, Shimizu H, Kawakami C, Koike K, Mitamura K, Kawaoka Y. 2007. Emergence of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. JAMA 297:1435–1442. doi: 10.1001/jama.297.13.1435. [DOI] [PubMed] [Google Scholar]
- 22.Sleeman K, Sheu TG, Moore Z, Kilpatrick S, Garg S, Fry AM, Gubareva LV. 2011. Influenza B viruses with mutation in the neuraminidase active site, North Carolina, USA, 2010-11. Emerg Infect Dis 17:2043–2046. doi: 10.3201/eid1711.110787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang D, Sleeman K, Huang W, Nguyen HT, Levine M, Cheng Y, Li X, Tan M, Xing X, Xu X, Klimov AI, Gubareva LV, Shu Y. 2013. Neuraminidase inhibitor susceptibility testing of influenza type B viruses in China during 2010 and 2011 identifies viruses with reduced susceptibility to oseltamivir and zanamivir. Antiviral Res 97:240–244. doi: 10.1016/j.antiviral.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Belser JA, Katz JM, Tumpey TM. 2011. The ferret as a model organism to study influenza A virus infection. Dis Model Mech 4:575–579. doi: 10.1242/dmm.007823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang SS, Banner D, Paquette SG, Leon AJ, Kelvin AA, Kelvin DJ. 2014. Pathogenic influenza B virus in the ferret model establishes lower respiratory tract infection. J Gen Virol 95(Pt 10):2127–2139. doi: 10.1099/vir.0.064352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YH, Kim HS, Cho SH, Seo SH. 2009. Influenza B virus causes milder pathogenesis and weaker inflammatory responses in ferrets than influenza A virus. Viral Immunol 22:423–430. doi: 10.1089/vim.2009.0045. [DOI] [PubMed] [Google Scholar]
- 27.Gubareva LV, Matrosovich MN, Brenner MK, Bethell RC, Webster RG. 1998. Evidence for zanamivir resistance in an immunocompromised child infected with influenza B virus. J Infect Dis 178:1257–1262. doi: 10.1086/314440. [DOI] [PubMed] [Google Scholar]
- 28.Mishin VP, Hayden FG, Gubareva LV. 2005. Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob Agents Chemother 49:4515–4520. doi: 10.1128/AAC.49.11.4515-4520.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pica N, Chou YY, Bouvier NM, Palese P. 2012. Transmission of influenza B viruses in the guinea pig. J Virol 86:4279–4287. doi: 10.1128/JVI.06645-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lai C, Struckhoff JJ, Schneider J, Martinez-Sobrido L, Wolff T, Garcia-Sastre A, Zhang DE, Lenschow DJ. 2009. Mice lacking the ISG15 E1 enzyme UbE1L demonstrate increased susceptibility to both mouse-adapted and non-mouse-adapted influenza B virus infection. J Virol 83:1147–1151. doi: 10.1128/JVI.00105-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, Garcia-Sastre A, Leib DA, Pekosz A, Knobeloch KP, Horak I, Virgin HW IV. 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc Natl Acad Sci U S A 104:1371–1376. doi: 10.1073/pnas.0607038104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fabian P, McDevitt JJ, DeHaan WH, Fung RO, Cowling BJ, Chan KH, Leung GM, Milton DK. 2008. Influenza virus in human exhaled breath: an observational study. PLoS One 3:e2691. doi: 10.1371/journal.pone.0002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin H, Li H, Cho HJ, Bian S, Roh HJ, Lee MK, Kim JS, Chung SJ, Shim CK, Kim DD. 2007. Air-liquid interface (ALI) culture of human bronchial epithelial cell monolayers as an in vitro model for airway drug transport studies. J Pharm Sci 96:341–350. doi: 10.1002/jps.20803. [DOI] [PubMed] [Google Scholar]
- 34.Roy CJ, Milton DK. 2004. Airborne transmission of communicable infection—the elusive pathway. N Engl J Med 350:1710–1712. doi: 10.1056/NEJMp048051. [DOI] [PubMed] [Google Scholar]
- 35.Burnham AJ, Baranovich T, Marathe BM, Armstrong J, Webster RG, Govorkova EA. 2014. Fitness costs for influenza B viruses carrying neuraminidase inhibitor-resistant substitutions: underscoring the importance of E119A and H274Y. Antimicrob Agents Chemother 58:2718–2730. doi: 10.1128/AAC.02628-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ilyushina NA, Govorkova EA, Gray TE, Bovin NV, Webster RG. 2008. Human-like receptor specificity does not affect the neuraminidase-inhibitor susceptibility of H5N1 influenza viruses. PLoS Pathog 4:e1000043. doi: 10.1371/journal.ppat.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson CI, Barclay WS, Zambon MC, Pickles RJ. 2006. Infection of human airway epithelium by human and avian strains of influenza A virus. J Virol 80:8060–8068. doi: 10.1128/JVI.00384-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoffmann E, Mahmood K, Yang CF, Webster RG, Greenberg HB, Kemble G. 2002. Rescue of influenza B virus from eight plasmids. Proc Natl Acad Sci U S A 99:11411–11416. doi: 10.1073/pnas.172393399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 40.Potier M, Mameli L, Belisle M, Dallaire L, Melancon SB. 1979. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-d-N-acetylneuraminate) substrate. Anal Biochem 94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 41.Marathe BM, Leveque V, Klumpp K, Webster RG, Govorkova EA. 2013. Determination of neuraminidase kinetic constants using whole influenza virus preparations and correction for spectroscopic interference by a fluorogenic substrate. PLoS One 8:e71401. doi: 10.1371/journal.pone.0071401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. 2003. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem 49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- 43.Marshall N, Priyamvada L, Ende Z, Steel J, Lowen AC. 2013. Influenza virus reassortment occurs with high frequency in the absence of segment mismatch. PLoS Pathog 9:e1003421. doi: 10.1371/journal.ppat.1003421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maree AF, Keulen W, Boucher CA, De Boer RJ. 2000. Estimating relative fitness in viral competition experiments. J Virol 74:11067–11072. doi: 10.1128/JVI.74.23.11067-11072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Holland JJ, de la Torre JC, Clarke DK, Duarte E. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J Virol 65:2960–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heldt FS, Frensing T, Pflugmacher A, Gropler R, Peschel B, Reichl U. 2013. Multiscale modeling of influenza A virus infection supports the development of direct-acting antivirals. PLoS Comput Biol 9:e1003372. doi: 10.1371/journal.pcbi.1003372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.WHO. 2013. Meeting of the WHO expert working group on surveillance of influenza antiviral susceptibility, Geneva, July 2013. Wkly Epidemiol Rec 88:477–482. [PubMed] [Google Scholar]
- 48.WHO. 2012. Laboratory methodologies for testing the antiviral susceptibility of influenza viruses: neuraminidase inhibitor (NAI). WHO, Geneva, Switzerland: www.who.int/influenza/gisrs_laboratory/antiviral_susceptibility/nai_overview/en/index.html Accessed 28 December 2014. [Google Scholar]
- 49.Pozo F, Lina B, Andrade HR, Enouf V, Kossyvakis A, Broberg E, Daniels R, Lackenby A, Meijer A, Community Network of Reference Laboratories for Human Influenza in Europe. 2013. Guidance for clinical and public health laboratories testing for influenza virus antiviral drug susceptibility in Europe. J Clin Virol 57:5–12. doi: 10.1016/j.jcv.2013.01.009. [DOI] [PubMed] [Google Scholar]
- 50.Marcus PI, Ngunjiri JM, Sekellick MJ. 2009. Dynamics of biologically active subpopulations of influenza virus: plaque-forming, noninfectious cell-killing, and defective interfering particles. J Virol 83:8122–8130. doi: 10.1128/JVI.02680-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nayak DP, Chambers TM, Akkina RK. 1985. Defective-interfering (DI) RNAs of influenza viruses: origin, structure, expression, and interference. Curr Top Microbiol Immunol 114:103–151. [DOI] [PubMed] [Google Scholar]
- 52.Saira K, Lin X, DePasse JV, Halpin R, Twaddle A, Stockwell T, Angus B, Cozzi-Lepri A, Delfino M, Dugan V, Dwyer DE, Freiberg M, Horban A, Losso M, Lynfield R, Wentworth DN, Holmes EC, Davey R, Wentworth DE, Ghedin E. 2013. Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J Virol 87:8064–8074. doi: 10.1128/JVI.00240-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Escuret V, Frobert E, Bouscambert-Duchamp M, Sabatier M, Grog I, Valette M, Lina B, Morfin F, Ferraris O. 2008. Detection of human influenza A (H1N1) and B strains with reduced sensitivity to neuraminidase inhibitors. J Clin Virol 41:25–28. doi: 10.1016/j.jcv.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 54.Monto AS, McKimm-Breschkin JL, Macken C, Hampson AW, Hay A, Klimov A, Tashiro M, Webster RG, Aymard M, Hayden FG, Zambon M. 2006. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob Agents Chemother 50:2395–2402. doi: 10.1128/AAC.01339-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hurt AC, Barr IG, Hartel G, Hampson AW. 2004. Susceptibility of human influenza viruses from Australasia and South East Asia to the neuraminidase inhibitors zanamivir and oseltamivir. Antiviral Res 62:37–45. doi: 10.1016/j.antiviral.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 56.Okomo-Adhiambo M, Sleeman K, Ballenger K, Nguyen HT, Mishin VP, Sheu TG, Smagala J, Li Y, Klimov AI, Gubareva LV. 2010. Neuraminidase inhibitor susceptibility testing in human influenza viruses: a laboratory surveillance perspective. Viruses 2:2269–2289. doi: 10.3390/v2102269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheu TG, Deyde VM, Okomo-Adhiambo M, Garten RJ, Xu X, Bright RA, Butler EN, Wallis TR, Klimov AI, Gubareva LV. 2008. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother 52:3284–3292. doi: 10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neuraminidase Inhibitor Susceptibility Network. 2007. Monitoring of neuraminidase inhibitor resistance among clinical influenza virus isolates in Japan during the 2003–2006 influenza seasons. Wkly Epidemiol Rec 82:149–150. [PubMed] [Google Scholar]
- 59.Tashiro M, McKimm-Breschkin JL, Saito T, Klimov A, Macken C, Zambon M, Hayden FG, Neuraminidase Inhibitor Susceptibility Network. 2009. Surveillance for neuraminidase-inhibitor-resistant influenza viruses in Japan, 1996–2007. Antivir Ther 14:751–761. doi: 10.3851/IMP1194. [DOI] [PubMed] [Google Scholar]
- 60.Jackson D, Barclay W, Zurcher T. 2005. Characterization of recombinant influenza B viruses with key neuraminidase inhibitor resistance mutations. J Antimicrob Chemother 55:162–169. doi: 10.1093/jac/dkh528. [DOI] [PubMed] [Google Scholar]
- 61.Staschke KA, Colacino JM, Baxter AJ, Air GM, Bansal A, Hornback WJ, Munroe JE, Laver WG. 1995. Molecular basis for the resistance of influenza viruses to 4-guanidino-Neu5Ac2en. Virology 214:642–646. doi: 10.1006/viro.1995.0078. [DOI] [PubMed] [Google Scholar]
- 62.Barnett JM, Cadman A, Burrell FM, Madar SH, Lewis AP, Tisdale M, Bethell R. 1999. In vitro selection and characterisation of influenza B/Beijing/1/87 isolates with altered susceptibility to zanamivir. Virology 265:286–295. doi: 10.1006/viro.1999.0058. [DOI] [PubMed] [Google Scholar]
- 63.Cheam AL, Barr IG, Hampson AW, Mosse J, Hurt AC. 2004. In vitro generation and characterisation of an influenza B variant with reduced sensitivity to neuraminidase inhibitors. Antiviral Res 63:177–181. doi: 10.1016/j.antiviral.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 64.Sheu TG, Deyde VM, Garten RJ, Klimov AI, Gubareva LV. 2010. Detection of antiviral resistance and genetic lineage markers in influenza B virus neuraminidase using pyrosequencing. Antiviral Res 85:354–360. doi: 10.1016/j.antiviral.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 65.Gubareva LV, Robinson MJ, Bethell RC, Webster RG. 1997. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J Virol 71:3385–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sleeman K, Guo Z, Barnes J, Shaw M, Stevens J, Gubareva LV. 2013. R292K substitution and drug susceptibility of influenza A(H7N9) viruses. Emerg Infect Dis 19:1521–1524. doi: 10.3201/eid1909.130724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang Q, Cheng F, Lu M, Tian X, Ma J. 2008. Crystal structure of unliganded influenza B virus hemagglutinin. J Virol 82:3011–3020. doi: 10.1128/JVI.02477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.von Itzstein M, Wu WY, Kok GB, Pegg MS, Dyason JC, Jin B, Van Phan T, Smythe ML, White HF, Oliver SW, Colman PM, Varghese JN, Ryan DM, Woods JM, Bethell RC, Hotham VJ, Cameron JM, Penn CR. 1993. Rational design of potent sialidase-based inhibitors of influenza virus replication. Nature 363:418–423. doi: 10.1038/363418a0. [DOI] [PubMed] [Google Scholar]
- 69.Higgins RR, Beniprashad M, Chong-King E, Li Y, Bastien N, Low DE, Gubbay JB. 2012. Recovery of influenza B virus with the H273Y point mutation in the neuraminidase active site from a human patient. J Clin Microbiol 50:2500–2502. doi: 10.1128/JCM.00682-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ilyushina NA, Seiler JP, Rehg JE, Webster RG, Govorkova EA. 2010. Effect of neuraminidase inhibitor-resistant mutations on pathogenicity of clade 2.2 A/Turkey/15/06 (H5N1) influenza virus in ferrets. PLoS Pathog 6:e1000933. doi: 10.1371/journal.ppat.1000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Duan S, Boltz DA, Seiler P, Li J, Bragstad K, Nielsen LP, Webby RJ, Webster RG, Govorkova EA. 2010. Oseltamivir-resistant pandemic H1N1/2009 influenza virus possesses lower transmissibility and fitness in ferrets. PLoS Pathog 6:e1001022. doi: 10.1371/journal.ppat.1001022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ives JA, Carr JA, Mendel DB, Tai CY, Lambkin R, Kelly L, Oxford JS, Hayden FG, Roberts NA. 2002. The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res 55:307–317. doi: 10.1016/S0166-3542(02)00053-0. [DOI] [PubMed] [Google Scholar]
- 73.Moscona A. 2009. Global transmission of oseltamivir-resistant influenza. N Engl J Med 360:953–956. doi: 10.1056/NEJMp0900648. [DOI] [PubMed] [Google Scholar]
- 74.Bloom JD, Gong LI, Baltimore D. 2010. Permissive secondary mutations enable the evolution of influenza oseltamivir resistance. Science 328:1272–1275. doi: 10.1126/science.1187816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rameix-Welti MA, Munier S, Le Gal S, Cuvelier F, Agou F, Enouf V, Naffakh N, van der Werf S. 2011. Neuraminidase of 2007-2008 influenza A(H1N1) viruses shows increased affinity for sialic acids due to the D344N substitution. Antivir Ther 16:597–603. doi: 10.3851/IMP1804. [DOI] [PubMed] [Google Scholar]
- 76.Hurt AC, Hardie K, Wilson NJ, Deng YM, Osbourn M, Leang SK, Lee RT, Iannello P, Gehrig N, Shaw R, Wark P, Caldwell N, Givney RC, Xue L, Maurer-Stroh S, Dwyer DE, Wang B, Smith DW, Levy A, Booy R, Dixit R, Merritt T, Kelso A, Dalton C, Durrheim D, Barr IG. 2012. Characteristics of a widespread community cluster of H275Y oseltamivir-resistant A(H1N1)pdm09 influenza in Australia. J Infect Dis 206:148–157. doi: 10.1093/infdis/jis337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Butler J, Hooper KA, Petrie S, Lee R, Maurer-Stroh S, Reh L, Guarnaccia T, Baas C, Xue L, Vitesnik S, Leang SK, McVernon J, Kelso A, Barr IG, McCaw JM, Bloom JD, Hurt AC. 2014. Estimating the fitness advantage conferred by permissive neuraminidase mutations in recent oseltamivir-resistant A(H1N1)pdm09 influenza viruses. PLoS Pathog 10:e1004065. doi: 10.1371/journal.ppat.1004065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pinilla LT, Holder BP, Abed Y, Boivin G, Beauchemin CA. 2012. The H275Y neuraminidase mutation of the pandemic A/H1N1 influenza virus lengthens the eclipse phase and reduces viral output of infected cells, potentially compromising fitness in ferrets. J Virol 86:10651–10660. doi: 10.1128/JVI.07244-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith AM, Perelson AS. 2011. Influenza A virus infection kinetics: quantitative data and models. Wiley Interdiscip Rev Syst Biol Med 3:429–445. doi: 10.1002/wsbm.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Matrosovich MN, Matrosovich TY, Gray T, Roberts NA, Klenk HD. 2004. Neuraminidase is important for the initiation of influenza virus infection in human airway epithelium. J Virol 78:12665–12667. doi: 10.1128/JVI.78.22.12665-12667.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]