Abstract
A considerable portion of vertebrate genomes are made up of endogenous retroviruses (ERVs). While aberrant or uncontrolled ERV expression has been perceived as a potential cause of disease, there is mounting evidence that some ERVs have become integral components of normal host development and physiology. Here, we revisit the longstanding concept that some of the gene products encoded by ERVs and other endogenous viral elements may offer to the host protection against viral infection. Notably, proteins produced from envelope (env) genes have been shown to act as restriction factors against related exogenous retroviruses in chickens, sheep, mice, and cats. Based on the proposed mode of restriction and the domain architecture of known antiretroviral env, we argue that many more env gene-derived restriction factors await discovery in vertebrate genomes, including the human genome.
INTRODUCTION
Scattered within vertebrate genomes are retroviral fossils called endogenous retroviruses (ERVs). The astonishing abundance and diversity of ERVs in a wide range of vertebrates is testimony to the ancient conflict and rich coevolutionary history of retroviruses and their hosts. For example, ∼8% of the human genome is comprised of ERV sequences, which can be classified into dozens of families that have been assimilated at different time points, ranging from over a hundred million years ago to perhaps less than a hundred thousand years ago (1). In any given species, the vast majority of ERV sequences appear to represent biochemically inert genomic material evolving under no selective constraint. Biochemical repression is imposed by host cellular machineries that appear to be dedicated to silencing the expression and proliferation of both exogenous and endogenous retroelements (2). Failure to muzzle ERV expression leads to rampant deregulation of the genome with likely deleterious consequences (2, 3). Indeed, aberrant overexpression of human ERVs has long been associated with various disease states, including cancers and autoimmune disorders, although the significance of ERV expression for the etiology and progression of these diseases remains unclear (1).
While these observations indicate that ERVs represent a threat to the integrity of genome function, there is also growing evidence that a fraction of this virus-derived genetic material has been adopted during evolution to serve beneficial functions for their host. In particular, it has come to light that ERVs have deposited a vast reservoir of prefunctional but generally latent cis-regulatory elements (e.g., promoters and transcription factor binding sites) that are occasionally recruited during evolution to become part of the “normal” regulation of adjacent host genes (3, 4). The protein-coding regions of ERVs also provide genetic material poised to be coopted by the host. The Syncytin genes of mammals represent the best-documented case of the molecular “domestication” of ERV gene products for cellular function. There is compelling evidence that Syncytin genes have been coopted from ERV-derived envelope (env) genes repeatedly and independently in various mammal lineages, where they appear to serve a physiological function in the placenta (5). Here, we focus on a distinct set of env-derived genes that appear to function in host antiviral defense in several vertebrate species.
Integration of the viral genome into the host genome is an obligatory step in the life cycle of an exogenous retrovirus. As such, integration of the proviral genome into the host germ line facilitates its vertical inheritance. The long-term persistence and potential fixation in the host genome is dependent on forces such as drift and natural selection (1, 6). Proviral genomes are delimited by regulatory regions identified as long terminal repeat (LTR)-flanking sequences, which code for three major polyproteins: the gag (group-specific antigen), pol (polymerase), and env (envelope) gene products. The env genes encode the surface (SU) and transmembrane (TM) subunits. The SU subunit determines cell specificity, dictating host and cellular tropism, while the TM subunit primarily drives fusion of viral and target cell surfaces. As Env facilitates targeting and entry to specific cells types, Env can also block further infection or “superinfection” (7). In this process, termed receptor interference, the Env of preinfected cells serves as a blockade to viruses that utilize the same host receptor. This speaks to a mechanism in which a retrovirus may “stake its claim” and monopolize the infected cell in an effort to eliminate competing viruses. Consequently, an inherited provirus expressing an Env that will confer protection to all cells in the next generation may rapidly sweep through a host population via natural selection (1, 6).
RECENT AND REPEATED EMERGENCE OF ANTIVIRAL ENDOGENOUS env GENES
Cases of endogenous env genes acting as a restriction factor have been documented for a wide range of vertebrates. One of the earliest reports was made for domestic chickens, for which several env genes derived from endogenous avian leukosis viruses (ALVs) were found to confer protection against exogenous ALV of the same subgroup by impeding retroviral entry (8). Similarly, in the domestic sheep, at least one copy of an endogenous Jaagsiekte sheep retrovirus (enJSRV56A1) expresses an Env protein that thwarts exogenous JSRV infection (9). At least 3 distinct antiviral env genes have been documented in mice. Friend virus susceptibility protein 4 (encoded by the Fv4 gene), also known as AKR virus restriction 1 (Akvr1 [10]), is derived from an endogenous ecotropic murine leukemia virus (MLV) and restricts ecotropic MLV (11). Resistance to mink cell focus-forming virus (Rmcf) is derived from an endogenous polytropic MLV and defends against polytropic mink cell focus-forming MLV (11). Rmcf2, derived from an endogenous xenotropic MLV, also confers resistance against polytropic MLV (11). Not to be outperformed by mice, domestic cats also carry a set of truncated env genes (ERV-DC7 and ERV-DC16) which encode the Refrex-1 restriction factors. These env genes are encoded by endogenous gammaretroviruses (group II) that protect against infection by exogenous feline leukemia virus subgroup D (FeLV-D) as well as endogenous group I gammaretroviruses (12). Each of the env genes listed above has been proposed to restrict via receptor interference, though there is still limited experimental evidence supporting a precise cellular mechanism. Nonetheless, these data indicate that in several vertebrate species, endogenous env genes derived from a variety of retroviruses can act as restriction factors against related retroviruses.
The aforementioned set of restriction env genes appear to be relatively recent additions to their host genomes. The sheep enJSRV56A1 insertion has been investigated throughout the Caprinae subfamily, is estimated to be ∼3 million years old, and was found to be fixed in the domestic sheep population (9). The ERVs encoding the Refrex-1 proteins also appear to be fixed in the domestic cat population (13), but their presence in other felid species has not been reported. Fv4 was first characterized in G-strain lab mice but was later found to be present in wild populations of Asian mice (11). Fv4 (Akvr1) was also characterized in a wild California mouse population descended from feral Asian mice; the California mice demonstrated resistance to naturally circulating ecotropic MLV but not amphotropic MLV (10). Wild populations of Mus musculus castaneus were found to carry Rmcf2, whereas Rmcf is carried by DBA/2 inbred mice (11). Population-level analyses of these mouse restriction env genes have been limited, but one study estimated the Fv4 locus to be ∼500,000 years old and largely confined to M. musculus castaneus and other related Asian wild mice (11). The ALV-restricting env genes of chickens were found to occur at low frequency in several Chinese chicken breeds and in two White Leghorn populations (14). Thus, all the known instances of vertebrate env genes with restriction activity appear to have originated relatively recently in evolution. More detailed evolutionary analyses are needed to reveal whether natural selection has acted to spread these genes within the population, preserving (purifying selection) or diversifying (positive selection) their coding sequence, as seen with other host restriction factors (15). It is also possible that the benefits of these sequences may be too transient to reach fixation or evince the telltale signatures of natural selection (6). Indeed, the longevity of ERV-derived restriction factors is contingent on many factors (3, 6). Unless env gene-derived restriction factors are continuously under the selective pressure of an evolutionary arms race or become coopted for alternative cellular functions (5), they are doomed to become pseudogenized and/or lost from the population.
MINIMAL ARCHITECTURE OF AN env GENE-DERIVED RESTRICTION FACTOR
Understanding the makeup of env genes operating via receptor interference may guide efforts toward uncovering additional restrictive env genes. Here, we focus on the specific interference mechanism of Env interacting with the host receptor, either during processing or at the cell surface, to serve as a blockade against exogenous retroviruses of the same interference group and the minimal domain architecture required for such restriction activity. Refrex-1 is the most recent addition to the set of env genes proposed to act via receptor interference (12). The Refrex-1 loci are unique compared to those previously described in that they encode truncated Envs that have retained their SU domain and signal peptide (SP) but lack TM domains (Fig. 1). As proposed by Ito et al. (12), these truncated Envs undergo processing and are secreted from cells, where they may then bind to receptors on adjacent host cells. Ito et al. (12) demonstrated that pretreating HEK293T cells with the supernatant of feline T cells (containing Refrex-1) restricts related exogenous retroviruses, suggesting that secreted Refrex-1 was able to protect surrounding cells from infection. Thus, Refrex-1 proteins may reveal the minimal domain architecture required for an env gene-derived restriction factor. Interestingly, these data echo those previously reported for Rmcf, whose restriction activity is maintained in the absence of the TM domain (11) (Fig. 1). Together, these observations substantiate the notion that restrictive env genes need not be full-length but that truncated env genes encoding only SP and SU are sufficient to be processed, secreted, and exert receptor interference activity.
FIG 1.
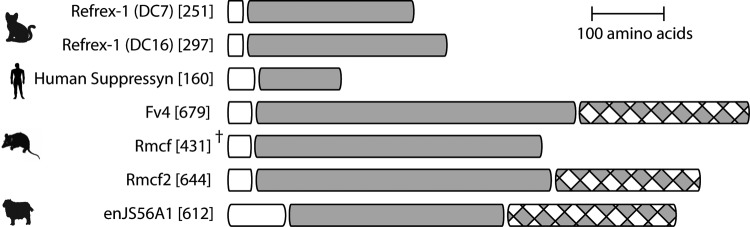
Domain comparison of endogenous Envs proposed to restrict via receptor interference. Domains were determined based on previous characterizations of Refrex-1 (12), Suppressyn (17), Fv4 (11), Rmcf (11), Rmcf2 (11), enJS56A1 (9), Phobius (http://phobius.sbc.su.se/), and UniProt (http://www.uniprot.org/). White boxes, signal peptide; gray boxes, surface subunit; gray and white checkered boxes, transmembrane subunit. †, complete Envs are not displayed. Numbers in brackets indicate length (in amino acids).
Suppressyn: A CANDIDATE env-GENE DERIVED RESTRICTION FACTOR IN HUMANS?
To date, all known env gene-derived restriction factors have been found outside primates. This raises the question of whether env genes coopted for host defense function can be found in humans. The human genome encodes about 30 full-length or near-full-length env genes but also thousands of loci potentially encoding env genes with various levels of truncation (16). These truncated env genes have historically been dismissed as decaying pseudogenes with no functional role. However, in light of the fact that an endogenous env gene need not be full-length to act as restriction factor (Fig. 1), it could well be that the human genome harbors many sequences encoding Env proteins with antiviral activity.
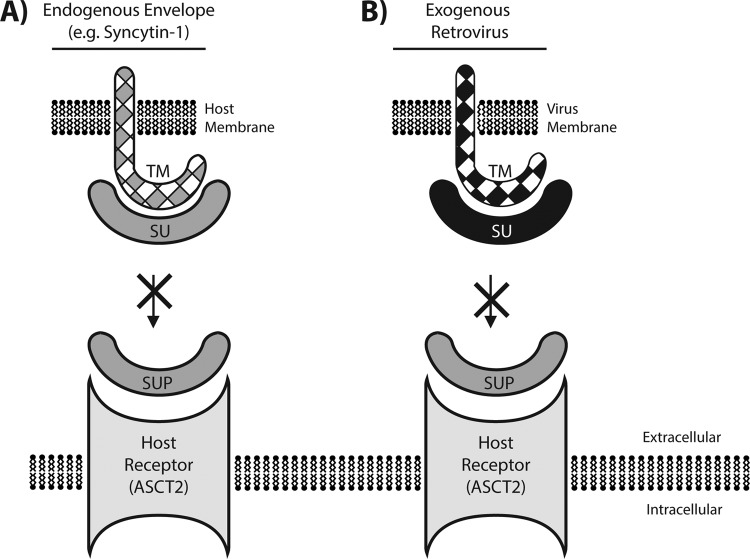
One recently emerged candidate is Suppressyn, a 160-amino-acid Env protein lacking a TM domain (Fig. 1). Suppressyn is encoded by a human endogenous retrovirus F family (HERV-F) element located on human chromosome 21 and expressed at high level in the placenta, where it is proposed to contribute to the regulation of Syncytin-1 (17), one of two full-length env gene-derived proteins thought to be coopted for human placentation (5). Suppressyn is secreted, and it competes with Syncytin-1 for binding to the cell receptor ASCT2, thereby potentially modulating Syncytin-1 function in the placenta (Fig. 2A). Thus, humans appear to encode a truncated env gene that functions via receptor interference to block another domesticated env gene. While a role for Suppressyn in controlling Syncytin-1, and thereby placentation, is plausible, the data also raise the possibility that Suppressyn acts as a restriction factor by preventing a range of exogenous retroviruses from binding to the ASCT2 receptor on placental cells (Fig. 2B). Indeed, several retroviruses are known to use the ASCT2 receptor: specifically, those belonging to the so-called type D interference group, including simian retroviruses 1 to 5, baboon endogenous virus, and feline RD114 (7). If Suppressyn is coopted for antiviral defense, then this activity may not be limited to humans. A cursory search of the University of California—Santa Cruz (UCSC) Genome Browser shows that Suppressyn can be found throughout hominoids and Old World monkeys but not in New World monkeys and prosimian primates, suggesting that the ancestral ERV inserted between 25 and 40 million years ago. It is of interest to note that the nonhuman Suppressyn orthologs were of nearly identical length to that found in humans. Thus, Suppressyn may represent not only an antiretroviral env gene-derived restriction factor in humans but one coopted and functional in a wide range of primate species.
FIG 2.
Suppressyn as a candidate restriction factor. (A) As reported by Sugimoto et al. (17), Suppressyn (SUP) is secreted to the extracellular surface to block Syncytin-1 from host receptor ASCT2. (B) This interaction between Sup and ASCT2 may also lead to a receptor interference-type defense of the host cell receptor from exogenous retroviruses of the same interference group. SU, surface subunit; TM, transmembrane subunit.
CONCLUSIONS
The recurrent appearance of endogenous env genes with antiretroviral activity suggests that this class of host defense genes may exist throughout many vertebrate lineages. These env genes further support a reoccurring theme of host genomes “fighting fire with fire” by tuning viral fossils for viral defense genes, which also encompasses sequences derived from other retroviral genes (e.g., Fv1 from gag [6]), and perhaps also nonvertebrate endogenous env genes (18) and nonretroviral elements (e.g., endogenous Borna-like virus N proteins [19]). The ancestral function of env genes is to facilitate specificity and entry of the virus to a host cell, though this can lead to territorial defense of the occupied host in the form of interference. In turn, this strategy opens the door for the cooption of ERV-encoded env genes for host cell protection. Considering that the human genome harbors hundreds of loci potentially encoding env genes of similar lengths and domain composition as some of the known restrictive env genes, it is tempting to speculate that humans and other primates encode a number of env gene-derived proteins with restriction activity. While additional work is required to further understand the role of Suppressyn, this truncated env gene product represents an excellent candidate as a restriction factor with broad antiretroviral activity encoded in human and other primates.
ACKNOWLEDGMENTS
We thank Jamie E. Henzy for comments on the manuscript. We apologize to colleagues who have produced primary research on the topic but could not be cited or discussed due to space limitations.
REFERENCES
- 1.Magiorkinis G, Belshaw R, Katzourakis A. 2013. ‘There and back again’: revisiting the pathophysiological roles of human endogenous retroviruses in the post-genomic era. Philos Trans R Soc Lond B Biol Sci 368:20120504. doi: 10.1098/rstb.2012.0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung DC, Lorincz MC. 2012. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci 37:127–133. doi: 10.1016/j.tibs.2011.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet 13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 4.Rebollo R, Romanish MT, Mager DL. 2012. Transposable elements: an abundant and natural source of regulatory sequences for host genes. Annu Rev Genet 46:21–42. doi: 10.1146/annurev-genet-110711-155621. [DOI] [PubMed] [Google Scholar]
- 5.Lavialle C, Cornelis G, Dupressoir A, Esnault C, Heidmann O, Vernochet C, Heidmann T. 2013. Paleovirology of ‘syncytins’, retroviral env genes exapted for a role in placentation. Philos Trans R Soc Lond B Biol Sci 368:20120507. doi: 10.1098/rstb.2012.0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aswad A, Katzourakis A. 2012. Paleovirology and virally derived immunity. Trends Ecol Evol 27:627–636. doi: 10.1016/j.tree.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Sommerfelt MA, Weiss RA. 1990. Receptor interference groups of 20 retroviruses plating on human cells. Virology 176:58–69. doi: 10.1016/0042-6822(90)90230-O. [DOI] [PubMed] [Google Scholar]
- 8.Robinson HL, Astrin SM, Senior AM, Salazar FH. 1981. Host susceptibility to endogenous viruses: defective, glycoprotein-expressing proviruses interfere with infections. J Virol 40:745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varela M, Spencer TE, Palmarini M, Arnaud F. 2009. Friendly viruses: the special relationship between endogenous retroviruses and their host. Ann N Y Acad Sci 1178:157–172. doi: 10.1111/j.1749-6632.2009.05002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gardner MB, Kozak CA, O'Brien SJ. 1991. The Lake Casitas wild mouse: evolving genetic resistance to retroviral disease. Trends Genet 7:22–27. doi: 10.1016/0168-9525(91)90017-K. [DOI] [PubMed] [Google Scholar]
- 11.Kozak CA. 2014. Origins of the endogenous and infectious laboratory mouse gammaretroviruses. Viruses 7:1–26. doi: 10.3390/v7010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ito J, Watanabe S, Hiratsuka T, Kuse K, Odahara Y, Ochi H, Kawamura M, Nishigaki K. 2013. Refrex-1, a soluble restriction factor against feline endogenous and exogenous retroviruses. J Virol 87:12029–12040. doi: 10.1128/JVI.01267-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anai Y, Ochi H, Watanabe S, Nakagawa S, Kawamura M, Gojobori T, Nishigaki K. 2012. Infectious endogenous retroviruses in cats and emergence of recombinant viruses. J Virol 86:8634–8644. doi: 10.1128/JVI.00280-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang J, Yu Y, Yao J, Chen Y, Xu G, Yang N, Sun D, Zhang Y. 2011. Molecular identification of avian leukosis virus subgroup E loci and tumor virus B locus in Chinese indigenous chickens. Poult Sci 90:759–765. doi: 10.3382/ps.2010-01133. [DOI] [PubMed] [Google Scholar]
- 15.Daugherty MD, Malik HS. 2012. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet 46:677–700. doi: 10.1146/annurev-genet-110711-155522. [DOI] [PubMed] [Google Scholar]
- 16.Villesen P, Aagaard L, Wiuf C, Pedersen FS. 2004. Identification of endogenous retroviral reading frames in the human genome. Retrovirology 1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugimoto J, Sugimoto M, Bernstein H, Jinno Y, Schust D. 2013. A novel human endogenous retroviral protein inhibits cell-cell fusion. Sci Rep 3:1462. doi: 10.1038/srep01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malik HS, Henikoff S. 2005. Positive selection of Iris, a retroviral envelope-derived host gene in Drosophila melanogaster. PLoS Genet 1:e44. doi: 10.1371/journal.pgen.0010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujino K, Horie M, Honda T, Merriman DK, Tomonaga K. 2014. Inhibition of Borna disease virus replication by an endogenous bornavirus-like element in the ground squirrel genome. Proc Natl Acad Sci U S A 111:13175–13180. doi: 10.1073/pnas.1407046111. [DOI] [PMC free article] [PubMed] [Google Scholar]