ABSTRACT
The expression of xenogeneic TRIM5α proteins can restrict infection in various retrovirus/host cell pairings. Previously, we have shown that African green monkey TRIM5α (AgmTRIM5α) potently restricts both human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus mac239 (SIVmac239) replication in a transformed human T-cell line (L. V. Coren, et al., Retrovirology 12:11, 2015, http://dx.doi.org/10.1186/s12977-015-0137-9). To assess AgmTRIM5α restriction in primary cells, we transduced AgmTRIM5α into primary rhesus macaque CD4 T cells and infected them with SIVmac239. Experiments with T-cell clones revealed that AgmTRIM5α could reproducibly restrict SIVmac239 replication, and that this restriction synergizes with an intrinsic resistance to infection present in some CD4 T-cell clones. AgmTRIM5α transduction of virus-specific CD4 T-cell clones increased and prolonged their ability to suppress SIV spread in CD4 target cells. This increased antiviral function was strongly linked to decreased viral replication in the AgmTRIM5α-expressing effectors, consistent with restriction preventing the virus-induced cytopathogenicity that disables effector function. Taken together, our data show that AgmTRIM5α restriction, although not absolute, reduces SIV replication in primary rhesus CD4 T cells which, in turn, increases their antiviral function. These results support prior in vivo data indicating that the contribution of virus-specific CD4 T-cell effectors to viral control is limited due to infection.
IMPORTANCE The potential of effector CD4 T cells to immunologically modulate SIV/HIV infection likely is limited by their susceptibility to infection and subsequent inactivation or elimination. Here, we show that AgmTRIM5α expression inhibits SIV spread in primary effector CD4 T cells in vitro. Importantly, protection of effector CD4 T cells by AgmTRIM5α markedly enhanced their antiviral function by delaying SIV infection, thereby extending their viability despite the presence of virus. Our in vitro data support prior in vivo HIV-1 studies suggesting that the antiviral CD4 effector response is impaired due to infection and subsequent cytopathogenicity. The ability of AgmTRIM5α expression to restrict SIV infection in primary rhesus effector CD4 T cells now opens an opportunity to use the SIV/rhesus macaque model to further elucidate the potential and scope of anti-AIDS virus effector CD4 T-cell function.
INTRODUCTION
The TRIM5α cellular protein is a well-studied resistance factor (1) that is a major contributor to the inability of human immunodeficiency virus type 1 (HIV-1) to replicate in Old World monkey CD4 T cells, especially those from rhesus macaque (2–6). While endogenous TRIM5α does not restrict permissive virus-cell pairings, expression of xenogeneic TRIM5α can make cells resistant to infection (7–11). Experiments with xenogeneic expression of TRIM5α have revealed a somewhat complicated pattern of restriction in a variety of virus-host pairings (5, 7–9, 11–14).
Cytoplasmic TRIM5α restricts infection rapidly after viral entry (15), disrupting reverse transcription (2–5, 16, 17) as well as later stages of the infection process (16, 17). During restriction, TRIM5α binds the retroviral capsid core, a capsid protein-coated structure which contains all of the viral molecules required for infection: the RNA genome, reverse transcriptase, and integrase. While the exact mechanism of restriction is not completely understood, two nonexclusive models posit that restricting TRIM5α binds the capsid core by forming a cage-like structure (18) that either causes the core to prematurely uncoat (16, 19–21), thereby interfering with reverse transcription, or engages the ubiquitin proteasome pathway through its ubiquitin ligase activity, causing the destruction of the caged core complex (10, 17, 22–24). Because TRIM5α binds cooperatively to the capsid core and its cytoplasmic concentration is limiting, restriction is saturable: increasing amounts of viral cores entering the cell from high multiplicities of infection (MOI) titrate out cytoplasmic TRIM5α, eliminating restriction (18, 25–28).
Conceptually, xenogeneic expression of rhesus macaque TRIM5α (rhTRIM5α) by gene transfer is an approach to protect primary human CD4 T cells from HIV-1 in vivo (29–32). However, in vitro experiments have found that, while rhTRIM5α-transduced cells protected human CD4 T cells in monoculture, there was no HIV-1 restriction in coculture with untransduced cells (33, 34) due to cell-to-cell infection (33). Similar results were observed with a stabilized human TRIM5α mutant that has a longer half-life (30). In contrast, our recent experiments found that near-physiological expression of African green monkey TRIM5α (AgmTRIM5α) in transformed human CD4 T cells provided potent restriction against both HIV-1 and simian immunodeficiency virus (SIV) in replication assays using both cell-free and cell-to-cell infection challenges (34). Thus, unlike rhTRIM5α with HIV-1, AgmTRIM5α could restrict both HIV-1 and SIV replication in the presence of infected cells. To extend our prior studies, we examined the ability of AgmTRIM5α to restrict SIVmac239 in primary rhesus macaque CD4 T cells as well as its impact on antiviral function. Our results found that AgmTRIM5α could effectively restrict SIVmac239 in primary CD4 T cells, and, importantly, augment the ability of SIV-specific T cells to suppress viral replication in autologous CD4 T cells.
MATERIALS AND METHODS
Retroviral vector production.
Retroviral vectors expressing AgmTRIM5α, gorilla TRIM5α (gorTRIM5α), and the CM9-6 rhesus macaque T-cell receptor (TCR), specific for the SIV CM9 epitope, were produced by transfecting the pSMS-Agm (34), pSMS-gor (34), and pCM9-6 (35) constructs, respectively, into Phoenix RD114 clone 22 packaging cells (kind gift of Hans-Peter Kleim, Fred Hutchinson Cancer Research Center, Seattle, WA) (36) using the TransIT-293 transfection agent (Mirus Bio). Phoenix RD114 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS), 10 mM HEPES buffer, 2 mM l-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin (all medium supplements were obtained from Life Technologies, Inc.). Vector supernatants were clarified by filtration though a 0.45-μm filter.
CD4 T cell transduction.
Primary rhesus macaque CD4 T-cell lines were isolated from peripheral blood of several rhesus macaques by magnetic sorting with anti-CD4 paramagnetic microbeads through an LS column (Miltenyi Biotech). CD4 clones generated by limiting dilution were tested for susceptibility to SIVmac239 prior to expansion and transduction with retroviral vectors. Except as noted, clones with median levels of infection susceptibility were selected for study. CD4 T-cell lines were cultured in RPMI 1640 media supplemented with 10% (vol/vol) FBS, 10 mM HEPES buffer, 2 mM glutamine, 50 U/ml interleukin-2 (IL-2; NIH AIDS Reference and Reagent Program). Primary cell cultures were expanded by biweekly stimulation by the addition of 30 ng/ml CD3 monoclonal antibody clone SP34-2 (BD Biosciences) and irradiated human peripheral blood mononuclear cells (PBMCs), as previously described (37–39). Transductions of primary cells were carried out as previously described (35). Briefly, non-tissue culture-treated plates were coated with 20 μg/ml RetroNectin (TaKaRa Bio) for 2 h, blocked with 2% (wt/vol) bovine serum albumin (BSA) for 30 min at room temperature, and washed with phosphate-buffered saline (PBS) prior to the addition of retrovirus supernatant. Plates with vector supernatants were centrifuged at 1,500 × g for 2 h at 32°C prior to the addition of 1 × 106 to 1.5 × 106 CD4 T cells in 500 μl RPMI and IL-2 to 40 U/ml (final concentration). Plates then were centrifuged at 1,500 × g for 1 h at 32°C. Cultures were analyzed for transductants by flow cytometry 48 to 72 h postransduction for either green fluorescent protein (GFP) fluorescence (TRIM5α vectors) or CM9 peptide-major histocompatibility complex class I (MHC-I)-phycoerythrin (PE) tetramer (MBL, Inc.) staining. Cell lines expressing AgmTRIM5α and gorTRIM5α were isolated with a BD FACSAria II flow cytometer (BD Biosciences). CM9-6 transductants were isolated using the CM9-MHC I-PE tetramer and anti-PE paramagnetic microbeads (Miltenyi Biotech). Postsort purity ranged between 95 and 99% (data not shown).
Direct infection of CD4 T-cell clones.
SIVmac239 stocks were produced by transfecting human embryonic kidney 293T cells with p239macSPXL (gift of Ronald Desrosiers) using the TransIT-293 transfection agent (40). Virus supernatants were collected at 48 and 72 h posttransfection and stored as aliquots at −80°C prior to use. For infection, 1 × 106 to 1.5 × 106 CD4 T cells were infected with 80 μl to 160 μl of the SIVmac239 virus stock in 1 ml of medium for a multiplicity of infection (MOI) of 1 with 2 μg/ml hexadimethrine bromide in a 15-ml tube (2058; BD Falcon).
Suppression of virus spread assay.
CD4 T-cell clones that were susceptible to infection and not SIV specific were infected with SIVmac239 at an MOI of >0.001, labeled with CellTrace violet (CTV) by using the provider's procedure (Life Technologies), and then cocultured with autologous SIV-specific effector CD4 T cells or those with an irrelevant specificity at a 1:5 (target-to-effector) ratio in a round-bottom tube (2058; BD Falcon). Cells were maintained in IL-2-supplemented RPMI 1640 media as described above. Suppression was analyzed by flow cytometry for infection by CD4 surface and intracellular Gag staining in the CTV-labeled population and by comparing the frequency of SIV-specific infected targets versus irrelevant CD4 T cell effector cocultures at multiple time points. Cultures with uninfected targets were included in each assay as negative controls.
Flow cytometry analysis of infected cells.
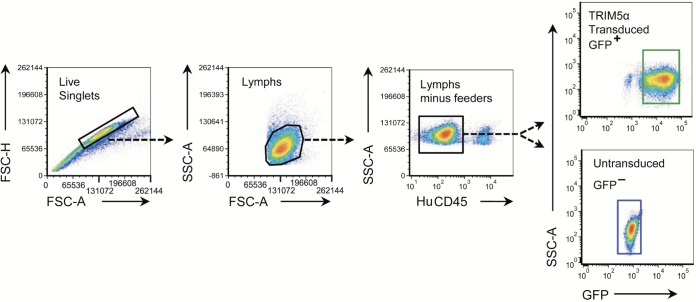
Cells (0.5 × 106 to 1 × 106) were analyzed for SIV infection by surface staining with CD4-peridinin chlorophyll protein (PerCP)-Cy5.5 (clone OKT4) and human CD45-allophycocyanin (APC) (BD Biosciences), followed by an intracellular stain with the p27CA-specfic 2F12-PE antibody (Quality Biological, Inc.). Staining for the CM9-6 TCR was carried out using CM9-MHC-I tetramer combined with CD3, CD4, and CD45 antibodies (BD Biosciences). The 2F12 antibody was labeled using the Zenon-PE antibody labeling kit (Life Technologies) by following the recommended protocol. Analyses were performed as previously described (39). Briefly, surface-labeled cells were washed prior to resuspension in the BD Cytofix/Cytoperm solution and incubated at room temperature for 30 min. Cells were fixed with 4% (wt/vol) paraformaldehyde in Dulbecco's PBS (D-PBS; Life Technologies) for 20 min at room temperature, followed by the addition of 3.5 ml of 0.1% (wt/vol) saponin (Sigma-Aldrich) in D-PBS and Zenon-PE-conjugated 2-F-12 for 30 min at room temperature in the dark and immediately analyzed by an LSRII flow cytometer (BD Biosciences). Data analysis was performed using FCS Express (De Novo Software, Los Angeles, CA). Cells were gated as follows (Fig. 1). Live single cells were gated by forward scatter height (FSC-H) and by forward side scatter area (FSC-A), then lymphocytes were gated by side scatter area (SSC-A) by forward side scatter area, then rhesus macaque T cells were gated from any remaining human feeder cells by human CD45 staining, and finally TRIM5α-expressing versus untransduced cells were gated by GFP fluorescence.
FIG 1.
Flow cytometry gating strategy used to study TRIM5α-transduced cells. Gated plots for analysis of infection in untransduced or TRIM5α-transduced cells are shown (left to right): cells gated for live singlets, live singlets gated for lymphocytes (Lymphs), human feeders removed from the lymphocyte population by gating for human CD45-negative cells, and TRIM5α-transduced cells from the GFP+ gate as well as untransduced cells from the GFP− gates.
RESULTS
AgmTRIM5α restricts SIVmac239 replication in primary rhesus CD4 T cells.
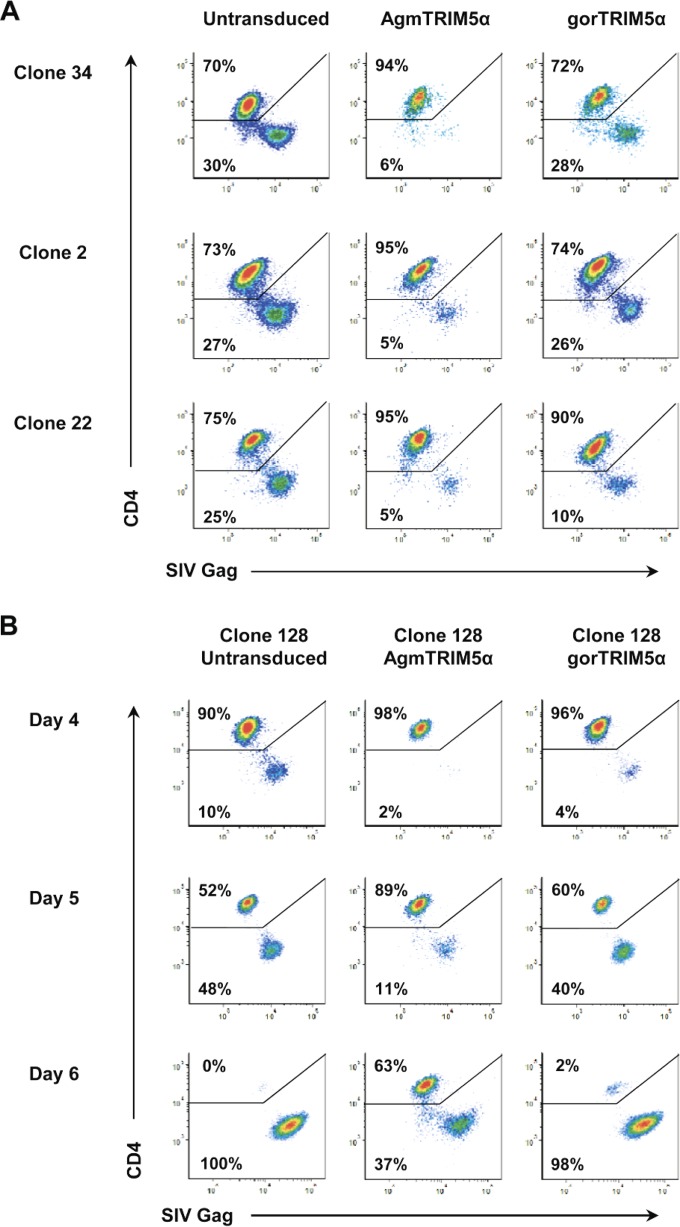
To examine the extent of AgmTRIM5α restriction of SIVmac239 replication in primary rhesus macaque CD4 T cells, we transduced T-cell clones with a retroviral vector containing both the AgmTRIM5α gene isolated from Tantalus African green monkey (9) and the eGFP gene. Based on our prior study, this vector produces nearly physiological levels of AgmTRIM5α and confers a 2- to 3-log level of restriction to HIV-1 and SIV replication in a transformed human CD4 T-cell line (34). To provide a negative control for ectopic expression of TRIM5α, we also transduced the primary CD4 T-cell clones with a gorilla TRIM5α gene. Our prior study found that gorTRIM5α expression induced only weak SIV restriction in the human cell system (34). GFP-positive AgmTRIM5α- and gorTRIM5α-transduced cells were exposed to SIVmac239 at an MOI of 0.1 and examined by flow cytometry for productively infected cells, i.e., the loss of surface CD4 and presence of intracellular Gag. At day 4 postinfection, three representative AgmTRIM5α-transduced clones, 34, 2, and 22, contained relatively low levels of CD4−/Gag+-infected cells (5 to 6%) compared to the levels present in the gorTRIM5α-transduced cells, 10 to 28%, and untransduced clones, 25 to 30% (Fig. 2A). Overall, the results from analyzing seven clones found AgmTRIM5α restriction to replication in primary CD4 T cells (data not shown).
FIG 2.
SIVmac239 restriction by AgmTRIM5α and gorTRIM5α. Flow cytometry analyses for SIV infection in untransduced CD4 T-cell clones and those expressing AgmTRIM5α or gorTRIM5α are presented. Plots of gated cells are shown for CD4 staining on the y axis and SIV Gag staining on the x axis. (A) SIVmac239 restriction in 3 representative clones (clones 34, 2, and 22) at day 5 postinfection. (B) SIVmac239 restriction in the representative clone 128 at days 4, 5, and 6 postinfection. Data presented are representative of three independent trials.
TRIM5α restriction can be overcome by exposure to high levels of virus (25–28), conditions that are present during cell-to-cell virus spread (41–50). To examine the durability of resistance over time as the exposure to cell-free and cell-associated virus increases, AgmTRIM5α-transduced cells were analyzed at 4, 5, and 6 days postinfection. The frequency of infected cells at all three time points was lower for the AgmTRIM5α-transduced cells, 2% at day 4, 11% at day 5, and 37% at day 6 (Fig. 2B), than that in either the gorTRIM5α (4%, 40%, and 98%, respectively) or untransduced cell cultures (10%, 48%, and 100%, respectively). The gorTRIM5α culture showed at most a slight restriction compared to the untransduced culture (4% versus 10%) at day 4 (Fig. 2B). However, by day 5 there was little difference between the two cultures (40% versus 48%), consistent with the weak SIV restriction by gorTRIM5α previously observed (34). Thus, xenogeneic expression of TRIM5α per se has little effect on SIV replication. Remarkably, by day 6 the majority (63%) of AgmTRIM5α-transduced T cells showed no signs of infection, being CD4+ and Gag−, while essentially all of the gorTRIM5α and untransduced control T cells were infected (98% and 100%, respectively). Thus, despite the high levels of SIVmac239, AgmTRIM5α expression was able to markedly restrict viral replication, protecting transduced cells from infection (Fig. 2A and B).
AgmTRIM5α restriction acts synergistically with natural resistance in CD4 T-cell clones.
We have observed that primary rhesus CD4 T-cell clones exhibit various degrees of susceptibility to SIVmac239 infection, with some having strong blocks to productive infection (51). For these experiments, we observed that out of 41 CD4 T-cell clones at day 5 postinfection, 17% were highly susceptible (>50% of cells infected) and 20% were highly resistant to infection (<5% of cells infected; M. T. Trivett and D. E. Ott, unpublished data). To determine if this intrinsic resistance to infection could augment AgmTRIM5α restriction, we combined these two factors. CD4 T-cell clones from the same donor animal were screened for their relative susceptibilities to SIVmac239 infection. Clones found to be exceptionally susceptible or resistant were used to produce AgmTRIM5α-transduced cells that were infected with our SIVmac239 stock and compared to infected, untransduced clones. Untransduced sensitive clones had significant levels of CD4−/Gag+-infected cells 5 days postinfection (24% in the representative data), which increased to essentially 100% after 13 days (Fig. 3A). Even though the resistant cells showed about half of the frequency of infected cells at the day 5 time point, few uninfected cells remained after 13 days (Fig. 3). Interestingly, there were some resistant cells in the culture that had downregulated CD4 yet remained Gag negative after 13 days. This is not due to CD4 antibody staining, because the OKT4 monoclonal antibody used is not blocked by gp120SU binding. Thus, the decrease in CD4 likely is due to infection. Perhaps these cells are in an earlier phase of virion production where Nef and Env downregulate CD4 surface expression before Gag accumulates to levels that are detectable (52). Indeed, CD4− cells are not substantially present in the resistant/AgmTRIM5α culture (Fig. 3B); thus, this population is induced by infection. Also, the Gag fluorescence intensity of the CD4− cells markedly shifts from day 5 to day 13 in all of the cultures, consistent with an increase in Gag with time (Fig. 3). Finally, the CD4− population is not discrete; rather, it is present as a continuum of Gag antibody fluorescence ranging from Gag− to Gag+, suggesting an accumulation of Gag in the cells over time. Taken together, the intrinsic resistance present in some CD4 T cells can be a relatively short-lived inhibitor of viral replication under these conditions.
FIG 3.
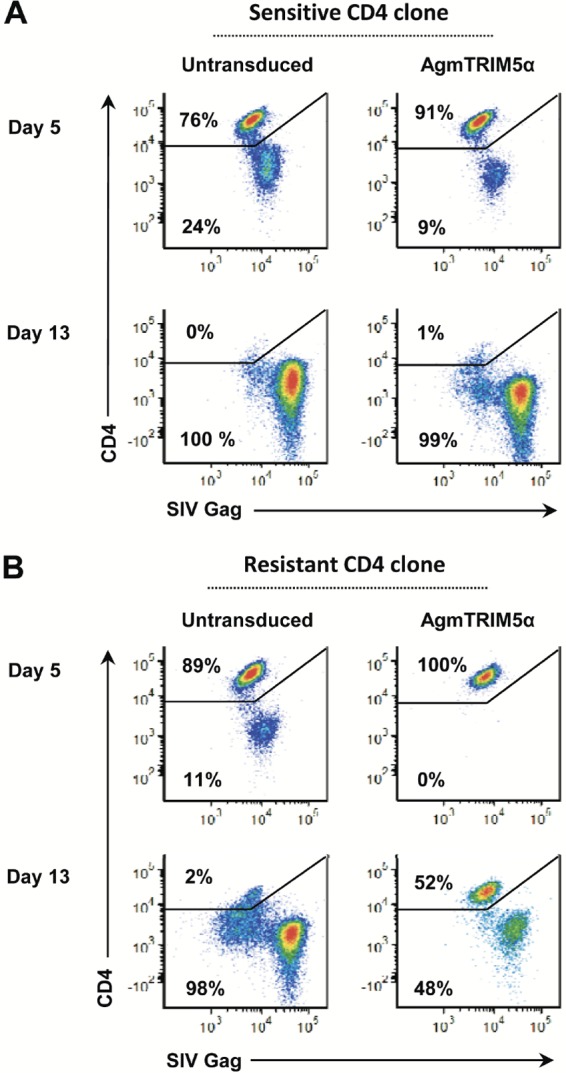
AgmTRIM5α restriction effect is enhanced in naturally resistant CD4 T-cell clones. Flow cytometry analyses for SIV infection in untransduced CD4 T-cell clones and those expressing AgmTRIM5α at days 5 and 13 postinfection are presented. Plots of gated cells are shown for CD4 staining on the y axis and SIV Gag staining on the x axis. (A) Relative SIVmac239 infection in a naturally susceptible CD4 T-cell clone. (B) Relative SIVmac239 infection in a naturally resistant clone. Data presented are representative of three independent trials.
The analysis of the sensitive cells expressing AgmTRIM5α cells (Fig. 3A) showed some restriction at day 5 postinfection (9% in the infected cells versus 24% in the untransduced cells), yet after 13 days, essentially all of the cells in the culture were CD4− and nearly all were Gag+. In contrast, the AgmTRIM5α-transduced resistant clone exhibited robust restriction; no detectable infected cells were observed at day 5, whereas 11% of infected cells were observed in the untransduced cells. Furthermore, this effect was durable, with 52% uninfected CD4+/Gag− cells remaining in the AgmTRIM5α culture at day 13 while essentially all of the untransduced cells were infected at that time point. Also, there were fewer of the CD4−/Gag− cells at day 13 in the AgmTRIM5α-transduced resistant clone culture than in the other cultures, suggesting fewer cells in early-stage infection. Therefore, AgmTRIM5α expression appears to act synergistically with this uncharacterized intrinsic cellular resistance factor to more effectively restrict SIV expression.
CD4 effector cells are susceptible to SIV infection in suppression assays.
In our studies of anti-SIV responses in CD8 T cells, we employ the retrovirus-mediated transfer of various SIV-specific TCR genes to evaluate effector functions of different TCR/CD8 T-cell combinations in a defined in vitro system (35). Of the parameters we evaluate, the most direct and physiologically relevant measure of antiviral function is the ability of SIV-specific CD8 T cells to specifically suppress SIV spread between autologous target CD4 T cells (35, 39). Applying this approach to study SIV-specific effector CD4 T cells is complicated by the possibility that the effectors themselves are substrates for infection. To study the extent of effector cell infection during suppression, CD4 T-cell clones transduced with the CM9-6 TCR were used as effectors in our suppression of the virus spread assay. This MHC class I-restricted TCR was chosen due to the lack of an appropriate MHC class II-restricted TCR and our prior successful transfer experiments with CM9-6 in CD8 T cells (35). Since both effectors and targets express CD4, the targets were labeled with the CTV fluorescent dye to differentiate them from the CVT− effectors in flow cytometry analyses (Fig. 4A). Suppression is evaluated by comparing the frequency of infected targets in the SIV-specific CD4 effector cell cocultures with that of the untransduced effector cocultures. In a representative flow cytometry analysis of SIV-specific clone A cells, the CTV-labeled targets at day 5 revealed strong suppression of SIV replication (Fig. 4B): 65% of the CD4 targets in the untransduced effector coculture were infected, while only 3% of those with the SIV-specific effectors were infected. Despite this initially robust suppression, at day 7, there was little difference between the two cultures: 68 to 78% of the target cells were infected (Fig. 4B). Thus, the suppression exhibited by the SIV-specific CD4 effectors, while initially effective, was not durable, potentially due to SIV infection of the effectors themselves.
FIG 4.
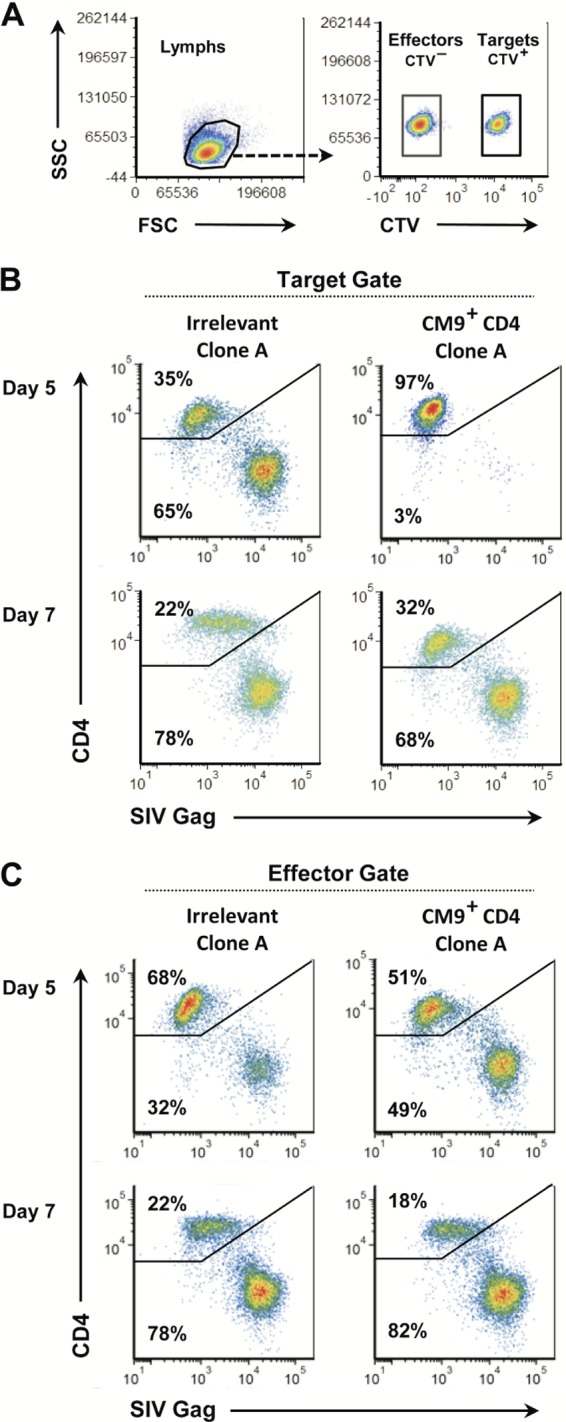
SIV-specific effector CD4 T cells exhibit a direct antiviral effect and are susceptible to infection during virus suppression. (A) Flow cytometry plots demonstrating CTV labeling differentiation between CTV− CD4+ effectors and CTV CD4 targets in coculture are presented. (B and C) Flow cytometry analyses for SIV infection in untransduced CD4 T-cell clones and those expressing the CM9 TCR at days 5 and 7 postinfection are presented. Plots of gated cells are shown for CD4 staining on the y axis and SIV Gag staining on the x axis. (B) Flow cytometry analysis gated on targets. (C) Flow cytometry analysis gated on targets. Data presented are representative of five independent trials.
To examine this possibility, we measured the infection status of the effectors. Flow cytometry analysis gated on CTV− revealed that both the transduced SIV-specific and untransduced effector cells were infected by virus produced from the target cells. Indeed, the amounts of infected effector cells were approximately half that of the target cells in the day 5 coculture with irrelevant effectors, with 65% of the target cells infected (Fig. 4B, upper left plot) and 32% to 49% of the effector cells infected (Fig. 4C). By day 7, the levels of infected cells rose in both the SIV-specific and irrelevant effector cultures to nearly that of their corresponding targets. These data show that, as expected, CD4 effectors are readily infected as they suppress viral replication in the target cells.
AgmTRIM5α restriction extends the durability of virus suppression by CD4 T-cell effectors.
The data described above suggest that the initially effective suppression by the SIV-specific effectors ultimately is lost as the effector population becomes infected (see day 5 versus 7 data in Fig. 4B and C). To examine whether reducing infection could increase the durability of suppression by CD4 effectors, we transduced AgmTRIM5α into genetically modified CM9-specific clones. Suppression assays with these transduced cell clones (representative results with clone B are shown in Fig. 5) showed that the SIV-specific cells expressing AgmTRIM5α maintained suppression of SIVmac239 in the CTV-labeled targets through day 7 (Fig. 5A), with only 6% of the targets showing signs of productive infection. In contrast, the SIV-specific cells without AgmTRIM5α showed only a modest level of suppression at day 5 (10% infected targets) compared to the irrelevant clone culture control (25% infected targets). In total, our experiments demonstrated that the relatively weak suppression of the SIV-specific CD4 effectors was not durable at day 7 without AgmTRIM5α.
FIG 5.
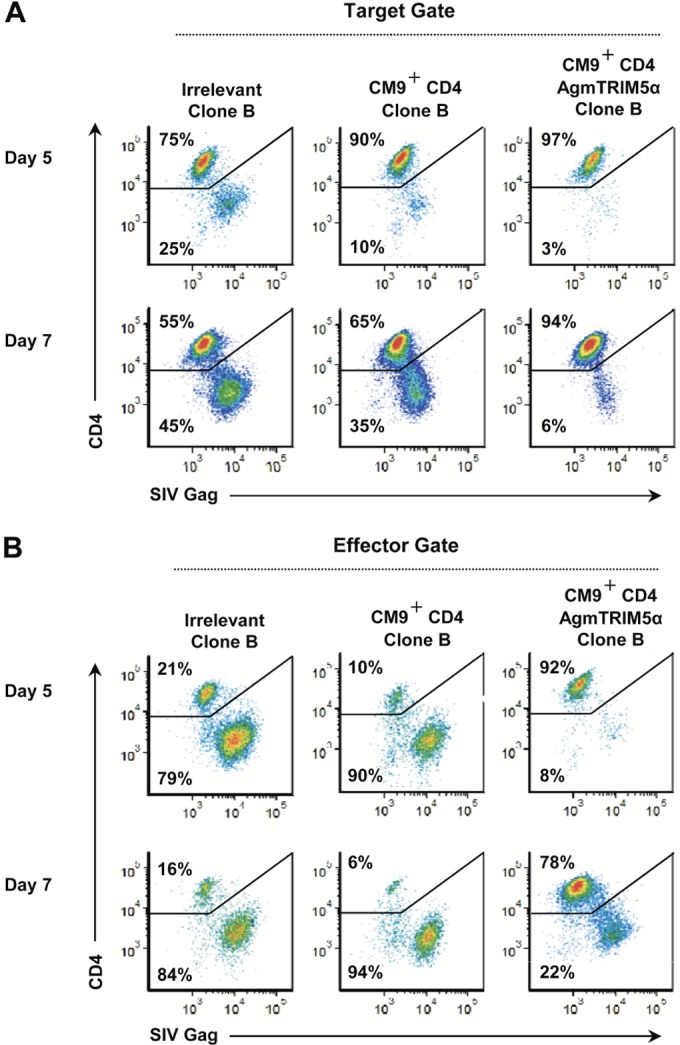
Virus-specific effector CD4 T cells, expressing AgmTRIM5α, are protected from infection during virus suppression and exhibit enhanced antiviral activity. Flow cytometry analyses for SIV infection in untransduced CD4 T-cell clones, those expressing CM9 TCR, and those expressing both CM9 TCR and AgmTRIM5α at days 5 and 7 postinfection are presented. Plots of gated cells are shown for CD4 staining on the y axis and SIV Gag staining on the x axis. (A) Flow cytometry analysis gated on targets. (B) Flow cytometry analysis gated on targets. Data presented are representative of three independent trials.
Examining the status of the CD4 effector populations revealed strong restriction of SIV infection in the AgmTRIM5α-transduced SIV-specific T cells at day 5, with only 8% productively infected cells versus 79% and 90% in the irrelevant and SIV-specific cells, respectively (Fig. 5B). While the frequency of infected effector cells in the AgmTRIM5α-transduced culture did rise to 22% at day 7, the non-AgmTRIM5α cultures showed no restriction at this time point, containing 94% and 85% infected cells. Taken together, these results show that SIV restriction induced by AgmTRIM5α preserves an uninfected pool of CD4 effector cells and provides for stronger and more durable viral suppression.
DISCUSSION
Here, we find that AgmTRIM5α-mediated restriction can both enhance and prolong the antiviral function of SIV-specific rhesus macaque CD4 T cells in our virus suppression assay. This effect correlates with reduced levels of infection in the AgmTRIM5α-expressing SIV-specific effector CD4 T-cell clones, suggesting that the infected effector cells lose their function by either cytopathogenicity or elimination by their fellow effector cells. Thus, the enhanced virus suppression activity in the transductants likely is due to AgmTRIM5α restriction protecting the effector T cells from the cell-free or cell-to-cell exposure to virus generated by infected target cells.
In addition to our suppression assays, we also observed AgmTRIM5α-mediated resistance to SIV replication in primary T-cell clones when directly challenged with virus. AgmTRIM5α reduced the extent of initial infection and reduced the kinetics of virus spread in the transduced T-cell clones compared to gorTRIM5α, a weakly restricting TRIM5α protein, or untransduced cells. It is important to consider that TRIM5α restriction inherently is not absolute: the effect is saturated by high levels of virus. Thus, in all of our studies, while AgmTRIM5α reduced the kinetics of SIV spread, it did not prevent infection of at least some of the cells. Our culture conditions, being a closed system with long-term contact with infected cells and virions, might present a higher effective MOI than physiologically encountered by many T cells in vivo. Thus, SIV restriction in AgmTRIM5α-transduced CD4 T cells might be more effective in vivo than in vitro. Nevertheless, the ability to prevent rapid infection of the CD4 effector cells, providing more time for them to exert their virus-specific function, should increase their contribution to the CD4-mediated antiviral response.
While less studied than their CD8 counterparts, generation of a strong virus-specific CD4 T-cell response has been closely correlated with less pathogenic disease outcomes and elite control in HIV-infected individuals (53, 54). In this context, our in vitro suppression/infection data support the concept that infection diminishes the contribution of the CD4 T-cell compartment to antiviral immunity in vivo (53–56). Therefore, providing resistance to CD4 T cells might allow this compartment of the immune response to contribute its full potential. The expression of restricting TRIM5α proteins has been proposed as a strategy to protect human CD4 T cells (29, 31–33, 50, 57–59). However, despite strong levels of HIV-1 restriction by rhTRIM5α and stabilized human TRIM5α in replication assays that contain only the modified TRIM5α-expressing cells, restriction failed to protect CD4 T cells in the presence of infected unmodified CD4 T cells (50, 59), conditions certainly encountered in vivo. Our results reveal that AgmTRIM5α can resist this type of infected-cell challenge, making it the only TRIM5α protein reported to date that provides such restriction. For this reason, using AgmTRIM5α restriction in the SIV/rhesus macaque model is an ideal system for studying the concept of TRIM5α-mediated restriction as a means to protect CD4 T-cell function in both virus suppression assays and adoptive transfer experiments. In light of our findings, selection of intrinsically resistant cells for these AgmTRIM5α experiments would provide for even greater resistance.
Even though AgmTRIM5α transduction holds promise as an approach to evaluate TRIM5α restriction, one caveat to using this protein in rhesus macaques is that it contains a 37-amino-acid region with a unique 20-amino-acid duplication within its B30.2/SPRY domain that is responsible for SIVmac restriction (9, 60). While the immunogenicity of AgmTRIM5α in rhesus macaques is unknown, short-term adoptive transfer experiments probably are feasible given its overall close relationship with rhTRIM5α. Nevertheless, future experiments will need to be designed to account for this possibility.
Our finding that AgmTRIM5α can enhance CD4-mediated viral suppression has important in vivo implications. Extrapolating our results in vivo predicts that any virus-specific CD4 effector response in an infected individual can be severely and preferentially hindered by cell-to-cell-mediated viral infection of CD4 effectors. Thus, introducing resistance into these cells should increase the efficacy of the CD4 effector response. This approach also could be used to more broadly protect the CD4 T-cell compartment as a whole to prevent immunodeficiency due to loss of helper T cells and their vital functions. While AgmTRIM5α restriction is a convenient and potentially powerful approach to experimentally study AIDS viruses in the rhesus macaque model, its clinical use is unlikely. However, the CCR5 gene inactivation CD4 T-cell therapy currently undergoing clinical trials (61) could be focused on protecting the SIV-specific CD4 effector cell compartment to implement this concept.
ACKNOWLEDGMENTS
We thank Theodora Hatziioannou for the African green monkey and gorilla TRIM5α genes and Hans-Peter Kleim for the Phoenix RD114 clone 22 packaging cell line. The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: 2F12 SIV p27 hybridoma (no. 1547), deposited by Niels Pedersen, and human recombinant IL-2 from Maurice Gately, Hoffmann-La Roche Inc.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, and mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. government.
REFERENCES
- 1.Sastri J, Campbell EM. 2011. Recent insights into the mechanism and consequences of TRIM5alpha retroviral restriction. AIDS Res Hum Retrovir 27:231–238. doi: 10.1089/aid.2010.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gartner S, Liu Y, Polonis V, Lewis MG, Elkins WR, Hunter EA, Miao J, Corts KJ, Eddy GA. 1994. Adaptation of HIV-1 to pigtailed macaques. J Med Primatol 23:155–163. doi: 10.1111/j.1600-0684.1994.tb00117.x. [DOI] [PubMed] [Google Scholar]
- 3.Himathongkham S, Luciw PA. 1996. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology 219:485–488. doi: 10.1006/viro.1996.0276. [DOI] [PubMed] [Google Scholar]
- 4.Shibata R, Kawamura M, Sakai H, Hayami M, Ishimoto A, Adachi A. 1991. Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J Virol 65:3514–3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 6.Hatziioannou T, Evans DT. 2012. Animal models for HIV/AIDS research. Nat Rev Microbiol 10:852–867. doi: 10.1038/nrmicro2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang F, Perez-Caballero D, Hatziioannou T, Bieniasz PD. 2008. No effect of endogenous TRIM5alpha on HIV-1 production. Nat Med 14:235–238. doi: 10.1038/nm0308-235. [DOI] [PubMed] [Google Scholar]
- 8.Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc Natl Acad Sci U S A 101:10774–10779. doi: 10.1073/pnas.0402361101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kratovac Z, Virgen CA, Bibollet-Ruche F, Hahn BH, Bieniasz PD, Hatziioannou T. 2008. Primate lentivirus capsid sensitivity to TRIM5 proteins. J Virol 82:6772–6777. doi: 10.1128/JVI.00410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson JL, Campbell EM, Wu X, Vandegraaff N, Engelman A, Hope TJ. 2006. Proteasome inhibition reveals that a functional preintegration complex intermediate can be generated during restriction by diverse TRIM5 proteins. J Virol 80:9754–9760. doi: 10.1128/JVI.01052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yap MW, Nisole S, Lynch C, Stoye JP. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci U S A 101:10786–10791. doi: 10.1073/pnas.0402876101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keckesova Z, Ylinen LM, Towers GJ. 2004. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc Natl Acad Sci U S A 101:10780–10785. doi: 10.1073/pnas.0402474101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song B, Javanbakht H, Perron M, Park DH, Stremlau M, Sodroski J. 2005. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J Virol 79:3930–3937. doi: 10.1128/JVI.79.7.3930-3937.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldschmidt V, Ciuffi A, Ortiz M, Brawand D, Munoz M, Kaessmann H, Telenti A. 2008. Antiretroviral activity of ancestral TRIM5alpha. J Virol 82:2089–2096. doi: 10.1128/JVI.01828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perez-Caballero D, Hatziioannou T, Zhang F, Cowan S, Bieniasz PD. 2005. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J Virol 79:15567–15572. doi: 10.1128/JVI.79.24.15567-15572.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kutluay SB, Perez-Caballero D, Bieniasz PD. 2013. Fates of retroviral core components during unrestricted and TRIM5-restricted infection. PLoS Pathog 9:e1003214. doi: 10.1371/journal.ppat.1003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu X, Anderson JL, Campbell EM, Joseph AM, Hope TJ. 2006. Proteasome inhibitors uncouple rhesus TRIM5alpha restriction of HIV-1 reverse transcription and infection. Proc Natl Acad Sci U S A 103:7465–7470. doi: 10.1073/pnas.0510483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganser-Pornillos BK, Chandrasekaran V, Pornillos O, Sodroski JG, Sundquist WI, Yeager M. 2011. Hexagonal assembly of a restricting TRIM5alpha protein. Proc Natl Acad Sci U S A 108:534–539. doi: 10.1073/pnas.1013426108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A 103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, Sodroski J. 2007. The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J Virol 81:2138–2148. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roa A, Hayashi F, Yang Y, Lienlaf M, Zhou J, Shi J, Watanabe S, Kigawa T, Yokoyama S, Aiken C, Diaz-Griffero F. 2012. RING domain mutations uncouple TRIM5alpha restriction of HIV-1 from inhibition of reverse transcription and acceleration of uncoating. J Virol 86:1717–1727. doi: 10.1128/JVI.05811-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J, Tipper C, Sodroski J. 2011. Role of TRIM5alpha RING domain E3 ubiquitin ligase activity in capsid disassembly, reverse transcription blockade, and restriction of simian immunodeficiency virus. J Virol 85:8116–8132. doi: 10.1128/JVI.00341-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatterji U, Bobardt MD, Gaskill P, Sheeter D, Fox H, Gallay PA. 2006. Trim5alpha accelerates degradation of cytosolic capsid associated with productive HIV-1 entry. J Biol Chem 281:37025–37033. doi: 10.1074/jbc.M606066200. [DOI] [PubMed] [Google Scholar]
- 24.Campbell EM, Perez O, Anderson JL, Hope TJ. 2008. Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5alpha. J Cell Biol 180:549–561. doi: 10.1083/jcb.200706154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatziioannou T, Cowan S, Goff SP, Bieniasz PD, Towers GJ. 2003. Restriction of multiple divergent retroviruses by Lv1 and Ref1. EMBO J 22:385–394. doi: 10.1093/emboj/cdg042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Besnier C, Takeuchi Y, Towers G. 2002. Restriction of lentivirus in monkeys. Proc Natl Acad Sci U S A 99:11920–11925. doi: 10.1073/pnas.172384599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cowan S, Hatziioannou T, Cunningham T, Muesing MA, Gottlinger HG, Bieniasz PD. 2002. Cellular inhibitors with Fv1-like activity restrict human and simian immunodeficiency virus tropism. Proc Natl Acad Sci U S A 99:11914–11919. doi: 10.1073/pnas.162299499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi J, Aiken C. 2006. Saturation of TRIM5 alpha-mediated restriction of HIV-1 infection depends on the stability of the incoming viral capsid. Virology 350:493–500. doi: 10.1016/j.virol.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Greene WC, Debyser Z, Ikeda Y, Freed EO, Stephens E, Yonemoto W, Buckheit RW, Este JA, Cihlar T. 2008. Novel targets for HIV therapy. Antiviral Res 80:251–265. doi: 10.1016/j.antiviral.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 30.Anderson JS. 2013. Using TRIM5alpha as an HIV therapeutic: the alpha gene? Expert Opin Biol Ther 13:1029–1038. doi: 10.1517/14712598.2013.779251. [DOI] [PubMed] [Google Scholar]
- 31.Chan E, Towers GJ, Qasim W. 2014. Gene therapy strategies to exploit TRIM derived restriction factors against HIV-1. Viruses 6:243–263. doi: 10.3390/v6010243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sloan RD, Wainberg MA. 2013. Harnessing the therapeutic potential of host antiviral restriction factors that target HIV. Expert Rev Anti Infect Ther 11:1–4. doi: 10.1586/eri.12.146. [DOI] [PubMed] [Google Scholar]
- 33.Anderson J, Akkina R. 2008. Human immunodeficiency virus type 1 restriction by human-rhesus chimeric tripartite motif 5alpha (TRIM 5alpha) in CD34(+) cell-derived macrophages in vitro and in T cells in vivo in severe combined immunodeficient (SCID-hu) mice transplanted with human fetal tissue. Hum Gene Ther 19:217–228. doi: 10.1089/hum.2007.108. [DOI] [PubMed] [Google Scholar]
- 34.Coren LV, Trivett MT, Jain S, Ayala VI, Del Prete GQ, Ohlen C, Ott DE. 2015. Potent restriction of HIV-1 and SIVmac239 replication by African green monkey TRIM5α. Retrovirology 12:11. doi: 10.1186/s12977-015-0137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barsov EV, Trivett MT, Minang JT, Sun H, Ohlen C, Ott DE. 2011. Transduction of SIV-specific TCR genes into rhesus macaque CD8+ T cells conveys the ability to suppress SIV replication. PLoS One 6:e23703. doi: 10.1371/journal.pone.0023703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neff T, Peterson LJ, Morris JC, Thompson J, Zhang X, Horn PA, Thomasson BM, Kiem HP. 2004. Efficient gene transfer to hematopoietic repopulating cells using concentrated RD114-pseudotype vectors produced by human packaging cells. Mol Ther 9:157–159. doi: 10.1016/j.ymthe.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 37.Riddell SR, Greenberg PD. 1990. The use of anti-CD3 and anti-CD28 monoclonal antibodies to clone and expand human antigen-specific T cells. J Immunol Methods 128:189–201. doi: 10.1016/0022-1759(90)90210-M. [DOI] [PubMed] [Google Scholar]
- 38.Berger C, Huang ML, Gough M, Greenberg PD, Riddell SR, Kiem HP. 2001. Nonmyeloablative immunosuppressive regimen prolongs in vivo persistence of gene-modified autologous T cells in a nonhuman primate model. J Virol 75:799–808. doi: 10.1128/JVI.75.2.799-808.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minang JT, Trivett MT, Coren LV, Barsov EV, Piatak M Jr, Ott DE, Ohlen C. 2009. Nef-mediated MHC class I down-regulation unmasks clonal differences in virus suppression by SIV-specific CD8+ T cells independent of IFN-gamma and CD107a responses. Virology 391:130–139. doi: 10.1016/j.virol.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kestler H, Kodama T, Ringler D, Marthas M, Pedersen N, Lackner A, Regier D, Sehgal P, Daniel M, King N, Desrosiers R. 1990. Induction of AIDS in rhesus monkeys by molecularly cloned simian immunodeficiency virus. Science 248:1109–1112. doi: 10.1126/science.2160735. [DOI] [PubMed] [Google Scholar]
- 41.Gousset K, Ablan SD, Coren LV, Ono A, Soheilian F, Nagashima K, Ott DE, Freed EO. 2008. Real-time visualization of HIV-1 Gag trafficking in infected macrophages. PLoS Pathog 4:e1000015. doi: 10.1371/journal.ppat.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearce-Pratt R, Malamud D, Phillips DM. 1994. Role of the cytoskeleton in cell-to-cell transmission of human immunodeficiency virus. J Virol 68:2898–2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sattentau QJ. 2011. The direct passage of animal viruses between cells. Curr Opin Virol 1:396–402. doi: 10.1016/j.coviro.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 44.Dimitrov DS, Willey RL, Sato H, Chang L-J, Blumenthal R, Martin MA. 1993. Quantitation of human immunodeficiency virus type 1 infection kinetics. J Virol 67:2182–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carr JM, Hocking H, Li P, Burrell CJ. 1999. Rapid and efficient cell-to-cell transmission of human immunodeficiency virus infection from monocyte-derived macrophages to peripheral blood lymphocytes. Virology 265:319–329. doi: 10.1006/viro.1999.0047. [DOI] [PubMed] [Google Scholar]
- 46.Chen P, Hubner W, Spinelli MA, Chen BK. 2007. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J Virol 81:12582–12595. doi: 10.1128/JVI.00381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sourisseau M, Sol-Foulon N, Porrot F, Blanchet F, Schwartz O. 2007. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J Virol 81:1000–1012. doi: 10.1128/JVI.01629-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong P, Agosto LM, Ilinskaya A, Dorjbal B, Truong R, Derse D, Uchil PD, Heidecker G, Mothes W. 2013. Cell-to-cell transmission can overcome multiple donor and target cell barriers imposed on cell-free HIV. PLoS One 8:e53138. doi: 10.1371/journal.pone.0053138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin N, Welsch S, Jolly C, Briggs JA, Vaux D, Sattentau QJ. 2010. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J Virol 84:3516–3527. doi: 10.1128/JVI.02651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richardson MW, Carroll RG, Stremlau M, Korokhov N, Humeau LM, Silvestri G, Sodroski J, Riley JL. 2008. Mode of transmission affects the sensitivity of human immunodeficiency virus type 1 to restriction by rhesus TRIM5alpha. J Virol 82:11117–11128. doi: 10.1128/JVI.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minang JT, Trivett MT, Barsov EV, Del Prete GQ, Trubey CM, Thomas JA, Gorelick RJ, Piatak M Jr, Ott DE, Ohlen C. 2011. TCR triggering transcriptionally downregulates CCR5 expression on rhesus macaque CD4(+) T-cells with no measurable effect on susceptibility to SIV infection. Virology 409:132–140. doi: 10.1016/j.virol.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klotman ME, Kim S, Buchbinder A, DeRossi A, Baltimore D, Wong-Staal F. 1991. Kinetics of expression of multiply spliced RNA in early human immunodeficiency virus type 1 infection of lymphocytes and monocytes. Proc Natl Acad Sci U S A 88:5011–5015. doi: 10.1073/pnas.88.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ranasinghe S, Flanders M, Cutler S, Soghoian DZ, Ghebremichael M, Davis I, Lindqvist M, Pereyra F, Walker BD, Heckerman D, Streeck H. 2012. HIV-specific CD4 T cell responses to different viral proteins have discordant associations with viral load and clinical outcome. J Virol 86:277–283. doi: 10.1128/JVI.05577-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soghoian DZ, Jessen H, Flanders M, Sierra-Davidson K, Cutler S, Pertel T, Ranasinghe S, Lindqvist M, Davis I, Lane K, Rychert J, Rosenberg ES, Piechocka-Trocha A, Brass AL, Brenchley JM, Walker BD, Streeck H. 2012. HIV-specific cytolytic CD4 T cell responses during acute HIV infection predict disease outcome. Sci Transl Med 4:123ra125. doi: 10.1126/scitranslmed.3003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brenchley JM, Ruff LE, Casazza JP, Koup RA, Price DA, Douek DC. 2006. Preferential infection shortens the life span of human immunodeficiency virus-specific CD4+ T cells in vivo. J Virol 80:6801–6809. doi: 10.1128/JVI.00070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Douek DC, Brenchley JM, Betts MR, Ambrozak DR, Hill BJ, Okamoto Y, Casazza JP, Kuruppu J, Kunstman K, Wolinsky S, Grossman Z, Dybul M, Oxenius A, Price DA, Connors M, Koup RA. 2002. HIV preferentially infects HIV-specific CD4+ T cells. Nature 417:95–98. doi: 10.1038/417095a. [DOI] [PubMed] [Google Scholar]
- 57.Sakuma R, Noser JA, Ohmine S, Ikeda Y. 2007. Inhibition of HIV-1 replication by simian restriction factors, TRIM5alpha and APOBEC3G. Gene Ther 14:185–189. [DOI] [PubMed] [Google Scholar]
- 58.Neagu MR, Ziegler P, Pertel T, Strambio-De-Castillia C, Grutter C, Martinetti G, Mazzucchelli L, Grutter M, Manz MG, Luban J. 2009. Potent inhibition of HIV-1 by TRIM5-cyclophilin fusion proteins engineered from human components. J Clin Investig 119:3035–3047. doi: 10.1172/JCI39354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Richardson MW, Guo L, Xin F, Yang X, Riley JL. 2014. Stabilized human TRIM5alpha protects human T cells from HIV-1 infection. Mol Ther 22:1084–1095. doi: 10.1038/mt.2014.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakayama EE, Miyoshi H, Nagai Y, Shioda T. 2005. A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5alpha determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J Virol 79:8870–8877. doi: 10.1128/JVI.79.14.8870-8877.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G, Holmes MC, Gregory PD, Ando DG, Kalos M, Collman RG, Binder-Scholl G, Plesa G, Hwang WT, Levine BL, June CH. 2014. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med 370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]